Abstract
MicroRNAs play pivotal roles in gene regulation. Despite various research efforts on microRNAs, how micro-RNA target genes are transcriptionally regulated and how the transcriptional regulation of microRNA target genes relates to that of the microRNA genes are not well studied. By investigating the transcriptional regulation of microRNA target genes, we found that different groups of target genes of the same microRNA are co-expressed under different conditions, and these groups rarely overlap with each other for the majority of microRNAs. We also discovered that co-expressed microRNA target genes are often co-regulated, and different groups of target genes of the same microRNA are often regulated differently. In addition, we observed that transcription factors regulating a microRNA gene often regulate its target genes. Our study sheds light on the regulation of microRNA target genes, which will facilitate the prediction of microRNA target genes and the understanding of the transcriptional regulation of microRNA genes.
Keywords: MicroRNA, Target gene, Co-expression, Co-regulation, Cis-regulatory module
1. Introduction
MicroRNAs (miRNAs) are members of a growing class of regulatory non-coding RNAs that play crucial roles in regulating expression of protein-coding genes [1–3]. MiRNAs can bind to mRNAs of their target genes to either induce mRNA degradation or repress translation [1,2]. Early studies have shown important regulatory roles of miRNAs in development timing control [4,5]. Later studies have implicated miRNAs as important regulators of more diverse vital programs including hematopoietic cell differentiation, apoptosis, cell proliferation, and organ development [1,6,7]. It is also known that miRNAs may regulate more than one-third of all protein-coding genes in humans and are conserved across a diverse array of plants and animals [1,8]. Because of their involvement in a wide variety of developmental and physiological processes, it is important to study miRNA function, miRNA target genes, and transcriptional regulation of miRNAs and their target genes.
Various aspects of miRNAs have been studied in the past decade. One central focus of these studies is to understand the biogenesis of miRNAs. Like protein-coding genes, miRNA genes are first transcribed as longer primary transcripts called primary miRNA (pri-miRNA). Most miRNAs are transcribed by the RNA polymerase II and a small number of miRNAs are produced by the RNA polymerase III [9–11]. The pri-miRNA transcripts are then cropped by ribonuclease into precursor-miRNA (pre-miRNA). The pre-miRNA is subsequently exported out of the nucleus for further cleavage into a 22-nucleotide duplex. The complementary strand becomes degraded, leaving one fully mature miRNA strand. Mature miRNAs then associate with several proteins to form the RNA-induced-silencing-complex that binds to specific mRNA transcripts of protein-coding genes, directing mRNA inactivation by translational repression, deadenylation, or degradation. Besides the above understanding of miRNA biogenesis, studies in the past decade have also discovered a large number of miRNA genes in different species [12–14]. For instance, there are over 17,000 distinct mature miRNA sequences in over 140 species annotated in the current miRBase database [15]. In addition, target genes of miRNAs have been predicted by different algorithms [6,8,16]. Finally, with next generation sequencing techniques, several studies have predicted transcription start sites of miRNAs in mammals [11,17,18].
Despite many studies on miRNAs and the existence of predicted miRNA target genes in different species, the transcriptional regulation of miRNA target genes has not been well studied [19]. For instance, to our knowledge, scientists have not addressed the following questions: Are the target genes of one miRNA co-expressed? What is the pattern of the co-expressed miRNA target genes? How are the co-expressed miRNA target genes transcriptionally regulated? What is the relationship between miRNA transcriptional regulation and that of their targets? And so on. In this paper, we attempt to address these questions.
In the following, we will first describe the data and methods used. We will then show that different groups of target genes of one miRNA are often co-expressed under different conditions and these groups from the same miRNA rarely overlap with each other for the majority of miRNAs. Next, we will show that co-expressed miRNA target genes are likely co-regulated and different groups of target genes of the same miRNA may be regulated differently. Finally, we will show that transcription factors (TFs) regulating a miRNA often regulates its target genes. Our study shed light on miRNA target gene regulation and may facilitate the understanding of the transcriptional regulation of miRNAs themselves as well.
2. Materials and methods
2.1. MiRNA target genes and expression data
We obtained all human miRNAs and their target genes from DIANA-microT 3.0 (http://diana.cslab.ece.ntua.gr/microT/) [20]. The DIANA-microT 3.0 is based on parameters calculated individually for each miRNA and combines conserved and non-conserved miRNA recognition elements into a final prediction score, which correlates with protein production fold changes. The miRNA target genes predicted by the DIANA-microT 3.0 were shown to have higher precision than or comparable precision as the most commonly used algorithms [21]. In total, we obtained 555 miRNAs and 8770 different human genes as their target genes. To study the transcriptional regulation of these target genes, we extracted the upstream 10,000 base pair (bp) sequences relative to their transcription start sites from the Ensembl Genome Browser [22]. We then masked repeats in these sequences using the RepeatMasker web server at http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker
We obtained gene expression data from the GEO database [23]. Since miRNAs are known to play critical roles in development, we downloaded all data series on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array) related to development by searching “development” at the GEO website. Data from the GPL570 platform is used because this microarray platform has the largest number of human samples. We further filtered data series that have fewer than 15 samples or have not been normalized by the RMA algorithm [24]. At least 15 samples are required because it helps to obtain co-expressed gene clusters more reliably. The requirement of normalization by RMA will be helpful for others to repeat this study. In total, we obtained 125 datasets and 39,629 samples. For each dataset, we calculated the Pearson’s correlation coefficient for each gene pair and performed hierarchical clustering with complete linkage to obtain co-expressed clusters. Thus, every pair of genes within each cluster has a Pearson’s correlation coefficient larger than 0.6 or smaller than −0.6. The cutoff of 0.6 with at least 15 samples represents a p-value smaller than 0.02. That is, fewer than 2% of gene pairs will have the Pearson’s correlation coefficient larger than 0.6 or smaller than −0.6 by chance. In total, we obtained 6685 clusters with at least 15 genes in each cluster from the 125 datasets. The series number of these datasets and the number of samples are listed in Section A of the supplementary file. All 6685 clusters are compared with miRNA target genes to define co-expressed miRNA target gene sets in Section 2.2. Besides using 0.6 as the cutoff, we also obtained clusters based on the cutoffs 0.7 and 0.8. The analysis in this paper is based on the clusters with the cutoff 0.6 without specification.
2.2. Significantly co-expressed miRNA target gene sets
For every miRNA target gene set, we calculated its significance of overlap with each co-expressed gene cluster obtained from the 125 microarray expression datasets, by using the hypergeometric test. Assume the set of all miRNA target genes that are included in the GPL570 platform is S and |S|, the number of genes in S is equal to N. Given a miRNA target gene set S1 and a co-expressed gene cluster S2, assume the number of genes in the intersection of the three sets S, S1, and S2 is |S1∩S| = n, |S2∩S| = M, and |S1∩S2| = m, respectively. Then the significance of the overlap of the miRNA target gene set S1 and the co-expressed gene cluster S2 is measured by the following p-value based on the hypergeometric test:
To determine whether the target gene set of a miRNA is significantly co-expressed, we applied the Q-Value software [25] to all p-values obtained above and found the p-value cutoff, p1, to control the false discovery rate at the level of 0.05. We then output each group of shared genes by the target genes of a miRNA and a co-expressed cluster as a co-expressed miRNA target gene group, if the p-value of the overlapping between this target gene set and this co-expressed cluster is smaller than p1.
2.3. TRANSFAC database, TFs regulating miRNAs, and others
All 522 vertebrate TF binding specificity patterns from the TRANSFAC 9.2 database [26] were extracted for this study. These TF binding specificity patterns, also called motifs, are represented as position weight matrices (PWMs) here. A PWM is a 4 by k matrix, where k is the length of the binding sites and the four numbers in each column represent the probabilities of A, C, G, and T occurring at one position of the binding sites of the TF under consideration, with the numbers in the i-th column for the i-th position of the binding sites. Pseudo counts are introduced to regularize these PWMs, as was used previously [27,28]. These PWMs will be used to study the transcriptional regulation of miRNA target genes by applying published computational methods.
To validate the computational prediction of transcriptional regulation, we downloaded the TFs regulating miRNAs from the TransmiR database [29]. There are regulatory TFs available for 140 human miRNAs, for about 86% (120/140) miRNAs we have the target genes predicted by the DIANA-microT 3.0. We also downloaded ChIP-seq data (Chromatin ImmunoPrecipitation followed by massively parallel DNA Sequencing) generated by Dr. Michael Snyder’s group at Yale University and Dr. Richard Myers’s Lab at the HudsonAlpha Institute for Biotechnology from the UCSC Genome Browser [30] for all TFs obtained from the TransmiR database. In total, there are 6 TFs (CTCF, E2F1, GATA1, SPI1, SP1 and YY1) with available ChIP-seq data at the UCSC Genome Browser and mentioned in the TransmiR database.
2.4. Putative TF combinations regulating co-expressed miRNA target genes
We want to study the transcriptional regulation of co-expressed miRNA target genes. In other words, we want to know which TFs have transcription factor binding sites (TFBSs) significantly shared by upstream sequences of co-expressed miRNA target genes. As it is known that TFBSs are short and degenerate [31], and one of the best computational approaches to identify TFBSs is through the identification of cis-regulatory modules (CRMs) [28,32–36]. CRMs are short DNA sequences of a few hundred bp, in which multiple TFBSs reside to coordinately regulate the expression of genes nearby. Because it is often the interplay of multiple TFBSs of different TFs, instead of a single TF, that determines the temporal and spatial expression patterns of genes [1,2], it is important to study CRMs for an understanding of gene regulation. In addition, the occurrence of a CRM by chance is much smaller than that of an individual TFBS, which makes the identification of a CRM more reliable than the identification of a single TFBS.
Many computational methods have been developed to predict CRMs [28,32–36]. We applied the MOPAT software [28] to predict CRMs here, because it is more reliable to identify CRMs composed of known motifs than to predict CRMs composed of new motifs [28,32]. In addition, MOPAT can predict CRMs using all known motifs from the TRANSFAC database [28]. We used the following parameters when applying the MOPAT software: the window size cutoff, w, is 400; and the gene number cutoff, g, is 50% of the input genes. MOPAT outputs TF combinations that are significantly shared by CRMs, and the corresponding CRMs.
3. Results
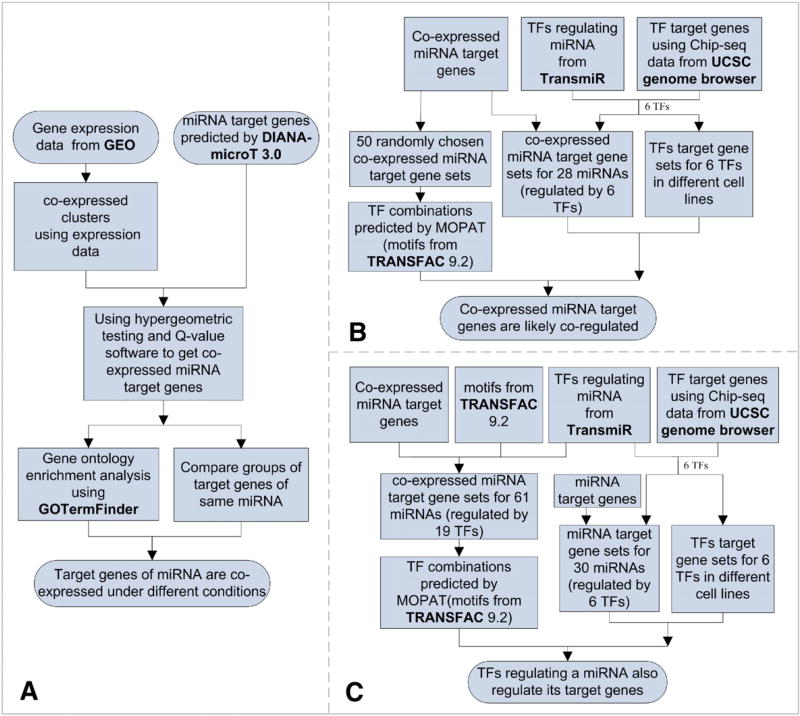
To study the transcriptional regulation of co-expressed miRNA target genes, we applied the methods described in the Materials and methods section to the above 8770 human miRNA target genes. We found that different groups of target genes of the same miRNA are often co-expressed under different conditions. In addition, co-expressed miRNA target genes are likely co-regulated. Finally, TFs regulating a miRNA often regulate its target genes. Fig. 1 illustrates the flowchart of the structure of our analysis. The details are in the following three subsections.
Fig. 1.
The flowcharts of the structure of our analyses. The flowcharts A, B and C describe the analyses in Sections 3.1, 3.2 and 3.3, respectively.
3.1. Different groups of target genes of the same miRNA are often co-expressed under different conditions
By comparing the miRNA target genes with the co-expressed gene clusters, with a false discovery rate of 0.05, we found that target genes of 90.27% (501/555) of miRNAs significantly overlap with at least one co-expressed cluster. See Section B of the supplementary file for groups of co-expressed miRNA target genes and the datasets in which they are co-expressed. In addition, the observation that miRNA target genes are significantly co-expressed is unchanged when we used different cutoffs to define co-expressed clusters. Since we only used 125 different microarray datasets related to development, with more microarray data under more experimental conditions, we may find an even higher percentage of miRNAs whose target genes are co-expressed. Note that it may be not surprising to observe target genes of 90.27% of miRNAs are co-expressed, as target genes of TFs have often been shown to be co-expressed [37].
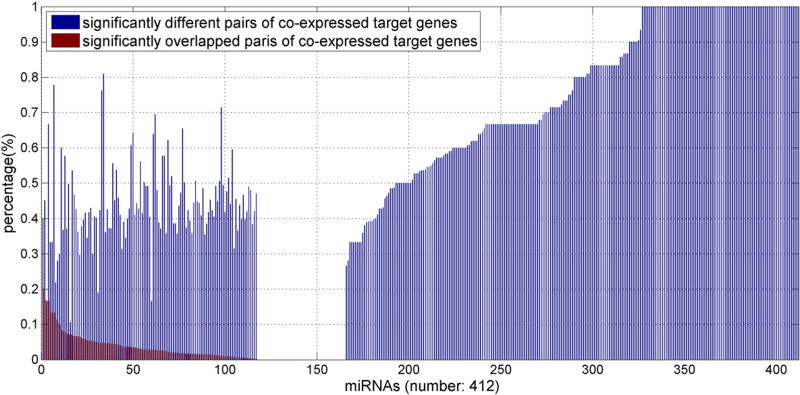
Since one miRNA target gene set may significantly overlap with several co-expressed clusters, we compared different groups of co-expressed target genes of the same miRNA. There are 412 miRNAs with target genes significantly overlap with at least two co-expressed clusters. Surprisingly, we found that for 12.62% (52/412) of miRNAs, at least two groups of co-expressed target genes do not share any target gene. To assess the significance of rare overlap between two groups of co-expressed target genes of the same miRNA, we applied the hypergeometric test to measure the chance that we could find two random subsets of target genes of the same miRNA (with the corresponding sizes) that have fewer shared genes than the observed number of genes shared by two co-expressed target gene groups. We found that for 88.35% (364/412) of miRNAs that have at least two co-expressed target gene groups, under the false discovery rate 0.05, the number of shared target genes by at least two groups of co-expressed target genes of these miRNAs is significantly small. However, we also noticed that for 28.4% (117/412) of miRNAs, there are at least two groups of co-expressed target genes significantly overlapped. That is, the two groups of co-expressed target genes of the same miRNA have almost exactly the same gene members, which essentially are one group of co-expressed target genes that are active under different conditions. Finally, we observed that for 11.65% (48/412) of miRNAs, there is no any pair of significantly overlapped (different) co-expressed miRNA target gene groups. We did not find any common characteristic shared by these 48 miRNAs (Section C of the supplementary file). See Fig. 2 for the number of significantly different (overlapped) co-expressed gene group pairs for each of the 412 miRNAs, which have at least two groups of co-expressed target genes. It is evident that the majority of miRNA genes may target different groups of target genes under different conditions and these groups rarely share target genes. In addition, a miRNA may target the same group of target genes under different conditions.
Fig. 2.
The percentage of the number of significantly different (blue bar) and significantly overlapped (red bar) pairs of co-expressed miRNA target gene groups for each of the 412 miRNAs. The miRNAs are sorted according to the percentages of significantly different pairs of miRNA target gene groups from smallest to largest first, then sorted according to the percentage of significantly overlapped pairs of co-expressed miRNA target gene groups.
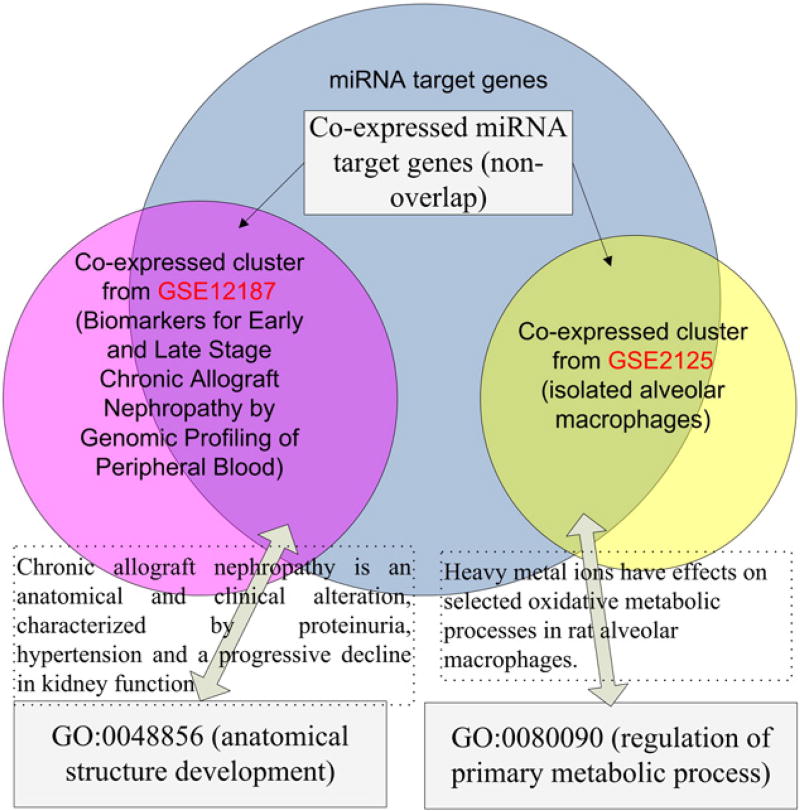
We next studied gene ontology terms shared by different groups of co-expressed target genes of one miRNA. For the above 364 miRNAs with at least two groups of rarely shared co-expressed target genes, we did gene ontology enrichment analysis for each group of co-expressed miRNA target genes by using GOTermFinder [38]. We found that different groups of co-expressed target genes of the same miRNA often have different functions. For instance, for the miRNA hsa-miR-203, we found its target genes are significantly co-expressed in the dataset GSE12187 (biomarkers for early and late stage chronic allograft nephropathy by genomic profiling of peripheral blood) and GSE2125 (isolated alveolar macrophages) (Fig. 3). The co-expressed hsa-miR-203 target gene group in GSE12187 is enriched with genes annotated with GO:0048856 (anatomical structure development). As chronic allograft nephropathy is an anatomical and clinical alteration, characterized by proteinuria, hypertension and a progressive decline in kidney function [39], the function of these co-expressed target genes is consistent with the experimental condition of GSE12187. Accordingly, the co-expressed hsa-miR-203 target gene group in GSE2125 is enriched with genes annotated with GO:0080090 (regulation of primary metabolic process). As heavy metal ions have effects on selected oxidative metabolic processes in rat alveolar macrophages [40], the function of these co-expressed target genes is also consistent with the experimental condition of the GSE2125.
Fig. 3.
A pair of significantly different miRNA target gene groups for the miRNA hsa-miR-203.
3.2. Co-expressed miRNA target genes are likely co-regulated based on computational prediction and ChIP-seq data
In the above, we showed that miRNA target genes can often be classified into several groups, genes in each of which are often co-expressed and share certain functions. Since it has been previously shown that co-expressed genes are often co-regulated [41], it is interesting to see whether there are common TFs regulating each group of the co-expressed miRNA target genes. We thus applied the MOPAT software [28] to identify significantly shared TF combinations by a group of co-expressed miRNA target genes. MOPAT searches the upstream sequences of a group of co-regulated genes for putative TFBSs of all input TF PWMs, and then discovers TF combinations whose TFBSs significantly co-occur in many short DNA regions (the length of these short regions is defined by users) in the upstream sequences.
We randomly chose 50 groups of co-expressed miRNA target genes. For all 50 groups, we identified significant TF combinations that have their TFBSs co-occurring in the upstream sequences of genes within one group. Here a significant TF combination means a group of cooperative TFs with multiple comparison corrected p-values smaller than 0.005 output from the MOPAT software [28]. The p-value of the most significant TF combinations from each of the 50 co-expressed target gene groups is much smaller than that from each of the 50 groups of random sequences of the same sizes. These significant TF combinations indicate that the co-expressed target genes may be co-regulated. The most significant TF combinations and their corrected p-values for each of the 50 miRNA target gene groups have been listed in Section D of the supplementary file. We also applied MOPAT to predict TF combinations for different groups of co-expressed target genes of the same miRNAs. We found that on average more than 90% of predicted TF combinations are different for different groups of co-expressed target genes of the same miRNAs. Here are two examples of the significant TF combinations predicted.
3.2.1. Example 1
One of the predicted TF combinations that regulate the target genes of the miRNA, hsa-miR-17, comprises motifs M00500 (STAT6), M00772 (IRF), and M00972 (IRF). The IL-4/IL-13/STAT6 signaling pathway promotes luminal mammary epithelial cell developments [42]. IRF (TRIM63) is important for microtubule, intermediate filament, and sarcomeric M-line maintenance in striated muscle development [43]. Thus, these TFs may work together in development. Consistently, hsamiR-17 family miRNAs are important for embryonic development. [44], which supports the functionality of the TFs in this TF combination. In addition, nearly 60% of target genes of this TF combination are shown to be related to development (see Section E of the supplementary file), which supports the function of this TF combination and that of hsa-miR-376a.
3.2.2. Example 2
The TF combination that regulates the target genes of the miRNA hsa-miR-224 comprises motifs M00143 (PAX5), M00800 (AP2), and M00982 (KROX). Among the three TFs, PAX5 expression correlates with increasing malignancy in human astrocytomas [45]. TFAP2A (AP-2) can inhibit cancer cell growth [46]; and EGR1 can promote growth and survival of prostate cancer cells [47]. Thus, these three TFs may work together to play a role in cancer. Consistently, the human miRNA hsa-miR-224 is associated with hepatocellular Carcinoma [48]. In addition, about 90% target genes of this TF combination are shown to be related to cancer (see Section E of the supplementary file). All these evidences support the functionality of this TF combination.
In addition to the above computational predictions, we also collected ChIP-seq data for 6 TFs from the UCSC Genome Browser [30], with a restriction date before December 31, 2010. These 6 TFs regulate 28 miRNAs that have co-expressed miRNA target gene groups, according to the TransmiR database [29] and http://diana.cslab.ece.ntua.gr/microT/. For each of the 6 TFs, we defined the target genes of this TF as the nearest gene to each ChIP-seq peak with q-value smaller than 0.001. The ChIP-seq peaks and their associated q-values have been provided by the UCSC Genome Browser. We then performed the hypergeometric test to indicate whether a significant number of genes in a group of co-expressed miRNA target genes are the target genes of a TF defined above. We found that for 75% (21/28) of miRNAs, their co-expressed target genes are co-regulated (see Table 1 and Section F of the supplementary file for details).
Table 1.
TF target genes defined from ChIP-seq data significantly overlap with co-expressed target genes of 21 miRNAs.
| miRNA | TF (p-value) |
|---|---|
| hsa-let-7a | E2F1 (7.65E-06), SP1 (6.05E-04) |
| hsa-let-7i | E2F1 (4.33E-06), SP1 (4.64E-04) |
| hsa-miR-106a | CTCF (2.04E-04), E2F1 (8.56E-07), SPI1 (1.59E-05), SP1 (1.24E-11), YY1 (1.47E-07) |
| hsa-miR-106b | E2F1 (2.79E-07), SP1 (3.26E-08), YY1 (1.92E-06) |
| hsa-miR-144 | E2F1 (3.67E-09), SPI1 (5.32E-09), SP1 (8.03E-08), YY1 (9.55E-12) |
| hsa-miR-15a | E2F1 (5.61E-06), SP1 (2.12E-04), YY1 (1.24E-03) |
| hsa-miR-15b | E2F1 (5.61E-06), SP1 (2.12E-04), YY1 (1.24E-03) |
| hsa-miR-16 | E2F1 (1.25E-05), SP1 (1.66E-04), YY1 (7.63E-04) |
| hsa-miR-17 | E2F1 (1.26E-04), SP1 (1.62E-05), YY1 (6.48E-06) |
| hsa-miR-195 | E2F1 (1.25E-05), SP1 (1.66E-04), YY1 (7.63E-04) |
| hsa-miR-19a | E2F1 (1.55E-04), SPI1 (6.00E-04), SP1 (2.99E-05), YY1 (6.16E-06) |
| hsa-miR-19b | SP1 (1.55E-03), YY1 (2.03E-04) |
| hsa-miR-20a | E2F1 (5.24E-07), SPI1 (6.91E-04), SP1 (9.00E-06), YY1 (3.06E-05) |
| hsa-miR-20b | E2F1 (5.38E-04), SP1 (8.66E-08), YY1 (5.12E-05) |
| hsa-miR-223 | YY1 (1.77E-03) |
| hsa-miR-23a | E2F1 (3.24E-07), SPI1 (5.17E-04), SP1 (2.83E-04), YY1 (3.22E-05) |
| hsa-miR-363 | E2F1 (4.16E-06), YY1 (5.45E-04) |
| hsa-miR-375 | E2F1 (2.01E-05), SPI1 (2.85E-04), SP1 (1.33E-04), YY1 (5.30E-05) |
| hsa-miR-449b | GATA (2.19E-03) |
| hsa-miR-92a | E2F1 (8.13E-04), SPI1 (1.26E-03) |
| hsa-miR-93 | E2F1 (1.11E-04), SP1 (7.39E-08), YY1(4.20E-05) |
When the target genes of one TF significantly overlap with several co-expressed target gene groups of a miRNA, only the smallest p-value of significant overlap for this TF is listed here.
3.3. TFs regulating a miRNA also regulate its target genes
How miRNA genes themselves are regulated is still not well studied [3]. It is difficult to study miRNA gene transcriptional regulation because the transcription start sites of the majority of miRNA genes have not been well defined [3]. With the observation that miRNAs and TFs co-regulate their target genes, it is reasonable to hypothesize that TFs regulating a miRNA may regulate its target genes, since miR-NAs and TFs must be active at the same time in order to coordinately regulate their target genes.
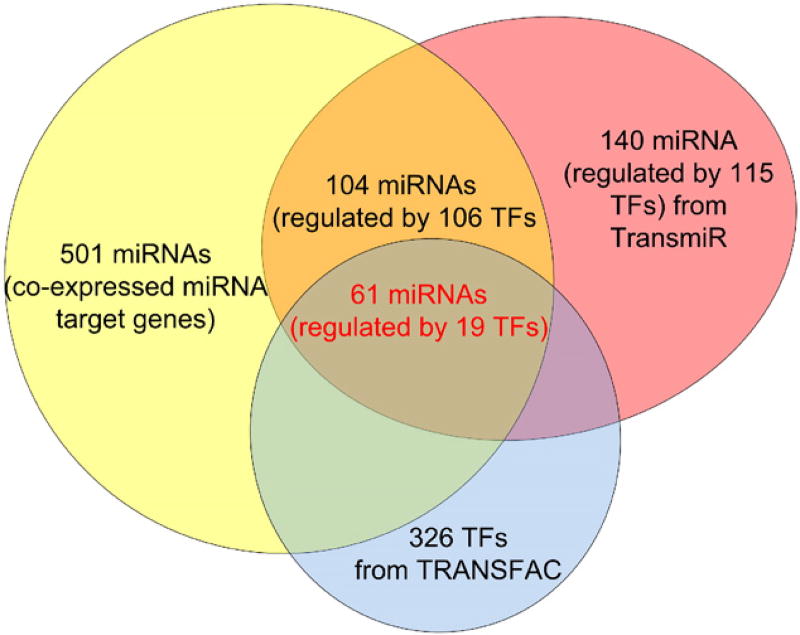
To test this hypothesis, we collected all miRNAs with known regulatory TFs from the TransmiR database [29], which collects such TF-miRNA regulation relationship from the literature. There are 104 miRNAs in the TransmiR database with co-expressed target genes identified above. These miRNAs are known to be regulated by 106 TFs. Among the 106 TFs, only 19 TFs (they regulate 61 miRNAs) are also included in the TRANSFAC database 9.2 we used (Fig. 4). We thus checked whether there is a significant TF combination predicted by MOPAT that contains the TFs regulating the miRNA under consideration. For each of the 61 miRNAs, we applied MOPAT to the group of co-expressed miRNA target genes that has the smallest overlapping p-value when we compared miRNA target genes with co-expressed clusters. From the analysis of MOPAT results, we found that about 36% (22/61) of miRNAs, the TF regulating the miRNA according to the TransmiR database also regulates at least one group of its co-expressed target genes. See Table 2 for the miRNAs, the TFs that regulate these miRNAs according to the TransmiR database [29], and the TF combinations that regulate target genes of these miRNAs from the MOPAT software. For about two thirds of the 61 miRNAs, we could not find any TF that regulates the miRNA in the MOPAT predictions, which could be due to the sequence range we considered (we only considered upstream 10,000 bp regions), the incomplete collection of TFs with known PWMs in TRANSFAC database, and the limitation of computational methods for motif identification.
Fig. 4.
61 miRNAs are regulated by 19 TFs shared by the TRANSFAC 9.2 database and the TransmiR database.
Table 2.
Target genes of 22 miRNAs are co-regulated based on computational prediction by the MOPAT software.
| miRNA | Regulatory TFs from TransmiR | Regulatory TFs from MOPAT |
|---|---|---|
| hsa-let-7a | E2F1, E2F3, EIF2C2, BRD2, LIN28, LIN28, MYC, TRIM32 | E2F1 |
| hsa-let-7i | E2F1, E2F3, EIF2C2, LIN28A, MYC, TRIM32 | E2F1 |
| hsa-miR | CEBPA, MYF6, MYF5, MYOD1, MYOG | CEBPA, MYOD1 |
| hsa-miR-106b | E2F1, E2F1, E2F3, MYC | E2F1 |
| hsa-miR-133a | PPP3R1, MYF6, MYF5, MYOD1, MYOG, TNFSF12 | MYOD1, MYOG |
| hsa-miR-144 | GATA1,GATA4 | GATA4 |
| hsa-miR-155 | AKT1, BCR, CAMP, FOXP3, FOXP3, NFKB1, SMAD4, SMG1, TGFB1, TP53 | FOXP3 |
| hsa-miR-15b | E2F1, E2F3, BRD2, STAT5 | E2F1 |
| hsa-miR-16 | E2F1, E2F3, NFKB1, STAT5 | E2F1 |
| hsa-miR-18a | E2F1, ERS1, MYC, MYCN, MYCN, NKX2-5, TLX1, TLX3 | E2F1, NKX2-5 |
| hsa-miR-195 | E2F1, E2F3, EGR3, EGR3, MYC, STAT5 | E2F1 |
| hsa-miR-19a | E2F1, ESR1, MYC, MYC, MYCN, MYCN, NKX2-5, PTEN, TLX1, TLX3 | E2F1 |
| hsa-miR-206 | MYF6, MYF5, MYOD1, MYOG TNFSF12 | MYOD1 |
| hsa-miR-20a | CCND1,E2F1,ESR1, MYC,MYCN, NKX2-5, TLX1, TLX3 | E2F1 |
| hsa-miR-20b | E2F1,ESR1, MYC | E2F1 |
| hsa-miR-29a | CEBPA, HMGA1, IL4,MYC, NFKB1, PDGF-B, TGFB1, YY1 | CEBPA, YY1 |
| hsa-miR-29b | CEBPA, NFKB1, YY1 | CEBPA, YY1 |
| hsa-miR-29c | MYC, NFKB1, YY1 | YY1 |
| hsa-miR-34a | CEBPA, CEBPA, MYC, NR1H4, NR1H4, TP53, TP53, TP53, TP53 | CEBPA |
| hsa-miR-363 | E2F1, ESR1 | E2F1 |
| hsa-miR-449a | CDK, E2F1, FOXJ1, RB1 | E2F1 |
| hsa-miR-92a | E2F1, ESR1, MYC, MYCN, NKX2-5, TLX1, TLX3 | E2F1 |
Since there could be false positive predictions in the MOPAT output, the above observation that TFs regulating a miRNA also regulate its target genes needs further validation. We thus also collected ChIP-seq data for TFs that regulate one of the miRNAs in the TransmiR database. In total, we obtained ChIP-seq data for 6 TFs that regulates at least one miRNA in the TransmiR database. These 6 TFs regulate 30 miRNAs according to the TransmiR database. We found that for 90% (27/30) of miRNAs, ChIP-seq data shows that the TF regulating miRNA also regulates its target genes. See Table 3 for the miRNAs, their regulatory TFs based on ChIP-seq data of 6 TFs, and the p-value of the overlapping between the TF target genes and the miRNA target genes. Thus, it is clear that for miRNAs, the TF regulates a miRNA may often regulate its target genes as well. This observation may be useful to facilitate the study of the transcriptional regulation of miRNA genes themselves, as it is often easier to know what TFs may regulate target genes of a miRNA.
Table 3.
TFs that regulate 27 miRNAs also regulate their target genes.
| miRNA | TF (p-value) |
|---|---|
| hsa-miR-106a | E2F1 (2.77E-08), SP1 (6.11E-09) |
| hsa-miR-106b | E2F1 (1.73E-08) |
| hsa-miR-144 | GATA (2.39E-02) |
| hsa-miR-146a | SPI1 (1.96E-04) |
| hsa-miR-15a | E2F1 (1.83E-06) |
| hsa-miR-15b | E2F1 (3.79E-07) |
| hsa-miR-16 | E2F1 (1.67E-06) |
| hsa-miR-17 | E2F1 (9.19E-07) |
| hsa-miR-18a | E2F1 (1.07E-03) |
| hsa-miR-18b | E2F1 (2.03E-03) |
| hsa-miR-195 | E2F1 (9.10E-07) |
| hsa-miR-19a | E2F1 (6.88E-06) |
| hsa-miR-19b | E2F1 (3.25E-06) |
| hsa-miR-20a | E2F1 (1.95E-08) |
| hsa-miR-20b | E2F1 (1.08E-06) |
| hsa-miR-223 | E2F1 (4.60E-04) |
| hsa-miR-23a | SPI1 (2.46E-10) |
| hsa-miR-25 | E2F1 (7.27E-03) |
| hsa-miR-29a | YY1 (3.29E-03) |
| hsa-miR-29b | YY1 (2.29E-03) |
| hsa-miR-29c | YY1 (4.36E-03) |
| hsa-miR-363 | E2F1 (3.98E-07) |
| hsa-miR-375 | CTCF (1.04E-04) |
| hsa-miR-449a | E2F1 (2.57E-02) |
| hsa-miR-451 | GATA (1.64E-02) |
| hsa-miR-92a | E2F1 (1.80E-05) |
| hsa-miR-93 | E2F1 (8.35E-07) |
Each p-value in the second column indicates the significance of the overlap between the TF target genes and the target genes of a miRNA.
4. Discussion
We used 125 microarray expression datasets related to development to study the expression patterns of miRNA target genes. With these 125 datasets, we have shown that target genes of 90.27% of miRNAs significantly overlap with at least one co-expressed gene cluster. In the future, more microarray expression datasets under conditions other than development could be used to study each miRNA and its target genes in detail. In this way, we may gain better understanding of the classification of the miRNA target genes and their functions for each miRNA.
Since a large number of miRNAs are transcribed by RNA polymerase II and several TFs are often needed to regulate RNA polymerase II transcribed genes, it is most likely that TF combinations regulating miRNAs will also regulate miRNA target genes of the miRNAs under consideration. In this study, we have only shown that individual TF regulating a miRNA often regulates its target genes. In the future, with more information about TFs that regulate miRNAs and with ChIP-seq data for more TFs, TF combinations regulating miRNAs can also be compared with TF combinations regulating miRNA target genes.
We applied the MOPAT software [28] to identify TF combinations that may regulate the miRNA target genes in this study. Other computational methods could be used here. We could also consider longer upstream sequences, the 5′ untranslated sequences, and the intronic sequences when applying computational methods for TF combination prediction. However, our conclusions should still hold with these alternatives, since comparisons of these predicted TF combinations with those in literature and those from ChIP-seq experiments supported our predictions. In the future, with more experimental information about transcriptional regulation of miRNA genes themselves and their target genes, we could extend the current study to have a deeper understanding of regulation of miRNA target genes, and the relationship between the regulation of miRNA genes and that of their targets.
5. Conclusions
We have shown that target genes of the same miRNA can be classified into several groups, and different groups of miRNA target genes are co-expressed under different conditions. These groups of miRNA target genes are often co-regulated. In addition, TFs regulating a miRNA often regulate its target genes as well. All these observations show different aspects of miRNA target genes, which have not been discovered before and will facilitate future studies of miRNA, miRNA target genes, and miRNA transcriptional regulation.
Supplementary Material
Acknowledgments
We apologize to colleagues whose work we could not cover due to space limitations. We thank Hou Lin at Beijing University for the co-expressed clusters generated. This project is supported by a National Institute of Health grant HG004359.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at doi:10.1016/j.ygeno.2011.09.004.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 3.Schanen BC, Li X. Transcriptional regulation of mammalian miRNA genes. Genomics. 2011;97:1–6. doi: 10.1016/j.ygeno.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Wang Y, Shen C, Huang YW, Huang K, Huang TH, Nephew KP, Li L, Liu Y. RNA polymerase II binding patterns reveal genomic regions involved in microRNA gene regulation. PLoS One. 2010;5:e13798. doi: 10.1371/journal.pone.0013798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Xuan Z, Zhao X, Li Y, Zhang MQ. High-resolution human core-promoter prediction with CoreBoost_HM. Genome Res. 2009;19:266–275. doi: 10.1101/gr.081638.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z. Insight into microRNA regulation by analyzing the characteristics of their targets in humans. BMC Genomics. 2009;10:594. doi: 10.1186/1471-2164-10-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maragkakis M, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinform. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25(23):3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 22.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claverie JM, Audic S. The statistical significance of nucleotide position-weight matrix matches. Comput. Appl. Biosci. 1996;12:431–439. doi: 10.1093/bioinformatics/12.5.431. [DOI] [PubMed] [Google Scholar]
- 28.Hu J, Hu H, Li X. MOPAT: a graph-based method to predict recurrent cis-regulatory modules from known motifs. Nucleic Acids Res. 2008;36:4488–4497. doi: 10.1093/nar/gkn407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchette M, Bataille AR, Chen X, Poitras C, Laganiere J, Lefebvre C, Deblois G, Giguere V, Ferretti V, Bergeron D, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai X, Hou L, Su N, Hu H, Deng M, Li X. Systematic identification of conserved motif modules in the human genome. BMC Genomics. 2010;11:567. doi: 10.1186/1471-2164-11-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frith MC, Hansen U, Weng Z. Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics. 2001;17:878–889. doi: 10.1093/bioinformatics/17.10.878. [DOI] [PubMed] [Google Scholar]
- 35.Gupta M, Liu JS. De novo cis-regulatory module elicitation for eukaryotic genomes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7079–7084. doi: 10.1073/pnas.0408743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Q, Wong WH. CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Luscombe NM, Qian J, Gerstein M. Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet. 2003;19:422–427. doi: 10.1016/S0168-9525(03)00175-6. [DOI] [PubMed] [Google Scholar]
- 38.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO:: TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scolari MP, Cappuccilli ML, et al. Therapy strategies in the prevention of chronic allograft nephropathy. G. Ital. Nefrol. 2005 Jan-Feb;22(Suppl 31):S36–S40. [PubMed] [Google Scholar]
- 40.Castranova V, Bowman L, Reasor MJ, Miles PR. Effects of heavy metal ions on selected oxidative metabolic processes in rat alveolar macrophages. Toxicol Appl Pharmacol. 1980 Mar 30;53(1):14–23. doi: 10.1016/0041-008x(80)90375-0. [DOI] [PubMed] [Google Scholar]
- 41.Allocco DJ, Kohane IS, Butte AJ. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinform. 2004;5:18. doi: 10.1186/1471-2105-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaled WT, Read EK, Nicholson SE, Baxter FO, Brennan AJ, Came PJ, Sprigg N, McKenzie AN, Watson CJ. The IL-4/IL-13/Stat6 signalling pathway promotes luminal mammary epithelial cell development. Development. 2007 Aug;134(15):2739–2750. doi: 10.1242/dev.003194. Epub 2007 Jul 4. [DOI] [PubMed] [Google Scholar]
- 43.McElhinny Abigail S, Perry Cynthia N, Witt Christian C, Siegfried Labeit, Gregorio Carol C. Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J. Cell. Sci. 2004 Jul 1;117:3175–3188. doi: 10.1242/jcs.01158. [DOI] [PubMed] [Google Scholar]
- 44.Foshay Kara M, Gallicano Ian G. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev. Biol. 2009 Feb 15;326(2):431–443. doi: 10.1016/j.ydbio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Stuart ET, Kioussi C, Aguzzi A, et al. PAX5 expression correlates with increasing malignancy in human astrocytomas. Clin. Cancer Res. 1995;1:207–214. [PubMed] [Google Scholar]
- 46.Zeng YX, Somasundaram K, el-Deiry WS. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet. 1997;15:78–82. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- 47.Virolle T, Krones-Herzig A, Baron V, De Gregorio G, Adamson ED, Mercola D. Egr1 promotes growth and survival of prostate cancer cells. Identification of novel Egr1 target genes. J Biol Chem. 2003 Apr 4;278(14):11802–11810. doi: 10.1074/jbc.M210279200. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 2008;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.