ABSTRACT
Nontuberculous mycobacteria (NTM) include species that colonize human epithelia, as well as species that are ubiquitous in soil and aquatic environments. NTM that primarily inhabit soil and aquatic environments include the Mycobacterium avium complex (MAC) (M. avium and Mycobacterium intracellulare) and the Mycobacterium abscessus complex (MABSC) (M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii) and can be free living, biofilm associated, or amoeba associated. Although NTM are rarely pathogenic in immunocompetent individuals, individuals who are immunocompromised, due to either an inherited or acquired immunodeficiency, are highly susceptible to NTM infection (NTMI). Several characteristics, such as biofilm formation and the ability of select NTM species to form distinct colony morphotypes, all may play a role in pathogenesis that is not observed in the related, well-characterized pathogen Mycobacterium tuberculosis. Different morphotypes of NTM have been recognized and characterized since the 1950s, but the mechanisms that underlie colony phenotype change and subsequent differences in pathogenicity are just beginning to be explored. Advances in genomic analysis have led to progress in identifying genes important to the pathogenesis and persistence of MAC disease as well as in illuminating genetic aspects of different colony morphotypes. Here we review recent literature regarding NTM ecology and transmission, as well as the factors which regulate colony morphotype and pathogenicity.
KEYWORDS: amoeba, M. abscessus, M. avium, M. intracellulare, morphotype, mycobacteria, nontuberculous mycobacteria, tuberculosis
TEXT
Nontuberculous mycobacteria (NTM) are a subset of Mycobacterium species that are found in many environmental niches in nature. Although NTM are harmless to most individuals, each year (at both the national and international levels) there are increasingly more individuals diagnosed with NTM infection (NTMI). Clinical presentations of NTMI include pulmonary infection, disseminated infection, skin disease, and lymphadenitis. Although risk factors for NTMI include immunodeficiency or underlying barrier dysfunction, most NTMI patients do not have any known risk factors. This is especially true of children with NTMI, in whom disease manifests primarily as a distressful and disfiguring cervical lymphadenitis. It is important to have a basic understanding of the host and bacterial factors that maintain human-NTM commensalism, as their perturbation may cause an infection to progress at the expense of the human host (1).
NTM include species that colonize human epithelia, as well as species that are found in soil and aquatic environments. The NTM species that colonize human epithelia are largely nonpathogenic and can be found on skin (2–4) and along the genitourinary (5–7), gastrointestinal (7), and respiratory (8–14) tracts. The NTM species that are found in soil and aquatic environments include Mycobacterium vaccae, the Mycobacterium avium complex (MAC), and the Mycobacterium abscessus complex (MABSC) (15, 16). Although most NTM are traditionally considered to be opportunistic pathogens, M. vaccae is unique in that it is also a transient human colonizer (17) and benefits the host in a manner that resembles ecological mutualism: M. vaccae inhibits pulmonary allergic inflammation in mice (18), as well as decreases anxiety in both mice (19) and humans (20), via an as-yet-undefined gut-brain-microbiota axis (21). MAC was first isolated from wood pigeons (22) but is now known to be ubiquitous in the environment and found in freshwater, salt water, soil, food, dust, and domestic and wild animals (23–26). In both water and soil, MAC and MABSC species (MAC/MAB) can be free living, biofilm associated, or amoeba associated (27, 28). Infection with MAC/MAB can follow exposure to aerosols of MAC/MAB-containing water while bathing (29, 30) or to aerosols of MAC/MAB-containing soil while gardening (31) or during a natural disaster (32). NTM therefore occupy a unique and broad ecological niche and necessarily exhibit a remarkable range of genetic adaptations to their varied environments.
With an ever-growing number of mycobacterial species being identified, the importance of understanding the interspecies relationships as well as the roles of individual species in human colonization and pathogenesis cannot be overlooked. Here we review the diversity, evolution, and genetic relationships between existing mycobacterial species, the environments in which they are found (specifically, their niche as aquatic organisms as well as their interactions with environmental amoebas), and the current research on NTM colony morphotypes.
THE DIVERSE LIVES OF NONTUBERCULOUS MYCOBACTERIA
The nomenclature and classification of mycobacteria has remained unchanged for most of its known history (33). Species are generally grouped into three major categories based on propensity for human infection: obligate pathogens (e.g., Mycobacterium leprae, Mycobacterium tuberculosis, Mycobacterium marinum, and Mycobacterium ulcerans), facultative or opportunistic pathogens (e.g., M. avium, Mycobacterium intracellulare, M. abscessus, and Mycobacterium kansasii), and strictly commensal or saprophytic bacilli (e.g., Mycobacterium smegmatis, Mycobacterium vanbaalenii, and Mycobacterium thermoresistibile) (34). There are more than 170 recognized species of mycobacteria, with more being added on a regular basis (35). Until recently, the phylogenetic relationships of mycobacterial species were based largely on 16S rRNA sequencing; however, the increased availability and cost-effectiveness of whole-genome sequencing (WGS) has led to more mycobacterial species being sequenced, as well as more robust comparative genomics on which to base phylogenetic relationships. Phylogenies based on WGS data are generally in concordance with those based on 16S rRNA data. Namely, rapid-growing mycobacteria (“rapid growers”) and slow-growing mycobacteria (“slow growers”) are clearly separated, with the rapid growers being more ancestral relative to the slow growers and the slow growers (i.e., more commonly pathogenic mycobacteria) being a distinct evolutionary branch. Where differences do exist between WGS- and 16S rRNA-based phylogenies is in regard to M. leprae (36). Some WGS-based phylogenetic trees resemble those of 16S rRNA-based trees, placing M. leprae more closely related to the NTM species M. avium (37), whereas others have placed it as a close sister clade to M. tuberculosis (38, 39). Many of these differences arise depending on the type of analysis used and the stringency of the thresholds used for software packages. Although these differences will likely continue to be a topic of debate, the overall structure of the mycobacterial phylogenetic tree is relatively accepted and well characterized (38).
Whole-genome sequencing of NTM has increased our understanding of the genetic evolution of the mycobacterial family. The recently sequenced Mycobacterium terrae complex has been placed as an intermediate group between rapid- and slow-growing mycobacteria, representing an evolutionary link for the growth rate shift (38). High gene turnover rates have been observed in the evolutionary timeline of NTM (39), and the number of 1:1 orthologs between sequenced NTM genomes appears to be very low (38). Even though NTM species may share a similar number of genes, the presence of species-specific genes appears to be very high and diverse (40). The role of horizontal gene transfer in mycobacteria is debated, with some studies reporting a very low impact on mycobacterial evolution, evidenced by the low number of transposable elements present in mycobacterial genomes (38). Other studies, however, report a larger role for horizontal gene transfer based on sequence similarities and genomic islands between species (41). A recent annotation of multiple NTM genomes demonstrated that a majority of predicted genes could not be assigned a specific function (38).
Importantly, mycobacterial genome comparisons have led to a novel model regarding the evolutionary divergence of NTM and obligate pathogenic mycobacteria: due to the relatively small genome sizes of the obligate pathogens M. leprae and M. tuberculosis, the evolution of human pathogenicity corresponded to a large loss of ancestral genes with a gain of several new genes more adapted to an obligate intracellular lifestyle (40, 42). M. leprae and M. tuberculosis are remarkably distinct from one another in terms of evolution: whereas M. leprae evolved ∼14 million years ago (43), the evolution of human-adapted M. tuberculosis was more recent, ∼10 thousand to 70 thousand years ago (44), and it was possibly dispersed to New World populations via migratory seals and sea lions (45). What M. leprae and M. tuberculosis have in common, however, is that in contrast to nearly all NTM, which must survive in soil and aquatic environments outside a living host, they do not exist as free-living organisms in nature. Based on their biologies, M. leprae and M. tuberculosis would therefore have less need for gene regulation and adaptive responses to the environment than NTM that are found in the environment. In addition to having smaller genome sizes, pathogenic mycobacteria also have genes enriched in DNA repair and recombination mechanisms, while opportunistic pathogens have an enrichment in membrane transport genes which aid in nutrient uptake and drug efflux (40). The genes responsible for energy metabolism appear to be more NTM species specific, owing to the fact that each NTM species must adapt to its own soil or aquatic environment (40).
THE AQUATIC LIVES OF NONTUBERCULOUS MYCOBACTERIA
Aquatic environments can be significant NTM reservoirs, and water is increasingly recognized as an important NTM transmission medium. The aquatic environments in which NTM reside can be as small as a showerhead or as large as a watershed (46). The aquatic microenvironments in which NTM have been found are largely of human origin: NTM-containing water supplies have been responsible for outbreaks of NTM disease in hospitals (47, 48), as well as for outbreaks among footbath customers (49, 50), metal workers (51, 52), and alternative-medicine users (53). Heating water can be insufficient to clear NTM from water supplies, as several NTM species resist water temperatures up to 55°C (a temperature at which Legionella pneumophila is heat susceptible), whereas other NTM species can resist water temperatures up to 70°C, with Mycobacterium xenopi being the most thermoresistant (54). Environmental sampling revealed that mycobacteria comprised 1/3 of all microbes in the water of an indoor pool and 8/10 of all microbes in the surrounding bioaerosol (55). The aquatic macroenvironments in which NTM have been found include those of human origin (e.g., municipal water supplies) (56–59) but also natural watersheds (46) and lakes (60, 61). Data from experimental models suggest that the water flow in aquatic micro- and macroenvironments is insufficient to prevent the formation of NTM biofilms (62), which facilitate NTM survival in numerous hostile environments (63).
Not surprisingly, NTM are well adapted to their aquatic niche and appear to have minimal requirements to survive as free-living organisms in water, as evidenced by the ability of NTM to survive in sterile deionized water for over 1 year (64), as well as in lake water under hypoxic conditions (60, 61). In their natural environment, NTM are often presumed to exist as free-living or biofilm-associated organisms; however, this seems at odds with the concept of mycobacteria being intramacrophagic pathogens. For this reason, increasing attention is being paid to the roles of environmental amoebas in sustaining NTM in the environment (28). The concept of intra-amoebae bacteria being a source of disease is accepted in the field of Legionnaires' disease and is increasingly recognized in the field of mycobacteriology since the first demonstrations that NTM-amoeba interactions promote NTM virulence (65, 66).
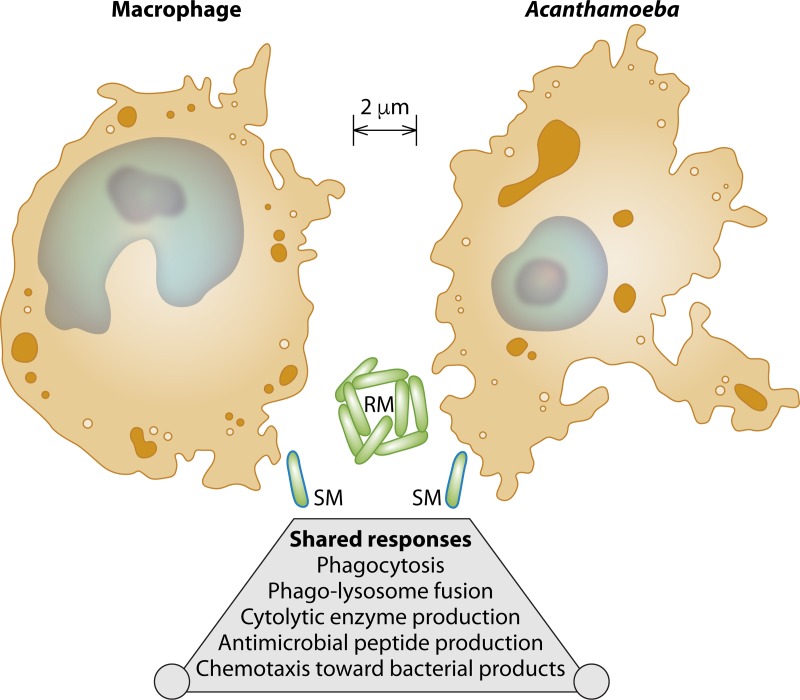
Acanthamoeba is a genus of free-living amoebas that have been found in a wide range of soil and aquatic environments, including city dust (67), bottled mineral water (68), eyewash stations (69), the waterlines of dental water flossers (70), and Antarctic soil and water (71). Acanthamoeba spp. are bacterivores, and their life cycle involves transitioning between encysted and trophozoite forms (72). Whereas the encysted form of Acanthamoeba resembles a capsule with a coarse and furrowed surface, the trophozoite form is oddly reminiscent of vertebrate macrophages, in terms of both morphology and cell biology (Fig. 1). Specifically, Acanthamoeba trophozoites chemotax toward bacterial products (73), release antimicrobial peptides (74), and engulf bacteria in phagosomes that subsequently undergo phagolysosomal fusion (75–77). Acanthamoeba trophozoites also affect bacterial lysis by expressing a broad range of cytolytic enzymes: lysozyme (78), serine and cysteine proteases that are active over a wide pH range (79), α- and β-glucosidases, β-galactosidase, β-N-acetylglucosaminidase, amylase, and peptidase (80). Also, similar to the case for vertebrate macrophages, the extent to which Acanthamoeba phagolysosomal fusion kills mycobacteria is mycobacterial species dependent (72, 81).
FIG 1.
Macrophages and Acanthamoeba species are similar in their morphology and responses to bacteria. Depicted are the cell morphologies and relative sizes of three cell types (macrophages, Acanthamoeba, and Mycobacterium), based on the electron microscopy images and measurements of Lei et al. (macrophages) (129), Gonzalez-Robles et al. (Acanthamoeba) (72), and Schoonmaker et al. (Mycobacterium) (130). Macrophages and Acanthamoeba spp. resemble one another in size and cell morphology: both are eukaryotes with cytoplasm (orange), intracellular vesicles (brown), a nucleus (light blue), and a nucleolus (dark blue). Smooth-morphotype (SM) mycobacteria (light green) can exist as singular bacilli due to the presence of surface glycopeptidolipids (GPLs) (dark blue outline); rough-morphotype (RM) mycobacteria (light green) can exist as multicellular aggregates due to loss of GPL transport and synthesis (represented by the absence of a dark blue outline). In M. abscessus, these GPL transport and synthesis genes include mps1, mbtH, and mmpL4b (120); however, there are also GPL-independent mechanisms underlying the smooth morphotype, as this phenotype and SM→RM transitions occur in NTM species that do not produce GPL (114). Upon encountering bacteria in their respective environments, macrophages and Acanthamoeba have shared responses: phagocytosis, phagolysosome fusion, production of cytolytic enzymes and antimicrobial peptides, and chemotaxis toward bacterial products.
Numerous NTM species can enter Acanthamoeba during the trophozoite phase and survive within cysts for prolonged periods (82). Specific Acanthamoeba species that are known to be parasitized by NTM are Acanthamoeba griffini, Acanthamoeba polyphaga, and Acanthamoeba castellanii. Acanthamoeba griffini is a halophilic species that was first isolated from seawater (83) and is now known to inhabit marine environments (84), hot springs (85), air conditioners (86), and contact lenses (79). In contrast, A. polyphaga is freshwater associated and was first isolated from a pond (83). NTM residing within A. polyphaga cysts can survive for over 2 weeks and are more resistant to the germicidal effects of chlorine (82). Acanthamoeba castellanii resides in soil, marine, and freshwater environments (83, 87). For any aquatic environment, the relative proportion of free versus amoeba-associated NTM is unknown; however, there are numerous reports demonstrating the ability of NTM to persist within Acanthamoeba in experimental systems (65, 81, 82, 88–91). These experimental data support the possibility that amoebal encystation allows NTM to persist in aquatic environments for extended periods. This is important, because growing M. avium in amoebas enhances their infectivity and virulence in a mouse infection model (compared to M. avium grown in the absence of amoebas) (65, 91).
Researchers are leveraging the possibility of an Acanthamoeba host to better understand how differences in bacterial genomes influence NTM survival. Acanthamoeba coculture systems have been used to collect virulent NTM isolates, identify conserved pathogenesis mechanisms, and make predictions regarding NTM transmission (81, 92–94). Just as the genetic tractability of another amoeba (Dictyostelium discoideum) has been used to discover how tubercular mycobacteria exit a cell (95), so too can novel methods of NTM transposon mutagenesis (96–99) be used to identify genes that augment or inhibit intra-Acanthamoeba survival. Once such genes have been identified, their presence in the genomes of virulent NTM isolates can be assayed and used to support a model wherein intra-amoeba survival is a prerequisite for intramacrophage survival. Collectively, existing data from Acanthamoeba coculture models support an intriguing concept that has already been applied to Legionella (100): that the resemblance of Acanthamoeba trophozoites to macrophages may naturally select for NTM bacilli that are adapted to survive within macrophages, thus increasing their fitness for intramacrophage survival in human hosts.
THE SMOOTH AND ROUGH LIVES OF NONTUBERCULOUS MYCOBACTERIA
Research on mycobacterial virulence factors has understandably focused on M. tuberculosis for many years; however, the emergence of NTM as globally significant pathogens (101–106) and their increasing prevalence in immunocompetent hosts (107, 108) have given rise to the need to further understand pathogenesis in a broader range of mycobacterial species. An excellent review of M. tuberculosis virulence factors and their corresponding roles in NTM was recently published (109). While the small genome size, large number of pseudogenes, and unculturable nature of M. leprae make it particularly hard to study, research elucidating the virulence factors of the culturable NTM species M. marinum and M. ulcerans is advancing at a high rate. Studying the virulence mechanisms of the more pathogenic species is a good starting point for deciphering pathogenesis in opportunistically pathogenic NTM, but previously mentioned genomic studies have found that a large number of species-specific genes are present in individual mycobacterial genomes, which may leave important gaps in our knowledge of novel virulence mechanisms. These genes are prime targets for functional studies so that we may better understand their place in the survival and possibly the pathogenic potential of mycobacteria. In order to further enhance our understanding of NTM in human infection, it is imperative that the species that are increasingly implicated in human disease, such as M. avium and M. abscessus, are studied further to define the functions of genes that are currently unclear.
In addition to potential differences in virulence factors, another important difference between M. tuberculosis and NTM species is the presence of dynamic colony morphotypes among NTM isolates. When plated on agar media, several NTM isolates form colonies with more than one morphology; the two most common morphologies are referred to as the smooth morphotype (SM) and the rough morphotype (RM). The SM is characterized by a uniform and glossy appearance, while the RM is characterized by an irregular, dry, and corded appearance (110). The manifestation of NTM species as SM or RM distinguishes NTM from their tuberculous counterparts, the colonies of which are predominantly rough. NTM species that exhibit both SM and RM colonies include Mycobacterium bolletii (111), M. kansasii (112), M. abscessus (113), M. vaccae (114), and M. avium (115). Mycobacterium avium colony description requires the use of additional qualifiers, as this species exhibits smooth opaque (SM-opaque), smooth transparent (SM-transparent), and RM colonies (116). Although SM and RM colony characteristics are stable over serial passages, spontaneous shifts of SM to RM and RM to SM have been reported (114, 117–119); even so, the SM is often treated in the literature as being “wild type.” A genetic basis for the SM→RM transition is the lost expression of genes that promote glycopeptidolipid (GPL) synthesis and transport (111, 120). GPLs are amphiphilic molecules that localize to the outermost layer of the mycobacterial cell wall (1). GPL localization to the mycobacterial cell wall is facilitated by several genes, including mps1, mbtH, and mmpL4b (120). In the presence of surface GPL, a dividing mycobacterium results in daughter cells that physically dissociate. In the absence of surface GPL, a dividing mycobacterium results in daughter cells that are attached end to end. After successive divisions, these attached cells form structures that resemble cords at microscopic and macroscopic levels (114, 118). However, different species without GPL can also form SM and RM, implicating other mechanisms and membrane-associated molecules in the morphotype (114). In addition to causing RM to form corded colonies on solid media, the lack of GPL on RM causes multibacillary aggregates in liquid media (121–123). Aggregate formation can impact NTM virulence by altering the phagolysosome composition and integrity (124–126), and the RMs of M. abscessus, M. avium, and M. kansasii are more virulent in experimental models (112, 127, 128). Collectively, the literature cited above demonstrates that SM and RM are two physical manifestations of a single NTM species and reflect the presence (SM) or absence (RM) of surface-associated GPL in the original CFU, as a result of mps1, mbtH, and mmpL4b gene activity.
CONCLUSION
Nontuberculous mycobacteria are a large group of organisms that include species able to colonize human epithelia and cause disease, as well as saprophytic species that are omnipresent in soil and aquatic environments. The taxonomic and evolutionary patterns elucidated by WGS and subsequent phylogenomic analysis reviewed here have allowed us a more complete picture of the genetic similarities between different mycobacterial species. These analyses have also shed light on some interesting evolutionary characteristics of the Mycobacterium family, such as gene gain and loss dynamics and the immense diversity and prevalence of species-specific genes with currently unknown functions. Further study of these genes presents us with an opportunity to advance our understanding of how these bacteria colonize and may cause disease. The already well-established ability of NTM to exist in many different environments is a testament to their remarkable adaptability, and their interesting association with environmental amoebas is giving rise to very exciting ideas about the evolution of mycobacterial pathogenesis and their ability to escape macrophage killing in NTM disease. This provides a thought-provoking model of how these bacteria, and perhaps even other intracellular pathogens, may have adapted to interact with the advanced immune systems of humans and potentially cause disease. We have also summarized the current research on the unique ability of NTM to form distinct colony morphotypes that seem to differ in their virulence and pathogenicity traits, as well as their ability to form biofilms and exhibit sliding motility, which may play important roles not only in the environmental survival of NTM but also in their ability to infect human hosts. The importance of continuing research on NTM and understanding the vast number of species belonging to this group is highlighted by the increasing emergence of facultative pathogenic mycobacteria and the increasing prevalence of NTMI in immunocompetent as well as immunocompromised humans.
ACKNOWLEDGMENTS
We thank Samuel Marshall (Department of Microbiology and Immunology, Medical College of Wisconsin, Milwaukee, WI) for his input and feedback during the preparation of the manuscript. We also thank Patrick Lane for his artistic services.
The work in our laboratory is supported by NIH R01 AI121212.
REFERENCES
- 1.Robinson RT, Huppler AR. 2017. The Goldilocks model of immune symbiosis with Mycobacteria and Candida colonizers. Cytokine 97:49–65. doi: 10.1016/j.cyto.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh AK, Marak RS, Maurya AK, Das M, Nag VL, Dhole TN. 2015. Mixed cutaneous infection caused by Mycobacterium szulgai and Mycobacterium intermedium in a healthy adult female: a rare case report. Case Rep Dermatol Med 2015:607519. doi: 10.1155/2015/607519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torkko P, Suomalainen S, Iivanainen E, Suutari M, Paulin L, Rudback E, Tortoli E, Vincent V, Mattila R, Katila ML. 2001. Characterization of Mycobacterium bohemicum isolated from human, veterinary, and environmental sources. J Clin Microbiol 39:207–211. doi: 10.1128/JCM.39.1.207-211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace RJ Jr, Nash DR, Tsukamura M, Blacklock ZM, Silcox VA. 1988. Human disease due to Mycobacterium smegmatis. J Infect Dis 158:52–59. doi: 10.1093/infdis/158.1.52. [DOI] [PubMed] [Google Scholar]
- 5.Alvarado-Esquivel C, Garcia-Corral N, Carrero-Dominguez D, Enciso-Moreno JA, Gurrola-Morales T, Portillo-Gomez L, Rossau R, Mijs W. 2009. Molecular analysis of Mycobacterium isolates from extrapulmonary specimens obtained from patients in Mexico. BMC Clin Pathol 9:1. doi: 10.1186/1472-6890-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon RE, Smith MM. 1953. Rapidly growing, acid fast bacteria. I. Species' descriptions of Mycobacterium phlei Lehmann and Neumann and Mycobacterium smegmatis (Trevisan) Lehmann and Neumann. J Bacteriol 66:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Springer B, Wu WK, Bodmer T, Haase G, Pfyffer GE, Kroppenstedt RM, Schroder KH, Emler S, Kilburn JO, Kirschner P, Telenti A, Coyle MB, Bottger EC. 1996. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol 34:1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirschner P, Teske A, Schroder KH, Kroppenstedt RM, Wolters J, Bottger EC. 1992. Mycobacterium confluentis sp. nov. Int J Syst Bacteriol 42:257–262. doi: 10.1099/00207713-42-2-257. [DOI] [PubMed] [Google Scholar]
- 9.Koukila-Kahkola P, Springer B, Bottger EC, Paulin L, Jantzen E, Katila ML. 1995. Mycobacterium branderi sp. nov., a new potential human pathogen. Int J Syst Bacteriol 45:549–553. doi: 10.1099/00207713-45-3-549. [DOI] [PubMed] [Google Scholar]
- 10.Lumb R, Goodwin A, Ratcliff R, Stapledon R, Holland A, Bastian I. 1997. Phenotypic and molecular characterization of three clinical isolates of Mycobacterium interjectum. J Clin Microbiol 35:2782–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meier A, Kirschner P, Schroder KH, Wolters J, Kroppenstedt RM, Bottger EC. 1993. Mycobacterium intermedium sp. nov. Int J Syst Bacteriol 43:204–209. doi: 10.1099/00207713-43-2-204. [DOI] [PubMed] [Google Scholar]
- 12.Reischl U, Emler S, Horak Z, Kaustova J, Kroppenstedt RM, Lehn N, Naumann L. 1998. Mycobacterium bohemicum sp. nov., a new slow-growing scotochromogenic mycobacterium. Int J Syst Bacteriol 48:1349–1355. doi: 10.1099/00207713-48-4-1349. [DOI] [PubMed] [Google Scholar]
- 13.van Ingen J, Boeree MJ, Stals FS, Pitz CC, Rooijmans-Rietjens JJ, van der Zanden AG, Dekhuijzen PN, van Soolingen D. 2007. Clinical Mycobacterium conspicuum isolation from two immunocompetent patients in The Netherlands. J Clin Microbiol 45:4075–4076. doi: 10.1128/JCM.00867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe J, Turenne C, Alfa M, Harding G, Thibert L, Kabani A. 2000. Mycobacterium branderi from both a hand infection and a case of pulmonary disease. J Clin Microbiol 38:3896–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pontiroli A, Khera TT, Oakley BB, Mason S, Dowd SE, Travis ER, Erenso G, Aseffa A, Courtenay O, Wellington EM. 2013. Prospecting environmental mycobacteria: combined molecular approaches reveal unprecedented diversity. PLoS One 8:e68648. doi: 10.1371/journal.pone.0068648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Zhai Y, Cao L, Tan H, Zhang R. 2016. Illumina-based analysis of core actinobacteriome in roots, stems, and grains of rice. Microbiol Res 190:12–18. doi: 10.1016/j.micres.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Rook GA, Brunet LR. 2005. Microbes, immunoregulation, and the gut. Gut 54:317–320. doi: 10.1136/gut.2004.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunt JR, Martinelli R, Adams VC, Rook GA, Brunet LR. 2005. Intragastric administration of Mycobacterium vaccae inhibits severe pulmonary allergic inflammation in a mouse model. Clin Exp Allergy 35:685–690. doi: 10.1111/j.1365-2222.2005.02239.x. [DOI] [PubMed] [Google Scholar]
- 19.Matthews DM, Jenks SM. 2013. Ingestion of Mycobacterium vaccae decreases anxiety-related behavior and improves learning in mice. Behav Processes 96:27–35. doi: 10.1016/j.beproc.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien ME, Anderson H, Kaukel E, O'Byrne K, Pawlicki M, Von Pawel J, Reck M, SR-ON-12 Study Group. 2004. SRL172 (killed Mycobacterium vaccae) in addition to standard chemotherapy improves quality of life without affecting survival, in patients with advanced non-small-cell lung cancer: phase III results. Ann Oncol 15:906–914. doi: 10.1093/annonc/mdh220. [DOI] [PubMed] [Google Scholar]
- 21.Mayer EA, Knight R, Mazmanian SK, Cryan JF, Tillisch K. 2014. Gut microbes and the brain: paradigm shift in neuroscience. J Neurosci 34:15490–15496. doi: 10.1523/JNEUROSCI.3299-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wayne LG, Sramek HA. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin Microbiol Rev 5:1–25. doi: 10.1128/CMR.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inderlied CB, Kemper CA, Bermudez LE. 1993. The Mycobacterium avium complex. Clin Microbiol Rev 6:266–310. doi: 10.1128/CMR.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolinsky E. 1979. Nontuberculous mycobacteria and associated diseases. Am Rev Respir Dis 119:107–159. [DOI] [PubMed] [Google Scholar]
- 25.Wolinsky E, Rynearson TK. 1968. Mycobacteria in soil and their relation to disease-associated strains. Am Rev Respir Dis 97:1032–1037. [DOI] [PubMed] [Google Scholar]
- 26.Yajko DM, Chin DP, Gonzalez PC, Nassos PS, Hopewell PC, Reingold AL, Horsburgh CR Jr, Yakrus MA, Ostroff SM, Hadley WK. 1995. Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J Acquir Immune Defic Syndr Hum Retrovirol 9:176–182. [PubMed] [Google Scholar]
- 27.Delafont V, Mougari F, Cambau E, Joyeux M, Bouchon D, Hechard Y, Moulin L. 2014. First evidence of amoebae-mycobacteria association in drinking water network. Environ Sci Technol 48:11872–11882. doi: 10.1021/es5036255. [DOI] [PubMed] [Google Scholar]
- 28.Drancourt M. 2014. Looking in amoebae as a source of mycobacteria. Microb Pathog 77:119–124. doi: 10.1016/j.micpath.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Thomson R, Tolson C, Carter R, Coulter C, Huygens F, Hargreaves M. 2013. Isolation of nontuberculous mycobacteria (NTM) from household water and shower aerosols in patients with pulmonary disease caused by NTM. J Clin Microbiol 51:3006–3011. doi: 10.1128/JCM.00899-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Reyn CF, Maslow JN, Barber TW, Falkinham JO III, Arbeit RD. 1994. Persistent colonisation of potable water as a source of Mycobacterium avium infection in AIDS. Lancet 343:1137–1141. doi: 10.1016/S0140-6736(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 31.De Groote MA, Pace NR, Fulton K, Falkinham JO 3rd. 2006. Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl Environ Microbiol 72:7602–7606. doi: 10.1128/AEM.00930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honda JR, Bernhard JN, Chan ED. 2015. Natural disasters and nontuberculous mycobacteria: a recipe for increased disease? Chest 147:304–308. doi: 10.1378/chest.14-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marks J. 1964. Nomenclature of the mycobacteria. Am Rev Respir Dis 90:278. [Google Scholar]
- 34.Rahman SA, Singh Y, Kohli S, Ahmad J, Ehtesham NZ, Tyagi AK, Hasnain SE. 2014. Comparative analyses of nonpathogenic, opportunistic, and totally pathogenic mycobacteria reveal genomic and biochemical variabilities and highlight the survival attributes of Mycobacterium tuberculosis. mBio 5:e02020. doi: 10.1128/mBio.02020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes BA. 2017. Mycobacterial taxonomy. J Clin Microbiol 55:380–383. doi: 10.1128/JCM.01287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith SE, Showers-Corneli P, Dardenne CN, Harpending HH, Martin DP, Beiko RG. 2012. Comparative genomic and phylogenetic approaches to characterize the role of genetic recombination in mycobacterial evolution. PLoS One 7:e50070. doi: 10.1371/journal.pone.0050070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biet F, Boschiroli ML, Thorel MF, Guilloteau LA. 2005. Zoonotic aspects of Mycobacterium bovis and Mycobacterium avium-intracellulare complex (MAC). Vet Res 36:411–436. doi: 10.1051/vetres:2005001. [DOI] [PubMed] [Google Scholar]
- 38.Fedrizzi T, Meehan CJ, Grottola A, Giacobazzi E, Fregni Serpini G, Tagliazucchi S, Fabio A, Bettua C, Bertorelli R, De Sanctis V, Rumpianesi F, Pecorari M, Jousson O, Tortoli E, Segata N. 2017. Genomic characterization of nontuberculous mycobacteria. Sci Rep 7:45258. doi: 10.1038/srep45258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Librado P, Vieira FG, Sanchez-Gracia A, Kolokotronis SO, Rozas J. 2014. Mycobacterial phylogenomics: an enhanced method for gene turnover analysis reveals uneven levels of gene gain and loss among species and gene families. Genome Biol Evol 6:1454–1465. doi: 10.1093/gbe/evu117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malhotra S, Vedithi SC, Blundell TL. 2017. Decoding the similarities and differences among mycobacterial species. PLoS Negl Trop Dis 11:e0005883. doi: 10.1371/journal.pntd.0005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reva O, Korotetskiy I, Ilin A. 2015. Role of the horizontal gene exchange in evolution of pathogenic mycobacteria. BMC Evol Biol 15(Suppl 1):S2. doi: 10.1186/1471-2148-15-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 43.Singh P, Benjak A, Schuenemann VJ, Herbig A, Avanzi C, Busso P, Nieselt K, Krause J, Vera-Cabrera L, Cole ST. 2015. Insight into the evolution and origin of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis. Proc Natl Acad Sci U S A 112:4459–4464. doi: 10.1073/pnas.1421504112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gagneux S. 2012. Host-pathogen coevolution in human tuberculosis. Philos Trans R Soc Lond B Biol Sci 367:850–859. doi: 10.1098/rstb.2011.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bos KI, Harkins KM, Herbig A, Coscolla M, Weber N, Comas I, Forrest SA, Bryant JM, Harris SR, Schuenemann VJ, Campbell TJ, Majander K, Wilbur AK, Guichon RA, Wolfe Steadman DL, Cook DC, Niemann S, Behr MA, Zumarraga M, Bastida R, Huson D, Nieselt K, Young D, Parkhill J, Buikstra JE, Gagneux S, Stone AC, Krause J. 2014. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature 514:494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lipner EM, Knox D, French J, Rudman J, Strong M, Crooks JL. 2017. A geospatial epidemiologic analysis of nontuberculous mycobacterial infection: an ecological study in Colorado. Ann Am Thorac Soc 14:1523–1532. doi: 10.1513/AnnalsATS.201701-081OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conger NG, O'Connell RJ, Laurel VL, Olivier KN, Graviss EA, Williams-Bouyer N, Zhang Y, Brown-Elliott BA, Wallace RJ Jr. 2004. Mycobacterium simae outbreak associated with a hospital water supply. Infect Control Hosp Epidemiol 25:1050–1055. doi: 10.1086/502342. [DOI] [PubMed] [Google Scholar]
- 48.Meyers H, Brown-Elliott BA, Moore D, Curry J, Truong C, Zhang Y, Wallace RJ Jr. 2002. An outbreak of Mycobacterium chelonae infection following liposuction. Clin Infect Dis 34:1500–1507. doi: 10.1086/340399. [DOI] [PubMed] [Google Scholar]
- 49.Vugia DJ, Jang Y, Zizek C, Ely J, Winthrop KL, Desmond E. 2005. Mycobacteria in nail salon whirlpool footbaths, California. Emerg Infect Dis 11:616–618. doi: 10.3201/eid1104.040936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winthrop KL, Abrams M, Yakrus M, Schwartz I, Ely J, Gillies D, Vugia DJ. 2002. An outbreak of mycobacterial furunculosis associated with footbaths at a nail salon. N Engl J Med 346:1366–1371. doi: 10.1056/NEJMoa012643. [DOI] [PubMed] [Google Scholar]
- 51.Wallace RJ Jr, Zhang Y, Wilson RW, Mann L, Rossmoore H. 2002. Presence of a single genotype of the newly described species Mycobacterium immunogenum in industrial metalworking fluids associated with hypersensitivity pneumonitis. Appl Environ Microbiol 68:5580–5584. doi: 10.1128/AEM.68.11.5580-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson RW, Steingrube VA, Bottger EC, Springer B, Brown-Elliott BA, Vincent V, Jost KC Jr, Zhang Y, Garcia MJ, Chiu SH, Onyi GO, Rossmoore H, Nash DR, Wallace RJ Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int J Syst Evol Microbiol 51:1751–1764. doi: 10.1099/00207713-51-5-1751. [DOI] [PubMed] [Google Scholar]
- 53.Galil K, Miller LA, Yakrus MA, Wallace RJ Jr, Mosley DG, England B, Huitt G, McNeil MM, Perkins BA. 1999. Abscesses due to Mycobacterium abscessus linked to injection of unapproved alternative medication. Emerg Infect Dis 5:681–687. doi: 10.3201/eid0505.990509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulze-Robbecke R, Buchholtz K. 1992. Heat susceptibility of aquatic mycobacteria. Appl Environ Microbiol 58:1869–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angenent LT, Kelley ST, St Amand A, Pace NR, Hernandez MT. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc Natl Acad Sci U S A 102:4860–4865. doi: 10.1073/pnas.0501235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaerewijck MJ, Huys G, Palomino JC, Swings J, Portaels F. 2005. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiol Rev 29:911–934. doi: 10.1016/j.femsre.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Torvinen E, Suomalainen S, Lehtola MJ, Miettinen IT, Zacheus O, Paulin L, Katila ML, Martikainen PJ. 2004. Mycobacteria in water and loose deposits of drinking water distribution systems in Finland. Appl Environ Microbiol 70:1973–1981. doi: 10.1128/AEM.70.4.1973-1981.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, Vincent V. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl Environ Microbiol 68:5318–5325. doi: 10.1128/AEM.68.11.5318-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Covert TC, Rodgers MR, Reyes AL, Stelma GN Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl Environ Microbiol 65:2492–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kirschner RA, Parker BC, Falkinham JO. 1999. Humic and fulvic acids stimulate the growth of Mycobacterium avium. FEMS Microbiol Ecol 30:327–332. doi: 10.1111/j.1574-6941.1999.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 61.Brooks RW, George KL, Parker BC, Falkinham JO III, Gruff H. 1984. Recovery and survival of nontuberculous mycobacteria under various growth and decontamination conditions. Can J Microbiol 30:1112–1117. doi: 10.1139/m84-174. [DOI] [PubMed] [Google Scholar]
- 62.Hall-Stoodley L, Lappin-Scott H. 1998. Biofilm formation by the rapidly growing mycobacterial species Mycobacterium fortuitum. FEMS Microbiol Lett 168:77–84. doi: 10.1111/j.1574-6968.1998.tb13258.x. [DOI] [PubMed] [Google Scholar]
- 63.Richards JP, Ojha AK. 2014. Mycobacterial biofilms. Microbiol Spectr doi: 10.1128/microbiolspec.MGM2-0004-2013. [DOI] [PubMed] [Google Scholar]
- 64.Archuleta RJ, Mullens P, Primm TP. 2002. The relationship of temperature to desiccation and starvation tolerance of the Mycobacterium avium complex. Arch Microbiol 178:311–314. doi: 10.1007/s00203-002-0455-x. [DOI] [PubMed] [Google Scholar]
- 65.Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun 65:3759–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tosetti N, Croxatto A, Greub G. 2014. Amoebae as a tool to isolate new bacterial species, to discover new virulence factors and to study the host-pathogen interactions. Microb Pathog 77:125–130. doi: 10.1016/j.micpath.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 67.Rivera F, Roy-Ocotla G, Rosas I, Ramirez E, Bonilla P, Lares F. 1987. Amoebae isolated from the atmosphere of Mexico City and environs. Environ Res 42:149–154. doi: 10.1016/S0013-9351(87)80016-6. [DOI] [PubMed] [Google Scholar]
- 68.Rivera F, Galvan M, Robles E, Leal P, Gonzalez L, Lacy AM. 1981. Bottled mineral waters polluted by protozoa in Mexico. J Protozool 28:54–56. doi: 10.1111/j.1550-7408.1981.tb02803.x. [DOI] [PubMed] [Google Scholar]
- 69.Paszko-Kolva C, Yamamoto H, Shahamat M, Sawyer TK, Morris G, Colwell RR. 1991. Isolation of amoebae and Pseudomonas and Legionella spp. from eyewash stations. Appl Environ Microbiol 57:163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbeau J, Buhler T. 2001. Biofilms augment the number of free-living amoebae in dental unit waterlines. Res Microbiol 152:753–760. doi: 10.1016/S0923-2508(01)01256-6. [DOI] [PubMed] [Google Scholar]
- 71.Brown TJ, Cursons RT, Keys EA. 1982. Amoebae from antarctic soil and water. Appl Environ Microbiol 44:491–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonzalez-Robles A, Salazar-Villatoro L, Omana-Molina M, Reyes-Batlle M, Martin-Navarro CM, Lorenzo-Morales J. 2014. Morphological features and in vitro cytopathic effect of Acanthamoeba griffini trophozoites isolated from a clinical case. J Parasitol Res 2014:256310. doi: 10.1155/2014/256310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schuster FL, Levandowsky M. 1996. Chemosensory responses of Acanthamoeba castellanii: visual analysis of random movement and responses to chemical signals. J Eukaryot Microbiol 43:150–158. doi: 10.1111/j.1550-7408.1996.tb04496.x. [DOI] [PubMed] [Google Scholar]
- 74.Michalek M, Sonnichsen FD, Wechselberger R, Dingley AJ, Hung CW, Kopp A, Wienk H, Simanski M, Herbst R, Lorenzen I, Marciano-Cabral F, Gelhaus C, Gutsmann T, Tholey A, Grotzinger J, Leippe M. 2013. Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba. Nat Chem Biol 9:37–42. doi: 10.1038/nchembio.1116. [DOI] [PubMed] [Google Scholar]
- 75.Oates PJ, Touster O. 1980. In vitro fusion of Acanthamoeba phagolysosomes. III. Evidence that cyclic nucleotides and vacuole subpopulations respectively control the rate and the extent of vacuole fusion in Acanthamoeba homogenates. J Cell Biol 85:804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oates PJ, Touster O. 1978. In vitro fusion of Acanthamoeba phagolysosomes. II. Quantitative characterization of in vitro vacuole fusion by improved electron microscope and new light microscope techniques. J Cell Biol 79:217–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oates PJ, Touster O. 1976. In vitro fusion of Acanthamoeba phagolysosomes. I. Demonstration and quantitation of vacuole fusion in Acanthamoeba homogenates. J Cell Biol 68:319–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Drozanski W, Drozanska D, Wicinska M. 1984. The cell wall of the obligate intracellular bacterial parasite of small free-living amoebae. I. Morphology and chemical composition of the rigid layer and peptidoglycan. Acta Microbiol Pol 33:195–206. [PubMed] [Google Scholar]
- 79.Heredero-Bermejo I, Criado-Fornelio A, De Fuentes I, Soliveri J, Copa-Patino JL, Perez-Serrano J. 2015. Characterization of a human-pathogenic Acanthamoeba griffini isolated from a contact lens-wearing keratitis patient in Spain. Parasitology 142:363–373. doi: 10.1017/S0031182014001140. [DOI] [PubMed] [Google Scholar]
- 80.Rosenthal S, Reed EJ, Weisman RA. 1969. Effect of lytic enzymes of Acanthamoeba castellanii on bacterial cell walls. J Bacteriol 98:182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bouam A, Ghigo E, Drancourt M. 2018. Intra-amoebal killing of Mycobacterium ulcerans by Acanthamoeba griffini: a co-culture model. Microb Pathog 114:1–7. doi: 10.1016/j.micpath.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 82.Adekambi T, Ben Salah S, Khlif M, Raoult D, Drancourt M. 2006. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl Environ Microbiol 72:5974–5981. doi: 10.1128/AEM.03075-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marciano-Cabral F, Cabral G. 2003. Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 16:273–307. doi: 10.1128/CMR.16.2.273-307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Todd CD, Reyes-Batlle M, Pinero JE, Martinez-Carretero E, Valladares B, Streete D, Lorenzo-Morales J, Lindo JF. 2015. Isolation and molecular characterization of Acanthamoeba genotypes in recreational and domestic water sources from Jamaica, West Indies. J Water Health 13:909–919. doi: 10.2166/wh.2015.232. [DOI] [PubMed] [Google Scholar]
- 85.Solgi R, Niyyati M, Haghighi A, Taghipour N, Tabaei SJ, Eftekhar M, Nazemalhosseini Mojarad E. 2012. Thermotolerant Acanthamoeba spp. isolated from therapeutic hot springs in Northwestern Iran. J Water Health 10:650–656. doi: 10.2166/wh.2012.032. [DOI] [PubMed] [Google Scholar]
- 86.Chan LL, Mak JW, Low YT, Koh TT, Ithoi I, Mohamed SM. 2011. Isolation and characterization of Acanthamoeba spp. from air-conditioners in Kuala Lumpur, Malaysia. Acta Trop 117:23–30. doi: 10.1016/j.actatropica.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anderson IJ, Watkins RF, Samuelson J, Spencer DF, Majoros WH, Gray MW, Loftus BJ. 2005. Gene discovery in the Acanthamoeba castellanii genome. Protist 156:203–214. doi: 10.1016/j.protis.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 88.Wheat WH, Casali AL, Thomas V, Spencer JS, Lahiri R, Williams DL, McDonnell GE, Gonzalez-Juarrero M, Brennan PJ, Jackson M. 2014. Long-term survival and virulence of Mycobacterium leprae in amoebal cysts. PLoS Negl Trop Dis 8:e3405. doi: 10.1371/journal.pntd.0003405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mba Medie F, Ben Salah I, Henrissat B, Raoult D, Drancourt M. 2011. Mycobacterium tuberculosis complex mycobacteria as amoeba-resistant organisms. PLoS One 6:e20499. doi: 10.1371/journal.pone.0020499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lahiri R, Krahenbuhl JL. 2008. The role of free-living pathogenic amoeba in the transmission of leprosy: a proof of principle. Lepr Rev 79:401–409. [PubMed] [Google Scholar]
- 91.Steinert M, Birkness K, White E, Fields B, Quinn F. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl Environ Microbiol 64:2256–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Azumah BK, Addo PG, Dodoo A, Awandare G, Mosi L, Boakye DA, Wilson MD. 2017. Experimental demonstration of the possible role of Acanthamoeba polyphaga in the infection and disease progression in Buruli ulcer (BU) using ICR mice. PLoS One 12:e0172843. doi: 10.1371/journal.pone.0172843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amissah NA, Gryseels S, Tobias NJ, Ravadgar B, Suzuki M, Vandelannoote K, Durnez L, Leirs H, Stinear TP, Portaels F, Ablordey A, Eddyani M. 2014. Investigating the role of free-living amoebae as a reservoir for Mycobacterium ulcerans. PLoS Negl Trop Dis 8:e3148. doi: 10.1371/journal.pntd.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kennedy GM, Morisaki JH, Champion PA. 2012. Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba Acanthamoeba castellanii. Appl Environ Microbiol 78:2049–2052. doi: 10.1128/AEM.06965-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hagedorn M, Rohde KH, Russell DG, Soldati T. 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rose SJ, Bermudez LE. 2016. Identification of bicarbonate as a trigger and genes involved with extracellular DNA export in mycobacterial biofilms. mBio 7:e01597-. doi: 10.1128/mBio.01597-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rathnaiah G, Bannantine JP, Bayles DO, Zinniel DK, Stabel JR, Grohn YT, Barletta RG. 2016. Analysis of Mycobacterium avium subsp. paratuberculosis mutant libraries reveals loci-dependent transposition biases and strategies to novel mutant discovery. Microbiology doi: 10.1099/mic.0.000258. [DOI] [PubMed] [Google Scholar]
- 98.Wang J, Moolji J, Dufort A, Staffa A, Domenech P, Reed MB, Behr MA. 2015. Iron acquisition in Mycobacterium avium subsp. paratuberculosis. J Bacteriol 198:857–866. doi: 10.1128/JB.00922-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khattak FA, Kumar A, Kamal E, Kunisch R, Lewin A. 2012. Illegitimate recombination: an efficient method for random mutagenesis in Mycobacterium avium subsp. hominissuis. BMC Microbiol 12:204. doi: 10.1186/1471-2180-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harb OS, Gao LY, Abu Kwaik Y. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ Microbiol 2:251–265. doi: 10.1046/j.1462-2920.2000.00112.x. [DOI] [PubMed] [Google Scholar]
- 101.Nunes-Costa D, Alarico S, Dalcolmo MP, Correia-Neves M, Empadinhas N. 2016. The looming tide of nontuberculous mycobacterial infections in Portugal and Brazil. Tuberculosis (Edinb) 96:107–119. doi: 10.1016/j.tube.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 102.Shah NM, Davidson JA, Anderson LF, Lalor MK, Kim J, Thomas HL, Lipman M, Abubakar I. 2016. Pulmonary Mycobacterium avium-intracellulare is the main driver of the rise in non-tuberculous mycobacteria incidence in England, Wales and Northern Ireland, 2007-2012. BMC Infect Dis 16:195. doi: 10.1186/s12879-016-1521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mirsaeidi M, Machado RF, Garcia JG, Schraufnagel DE. 2014. Nontuberculous mycobacterial disease mortality in the United States, 1999-2010: a population-based comparative study. PLoS One 9:e91879. doi: 10.1371/journal.pone.0091879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Billinger ME, Olivier KN, Viboud C, de Oca RM, Steiner C, Holland SM, Prevots DR. 2009. Nontuberculous mycobacteria-associated lung disease in hospitalized persons, United States, 1998-2005. Emerg Infect Dis 15:1562–1569. doi: 10.3201/eid1510.090196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. 2012. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med 185:881–886. doi: 10.1164/rccm.201111-2016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wentworth AB, Drage LA, Wengenack NL, Wilson JW, Lohse CM. 2013. Increased incidence of cutaneous nontuberculous mycobacterial infection, 1980 to 2009: a population-based study. Mayo Clin Proc 88:38–45. doi: 10.1016/j.mayocp.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lopez-Varela E, Garcia-Basteiro AL, Santiago B, Wagner D, van Ingen J, Kampmann B. 2015. Non-tuberculous mycobacteria in children: muddying the waters of tuberculosis diagnosis. Lancet Respir Med 3:244–256. doi: 10.1016/S2213-2600(15)00062-4. [DOI] [PubMed] [Google Scholar]
- 108.Wu UI, Holland SM. 2015. Host susceptibility to non-tuberculous mycobacterial infections. Lancet Infect Dis 15:968–980. doi: 10.1016/S1473-3099(15)00089-4. [DOI] [PubMed] [Google Scholar]
- 109.Bottai D, Stinear TP, Supply P, Brosch R. 2014. Mycobacterial pathogenomics and evolution. Microbiol Spectr 2:MGM2-0025–2013. doi: 10.1128/microbiolspec.MGM2-0025-2013. [DOI] [PubMed] [Google Scholar]
- 110.Fregnan GB, Smith DW. 1962. Description of various colony forms of mycobacteria. J Bacteriol 83:819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bernut A, Viljoen A, Dupont C, Sapriel G, Blaise M, Bouchier C, Brosch R, de Chastellier C, Herrmann JL, Kremer L. 2016. Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol Microbiol 99:866–883. doi: 10.1111/mmi.13283. [DOI] [PubMed] [Google Scholar]
- 112.Belisle JT, Brennan PJ. 1989. Chemical basis of rough and smooth variation in mycobacteria. J Bacteriol 171:3465–3470. doi: 10.1128/jb.171.6.3465-3470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ruger K, Hampel A, Billig S, Rucker N, Suerbaum S, Bange FC. 2014. Characterization of rough and smooth morphotypes of Mycobacterium abscessus isolates from clinical specimens. J Clin Microbiol 52:244–250. doi: 10.1128/JCM.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Agusti G, Astola O, Rodriguez-Guell E, Julian E, Luquin M. 2008. Surface spreading motility shown by a group of phylogenetically related, rapidly growing pigmented mycobacteria suggests that motility is a common property of mycobacterial species but is restricted to smooth colonies. J Bacteriol 190:6894–6902. doi: 10.1128/JB.00572-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Prinzis S, Rivoire B, Brennan PJ. 1994. Search for the molecular basis of morphological variation in Mycobacterium avium. Infect Immun 62:1946–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Torrelles JB, Ellis D, Osborne T, Hoefer A, Orme IM, Chatterjee D, Brennan PJ, Cooper AM. 2002. Characterization of virulence, colony morphotype and the glycopeptidolipid of Mycobacterium avium strain 104. Tuberculosis (Edinb) 82:293–300. doi: 10.1054/tube.2002.0373. [DOI] [PubMed] [Google Scholar]
- 117.Park IK, Hsu AP, Tettelin H, Shallom SJ, Drake SK, Ding L, Wu UI, Adamo N, Prevots DR, Olivier KN, Holland SM, Sampaio EP, Zelazny AM. 2015. Clonal diversification and changes in lipid traits and colony morphology in Mycobacterium abscessus clinical isolates. J Clin Microbiol 53:3438–3447. doi: 10.1128/JCM.02015-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Howard ST, Rhoades E, Recht J, Pang X, Alsup A, Kolter R, Lyons CR, Byrd TF. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152:1581–1590. doi: 10.1099/mic.0.28625-0. [DOI] [PubMed] [Google Scholar]
- 119.Byrd TF, Lyons CR. 1999. Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect Immun 67:4700–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pawlik A, Garnier G, Orgeur M, Tong P, Lohan A, Le Chevalier F, Sapriel G, Roux AL, Conlon K, Honore N, Dillies MA, Ma L, Bouchier C, Coppee JY, Gaillard JL, Gordon SV, Loftus B, Brosch R, Herrmann JL. 2013. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol Microbiol 90:612–629. doi: 10.1111/mmi.12387. [DOI] [PubMed] [Google Scholar]
- 121.Brambilla C, Sanchez-Chardi A, Perez-Trujillo M, Julian E, Luquin M. 2012. Cyclopropanation of alpha-mycolic acids is not required for cording in Mycobacterium brumae and Mycobacterium fallax. Microbiology 158:1615–1621. doi: 10.1099/mic.0.057919-0. [DOI] [PubMed] [Google Scholar]
- 122.Sanchez-Chardi A, Olivares F, Byrd TF, Julian E, Brambilla C, Luquin M. 2011. Demonstration of cord formation by rough Mycobacterium abscessus variants: implications for the clinical microbiology laboratory. J Clin Microbiol 49:2293–2295. doi: 10.1128/JCM.02322-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Julian E, Roldan M, Sanchez-Chardi A, Astola O, Agusti G, Luquin M. 2010. Microscopic cords, a virulence-related characteristic of Mycobacterium tuberculosis, are also present in nonpathogenic mycobacteria. J Bacteriol 192:1751–1760. doi: 10.1128/JB.01485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Llorens-Fons M, Perez-Trujillo M, Julian E, Brambilla C, Alcaide F, Byrd TF, Luquin M. 2017. Trehalose polyphleates, external cell wall lipids in Mycobacterium abscessus, are associated with the formation of clumps with cording morphology, which have been associated with virulence. Front Microbiol 8:1402. doi: 10.3389/fmicb.2017.01402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brambilla C, Llorens-Fons M, Julian E, Noguera-Ortega E, Tomas-Martinez C, Perez-Trujillo M, Byrd TF, Alcaide F, Luquin M. 2016. Mycobacteria clumping increase their capacity to damage macrophages. Front Microbiol 7:1562. doi: 10.3389/fmicb.2016.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roux AL, Viljoen A, Bah A, Simeone R, Bernut A, Laencina L, Deramaudt T, Rottman M, Gaillard JL, Majlessi L, Brosch R, Girard-Misguich F, Vergne I, de Chastellier C, Kremer L, Herrmann JL. 2016. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol 6:160185. doi: 10.1098/rsob.160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Catherinot E, Clarissou J, Etienne G, Ripoll F, Emile JF, Daffe M, Perronne C, Soudais C, Gaillard JL, Rottman M. 2007. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect Immun 75:1055–1058. doi: 10.1128/IAI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kansal RG, Gomez-Flores R, Mehta RT. 1998. Change in colony morphology influences the virulence as well as the biochemical properties of the Mycobacterium avium complex. Microb Pathog 25:203–214. doi: 10.1006/mpat.1998.0227. [DOI] [PubMed] [Google Scholar]
- 129.Lei L, Tzekov R, Tang S, Kaushal S. 2012. Accumulation and autofluorescence of phagocytized rod outer segment material in macrophages and microglial cells. Mol Vis 18:103–113. [PMC free article] [PubMed] [Google Scholar]
- 130.Schoonmaker MK, Bishai WR, Lamichhane G. 2014. Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to beta-lactams. J Bacteriol 196:1394–1402. doi: 10.1128/JB.01396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]