ABSTRACT
The type VI secretion system (T6SS) inhibits the growth of neighboring bacterial cells through a contact-mediated mechanism. Here, we describe a detailed characterization of the protein localization dynamics in the Pseudomonas aeruginosa T6SS. It has been proposed that the type VI secretion process is driven by a conformational-change-induced contraction of the T6SS sheath. However, although the contraction of an optically resolvable TssBC sheath and the subsequent localization of ClpV are observed in Vibrio cholerae, coordinated assembly and disassembly of TssB and ClpV are observed without TssB contraction in P. aeruginosa. These dynamics are inconsistent with the proposed contraction sheath model. Motivated by the phenomenon of dynamic instability, we propose a new model in which ATP hydrolysis, rather than conformational change, generates the force for secretion.
IMPORTANCE The type VI secretion system (T6SS) is widely conserved among Gram-negative bacteria and is a central determinant of bacterial fitness in polymicrobial communities. The secretion system targets bacteria and secretes effectors that inhibit the growth of neighboring cells, using a contact-mediated-delivery system. Despite significant homology to the previously characterized Vibrio cholerae T6SS, our analysis reveals that effector secretion is driven by a distinct force generation mechanism in Pseudomonas aeruginosa. The presence of two distinct force generation mechanisms in T6SS represents an example of the evolutionary diversification of force generation mechanisms.
KEYWORDS: subcellular dynamics, T6SS, bacterial cell biology, quantitative fluorescent imaging
INTRODUCTION
Bacteria compete against neighboring cells for limited resources. Many bacterial species have consequently developed mechanisms to increase their relative fitness. One prevalent mechanism is the type VI secretion system (T6SS), which is found in about a quarter of Gram-negative bacterial species, including Pseudomonas aeruginosa and Vibrio cholerae. This system secretes toxic effectors by a contact-mediated-mechanism into neighboring bacteria, causing cell lysis or cessation of growth (1, 2). In the V. cholerae T6SS, two proteins (TssB and TssC) assemble into a tubular sheath in the donor cell cytosol. Effector delivery is driven by a conformational-change-induced contraction of the protein sheath, which drives the shaft through the outer membranes of neighboring recipient cells, delivering the effectors (3). The T6SS is then disassembled in a ClpV-dependent process. This duty cycle of assembly contraction and disassembly is typically observed only a single time in a given location. Subsequent events typically occur elsewhere in the cell (3, 4). In in vitro biochemical reconstitution in V. cholerae, ClpV interacts through its N-terminal domain with only the contracted form of the TssBC complex, disassembling the tubules formed by that complex (4, 5). V. cholerae ΔclpV strains have a partial loss of function of T6SS, demonstrating ClpV to be an important yet nonessential component of T6SS in V. cholerae (5, 6, 7). These observations have led to a syringe model of force generation for toxin delivery, wherein TssBC sheath contraction provides the force to push the inner needle out of the cell and the role of ClpV is to disassemble the sheath after contraction (Fig. 1).
FIG 1.
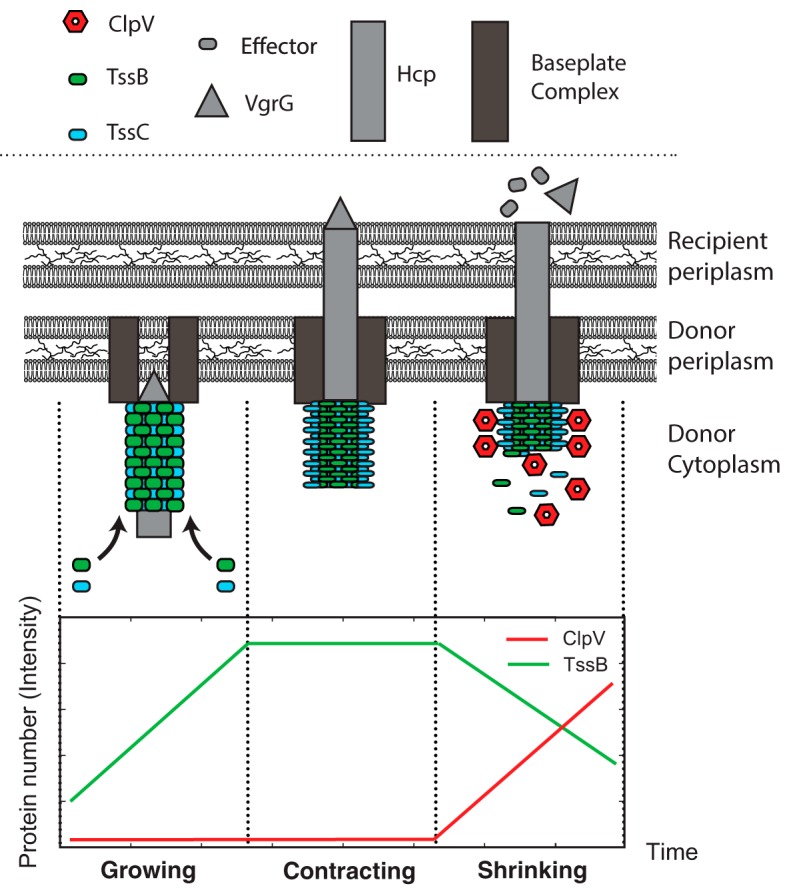
Schematic model for V. cholerae T6SS structure and secretion mechanism. In the growing phase of the duty cycle, the sheath and shaft assemble at the membrane. In the contraction phase of the duty cycle, the TssBC sheath undergoes conformational change, contracting the sheath and driving the Hcp/VgrG shaft through the recipient cell membrane. In the shrinking phase of the duty cycle, ClpV binds the N terminus of TssC which is exposed only in the contracted conformation. The ATPase activity of the TssC-bound ClpV then proceeds to disassemble the sheath.
In contrast, previous research shows that in the Hcp secretion island I-encoded T6SS (H1-T6SS) of P. aeruginosa and the T6SS of Serratia marcescens, both TssB and ClpV repeatedly localize to form punctate foci in the same location before final disassembly, consistent with significant functional differences between these T6SSs and that of V. cholerae (8). Significant structural differences also exist: the N terminus of ClpV in P. aeruginosa typically does not interact directly with TssC, as it does in V. cholerae, but there is instead a preferential interaction between ClpV and TssB that is mediated by TagJ (8, 9, 10). These differences motivated our investigation into the function of the T6SS system in P. aeruginosa.
To characterize the T6SS in P. aeruginosa, we investigated the dynamics of three important H1-T6SS components—ClpV, TssB, and Fha—by imaging fluorescent fusions to these T6SS proteins to visualize protein localization and dynamics in vivo. Although the localization dynamics of these proteins have not previously been quantified in vivo, these proteins are each known to play key roles in the P. aeruginosa T6SS. The ATPase ClpV is responsible for sheath disassembly, ClpV is required for Hcp secretion, and ClpV foci have been shown to correlate with T6SS-dependent cell killing (6, 11). The sheath protein TssB can be visualized to characterize the sheath dynamics (4). Fha has been shown to recruit downstream proteins to the H1-T6SS apparatus (12, 13), and it is hypothesized to be a component of the T6SS baseplate, arriving upstream of TssB and ClpV in T6SS assembly (11) (in this study, we present the first visualization of T6SS Fha dynamics). The observed dynamics of these proteins is inconsistent with those in the proposed V. cholerae model. We therefore present a new pump model for T6SS toxin delivery, in which sheath growth and disassembly drive the secretion process.
RESULTS
ClpV assembles and disassembles repeatedly and rapidly at the same location.
We initially focused on ClpV localization dynamics, since ClpV has been widely studied in both P. aeruginosa and V. cholerae and is thus a well-established comparison protein for the two species (3, 4, 5, 7, 11, 12, 14). ClpV is a member of the AAA+ ATPase protein family, which is an essential component of the T6SS (15). AAA+ proteins typically assemble into oligomeric ring-like enzyme machines that generate mechanical force by ATP hydrolysis and are known to remodel a variety of substrates (16). In V. cholerae, ClpV is known to disassemble the TssBC sheath after sheath contraction (4).
It is already known that the activation and dynamics of ClpV in P. aeruginosa are qualitatively different from those in V. cholerae. Although the T6SS is constitutively active in V. cholerae, as identified by the localization of T6SS proteins (4), in P. aeruginosa the ClpV of H1-T6SS rarely localizes unless it is specifically activated (17). ClpV is a nonessential protein in V. cholerae T6SS but is required in P. aeruginosa H1-T6SS (5, 6, 7, 11). In P. aeruginosa, ClpV forms a focus that shows repeated spiking of intensity in the same location before final disassembly (see Fig. 2). Additionally, data from Basler et al. demonstrate that labeled V. cholerae ClpV foci can move in the cell prior to disassembly, while assembled P. aeruginosa ClpV foci remain stationary (4). Although P. aeruginosa ClpV foci can appear in new locations in the cell, in ≈2,000 observations of P. aeruginosa ClpV foci, these foci were always assembled on location, rather than moving as an assembled complex as observed in V. cholerae (4).
FIG 2.
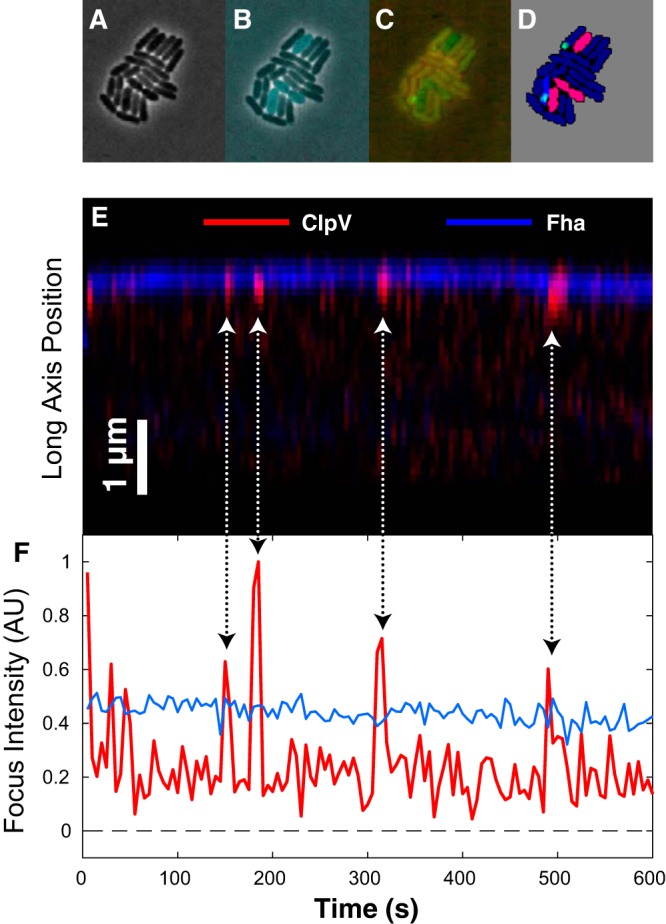
(A) Phase-contrast image of a polymicrobial microcolony consisting of both P. aeruginosa and Burkholderia thailandensis cells. (B) Cell identity is inferred by cytoplasmic fluorescence of cyan fluorescent protein (CFP) expressed in B. thailandensis cells. (C) ClpV-mCherry and Fha-GFP visualized by fluorescence microscopy in a single frame. (D) Cells are segmented from the phase-contrast image. Cell identity is inferred from CFP fluorescence and foci are detected in mCherry and GFP channels. B. thailandensis and P. aeruginosa cells are pink and blue, respectively. Foci with neighboring B. thailandensis cells are green; those without are blue. (E) Kymograph showing fluorescence intensity of Fha (blue) and ClpV (red) long-axis dynamics over a 4-min interval. (F) Fha (blue) and ClpV (red) focus intensity dynamics. Arrows show the corresponding events in the kymograph and intensity plots.
To make a quantitative comparison between ClpV dynamics in P. aeruginosa and dynamics reported for V. cholerae, we measured ClpV focus intensity over time (Fig. 2 and 3). We observed repeated ClpV localization-delocalization events in quick succession in the same location, followed by long pauses. We call individual events spikes and a cluster of spikes a firing event (Fig. 3, top). To analyze these intensity traces, we segmented the trace into two states based on the focus intensity: assembled, corresponding to a ClpV focus, and disassembled, corresponding to background intensity (see the supplemental material; Fig. 2). We then quantified the lifetimes of the observed states and the number of spikes per firing event. A histogram of both the spike lifetimes and the pauses (duration of time between spikes) is shown in Fig. 3 (bottom).
FIG 3.
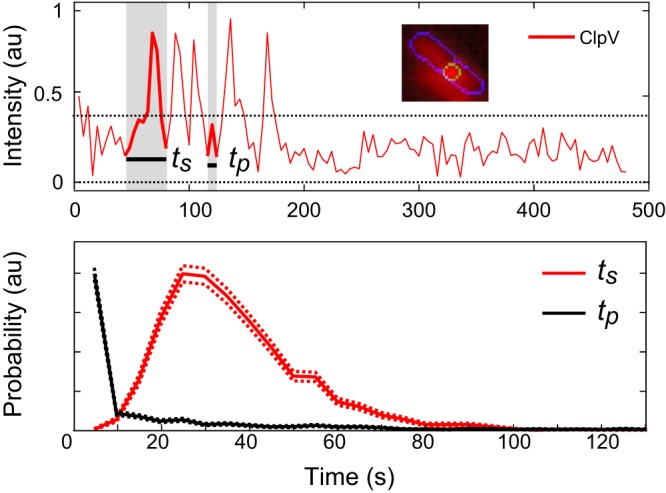
Example data demonstrating measurement of ClpV firing lifetimes (ts) and time between spikes (tp). There are five spikes in this trace. (Top) Representative ClpV firing for a single cell at one focus region. Inset shows image of cell with focus area indicated by outline. Here and in subsequent figures, each trace is representative of one focus area in one cell. (Bottom) Comparisons of ClpV lifetimes. Mean lifetime of the ClpV spike is 37 s, n ≈ 2,200; measurement of time between spikes, n ≈ 1,100.
In P. aeruginosa, ClpV firing consisted of a group of one to eight spikes, with a mean of 2.0 (standard deviation [SD] = 1.2, n = 900) spikes per firing event. The ClpV foci remained relatively stationary, and the change in focus displacement was not observed to surpass 500 nm during firing. The spikes had a mean lifetime of 37 s (SD = 16 s, n = 1,960) and a median pause of 33 s (SD = 42 s, n = 380) between ungrouped spikes. Grouped spikes had a median pause of shorter duration than our frame rate of 5 s. After the last delocalization event, the intensity returned to cell background intensity, as seen in Fig. 3 (top). The maximum focus intensity was approximately constant from spike to spike in a firing event. We hypothesize that this maximum intensity is regulated by the depletion of cytoplasmic protein. In V. cholerae, firing events correspond to a single spike and therefore the repeated spiking at a fixed location observed in P. aeruginosa is an important qualitative distinction between the two T6SSs (4). Interestingly, in spite of this qualitative difference, the average lifetime of one ClpV spike in V. cholerae and P. aeruginosa is nearly identical (34 s, SD = 6.2 s, n = 10 for ClpV recruitment and n = 40 for the rest of ClpV lifetime, and 37 s, SD = 16 s, n = 1,960, respectively) (Fig. 3, bottom).
To compare the activity of the T6SS between V. cholerae and P. aeruginosa, we estimated the overall firing rate between the two species. In P. aeruginosa cells, we observed that a ClpV focus appeared in just 11.3% of cells over a 7-min experiment, consistent with a firing rate of 0.01 spikes/cell · second. At any time, 1.4% of cells had a ClpV focus (n = 15,600 cells; n = 1,760 for cells with ClpV foci). In P. aeruginosa cells, 0.3% of cells had two or more firing locations, but in 11% of cells, a second firing event occurred in the same location as in the original, as measured by a firing event occurring a minute or more after firing had ceased. This suggests that the T6SS apparatus does not always fully disassemble after firing but can sometimes reinitiate T6SS activity at that same location. In contrast, >90% of all V. cholerae cells were observed to have a T6SS focus, and many cells were observed to have two or more firing events, although there was only one firing event at each location (4, 6).
TssB contraction events cannot be resolved in P. aeruginosa.
Having characterized ClpV dynamics, we next performed a detailed analysis of the dynamics of the TssB protein, a component of the T6SS sheath (5). TssB is an essential component of T6SS, and in V. cholerae it is known to assemble into tubules with TssC (5). In V. cholerae, these tubules have both an extended and a contracted form; they assemble in the extended form and then spontaneously contract (3, 5). This contractile tubule sheath has been hypothesized to be the effector delivery mechanism. After effector delivery, the contracted tubule is depolymerized by ClpV, and the sheath components are recycled (3, 5). However, analogous contraction has not been observed in all T6SSs. For instance, although the sheath has been visualized by fluorescence microscopy in S. marcescens, contraction has not been observed, since the sheath focus is always below the diffraction limit (4, 8). In close analogy to the observations in S. marcescens, we observed TssB foci in wild type P. aeruginosa and determined that these foci were below the diffraction limit (Fig. 4A), and therefore no contraction events could be directly resolved.
FIG 4.
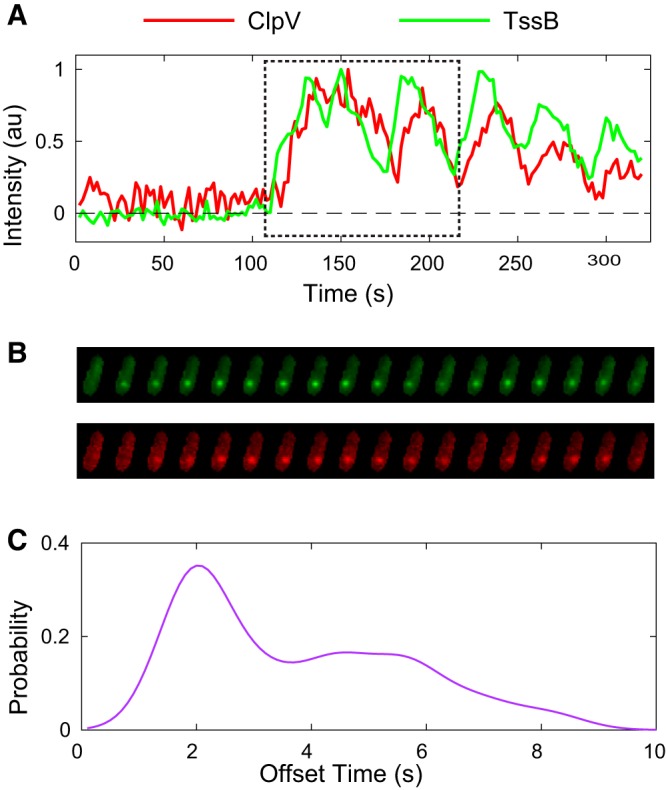
(A) ClpV and TssB normalized fluorescence intensities over time in single cell (2-s frame rate). Fluorescence intensity fluctuates, and focus motion is observed, but the focus remains diffraction limited, as seen in panel B. Cell images for the region indicated by the dashed boundary are shown in panel B. (B) Typical single-focus data show a representative coordinated ClpV and TssB fluorescence intensity traces for one focus region in ΔretS background. (C) Population-level analysis reveals average lag times between ClpV and TssB traces. n ≈ 280.
TssB and ClpV localization dynamics are coordinated, not sequential.
To test the proposed mechanism of T6SS dynamics, we simultaneously visualized the sheath (TssBC) and the ATPase that catalyzes its disassembly (ClpV). In V. cholerae, Basler et al. observed the sequential processes of sheath assembly (TssB), contraction, ClpV localization, and disassembly (4), which motivated the proposal of the syringe model. To test whether the dynamics were also sequential in P. aeruginosa, we visualized the T6SS dynamics in a clpV-mCherry tssB-GFP strain.
In this dual-label strain, the behavior of ClpV was markedly different from that in the single-label strain (clpV-mCherry): although the ClpV-mCherry focus lifetime was identical in both strains, the clpV-mCherry tssB-GFP strain had a significantly diminished firing rate compared to that of the clpV-mCherry strain, with at most one ClpV/TssB spike seen over the 7- to 9-min experiment and with a spike lifetime five times as long as that of wild-type ClpV-mCherry (see the supplemental material; Fig. 1). However, this loss of function appears to be circumvented by an increase in T6SS protein expression. In the ΔretS background, in which H1-T6SS is constitutively active, ΔretS clpV-mCherry tssB-GFP ClpV behavior was qualitatively and quantitatively similar to that of ClpV behavior in the wild type (i.e., the observed shape of the intensity trace and dynamics in the clpV-mCherry strain). Additionally, although both spike frequency and lifetime were affected by the dual label in the wild type, the lag time between ClpV and TssB spiking was the same in both the dual-label wild-type and ΔretS backgrounds. We shall therefore assume that the dynamics observed in the ΔretS clpV-mCherry tssB-GFP strain are at least a qualitative approximation for the dynamics in unlabeled wild-type cells. All subsequent ClpV-mCherry TssB-GFP experiments were performed in this strain.
Both the ClpV-mCherry and TssB-GFP fusions formed foci that were observed to colocalize and to undergo very similar (and correlated) dynamics, as shown in Fig. 4A and B. Consistent with the syringe model, in which ClpV disassembles the TssBC sheath, the ClpV intensity profile (localization) in V. cholerae was observed to lag behind the TssB intensity profile: ClpV localized to the TssBC sheath 5 to 10 s after sheath contraction (3, 4). In P. aeruginosa, we observed that the ClpV and TssB intensity profiles were nearly identical, except for a short lag in the ClpV localization profile, as shown in Fig. 4B. To quantify the lag time, we calculated the cross-correlation between the ClpV-mCherry and TssB-GFP fluorescence. The mean lag time was found to be 2.7 s (SD = 1.6, n = 280) (Fig. 4C).
In contrast to the syringe model, which predicts sequential dynamics, the similarity between the TssB and ClpV localization profiles suggests coordinated dynamics or a leader-follower mechanism. An interesting contrast between the dynamics in P. aeruginosa and V. cholerae is the length of the lag time; in V. cholerae there is a 100- to 200-s lag between TssB assembly and ClpV localization, in contrast to our observed 2.7-s lag in P. aeruginosa TssB and ClpV localization. Furthermore, in V. cholerae, the localization profiles of TssB and ClpV are distinctly different (4).
Fha localization precedes and remains throughout ClpV activity.
After observing the close coordination between TssB and ClpV, we wished to determine whether this coordination was specific to sheath and sheath-associated proteins or representative of all T6SS proteins. Another key difference between P. aeruginosa and V. cholerae T6SS function lies in the requirement of Fha. Fha is required for P. aeruginosa T6SS functionality, likely as a component of the T6SS baseplate, and it precedes ClpV recruitment, but there is no similar protein in V. cholerae (11, 12, 13, 18). We therefore investigated the localization dynamics of the upstream T6SS-essential protein Fha. We constructed a double-labeled ClpV-mCherry/Fha-sfGFP strain and observed the localization dynamics of both proteins (Fig. 5). Fha has qualitatively different localization dynamics than those of either ClpV or TssB. Fha was observed to localize and delocalize gradually, making it difficult to consistently classify the dynamics using a method analogous to that used for ClpV and TssB. Although Fha did not spike, its persistent localization appears to be essential for ClpV localization, since in observations of ≈10,000 cells, ClpV firing was never observed without Fha localization and on average Fha appeared ≈20 s before ClpV began to spike. Furthermore, Fha enveloped the ClpV spikes; out of approximately 1,800 observations, Fha always appeared before ClpV firing and was retained throughout and disassembled after the end of the firing event, consistent with its proposed role as part of the T6SS baseplate complex (11). Interestingly, the Fha focus sometimes persisted minutes beyond the cessation of ClpV firing, indicating that Fha appears to be required for firing to occur, but the presence of Fha does not appear to be sufficient to generate a firing event. Therefore, the close coordination between ClpV and TssB localization patterns appears to be specific to the sheath and not to H1-T6SS proteins in general.
FIG 5.
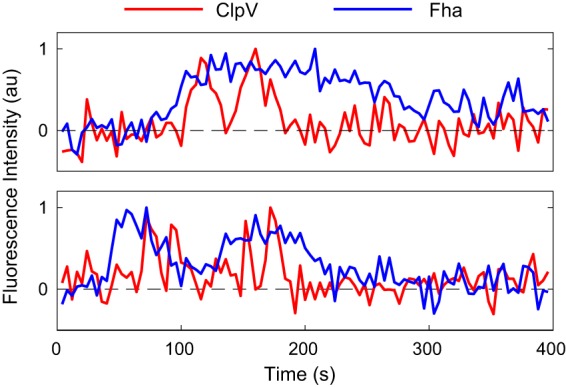
Representative intensity traces of ClpV and Fha. (Top) Fha levels start increasing a few seconds (2 to 60 s) before ClpV begins to fire. After the firing has terminated, Fha returns to a baseline level. (Bottom) Fha disappears and returns. Due to the diffuse nature of the Fha focus, an automated approach to determining lifetime and lag time was not reliable. The intensity traces shown above were representative of the traces examined by eye (n ≈ 1,760).
Focus movement is linear and normal to the cell membrane.
Cryogenic electron microscopy (CryoEM) images have shown that the T6SS in V. cholerae forms a sheath, which is large enough to be resolved by fluorescence microscopy, roughly perpendicular to the cell membrane (3). The sheath contraction can also be directly visualized by fluorescence microscopy. Due to the small size of the P. aeruginosa TssB and ClpV foci, no direct motion or TssBC contraction has yet been observed in P. aeruginosa, but the movement of the focus can be tracked to a higher resolution than the diffraction limit, using Gaussian fitting to the focus location. We therefore tracked ClpV foci in P. aeruginosa H1-T6SS with 5-s resolution time to determine whether the observed motions were consistent with sheath contraction. The resolution for tracking foci was determined to be 50 nm, limited by the nonuniform fluorescence background due to the cytoplasmic fluorescence. (We chose to track ClpV foci rather than TssB foci because we had determined that ClpV and TssB foci have nearly identical behavior, but the fluorescent labeling of TssB tended to disrupt H1-T6SS dynamics to a greater extent than the labeling of ClpV, as discussed previously.) Thus, investigating the dynamics of ClpV foci minimized the disruption to the system caused by fluorescent labeling, while still being representative of key dynamic components of the H1-T6SS.
To probe whether the motion was consistent with a sheath having a length that varied over time, we first investigated whether the observed motions of the protein foci were better modeled by isotropic or anisotropic random motion. Sheath contraction would result in anisotropic motion, due to the increased variance along the axis of sheath due to contraction. To measure the anisotropy, we calculated an anisotropy score for each trace by finding the eigenvectors of the covariance matrix of the focus position and then determining the ratio of the maximum and minimum eigenvalues. A high ratio would indicate strong anisotropy, while a ratio close to one would indicate isotropy. To determine the significance of the anisotropy, we computed the test statistic by simulating the distribution of the anisotropy score for a two-dimensional normal distribution using an isotropic covariance matrix with the same number of observations as the focus trajectory. An example of measured and random data in the context of a cell is shown in Fig. 6B.
FIG 6.
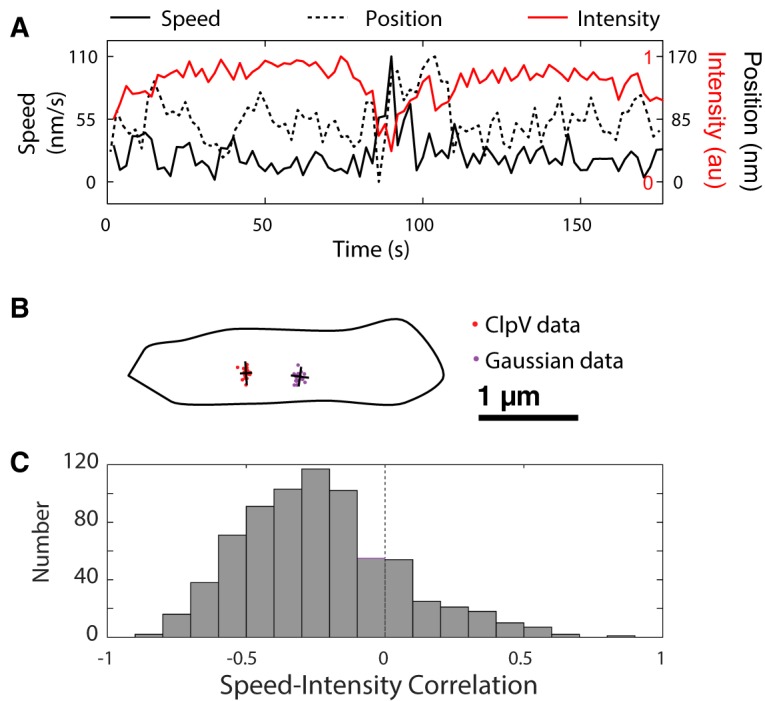
(A) Representative intensity/position/speed graph for one region. Frame rate, 2 s. Position is measured by projecting coordinates onto best-fit line. (B) Example spread of ClpV data and Gaussian-distributed data. Major and minor axes are indicated in black. (C) Correlation coefficients between speed and intensity in wild-type cells (n ≈ 650). There is generally a negative correlation between speed and intensity. Note that the right tail of the distribution is primarily due to uncertainty in resolving the focus centroid at lower intensities, leading to imperfections in calculating the speed.
If the anisotropy score were due to statistical fluctuations, we would expect just 5% of scores to exceed the 95% confidence level. However, we found that the anisotropy scores were too large to be described by isotropic random motion: 45% of the measured data had anisotropy scores higher than the 95% confidence level. Using the Fisher exact test, we combined these measurements to generate a single P value, which was less than 10−4, allowing us to reject the null hypothesis that the distribution of positions is isotropic. Although the anisotropy of the motion is clearly statistically significant, it is not large. The mean range of ClpV movements was 300 nm.
Finally, we investigated the orientation of ClpV movement relative to the cell membrane. For each focus, we determined the normal line to the cell membrane and the best-fit line of ClpV and calculated the angle between them. We discovered that, compared to random ClpV movement orientation, the slope of the ClpV best-fit line was about six times more likely to be within 10° of the normal line, indicating strong perpendicularity. Like the V. cholerae syringe model, this apparatus is also oriented to deliver toxins outside the cell.
ClpV motion correlates with ClpV intensity.
We hypothesized that a change in the length of the sheath would correspond with focus movement, as well with as a change in fluorescence intensity. We therefore posited that focus motion corresponds with changes in the length of the sheath and that rapid changes in motion correspond to fluctuations in intensity. In order to calculate the significance of this relationship, we calculated Pearson's correlation coefficient for each data set. We noted that significant increases in speed corresponded with both increases and decreases in fluorescence intensity (Fig. 6A and C).
This result is clearly inconsistent with the V. cholerae syringe model. In the syringe model, effector delivery is driven by sheath contraction, which occurs due to a conformational change in TssBC. The length of the sheath is reduced without reducing the number of proteins. Therefore, contraction is expected to lead to a rapid change in the focus position, without a change in the focus intensity. However, in contrast, manual and automated inspection of the intensity and position traces in P. aeruginosa demonstrated that significant changes in position of the TssB focus were accompanied by changes in intensity. Therefore, even with sub-diffraction-limited tracking of the centroid of the sheath, we found no clear evidence for contraction events in P. aeruginosa.
DISCUSSION
Evidence for a novel secretion mechanism.
Our analysis reveals striking quantitative and qualitative differences between V. cholerae and P. aeruginosa T6SSs in both TssB and ClpV dynamics and structure. In P. aeruginosa, ClpV intensity traces resemble triangle waves; that is, the intensity first undergoes steady growth, followed by steady decay, and closely follows an analogous pattern in the TssB trace. Several of these spiking events may occur in close succession. In contrast, in V. cholerae, the ClpV intensity resembles a single square wave; that is, ClpV rapidly localizes and then delocalizes, but only after TssBC contraction is observed (4). In this case, ClpV and TssB localization is sequential, rather than coordinated, and only a single spiking event is observed during firing. In P. aeruginosa, the rapid localization of ClpV to the assembling sheath, the repeated spiking, and the absence of detectable sheath contraction all conflict with the proposed syringe model and the observed dynamics in V. cholerae.
In addition to the differences in dynamics, there are cellular-scale differences in the T6SS structure. In P. aeruginosa, the TssB focus is diffraction limited, but it is optically resolvable in V. cholerae. The greatly reduced size of the P. aeruginosa H1-T6SS may require repeated secretion events to secrete the same number of effectors as the V. cholerae T6SS, consistent with our observations of multiple spikes per firing event. There is also precedent for mechanistic distinctions in the constituent proteins and characterized protein interactions. In V. cholerae, there is a well-characterized and direct interaction between ClpV and TssC: once the TssBC sheath contracts, the N-terminal ClpV-binding domain of TssC is exposed, facilitating sheath disassembly (4). In P. aeruginosa, no such direct interaction between ClpV and TssC has been observed, and the conserved TssC binding groove in ClpV is only partially conserved in the ClpV of P. aeruginosa (4, 9, 10). This partial conservation decreases the hydrophobicity of the binding site and is inconsistent with the binding of the alpha helix that is seen in V. cholerae. Free ClpV and TssC can still bind, but this binding no longer depends on TssC conformational change (9). Rather, in P. aeruginosa, ClpV and TssB interactions are more plausibly mediated by TagJ, and they do not appear to be dependent on sheath contraction (9, 10). Together, the differences in T6SS dynamics, sheath size, and protein function are consistent with the existence of a distinct secretion mechanism in P. aeruginosa.
The pump model and a dynamic instability.
The mismatch between the dynamics predicted by the syringe model and the observed phenomenology motivates the consideration of alternative models for secretion and force generation in the T6SS. In particular, the sheath contraction of the V. cholerae T6SS is hypothesized to be a principal mechanism responsible for puncturing the cell wall of the neighboring cell. During this contraction, the V. cholerae sheath contracts to half of its length without depolymerization, but we could not detect sheath contraction in P. aeruginosa. Instead, the P. aeruginosa sheath alternates between growth and decay states (i.e., it changes in length due to changes in the number of protein subunits). We propose that this observed growth and decay play a central role in the secretion process. It is interesting to speculate that the disassembly of the TssBC sheath, driven by ClpV, could provide the force required to secrete the effectors. We therefore call this model a pump model, which is capable of delivering sequential secretions of effectors.
It is known that the assembly and disassembly of cytoskeletal filaments can generate force (19). For instance, microtubule disassembly is responsible for the generation of force on chromosome centromeres during metaphase (20). Even in in vitro reconstitution, purified tubulin can assemble into microtubules that transition between growth and rapid decay due to a phenomenon called dynamic instability, in which the GTP-hydrolysis front catches the filament tip (20).
The similarities between the observed dynamics of the P. aeruginosa H1-T6SS and microtubules suggest that there may be mechanistic similarities between the two processes. For instance, the growth rate of the TssBC filament is observed to transition back and forth between growth and decay states. Since the filaments do not decay in the absence of ClpV ATPase activity, ClpV is known to bind to the TssBC sheath, and ClpV is required for secretion in H1-T6SS, it is natural to assume that ClpV modulates filament growth (11). The switch between growth and decay states, causing the ClpV and TssB foci to appear and disappear, is caused by the GTP-hydrolysis front in the context of microtubules. In P. aeruginosa H1-T6SS, this switch could be caused by a number of putative mechanisms, as follows. (i) The ClpV intensity trace lags the TssB intensity trace, motivating a model in which the ClpV coat is the analogue to the hydrolysis front. When the ClpV coat catches the tip of the assembling sheath, the sheath transitions to a decaying state, and TssBC subunits are depolymerized by ClpV ATP hydrolysis. (ii) Alternatively, an analogous ClpV ATP-hydrolysis front that propagates through the sheath may exist in TssBC-bound ClpV. Disassembly may be sterically blocked except at the tip. When the hydrolysis front catches the tip, the sheath transitions to the decaying state. This model could result in consistent maximum spike intensity, as shown in Fig. 3A, if the assembly is limited by the depletion of cytoplasmic TssBC. The proposed pump and syringe models have distinct mechanisms of force generation but many similar features. In both models, it is hypothesized that changes in the sheath length drive the internal shaft that carries the effectors into the neighboring cell. In the V. cholerae syringe model, the change in the sheath length (and force) required for effector delivery is generated by the conformational change of TssBC, whereas the pump model depends directly on ATP hydrolysis to shorten the filament and generate force, as was originally proposed (17). Both models predict that clpV mutants would significantly compromise secretion, as observed previously (5, 6, 7). Our proposed model is described in Fig. 7.
FIG 7.
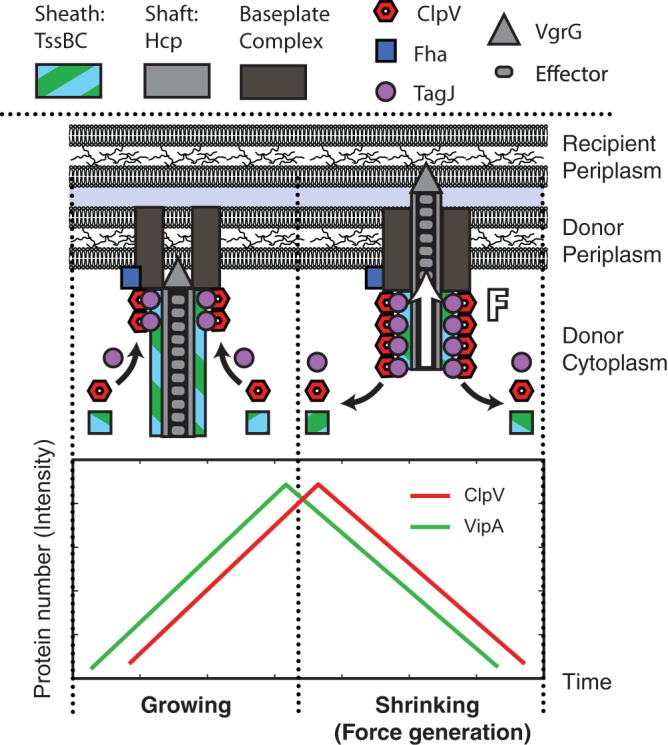
Schematic model for P. aeruginosa T6SS structure and secretion mechanism. As the TssBC sheath begins formation, TagJ associates with the sheath. TagJ in turns complexes with ClpV, which allows ClpV to associate with the TssBC sheath without any sheath conformational changes. ClpV can thus localize to the T6SS apparatus within a few seconds after the initiation of TssB localization. ClpV then disassembles the TssBC sheath, generating a force similar to that seen in microtubule growth and decay, which is used to deliver effectors to the neighboring cell (20). This is in direct contrast to ClpV and TssBC in the T6SS of V. cholerae, which require a conformational change to bind (4).
We are not necessarily questioning the applicability of the syringe model to V. cholerae, which appears to have distinct localization dynamics. However, it is important to note that many experiments in V. cholerae have been performed in a mutant with increased expression and activity of T6SS (3, 4, 21). It may be necessary to reevaluate the syringe model in V. cholerae under conditions where the T6SS proteins are not overexpressed.
T6SS diversity in bacteria.
Evidence for distinct mechanisms in P. aeruginosa and V. cholerae naturally motivate the questions about the diversity of the T6SS mechanisms in T6SS+ Gram-negative bacteria as a whole. Are there two or more classes of T6SS mechanisms? If so, what are the key genetic signatures of these families? To explore the diversity of the T6SS, we considered another characterized system, the S. marcescens T6SS. The observed diffraction-limited protein localization of TssB and ClpV and genetic evidence (the presence of both Fha and TagJ, the latter of which mediates the interaction between TssB and ClpV) suggest that the S. marcescens T6SS mechanism is more consistent with that of the P. aeruginosa T6SS than that of the V. cholerae T6SS (8, 10). Consistent with these observations, Forster et al. suggested that ClpV, TssB, TssC, and TagJ (when present) have coevolved with each other, creating at least two separate classes of ClpV/TssBC binding proteins (9). Initially, we hypothesized that TagJ and Fha might also be phylogenetically linked and a proxy for the P. aeruginosa-like T6SS mechanism, since both genes are absent from V. cholerae and present in P. aeruginosa and S. marcescens. However, the Agrobacterium tumefaciens T6SS requires Fha phosphorylation but has no tagJ homolog (10, 13). None of these T6SSs appear to be exceptional, as 30% of T6SSs have an fha homolog and 50% of T6SSs have a tagJ homolog (18). Additionally, TssB without ClpV localization appears to be present significantly more frequently in both V. cholerae and S. marcescens than in P. aeruginosa. These distinctions suggest that T6SS function may be diversified beyond the distinct mechanisms observed in P. aeruginosa and V. cholerae.
Conclusion.
We report on a fine-scale analysis of the dynamics of the T6SS in P. aeruginosa, using quantitative image analysis of single cells. The simultaneous imaging of TssB and ClpV revealed that the localization of these two proteins in P. aeruginosa is synchronized rather than sequential, unlike that observed in V. cholerae. To accommodate these observations, we proposed a pump model in which effector delivery is powered by sheath depolymerization in analogy to force generation by cytoskeletal filaments. Structural differences between the TssC proteins in V. cholerae and P. aeruginosa (and their concomitant interactions with ClpV) is consistent with the view that the functional mechanism may be distinct in the two organisms. The genetic similarity between the T6SS in P. aeruginosa and those of other Gram-negative bacteria suggests that this pump mechanism may describe the T6SS function in a significant number of other bacteria.
MATERIALS AND METHODS
Strains.
Strains used in this study are described in Table 1. Strains were derived from P. aeruginosa PAO1 and Burkholderia thailandensis E264 (22, 23). Functional translational fluorescent C-terminal fusions of either superfolder green fluorescent protein (sfGFP) or monomeric mCherry (mCherry) were generated to TssB, ClpV, and Fha at their endogenous chromosomal loci by allelic exchange, as described in the supplemental material (14). To ascertain the functionality of these strains, we performed bacterial growth competition experiments using fluorescently labeled and unlabeled strains with B. thailandensis recipients under comparable conditions. T6SS-dependent fitness was determined by comparing the ratio of P. aeruginosa to B. thailandensis before and after 7 h of coculture. We found that all strains reduced populations of B. thailandensis in a T6S-dependent manner by an order of magnitude.
TABLE 1.
Strains used in this study
Growth conditions and microscopy.
All strains were grown overnight from a single colony in Luria broth (LB) medium. Doubling time in liquid medium was roughly 30 min. Overnight cultures were diluted 1:10 into fresh medium and grown to an optical density at 600 nm (OD600) of ≈0.8 (the T6SS is less active in log-phase growth) (14). Agarose pads were prepared by pouring 1 ml of growth medium and 2% agarose into 2- by 2-cm wells cut into a rubber gasket sealed onto a standard microscope slide. We spotted 2 μl of cells onto dried pads, and a coverslip was placed on top of each pad. The entire slide was sealed with VaLP (1:1:1 vaseline-lanolin-paraffin).
To study interactions with a second species, we introduced B. thailandensis. B. thailandensis was distinguished from P. aeruginosa by the expression of cytoplasmic cyan fluorescent protein (CFP) and gating cells on fluorescence levels in the CFP channel. Introducing B. thailandensis also had the advantage of activating the T6SS in wild-type P. aeruginosa (14). Under normal growth conditions, wild-type P. aeruginosa has about 1 to 2% of cells with an active T6SS, as identified by the presence of a ClpV focus, but upon exposure to B. thailandensis, which activates T6SS via the P. aeruginosa response to antagonism (PARA), about 10% of cells have a ClpV focus (14). All wild-type cells in this study were activated via PARA to achieve a higher percentage of cells with foci. To activate P. aeruginosa via PARA, immediately prior to spotting cells on the slide, we added 100 μl of B. thailandensis culture to 100 μl of P. aeruginosa culture and spun down (3 min at 8,000 rpm), removed the supernatant, and resuspended the pellet in 40 μl of fresh LB before spotting. The V. cholerae strain, to which we compared P. aeruginosa T6SS dynamics, carries a mutation which increases T6SS activity.
Under imaging conditions, the average doubling time for cells was ≈40 min. Fluorescence microscopy was performed on a Nikon-TiE instrument (Nikon, Tokyo, Japan) with an environmental chamber, using NIS-Elements software. Images were captured on an Andor Neo sCMOS camera (Belfast, Northern Ireland) at either 2-s or 5-s intervals.
Quantitative image analysis.
Cells were segmented and linked from phase-contrast images using our SuperSegger segmentation engine (24). All time-lapse images were aligned to compensate for stage drift. Cell growth, as observed by phase-contrast microscopy, was not appreciable over time-lapse experiments. Although SuperSegger can identify and track foci, the current implementation identifies foci in each frame and links foci between frames. This approach is problematic for foci that repeatedly appear and disappear, since it can be difficult to ascertain whether a focus is returning to precisely the same location or not. In the current context, foci are imaged over a sufficiently short period, such that the movement of foci in this system is small (<500 nm). We therefore exploited these slow dynamics to track foci which reappear over time. We first averaged over all frames to generate a summed image and then identified loci in this summed image and defined a circular region (radius, 500 nm around the focus position) where loci were allowed to move. To compute the focus intensity in each frame, we convolved each image with a disk (radius, 300 nm), and defined the focus intensity as the maximum pixel value in the allowed region of the convolved image minus the background intensity. We computed the background intensity by averaging over the fluorescence intensity in the cell, having first masked out the focus using the 300-nm-radius disk.
Supplementary Material
ACKNOWLEDGMENTS
P.A.W. and J.C. acknowledge Robin Kirkpatrick for edits to the image analysis code and Brandon Hoang for performing the initial Fha experiments.
Support for this research was graciously provided by National Science Foundation Graduate Research Fellowship DGE-1256082 and by the National Institutes of Health under award number R21 AI 105268-01.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00744-17.
REFERENCES
- 1.Cianfanelli FR, Monlezun L, Coulthurst SJ. 2016. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Micro 12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler M, Pilhofer M, Henderson G, Jensen G, Mekalanos J. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler M, Mekalanos JJ. 2012. Type 6 secretion dynamics within and between bacterial cells. Science 337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapitein N, Bönemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. 2013. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient Type VI protein secretion. Mol Microbiol 87:1013–1028. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- 7.LeRoux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. 2012. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci U S A 109:19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerc AJ, Diepold A, Trunk K, Porter M, Rickman C, Armitage JP, Stanley-Wall NR, Coulthurst SJ. 2015. Visualization of the Serratia type VI Secretion system reveals unprovoked attacks and dynamic assembly. Cell Rep 12:2131–2142. doi: 10.1016/j.celrep.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Förster A, Planamente S, Manoli E, Lossi NS, Freemont PS, Filloux A. 2014. Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes. J Biol Chem 289:33032–33043. doi: 10.1074/jbc.M114.600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lossi N, Manoli E, Simpson P, Jones C, Hui K, Dajani R, Coulthourst SJ, Freemont P, Filloux A. 2012. The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel subcomplex in the bacterial type VI secretion system. Mol Microbiol 86:437–456. doi: 10.1111/j.1365-2958.2012.08204.x. [DOI] [PubMed] [Google Scholar]
- 11.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 12.Hsu F, Schwarz S, Mougous JD. 2009. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol 72:1111–1125. doi: 10.1111/j.1365-2958.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin JS, Wu HH, Hsu PH, Ma LS, Pang YY, Tsai MD, Lai EM. 2014. Fha interaction with phosphothreonine of TssL activates type VI secretion in Agrobacterium tumefaciens. PLoS Pathog 10:e1003991. doi: 10.1371/journal.ppat.1003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, Goodlett DR, Wiggins PA, Mougous JD. 2015. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 4:e05701. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlieker C, Zentgraf H, Dersch P, Mogk A. 2005. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem 386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- 16.Hanson PI, Whiteheart SW. 2005. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 17.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner MK, Zanic M, Howard J. 2013. Microtubule catastrophe and rescue. Curr Opin Cell Biol 25:14–22. doi: 10.1016/j.ceb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoué S, Salmon ED. 1995. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell 6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GKS, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, Derr A, Engels R, DeShazer D, Birren B, Nierman WC, Tan P. 2006. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol 6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stylianidou S, Brennan C, Nissen SB, Kuwada NJ, Wiggins PA. 2016. SuperSegger: robust image segmentation, analysis and lineage tracking of bacterial cells. Mol Microbiol 102:690–700. doi: 10.1111/mmi.13486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


