Abstract.
Obtaining RNA from clinical samples collected in resource-limited settings can be costly and challenging. The goals of this study were to 1) optimize messenger RNA extraction from dried blood spots (DBS) and 2) determine how transcriptomes generated from DBS RNA compared with RNA isolated from blood collected in Tempus tubes. We studied paired samples collected from eight adults in rural Tanzania. Venous blood was collected on Whatman 903 Protein Saver cards and in tubes with RNA preservation solution. Our optimal DBS RNA extraction used 8 × 3-mm DBS punches as the starting material, bead beater disruption at maximum speed for 60 seconds, extraction with Illustra RNAspin Mini RNA Isolation kit, and purification with Zymo RNA Concentrator kit. Spearman correlations of normalized gene counts in DBS versus whole blood ranged from 0.887 to 0.941. Bland–Altman plots did not show a trend toward over- or under-counting at any gene size. We report a method to obtain sufficient RNA from DBS to generate a transcriptome. The DBS transcriptome gene counts correlated well with whole blood transcriptome gene counts. Dried blood spots for transcriptome studies could be an option when field conditions preclude appropriate collection, storage, or transport of whole blood for RNA studies.
INTRODUCTION
Next-generation sequencing in translational research has expanded dramatically as costs have decreased and technology has improved. Although greater quantities of high-quality RNA (RNA Integrity Number [RIN] > 7) are ideal, intermediate quality RNA has been used to generate reliable transcriptome data.1,2 Dried blood spot (DBS) collection is a widely used method for blood collection and specimen preservation. Collection of blood from a prick of the finger is less invasive for the patient, can be easily performed in field settings, is more acceptable than larger volume collection, particularly for pediatric populations, does not need immediate refrigeration, and can be stored indefinitely.3 Methods to isolate RNA of acceptable integrity and quantity from DBS have been explored. Several studies have used DBS-sourced messenger RNA (mRNA) or micro RNA for targeted gene expression studies4–6 or microarray.7,8 For next-generation sequencing, some studies have used a pre-amplification step to generate sufficient mRNA for array studies,9–11 although pre-amplification steps can introduce bias. As Grauholm et al.10,12 demonstrated, different types of pre-amplification protocols generate different gene expression profiles. One study reported RNA sequencing data generated from archived DBS collected as part of neonatal screening12 and demonstrated the ability to differentiate males from females by expression of Y-chromosome genes but did not have whole blood–derived RNA available to compare DBS transcriptomes with transcriptomes from whole blood.
We conducted this study to optimize RNA quality and yield from DBS to generate RNA for next-generation RNA sequencing studies. Our goal was to use RNA directly after DBS extraction without a pre-amplification step because of the bias of pre-amplification of RNA. We also compared transcriptome data from DBS-derived RNA with whole blood–derived RNA.
MATERIALS AND METHODS
Sample collection.
Specimens included in this study were collected from eight adults in a rural area of northwest Tanzania in November 2015. The adults were enrolled in a larger study of schistosomiasis in the region and provided written informed consent for participation. Peripheral blood was collected into a sterile syringe. On completion of the blood collection, 3 mL of blood were placed into each of two Tempus Blood RNA (Invitrogen, Waltham, MA) tubes that contained 6 mL of stabilizing reagent and the tubes were shaken vigorously for 15 seconds according to the manufacturer’s instructions. Blood was expelled from the tip of the syringe to fill each of five spots on one Whatman Protein Saver 903 card (GE Healthcare, Chicago, IL). The cards were dried out of direct sunlight and then sealed in an impermeable zip bag with a desiccant. They were stored and transported at room temperature for 2 weeks and frozen at −30°C on arrival at Weill Cornell in New York.
Ethics.
Ethical permission for the conduct of this study was obtained from Bugando Medical Center and the National Institute for Medical Research (both in Tanzania) and from Weill Cornell Medical College in New York.
Optimization of RNA quality and yield from DBSsamples.
DBS were sampled using 3 or 6 mm punches (Uni-Core Punch; GE Healthcare Life Sciences, Marlborough, MA). If an experiment used a “full” spot, it was removed using sterile scissors. All RNA extraction protocols included on-column DNase digestion. All RNA extracts were purified and concentrated using the Zymo RNA Purification and Concentration kit (Zymo Research Corp., Irvine, CA) before assessing RNA integrity. RNA concentrations and 260/280 ratios were measured using NanoDrop 8000 (Thermo Fisher Scientific, Inc., Waltham, MA). RNA integrity was evaluated using RNA 6000 pico assay kit (on the Agilent 2100 Bioanalyzer, Santa Clara, CA). The optimization steps are summarized in Supplemental Figure 1.
RNA extraction method.
The starting material for the comparison of three RNA extraction methods was three 3-mm DBS punches per extraction. The methods were as follows: 1) Extraction was performed according to the kit instructions for the Illustra RNAspin Mini RNA Isolation Kit (GE Healthcare Life Sciences) with the agitation method for sample disruption (1 hour incubation in lysis buffer with every 15-minute vortexing). 2) RNA was extracted using the RNeasy Isolation Kit (Qiagen, Hilden, Germany) with sample disruption using a Thermomixer Compact (Eppendorf, Hamburg, Germany) at 37°C and 1,000 rpm for 30 minutes. 3) RNA was extracted as in Method 2, with an additional sample disruption step of centrifugation through a QIAshredder column (Qiagen).
Sample disruption.
We compared agitation and homogenization methods for the initial step of RNA extraction from DBS using a blood spot in 350 μL Buffer RA1 (Illustra Kit, GE Healthcare) and 3.5 μL 2-mercaptoethanol (Sigma-Aldrich Corp, St. Louis, MO). For the agitation method, the blood spot was incubated for 1 hour with 30 seconds of vortexing at high speed every 15 minutes. For the homogenization, the blood spot was homogenized with a homogenizer (Bead Mill 4; Thermo Fisher Scientific, Inc.) using 2.38-mm metal beads in RNase-free 2-mL screw-cap tubes. Homogenization protocol was two rounds of 30 seconds at 5 m/s, the highest setting. For both homogenization methods, RNA was isolated using the Illustra kit.
Optimization of homogenizer settings and input DBS.
Homogenization was in 350 μL Buffer RA1 and 3.5 μL 2-mercaptoethanol. Starting material was three 3-mm punches. Homogenization time was compared at 5 m/s (maximum speed) for 30 seconds versus 60 seconds at 5 m/s. Homogenization speeds were compared for 60 seconds at 1, 3, and 5 m/s. After homogenization, the settings were optimized for three, five, and eight 3-mm punches (Supplemental Figure 1).
Preparation of RNA for transcriptome studies.
RNA from DBS was extracted using eight 3-mm punches for each reaction tube. The punches were homogenized using the Bead Mill at 5 m/s for 60 seconds, extracted using the Illustra kit, and purified using Zymo concentration. RNA from whole blood collected into Tempus RNA isolation tubes (Invitrogen) was extracted with the Tempus Spin RNA Isolation Kit (Invitrogen) according to the manufacturer’s instructions with on-column DNase digestion.
RNA quality assessment.
Following RNA isolation, total RNA integrity was checked using an Agilent Technologies 2100 Bioanalyzer. Amount of RNA present for each patient sample was measured using the NanoDrop 8000 (Thermo Fisher Scientific, Inc.).
RNA sample library preparation.
Preparation of RNA sample library and next-generation sequencing was performed by the Genomics Core Laboratory at Weill Cornell Medicine. Messenger RNA was prepared using TruSeq (Illumina, San Diego, CA), according to the manufacturer’s instructions. Messenger RNA was purified using magnetic beads for six of the whole blood–generated RNA with higher RNA integrity value. For two of the whole blood–generated RNA and all of the DBS-generated RNA, mRNA was purified using biotinylated, target-specific oligos combined with RiboZero rRNA removal beads provided in the Illumina TruSeq Stranded Total RNA Sample Preparation kits.
Sequencing with Illumina HiSeq 4000.
Before performing a sequencing run on the HiSeq 4000, the cBot (Illumina), a fluidics device that hybridizes samples onto a patterned flow cell and amplifies them for later sequencing, was used. The patterned flow cell was sequenced on a HiSeq 4000 sequencer (Illumina) with single-stranded 50-bp cycles. Sequencing quality was assessed using FastQC (Babraham Bioinformatics, Cambridge, United Kingdom). Reads were aligned to the human hg19 reference genome using Tophat213 and counts data were generated using HTSeq-count.14
Statistical methods.
Transcript count data were normalized by library size using DESeq2 (v. 1.161 from Bioconductor 3.4).15 Median count and trimmed mean of M values normalizations were included as part of the analysis, but results did not differ significantly from DESeq2 normalization (data not shown).16 To assess agreement between transcriptomes derived from DBS versus blood RNA, we compared DBS and whole blood RNA transcript counts from the same participants with a paired analysis. Spearman correlations were generated to quantify the strength of the relationship between DBS and whole blood counts. To identify the association between DBS and whole blood counts visually, scatter plots of the two log2 count data were generated. Bland–Altman, or minus versus average (MA), plots were used to visually assess fold changes between DBS and whole blood compared with the average size.17 For each person, difference in log2 counts between DBS and whole blood RNA were plotted against the average of log2 counts. To compare gene lengths for genes with greatest differences in counts between DBS and whole blood RNA, the rank sum test was used. Statistical analysis was performed in R (v. 3.3.2).
RESULTS
Participant characteristics.
The people whose blood was included in this study are summarized in Table 1. Two of the eight people included were men (25%) and the other six were women. The median and interquartile range for age were 33 (27.5–37.5) years. Six of the eight participants were human immunodeficiency virus (HIV)-infected (75%), one had Schistosoma haematobium infection (12.5%), and one had syphilis infection (12.5%). These people are from a cohort described in Downs et al. (manuscript in preparation). Library size was greater in whole blood transcriptomes than in DBS transcriptomes which reflects lower RIN obtainable from RNA extracted from DBS. Library sizes were accounted for as part of normalization for differential expression analyses.
Table 1.
Demographics of study participants
| DBS transcriptome | Whole blood transcriptome | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | Gender | Age (years) | HIV infection | RIN | Total reads | Library size | RIN | Total reads | Library size |
| 1 | Male | 40 | Yes | 1.1 | 19,988,831* | 2,468,097 | 8.2 | 24,151,082 | 10,536,497 |
| 2 | Male | 31 | No | 1.1 | 26,350,541* | 800,788 | 7.5 | 22,607,544 | 14,158,669 |
| 3 | Female | 30 | No | 2.3 | 20,713,021* | 1,667,781 | 6.9 | 29,685,995 | 19,030,425 |
| 4 | Female | 38 | Yes | 1.7 | 22,426,091* | 1,960,781 | 6.3 | 21,246,880 | 10,211,765 |
| 5 | Female | 37 | Yes | 2.4 | 26,206,356* | 4,573,521 | 6.2 | 21,190,805 | 9,636,586 |
| 6 | Female | 20 | Yes | 1.8 | 21,902,329* | 2,335,387 | 7.4 | 21,481,941 | 11,082,507 |
| 7 | Female | 25 | Yes | 1.3 | 23,106,327* | 964,788 | 2.9 | 27,182,033* | 2,205,173 |
| 8 | Female | 35 | Yes | 2.4 | 20,416,063* | 1,469,987 | 2.2 | 26,095,689* | 1,299,210 |
DBS = dried blood spot; RIN = RNA integrity number.
Ribosomal RNA removal by RiboZero. All other libraries were generated from RNA which had been polyA selected.
Optimization of RNA extraction from DBS.
The Illustra RNA MiniSpin Mini RNA isolation kit and the Qiagen RNeasy Micro Kit with additional QIAshredder homogenization step gave comparable RNA concentrations and 260/280 ratios when tested on 3-mm blood spots. Sample disruption using beads and a Bead Mill yielded more RNA than agitation methods, such that there was a visible band on the Agilent electropherogram. The optimal homogenization setting using Bead Mill was one round of 60 seconds at 5 m/s. The number of input DBS that yielded the greatest concentration of RNA was eight 3-mm punches. The Agilent results from eight 3-mm DBS, homogenized using the Bead Mill at 5 m/s for 60 seconds and extracted using the Illustra kit, are shown in Supplemental Figure 1.
Comparison of DBS and Tempus whole blood extractions.
RNA was extracted from DBS using the optimized method described previously, and from whole blood using the Tempus Spin protocol. The RIN reported by Agilent for DBS RNA and whole blood RNA are shown in Table 1. The library sizes were larger for whole blood transcriptome than for DBS transcriptome in seven of eight people (Table 1).
Despite the differences in input RNA concentration and quality, both DBS and whole blood–derived RNA yielded comparable numbers of aligned reads. The average number of reads generated for the DBS-derived transcriptome was 22,638,695, smaller than for blood-derived transcriptome (mean 24,205,246). More than 99.8% of reads passed quality control for both DBS and blood-derived transcriptomes, and the alignment rate for both DBS and blood-derived transcriptomes was 97%.
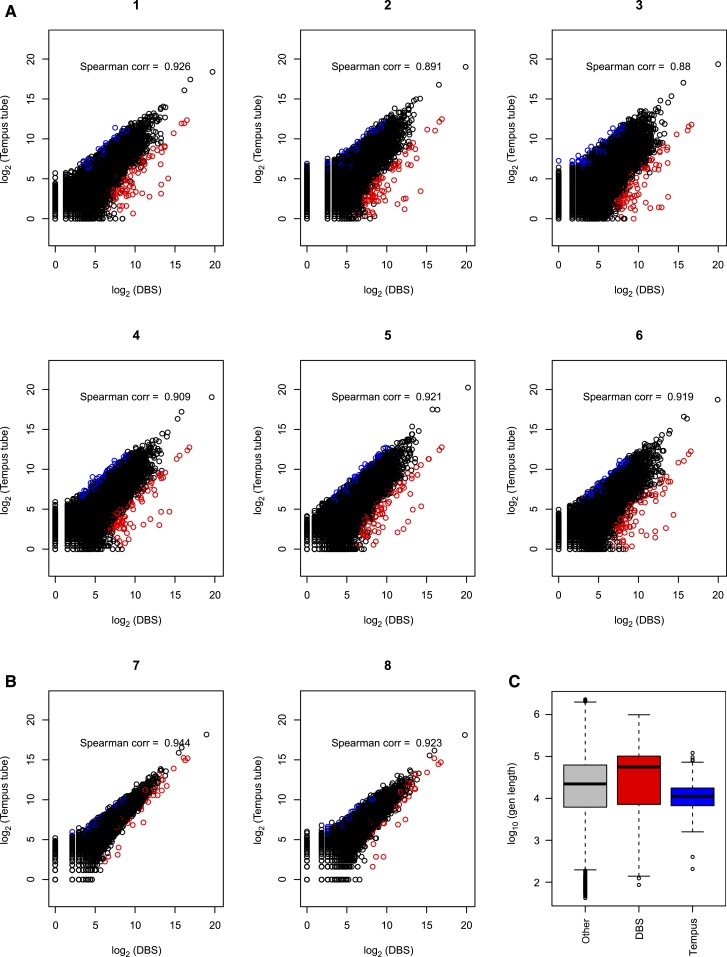
To determine the comparability of transcriptomes derived from DBS compared with blood RNA, we completed a correlation analysis comparing log2-adjusted gene counts in DBS and blood transcriptomes (Figure 1). If the order X is completely preserved in Y, then the Spearman correlation is 1. In Figure 1, each panel corresponds to an individual person with DBS count (X) plotted with whole blood RNA count (Y) for each gene. The sample-specific Spearman correlation coefficients ranged from 0.88 to 0.94. In each scatter plot, larger variation in counts is seen in the low-count area (points in widely stretched band in bottom left corner), whereas strong correlation between two measures is pronounced in the higher counts area (points tightly gathered along 45° line toward upper right corner). Using the rank sum test, the top 1% of genes with increased expression in DBS (Supplemental Table 1) were longer than the genes with increased expression in whole blood (Figure 1; Supplemental Table 2). Genes with increased counts in DBS (median = 61,533 bp, 95% confidence interval [CI] = [45,697, 78,278]) are longer than other genes (median = 32,969 bp, 95% CI = [32,249, 33,718]) by rank sum test (P = 0.003). Genes with increased counts in whole blood (median = 12,010 bp, 95% CI = [10,175, 13,986]) are shorter than other genes by rank sum test (P value < 0.001).
Figure 1.
Correlation of counts for gene expression data from dried blood spot (DBS) RNA and whole blood RNA. The gene counts were plotted and Spearman correlation calculated for six subjects who had polyA selection of non-ribosomal RNA from whole blood (A) and two subjects who had rRNA depletion from whole blood (B). All DBS-derived RNA had rRNA depletion before RNA-seq. Points in red on the graphs are those genes that have average fold changes (fold change in each subject is defined as counts from DBS over whole blood) in the top 1% and average normalized log2 counts greater than five. Points in blue are those genes whose average fold changes fall in the bottom 1% (genes with increased expression in whole blood compared with DBS) and average normalized log2 counts greater than five. Lengths of genes in the top and bottom 1% fold-change groups are compared with the length of remaining 98% genes (taking this as the reference gene set). (C) Genes with increased counts in DBS are longer than other genes by rank sum test (P = 0.003). Genes with increased counts in whole blood are shorter than other genes by rank sum test (P value < 0.001).
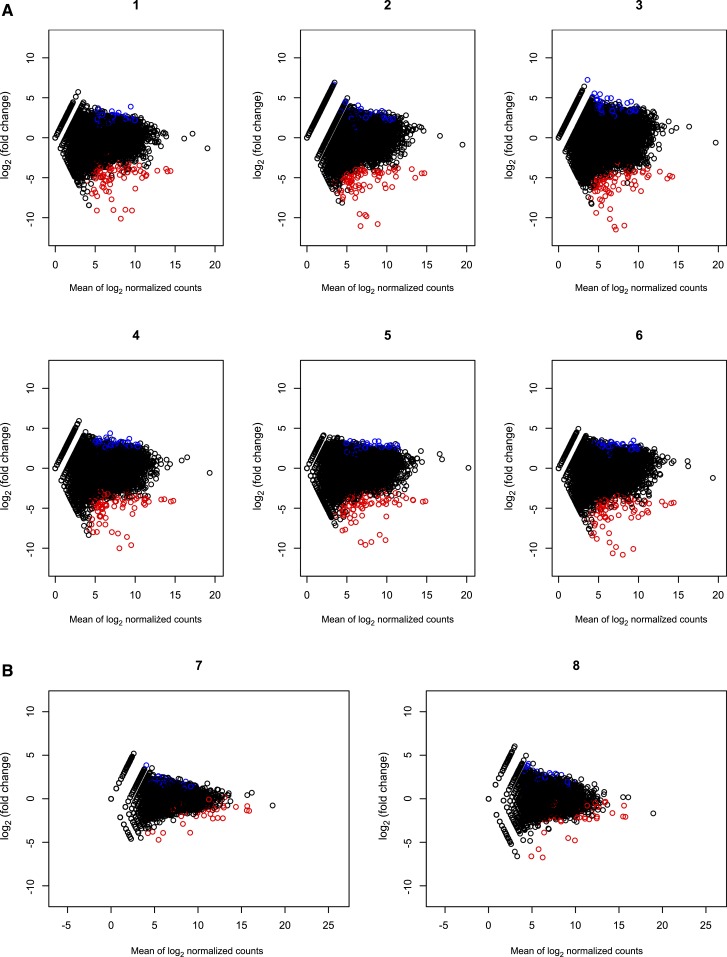
To look at whether there was a trend toward over- or under-estimation of gene counts in DBS and blood transcriptomes, we analyzed the count data agreement between the two RNA sources with MA plots (Figure 2). For each person, difference in log2 counts between DBS and whole blood RNA were plotted against the average of log2 counts. Genes with low average counts, between zero and five on horizontal axis, show higher dispersion on the vertical axis. This higher dispersion indicates that low-count genes have larger discrepancy between DBS versus whole blood readings, and the difference decreases for genes with larger counts. These plots show that there is no bias toward increased or decreased counts when comparing DBS and whole blood gene expression at the transcriptome level. To examine whether the lack of exact correlation was related to the different mRNA selection techniques (polyA selection versus rRNA control) versus to the different starting materials (DBS versus whole blood), we examined the inter-person correlation for each individual. The inter-person correlation for whole blood extractions was 0.97–0.99 when both samples had polyA selection and 0.91–0.94 when the mRNA isolation methods were discordant (Supplemental Figure 2). The inter-sample correlations from DBS were higher than those of whole blood sample pairs with disparate mRNA selection methods, which likely reflects the use of the same mRNA selection technique (Supplemental Figure 3).
Figure 2.
Count data agreement between the dried blood spot (DBS) and whole blood RNA sources were assessed with Bland–Altman, also known as minus vs. average, plots. Genes with lower average counts, between 0 and 5 on the horizontal axis, show higher dispersion on the vertical axis. This higher dispersion indicates that low-count genes have larger discrepancy between DBS vs. whole blood readings. Dispersion was lower when whole blood RNA was rRNA depleted (B) compared with polyA selected (A) before RNA-seq.
DISCUSSION
Our study demonstrates a positive correlation between RNA transcriptomes in paired whole blood and DBS samples. Our work has important practical implications for evaluation of gene expression in studies in which obtaining whole blood RNA is not feasible or practical. The ongoing advancement of RNA sequencing methodologies and ribosomal RNA depletion methods has enabled researchers to obtain usable transcriptome data from increasingly small quantities of RNA that were previously insufficient for transcriptome studies, and from RNA of lower quality than was previously required.18 A recent study reported that heat-degraded RNA could give comparable transcriptome results to non-degraded RNA.2 Our optimization of a protocol that yields RNA transcriptomes from DBS samples that correlate with whole blood RNA transcriptomes is timely and practical, opening possibilities for RNA sequencing in research settings in which gene expression studies were previously thought to be unfeasible because of sample collection and storage challenges.
The whole blood transcriptome, generated from RNA with RIN > 6 with polyA selection for rRNA depletion, yielded larger library sizes. Count data were adjusted for library size, as is the default in multiple gene expression analysis programs, including in DESeq2.8 The differences between transcriptomes from DBS versus whole blood were significant enough that DBS and whole blood–generated RNA transcriptomes are not directly comparable to each other. However the normalized counts data from DBS and whole blood RNA were correlated and suggest that DBS-generated transcriptomes could be used in settings where whole blood collection for RNA studies was not feasible, but gene expression profiling is desired. Studies on deparaffinized biopsy samples have shown similar results, again validating that RNA profiles from suboptimal quality RNA can yield usable RNA transcription profiles.1,19
One limitation of this study is that we did not generate transcriptomes from all stages of optimization of RNA extraction. The projected costs for this were outside the scope of our project, with primary comparison being transcriptomes of optimized RNA from DBS compared with whole blood. To compare directly the method of storage of blood (on filter paper versus frozen in RNA preservation solution), we used DBS where blood was collected from the same venous phlebotomy as the whole blood. One of the advantages of DBS is that blood can be collected from a finger prick, which can be a more feasible method for blood collection in the field. Of note, concentration gradients may exist between capillary blood collected from finger stick and venous blood from phlebotomy, as seen with HIV viral load.20 Because our DBS comparison was performed using DBS with blood spotted from a venous phlebotomy, this likely improves the correlation of counts between DBS and whole blood transcriptome.
In conclusion, we provide a protocol and evidence of correlation between DBS samples and whole blood for RNA gene expression studies. Our work represents an important and practical step forward for broadening the applicability of RNA-seq to DBS samples—which were previously considered to yield insufficient quality and quantity of RNA. Populations for whom DBSs are typically collected include harder-to-reach groups, including pediatric patients, those in resource-limited settings without a cold-storage chain, and studies for which blood spots but not whole blood were archived. Our work opens new avenues for studies of gene expression and highlights the need for additional investigation to confirm and extend our findings.
Supplementary Material
Supplemental Figures and Tables
Acknowledgments:
We would like to thank the Mwanza, Tanzania-based team for their field work and blood collection, especially Donald Miyaye, Ruth Magawa, Jane Mlingi, Ndalloh Paul, and Inobena Tosiri. We also recognize the Genomics Core laboratory at Weill Cornell Medical College for generating RNA-seq data.
Note: Supplemental figures and tables appear at www.ajtmh.org.
REFERENCES
- 1.Hedegaard J, et al. 2014. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 9: e98187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuierer S, Carbone W, Knehr J, Petitjean V, Fernandez A, Sultan M, Roma G, 2017. A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples. BMC Genomics 18: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patton JC, Akkers E, Coovadia AH, Meyers TM, Stevens WS, Sherman GG, 2007. Evaluation of dried whole blood spots obtained by heel or finger stick as an alternative to venous blood for diagnosis of human immunodeficiency virus type 1 infection in vertically exposed infants in the routine diagnostic laboratory. Clin Vaccine Immunol 14: 201–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gauffin F, Nordgren A, Barbany G, Gustafsson B, Karlsson H, 2009. Quantitation of RNA decay in dried blood spots during 20 years of storage. Clin Chem Lab Med 47: 1467–1469. [DOI] [PubMed] [Google Scholar]
- 5.Ponnusamy V, Kapellou O, Yip E, Evanson J, Wong LF, Michael-Titus A, Yip PK, Shah DK, 2016. A study of microRNAs from dried blood spots in newborns after perinatal asphyxia: a simple and feasible biosampling method. Pediatr Res 79: 799–805. [DOI] [PubMed] [Google Scholar]
- 6.Maeno Y, Nakazawa S, Nagashima S, Sasaki J, Higo KM, Taniguchi K, 2003. Utility of the dried blood on filter paper as a source of cytokine mRNA for the analysis of immunoreactions in Plasmodium yoelii infection. Acta Trop 87: 295–300. [DOI] [PubMed] [Google Scholar]
- 7.Haak PT, Busik JV, Kort EJ, Tikhonenko M, Paneth N, Resau JH, 2009. Archived unfrozen neonatal blood spots are amenable to quantitative gene expression analysis. Neonatology 95: 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khoo SK, Dykema K, Vadlapatla NM, LaHaie D, Valle S, Satterthwaite D, Ramirez SA, Carruthers JA, Haak PT, Resau JH, 2011. Acquiring genome-wide gene expression profiles in Guthrie card blood spots using microarrays. Pathol Int 61: 1–6. [DOI] [PubMed] [Google Scholar]
- 9.Ho NT, et al. 2013. Gene expression in archived newborn blood spots distinguishes infants who will later develop cerebral palsy from matched controls. Pediatr Res 73: 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grauholm J, Khoo SK, Nickolov RZ, Poulsen JB, Baekvad-Hansen M, Hansen CS, Hougaard DM, Hollegaard MV, 2015. Gene expression profiling of archived dried blood spot samples from the Danish Neonatal Screening Biobank. Mol Genet Metab 116: 119–124. [DOI] [PubMed] [Google Scholar]
- 11.McDade TW, Ross K, Fried R, Arevalo JM, Ma J, Miller GE, Cole SW, 2016. Genome-wide profiling of RNA from dried blood spots: convergence with bioinformatic results derived from whole venous blood and peripheral blood mononuclear cells. Biodemography Soc Biol 62: 182–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bybjerg-Grauholm J, Hagen CM, Khoo SK, Johannesen ML, Hansen CS, Baekvad-Hansen M, Christiansen M, Hougaard DM, Hollegaard MV, 2017. RNA sequencing of archived neonatal dried blood spots. Mol Genet Metab Rep 10: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anders S, Pyl PT, Huber W, 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson MD, Oshlack A, 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altman DG, Bland JM, 1983. Measurement in medicine—the analysis of method comparison studies. Statistician 32: 307–317. [Google Scholar]
- 18.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T, 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webster AF, Zumbo P, Fostel J, Gandara J, Hester SD, Recio L, Williams A, Wood CE, Yauk CL, Mason CE, 2015. Mining the archives: a cross-platform analysis of gene expression profiles in archival formalin-fixed paraffin-embedded tissues. Toxicol Sci 148: 460–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fajardo E, Metcalf CA, Chaillet P, Aleixo L, Pannus P, Panunzi I, Triviño L, Ellman T, Likaka A, Mwenda R, 2014. Prospective evaluation of diagnostic accuracy of dried blood spots from finger prick samples for determination of HIV-1 load with the NucliSENS Easy-Q HIV-1 version 2.0 assay in Malawi. J Clin Microbiol 52: 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures and Tables