Abstract
The selectivity of an enzyme inhibitor is a key determinant of its usefulness as a tool compound or its safety as a drug. Yet selectivity is never assessed comprehensively in the early stages of the drug discovery process, and only rarely even in the later stages, because technical limitations prohibit doing otherwise. Here, we report EnPlex, an efficient, high-throughput method for simultaneously assessing inhibitor potency and specificity, and pilot its application to 96 serine hydrolases. EnPlex analysis of widely used serine hydrolase inhibitors revealed numerous previously unrecognized off-target interactions, some of which may help to explain previously confounding adverse effects. In addition, EnPlex screening of a hydrolase-directed library of boronic acid- and nitrile-containing compounds provided dual potency/selectivity structure-activity relationships from which lead candidates could be more effectively prioritized. Follow-up of a series of dipeptidyl peptidase 4 (DPP4) inhibitors showed that EnPlex indeed predicted efficacy and safety in animal models. These results demonstrate the feasibility and value of high-throughput, superfamily-wide selectivity profiling, and suggest such profiling can be incorporated into the earliest stages of drug discovery.
INTRODUCTION
Enzyme inhibitor discovery has typically followed a sequential process in which inhibitors for a chosen target are first identified, optimized for potency, and then checked for selectivity1, 2. Greater effort is typically devoted to addressing potency, with selectivity analysis often limited to testing a handful of lead candidates against closely related enzymes. As a result, off-target effects are often discovered only in the late stages of drug development, in many cases resulting in clinical failure because of unanticipated off-target toxicity. Meanwhile, potentially highly selective inhibitors may be discarded early in the course of discovery because they are slightly less potent than others and there is no systematic way to recognize their specificity. An alternative, perhaps more efficient and productive strategy might be one where compound libraries are screened against a large panel of related enzymes from the outset 2. In principle, this approach would simultaneously identify hits for many enzymes, and would enable lead inhibitor selection and medicinal chemistry optimization for each enzyme of interest to be based on both potency and selectivity. In practice, however, there is no method able to accomplish these goals. Some progress has recently been made in the family-wide profiling of kinase inhibitors3–5, although the throughput of such assays remains modest. Unfortunately, high-throughput, family-wide assays are entirely lacking for all other enzyme families.
For example, the serine hydrolases are one of the largest enzyme superfamilies in Nature, with ~240 members in humans alone6. They play key roles in diverse biological processes such as blood clotting, glucose homeostasis, neural signaling, and bacterial and viral infection. Members of this superfamily, including ones of human, viral, and bacterial origin, are validated targets for more than a dozen FDA-approved drugs6. Many others are the targets of inhibitor discovery efforts where the objective is first to use the inhibitors as chemical probes of the hydrolase’s biological function, and then ultimately as a lead candidate for clinical development 6, 7. The structural and mechanistic characteristics of the serine hydrolases make off-target interactions far more likely to occur within rather than outside the superfamily. For example, all serine hydrolases share a catalytic mechanism featuring an usually reactive serine hydroxyl group in their active sites. As a result, electrophillic groups are widely employed in designing inhibitors targeting these enzymes, thereby dramatically increasing the probability of intra-superfamily cross-reactivities. Unfortunately, screening even a single serine hydrolase inhibitor against the entire superfamily, let alone hundreds or thousands of compounds, is not feasible with current technologies.
We therefore sought to develop a method for high-throughput, superfamily-wide serine hydrolase activity profiling, reasoning that if the approach was successful, it could be subsequently expanded to other enzyme families. We believed that such a technology would enable not only 1) the rapid selectivity profiling of the many existing serine hydrolase drugs and chemical probes, but also 2) enable a large-scale, superfamily-wide screening approach for the development of new inhibitors. We recognized that competitive activity-based protein profiling (ABPP) had the potential to form the basis of this technology8. Activity-based probes typically possess a reactive chemical group that covalently interacts with the active-site residues of a large number of mechanistically related enzymes, and a tag (for example, a biotin or fluorophore) that enables the detection and/or enrichment of probe-labeled enzymes. The prototypical activity-based probe for serine hydrolases relies on a fluorophosphonate chemical group that reacts with their active site serine nucleophiles9. ABPP is typically used for inhibitor identification by assessing the ability of compounds to block the labeling of enzymes with an activity-based probe directly in a complex cell or tissue lysate10. However, current ABPP methodologies permit screening only a limited number of compounds against a small fraction of the serine hydrolase superfamily. The number of compounds is limited by the low-throughput of the gel or mass-spectrometry (MS) readouts used to analyze the probe-labeled enzymes, and the number of enzymes is limited by the small fraction of active enzymes present in any particular lysate. A recently described method to monitor the enzyme-probe reaction by fluorescence polarization (fluopol-ABPP) dramatically increases throughput, but can evaluate only a single enzyme at a time and requires large amounts of purified protein, thereby restricting its utility to those proteins easily produced in high yields11. Therefore, despite a number of recent advances, current ABPP methods cannot yet screen large numbers of compounds across entire enzyme families (see Supplementary Results, Supplementary Table 1 for a comparison of competitive ABPP methods).
Here we report a method, called EnPlex, that enables multiplexed, high-throughput serine hydrolase inhibitor screening, yet only requires small (picomolar) quantities of protein. We used EnPlex to simultaneously screen >90 serine hydrolases in each well of a high-throughput screening plate against a panel of widely used inhibitors at multiple doses—thereby instantaneously providing half-maximal inhibitory concentration (IC50) values for every hit. The results revealed hundreds of previously unknown enzyme-inhibitor interactions, including several for FDA approved drugs. EnPlex was also used to screen a library of ~300 boronic acid- and nitrile-containing compounds, identifying lead inhibitors for many enzymes on the basis of both selectivity and potency. Among the many notable examples was a remarkably selective boronic acid-based DPP4 inhibitor that exhibited both efficacy and safety in animal models.
RESULTS
Development of a multiplexed, high-throughput assay
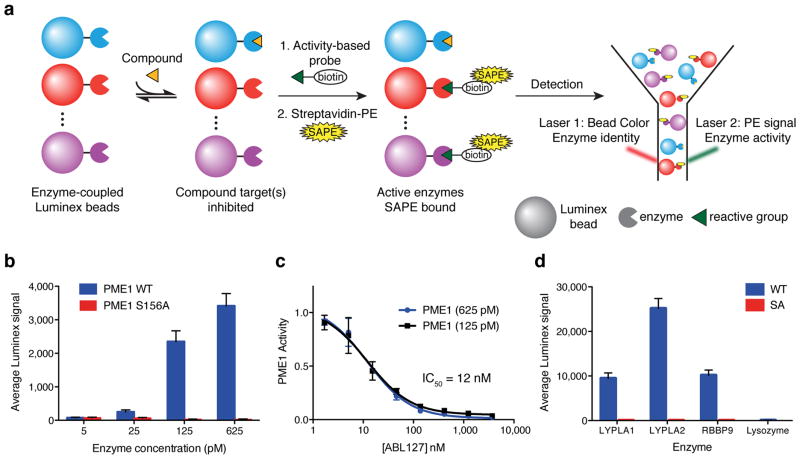
We hypothesized that a high-throughput, superfamily-wide multiplexed profiling method could be developed by coupling serine hydrolases to polystyrene microspheres (Luminex beads), each of which is dyed a different color (Fig. 1a). Performing competitive ABPP experiments on the Luminex bead-bound enzymes, where inhibitors compete with a biotinylated activity-based probe for binding the enzyme active sites, then permits a two-laser flow cytometer to simultaneously identify each enzyme (by the bead color) and its degree of inhibition (by streptavidin-phycoerythrin staining).
Figure 1.
The EnPlex platform. (a) Schematic representation. Purified enzymes are coupled to Luminex microspheres, with a different bead color for each enzyme. Multiplexed bead complexes are incubated with a compound before being treated with a biotinylated activity-based probe and a streptavidin R-phycoerythrin conjugate (SAPE). The mixtures are scanned on a Luminex flow cytometer, where one laser detects the bead color (enzyme identity) and a second laser detects the R-phycoerythrin signal (enzyme activity). (b) A strong Luminex signal was observed with wild-type (WT) bead-coupled PME1 at low enzyme concentrations. No signals were observed with the catalytically dead S156A mutant PME1. Error bars represent the s.d of three independent experiments. The enzyme concentration was calculated assuming 100% of the protein was coupled to the beads. (c) IC50 curve for the ABL127-PME1 interaction determined by EnPlex. Note that the exact enzyme concentration is not important for EnPlex, as both PME1 concentrations gave identical IC50 values. Data are means ± s.e.m of three independent experiments. (d) Luminex signals for WT and inactive (SA) LYPLA1, LYPLA2, and RBBP9, and for WT lysozyme (1 nM of each enzyme was used). Error bars represent the s.d of three independent experiments.
We first tested the EnPlex idea with the serine hydrolase protein phosphatase methylesterase 1 (PME1), which was recently screened by conventional ABPP methods 12, 13. We coupled varying amounts of purified PME1 to equivalent numbers of microspheres (Fig. 1b) and observed robust signal with only 125 pM of PME1, a concentration 8,000-fold below that required for fluopol-ABPP 13. Mutation of the PME1 serine nucleophile to alanine completely abolished the signal, demonstrating that it truly reflects enzyme activity and not simply abundance (Fig. 1b). We next incubated beads with the PME1 inhibitor ABL127 before probe labeling, and observed dose-dependent inhibition of PME1 activity with an IC50 value of 12 nM, nearly identical to previously reported IC50 measurements (Fig. 1c) 13. Testing of three additional pairs of wild-type and catalytically inactive mutant serine hydrolases [acyl-protein thioesterase 1 (LYPLA1), acyl-protein thioesterase 2 (LYPLA2), and retinoblastoma-binding protein 9 (RBBP9)] all yielded similar activity-based signals (Fig. 1d). In contrast, the enzyme lysozyme, which is not a serine hydrolase, generated no signal (Fig. 1d). These results indicate that the EnPlex method is sensitive, specific and in principle scalable to all members of the serine hydrolase superfamily in a single, multiplexed assay.
To test the feasibility of large-scale screening, we next produced a diverse panel of 103 of the ~240 serine hydrolases (Supplementary Fig. 1 and Supplementary Table 2). This enzyme panel spans the entire serine hydrolase superfamily, and notably includes active, soluble forms of membrane-associated and transmembrane domain-containing enzymes [e.g., fatty-acid amide hydrolase 1 (FAAH), monoglyceride lipase (MGLL), and monoacylglycerol lipase ABHD6 (ABHD6)]. After purification, the enzymes were incubated with the FP-biotin probe and analyzed by Western blotting with a strepavidin-conjugated infrared dye to confirm probe-labeling (Supplementary Fig. 2). Only two enzymes, tripeptidyl-peptidase 2 (TPP2) and valacyclovir hydrolase (BPHL), did not detectably label with the probe, indicating that they were either inactive or were not sensitive to this reagent. Of the 101 active, purified enzymes, 94 (93%) also generated strong signals with high reproducibility in the Luminex format (Supplementary Fig. 3). Importantly, we found that pure proteins were not required for this assay; they only need to be separated from other FP-sensitive enzymes. Accordingly, most proteins needed only a single purification step (Supplementary Fig. 2c). The results demonstrate the feasibility of profiling the activity of ~ 40% of the entire serine hydrolase superfamily in a single well of a 384-well plate – by far the most comprehensive such assay reported to date.
Serine hydrolase inhibitor profiling by EnPlex
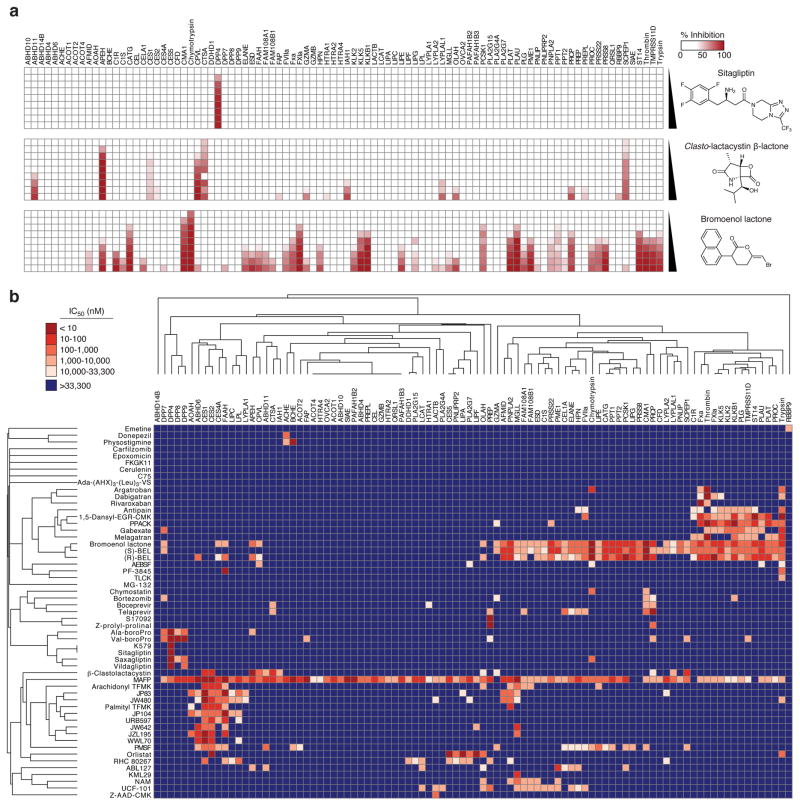
Having established an assay capable of superfamily-wide activity profiling, we next asked whether EnPlex could rapidly recapitulate what has taken years to discover using traditional methods, and perhaps also yield new insights. To do this, we assembled a panel of well-characterized serine hydrolase inhibitors (and related proteasome inhibitors) (Supplementary Table 3). These 55 small-molecules span multiple mechanisms of inhibition (reversible, covalent-reversible, and irreversible) with varying degrees of potency. Replicate profiling of each compound was performed at 9 doses, yielding a total of ~110,000 enzyme-compound interactions in a single experiment (Fig. 2a, Supplementary Fig. 4, and Supplementary Fig. 5). We calculated IC50 values for each compound and enzyme tested (Supplementary Data Set 1), and observed that our values for known enzyme-inhibitor interactions, whether reversible or irreversible, were largely concordant with published measurements (Supplementary Table 4). That reversible interactions, for example dipeptidyl peptidase 4 (DPP4) with sitagliptin (Fig. 2a, top panel), thrombin with argatroban, melagatran, and dabigatran (Supplementary Fig. 4d), and RBBP9 with emetine (Supplementary Fig. 4i), were readily identified underscores the versatility of the EnPlex method (and ABPP in general). In theory, reversible inhibitors could be outcompeted by covalent activity-based probes over time, but performing experiments using this probe concentration and labeling time can clearly reveal reversible inhibitors without requiring specifically optimized assay conditions11. In order to take a global view of serine hydrolase inhibitory activity, we next performed hierarchical clustering of these IC50 values (Fig. 2b) As expected, structurally related compounds (e.g., O-aryl carbamates and compounds containing arginine mimetics) generally clustered together, as did enzymes with high sequence similarity and good pharmacological coverage (e.g., the dipeptidyl peptidases). Importantly, we also observed shared pharmacological sensitivities between unrelated enzymes, demonstrating that structural relationships between active sites often cannot be predicted from protein sequences alone.
Figure 2.
Large-scale profiling of serine hydrolase-inhibitor interactions. (a) Complete dose-response profiles of sitagliptin, clasto-lactacystin β-lactone, and bromoenol lactone, which represent varying degrees of selectivity. Compounds were assayed at nine doses (from 5 nM to 33 μM) in duplicate or triplicate. The percent inhibition at each concentration relative to DMSO controls is shown. (b) Two-dimensional hierarchical cluster analysis of IC50 values obtained by EnPlex. Values are listed in Supplementary Data Set 1.
Revealing off-targets of probes and drugs
Even a cursory inspection of the EnPlex data set demonstrates that it efficiently identified previously unrecognized targets of commonly used drugs and tool compounds. For example, clasto-lactacystin β-lactone, the active metabolite of lactacystin, was originally discovered as a proteasome inhibitor 14 with its only other known target being lysosomal protective protein (CTSA)15, 16. EnPlex not only confirmed inhibition of CTSA, but also revealed strong inhibition of its closest two homologs [probable serine carboxypeptidase CPVL (CPVL) and retinoid-inducible serine carboxypeptidase (SCPEP1)], as well as of the evolutionarily distant enzymes acylamino-acid-releasing enzyme (APEH) and liver carboxylesterase 1 (CES1) (Fig. 2a, middle panel). The biological effects of lactacystin have long been presumed to be mediated exclusively through proteasome inhibition 14, 17, but these results raise the possibility that some of its biological effects may be mediated at least in part by inhibition of one or more of these newly identified targets, or that lactacystin may have other biological effects hitherto not recognized. Similarly, EnPlex revealed new targets of the proteasome inhibitor bortezomib, a drug used to treat the B-cell malignancy multiple myeloma18. The principal side effect of bortezomib is peripheral neuropathy, which is reduced in the second-generation proteasome inhibitor carfilzomib19. Bortezomib, but not carfilzomib, has been reported to inhibit seven serine proteases in addition to the proteasome, possibly explaining their different toxicity profiles 19. EnPlex confirmed all of these interactions with the exception of serine protease HTRA2 (HTRA2), but follow up with a traditional substrate assay confirmed that the EnPlex HTRA2 result was correct: bortezomib did not significantly inhibit HTRA2 activity (Supplementary Fig. 6a). Moreover, EnPlex revealed that bortezomib inhibits seven additional enzymes not previously recognized as targets, including lysosomal Pro-X carboxypeptidase (PRCP), granzyme A (GZMA), and plasma kallikrein (KLKB1) (Supplementary Table 5 and Supplementary Fig. 6b). In contrast, carlfilzomib did not interact with any enzymes in the panel. Notably, the serine protease targets of bortezomib share little sequence identity or substrate specificity, making it unlikely that these off-target hits could have been predicted computationally. Thus, EnPlex not only comprehensively confirmed the known inhibitory profile of well-characterized inhibitors, but it also efficiently revealed additional targets not previously known. We note, however, that while the bortezomib results provide a good example of the comprehensiveness of EnPlex, it remains to be determined whether these newly discovered off-target effects explain the peripheral neuropathy that often limits bortezomib’s clinical use.
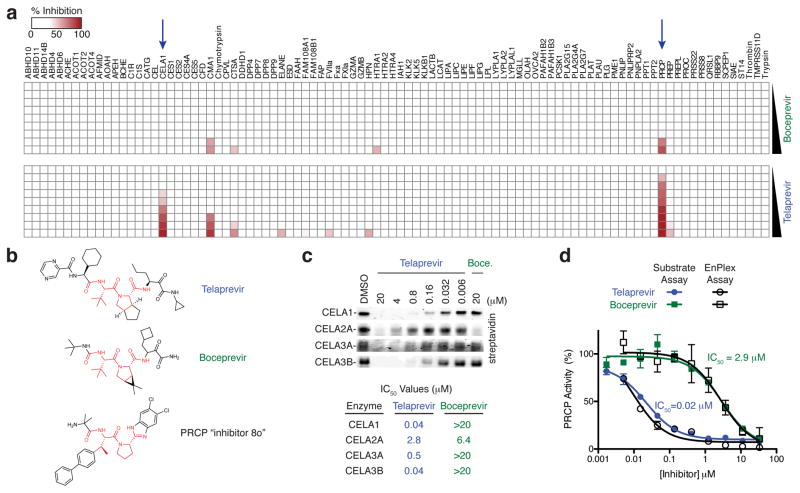
EnPlex also revealed new insights into the drug telaprevir, which was recently FDA-approved for the treatment of hepatitis C virus (HCV) infection. Telaprevir is an effective inhibitor of the HCV NS3 serine protease, but it is associated with serious, sometimes fatal, skin reactions20, 21, presumed to be mediated by off-target inhibition of a human protease(s). Unlike telaprevir, the structurally similar NS3 protease inhibitor boceprevir does not induce such rash, and therefore presumably does not engage the same off-target human enzyme (Fig. 3a,b). EnPlex identified two serine hydrolases, chymotrypsin-like elastase family member 1 (CELA1) and PRCP, that were potently inhibited by telaprevir but not boceprevir (Fig. 3a). Intriguingly, CELA1 is exclusively expressed in skin 22, making it an attractive candidate for mediating telaprevir’s skin toxicity, but its physiological functions have yet to be determined. Given the CELA1 result, we extended EnPlex analysis to the related chymotrypsin-like elastase family members CELA2A, CELA3A and CELA3B (which were not among the 94 enzymes screened initially). Both compounds inhibited CELA2A with similar potencies, but only telaprevir inhibited CELA3A and CELA3B (Fig. 3c). However, as CELA3A and CELA3B are expressed only in the pancreas 23, it seems unlikely that their inhibition contributes to telaprevir-associated rash. We note that whereas telaprevir’s inhibition of CELA1 might have been anticipated because both CELA1 and NS3 protease have chymotrypsin-like folds and similar substrate specificities 24–26, such is not the case with PRCP (validation of which was confirmed using a fluorometric substrate assay (Fig. 3d)), which has an α/β hydrolase fold and cleaves C-terminal amino acids after proline27. Interestingly, both boceprevir and telaprevir share a structural feature with a recently described PRCP inhibitor, compound “8o” 28, that is striking: a central proline-like residue (Fig. 3b, colored red). Whether PRCP, whose function has been implicated in hypertension and appetite regulation 28–30, contributes to the telaprevir-induced rash remains to be established. Regardless, this result further highlights the power of EnPlex to detect off-target interactions that are not easily predicted by enzyme sequence, structural, or known substrate specificity similarities.
Figure 3.
Comparative selectivity profiling of telaprevir and boceprevir. (a) Complete EnPlex profiles. The blue arrows indicate the enzymes inhibited by telaprevir. (b) Structures of telaprevir, boceprevir, and PRCP inhibitor “compound 8o”. Similar portions of these compounds are colored red. (c) Gel-based competitive ABPP analysis with the FP-biotin probe against the chymotrypsin-like elastases. Full gel images are shown in Supplementary Figure 11. IC50 values determined from three independent experiments are shown. (d) PRCP IC50 curves obtained by EnPlex and by a fluorometric substrate assay. Data are means ± s.e.m of three independent experiments.
Lead inhibitor discovery by EnPlex
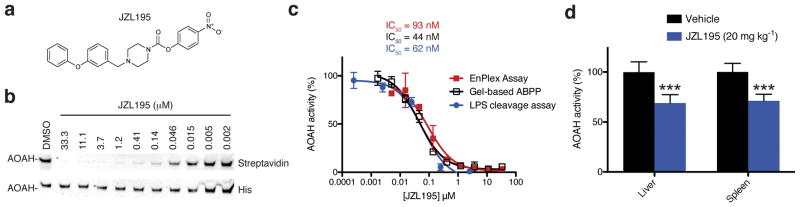
We next asked whether EnPlex could identify inhibitors for enzymes for which inhibitors have not yet been reported. Many such hits were identified, seven of which were confirmed in secondary assays (Fig. 4a,b and Supplementary Fig. 7). For example, EnPlex indicated that JZL195 (originally developed as a dual FAAH/MGLL inhibitor)31 potently inhibits acyloxyacyl hydrolase (AOAH), which cleaves secondary acyl chains from bacterial lipopolysaccharides (LPS) 32. JZL195 has been previously characterized by gel- and MS-based ABPP, but the AOAH interaction was repeatedly missed because AOAH was not expressed in the mouse tissues evaluated 31, 33. Genetic inactivation of AOAH alters innate antibody responses to Gram-negative bacteria and prolongs LPS-induced immune tolerance and immunosuppression 34, 35, but AOAH inhibitors have yet to be reported. We confirmed that JZL195 indeed inhibits deacylation of LPS by purified AOAH in a radioactivity-based assay (Fig. 4c) 36. Moreover, in vivo treatment of mice with JZL195 (20 mg kg−1, i.p.), resulted in inhibition of liver and spleen AOAH activity, as detected with a radioactivity-based LPS deacylation assay (Fig. 4d). This experiment clearly showed that JZL195 inhibits AOAH in vivo, demonstrating the power of EnPlex to function as an unbiased approach to discovering new uses for existing drugs.
Figure 4.
JZL195 inhibits AOAH deacylation of LPS in vitro and in vivo. (a) Structure of JZL195. (b) Confirmation of the interaction between JZL195 and AOAH by gel-based competitive ABPP. Full gel images are shown in Supplementary Figure 11. (c) IC50 curves for the JZL195-AOAH interaction as determined by EnPlex, gel-based ABPP, and in a radioactivity-based LPS deacylation assay. Data are means ± s.e.m of three independent experiments. (d) Mice treated with vehicle or JZL195 (20 mg kg−1, i.p., 6 h) were sacrificed and AOAH activity was measured in the indicated tissue lysates using a LPS deacylation assay. ***p < 0.001 for vehicle versus JZL195 groups. Data are presented as mean values ± s.e.m.; n = 4–6/group.
Incorporating specificity testing into primary screening
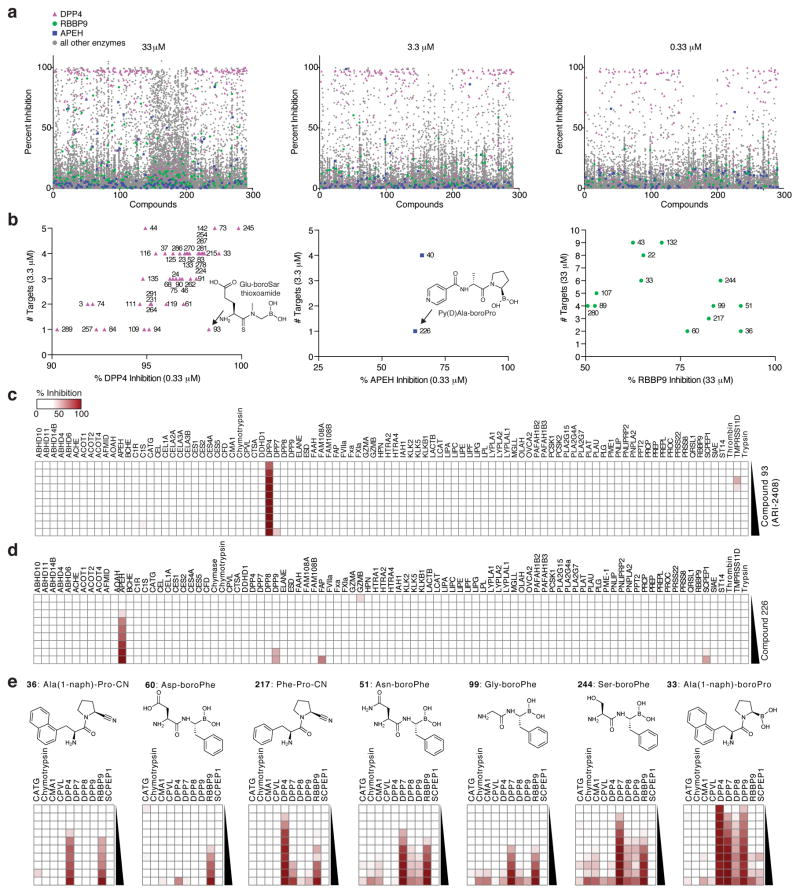
The experiments described above involved the EnPlex profiling of compounds that had been optimized for a particular serine hydrolase target(s), and our results showed that even for such optimized compounds, previously unrecognized off-target effects abound. We therefore asked whether such specificity testing might be incorporated into the earliest stages of drug discovery, rather than only late in optimization process. To address this, we turned to a library of ‘hydrolase-directed’ electrophilic compounds. Electrophilic groups often drive potent enzyme inhibition (as is the case with the FDA-approved drugs saxagliptin and bortezomib), but this same electrophilicity often leads to lack of specificity 6, 12. We hypothesized that EnPlex could prioritize compounds based on their potency and specificity directly in the primary screen. To test this premise, we synthesized 291 electrophiles (261 boronic acids and 30 nitriles (Supplementary Table 6)) and screened these compounds at three doses (Fig. 5a, Supplementary Fig. 8), essentially performing 96 primary screens simultaneously, one for each enzyme. However, unlike traditional primary screens, EnPlex reported both the potency and the selectivity of each compound for each enzyme. Moreover, since the compounds shared structural features, the results also revealed structure-activity relationships (SAR) in both potency and selectivity dimensions.
Figure 5.
EnPlex analysis of a boronic acid and nitrile compound library. (a) Primary screening of all compounds (numbers correspond to Supplementary Table 6) against all enzymes at three concentrations. DPP4, RBBP9, and APEH are colored as indicated, and all other enzymes are colored gray. (b) Hit compounds for DPP4, APEH, and RBBP9 are displayed according to potency (x-axis) and selectivity (y-axis). “# Targets” refers to the number of enzymes inhibited >50% at the indicated concentration. Complete EnPlex profiles for (c) the DPP4 inhibitor ARI-2408 (from 1.7 nM to 33 μM) and (d) the APEH inhibitor 226 (from 5 nM to 33 μM) are shown. (e) EnPlex profiles for the enzymes inhibited by the most potent RBBP9 inhibitors. These inhibitors have similar potencies against RBBP9, but widely varying selectivities.
For example, EnPlex identified two inhibitors of APEH, a serine peptidase that cleaves acetylated N-terminal amino acids from proteins 37 (Fig. 5b, middle panel). Only one of these, compound 226 (Py(D)Ala-boroPro), inhibited APEH selectively (Fig. 5b, d). We confirmed this interaction in a substrate assay (IC50 = 104 nM) (Supplementary Fig. 9). Compound 226 thus represents the more attractive starting point for further optimization. Similarly, EnPlex identified numerous inhibitors of the enzyme RBBP9 (Fig. 5b, right panel), which confers resistance to the growth inhibitory effects of TGF-β38, binds the tumor suppressor RB138, and contributes to pancreatic neoplasia39, but whose endogenous substrate(s) remains unknown and lacks useful inhibitors despite considerable prior efforts11, 40. The EnPlex hit compounds belong to two structural classes: Xaa-Pro-CN and Xaa-boroPhe (Fig. 5e). Compounds 36 (Ala(1-naph)-Pro-CN, IC50 = 0.88 μM) and 60 (Asp-boroPhe, IC50 = 1.08 μM) are the most selective of these compounds, compared to others that inhibit as many as 8 other enzymes in the assay. Compounds 36 and 60, even before any optimization, represent the best RBBP9 inhibitors known to date and give insights for further medicinal chemistry optimization. For example, the increased size of the aromatic group on 36 compared with 217 increases potency against RBBP9 and decreases potency against DPP4 (Fig. 5e), suggesting further derivatization of this part of the molecule may give even greater improvements.
Next, we focused on DPP4, the key enzyme that cleaves and inactivates the incretin glucagon-like peptide 1 (GLP-1) 41, and, owing to this activity, is now a validated target for inhibitors for the treatment of type 2 diabetes42, 43. Dipeptide boronic acids, especially those with a Xaa-boroPro motif, were among the first and most potent inhibitors of DPP4, but perplexing adverse effects in animals hampered their development as clinical candidates44, 45. Off target inhibition of dipeptidyl peptidases 8 and 9 (DPP8 and DPP9) was proposed to explain the adverse effects, but this hypothesis was never fully established and remains controversial even to this day 46. EnPlex confirmed that many boroPro-based compounds are indeed potent inhibitors of DPP4, but also revealed, not surprisingly, that a great many of these are also highly promiscuous, inhibiting multiple other serine hydrolases (Fig. 5b, left panel). For example, compound 24 (Ala-boroPro thioxoamide, also known as ARI-2243) potently inhibited DPP4 (IC50 = 0.7 nM), but also inhibited dipeptidyl peptidase 7 (DPP7), DPP8, DPP9, PRCP, and APEH (Supplementary Fig. 8 and Supplementary Table 7). However, EnPlex also revealed that several boronic acid-based compounds are both highly potent and specific inhibitors of DPP4. For example, compound 93 (Glu-boroSar thioxoamide, also known as ARI-2408), inhibits DPP4 with an IC50 of 1.7 nM and shows no significant off-target interaction with any of the other 95 serine hydrolases assayed (Fig. 5b, c). The EnPlex results indicate that compounds ARI-2243 and ARI-2408 should have essentially equal anti-hyperglycemic activity, but that ARI-2243 presents far greater risks of adverse effects from off-target interactions. Prior to EnPlex profiling and therefore entirely independent of these results, ARI-2243 and ARI-2408 were part of a DPP4 drug discovery effort at Arisaph Pharmaceuticals, and consistent with the EnPlex results, both exhibited highly effective and essentially equal anti-hyperglycemic glycemic activity in mice (Supplementary Fig. 10). More importantly, whereas both ARI-2408 and ARI-2243 exhibited no significant adverse effects in mice, only ARI-2408 proved completely safe in non-human primate studies; ARI-2243 exhibited serious adverse effects even at low doses (Supplementary Table 8 and Supplementary Note 1). These data not only confirm that boronic acids can indeed inhibit an enzyme with high specificity, but also demonstrate the power of incorporating EnPlex specificity testing into the earliest stages of drug discovery. Had the EnPlex results been available earlier, ARI-2408 could have been immediately prioritized over other candidate DPP4 inhibitors.
DISCUSSION
Here we have described a first-ever multiplexed, high-throughput enzyme inhibitor screening method, and have demonstrated its power by profiling entire small-molecule libraries at multiple doses against a significant portion of the human serine hydrolases. The results revealed numerous previously unknown off-target interactions for well characterized and widely used drugs, and identified new lead inhibitors for enzymes for which no inhibitors have previously been reported. Many of the newly discovered interactions were not predictable based on protein structure or enzyme specificity, underscoring the importance of superfamily-wide inhibitor profiling.
EnPlex, inspired by the gel- and MS-based competitive ABPP methods, represents a significant advance over these methods in terms of enzyme superfamily coverage, throughput, reproducibility, and ease of use (Supplementary Table 1). In the largest screen prior to this work, gel-based competitive ABPP was used to assay ~140 inhibitors against 72 serine hydrolases 12. However, that effort included only one replicate at a single compound dose, required the separation of enzymes into many gel-resolvable groups (~4–6 enzymes in ~15 separate groups), took months to perform, and the assay output was a large number of gel images that were difficult to quantify. In contrast, EnPlex is capable of screening orders of magnitude more compounds, all in a single day, and yields quantitative, easily analyzable results. Further, EnPlex is substantially more sensitive than the high-throughput, single-plex fluopol-ABPP platform (~1 μg versus ~1 mg of protein for every 1,000 wells for EnPlex and fluopol-ABPP, respectively). This advantage is particularly important because many enzymes are difficult to express and purify in large quantities. As a result, EnPlex permits the screening of many enzymes previously inaccessible to high-throughput analysis, including for example abhydrolase domain-containing 4 (ABHD4), ABHD6, and serine beta-lactamase-like protein LACTB (LACTB), which we were only able to isolate in microgram quantities despite considerable efforts aimed at optimizing expression.
We should note that EnPlex has limitations that merit further discussion. Most notably, even though EnPlex only requires small amounts of purified enzyme, it nonetheless requires enzyme purification and immobilization. Some enzymes are challenging to purify (e.g., those with transmembrane domains), and even those that are easily obtainable might have their catalytic activity affected by the appendage of epitope tags, removal from their endogenous settings, and immobilization on beads. Encouragingly, the vast majority of the enzymes we purified retained their activity on beads (93%), including soluble forms of several transmembrane domain-containing and membrane-associated enzymes. Moreover, virtually all of the compounds that had been previously profiled by low-throughput competitive ABPP in proteomes gave strikingly similar profiles by EnPlex (e.g., emetine11, WWL7047, PF-384548, URB59749, and orlistat50, Supplementary Fig. 4), suggesting EnPlex corresponds well to endogenous settings. In fact, in many cases EnPlex was the more comprehensive ABPP technology, as described in detail for JZL195 and bortezomib (Supplementary Table 5). Regardless, we believe that EnPlex and competitive ABPP in proteomes will serve complementary roles in the future; EnPlex can rapidly and comprehensively profile large compound collections, and lower throughput, proteomic ABPP methods can confirm the potency and selectivity of select compounds in situ.
Until now, technical limitations have generally limited small-molecule selectivity profiling to the late stages of drug development. Even then, a handful of optimized compounds are usually only tested against a relatively small number of enzymes structurally related to the intended target. EnPlex now makes it possible to perform high-throughput, superfamily-wide specificity profiling early in the drug discovery process. Used in this way, EnPlex could be used throughout primary screening and optimization to prioritize and modify compounds based not only on their on-target potency, but also on their degree of specificity. While we have demonstrated the feasibility of EnPlex for serine hydrolases, the approach can be extended in principle to any of the more than a dozen enzyme classes for which activity-based probes have been developed8. Moreover, we speculate that EnPlex may even be compatible with probes beyond those that work in traditional ABPP formats; for example, as EnPlex is not performed in complex lysates, EnPlex probes may not require the same levels of biological stability and enzyme specificity as existing activity-based probes. Regardless, this high-throughput profiling method is a powerful new approach for inhibitor selectivity profiling and should further motivate the development of additional activity-based probes.
ONLINE METHODS
Cloning
Open-reading frames (ORFs) encoding serine hydrolases were either obtained in a pDONR223 vector from The Broad Institute’s ORFeome 51, or were cloned from cDNAs obtained from Open Biosystems into a pDONR221 vector after performing PCR with primers flanked by attB sequences. For expression in E. coli, ORFs were transferred into either a pET-DEST42 or a pTrcHisB (modified for Gateway compatibility) vector, both of which encode C-terminal epitope tags. For expression in HEK 293T cells, ORFs were transferred into a Gateway-compatible version of the pCLNCX vector (Imgenex) containing C-terminal FLAG and His tags 52, and cells were infected according to the manufacturer’s instructions.
Protein expression in E. coli
Rosetta 2 (DE3) cells (Novagen) were grown in LB media containing 75 mg/L carbenicillin with shaking at 37 °C to an OD600 of 0.5. The cells were then induced with 1 mM IPTG and harvested 4 hours later by centrifugation. Cells were lysed by stirring for 20 mins at 4 °C in PBS (pH 7.4) supplemented with 1 mg/mL lysozyme. The lysate was then sonicated and centrifuged at 10,000 × g for 10 min. Talon cobalt affinity resin (Clontech; 400 μL of slurry/g of cell paste) was added to the supernatant, and the mixture was rotated at room temperature for 1 h. Beads were collected by centrifugation at 700 × g for 3 min, washed twice with PBS, and applied to a 1 cm column. The column was washed twice with PBS buffer (10 mL/400 μL of resin slurry). The bound protein was eluted by the addition of 100 mM imidazole (2 mL/400 μL of resin). Imidazole was removed by passage over a Sephadex G-25M column (GE Healthcare). Protein concentrations were determined using the Bio-Rad DC Protein Assay kit. Glycerol was added to a final concentration of 10%, and proteins were aliquoted stored at −80 °C until use.
Protein Expression in HEK 293T cells
HEK 293T cells were grown in DMEM with 10% fetal bovine serum at 37 °C with 5% CO2. Infected cells were selected with media containing hygromycin (100 μg/mL) and grown to 100% confluency (typically 20 × 15 cm plates per protein).
For isolation of intracellular proteins, cells were washed two times with PBS and scraped. Cell pellets were then isolated by centrifugation at 1,400 × g for 3 min. The pellets were resuspended in PBS, sonicated, and debris was removed by centrifugation at 12,000 × g for 45 min. Proteins were then bound by incubation overnight with 500 μL of Anti-FLAG M2 affinity gel (Sigma). The agarose gel was washed three times with PBS (5 mL), and proteins were eluted with 1 mL of 150 ng/μL solution of 3X FLAG peptide (Sigma) in PBS. Proteins were then concentrated using an ultra centrifugal filter unit (Amicon) according to manufacturer’s instructions. If the protein was larger than 30 kDa, the 3X FLAG peptide was removed by using a filter unit with a 30 kDa cut-off; for smaller proteins, a 10 kDa cut-off filter was used and some peptide was retained. Protein concentrations were determined using a protein assay kit (Bio-Rad). Glycerol was added to a final concentration of 10%, and proteins were aliquoted stored at −80 °C until use.
For isolation of secreted proteins, cells were washed three times with PBS and incubated in serum free DMEM for 48 h. Media was collected from these plates and spun at 2,500 × g for 5 minutes to remove debris. Ammonium sulfate (80% cut) was then added to the media. After incubation overnight on ice, protein was precipitated by spinning for 30 min at 28,000 × g. After resuspension in 10 mL PBS, proteins were isolated using Anti-FLAG M2 affinity gel (Sigma) as described above.
Buffer exchange of purchased proteins
Purchased proteins suspended in amine-containing solutions were buffer exchanged into PBS using protein desalting columns (Pierce) according to manufacturer’s instructions. Glycerol was then added to a final concentration of 10%, and proteins were aliquoted and stored at −80 °C until use.
Protein coupling to Luminex beads
MagPlex microspheres (Luminex) with different spectral properties were each coupled separately to purified proteins. 200 μL of microspheres (~2.5 million beads) of each type were aliquoted into separate wells of a 96-well plate (Costar, 3960) and pelleted on a magnetic separator for 2 min. Supernatant was removed, beads were washed with 200 μL dH2O, and resuspended in 80 μL 100 mM NaH2PO4 (pH 6.2). 10 μL 50 mg/ml Sulfo-NHS (Pierce) and 10 μL 50 mg/ml EDC (Pierce) were added and incubated at 25 °C for 20 min while shaking. The activated beads were washed with PBS (300 μL) three times and then resuspended in PBS (300 μL). ~10 μL of purified protein in PBS containing 10% glycerol was added to each well (typically amounting to ~1–10 μg of protein/well). The mixtures were incubated at 25 °C for 2 h while shaking in dark. The protein-coupled beads were washed twice with PBS (300 μL), once with 300 μL 50 mM Tris, 150 mM NaCl (pH 8.0), and resuspended in PBS (500 μL). Microspheres coupled at the same time were pooled together, glycerol was added to a final concentration of 10%, and these mixtures were aliquoted and frozen at −80 °C until use. Due to concerns about potential degradation of other enzymes, trypsin and chymotrypsin were specifically stored in a separate pool. All other groups were pooled based solely on which proteins were available for coupling at that time. In total, we made 11 different groups: Group 1: ABHD14B, ACOT1, ACOT4, AFMID, FAAH, FAM108B1, LYPLA1 (SA), LYPLA1(WT), LYPLA2(SA), LYPLA2(WT), LYPLAL1, lysozyme, MGLL, PREP, PREPL, RBBP9(SA), RBBP9(WT); Group 2: ABHD10, ABHD4, DPP9, ESD, FAM108A1, FAM108C1, IAH1, LACTB, OVCA2, PAFAH1B2, PAFAH1B3, PME1(SA), PME1(WT), PRCP; Group 3: ABHD11, ABHD6, APEH, CEL, CES1, CPVL, DDHD1, DPP4, DPP7, DPP8, LIPC, LIPG, OLAH, PLA2G7, PPT2, SCPEP1, SIAE; Group 4: ABHD2, ACHE, ACOT2, AOAH, BCHE, CES2, CES3, CES4A, CES5, CTSA, FAP, LCAT, LIPA, LIPF, LPL, PLA2G15, PNLIP, PNLIPRP2, PNPLA2, QRSL1; Group 5: HTRA1, HTRA4, PCSK1, PLAT; Group 6: BPHL, PPT1, TPP2; Group 7: C1R, C1S, C2, CFD, CMA1, CTSG, ELANE, F10a, F2a (thrombin), F7a, GZMA, GZMB, HPN, HTRA2, KLK2, KLK5, PCSK2, PLA2G4A, PLAU, PROC, PRSS8, ST14, TMPRSS11D; Group 8: LIPE, PRSS22, PRSS27; Group 9: F11a, KLKB1, PLG; Group 10: KLK1, CELA1; Group 11: CTRC, PRSS1; Group 12: CELA2A, CELA3A, CELA3B. Frozen mixtures maintained their activity for >1 year. On average, enzymes thawed one year after freezing retained >90% of their median Luminex signal (an average of 90.8% with a standard deviation of 9.8%). Importantly, all enzymes retained >50% of their activity; all but two [SCPEP1 (61%) and PREP (56%)] retained >70% of their activity. Aliquots were used for only one experiment and were not refrozen after use.
EnPlex assay
Beads mixtures were thawed on ice and pooled in PBS (pH 7.4) containing 0.01% Pluronic F127 (Invitrogen) with ~7,500 beads of each type per mL of solution. 30 μL of this microsphere mixture was transferred into each well (~225 beads of each type/well) of a 384-well twin.tec PRC plate (Eppendorf). Compounds or DMSO were then added (100 nL) at the indicated final concentrations, and plates were incubated at 25 °C for 30 min while shaking. For the boronic acids, compounds were dissolved in DMSO containing 0.1% TFA to prevent cyclization. Plates were spun (1 min, 1800 × g), and FP-biotin was added (100 nL) to a final concentration of 1 μM (ten control wells on each plate did not receive FP-probe). Plates were incubated at 25 °C for 1 hour with shaking. The plates were then spun (1 min, 1800 × g), placed in a magnetic separator, and buffer was removed and replaced with PBS containing 1% SDS. Plates were incubated at 95 °C for 10 min, spun (1 min, 1800 × g), and the media was removed. 30 μL of PBS plus 1% BSA containing 20 μg/mL R-phycoerythrin streptavidin conjugate (Molecular Probes) was added, and plates were incubated at 25 °C for 30 min. The samples were washed with 30 μl PBS plus 1% BSA three times and resuspended in 30 μl PBS plus 1% BSA.
The data were then acquired on a Luminex FLEXMAP 3D instrument according to manufacturer’s instructions. The background for each bead, determined from wells to which no FP-probe was added, was subtracted from each measurement. Percent activity was then determined relative to DMSO controls and plotted using GENE-E software from The Broad Institute (http://www.broadinstitute.org/cancer/software/GENE-E/dev/GENE-E-dev.jnlp). Two-dimensional hierarchical clustering was performed based on the Pearson correlation. IC50 values were determined from dose-response curves generated using Prism software (GraphPad).
Gel-based competitive ABPP assays
Enzymes were diluted in PBS containing 0.01% Pluronic F127 (Invitrogen) and incubated with DMSO or compound for 30 min at 25 °C (25 μL total reaction volume). FP-biotin was then added at a final concentration of 1 μM. After 1 h, the reactions were quenched with 2′ SDS-PAGE loading buffer (reducing), separated by SDS-PAGE (10% acrylamide), and transferred to a nitrocellulose membrane. Blots were probed using the indicated antibodies and/or a streptavidin-coupled IR dye following manufacturers’ instructions, and were visualized and quantified using the Odyssey Imaging System (Li-Cor). IC50 values were determined from dose-response curves generated using Prism software (GraphPad).
Enzyme substrate assays
PRCP
The PRCP substrate assay was performed essentially as described previously 28. Briefly, PRCP (1 nM final concentration) was added a 96-well black, clear-bottom plate (Costar, 3603) in 100 μL buffer (10 mM sodium acetate, 100 mM NaCl, 25 μg/mL BSA, pH 5.5). Compound was added at the indicated concentration (1% final DMSO) and incubated for 30 min at 25 °C. Substrate Mca-Ala-Pro-Lys(Dnp)-OH (Anaspec) was then added at a final concentration of 25 μM. Reactions were incubated for 1 h at 25 °C and then read on a Spectramax M5 plate reader (Molecular Devices) using an excitation wavelength of 320 nm and an emission wavelength of 405 nm.
AOAH
The AOAH assay was performed following described previously protocols 36, 53. Briefly, JZL195 was incubated with purified AOAH (5 ng) in PBS for 30 min before adding 3H/14C labeled LPS (1 μg). After 4 h, intact LPS was precipitated with ethanol and 3H in the ethanol supernatant was counted. To assay for AOAH inhibition by JZL195 in vivo, C57BL/6J mice were injected were treated intraperitoneally with vehicle (18:1:1 Saline:Emulphor:DMSO) or JZL195 (20 mg kg−1) in vehicle. After 6 hours, the animals were sacrificed and the livers and spleens were harvested, weighed, and sonicated in PBS containing 0.1% Triton X-100. Tissue lysates (50 μL containing 5–10 mg tissue [wet weight]) were then incubated with 3H/14C labeled LPS in 0.5 mL AOAH reaction mixture 36 overnight and AOAH activity was analyzed as described above. The Institutional Animal Care and Use Committee of the National Institutes of Allergy and Infectious Diseases approved the Animal Study Protocol.
HTRA2
3x stocks HTRA2, β-casein substrate (Sigma), and inhibitor were prepared in assay buffer (50 mM Tris, pH 8.0). 20 μL each of enzyme and inhibitor were mixed and incubated for 30 min at 45 °C. 20 μL of substrate was added and incubated for an additional hour at 45 °C. Each sample was quenched, separated by SDS-PAGE, and analyzed by silver staining (Thermo Scientific).
APEH
APEH (1 mg/mL) was diluted 1:20,000 in assay buffer (50 mM Tris, pH 7.5). 180 μL was added to each well on a 96-well clear bottom plate (Costar, Cat. No. 3603). 20 μL of inhibitor (diluted previously to 100x in assay buffer) was added to each sample and incubated for 10 min at 25 °C. 10 μL of 20X Ac-Ala-AMC substrate (Bachem, Cat. No. I-1830) in assay buffer was added and incubated for an additional 30 min at 25 °C, shaking the plate for the first two minutes. Fluorescence was read using 380 nm excitation and 460 nm emission wavelengths.
DPP4, DPP8, and DPP9
Compounds were dissolved in DMSO at a concentration of 100 mM. A 1 mM compound stock at pH 2.0 (1:100 dilution into 0.01 N HCl) was then prepared and incubated 4 h to overnight at room temperature. Compounds were then serially diluted in assay buffer (25 mM Tris, pH 8.0 for DPP4 and DPP9; 50 mM Tris, pH 7.5 for DPP8) to the appropriate concentrations. A 4000x substrate solution (100 mM Gly-Pro-AMC, VWR Cat. No. 100042-646) was prepared in DMSO and diluted to 20x in each enzyme assay buffer. Enzymes were then also diluted in the appropriate assay buffer at a final concentration determined specifically for each enzyme lot. 180 uL of diluted enzyme was added to each well of a 96-well black clear-bottom plate (Costar, Cat. No. 3603). 20 uL of compound was added and incubated for 10 min at room temperature with shaking for the first two minutes. 10 uL of 20x substrate was added and incubated for 25 min at room temperature with shaking for the first two minutes. Fluorescence was read using 380 nm excitation and 460 nm emission wavelengths.
Oral glucose tolerance test
Male C57BL/6 mice were fasted overnight prior to administration of vehicle (water with 0.25% methylcellulose) or drug at the indicated dosages by oral gavage. One hour after drug administration the animals were given dextrose at 5 g/kg body weight by oral gavage. Blood glucose was measured with a hand-held glucometer before dosing with the drug, before the glucose challenge and at 20, 40, 60 and 120 minutes after the glucose challenge. The sample size was 7 animals per treatment group. All animal procedures were approved by the Tufts University Institutional Animal Care and Use Committee (IACUC).
Compound toxicity studies in monkeys
ARI-2408
Six cynomolgus monkeys (3 male and 3 female) were given a single dose of 300 mg/kg via naso-gastric gavage. Four cynomolgus monkeys (2 male and 2 female) were administered 100 mg/kg dose daily for 7 days by naso-gastric gavage. Starting on the first day of dosing, detailed clinical observations (i.e. - monkeys removed from their cages and observed for clinical signs of toxicity) were conducted on monkeys for 7 days (from Day 1 through 8). Standard clinical observations (i.e. - observed while in their cages) were then conducted for seven days (from Day 8 through Day 14). On the final day of the study (Day 15) detailed clinical observations were performed. Body weights were taken on the day of dosing, prior to dosing, for calculation of dose amount. Hematology and clinical chemistry parameters were evaluated pre-dose on Day 1, approximately 24 hours following the initial dose and on Day 7. These studies were carried out by BASi (Study 0807-09031).
ARI-2243
Six cynomolgus monkeys/sex/group were given daily oral doses of ARI-2243 or control article for two days at 0, 0.3, 1.0, and 3.0 mg/kg. One monkey (male, 3 mg/kg) was observed to be in overtly poor health on Day 2, and so was not given a second dose. Two monkeys that died or were euthanized early were given postmortem examinations. Remaining monkeys were observed for 12 days after the last dose and then were returned to the stock colony. In life, monkeys were observed for clinical signs of toxicity and changes in body weight, food consumption, hematology, coagulation, and clinical chemistry parameters. At necropsy, macroscopic pathologic findings were recorded and tissues were collected and fixed for subsequent processing and examination by light microscopy for histopathologic findings. These studies were carried out by BASi (Study 0807-08269).
Synthetic methods
See Supplementary Note 2.
Supplementary Material
Acknowledgments
We thank B. Cravatt for the FP serine hydrolase probe, B. Martin, J. Long, A. Lone, and A. Saghatelian for constructs, J. Davis and D. Peck for technical assistance, and D. Gray and C. Yu for helpful discussions. This work was supported by the NCI (grant no. U54CA112962 to T.R.G.), HHMI (T.R.G.), the NIH (grant R01 CA163930 to WWB), Arisaph Pharmaceuticals (W.W.B), and in part by the Division of Intramural Research, NIAID, NIH (R. S. M.).
Footnotes
AUTHOR CONTRIBUTIONS
D.A.B. conceived and developed the EnPlex assay, performed experiments, and analyzed data; L.W.K. assisted with enzyme cloning and expression; W.W., Y.Liu, Y.Li, P.Z, I.W., and Y.S. synthesized and characterized boronic acid and nitrile-based compounds; J.H.L. directed the synthetic efforts; S.E.P. carried out standard enzymatic substrate assays (HTRA2, DPPs); C.K. directed monkey toxicity studies; S.E.H. and M.D. carried out mouse OGTT studies; D.G.S. directed OGTT studies; R.S.M. performed AOAH experiments; W.W.B. directed synthesis, enzymatic substrate assays, and OGTT experiments; T.R.G. directed the project; D.A.B. and T.R.G. wrote the paper.
COMPETING FINANCIAL INTERESTS
William W. Bachovchin is a co-founder, advisor and Board member of Arisaph Pharmaceuticals, a biotechnology company interested in developing boronic acid-based inhibitors of serine hydrolases as therapeutics. Christopher Kiritsy is a co-founder, CEO, and Board member of Arisaph Pharmaceuticals.
References
- 1.Collins I, Workman P. New approaches to molecular cancer therapeutics. Nat Chem Biol. 2006;2:689–700. doi: 10.1038/nchembio840. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein DM, Gray NS, Zarrinkar PP. High-throughput kinase profiling as a platform for drug discovery. Nat Rev Drug Discov. 2008;7:391–397. doi: 10.1038/nrd2541. [DOI] [PubMed] [Google Scholar]
- 3.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 4.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 5.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 11.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes. Nat Biotechnol. 2009;27:387–394. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachovchin DA, et al. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc Natl Acad Sci USA. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachovchin DA, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proc Natl Acad Sci USA. 2011;108:6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenteany G, et al. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 15.Ostrowska H, Wojcik C, Omura S, Worowski K. Lactacystin, a specific inhibitor of the proteasome, inhibits human platelet lysosomal cathepsin A-like enzyme. Biochem Biophys Res Commun. 1997;234:729–732. doi: 10.1006/bbrc.1997.6434. [DOI] [PubMed] [Google Scholar]
- 16.Ostrowska H, et al. Separation of cathepsin A-like enzyme and the proteasome: evidence that lactacystin/beta-lactone is not a specific inhibitor of the proteasome. Int J Biochem Cell Biol. 2000;32:747–757. doi: 10.1016/s1357-2725(00)00021-2. [DOI] [PubMed] [Google Scholar]
- 17.Fenteany G, Schreiber SL. Lactacystin, proteasome function, and cell fate. J Biol Chem. 1998;273:8545–8548. doi: 10.1074/jbc.273.15.8545. [DOI] [PubMed] [Google Scholar]
- 18.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 19.Arastu-Kapur S, et al. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: a link to clinical adverse events. Clin Cancer Res. 2011;17:2734–2743. doi: 10.1158/1078-0432.CCR-10-1950. [DOI] [PubMed] [Google Scholar]
- 20.Roujeau JC, et al. Telaprevir-Related Dermatitis. Arch Dermatol. 2012;149:1–7. doi: 10.1001/jamadermatol.2013.938. [DOI] [PubMed] [Google Scholar]
- 21.Schlutter J. Therapeutics: new drugs hit the target. Nature. 2011;474:S5–7. doi: 10.1038/474S5a. [DOI] [PubMed] [Google Scholar]
- 22.Talas U, Dunlop J, Khalaf S, Leigh IM, Kelsell DP. Human elastase 1: evidence for expression in the skin and the identification of a frequent frameshift polymorphism. J Invest Dermatol. 2000;114:165–170. doi: 10.1046/j.1523-1747.2000.00825.x. [DOI] [PubMed] [Google Scholar]
- 23.Tani T, Ohsumi J, Mita K, Takiguchi Y. Identification of a novel class of elastase isozyme, human pancreatic elastase III, by cDNA and genomic gene cloning. J Biol Chem. 1988;263:1231–1239. [PubMed] [Google Scholar]
- 24.Sawyer L, et al. The atomic structure of crystalline porcine pancreatic elastase at 2.5 A resolution: comparisons with the structure of alpha-chymotrypsin. J Mol Biol. 1978;118:137–208. doi: 10.1016/0022-2836(78)90412-6. [DOI] [PubMed] [Google Scholar]
- 25.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 26.Liverton NJ, et al. MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob Agents Chemother. 2010;54:305–311. doi: 10.1128/AAC.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soisson SM, et al. Structural definition and substrate specificity of the S28 protease family: the crystal structure of human prolylcarboxypeptidase. BMC Struct Biol. 2010;10:16. doi: 10.1186/1472-6807-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C, et al. Design and synthesis of prolylcarboxypeptidase (PrCP) inhibitors to validate PrCP as a potential target for obesity. J Med Chem. 2010;53:7251–7263. doi: 10.1021/jm101013m. [DOI] [PubMed] [Google Scholar]
- 29.Wallingford N, et al. Prolylcarboxypeptidase regulates food intake by inactivating alpha-MSH in rodents. J Clin Invest. 2009;119:2291–2303. doi: 10.1172/JCI37209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallela J, Yang J, Shariat-Madar Z. Prolylcarboxypeptidase: a cardioprotective enzyme. Int J Biochem Cell Biol. 2009;41:477–481. doi: 10.1016/j.biocel.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Long JZ, et al. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci USA. 2009;106:20270–20275. doi: 10.1073/pnas.0909411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munford RS, Hall CL. Purification of acyloxyacyl hydrolase, a leukocyte enzyme that removes secondary acyl chains from bacterial lipopolysaccharides. J Biol Chem. 1989;264:15613–15619. [PubMed] [Google Scholar]
- 33.Long JZ, Jin X, Adibekian A, Li W, Cravatt BF. Characterization of tunable piperidine and piperazine carbamates as inhibitors of endocannabinoid hydrolases. J Med Chem. 2010;53:1830–1842. doi: 10.1021/jm9016976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu M, Varley AW, Ohta S, Hardwick J, Munford RS. Host inactivation of bacterial lipopolysaccharide prevents prolonged tolerance following gram-negative bacterial infection. Cell Host Microbe. 2008;4:293–302. doi: 10.1016/j.chom.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu M, et al. Lipopolysaccharide deacylation by an endogenous lipase controls innate antibody responses to Gram-negative bacteria. Nat Immunol. 2005;6:989–994. doi: 10.1038/ni1246. [DOI] [PubMed] [Google Scholar]
- 36.Munford RS, Erwin AL. Eukaryotic lipopolysaccharide deacylating enzyme. Methods Enzymol. 1992;209:485–492. doi: 10.1016/0076-6879(92)09059-c. [DOI] [PubMed] [Google Scholar]
- 37.Perrier J, Durand A, Giardina T, Puigserver A. Catabolism of intracellular N-terminal acetylated proteins: involvement of acylpeptide hydrolase and acylase. Biochimie. 2005;87:673–685. doi: 10.1016/j.biochi.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Woitach JT, Zhang M, Niu CH, Thorgeirsson SS. A retinoblastoma-binding protein that affects cell-cycle control and confers transforming ability. Nat Genet. 1998;19:371–374. doi: 10.1038/1258. [DOI] [PubMed] [Google Scholar]
- 39.Shields DJ, et al. RBBP9: a tumor-associated serine hydrolase activity required for pancreatic neoplasia. Proc Natl Acad Sci USA. 2010;107:2189–2194. doi: 10.1073/pnas.0911646107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachovchin DA, et al. Oxime esters as selective, covalent inhibitors of the serine hydrolase retinoblastoma-binding protein 9 (RBBP9) Bioorg Med Chem Lett. 2010;20:2254–2258. doi: 10.1016/j.bmcl.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mentlein R, Gallwitz B, Schmidt WE. Dipeptidyl-peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon-like peptide-1(7–36)amide, peptide histidine methionine and is responsible for their degradation in human serum. Eur J Biochem. 1993;214:829–835. doi: 10.1111/j.1432-1033.1993.tb17986.x. [DOI] [PubMed] [Google Scholar]
- 42.Choy M, Lam S. Sitagliptin: a novel drug for the treatment of type 2 diabetes. Cardiol Rev. 2007;15:264–271. doi: 10.1097/CRD.0b013e318123f771. [DOI] [PubMed] [Google Scholar]
- 43.Thareja S, et al. Saxagliptin: a new drug for the treatment of type 2 diabetes. Mini Rev Med Chem. 2010;10:759–765. doi: 10.2174/138955710791572424. [DOI] [PubMed] [Google Scholar]
- 44.Flentke GR, et al. Inhibition of dipeptidyl aminopeptidase IV (DP-IV) by Xaa-boroPro dipeptides and use of these inhibitors to examine the role of DP-IV in T-cell function. Proc Natl Acad Sci USA. 1991;88:1556–1559. doi: 10.1073/pnas.88.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connolly BA, et al. Dipeptide boronic acid inhibitors of dipeptidyl peptidase IV: determinants of potency and in vivo efficacy and safety. J Med Chem. 2008;51:6005–6013. doi: 10.1021/jm800390n. [DOI] [PubMed] [Google Scholar]
- 46.Lankas GR, et al. Dipeptidyl peptidase IV inhibition for the treatment of type 2 diabetes: potential importance of selectivity over dipeptidyl peptidases 8 and 9. Diabetes. 2005;54:2988–2994. doi: 10.2337/diabetes.54.10.2988. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Blankman JL, Cravatt BF. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J Am Chem Soc. 2007;129:9594–9595. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]
- 48.Ahn K, et al. Discovery and characterization of a highly selective FAAH inhibitor that reduces inflammatory pain. Chem Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander JP, Cravatt BF. Mechanism of carbamate inactivation of FAAH: implications for the design of covalent inhibitors and in vivo functional probes for enzymes. Chem Biol. 2005;12:1179–1187. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorg Med Chem Lett. 2008;18:5838–5841. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 53.Munford R, Lu M, Varley A. Chapter 2: Kill the bacteria...and also their messengers? Adv Immunol. 2009;103:29–48. doi: 10.1016/S0065-2776(09)03002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.