Abstract
Few techniques are suited to probe the structure and dynamics of molecular complexes at the mesoscale level (100–1000 nm). We have developed a single-molecule technique that uses tracking fluorescence correlation spectroscopy (tFCS) to probe the conformation and dynamics of mesoscale molecular assemblies. tFCS measures the distance fluctuations between two fluorescently labeled sites within an untethered, freely diffusing biomolecule. To achieve subdiffraction spatial resolution, we developed a feedback scheme that allows us to maintain the molecule at an optimal position within the laser intensity gradient for fluorescence correlation spectroscopy. We characterized tFCS spatial sensitivity by measuring the Brownian end-to-end dynamics of DNA molecules as short as 1000 bp. We demonstrate that tFCS detects changes in the compaction of reconstituted nucleosome arrays and can assay transient protein-mediated interactions between distant sites in an individual DNA molecule. Our measurements highlight the applicability of tFCS to a wide variety of biochemical processes involving mesoscale conformational dynamics.
Introduction
The regulation of gene expression, intracellular transport, replication, and many intracellular processes is mediated by the dynamics of molecular systems occurring at the mesoscale level (100–1000 nm). Yet, methods for probing dynamics of mesoscale molecules are sparse. Super-resolution microscopy techniques (1) and biochemical methods to identify proximal interactions in vivo such as Hi-C (2, 3), proximity-dependent biotin identification (4), or spatially restricted enzymatic tagging (5) are powerful tools that can provide a static view of mesoscale organization inside the cell. However, it would be highly valuable to complement these methods with approaches that can 1) report on dynamic changes in the conformation of mesoscale systems and 2) be applied to reconstituted systems, in which the dynamics of individual molecules can be measured in isolation or with selected partners and biochemical and biophysical models can be directly tested. Single-molecule methods that have been used to study mesoscale systems include tethered particle motion (6, 7) and force spectroscopy (8, 9). To our knowledge, we have developed a new approach that enables the measurement of conformational fluctuations in mesoscale biological molecules in tether-free and force-free conditions.
Tracking fluorescence correlation spectroscopy (tFCS) is a method that combines confocal microscopy, feedback-based single-molecule tracking, and fluorescence correlation spectroscopy (FCS) to measure the conformational dynamics of individual molecules without the need for mechanical tethering (10, 11, 12, 13, 14). The key element in tFCS is the use of active feedback to compensate for center of mass diffusion by repositioning the microscope stage in real time so that the fluorescence from an individual molecule can be monitored over a long period. However, tFCS has previously been restricted to very large molecules such as λ-phage DNA (15) (radius of gyration 1 μm) because of spatial resolution limitations. Other active tracking methods (16, 17, 18) have been developed to measure hydrodynamic mobilities (19, 20), stoichiometry of molecular complexes (21), or nanoscale conformational dynamics (22, 23), but none has permitted mesoscale intramolecular dynamics measurements.
Here, we developed a dual-color tFCS method that specifically assays the distance fluctuations between two discrete sites within a single macromolecule with ∼100–150 nm spatial resolution and sub-millisecond temporal resolution. By optimizing the focal position of the lasers used for tracking and for intramolecular dynamics detection, we showed that tFCS has sufficient spatial and temporal resolution to probe the relative motion between two sites in freely diffusing DNA molecules separated by as little as 1000 bp. Additionally, we showed that tFCS can detect conformational transitions in diffusing chromatin fibers and DNA-looping processes induced by lac repressor. These measurements provide, to our knowledge, the first demonstration that tFCS can measure dynamic biophysical processes occurring on a subdiffraction scale, which has important implications for many molecular systems, particularly the dynamics of nucleoprotein systems.
Materials and Methods
Single-molecule tracking microscope
To track individual molecules, we used a custom-built confocal microscope augmented with a feedback loop as well as beam-steering capabilities that enabled high-bandwidth electronic control of the position of the focus of each laser beam in the sample (details in Supporting Materials and Methods). Briefly, to achieve all-optical sensing of the three-dimensional (3D) position of the diffusing particle and real-time tracking, we set up the tracking laser according to the optical and feedback scheme previously described (13). To increase the tracking bandwidth of the instrument, we used two pairs of acousto-optic deflectors (AODs) placed in the path of the tracking and probe beams (Fig. 1 A) to serve as fast actuators, with microsecond-scale response time. This strategy allowed us to apply feedback faster than the mechanical resonance frequency of the piezo stage (100 Hz) and minimize the tracking localization error, which is key to resolving intramolecular dynamics at the subdiffraction limit scale.
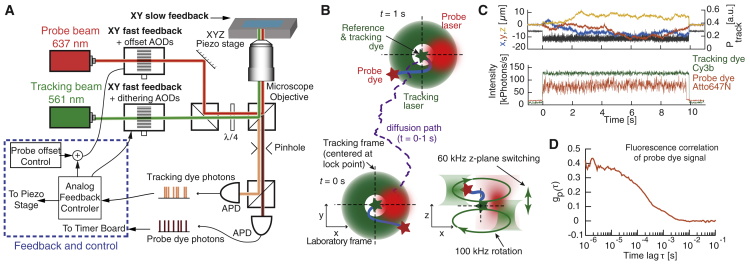
Figure 1.
Instrumentation and concept of the tFCS assay. (A) Key optical and electronic elements of the tFCS microscope are shown. The feedback is applied by an analog controller (blue box), which drives the microscope piezo stage and the acousto-optic deflectors (AODs) controlling the position of the tracking and probe beams. The optical elements used to dither the tracking laser axially are omitted for simplification (Supporting Materials and Methods). (B) A schematic of the tracking and probe beam motions during tracking of a freely diffusing molecule is shown (blue). The trajectory (purple line) of the green-labeled site on the molecule (green star, reference dye) is followed by the tracking laser (green beam), whereas the probe laser (red beam) is maintained at a fixed position with respect to the tracking laser. The green ring indicates circular dithering of the tracking beam used to locate the molecule in XY (13). The focus of the rotating tracking laser is also axially dithered between two transverse planes to locate the molecule along the z axis (bottom right) (13). The green-labeled site is immobile with respect to the probe laser (at the tracking/reference lock point, dashed crosshair), whereas the red-labeled site (red star, probe dye) moves because of intramolecular dynamics. (C) Single-molecule signals recorded in tFCS are shown, including the probe fluorescence (red), the stage trajectory, and the tracking laser power (black), which is under feedback control to keep the tracking dye fluorescence at a fixed value (green). (D) A single-molecule fluorescence correlation function from the probe signal in (C) is shown. To see this figure in color, go online.
Measurements conditions
All tFCS measurements were taken with molecules at concentration so that one individual molecule drifted in the microscope confocal volume every 15–30 s, making it unlikely that two molecules were tracked at once. All of the DNA and LacI experiments were done in 10 mM Tris (pH 7.4), 50 mM KCl. The arrays data were taken in 10 mM Tris (pH 7.4), 25 mM NaCl, and either 2 mM MgCl (MgCl2 conditions) or 0.5 mM EDTA (no MgCl2 conditions). All buffers were complemented with an oxygen-scavenging system consisting of 0.01 units/mL of protocatechuate-dioxygenase, 2 mM of protocatechuic acid, and 1 mM of aged Trolox to serve as a reducing oxidizer system (24, 25). For tFCS, 25 μL of sample was loaded into a microfluidic channel formed by two microscope cover slips (40 × 20 #1) separated by two overlaid layers of double-sided tape (3 M) positioned to create a narrow channel (3 × 20 × 0.3 mm, channel volume 15–20 μL). For the LacI and chromatin arrays experiments, the channels were passivated by incubation of a 1 mg mL-1 solution of casein for 5 min and rinsed with 100 μL of buffer before sample loading.
tFCS data recording and preprocessing
For each experiment, we collected data in continuous mode for 10–30 min, during which we recorded the fluorescence signal from the reference and probe dyes using the time-interval-analyzer board (GT653; GuideTech, Sunnyvale, CA) operating in time-tagged mode. The microscope stage trajectory and the tracking laser power were digitized at 1 kHz (National Instruments, Austin, TX). The reference-dye fluorescence was locked at , and the red laser power was chosen to achieve a probe-dye fluorescence rate of . The raw signals were semiautomatically preprocessed offline to detect and classify individual molecules, compute the fluorescence correlation functions, and correct for background effects (Supporting Materials and Methods).
Compaction scores and clustering of conformational states
To compare molecular macrostates, tFCS data were quantified using a simple metric of molecular compaction. We defined the relative compaction between two molecules as the ratio of the inverse of the integral of the correlation function of the two molecules, computed over the interval of time lags from 5 ms to 5 μs. These bounds were chosen empirically to encompass the region of the correlation signal, which differs between the macrostates of interest, while avoiding introduction of noise from the shorter (5 μs) and longer (5 ms) timescales.
DNA constructs labeling and nucleosome arrays reconstitution
The DNA molecules used in all the experiments were fluorescently labeled with a single probe dye and a single reference dye by ligation or polymerase chain reaction. Nucleosome arrays were reconstituted using the salt dialysis method with purified H3/H4/H2A/H2B octamers, as previously described (26) (Supporting Materials and Methods).
Results
Design of the tFCS assay to measure the intramolecular dynamics of freely diffusing single molecules
To measure the conformation of biological macromolecules in the absence of mechanical perturbation, we custom-designed a confocal microscope with a feedback system that compensates for the motion of individual molecules as they freely diffuse (13). The feedback system allows us to track single molecules for several seconds in 3D space while simultaneously observing their fluorescence emissions (Fig. 1 A). We tailored the system to specifically assay the distance and distance fluctuations between two parts of a single molecule or complex. To accomplish this, we used a green reference dye (Cy3b) as a reference point for tracking the movement of the molecule in three dimensions and a red probe dye (Atto647N) as a reporter of the intramolecular dynamics. We then controlled, via feedback, the position of two lasers in real time to simultaneously excite the reference tracking and probe dyes. To follow the diffusive path of the reference dye in the transverse plane (XY, perpendicular to the propagation axis), we dithered the tracking laser beam around a circular orbit at 100 kHz to generate a particle-position-dependent modulation of the dye fluorescence signal (Supporting Material; (11, 27)). To track the displacement of the particle in the axial direction (Z), we dithered the axial focus of the rotating tracking laser at 65 kHz between two planes slightly above and below the focal plane of the probe laser (Supporting Material; (13)). Importantly, we coordinated the positions of the probe and tracking lasers in the XY plane (Fig. 1 B) to effectively maintain the probe laser focus at a fixed point relative to the reference site of the molecule. With this tFCS assay, which uses a dual-focus orbital tracking system and a two-fluorophore labeling scheme, we were able to collect 1 million photons per molecule in both fluorescence channels (Fig. S1; Table S2).
Readout of the intramolecular dynamics
The schematic representation of a typical trajectory of the molecule and the focal positions of the lasers (Fig. 1 B) highlights how intramolecular dynamics results in a motion of the probe dye within the probe beam. Fluctuations of the probe fluorescence thus encode the 3D motion of the probe dye. Because the probe laser is spatially locked with respect to the reference dye, the tFCS measurement is only sensitive to the relative motion of the probe dye with respect to the reference dye, not to the overall center of mass diffusion nor to a static configuration in which the reference-to-probe dye vector is frozen (for example, if the molecule is immobilized).
However, because the molecule is free to tumble and rotate around the reference dye during tracking, the orientation of the reference-to-probe dye vector is never static in tFCS; therefore, even a rigid molecule with a fixed reference-to-probe dye distance exhibits fluorescence fluctuations that contain information about this distance (Fig. S2).
To quantify the reference-to-probe dye dynamics, we computed the autocorrelation of the probe dye fluorescence signal, as is standard in conventional FCS (Fig. 1, C and D). Given a model describing the motion of the probe dye and its blinking dynamics, it is theoretically possible to fit the tFCS signal and quantitatively estimate the model parameters, such as the amplitude and relaxation time of the intramolecular dynamics, but this approach is challenging (Discussion and Supporting Material Appendix). In this manuscript, we examined the amplitude of the tFCS signals of individual molecules as an uncalibrated readout of the amplitude of the reference-to-probe dye dynamics (Supporting Material Appendix; Fig. S6). Additionally, from the 3D movement of the molecule over time provided by the X, Y, and Z stage trajectory (Fig. 1 C), we obtained an independent and calibrated readout of the diffusion coefficient. Our approach is generalizable to any biological macromolecule that can be labeled with two different fluorophores. The labeling sites can be located on the same individual molecule or on two separate binding partners. Thus, by selecting the appropriate position of the reference and probe dyes, we can measure the dynamics between any pairs of sites within a macromolecular complex or even the distance between the sites of a rigid molecule by leveraging the rotational diffusion of the molecule.
Optimization of the spatial sensitivity of tFCS through optimization of the illumination geometry
A major challenge for mesoscale measurements is that the target resolution (100 nm) is smaller than the size of the diffraction-limited probe beam. In prior implementations of tFCS, the intramolecular motion was larger than the beam dimensions, and the fluorescence fluctuations stemmed from large excursions of the probe dye in and out of the entire Gaussian beam profile (15). In this new tFCS assay, the trajectory of the probe dye samples only a small region of the illumination volume. To resolve intramolecular dynamics in the subdiffraction-limit regime, we increased the spatial sensitivity of the assay by configuring the feedback to maintain the molecule at the edge of the Gaussian profile of the probe laser, where the local intensity gradient is large (Fig. 2 B). If the reference and probe beams were co-linear (centered-illumination), the local intensity gradient seen by the probe fluorophore would be small, and intramolecular dynamics would give rise to small fluorescence fluctuations (Fig. 2 A). Therefore, we modified the microscope optical path so that the probe beam was laterally offset with respect to the tracking beam. We tuned the distance between the probe and tracking beam electronically using the AODs in the probe beam path (Fig. 1 A). With this “side-illumination” configuration, even small displacements of the probe dye result in large changes in the laser intensity seen by the probe dye, which provides a sensitive way to convert molecular motion into fluorescence fluctuations (Fig. S3).
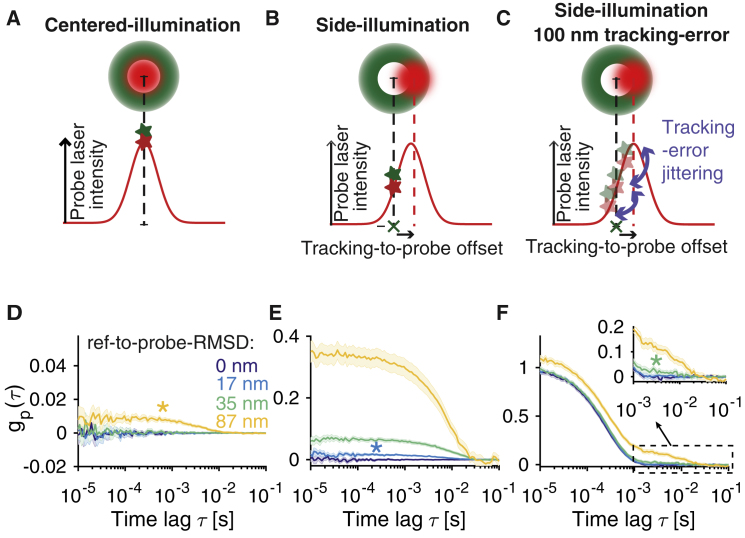
Figure 2.
Optimization of tFCS spatial sensitivity with side-illumination, and expected resolution in presence of tracking localization error. (A and B) A schematic of the centered- (A) and side-illumination (B) configurations is shown. Top: the relative positions of the tracking (green) and probe (red) beams are shown. Bottom: the X axis cross-section of the probe laser intensity profile seen by the probe dye is shown. The green cross represents the lock point of the feedback (at the center of tracking laser circular orbit) with respect to which the reference dye is immobile. The green and red stars represent the average position of the reference and probe dyes in the beam, respectively. (C) The illustration shows the effect of finite-bandwidth tracking, in which the reference dye is imperfectly maintained at the lock point, which results in a residual jitter of the probe dye in the probe beam frame. (D and E) Simulated tFCS signals (mean 95% confidence interval) of molecules with a reference-to-probe dye root mean-square distance (RMSD) of 0, 17, 35, or 87 nm are shown, using either the centered- (D) or the side-illumination (E) configuration. In the side-illumination geometry, the reference-to-probe beam offset was set to , where . Asterisk color indicates the shortest resolvable reference-to-probe RMSD. (F) The same simulation as that in (E) is shown, but this time taking into account imperfect feedback (100 nm RMS localization error). To see this figure in color, go online.
Effect of tracking-localization error and experimental considerations for fast feedback
To obtain a high spatial resolution, it is essential that the microscope feedback loop tracks the displacement of the reference dye as accurately as possible. In practice, the actuator bandwidth and the finite dye brightness set limits on the feedback bandwidth and tracking accuracy (11). This leads to a residual motion of the probe dye in the probe beam, which impairs spatial sensitivity, as this is equivalent to random displacements of the probe beam away from its optimal position and reduces the local intensity gradient seen by the probe dye (Fig. 2 C).
To maximize tracking accuracy, we improved the feedback architecture so that we could track the molecule with a bandwidth larger than the resonance frequency of the piezo stage (100 Hz). To do so, we implemented in the dimensions a feedback loop with two branches. We corrected for low-frequency components of the particle motion via feedback on the microscope piezo stage, whereas higher bandwidth components were canceled via feedback on the laser position, controlled with AODs. With this scheme, we were able to feedback at 1 kHz bandwidth for tracking and maintain the reference dye within better than 100 nm of the desired lock point (root mean-square (RMS) error) for molecules diffusing up to 15 μm2/s.
Numerical estimation of the spatial resolution of tFCS
To estimate the smallest distance fluctuations that can be resolved by tFCS and obtain a theoretical lower-bound on the tFCS resolution, we first conducted numerical simulations of the assay. We simulated tFCS signals for intramolecular dynamics, where the reference-to-probe dye distance fluctuates with a root mean-square distance (RMSD) ranging from 17 to 87 nm (Fig. 2, D–F) and a characteristic timescale of 10 ms. The number of photons collected per molecule , signal-to-noise , and duration of tracking trace were chosen to represent typical experimental values (Fig. S1; Table S2). We then compared the tFCS signal obtained for each reference-to-probe RMSD with that of control molecules where the reference and probe sites were colocalized (i.e., reference-to-probe RMS distance equals 0). The simulated tFCS signals matched well with the theoretical prediction (Supporting Material Appendix Eq. 35) and allowed us to characterize the measurement noise and the resolution. In the ideal case of perfect tracking, the side-illumination configuration was able to resolve the intramolecular dynamics of all the molecules ranging from 17 nm and upward (Fig. 2 E), compared with the centered-illumination geometry, where only molecules with a reference-to-probe RMS distance of 87 nm were resolved (Fig. 2 D). When we accounted for a realistic tracking localization RMS error of 100 nm, the resolution was reduced to 35 nm with the side-illumination configuration (Fig. 2 F). Simulations with different intramolecular timescales of motion (1 and 10 ms) yielded similar results (Fig. S4), indicating that the resolution is not strongly dependent on the dynamics timescale.
To characterize the performance of tFCS in measuring the intramolecular dynamics of larger molecules, we next simulated signals from molecules with an RMSD ranging from 100 to 1000 nm. The time intervals in which the probe dye explores the focus of the probe beam become sparse, but the tFCS signal remains quantifiable (Fig. S5). Theoretical analysis of the tFCS amplitude indicates that as the end-to-end dynamics amplitude reaches the order of the beam dimension, the illumination geometry becomes irrelevant, and centered- and side-illumination geometry lead to similar correlation signals (Fig. S6).
We then examined the ability of tFCS to measure the distance between two sites in a rigid molecule. To do so, we simulated a rotational diffusion such that the probe dye moved stochastically on the surface of a sphere and at a fixed distance from the reference dye. We found that increasing the distance between the sites gave rise to tFCS signals of increasing amplitude and that these signals were above the noise floor for molecules longer than or , depending on the tracking localization accuracy (Fig. S7).
Together, these results show that with a tracking localization accuracy of 100 nm, the theoretical detection limit in tFCS is on the order of and that tFCS can measure both distance fluctuations and static distances between two sites at this length scale. Importantly, these simulations highlight how technical limitations affect the spatial resolution and indicate that the spatial resolution could be pushed near the maximal range for fluorescence resonance energy transfer (FRET) with a smaller tracking localization error.
Measurement of the intramolecular dynamics of DNA
Decades of experimental and theoretical work have led to a deep understanding of the polymer dynamics of double-stranded DNA (dsDNA). Static and dynamical properties of DNA such as radius of gyration, diffusivity, and relaxation timescale have been previously measured using dynamic light scattering (28, 29), FCS (30, 31, 32), and fluorescence microscopy (33, 34). The intramolecular Brownian dynamics of DNA has also been studied by fluorescence microscopy, FCS, and other methods (15, 35, 36). Here, we used DNA as a benchmark system to experimentally characterize the spatial resolution of our tFCS approach. We labeled dsDNA of 0.5, 1, and 3.9 kbp with a single Cy3b and a single Atto647N at opposite ends (OE) of the molecule. As a control, dsDNA of the same three lengths were labeled with the two dyes placed on the same end (SE) of the duplex, separated by only 31 bp.
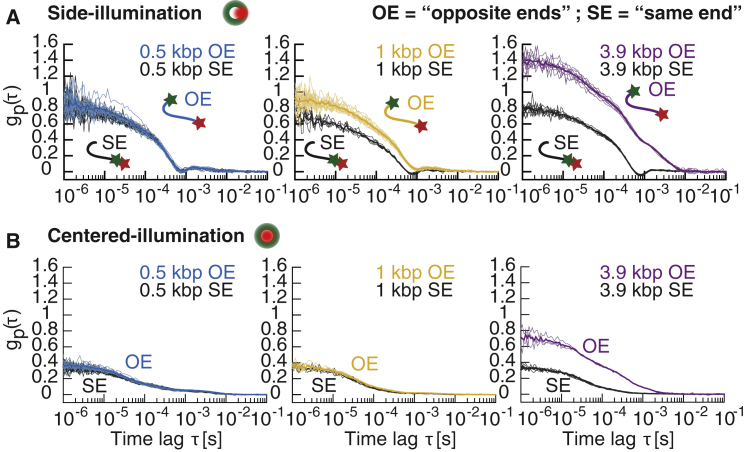
We clearly resolved the end-to-end dynamics of both the 1 and 3.9 kbp DNA when we used the side-illumination geometry (Fig. 3 A). Conversely, we found that the centered-illumination geometry with near-perfect alignment of the probe and tracking beams resulted in poorer sensitivity and could only resolve the 3.9 kbp DNA (Fig. 3 B). The 0.5 kbp DNA fragment was below the resolution threshold for both configurations, as suggested by the overlap of the correlation signal of the SE- and OE-labeled molecules. When observed with the side-illumination geometry, the 3.9 kbp fragments exhibited fluorescence fluctuations with larger correlation amplitude at all time lags and slower timescales of motion compared with the 1 kbp molecules, which was consistent with a larger radius of gyration and slower polymer relaxation modes. Even though the reference and probe sites were nearly colocalized in the SE-labeled DNA molecules, they exhibited a nonzero fluorescence correlation signal (Fig. 3, A and B), indicating the presence of systematic noise in the tFCS assay. This systematic noise likely stems from imperfect localization of the reference dye, as inferred from our previous simulations (Fig. 2 F) and from the triplet-state dynamics of the Atto647N dye, which is also observed in conventional FCS measurements (37, 38). Importantly, the assay separates molecules that differ only in the distance between the reference and probe sites (SE and OE constructs) and not in their overall dimensions, indicating that this is an appropriate tool for measuring macromolecular conformation in addition to its ability to measure hydrodynamic properties.
Figure 3.
tFCS measurement of the end-to-end dynamics of freely diffusing DNA molecules. (A and B) Fluorescence correlation signals of DNA molecules using the side- (A) or centered-illumination (B) configuration are shown. DNA were labeled with Cy3b and Atto647N either on opposite ends (OE) or on the same end (SE) of the molecule. DNA lengths were 0.5 kbp (blue OE, black SE), 1 kbp (yellow OE, black SE), and 3.9 kbp (purple OE, black SE). Correlation signals from individual molecules (thin lines) and population averages (thick lines) are shown. To see this figure in color, go online.
To test the effect of changing the relative position of the reference and probe lasers, which results in a change in the local laser intensity gradient seen by the probe dye, we repeated the measurement with varying tracking-to-probe beam offsets. We found that the changes in the amplitude of the correlation signal and of the mean fluorescence signal had a quadratic dependency in the offset, as predicted analytically (Fig. S8; Supporting Material Appendix Eq. 43). Together, these results demonstrate that the tFCS assay detects the end-to-end dynamics of DNA constructs of 1 kbp and longer. Based on prior tethered particle motion (39, 40) and atomic force microscopy (41) measurements of the RMS distance between the two ends of 1-kbp-long DNA molecules, these data indicate that we achieved an experimental resolution of 100–150 nm.
Measuring chromatin compaction in freely diffusing molecules
Changes in the local compaction of chromatin regulate multiple essential processes, including gene expression, mitotic chromosome condensation, and heterochromatin formation (42, 43, 44). Reconstituted arrays of nucleosomes (45) can recapitulate the mechanics of chromatin condensation in response to electrostatic forces (46, 47), chromatin-binding proteins (48), and histone modifications or variants (49). Traditionally, measurements of chromatin compaction in solution use sedimentation assays or analytical ultracentrifugation (AUC) to differentially fractionate compact chromatin (46). However, these assays require a large amount of material and are not typically compatible with complex biochemical systems such as cellular extracts. FRET has been used to study the dynamics of nucleosome arrays but can only probe local conformational changes at the scale of a single nucleosome or a pair of adjacent nucleosomes (50, 51, 52). We investigated the applicability of tFCS in characterizing the conformational state of single reconstituted nucleosome arrays and as an alternative approach to studying chromatin structure and dynamics.
We reconstituted nucleosomes on a tandem DNA array of 19 copies of a high-affinity nucleosome-positioning sequence (19×601) (53) labeled with Cy3b and Atto647N on OE. To test whether tFCS could measure changes in the end-to-end distance of individual DNA molecules upon histone assembly, we prepared arrays with increasing histone saturation ratios, defined as the fraction of positioning sites occupied by a histone octamer. Different saturation ratios were obtained by adjusting the relative stoichiometry of histone octamers and positioning sites during assembly, and the ratios were measured at the population level by electrophoresis (Fig. S9 A). The amplitude of the side-illumination tFCS signals decreased with the saturation of the arrays, indicating that the assay could detect the reduction in the distance between the two ends of the molecule as a result of DNA wrapping by histones (Fig. 4 A). To further quantify the folding state of individual arrays, we computed for each molecule a compaction score quantifying the reduction in amplitude of its correlation signal with respect to the bare 19×601 DNA (Materials and Methods). We found that both the compaction score and the diffusivity of the arrays increased with the histone occupancy level (Figs. 4 B and S9, B and C), confirming that both quantities reflect on the folding state of the molecule. Despite the low concentration of molecules during tracking, the arrays were stable, and it is unlikely that significant nucleosome dissociation occurred during the measurement because the tFCS signals were unchanged even 2 h after dilution of the arrays for tFCS (Fig. S9 E).
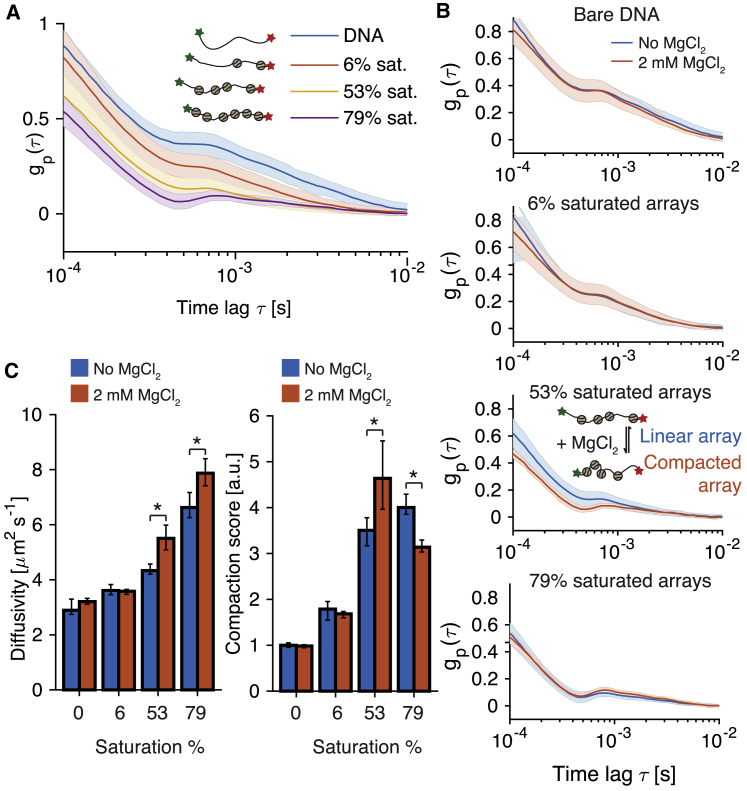
Figure 4.
Measurement of the compaction state of nucleosome arrays. (A) tFCS signals (population median standard deviation) for 19×601 DNA molecules before nucleosome assembly (blue) and for chromatinized 19×601 molecules with increasing histone occupancy are shown. Data were taken using a side-illumination configuration. (B) tFCS measurement of the MgCl2-induced nucleosome arrays compaction is shown as a function of histone occupancy (top to bottom: bare DNA, 6, 53, and 79% saturated arrays). (C) The median (95% bootstrap confidence intervals) diffusivity (left) and compaction score (right) of bare DNA and nucleosome arrays before and after addition of 2 mM MgCl2 are shown. The asterisk indicates statistical significance (two-sided Wilcoxon rank sum test, p-value0.01). To see this figure in color, go online.
We next tested whether tFCS could measure the compaction of nucleosome arrays in the presence of divalent cations (46, 54). Upon addition of 2 mM MgCl2, we observed a reduction in the tFCS signal amplitude for the 53% saturated arrays along with an increase in diffusivity. For the 6% saturated arrays, neither the tFCS signals nor the diffusion coefficients were significantly affected by the addition of magnesium, suggesting that the low nucleosome density in these molecules precludes compaction, which is mediated by intranucleosomal interactions. For the 79% saturated arrays, the addition of 2 mM MgCl2 resulted in a significant increase in their diffusion coefficient, as extracted from the molecule trajectory, indicating that these molecules were able to compact. However, the compaction was not detected in the fluorescence correlation signals. These data can be explained by the fact that the 79% saturated arrays have dimensions on the order of the spatial resolution of the tFCS assay because their diffusion coefficient was close to that of the 1000 bp DNA molecules ( ), which are the shortest DNA we could resolve. Therefore, further compaction upon magnesium addition cannot be detected. To confirm this hypothesis, we measured the tFCS signals of similarly saturated arrays, but where the probe and reference dye were placed on the SE of the molecule, and we found that indeed the signals of these arrays nearly overlapped with those where the two dyes were on OE of the DNA (Fig. S9 D).
Together, these data show that the tFCS approach, when applied within its resolution limits, allows us to measure changes in the compaction of freely diffusing nucleosome arrays at the single-molecule level.
Measurement of repressor-induced DNA-looping dynamics
Control of bacterial operons (55, 56), promoter-enhancer interactions in eukaryotes (57), and the large-scale organization of topological domains in metazoan chromosomes (58) are all regulated by chromatin folding to juxtapose DNA sites separated by tens to millions of DNA bases. To demonstrate the ability of tFCS to monitor transient protein-mediated interactions between remote DNA loci, we tested whether we could detect the formation of looped states in freely diffusing DNA induced by lac repressor binding (6, 55, 59) (Fig. 5).
Figure 5.
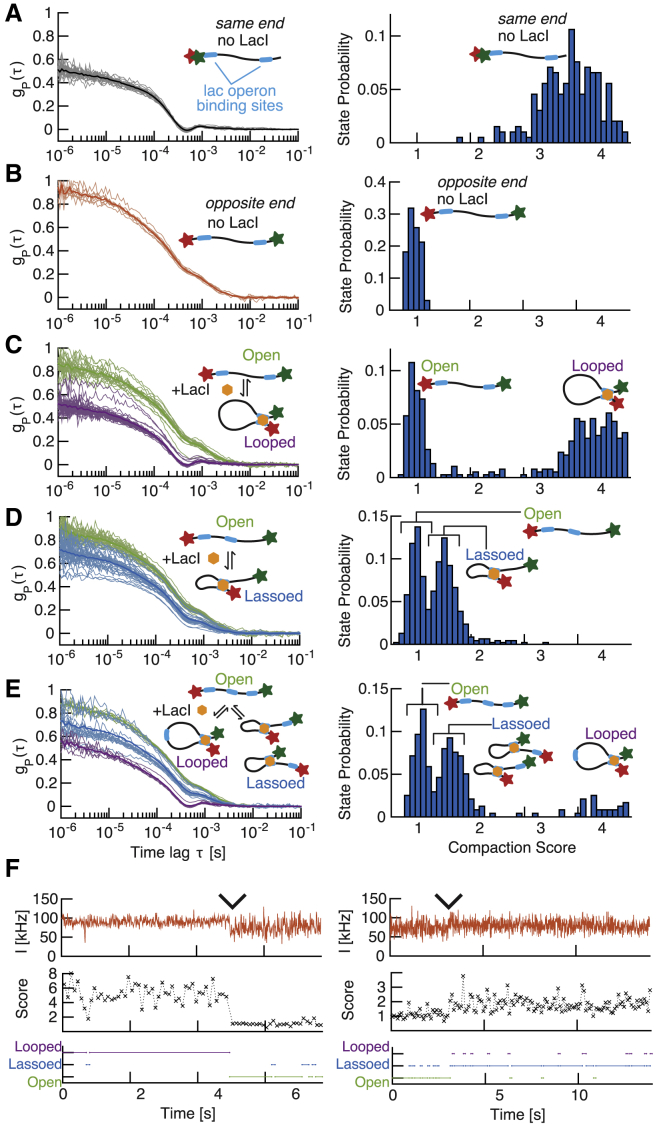
Detection of protein-mediated conformational changes. (A and B) Left: single-molecule tFCS signals (thin lines) and population mean (thick lines) of the looping DNA construct bearing two lac operators are shown. The molecules were labeled with Atto647N and Cy3b either at the same end (A) or on opposite ends (B), and data are shown in the absence of LacI. Right: the distribution of compaction scores for the 100 ms subtraces of all the individual molecules is shown. (C–E) Left: tFCS signals of individual DNA molecules after addition of 64 nM LacI are shown. The DNA used were the looping construct in (C), the lasso construct in (D), and the construct with three LacO sites in (E). The conformational state inferred by clustering the tFCS signals are categorized as the following: open in green (no LacI bound or LacI bound at a single site), lassoed in blue (LacI-mediated short loop), or looped in purple (LacI-mediated long loop). Cluster averages (thick lines) and individual molecules (thin lines) are shown for each of these three groups. Right: the distribution of compaction scores for the 100 ms subtraces of all the individual molecules is shown. (F) A representative example of a conformational transition between a looped and an open state (left) or an open and a lassoed state (right), detected during the tracking of an individual molecule. Probe dye fluorescence trace (top), 100 ms binned compaction score (middle), and inferred conformational state (bottom) are shown. Wedges indicate time of transition between the two conformations. To see this figure in color, go online.
We generated two 2.6 kbp DNA substrates containing two lac operator sites spaced at specific distances (Table S1): a “looping” construct with the two binding sites at each end of the molecule and a “lasso” construct with one operator site moved to the middle of the molecule (1.3 kbp between the operator sites). For both constructs, we found that addition of lac repressor resulted in the appearance of two populations of molecules characterized by distinct correlation functions (Fig. 5, C and D). The tFCS signal of the population with the largest correlation amplitude matched the signal measured before the addition of LacI (Fig. 5 B), which is consistent with molecules in an unlooped conformation (with or without LacI binding). In addition, for the “looping” construct, the tFCS signal of the molecules with the smallest amplitude overlapped with the signal obtained from control DNA molecules in which the two dyes were juxtaposed, validating that these molecules were in a state with the Cy3b and Atto647N dyes in close proximity (Fig. 5 A). For the lasso construct, we found that the folded molecules exhibited a signal of intermediate amplitude, which was consistent with the formation of a smaller loop in which the dyes are separated by about half the length of the molecule. Importantly, the addition of 2 mM of the LacI inhibitor isopropyl β-D-thiogalactoside efficiently destabilized the loops (Fig. S10).
To test whether we could distinguish more than two conformational states for a given molecule, we then generated a construct containing three lac operator sites: one at each end of the DNA, as in the looping construct, and one in the middle of the molecule, as in the lasso construct. Because three different pairs of LacO sites can be brought into contact by a LacI tetramer, this construct can adopt four distinct conformational states: 1) an open configuration in which none of the LacO sites are bridged together, 2) a long loop state in which LacI maintains the two terminal LacO sites in contact, 3) a short loop state in which LacI maintains the Cy3b-labeled terminal site in contact with the internal site, and 4) a short loop state in which LacI maintains the Atto647N-labeled terminal site in contact with the internal site. In the presence of LacI, we clearly detected three clusters of tFCS traces. The tFCS signals in the clusters with low, medium, and high amplitudes overlapped with those of the looped molecules from the looping construct, the looped molecules from the lasso construct, and the molecules in the absence of LacI, respectively, indicating that these clusters correspond to the open, short loop, and long loop configurations. However, because the internal site was roughly in the middle of the molecule, we were unable to distinguish between the short loop configurations in 3) and 4), which have almost identical Cy3b-Atto647N distance. Altogether, these measurements demonstrate that the tFCS signal provides a single-molecule readout of the long-range interactions between specific sites, induced by LacI.
Finally, we asked whether we could detect transitions between looped and unlooped states in real time. Because the LacI-mediated loop lifetime (tens of seconds) is long compared with the duration of the tracking traces (6, 59), each trace is expected to contain at most one transition. In contrast with the DNA and nucleosome arrays, in which the dynamics is fast ( relaxation timescale) and in which each molecule explores its full conformational space over the course of the measurement, the lac operator/repressor looping dynamics is not at equilibrium during a tracking trace, and it is therefore not meaningful to compute the fluorescence correlation function over the full trace. To scan for the presence of transitions, we segmented each tracking trajectory into 100 ms time bins and computed the fluorescence correlation signal within each bin. We compared the sliding tFCS signal with the average correlation signal of the molecules in the open, folded loop and folded lasso state, and we inferred the state of the molecule at each time point. We inferred transitions between states (looped/unlooped) from changes in the fluorescence fluctuation spectrum across time bins. Representative examples of an unlooping event in the loop construct and a looping event in the lasso construct showed that the transition points uncovered by classifying the 100 ms tFCS signals aligned as expected with abrupt changes in the compaction score and with visible transitions in the amplitude of fluorescence fluctuations (Figs. 5 F and S11). These data show that slow conformational dynamics between discrete states can be extracted by segmentation of the tFCS data.
Discussion
A major challenge for studying subcellular biological processes is that few methods can interrogate the structure and dynamics of large macromolecules and complexes at the distance and timescales relevant to their function. We show here that tFCS enables measurement of the internal dynamics of freely diffusing mesoscale molecules in three different experimental systems: DNA molecules in solution, nucleosome arrays, and repressor-protein-induced DNA looping. Because tFCS can be applied to freely diffusing molecules, it is an alternative to trapping-based single-molecule methods to study molecular dynamics in a zero-force regime. The assay is, in its aim, similar to single-molecule FRET, but whereas FRET is powerful for studying fast structural dynamics at the nanometer scale, it cannot be applied to distances greater than nm because of intrinsic limitations in the energy-transfer mechanism (60).
Using DNA of various lengths to benchmark tFCS, we demonstrated that we achieved a spatial resolution of . However, this is not a fundamental limit for the technique but the result of practical limitations in the implementation. Even with a 100 nm tracking localization RMS error, our simulations suggest a spatial resolution of . Anisotropic tracking accuracy—where the axial particle localization is poorer than the lateral localization, resulting in suboptimal positioning of the molecule within the probe beam—likely accounts for much of the discrepancy between the numerical and experimental resolution. This could be overcome with alternative optical designs, such as using a pair of perpendicularly positioned objectives, which would permit the use of AODs to displace the beam along three directions of space and increase the feedback bandwidth. Imperfect coordination in the motion of the tracking and probe beams may also explain some of the discrepancy, but this could be resolved with improved feedback schemes.
Fundamentally, tFCS spatial and temporal resolutions are limited by photon counting in two ways: 1) the fluorescence intensity of the probe dye determines the level of shot noise in the correlation signal, and 2) the count rate of the tracking dye fluorescence defines the maximal feedback bandwidth that sets the tracking accuracy. tFCS would therefore doubly benefit from brighter and more photostable tags, which are actively being developed (61, 62). Such improvements would likely enable the use of tFCS in more complex biochemical environments, such as in cellular extracts. In addition, because tFCS does not require surface immobilization, can track molecules in three dimensions over several tens of micrometers, and operates in a confocal microscopy setting, applications in living cultured cells can be foreseen. The key challenges toward such applications are the development of fluorophores that allow stable imaging of individual molecules within cells with sufficient photon flux and of methods to control the specificity and sparsity of the labeling.
In contrast with traditional localization microscopy techniques, spatial resolution in tFCS is not directly affected by the dimensions of the point spread function but rather by the steepness of the laser intensity gradient at the tracking lock point. This specificity defines an optimization problem for achieving high-resolution imaging, which is distinct from conventional point spread function engineering. It would be interesting to investigate tFCS signals under different excitation profiles, such as those used in light-sheet microscopy (63) or stimulated emission depletion microscopy (64).
The fluorescence fluctuations in tFCS derive from the intramolecular dynamics but also originate from the triplet-state dynamics of the probe dye and from the residual motion of the reference dye in the probe beam frame because of tracking localization error. The convolution of these three processes makes direct fitting of the fluorescence correlation function, as done in conventional FCS, challenging. Future work will focus on deriving methods to better quantify tFCS data. We presented a detailed mathematical analysis of the tFCS signals (Supporting Material Appendix), which suggests that converting the fluorescence correlation into a distance correlation signal to infer the statistics of the relative motion between two sites in calibrated length units is within reach.
We showed that tFCS can measure the Brownian end-to-end dynamics of DNA molecules ranging from , but our simulations (Fig. S5) and previous studies indicate that the method can be applied to probe DNA molecules that are several tens of kilobase pairs long (15). Our current experimental resolution is lower than that of tethered particle motion (TPM), which is a standard approach used for studying the time-dependent motion of DNA (6, 40). However, tFCS could be done in an immobilized setting with the reference dye tethered to a cover slip, in which case the tracking feedback error would be reduced to zero, and the tFCS resolution would be comparable to the TPM resolution, as suggested by our simulations (Fig. S4). The key advantage of tFCS for the study of biopolymers is that the end-to-end motion can be measured in diffusion and force-free conditions. The volume exclusion effects due to the bead and surface proximity in TPM remain difficult to model and account for and are completely bypassed in tFCS (65). More importantly, the time resolution of TPM is intrinsically limited by the hydrodynamic friction of the bead, which slows down the DNA relaxation and by the camera frame rate (66). Because tFCS is bead-free and the signal is collected by an avalanche photodiode with resolution, the time resolution in tFCS is orders of magnitude larger and only limited by the total number of photons collected. tFCS may thus find interesting applications in the measurement of fast DNA relaxation modes, which is important to test polymer models, or in the study of rapid conformational transitions resulting from transient DNA-protein interactions.
Our experiments demonstrate the applicability of tFCS to characterize the compaction state of nucleosome arrays. AUC has been traditionally used to study the response of chromatin to various biochemical perturbations, such as changes in histone composition, salt concentration, or protein activity (46). However, AUC readout is limited to sedimentation coefficients, which are not sufficient to construct precise structural models or characterize the polymer properties of the arrays. Computational studies have proposed different classes of chromatin-fiber structures (67), but we lack experimental measurements to test these models. The end-to-end distance fluctuation spectrum measured by tFCS contains information on the polymer persistence length and relaxation timescales and is amenable to polymer model fitting. We expect that with improved quantification schemes, tFCS of nucleosome arrays will provide a key data set to characterize the polymer properties and structural organization of chromatin under different conditions. Because chromatin organization is thought to regulate genome accessibility and gene expression, we anticipate that tFCS will be impactful in understanding dynamic chromatin processes within the nucleus.
More broadly, tFCS is applicable to probe conformational changes within any large molecular system—such as the macromolecular machines involved in transcription, replication, or chromosome segregation—or within the large elongated proteins implicated in membrane transport. tFCS has the potential to play an important role in the study of these systems, which is similar to the role played by single-molecule FRET in the realm of small individual molecules (15 nm (68)). We envision that tFCS will be powerfully combined with biochemical manipulations and possibly applied inside living cells to tease apart the basic mechanisms that drive organization at the mesoscale.
Author Contributions
H.M. and A.F.S. supervised the project. C.L., A.F.S., and H.M. designed the instrumentation and the method. C.L. built the instrument, did the tFCS experiments, and processed the data. C.L., A.F.S., and H.M. designed and interpreted the DNA and chromatin dynamics experiments. C.L., J.C.B., A.F.S., and H.M. designed and interpreted the LacI experiments. C.L. prepared the DNA and nucleosome arrays constructs, J.C.B. purified the lac repressor, and C.J.F. purified the recombinant histones. C.L. developed the analysis pipeline and wrote the code. C.L., J.C.B., A.F.S., and H.M. wrote and/or commented on the manuscript.
Acknowledgments
We thank Michael Armen for his help with the instrumentation development and his technical assistance, and we thank Andrew Spakowitz, Tom Lampo, Dmitri Pavlichin, and Peter McMahon for insightful discussions on polymer dynamics and stochastic processes. We also thank Dodd Gray for his help with the manuscript; Whitney Johnson, Bradley French, and Frederic Westhorpe for their feedback on the figures; and the members of the Mabuchi and Straight Labs for helpful discussions.
This work was supported by National Institutes of Health grants R01 GM106005 (to A.F.S. and H.M.), R01 GM074728 (to A.F.S.), and by National Science Foundation grant CMMI-0856205 (to H.M.).
Editor: Anatoly Kolomeisky.
Footnotes
Supporting Materials and Methods, eleven figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30254-6
Contributor Information
Aaron F. Straight, Email: astraigh@stanford.edu.
Hideo Mabuchi, Email: hmabuchi@stanford.edu.
Supporting Material
References
- 1.Huang B., Bates M., Zhuang X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J., Rippe K., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 3.de Wit E., de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roux K.J., Kim D.I., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee H.W., Zou P., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanzi F., Broggio C., Pavone F.S. Lac repressor hinge flexibility and DNA looping: single molecule kinetics by tethered particle motion. Nucleic Acids Res. 2006;34:3409–3420. doi: 10.1093/nar/gkl393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han L., Garcia H.G., Phillips R. Concentration and length dependence of DNA looping in transcriptional regulation. PLoS One. 2009;4:e5621. doi: 10.1371/journal.pone.0005621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monico C., Tempestini A., Capitanio M. Mapping DNA-LAC repressor interaction with ultra-fast optical tweezer. 2015 Fotonica AEIT Italian Conference on Photonics Technologies. 2015:1–3. [Google Scholar]
- 9.Ha T. Single-molecule methods leap ahead. Nat. Methods. 2014;11:1015–1018. doi: 10.1038/nmeth.3107. [DOI] [PubMed] [Google Scholar]
- 10.Berglund A., Mabuchi H. Tracking-FCS: fluorescence correlation spectroscopy of individual particles. Opt. Express. 2005;13:8069–8082. doi: 10.1364/opex.13.008069. [DOI] [PubMed] [Google Scholar]
- 11.Berglund A.J., McHale K., Mabuchi H. Feedback localization of freely diffusing fluorescent particles near the optical shot-noise limit. Opt. Lett. 2007;32:145–147. doi: 10.1364/ol.32.000145. [DOI] [PubMed] [Google Scholar]
- 12.Berglund A.J., McHale K., Mabuchi H. Fluctuations in closed-loop fluorescent particle tracking. Opt. Express. 2007;15:7752–7773. doi: 10.1364/oe.15.007752. [DOI] [PubMed] [Google Scholar]
- 13.McHale K., Berglund A.J., Mabuchi H. Quantum dot photon statistics measured by three-dimensional particle tracking. Nano Lett. 2007;7:3535–3539. doi: 10.1021/nl0723376. [DOI] [PubMed] [Google Scholar]
- 14.McHale K., Mabuchi H. Intramolecular fluorescence correlation spectroscopy in a feedback tracking microscope. Biophys. J. 2010;99:313–322. doi: 10.1016/j.bpj.2010.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHale K., Mabuchi H. Precise characterization of the conformation fluctuations of freely diffusing DNA: beyond Rouse and Zimm. J. Am. Chem. Soc. 2009;131:17901–17907. doi: 10.1021/ja906979j. [DOI] [PubMed] [Google Scholar]
- 16.Cohen A.E., Moerner W.E. Suppressing Brownian motion of individual biomolecules in solution. Proc. Natl. Acad. Sci. USA. 2006;103:4362–4365. doi: 10.1073/pnas.0509976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levi V., Ruan Q., Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama Y., Burkacky O., Lamb D.C. Real-time nanomicroscopy via three-dimensional single-particle tracking. Chemphyschem. 2009;10:2458–2464. doi: 10.1002/cphc.200900436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardarelli F., Lanzano L., Gratton E. Fluorescence correlation spectroscopy of intact nuclear pore complexes. Biophys. J. 2011;101:L27–L29. doi: 10.1016/j.bpj.2011.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q., Moerner W.E. An adaptive anti-Brownian electrokinetic trap with real-time information on single-molecule diffusivity and mobility. ACS Nano. 2011;5:5792–5799. doi: 10.1021/nn2014968. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y., Wang Q., Moerner W.E. Hardware-based anti-Brownian electrokinetic trap (ABEL trap) for single molecules: control loop simulations and application to ATP binding stoichiometry in multi-subunit enzymes. Proc SPIE Int Soc Opt Eng. 2008;7038:1–12. doi: 10.1117/12.798093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlau-Cohen G.S., Yang H.Y., Moerner W.E. Single-molecule identification of quenched and unquenched states of LHCII. J. Phys. Chem. Lett. 2015;6:860–867. doi: 10.1021/acs.jpclett.5b00034. [DOI] [PubMed] [Google Scholar]
- 23.Bockenhauer S.D., Duncan T.M., Börsch M. Proceedings of SPIE 8950, 89500H. International Society for Optical Engineering; 2014. The regulatory switch of F1-ATPase studied by single-molecule FRET in the ABEL Trap. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aitken C.E., Marshall R.A., Puglisi J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordes T., Vogelsang J., Tinnefeld P. On the mechanism of Trolox as antiblinking and antibleaching reagent. J. Am. Chem. Soc. 2009;131:5018–5019. doi: 10.1021/ja809117z. [DOI] [PubMed] [Google Scholar]
- 26.Guse A., Fuller C.J., Straight A.F. A cell-free system for functional centromere and kinetochore assembly. Nat. Protoc. 2012;7:1847–1869. doi: 10.1038/nprot.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enderlein J. Tracking of fluorescent molecules diffusing within membranes. Appl. Phys. B. 2000;71:773–777. [Google Scholar]
- 28.Sorlie S.S., Pecora R. A dynamic light scattering study of four DNA restriction fragments. Macromolecules. 1990;23:487–497. [Google Scholar]
- 29.Smith D.E., Perkins T.T., Chu S. Dynamical scaling of DNA diffusion coefficients. Macromolecules. 1996;29:1372–1373. [Google Scholar]
- 30.Petrov E.P., Ohrt T., Schwille P. Diffusion and segmental dynamics of double-stranded DNA. Phys. Rev. Lett. 2006;97:258101. doi: 10.1103/PhysRevLett.97.258101. [DOI] [PubMed] [Google Scholar]
- 31.Robertson R.M., Laib S., Smith D.E. Diffusion of isolated DNA molecules: dependence on length and topology. Proc. Natl. Acad. Sci. USA. 2006;103:7310–7314. doi: 10.1073/pnas.0601903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler R.G., Keller S., Rädler J.O. Intramolecular dynamics of linear macromolecules by fluorescence correlation spectroscopy. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;73:041919. doi: 10.1103/PhysRevE.73.041919. [DOI] [PubMed] [Google Scholar]
- 33.Perkins T.T., Quake S.R., Chu S. Relaxation of a single DNA molecule observed by optical microscopy. Science. 1994;264:822–826. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 34.Lumma D., Keller S., Rädler J.O. Dynamics of large semiflexible chains probed by fluorescence correlation spectroscopy. Phys. Rev. Lett. 2003;90:218301. doi: 10.1103/PhysRevLett.90.218301. [DOI] [PubMed] [Google Scholar]
- 35.Shusterman R., Alon S., Krichevsky O. Monomer dynamics in double- and single-stranded DNA polymers. Phys. Rev. Lett. 2004;92:048303. doi: 10.1103/PhysRevLett.92.048303. [DOI] [PubMed] [Google Scholar]
- 36.Cohen A.E., Moerner W.E. Principal-components analysis of shape fluctuations of single DNA molecules. Proc. Natl. Acad. Sci. USA. 2007;104:12622–12627. doi: 10.1073/pnas.0610396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Widengren J., Mets U., Rigler R. Fluorescence correlation spectroscopy of triplet states in solution: a theoretical and experimental study. J. Phys. Chem. 1995;99:13368–13379. [Google Scholar]
- 38.Widengren J., Schwille P. Characterization of photoinduced isomerization and back-isomerization of the cyanine dye Cy5 by fluorescence correlation spectroscopy. J. Phys. Chem. A. 2000;104:6416–6428. [Google Scholar]
- 39.Nelson P.C., Zurla C., Dunlap D. Tethered particle motion as a diagnostic of DNA tether length. J. Phys. Chem. B. 2006;110:17260–17267. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 40.Pouget N., Dennis C., Salomé L. Single-particle tracking for DNA tether length monitoring. Nucleic Acids Res. 2004;32:e73. doi: 10.1093/nar/gnh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valle F., Favre M., Dietler G. Scaling exponents and probability distributions of DNA end-to-end distance. Phys. Rev. Lett. 2005;95:158105. doi: 10.1103/PhysRevLett.95.158105. [DOI] [PubMed] [Google Scholar]
- 42.Ho L., Crabtree G.R. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chambeyron S., Bickmore W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hübner M.R., Eckersley-Maslin M.A., Spector D.L. Chromatin organization and transcriptional regulation. Curr. Opin. Genet. Dev. 2013;23:89–95. doi: 10.1016/j.gde.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen J.C., Ausio J., van Holde K.E. Homogeneous reconstituted oligonucleosomes, evidence for salt-dependent folding in the absence of histone H1. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz P.M., Hansen J.C. Formation and stability of higher order chromatin structures. Contributions of the histone octamer. J. Biol. Chem. 1994;269:16284–16289. [PubMed] [Google Scholar]
- 47.Schwarz P.M., Felthauser A., Hansen J.C. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 48.Georgel P.T., Horowitz-Scherer R.A., Hansen J.C. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 49.Fan J.Y., Rangasamy D., Tremethick D.J. H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 50.Torres T., Levitus M. Measuring conformational dynamics: a new FCS-FRET approach. J. Phys. Chem. B. 2007;111:7392–7400. doi: 10.1021/jp070659s. [DOI] [PubMed] [Google Scholar]
- 51.Koopmans W.J., Buning R., van Noort J. spFRET using alternating excitation and FCS reveals progressive DNA unwrapping in nucleosomes. Biophys. J. 2009;97:195–204. doi: 10.1016/j.bpj.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poirier M.G., Oh E., Widom J. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 2009;16:938–944. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowary P.T., Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 54.Finch J.T., Klug A. Solenoidal model for superstructure in chromatin. Proc. Natl. Acad. Sci. USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krämer H., Niemöller M., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman A.M., Fischmann T.O., Steitz T.A. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 1995;268:1721–1727. doi: 10.1126/science.7792597. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y., Wong C.H., Wei C.L. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao S.S., Huntley M.H., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y.J., Johnson S., Phillips R. Modulation of DNA loop lifetimes by the free energy of loop formation. Proc. Natl. Acad. Sci. USA. 2014;111:17396–17401. doi: 10.1073/pnas.1415685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krainer G., Hartmann A., Schlierf M. farFRET: extending the range in single-molecule FRET experiments beyond 10 nm. Nano Lett. 2015;15:5826–5829. doi: 10.1021/acs.nanolett.5b01878. [DOI] [PubMed] [Google Scholar]
- 61.Fouz M.F., Mukumoto K., Das S.R. Bright fluorescent nanotags from bottlebrush polymers with DNA-tipped bristles. ACS Cent. Sci. 2015;1:431–438. doi: 10.1021/acscentsci.5b00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grimm J.B., English B.P., Lavis L.D. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen B.C., Legant W.R., Betzig E. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hell S.W., Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 65.Kumar S., Manzo C., Dunlap D. Enhanced tethered-particle motion analysis reveals viscous effects. Biophys. J. 2014;106:399–409. doi: 10.1016/j.bpj.2013.11.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manghi M., Tardin C., Destainville N. Probing DNA conformational changes with high temporal resolution by tethered particle motion. Phys Biol. 2010;7:046003. doi: 10.1088/1478-3975/7/4/046003. [DOI] [PubMed] [Google Scholar]
- 67.Koslover E.F., Fuller C.J., Spakowitz A.J. Local geometry and elasticity in compact chromatin structure. Biophys. J. 2010;99:3941–3950. doi: 10.1016/j.bpj.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.