Abstract
There is emerging evidence that the immune biology associated with lung and other solid tumors, as well as patient immune genetic traits, contributes to individual survival. At this time, dramatic advances in immunologic approaches to the study and management of human cancers are taking place, including lung and head and neck squamous cell carcinoma. However, major obstacles for therapies are the profound immune alterations in blood and in the tumor microenvironment that arise in tandem with the cancer. Although there is a significant current effort underway across the cancer research community to probe the tumor environment to uncover the dynamics of the immune response, little similar work is being done to understand the dynamics of immune alterations in peripheral blood, despite evidence showing the prognostic relevance of the neutrophil/lymphocyte ratio for these cancers. A prominent feature of cancer-associated inflammation is the generation of myeloid-derived suppressor cells, which arise centrally in bone marrow myelopoiesis and peripherally in response to tumor factors. Two classes of myeloid-derived suppressor cells are recognized: granulocytic and monocytic. To date, such immune factors have not been integrated into molecular classification or prognostication. Here, we advocate for a more complete characterization of patient immune profiles, using DNA from archival peripheral blood after application of methylation profiling (immunomethylomics). At the heart of this technology are cell libraries of differentially methylated regions that provide the “fingerprints” of immune cell subtypes. Going forward, opportunities exist to explore aberrant immune profiles in the context of cancer-associated inflammation, potentially adding significantly to prognostic and mechanistic information for solid tumors.
Keywords: immunology, cancer, methylation, epidemiology
It has long been known that DNA methylation patterns are completely erased and reprogramed in preimplantation embryos, enabling the developmental differentiation processes associated with the genesis of somatic lineages (1). In general, genomic methylation patterns in somatic differentiated cells are stable and heritable (1) and include such generally irreversible states as those associated with genomic imprinting and X-inactivation. Our early data (2) are consistent with those of numerous other labs showing that patterns of methylation in promoters, enhancers, and gene bodies strongly correlate with expression (3) and define essentially invariant, stable differentially methylated regions (DMRs) that can be used to predict lineage (4, 5). These DNA methylation changes act to form barriers to ensure that cell-type specification, within the context of the normal development of an organism, is a one-way street. We note that there is some evidence of somatic cell plasticity, but this has been observed under unusual circumstances (e.g., in cancers or in inducing pluripotent cells) (6–8). The Epigenome Roadmap Consortia recently confirmed this, noting that DMRs define lineage (in the developmental context) and that the environment has little (if any) influence on this process (9). Of course, the “environment” does influence cellular lineage, but it does so with some specificity, in the sense that it modifies the major differentiation pathways so as to hone the genome to best respond to the anticipated challenges for the individual. Finally, DNA methylation has been explored extensively in hematopoietic lineage differentiation. In the case of T-cell differentiation, stable and heritable changes in DNA methylation, impervious to minimally responsive environmental perturbations, define the major lineage (10), although it must be noted that the environment can alter the relative size and nature of the differing cell lineages and the pool of cells in each lineage. Regulatory T cells, as one example of a lineage, have been exhaustively shown to have stable and invariant methylation marks that define their phenotype (11, 12). As a consequence, methylation marks with lineage specificity have a potentially profound utility as biomarkers, signaling programmed cellular responses to regulatory stimuli that are distinct from environmental influences upon methylation. Thus, it is now clear that DMRs are specific, genetically determined, developmentally programmed, invariant marks of lineage. Hence, we have applied these methylation biomarkers to probe the immune response status of patients with nonhematopoietic cancer, seeking to enhance our understanding of the normal immune response and devise predictors of disease outcome.
DNA Methylation Data Can Be Used to Define the Immune Profile in Peripheral Blood
Epigenetic modifications, specifically DNA methylation, dictate programmed lineage differentiation within the immune system (13–16). Remodeling the epigenome during development leads to progressively restricted immune subtypes, and DNA methylation provides a chemically stable mark for these cell fate decisions that are immutable and unchanged by lifestyle or exposures (17). This fact, in conjunction with our empirical observations of isolated leukocyte DNA methylation array data, led our group to hypothesize that all lineage-specific immune cells in the peripheral blood could be distinguished by a signature or “fingerprint” of DMRs. In 2012, with our group, Dr. Houseman developed the first statistical algorithm for estimating leukocytes solely by reference to DNA methylation data (18). We have since performed several more extensive validation experiments and continue to evolve ever more sophisticated and accurate bioinformatic methods for immunomethylomics; that is, immune cell typing using methylation (19–22). Using pure cell type reference DNA methylation data (cell type libraries), we deconvolute separate target DNA methylation data sets into constituent cell-type proportions. At this time, there are DMR libraries based on the Illumina 450K methylation platform (Illumina) for normal leukocyte subtypes, including CD4, CD8 T-cells, B cells, natural killer cells, dendritic cells, monocytes, neutrophils, basophils, eosinophils (23), activated natural killer cells (24), and cord blood (25).
Host Immunity Has a Significant Effect on Cancer Survival
Although the classification of tumors has improved our understanding of lung cancer prognosis, immune factors are notably absent in existing prognostic models of lung cancer (26). This omission is significant both because immune evasion is a recognized hallmark of cancer (27) and because of the abundant evidence that patients with lung and aerodigestive cancer suffer systemic immune defects (28–46), a portion of which are now known to respond to immunotherapy (47, 48). Blood lymphocyte counts (particularly CD4 T-cells) and T-cell function are altered in patients with cancer, with T-regulatory cells having been of significant interest to date in the literature (30, 38, 40, 42, 45, 46). Several studies have reported that the increased frequency of T-regulatory CD4+ lymphocytes in the peripheral circulation correlates with prognosis (49–51). In addition to the inhibitory signaling alterations mediated by T-regulatory cells that are associated with cancers, it is also clear that natural killer cells play an important role in aerodigestive cancers (32, 38, 52). In addition to mediating direct cytotoxicity, they participate in the regulation of the antitumorous adaptive immune response, as they produce cytokines such as interferon-γ, tumor necrosis factor-α, interleukin-10, several chemokines, and growth factors. Thus, natural killer cells exert an influence on macrophages, neutrophils, and dendritic cells during the immune response (53). We demonstrated depressed natural killer cell numbers in patients with head and neck squamous cell carcinoma, consistent with their importance in the immune response to head and neck squamous cell carcinoma (54). In addition, the solid tumor microenvironment is highly immunosuppressive (30, 33, 37, 40, 44, 52, 53) through secretion of soluble factors, most notably transforming growth factor-β, interleukin-4, interleukin-10, interleukin-13, and other mechanisms. At the same time, polymorphonuclear granulocytes have been shown to play an important role in the immune and inflammatory responses in head and neck squamous cell carcinoma and lung cancers (28, 29, 41–43, 52, 53).
Neutrophil–Lymphocyte Ratio as a Prognostic Biomarker
Shifts in the distribution of blood leukocytes are important predictors of cancer patient survival. The neutrophil–lymphocyte ratio (NLR) in whole blood has received a great deal of attention as a marker of cancer inflammation (55). The NLR can be derived using the common five-part white blood cell differential (neutrophil, basophil, eosinophil, monocytes, and lymphocytes) from automated cell analyzers. Because the NLR reflects the relative balance of the myeloid and lymphocytic lineages in peripheral blood, it is sensitive to the altered myelopoiesis arising in chronic inflammation and cancer. Extensive studies show that the NLR is a remarkably consistent prognostic factor for survival in malignant and cardiovascular disease (56–61). An NLR <3 is widely considered a favorable predictor for solid tumors as well as related disease mortalities, and an NLR >5 has often been used as the threshold that predicts poor outcome (62). A recent meta-analysis of solid tumor prognosis including 100 studies and 40,559 subjects showed that a higher NLR was significantly associated with reduced overall survival, reduced cancer-specific survival, and reduced progression-free and disease-free survival (55).
There are now numerous studies that all show shorter survival times in patients with lung cancer with an elevated NLR, and a recent meta-analysis confirms the data are consistent (63). Although the thresholds for defining an elevated NLR were somewhat different in these studies, an NLR >5 was associated with poor prognosis independent of known risk factors (e.g., age, stage). We have devised an algorithm using DNA methylation to estimate (64) NLR from 27K and 450K methylation data, and our approach is easily adaptable to the new 850K array platform. In published studies, we found that this DNA methylation-derived NLR at values >5 was independently associated with significantly shorter survival time in studies of multiple solid tumors (65). Because the conventional NLR is based on simple normal cell morphology, the presumed pathologic cell types within the blood cannot be phenotypically (or otherwise) distinguished in blood smears or automated differential counters. Similarly, the current epigenetic methylation-derived NLR measure is based on the methylomes of normal mature leukocyte populations. We have also created an algorithm that estimates the common clinical NLR parameter using only DMR information from normal leukocyte libraries. Because cancer inflammation leads to aberrant myeloid populations in the blood and associated shifts toward higher values of the NLR, immune biomarkers specific to these pathologic cell types (driving immunosuppression) will provide the greatest power to evaluate the role of cancer inflammation in lung cancer survival. Importantly, new preliminary data are poised to answer the obvious question about the predictive power of the NLR. Using either complete blood cell counts or blood methylation data, emerging data suggest the NLR can also prospectively predict solid tumor risk (65). Confirmation of these results awaits further studies.
Mechanistic Considerations
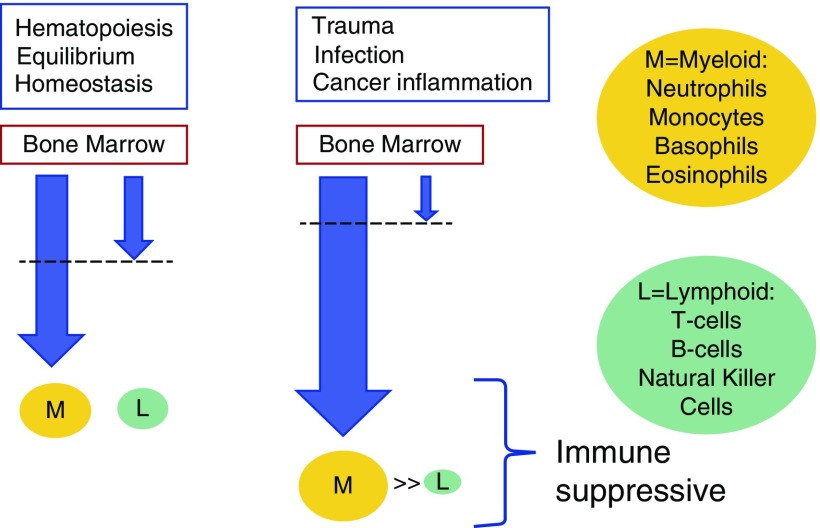
As a result of the overwhelming concordance of this body of literature, there is now an urgent need to investigate the molecular drivers of this phenotype. Researchers long ago observed that tumors affect the host’s hematopoietic progenitor cells, resulting in expansion of myeloid lineage populations and a decrease in circulating lymphoid cells (66). Chronic inflammation, infection, and aging lead to the same reciprocal dynamic between myeloid and lymphoid lineages (67, 68). Today, cancer-associated shifts in myelopoiesis are actively studied, fueled by the realization that inflammation-induced myeloid cells suppress host immune cells (69–71). These myeloid-derived suppressor cells (MDSCs) (72) suppress antigen-specific CD8+ T-cell activity via production of reactive oxygen species and nitric oxide (73, 74), and increase L-arginine metabolism via arginase secretion (75), leading to arginine depletion. These effects lead to downregulation of crucial T-cell receptor components (76), as well as natural killer cell–suppression and cytokine secretion (77). MDSCs also downregulate the NKG2D gene (an activation receptor on natural killer cells), rendering them ineffective in attacking malignant cells (78). Importantly, MDSCs contribute to maternofetal tolerance, modulating immune response via a presence in cord blood (79). Although much research has focused on the effects cancer cells have on bone marrow MDSC precursors, compelling evidence exists that circulating MDSCs may arise from cancer-associated normal blood monocytes and bone marrow precursors, likely via tumor-derived soluble factors such as lactate dehydrogenase and prostaglandin E2 (80, 81). As the myeloid cascade is induced by the presence of a cancer, there exists an opportunity to capture these cells in the peripheral blood; this is a particularly attractive opportunity to use immunomethylomic methods (see Figure 1). The role of MDSCs in lung cancer has been studied, with recent work highlighting the potentially crucial role they might play in personalized medicine (82–85).
Figure 1.
Immune suppression detectable in peripheral blood. Myeloid-derived neutrophil suppressors diminish lymphocytes in many inflammatory conditions, including cancer. The dashed lines indicate cells leaving the bone marrow.
Challenges and Opportunities
Immunomethylomic approaches have demonstrated a completely novel approach to the interrogation of the peripheral blood immune profile. This can be accomplished using archived DNA from blood and does not require flow cytometry. As a consequence, new opportunities have arisen for application of epidemiologic techniques to study of the immune response. At this time, there are few quantitative data enumerating the immune subtype response to environmental insult; these new tools offer promise for in-depth studies of the effects of the environment on the immune response. There are opportunities for assessing the immune profile prospectively, as well as the immune correlates of treatment. Application of this tool will require building additional immune subtype libraries and devising rich quantitative approaches to detection of more rare cell subtypes. Using the tools derived from the stable, developmental methylome, a rich new array of biomarkers and mechanistically based epidemiologic assessments of the immune response is likely to be discovered moving ahead.
Supplementary Material
Footnotes
Supported by National Institutes of Health grants 1R01CA207360 (J.K.W.) and CA207110 (K.T.K.).
Author Contributions: K.T.K. and J.K.W.: Substantial contributions to the conception or design of the work; drafting the work and revising it critically for important intellectual content; and final approval of the version submitted for publication. K.T.K.: Accountability for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 2.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland MJ, Nazor KL, Loring JF. Epigenetic regulation of pluripotency and differentiation. Circ Res. 2014;115:311–324. doi: 10.1161/CIRCRESAHA.115.301517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy S, Gascard P, Dumont N, Zhao J, Pan D, Petrie S, et al. Rare somatic cells from human breast tissue exhibit extensive lineage plasticity. Proc Natl Acad Sci USA. 2013;110:4598–4603. doi: 10.1073/pnas.1218682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan D, Roy S, Gascard P, Zhao J, Chen-Tanyolac C, Tlsty TD. SOX2, OCT3/4 and NANOG expression and cellular plasticity in rare human somatic cells requires CD73. Cell Signal. 2016;28:1923–1932. doi: 10.1016/j.cellsig.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lay FD, Triche TJ, Jr, Tsai YC, Su SF, Martin SE, Daneshmand S, et al. Reprogramming of the human intestinal epigenome by surgical tissue transposition. Genome Res. 2014;24:545–553. doi: 10.1101/gr.166439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naito T, Taniuchi I. Roles of repressive epigenetic machinery in lineage decision of T cells. Immunology. 2013;139:151–157. doi: 10.1111/imm.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toker A, Engelbert D, Garg G, Polansky JK, Floess S, Miyao T, et al. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J Immunol. 2013;190:3180–3188. doi: 10.4049/jimmunol.1203473. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Zheng Y. Regulatory T cell identity: formation and maintenance. Trends Immunol. 2015;36:344–353. doi: 10.1016/j.it.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–e189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 15.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calvanese V, Fernández AF, Urdinguio RG, Suárez-Alvarez B, Mangas C, Pérez-García V, et al. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic Acids Res. 2012;40:116–131. doi: 10.1093/nar/gkr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 18.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Accomando WP, Wiencke JK, Houseman EA, Nelson HH, Kelsey KT. Quantitative reconstruction of leukocyte subsets using DNA methylation. Genome Biol. 2014;15:R50. doi: 10.1186/gb-2014-15-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koestler DC, Christensen B, Karagas MR, Marsit CJ, Langevin SM, Kelsey KT, et al. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8:816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houseman EA, Kelsey KT, Wiencke JK, Marsit CJ. Cell-composition effects in the analysis of DNA methylation array data: a mathematical perspective. BMC Bioinformatics. 2015;16:95. doi: 10.1186/s12859-015-0527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houseman EA, Kim S, Kelsey KT, Wiencke JK. DNA methylation in whole blood: uses and challenges. Curr Environ Health Rep. 2015;2:145–154. doi: 10.1007/s40572-015-0050-3. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Eliot M, Koestler DC, Houseman EA, Wetmur JG, Wiencke JK, et al. Enlarged leukocyte referent libraries can explain additional variance in blood-based epigenome-wide association studies. Epigenomics. 2016;8:1185–1192. doi: 10.2217/epi-2016-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiencke JK, Butler R, Hsuang G, Eliot M, Kim S, Sepulveda MA, et al. The DNA methylation profile of activated human natural killer cells. Epigenetics. 2016;11:363–380. doi: 10.1080/15592294.2016.1163454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, L McKenney S, et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics. 2016;11:354–362. doi: 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashworth AB, Senan S, Palma DA, Riquet M, Ahn YC, Ricardi U, et al. An individual patient data metaanalysis of outcomes and prognostic factors after treatment of oligometastatic non-small-cell lung cancer. Clin Lung Cancer. 2014;15:346–355. doi: 10.1016/j.cllc.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Trellakis S, Bruderek K, Dumitru CA, Gholaman H, Gu X, Bankfalvi A, et al. Polymorphonuclear granulocytes in human head and neck cancer: enhanced inflammatory activity, modulation by cancer cells and expansion in advanced disease. Int J Cancer. 2011;129:2183–2193. doi: 10.1002/ijc.25892. [DOI] [PubMed] [Google Scholar]
- 29.Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123:1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chikamatsu K, Sakakura K, Toyoda M, Takahashi K, Yamamoto T, Masuyama K. Immunosuppressive activity of CD14+ HLA-DR- cells in squamous cell carcinoma of the head and neck. Cancer Sci. 2012;103:976–983. doi: 10.1111/j.1349-7006.2012.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turksma AW, Bontkes HJ, van den Heuvel H, de Gruijl TD, von Blomberg BM, Braakhuis BJ, et al. Effector memory T-cell frequencies in relation to tumour stage, location and HPV status in HNSCC patients. Oral Dis. 2013;19:577–584. doi: 10.1111/odi.12037. [DOI] [PubMed] [Google Scholar]
- 32.Böttcher A, Ostwald J, Guder E, Pau HW, Kramp B, Dommerich S. Distribution of circulating natural killer cells and T lymphocytes in head and neck squamous cell carcinoma. Auris Nasus Larynx. 2013;40:216–221. doi: 10.1016/j.anl.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Green VL, Michno A, Stafford ND, Greenman J. Increased prevalence of tumour infiltrating immune cells in oropharyngeal tumours in comparison to other subsites: relationship to peripheral immunity. Cancer Immunol Immunother. 2013;62:863–873. doi: 10.1007/s00262-013-1395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czystowska M, Gooding W, Szczepanski MJ, Lopez-Abaitero A, Ferris RL, Johnson JT, et al. The immune signature of CD8(+)CCR7(+) T cells in the peripheral circulation associates with disease recurrence in patients with HNSCC. Clin Cancer Res. 2013;19:889–899. doi: 10.1158/1078-0432.CCR-12-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Costa AM, Schuyler CA, Walker DD, Young MR. Characterization of the evolution of immune phenotype during the development and progression of squamous cell carcinoma of the head and neck. Cancer Immunol Immunother. 2012;61:927–939. doi: 10.1007/s00262-011-1154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romano E, Romero P. The therapeutic promise of disrupting the PD-1/PD-L1 immune checkpoint in cancer: unleashing the CD8 T cell mediated anti-tumor activity results in significant, unprecedented clinical efficacy in various solid tumors. J Immunother Cancer. 2015;3:15. doi: 10.1186/s40425-015-0059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oguejiofor K, Hall J, Slater C, Betts G, Hall G, Slevin N, et al. Stromal infiltration of CD8 T cells is associated with improved clinical outcome in HPV-positive oropharyngeal squamous carcinoma. Br J Cancer. 2015;113:886–893. doi: 10.1038/bjc.2015.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linsley PS, Chaussabel D, Speake C. The relationship of immune cell signatures to patient survival varies within and between tumor types. PLoS One. 2015;10:e0138726. doi: 10.1371/journal.pone.0138726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian X, Ma C, Nie X, Lu J, Lenarz M, Kaufmann AM, Albers AE. Biology and immunology of cancer stem(-like) cells in head and neck cancer. Crit Rev Oncol Hematol. 2015;95:337–345. doi: 10.1016/j.critrevonc.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Partlová S, Bouček J, Kloudová K, Lukešová E, Zábrodský M, Grega M, et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. OncoImmunology. 2015;4:e965570. doi: 10.4161/21624011.2014.965570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumitru CA, Fechner MK, Hoffmann TK, Lang S, Brandau S. A novel p38-MAPK signaling axis modulates neutrophil biology in head and neck cancer. J Leukoc Biol. 2012;91:591–598. doi: 10.1189/jlb.0411193. [DOI] [PubMed] [Google Scholar]
- 42.Schaefer C, Kim GG, Albers A, Hoermann K, Myers EN, Whiteside TL. Characteristics of CD4+CD25+ regulatory T cells in the peripheral circulation of patients with head and neck cancer. Br J Cancer. 2005;92:913–920. doi: 10.1038/sj.bjc.6602407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dumitru CA, Lang S, Brandau S. Modulation of neutrophil granulocytes in the tumor microenvironment: mechanisms and consequences for tumor progression. Semin Cancer Biol. 2013;23:141–148. doi: 10.1016/j.semcancer.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Varilla V, Atienza J, Dasanu CA. Immune alterations and immunotherapy prospects in head and neck cancer. Expert Opin Biol Ther. 2013;13:1241–1256. doi: 10.1517/14712598.2013.810716. [DOI] [PubMed] [Google Scholar]
- 45.Drennan S, Stafford ND, Greenman J, Green VL. Increased frequency and suppressive activity of CD127(low/-) regulatory T cells in the peripheral circulation of patients with head and neck squamous cell carcinoma are associated with advanced stage and nodal involvement. Immunology. 2013;140:335–343. doi: 10.1111/imm.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukesova E, Boucek J, Rotnaglova E, Salakova M, Koslabova E, Grega M, et al. High level of Tregs is a positive prognostic marker in patients with HPV-positive oral and oropharyngeal squamous cell carcinomas. BioMed Res Int. 2014;2014:303929. doi: 10.1155/2014/303929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 48.Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen C, Chen D, Zhang Y, Chen Z, Zhu W, Zhang B, et al. Changes of CD4+CD25+FOXP3+ and CD8+CD28- regulatory T cells in non-small cell lung cancer patients undergoing surgery. Int Immunopharmacol. 2014;18:255–261. doi: 10.1016/j.intimp.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Wang WJ, Tao Z, Gu W, Sun LH. Variation of blood T lymphocyte subgroups in patients with non- small cell lung cancer. Asian Pac J Cancer Prev. 2013;14:4671–4673. doi: 10.7314/apjcp.2013.14.8.4671. [DOI] [PubMed] [Google Scholar]
- 51.Zhao L, Yang J, Wang HP, Liu RY. Imbalance in the Th17/Treg and cytokine environment in peripheral blood of patients with adenocarcinoma and squamous cell carcinoma. Med Oncol. 2013;30:461. doi: 10.1007/s12032-013-0461-7. [DOI] [PubMed] [Google Scholar]
- 52.Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol. 2015;33:3293–3304. doi: 10.1200/JCO.2015.61.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Remark R, Becker C, Gomez JE, Damotte D, Dieu-Nosjean MC, Sautès-Fridman C, et al. The non-small cell lung cancer immune contexture: a major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191:377–390. doi: 10.1164/rccm.201409-1671PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Accomando WP, Wiencke JK, Houseman EA, Butler RA, Zheng S, Nelson HH, et al. Decreased NK cells in patients with head and neck cancer determined in archival DNA. Clin Cancer Res. 2012;18:6147–6154. doi: 10.1158/1078-0432.CCR-12-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Templeton AJ, McNamara MG, Šeruga B, Vera-Badillo FE, Aneja P, Ocaña A, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and meta-analysis. J Natl Cancer Inst. 2014;106:dju124. doi: 10.1093/jnci/dju124. [DOI] [PubMed] [Google Scholar]
- 56.Avci A, Alizade E, Fidan S, Yesin M, Guler Y, Kargin R, Esen AM. Neutrophil/lymphocyte ratio is related to the severity of idiopathic dilated cardiomyopathy. Scand Cardiovasc J. 2014;48:202–208. doi: 10.3109/14017431.2014.932922. [DOI] [PubMed] [Google Scholar]
- 57.Shao Q, Chen K, Rha SW, Lim HE, Li G, Liu T. Usefulness of neutrophil/lymphocyte ratio as a predictor of atrial fibrillation: a meta-analysis. Arch Med Res. 2015;46:199–206. doi: 10.1016/j.arcmed.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Saliba W, Barnett-Griness O, Elias M, Rennert G. Neutrophil to lymphocyte ratio and risk of a first episode of stroke in patients with atrial fibrillation: a cohort study. J Thromb Haemost. 2015;13:1971–1979. doi: 10.1111/jth.13006. [DOI] [PubMed] [Google Scholar]
- 59.Bozbay M, Uyarel H. Neutrophil-to-lymphocyte ratio: a novel and simple prognostic marker for infective endocarditis. J Crit Care. 2015;30:822. doi: 10.1016/j.jcrc.2015.04.115. [DOI] [PubMed] [Google Scholar]
- 60.Verdoia M, Barbieri L, Di Giovine G, Marino P, Suryapranata H, De Luca G Novara Atherosclerosis Study Group (NAS) Neutrophil to lymphocyte ratio and the extent of coronary artery disease: results from a large cohort study. Angiology. 2016;67:75–82. doi: 10.1177/0003319715577529. [DOI] [PubMed] [Google Scholar]
- 61.Verdoia M, Schaffer A, Barbieri L, Aimaretti G, Marino P, Sinigaglia F, et al. Novara Atherosclerosis Study Group (NAS) Impact of diabetes on neutrophil-to-lymphocyte ratio and its relationship to coronary artery disease. Diabetes Metab. 2015;41:304–311. doi: 10.1016/j.diabet.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 62.Guthrie GJ, Charles KA, Roxburgh CS, Horgan PG, McMillan DC, Clarke SJ. The systemic inflammation-based neutrophil-lymphocyte ratio: experience in patients with cancer. Crit Rev Oncol Hematol. 2013;88:218–230. doi: 10.1016/j.critrevonc.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 63.Peng B, Wang YH, Liu YM, Ma LX. Prognostic significance of the neutrophil to lymphocyte ratio in patients with non-small cell lung cancer: a systemic review and meta-analysis. Int J Clin Exp Med. 2015;8:3098–3106. [PMC free article] [PubMed] [Google Scholar]
- 64.Perisanidis C, Kornek G, Pöschl PW, Holzinger D, Pirklbauer K, Schopper C, et al. High neutrophil-to-lymphocyte ratio is an independent marker of poor disease-specific survival in patients with oral cancer. Med Oncol. 2013;30:334. doi: 10.1007/s12032-012-0334-5. [DOI] [PubMed] [Google Scholar]
- 65.Koestler DC, Usset J, Christensen BC, Marsit CJ, Karagas MR, Kelsey KT, et al. DNA methylation-derived neutrophil-to-lymphocyte ratio: an epigenetic tool to explore cancer inflammation and outcomes. Cancer Epidemiol Biomarkers Prev. 2017;26:328–338. doi: 10.1158/1055-9965.EPI-16-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee MY, Lottsfeldt JL. Augmentation of neutrophilic granulocyte progenitors in the bone marrow of mice with tumor-induced neutrophilia: cytochemical study of in vitro colonies. Blood. 1984;64:499–506. [PubMed] [Google Scholar]
- 67.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ueda Y, Kondo M, Kelsoe G. Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J Exp Med. 2005;201:1771–1780. doi: 10.1084/jem.20041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagerling C, Casbon AJ, Werb Z. Balancing the innate immune system in tumor development. Trends Cell Biol. 2015;25:214–220. doi: 10.1016/j.tcb.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-derived suppressor cells: critical cells driving immune suppression in the tumor microenvironment. Adv Cancer Res. 2015;128:95–139. doi: 10.1016/bs.acr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmielau J, Finn OJ. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001;61:4756–4760. [PubMed] [Google Scholar]
- 74.Serafini P, Borrello I, Bronte V. Myeloid suppressor cells in cancer: recruitment, phenotype, properties, and mechanisms of immune suppression. Semin Cancer Biol. 2006;16:53–65. doi: 10.1016/j.semcancer.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 75.Munder M, Schneider H, Luckner C, Giese T, Langhans CD, Fuentes JM, et al. Suppression of T-cell functions by human granulocyte arginase. Blood. 2006;108:1627–1634. doi: 10.1182/blood-2006-11-010389. [DOI] [PubMed] [Google Scholar]
- 76.Rodriguez PC, Ernstoff MS, Hernandez C, Atkins M, Zabaleta J, Sierra R, et al. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009;69:1553–1560. doi: 10.1158/0008-5472.CAN-08-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oberlies J, Watzl C, Giese T, Luckner C, Kropf P, Müller I, et al. Regulation of NK cell function by human granulocyte arginase. J Immunol. 2009;182:5259–5267. doi: 10.4049/jimmunol.0803523. [DOI] [PubMed] [Google Scholar]
- 78.Crane CA, Austgen K, Haberthur K, Hofmann C, Moyes KW, Avanesyan L, et al. Immune evasion mediated by tumor-derived lactate dehydrogenase induction of NKG2D ligands on myeloid cells in glioblastoma patients. Proc Natl Acad Sci USA. 2014;111:12823–12828. doi: 10.1073/pnas.1413933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rieber N, Gille C, Köstlin N, Schäfer I, Spring B, Ost M, et al. Neutrophilic myeloid-derived suppressor cells in cord blood modulate innate and adaptive immune responses. Clin Exp Immunol. 2013;174:45–52. doi: 10.1111/cei.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Califano JA, Khan Z, Noonan KA, Rudraraju L, Zhang Z, Wang H, et al. Tadalafil augments tumor specific immunity in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2015;21:30–38. doi: 10.1158/1078-0432.CCR-14-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weed DT, Vella JL, Reis IM, De la Fuente AC, Gomez C, Sargi Z, et al. Tadalafil reduces myeloid-derived suppressor cells and regulatory T cells and promotes tumor immunity in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2015;21:39–48. doi: 10.1158/1078-0432.CCR-14-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koinis F, Vetsika EK, Aggouraki D, Skalidaki E, Koutoulaki A, Gkioulmpasani M, et al. Effect of first-line treatment on myeloid-derived suppressor cells’ subpopulations in the peripheral blood of patients with non-small cell lung cancer. J Thorac Oncol. 2016;11:1263–1272. doi: 10.1016/j.jtho.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 83.Eruslanov EB. Phenotype and function of tumor-associated neutrophils and their subsets in early-stage human lung cancer. Cancer Immunol Immunother. 2017;66:997–1006. doi: 10.1007/s00262-017-1976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pogoda K, Pyszniak M, Rybojad P, Tabarkiewicz J. Monocytic myeloid-derived suppressor cells as a potent suppressor of tumor immunity in non-small cell lung cancer. Oncol Lett. 2016;12:4785–4794. doi: 10.3892/ol.2016.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adah D, Hussain M, Qin L, Qin L, Zhang J, Chen X. Implications of MDSCs-targeting in lung cancer chemo-immunotherapeutics. Pharmacol Res. 2016;110:25–34. doi: 10.1016/j.phrs.2016.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.