ABSTRACT
Candida tropicalis is one of the most important human fungal pathogens causing superficial infections in locations such as the oral mucosa and genital tract, as well as systemic infections with high mortality. In its sister species Candida albicans, the cyclic AMP/protein kinase A (cAMP/PKA) pathway regulates fungal adhesion and dimorphism, both of which correlate closely with virulence. CaTpk1 and CaTpk2, the catalytic subunits of PKA, not only share redundant functions in hyphal growth, adhesion, and biofilm formation, but also have distinct roles in stress responses and pathogenesis, respectively. However, studies on PKA in the emerging fungal pathogen C. tropicalis are limited. Our results suggest that Tpk1 is involved in cell wall integrity and drug tolerance. The tpk2/tpk2 mutants, which have no protein kinase A activity, have reduced hyphal growth and adhesion. In addition, the tpk1/tpk1 tpk2/tpk2 double deletion mutant demonstrated delayed growth and impaired hyphal formation. In a murine model of systemic infection, both TPK1 and TPK2 were required for full virulence. We further found that EFG1 and HWP1 expression is regulated by PKA, while BCR1, FLO8, GAL4, and RIM101 are upregulated in the tpk1/tpk1 tpk2/tpk2 mutant. This study demonstrates that Tpk1 is involved in drug tolerance and cell wall integrity, while Tpk2 serves as a key regulator in dimorphism and adhesion. Both Tpk1 and Tpk2 are required for growth and full virulence in C. tropicalis.
KEYWORDS: adhesion, C. tropicalis, hyphal growth, PKA, virulence
Introduction
Superficial and invasive fungal infections in humans are an emerging problem worldwide. Although the incidence of superficial infections is much higher than invasive infections, invasive infections are studied more frequently due to higher mortality rates [1]. Of the human pathogenic fungi, the genus Candida is the first or second most frequently isolated group from patients with invasive fungal infections [1]. C. albicans is the most prevalent causative agent of all forms of candidiasis, but more than 40% of Candida infections are caused by non-albicans Candida species (NACs) such as C. tropicalis, C. glabrata, and C. parapsilosis [2, 3]. Previous studies indicated that C. tropicalis, an evolutionary counterpart of C. albicans, is the most frequently isolated NAC from patients with candidiasis [2, 4]. Furthermore, the mortality rate of infections caused by C. tropicalis is higher than that of infections caused by other NACs or C. albicans in Brazil [5]. Recently, drug-tolerant or -resistant C. tropicalis isolates have been isolated from patients and environmental samples worldwide [6–11]. However, the molecular mechanisms of drug tolerance and pathogenesis in C. tropicalis remain elusive.
Among the human fungal pathogens causing invasive infections, Histoplasma capsulatum as well as dimorphic fungi in general, undergo transitions between filamentous and yeast growth [12]. The morphological transition enables pathogens to adapt to the harsh environmental conditions within the host [12, 13]. In C. albicans and C. tropicalis, dimorphism is involved in the regulation of adhesion, biofilm formation, invasion, and virulence [12, 14–16]. Within the host, various environmental cues, such as nutrients (carbon, ammonium, or amino acids), N-acetylglucosamine (GlcNAc), pH, temperature, serum, and reactive oxygen species, trigger the morphological change between C. albicans yeast and hyphal forms via various signaling pathways, including the cyclic adenosine monophosphate/protein kinase A (cAMP/PKA), calcineurin, HOG, and MAPK pathways [12, 13].
The cAMP/PKA pathway receives much attention due to its crucial roles in morphological development and pathogenesis, not only in human fungal pathogens such as C. albicans, Cryptococcus neoformans, and Aspergillus fumigatus, but also in plant pathogenic fungi such as Magnaporthe oryzae and Ustilago maydis [17–21]. Upon receiving environmental cues, activated adenylyl cyclase converts ATP to cAMP. Being a secondary messenger, cAMP binds to the regulatory subunits of PKA and causes a conformational change that releases the PKA catalytic subunits. These then activate downstream transcription factors such as Efg1 and Flo8 in C. albicans and Saccharomyces cerevisiae via phosphorylation [18, 20, 22].
In the human fungal pathogen C. albicans, the cAMP/PKA pathway is activated by various environmental cues, such as serum and GlcNAc, and controls the morphological transition between the yeast and hyphal forms, which is important for pathogenesis [13, 15, 23]. In C. albicans, the cAMP/PKA pathway comprises adenylyl cyclase (CYR1/CDC35), two PKA catalytic subunits (TPK1 and TPK2), and a PKA regulatory subunit (BCY1) [18, 23]. Ras1 and Cyr1 have been shown to play important roles in sensing environmental cues, regulating cAMP levels, and governing the expression of Efg1, which is one of the major regulators of filamentation [24]. CaTpk1 and CaTpk2 have redundant roles in growth and dimorphic switching, but each has separate roles in filamentous growth in various filamentation-inducing conditions [25–29]. CaTpk1 contributes to filamentation on solid medium, while CaTpk2 is involved in hyphal growth in liquid medium. Interestingly, only CaTpk2 is required for virulence in murine models of systemic infection [29, 30]. In C. albicans, PKA activates downstream targets, for example Efg1, via phosphorylation, leading to a transition from yeast to pseudohyphal or hyphal growth, a key virulence factor [14]. Morphological switching also plays an important role in biofilm formation [30], which contributes to antifungal drug tolerance [31, 32]. As an evolutionary relative of C. albicans, C. tropicalis was reported to produce pseudohyphae in addition to true hyphae in culture media [2, 4, 16], suggesting that the cAMP/PKA pathway may also be involved in the morphological switching and pathogenesis of C. tropicalis. C. tropicalis PKA comprises two catalytic subunits (Tpk1 and Tpk2) and a regulatory subunit (Bcy1). Recently, Zhang et al. showed that loss of either TPK1 or TPK2 results in normal glucose-induced filamentation in C. tropicalis, indicating that the two catalytic isoforms have a redundant function in morphological transition [33]. However, the impact of PKA signaling pathway-regulated dimorphic switching on growth, drug tolerance, biofilm formation, and virulence remain unclear.
In the present study, we demonstrate that Tpk1 and Tpk2 not only have separate functions in hyphal growth, but also play distinct roles in stress tolerance. Tpk1 is required for cell wall integrity and drug tolerance, while Tpk2 is involved in flocculation and biofilm formation. Furthermore, the tpk1/tpk1 tpk2/tpk2 mutant exhibits severe defects in growth and morphological transition compared to the wild type and tpk1/tpk1 or tpk2/tpk2 mutants. Moreover, Efg1, Brg1, Hwp1, and Als3 might be involved in biofilm formation. Tpk1 and Tpk2 are redundant in pathogenesis, while the tpk1/tpk1 tpk2/tpk2 mutant is avirulent in a murine model of systemic infection.
Results
Identification of Tpk1 and Tpk2 in C. tropicalis
To identify the genes encoding the PKA subunits in C. tropicalis, the amino acid sequences of C. albicans PKA catalytic Tpk1 and Tpk2 and S. cerevisiae Tpk1-3 were used for reciprocal BLAST searches. Each search resulted in the identification of the C. tropicalis TPK1 (CTRG_03315, 405 amino acids) and TPK2 (CTRG_01736, 425 amino acids) genes [34]. Sequence alignments showed that C. tropicalis Tpk1 shares 92% and 68% identity with the corresponding proteins in C. albicans and S. cerevisiae, respectively. Similarly, C. tropicalis Tpk2 has 89% and 78% identity over the full-length proteins with CaTpk2 and ScTpk2, respectively (Fig. 1A, above the diagonal line). Further analysis of the conserved PKA domain (accession: cd05580) revealed that the PKA domains of C. tropicalis Tpk1 and Tpk2 each share 99% homology with the corresponding proteins in C. albicans (Fig. 1A, below the diagonal line). A phylogenetic tree derived from the full-length proteins suggested that Candida Tpk2 proteins are more closely related to ScTpk2 than the Tpk1 proteins of C. albicans and C. tropicalis; however, the Candida PKA catalytic isoforms are more distantly related to ScTpk1 and ScTpk3 (Fig. 1B). These relationships suggest that the C. tropicalis catalytic isoforms (Tpk1 and Tpk2) might have similar functions in filamentation as the corresponding proteins in C. albicans and Tpk2 of S. cerevisiae.
Figure 1.
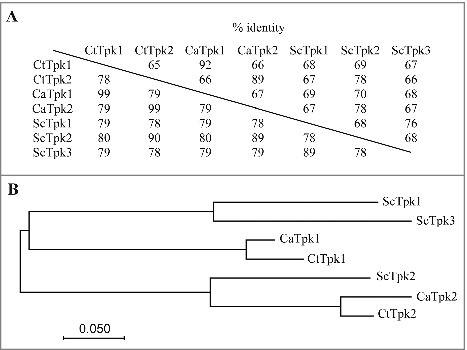
Identification of C. tropicalis PKA catalytic subunits (Tpk1 and Tpk2). (A) The amino acid identity of Tpk proteins from C. albicans (Ca), C. tropicalis (Ct), and S. cerevisiae (Sc). Numbers above the diagonal line indicate sequence identity between the full-length proteins, while numbers below represent the identity of PKA domain (accesion:cd05580). (B) Phylogenetic tree of the PKA catalytic subunits of C. albicans, C. tropicalis, and S. cerevisiae. The amino acid identity and the multiple alignment were analyzed and constructed using the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/). The phylogenetic tree was constructed in MEGA7 [68] software using Maximum Likelihood method based on the JTT matrix-based model [69]. The black bar indicates an evolutionary distance of 0.05 substitutions per site. Proteins and GenBank accession numbers: CaTpk1, XP_723574.1; CaTpk2, XP_714866.2; CtTpk1, XP_002549018.1; CtTpk2, XP_002547429.1; ScTpk1, NP_012371.2; ScTpk2, NP_015121.1; ScTpk3, NP_012755.1.
Tpk1 and Tpk2 have redundant functions in cell growth
The cAMP/PKA signaling pathway regulates a variety of cellular functions, and C. albicans PKA catalytic isoforms contribute to growth fitness [28]. To investigate whether C. tropicalis Tpk1 and Tpk2 are involved in growth fitness, we disrupted the TPK1 and TPK2 genes in C. tropicalis MYA3404 [34], resulting in two independent tpk1/tpk1 (YC710 and YC757) and two independent tpk2/tpk2 (YC681 and YC711) mutants. Both the tpk1/tpk1 and tpk2/tpk2 mutants grew as fast as wild type at 30°C in liquid YPD medium (Fig. 2A). Furthermore, these mutants showed similar growth that was indistinguishable from wild type on solid YPD medium at various temperatures (Fig. 2B). These data suggest that neither Tpk1 nor Tpk2 alone are required for growth fitness.
Figure 2.
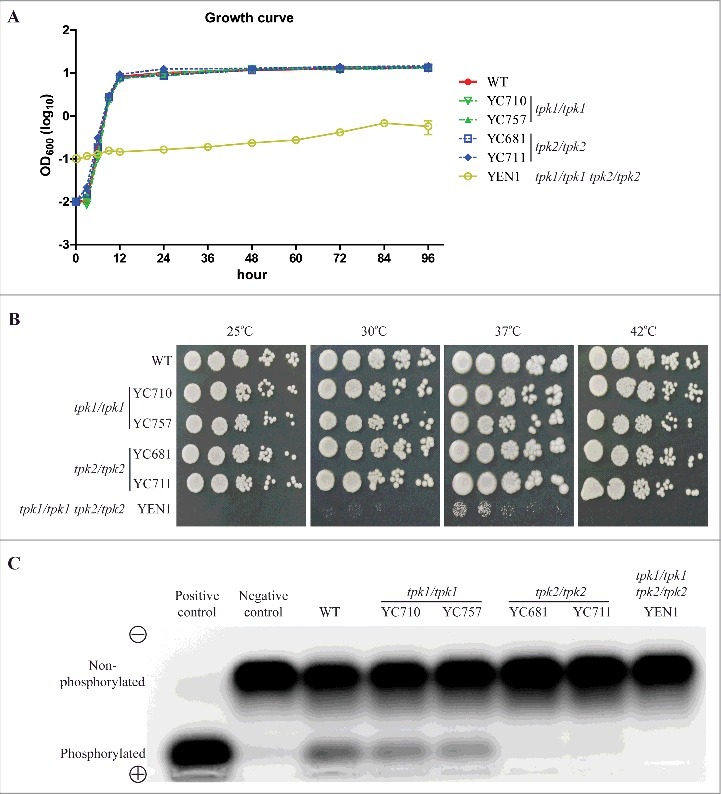
Protein kinase A is required for growth. (A) Growth curves of C. tropicalis wild type and mutants. Cells were grown overnight in YPD at 30°C, washed twice with dH2O, diluted to 0.01 OD600, except for the tpk1/tpk1 tpk2/tpk2 mutant (YEN1), which was diluted to 0.1 OD600 with fresh YPD medium, and incubated at 30°C and 200 rpm for four days. The OD600 of strains was measured via microplate spectrophotometer at the indicated time. The experiments were performed in triplicate, and the values represent the mean ± the standard error of the mean. (B) Spot assay of the indicated strains at different temperatures. Cells were grown overnight in YPD at 30°C (except the tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, and diluted to 0.2 OD600 as the starting concentration. Cells were then five-fold serially diluted, spotted onto YPD medium, and incubated at 25°C, 30°C, 37°C, or 42°C for 48 h and photographed. (C) Protein kinase A activity of the indicated strains. Cells were grown overnight in YPD at 30°C (except the tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days) and washed twice with dH2O. Crude protein extracts of the samples were isolated. PKA activity assays were carried out using the PepTag assay for non-radioactive detection of cAMP-dependent protein kinase kit.
Previous studies indicated that the PKA catalytic subunits are essential in yeast growth, as loss of all catalytic isoforms in S. cerevisiae or C. albicans result in loss of viability [28, 35–37]. However, PKA catalytic subunit-null mutants in C. neoformans and A. fumigatus are viable [38, 39]. To determine whether disruption of both TPK1 and TPK2 results in a growth defect or lethality in C. tropicalis, we deleted the TPK2 gene in the tpk1/tpk1 mutant (YC914) background (Table 1). In contrast to previous attempts by other laboratories [33], we obtained the tpk1/tpk1 tpk2/tpk2 double deletion mutant (YEN1) and found that this mutant exhibits severely impaired growth compared to the wild type or tpk1/tpk1 or tpk2/tpk2 mutants at 30°C (Fig. 2A). In addition, the loss of both PKA catalytic isoforms abolished fungal growth at 25°C and 42°C and drastically reduced growth at 30°C and 37°C (Fig. 2B). The growth defect of the tpk1/tpk1 tpk2/tpk2 mutant was partially rescued by reintroducing either the TPK1 or the TPK2 gene (Figure S1). These results suggest that Tpk1 and Tpk2 have redundant function in fungal growth and temperature sensing.
Table 1.
C. tropicalis strains used in this study.
| Strain | Genotype | Background | Reference |
|---|---|---|---|
| MYA3404 | Prototrophic wild type | Clinical isolate | [34] |
| YC680 | tpk1Δ::SAT1-FLP/TPK1 | MYA3404 | This study |
| YC683 | tpk1Δ::FRT/TPK1 | YC680 | This study |
| YC710aa | tpk1Δ::FRT/tpk1Δ::SAT1-FLP | YC683 | This study |
| YC914 | tpk1Δ::FRT/tpk1Δ::FRT | YC710 | This study |
| YC708 | tpk1Δ::SAT1-FLP/TPK1 | MYA3404 | This study |
| YC718 | tpk1Δ::FRT/TPK1 | YC708 | This study |
| YC757aa | tpk1Δ::FRT/tpk1Δ::SAT1-FLP | YC718 | This study |
| YEN10 | tpk1Δ::FRT/tpk1Δ::FRT | YC757 | This study |
| YEN43 | tpk1Δ::FRT/tpk1Δ::TPK1-SAT1-FLP | YEN10 | This study |
| YC650 | tpk2Δ::SAT1-FLP/TPK2 | MYA3404 | This study |
| YC670 | tpk2Δ::FRT/TPK2 | YC650 | This study |
| YC711bb | tpk2Δ::FRT/tpk2Δ::SAT1-FLP | YC670 | This study |
| YC653 | tpk2Δ::SAT1-FLP/TPK2 | MYA3404 | This study |
| YC672 | tpk2Δ::FRT/TPK2 | YC653 | This study |
| YC681bb | tpk2Δ::FRT/tpk2Δ::SAT1-FLP | YC672 | This study |
| YEN20 | tpk2Δ::FRT/tpk2Δ::FRT | YC681 | This study |
| YEN26 | tpk2Δ::FRT/tpk2Δ::TPK2-SAT1-FLP | YEN20 | This study |
| YC928 | tpk1Δ::FRT/tpk1Δ::FRT tpk2Δ::SAT1-FLP/TPK2 | YC914 | This study |
| YSJ13 | tpk1Δ::FRT/tpk1Δ::FRT tpk2Δ::FRT/TPK2 | YC928 | This study |
| YEN1 | tpk1Δ::FRT/tpk1Δ::FRT tpk2Δ::FRT/tpk2Δ::SAT1-FLP | YSJ13 | This study |
| YEN38 | tpk1Δ::FRT/tpk1Δ::FRT tpk2Δ::FRT/tpk2Δ::FRT | YEN1 | This study |
| YEN40 | tpk1Δ::FRT/tpk1Δ::TPK1-SAT1-FLP tpk2Δ::FRT/tpk2Δ::FRT | YEN38 | This study |
| YEN42 | tpk1Δ::FRT/tpk1Δ::FRT tpk2Δ::FRT/tpk2Δ::TPK2-SAT1-FLP | YEN38 | This study |
Two independent tpk1/ tpk1 mutants.
Two independent tpk2/ tpk2 mutants.
In C. albicans, Tpk2 contributes more to PKA activity than the Tpk1 isoform [27, 40]. To investigate whether differential PKA activity is conserved in C. tropicalis, we extracted total protein from the wild type and mutants to determine PKA activity. PKA activity of the tpk1/tpk1 mutants was slightly reduced compared to the wild type (Figure S2), and levels of PKA activity of tpk2/tpk2 and tpk1/tpk1 tpk2/tpk2 mutants were strongly reduced, indicating that Tpk2 contributes more to PKA activity than Tpk1 in C. tropicalis (Fig. 2C).
Tpk1 plays a key role in stress responses
To determine the roles of the C. tropicalis PKA subunits in the stress response, we first screened the tpk1/tpk1 and tpk2/tpk2 mutants on solid media containing various stressors. Loss of the TPK1 gene resulted in impaired growth when exposed to antifungal drugs (micafungin, anidulafungin, or posaconazole) or cell wall-perturbing agents (sodium dodecyl sulfate [SDS] or dithiothreitol [DTT]) (Fig. 3). The TPK1 complemented strain showed tolerance to SDS and antifungal drugs (Figure S3). In contrast, the tpk2/tpk2 mutants showed similar growth to the wild type under various stresses except when exposed to 0.85 M MgCl2, and growth of the tpk1/tpk1 tpk2/tpk2 mutant was completely abolished (Fig. 3). Our results suggest that Tpk1 plays a major role in response to stresses in C. tropicalis, while Tpk2 might be involved in magnesium homeostasis.
Figure 3.
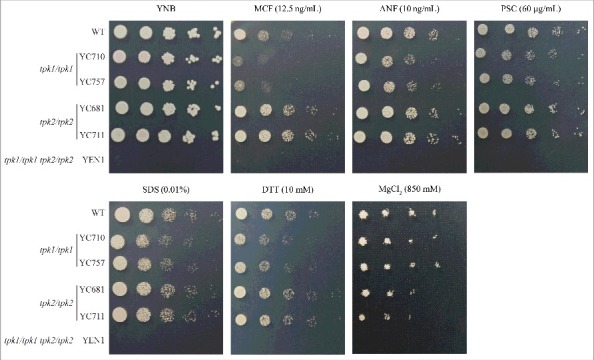
Tpk1 contributes to cell wall integrity and drug tolerance. Growth of the indicated strains exposed to cell wall-perturbing agents, antifungal drugs, and various stresses. Cells were grown overnight in YPD at 30°C (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, and diluted to 0.2 OD600 as the initial concentration. Samples were then five-fold serially diluted, spotted onto YNB medium containing the indicated chemicals, and incubated at 30°C for 48 h. MCF, micafungin; ANF, anidulafungin; PSC, posaconazole; SDS, sodium dodecyl sulfate; DTT, dithiothreitol.
PKA catalytic subunits play different roles in morphogenesis and adhesion
Tpk2 is involved in the regulation of cell-cell and cell-surface adherence in S. cerevisiae [22] and loss of the TPK2 gene in C. albicans results in defects in hyphal growth, adherence, and biofilm formation [27]. To determine the roles of C. tropicalis Tpk1 and Tpk2 in morphogenesis and adhesion, we performed a set of hyphal growth and adhesive assays. Hyphal growth was reduced in tpk2/tpk2 mutants, but not tpk1/tpk1 mutants, when grown in synthetic complete (SC) liquid medium with or without 10 mM GlcNAc or 10 mM 3AT (Fig. 4A). Interestingly, cell clumps and reduced hyphal growth were observed in the tpk1/tpk1 tpk2/tpk2 mutant (Fig. 4A). On nitrogen source starvation (SLAD) solid medium, loss of TPK2 resulted in delayed hyphal growth compared with the wild type and tpk1/tpk1 mutants (Fig. 4B, upper panel). In contrast, the tpk2/tpk2 mutants showed hyper-filamentation on the carbon source starvation medium (Spider) compared with the wild type and tpk1/tpk1 mutants (Fig. 4B, lower panel). As in the liquid medium, the tpk1/tpk1 tpk2/tpk2 mutant exhibited no filamentation on either solid SLAD or Spider media (Fig. 4B).
Figure 4.
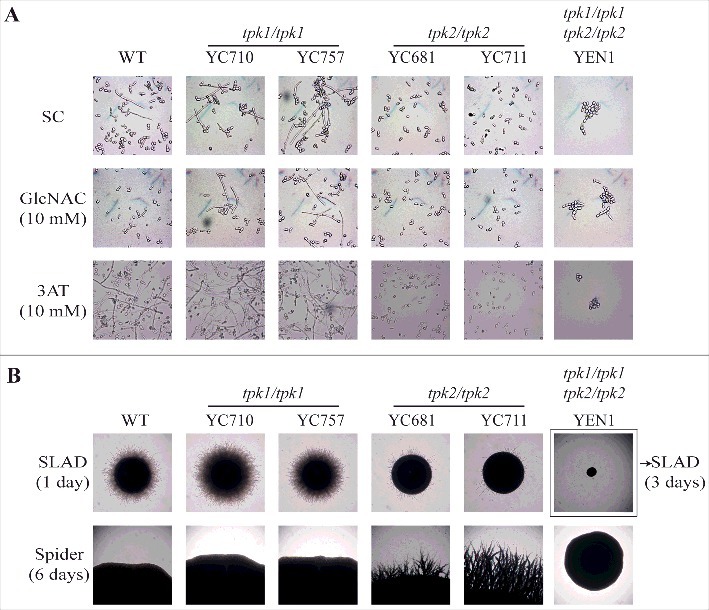
Tpk2, but not Tpk1, regulates hyphal growth. (A) Hyphal growth of the indicated strains in liquid media. Cells were grown overnight in YPD at 30°C (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, diluted to 0.2 OD600 with fresh SC medium or SC medium supplemented with 10 mM of GlcNAC or 3AT, and incubated at 37°C and 200 rpm. (B) Hyphal growth of the indicated strains on solid media. Cells were grown overnight in YPD at 30°C (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, and diluted to 103 cells/mL. Then, 50 μL containing ∼50 cells were spread onto filament-inducing SLAD or Spider plates and incubated at 37°C for the indicated time.
In S. cerevisiae and C. albicans, hyphal growth is correlated with cell-cell and cell-surface adherence [27, 41]. We showed that in C. tropicalis, Tpk1 and Tpk2 play specific roles in filamentation under different conditions (Fig. 4). Therefore, we hypothesized that in C. tropicalis these two proteins may play distinct roles in flocculation, adherence, and biofilm formation, which all require cell-cell or cell-surface adhesion. The tpk2/tpk2 mutants had flocculation defects compared to the wild type and tpk1/tpk1 mutants (Fig. 5A). Interestingly, the tpk1/tpk1 tpk2/tpk2 mutant also exhibited sedimentation at the bottom of the glass tube, indicating that the tpk1/tpk1 mutation is epistatic to the tpk2/tpk2 mutation regarding the flocculation phenotype (Fig. 5A). In contrast to cell-cell adherence, loss of either PKA subunit has no effect on yeast adherence to plastic surfaces (Figure S4). However, the tpk2/tpk2 mutants showed reduced biofilm formation on a polystyrene 12-well plate compared to the wild type and tpk1/tpk1 mutants, whereas no biofilm formation was observed for the tpk1/tpk1 tpk2/tpk2 mutant (Fig. 5B). Furthermore, the hyphal growth and biofilm defects of the tpk2/tpk2 mutant were rescued by reintroducing the TPK2 gene (Figure S5), suggesting that Tpk2 is required for hyphal growth in liquid medium and biofilm formation.
Figure 5.
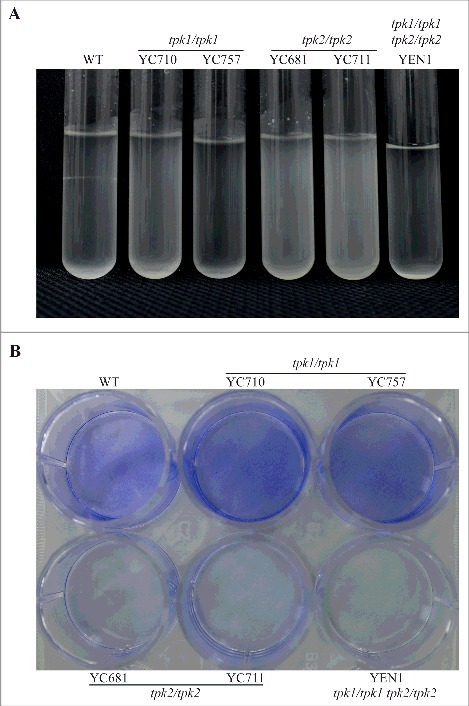
Tpk2 is involved in flocculation and biofilm formation. (A) Tpk2 is required for flocculation. Cells were grown overnight in SC medium containing 10 mM GlcNAc at 30°C (except the tpk1/tpk1 tpk2/tpk2 mutant, which was grown 2 days in YPD, washed twice with dH2O, transferred to SC medium containing 10 mM GlcNAc and incubated at 30°C overnight). The cells were then vortexed, centrifuged at 500 rpm for 1 min, and photographed. (B) Tpk2 is critical for biofilm formation. Cells were grown in YPD medium overnight at 30°C (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, and diluted to 0.5 OD600 in SC medium. Then, 2 mL of each sample were inoculated into a 12-well plate for 90 min at 37°C and 200 rpm. The wells were then washed twice with PBS buffer, inoculated with 2 mL of fresh SC medium, and incubated for 24 h at 37°C and 200 rpm. The wells were then washed twice with PBS, stained with 0.05% crystal violet, and photographed.
Both Tpk1 and Tpk2 are required for full virulence
The adherence of fungal pathogens to host cells is a key step in the establishment of infection [42, 43]. In Candida species, adherence often leads to invasion and biofilm formation [15, 43], and the cAMP/PKA pathway regulates the expression of adherence-associated genes in yeast cells [12, 44]. Our data show that both Tpk1 and Tpk2 are required for hyphal growth, supported by our findings that loss of either TPK1 or TPK2 does not affect yeast adherence to plastic surfaces and that tpk2/tpk2 mutants have a defect in biofilm formation (Fig. 5B and S4). Hence, we analyzed whether loss of hyphal growth or biofilm formation of the PKA null mutants is associated with attenuated virulence in C. tropicalis in a murine model of systemic infection. The tpk1/tpk1 (YC710 and YC757) and tpk2/tpk2 (YC681 and YC711) mutants exhibited similar virulence to wild type, while the tpk1/tpk1 tpk2/tpk2 mutant was avirulent in this infection model (P <0.001, log-rank test) (Fig. 6A).
Figure 6.
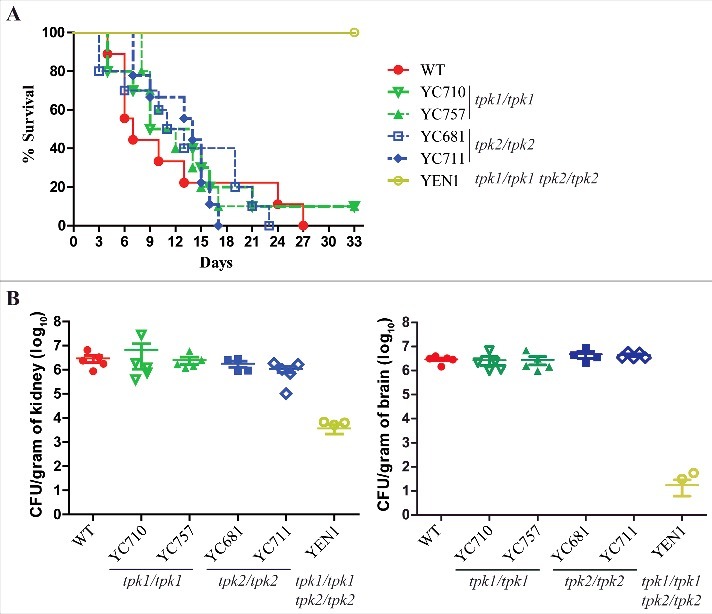
Both Tpk1 and Tpk2 are both required for full virulence. (A) Survival curves of mice infected with the indicated C. tropicalis strains. Cells were grown overnight in YPD medium (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed three times with PBS buffer (pH 7.4), resuspended in PBS buffer, and diluted to a concentration equal to 2.5 × 107 CFU/mL. Mice were inoculated with with 5 × 106 C. tropicalis cells in 200 µL and were monitored for 33 days. Ten mice per strain were used, except for the wild-type strain for which nine were used because one mouse died during the infection process. (B) The fungal burden in the kidneys and brains was measured on day 3 post-C. tropicalis infection in five mice per strain.
To analyze the colonization ability of C. tropicalis PKA mutants, we performed fungal burden analyses in the murine kidneys and brains. The tpk1/tpk1 or tpk2/tpk2 mutants had fungal burdens similar to wild-type, whereas the tpk1/tpk1 tpk2/tpk2 mutant exhibited a significantly decreased fungal burden in the kidneys (P = 0.016) and brains (P < 0.001) compared to the wild type (Fig. 6B).
In the histopathological examination, infection with wild type, tpk1/tpk1 mutants, or tpk2/tpk2 mutants resulted in hyphal growth in the periodic acid-Schiff (PAS)-stained kidney tissues (Fig. 7). However, fungal cells were barely observed in the PAS-stained kidney tissues from mice infected with the tpk1/tpk1 tpk2/tpk2 mutant. Similarly, necrotic tissues observed with hematoxylin-eosin (H&E) staining were only observed in mice infected with the wild type, tpk1/tpk1 mutant, or tpk2/tpk2 mutant, but not in those infected with the tpk1/tpk1 tpk2/tpk2 mutant (Fig. 7). Similarly, brain tissue necrosis was observed in mice infected with the wild type, tpk1/tpk1 mutant, or tpk2/tpk2 mutant, while few yeast cells and no obvious necrosis were observed in the brains of mice infected with the tpk1/tpk1 tpk2/tpk2 mutant (Fig. 7).
Figure 7.
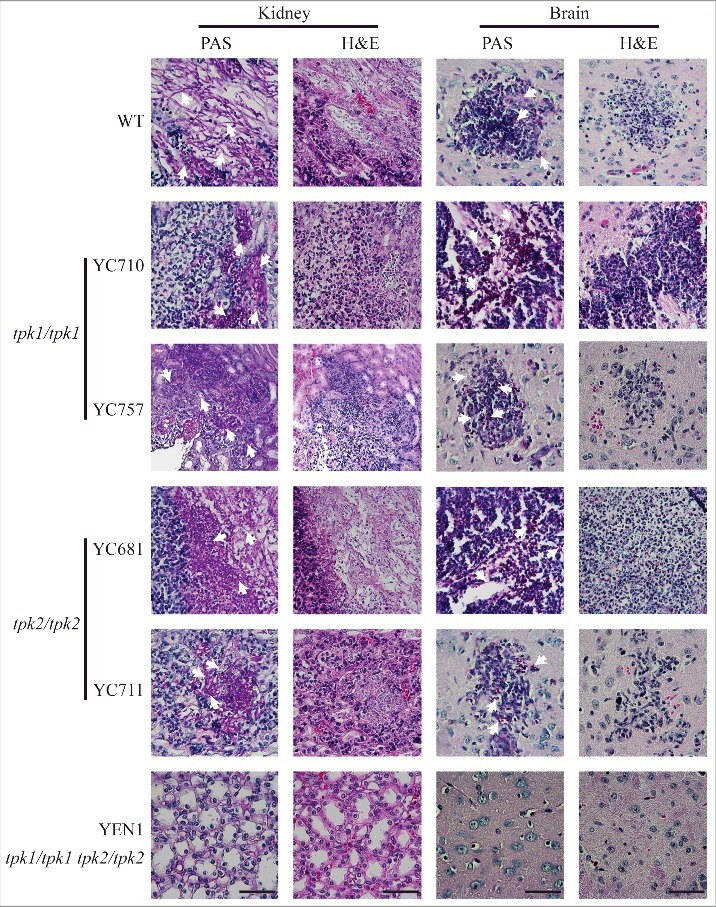
Both Tpk1 and Tpk2 are required for colonization and necrosis in kidneys and brains. Histopathological analyses of mouse kidney and brain organs obtained from C. tropicalis-infected mice at day 3. Organs were fixed in 10% formaldehyde solution, paraffin-embedded, and stained with PAS and H&E to observe C. tropicalis colonization and tissue necrosis, respectively. White arrowheads indicate fungal cells (hyphae or yeasts). Scale bar = 50 µm.
PKA regulates the expression of virulence-related genes
In the present study, we showed that the tpk2/tpk2 mutants exhibited increased filamentation on solid Spider media (Fig. 4B), which is a key feature for invasion and biofilm formation. To test whether hyperfilamentation of the tpk2/tpk2 mutant is due to differential expression of key transcription factors, we performed qRT-PCR with the wild type and PKA null mutants with RNA isolated after incubation in Spider medium for 4 h. Expression of EFG1 and HWP1 was downregulated in tpk1/tpk1, tpk2/tpk2, and tpk1/tpk1 tpk2/tpk2 mutants compared to the wild type (Fig. 8). In addition, loss of TPK2 resulted in reduced expression of the major adhesin ALS3 and transcription factor BRG1 compared with the wild type and tpk1/tpk1 mutants. Interestingly, the tpk1/tpk1 tpk2/tpk2 mutant showed upregulation of four transcription factors (BCR1, FLO8, GAL4, and RIM101) in C. tropicalis (Fig. 8), all of which have demonstrated roles in adherence or biofilm formation in C. albicans [45-48].
Figure 8.
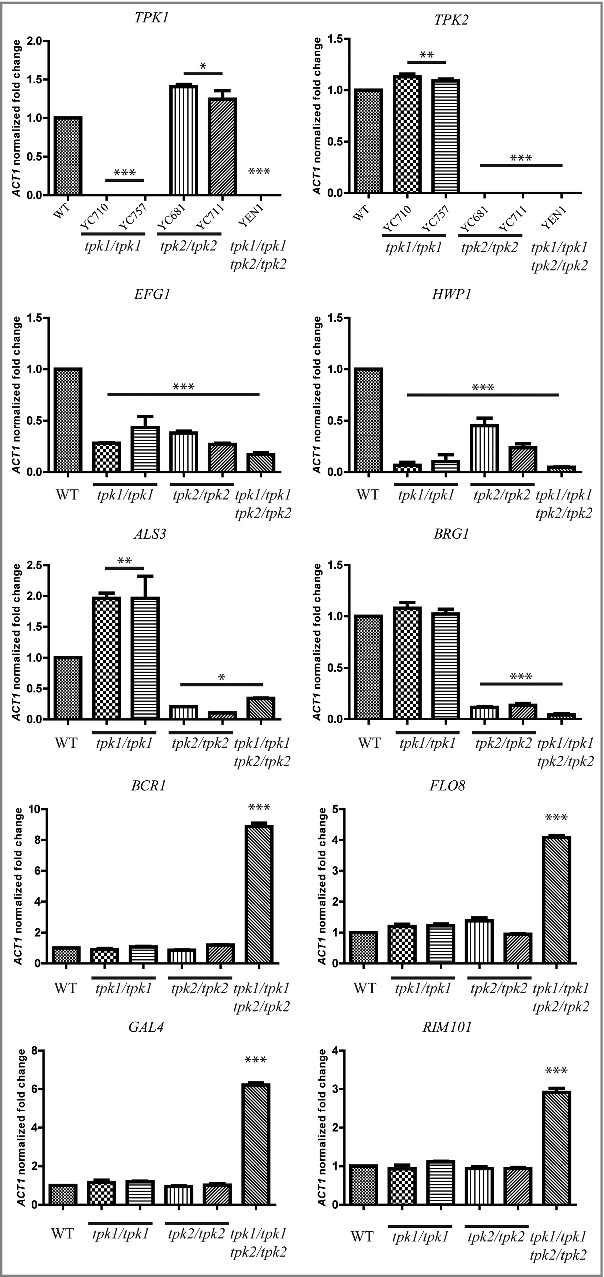
Tpk1 and Tpk2 are involved in the regulation of virulence-related gene expression. Cells were grown overnight in YPD at 30°C, washed twice with dH2O, diluted to 0.1 OD600 in 10 mL of Spider medium, and incubated for 4 h at 37°C. Total RNA was extracted using TRIzol and treated with the TURBO DNA-free™ Kit to remove genomic DNA. cDNA synthesis was carried out using 200 ng of RNA with the high-capacity cDNA reverse transcription kit. Transcript expression levels were analyzed by SYBR® Green PCR Master Mix and normalized to the C. tropicalis ACT1 gene using method. Asterisks indicate statistically significant differences compared with the wild type using Dunnett's multiple comparison test compared to wild type (*P < 0.05; **P < 0.01; ***P < 0.001).
Discussion
Roles of PKA in growth and stress responses of C. tropicalis
In the model organism S. cerevisiae, the cAMP/PKA signaling pathway is involved in the regulation of cell growth and cellular differentiation upon sensing nutrients and stressors from the environment [44, 49–51]. The PKA catalytic subunits (Tpk1-3) of S. cerevisiae have overlapping functions in fungal growth, but a triple deletion mutant was not viable [35] with the exception of a temperature-sensitive tpk2 mutant in the tpk1 tpk3 mutant background [52]. In C. albicans, loss of either catalytic subunit (Tpk1 or Tpk2) resulted in growth similar to the wild type and, as in S. cerevisiae, the tpk1/tpk1 tpk2/tpk2 mutant was not viable [28]. However, the conditional tpk1/tpk1 tpk2/tpk2 mutant was viable but had severe growth defects [28]. Recently, Zhang et al. showed that C. tropicalis Tpk1 and Tpk2 had redundant functions in growth and, as previously reported, that the PKA catalytic null mutant could not grow. Here, our results showed that the tpk1/tpk1 tpk2/tpk2 mutant was viable despite severe growth defects (Fig. 1), which is in contrast to the C. albicans tpk1/tpk1 tpk2/tpk2 mutant, but similar to the corresponding tpk1/PCK1p::TPK1 tpk2/tpk2 conditional mutant in C. albicans [28]. Our results are in line with previous studies in other fungi, such as C. neoformans or A. fumigatus, showing that PKA is not essential [38, 39]. Ding et al. identified a C. albicans bcy1/bcy1 mutant, which had previously been attempted and unsuccessful [53], that is viable, and Bcy1 was found to play a role in filamentation [54]. Taken together, our findings suggest that PKA is not essential, at least in our tested conditions, in C. tropicalis.
The phylogenetic analysis of S. cerevisiae, C. albicans, and C. tropicalis PKA catalytic subunits further supports that CtTpk1 and CaTpk1 are evolutionary similar compared to ScTpk1 or ScTpk3. These data support findings from a previous study performed in C. albicans [28] and suggest that the cAMP/PKA pathway of C. albicans and C. tropicalis might have conserved functions in the regulation of morphogenesis and adhesion. In C. tropicalis, only the tpk1/tpk1 mutant is sensitive to various stresses, such as cell wall-perturbing agents, amino acid starvation, and exposure to antifungal drugs. In contrast to C. albicans in which loss of TPK1 results in increased susceptibility to osmotic, oxidative, and thermal stresses [26], C. tropicalis tpk1/tpk1 and tpk2/tpk2 mutants showed similar growth compared to wild type under osmotic, oxidative, and thermal stress (Fig. 2B and S6). Furthermore, S. cerevisiae PKA catalytic isoforms have overlapping functions in stress responses [52, 55]. CaTpk1 and CtTpk1 showed distinct phenotypes against various stresses, indicating that these two proteins might activate different downstream targets in order to survive under harsh conditions.
Roles of PKA in C. tropicalis hyphal growth and adhesion
The current understanding of the roles of the cAMP/PKA signaling pathway in filamentous growth and adhesion is based on studies of the S. cerevisiae and C. albicans dimorphic lifestyle [18, 22, 44]. In C. albicans, PKA activates downstream targets, for example Efg1, via phosphorylation, leading to a transition from yeast to pseudohyphal or hyphal growth, a key virulence factor [14]. Morphological switching also plays an important role in biofilm formation [30], which contributes to antifungal drug tolerance [31, 32]. A recent study in C. tropicalis suggested that the Efg1 transcription factor mediates hyphal growth as its counterpart does in C. albicans [56], indicating that C. tropicalis and C. albicans PKA, which is upstream of Efg1, may have conserved functions in hyphal growth and adhesion. Our results showed that C. tropicalis Tpk1 and Tpk2 have distinct effects on filamentation in liquid or on solid media (Fig. 4); however, the roles of C. tropicalis PKA in dimorphic switching are distinct from its relative C. albicans. Sonneborn et al. found that hyphal growth of a tpk2/tpk2 mutant in C. albicans was compromised at 30°C but not at 37°C, suggesting that dimorphic switching on solid medium via Tpk2 is temperature-dependent [29]. The same group further showed that loss of TPK1 resulted in abolishment of filamentation at 37°C and that TPK2 is required for hyphal growth in liquid medium [28]. In C. tropicalis, we showed that the tpk2/tpk2 mutant had a filamentation defect both in liquid and on solid media at 37°C, while the loss of TPK1 did not affect hyphal growth at 37°C compared with the wild type (Fig. 4). These results suggest that Tpk1 plays a divergent role in the yeast-hyphal transition of C. tropicalis and C. albicans upon sensing various environmental cues.
Dimorphism, biofilm formation, and adhesion are associated with pathogenicity in Candida species [15]. Candida biofilms consist of yeast cells attached to surfaces, hyphae or pseudohyphae and an extracellular matrix secreted by the cells [57]. Hence, both adherence and dimorphism are key determinants of biofilm formation in C. albicans [57]. Our data showed that the C. tropicalis tpk2/tpk2 mutant had reduced ability to undergo dimorphic switching in specific conditions and subsequently exhibited defects in flocculation and biofilm formation, although the tpk2/tpk2 mutant showed similar adherence to plastic surfaces as the wild type and tpk1/tpk1 mutants. To date, the understanding of how PKA regulates biofilm formation in C. tropicalis is limited. C. albicans biofilm formation has been well characterized, and several transcription factors, including Efg1, Flo8, Tec1, Ndt80, Rob1, Brg1, and Bcr1, are known to be involved in this process [48]. Previous studies showed that Efg1 is a direct target of the PKA signaling pathway in C. albicans and that it governs biofilm formation and adherence via upregulation ALS3 [58, 59]. In addition, loss of EFG1 results in abolishment of biofilm formation on plastic surfaces in C. tropicalis [56]. Our data showed that tpk2/tpk2 mutants were unable to form a biofilm, indicating possible reduced expression of EFG1 and ALS3 as well as other adhesion-related genes. Although the two independent tpk2/tpk2 mutants showed slightly different expressions of EFG1 and HWP1, it is clear that loss of TPK2 resulted in low-level expression of EFG1, BRG1, ALS3, and HWP1 in comparison to the wild type, indicating that C. tropicalis Tpk2 may regulate biofilm formation and adhesion via the transcription factors Efg1 or Brg1 and adhesins Als3 or Hwp1. The different expression levels of EFG1 and HWP1 might be due to experimental issues, such as a fitness change during the transformation procedure in one of the independent tpk2/tpk2 mutants, as the transcription levels of other genes were similar between the two independent mutants. Interestingly, the tpk1/tpk1 tpk2/tpk2 mutant exhibited cell aggregation but neither hyphal growth nor biofilm formation in the inducing conditions (Fig. 4A and 5B). We found high-level expression of adherence-related transcription factors such as Bcr1, Flo8, Gal4, and Rim101 (Fig. 8), indicating that other signaling pathways, such as MAPK or RIM101, may be activated to compensate for the loss of the cAMP/PKA pathway in the tpk1/tpk1 tpk2/tpk2 mutant. The upregulation of these transcription factors might serve as a partial explanation for the aggregation observed in the tpk1/tpk1 tpk2/tpk2 mutant.
Roles of PKA in C. tropicalis virulence
Previous studies of the PKA signaling pathway and virulence focused on C. albicans, A. fumigatus, and C. neoformans [17]. Though C. albicans Tpk1 and Tpk2 are involved in hyphal growth, only Tpk2 is required for virulence in a murine model of systemic infection [29, 60]. Our study indicated that in C. tropicalis, the PKA catalytic subunits play redundant roles in virulence. Loss of either Tpk1 or Tpk2 resulted in virulence similar to the wild type, while the tpk1/tpk1 tpk2/tpk2 mutant was avirulent. Previous studies in C. albicans indicated that mutants with filamentation defects were avirulent [61] and found that a tpk2/tpk2 mutant exhibited reduced hyphal growth and attenuated virulence [29]. A possible explanation for the full virulence of the C. tropicalis tpk2/tpk2 mutants is that these mutants have normal expression of hyphal-related genes such as BCR1, FLO8, and RIM101 compared with the wild type and tpk1/tpk1 mutants. In addition, tpk2/tpk2 mutants exhibited similar tolerance as the wild type to various stresses such as cell wall perturbation and exposure to antifungal drugs, indicating that these mutants might survive under relatively harsh conditions within the host. The tpk1/tpk1 mutants exhibited virulence similar to that of wild-type and the tpk2/tpk2 mutants, but had increased susceptibility to the stressors mentioned above. Dimorphic switching and biofilm formation, which are critical for pathogenesis, may contribute to the virulence of the tpk1/tpk1 mutant. Interestingly, the tpk1/tpk1 tpk2/tpk2 mutant is avirulent, which may be explained by its severe growth defects. Taken together, our data suggest that PKA isoforms have distinct roles in stress responses, hyphal growth, and biofilm formation, but Tpk1 and Tpk2 coordinately regulate growth and virulence of the human fungal pathogen C. tropicalis.
In this study, we summarized and proposed a model to illustrate the distinct roles of Tpk1 and Tpk2 in different cellular process upon encountering various environmental conditions (Fig. 9). In contrast to CaTpk1, which regulates salt and oxidative stresses [26], CtTpk1 regulates cell wall integrity and drug tolerance. CtTpk2 has, at least in part, similar functions to CaTpk2 in the regulation of morphological switching under filamentation-inducing media in liquid or on solid media. CtTpk2 regulates flocculation and biofilm formation via expression of transcription factors and adhesins. However, unlike CaTpk2, CtTpk1 works together with CtTpk2 to reach full virulence in a murine model of systemic infection (Fig. 9).
Figure 9.
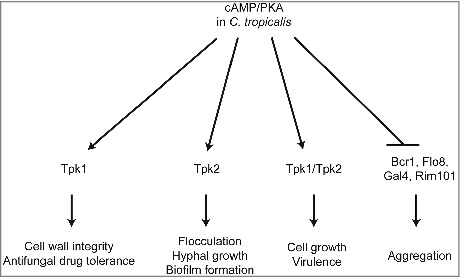
Proposed roles of PKA in C. tropicalis. The C. tropicalis PKA catalytic subunit Tpk1 controls cell wall integrity and drug tolerance, while Tpk2 is required for flocculation, hyphal growth, and biofilm formation. In addition, both proteins are not essential in C. tropicalis and have redundant roles in growth and virulence; loss of either gene results in growth and virulence similar to the wild type. In contrast, Tpk1 and Tpk2 negatively regulate expression of adherence-associated transcription factors (Bcr1, Flo8, Gal4, and Rim101), which were upregulated in the tpk1/tpk1 tpk2/tpk2 mutant that showed cell aggregation.
Materials and methods
Yeast strains, media, and chemicals
C. tropicalis strains used in this study are listed in Table 1. The following media were used in this study: YPD (1% yeast extract, 2% peptone, 2% glucose); YPM (1% yeast extract, 2% peptone, 2% maltose); YNB (0.17% yeast nitrogen base without amino acids and (NH4)2SO4, 0.5% (NH4)2SO4, 2% glucose); SC (YNB with amino acids); SLAD (0.17% yeast nitrogen base without amino acids and (NH4)2SO4, 50 µM (NH4)2SO4, 2% glucose); Spider medium (1% nutrient broth, 1% mannitol, 0.4% K2HPO4, pH 7.2 with H3PO4). Agar (2%) was added to all solid media with the exception of Spider medium, which contained 1.4% agar. For selection assays, YPD medium was supplemented with 100 µg/mL nourseothricin (Werner BioAgents, Jena, Germany). The following supplements were added to media at the indicated concentrations: 0.01% SDS (Bioman, New Taipei City, Taiwan), 60 µg/mL posaconazole (Merck, Rahway, NJ, USA), 12.5 ng/mL micafungin (Astellas Pharma Inc., Deerfield, IL, USA), 10 ng/mL anidulafungin (Selleckchem, Houston, TX, USA), 10 mM DTT (Bioshop, Ontario, Canada), 10 mM 3AT (Sigma, St. Louis, MO, USA), 750 mM NaCl (Bioshop, Ontario, Canada), 750 mM KCl (Bioshop, Ontario, Canada), 500 mM CaCl2 (MDBio, New Taipei City, Taiwan), 100 mM LiCl (MDBio, New Taipei City, Taiwan), 850 mM MgCl2 (MDBio, New Taipei City, Taiwan), 0.005% H2O2 (Sigma, St. Louis, MO, USA), and 10 mM GlcNAc (Sigma, St. Louis, MO, USA).
Strain construction
Both alleles of the C. tropicalis TPK1 and TPK2 genes were disrupted with the SAT1 flipper cassette [62] using the Frozen-EZ Yeast Transformation Kit (Zymo Research, Orange, CA, USA) or electroporation method [63]. To disrupt the TPK1 gene, approximately 1-kb 5′ (amplified with primers JC513/JC514, Table S1) and 3′ (amplified with primers JC515/JC516) noncoding regions (NCRs) of TPK1 were PCR-amplified from genomic DNA of the wild type strain MYA3404 [34]. The 4.2-kb SAT1 flipper sequence was amplified from plasmid pSFS2A [62] with primers JC17/JC18. These three PCR products were treated with ExoSAP-IT (USB corp., Ohio, USA) to remove additional primers and dNTPs, and then combined in a 1:3:1 molar ratio (5′ TPK1NCR :SAT1 flipper:3′ TPK1NCR) to generate the disruption allele by overlap PCR using nesting primers JC517/JC518 (∼100 bp closer to TPK1 compared with JC513/JC516, respectively), resulting in an ∼6-kb TPK1 disruption fragment with 5′ TPK1NCR-SAT1 flipper-3′ TPK1NCR.
The first allele of the TPK1 gene was disrupted in the wild-type strain MYA3404 by transformation with 0.2 to 1 µg of gel-purified disruption DNA using the Frozen-EZ Yeast Transformation Kit. Two independent heterozygous nourseothricin-resistant mutants (YC680 and YC708, Table 1) were obtained from two separate transformations. Liquid YPM medium was used to activate the expression of the FLP recombinase under the control of the C. albicans MAL2 promoter. The SAT1 flipper was then excised, leaving a FLP recombination target (FRT) sequence. This resulted in nourseothricin-sensitive TPK1/tpk1 mutant strains (YC683 and YC718). Despite multiple attempts, the second allele of the TPK1 gene could not be disrupted with the same overlap-PCR disruption allele. Therefore, we amplified 5′ TPK1NCR with JC513/555 and 3′ TPK1NCR with JC556/516 from the second wild-type TPK1 allele of 683 and YC718 and mixed these with the SAT1 flipper to produce an overlap-PCR TPK1 disruption allele specific for the second allele of the TPK1 gene. Eventually, two independent nourseothricin-resistant homozygous tpk1/tpk1 mutants (YC710 and YC757) derived from two separate transformations were obtained.
A similar approach was employed to disrupt the TPK2 gene, with ∼1-kb 5′ and 3′ NCRs for homologous recombination. To generate the ∼6.0-kb TPK2 disruption allele, the overlap PCR DNA products 5′ TPK2NCR (amplified with primers JC521/JC522), SAT1 flipper (amplified with primers JC17/JC18), and 3′ TPK2NCR (amplified with primers JC523/JC524) were mixed in a 1:3:1 molar ratio and amplified with primers JC525/JC526 (∼100 bp closer to the TPK2 ORF compared with JC521/JC524, respectively). A similar approach was used to disrupt the second allele of the TPK2 gene. We amplified the 5′ TPK2NCR with JC521/JC545 and 3′ TPK2 NCR with JC546/524 from the second TPK2 allele of the heterozygous TPK2/tpk2 mutant. We obtained two independent nourseothricin-resistant tpk2/tpk2 mutants (YC711 and YC681, Table 1) from two separate transformations. Correct integration of the disruption cassettes was confirmed by PCR using primers for the ORF and the 5′/3′ NCRs.
To obtain the tpk1/tpk1 tpk2/tpk2 double deletion mutant, YC710 mutant (tpk1/tpk1::SAT1-FLP mutant) was cultured in YPM media to excise the SAT1 flipper, leaving an FRT sequence and resulting in a nourseothricin-sensitive tpk1/tpk1 mutant (YC914). In contrast to the fusion PCR method, the plasmid pYSJ10 was constructed to carry the TPK2 disruption cassette. A ∼1-kb 5′ TPK2NCR containing KpnI and ApaI enzyme cutting sites with primer JC758/759 and ∼1 kb 3′ TPK2NCR containing SacII and SacI enzyme cutting sites with JC760/761 were amplified. Then, a SacII/SacI-digested 3′ TPK2NCR fragment was cloned into the SacII- and SacI-digested pSFS2A to generate pYSJ6. The 5′ TPK2NCR fragment was then subcloned into KpnI- and ApaI-digested pYSJ6 to generate pYSJ10 (Table S2).
The TPK2 disruption cassette isolated from pYSJ10 by SacI and KpnI digestion was transformed into the tpk1/tpk1 mutant (YC914) using electroporation to obtain the tpk1/tpk1 tpk2::SAT1-FLP/TPK2 mutant (YC928). As described previously, the SAT1 flipper of YC928 was excised to generate a nourseothricin-sensitive tpk1/tpk1 tpk2/TPK2 mutant (YSJ13). Because the second TPK2 allele could not be deleted using the same disruption fragment, we amplified ∼700 bp of the 5′ TPK2NCR containing KpnI and ApaI enzyme cutting sites with JC782/JC783 and ∼1 kb of the 3′ TPK2NCR containing SacII and SacI enzyme cutting sites with JC784/JC785 from the YSJ13 mutant. Afterwards, a SacII/SacI-digested 3′ TPK2NCR fragment was cloned into the SacII- and SacI-digested pSFS2A to generate pYSJ25 and the 5′ TPK2NCR fragment was then subcloned into KpnI- and ApaI-digested pYSJ25 to generate pYSJ29 (Table S2). The tpk1/tpk1 tpk2/tpk2 double mutant (YEN1) was obtained by transforming the TPK2 disruption cassette from pYSJ29 into the tpk1/tpk1 tpk2/TPK2 mutant (YSJ13). All strains were confirmed by amplifying the ORF and 5′ and 3′ NCR integrations of the disruption cassettes, as well as by qRT-PCR to detect expression of TPK1 or TPK2 ORFs.
Similar to the deletion cassette, the SAT1-flipper marker was also used to obtain the revertant construct. First, a ∼1.8-kb fragment carrying the 5′ NCR and ORF of TPK1 with ApaI and XhoI restriction sites and a ∼1-kb fragment with the 3′ NCR of TPK1 containing SacII and SacI sites were amplified with primers JC1006/1007 and JC1004/1023, respectively. Then, a SacII/SacI-digested 3′ TPK1NCR fragment was cloned into pSFS2A and digested with the same enzymes to generate pYEN37 (Table S2). Similarly, the TPK1 5′ NCR and ORF were cloned into ApaI and XhoI-digested pYEN37 to generate pYSJ122. The TPK1 complementary cassette containing the ∼7-kb 5′NCR-ORF-SAT1-FLP-3′NCR digested with ApaI and SacI from pYSJ112 was transformed into the tpk1/tpk1 pop-out mutant (YEN10) to generate the tpk1/tpk1::TPK1-SAT1 complemented strain (YEN43) and into the tpk1/tpk1 tpk2/tpk2 pop-out strain (YEN38) to obtain the tpk1/tpk1::TPK1-SAT1 tpk2/tpk2 complemented strain (YEN40).
The same procedure was also used to construct the TPK2 revertant construct. The TPK2 complementary cassette was obtained by cloning a ∼2.3 kb of the 5′ NCR and ORF of TPK2 amplified with primers JC998/999 into KpnI- and ApaI-digested pYSJ6 to generate pYEN2. The TPK2 revertants tpk2/tpk2::TPK2-SAT1 (YEN26) and tpk1/tpk1 tpk2/tpk2::TPK2-SAT1 (YEN42) were obtained by transforming the TPK2 complementary cassette from pYEN2 into the tpk2/tpk2 pop-out mutant (YEN20) and tpk1/tpk1 tpk2/tpk2 pop-out mutant (YEN38), respectively.
Cell adhesion assay
The cell adhesion assay was performed as previously described with modifications to mimic the adherence step of the biofilm assay [64]. Briefly, cells grown in YPD medium overnight were washed twice with PBS and then adjusted to a concentration of OD600 = 0.5 with SC medium. Then, 200 µL of cells were inoculated into a 96-well plate and incubated for 90 min (37°C and 200 rpm). Afterwards, the wells were washed twice with PBS to remove non-adherent cells. The adhered cells were vigorously resuspended in water, serially diluted (2,000-fold), plated on YPD medium, and incubated for two days at 30°C. The colony forming units (CFU) were then determined.
Serial dilution growth assays
Cells were grown in YPD broth at 30°C overnight (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, and diluted with dH2O to an OD600 of 0.2 as the starting concentration. Then, 3 μL of five-fold serial dilutions in dH2O of the indicated strains were spotted onto solid agar plates with a multichannel pipette, incubated at 25°C, 30°C, 37°C, or 42°C for 48 h, and photographed.
Growth kinetics and protein kinase A activity assays
For growth kinetics assays, cells were grown in YPD broth at 30°C overnight (except the tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, diluted to 0.01 OD600 (0.1 OD600 for the tpk1/tpk1 tpk2/tpk2 mutant) in fresh YPD medium, and incubated at 30°C with shaking at 200 rpm. The OD600 of cultures was measured at 0, 3, 6, 9, 12, 24, 48, 72, and 96 h via microplate spectrophotometer (Spectra MAX 190, Molecular Devices, CA, USA). The experiments were performed in triplicate, and the data were plotted using Prism 5.03 (GraphPad Software, CA, USA).
The PKA activity assay was carried out using PepTag Assay for non-radioactive detection of cAMP-dependent protein kinase kit (Promega, Madison, WI, USA) according to the user manual. Briefly, strains were grown in 10 mL of YPD broth at 30°C overnight (except the tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days) and washed twice with dH2O. Crude protein extracts were purified using 500 µL of PKA extraction buffer (0.5 mM EDTA, 0.5 mM EGTA, 10 mM β-mercaptoethanol, 25 mM Tris-HCl [pH 7.4], and 1 µg/mL leupeptin and aprotinin), and the total protein concentration was measured by Bradford assay [65]. Then, 4 µg of the samples were mixed with PKA reaction buffer (1 µM cAMP, 1 mM ATP, 10 mM MgCl2, and 20 mM Tris-HCl [pH 7.4]) and incubated at room temperature for 30 min. The reaction was stopped by incubating the tube at 95°C for 10 min. The samples were loaded and separated on a 0.8% agarose gel in 50 mM Tris-HCl (pH 8.0) at 100 V for 25 min, and the gel was photographed.
Flocculation and biofilm formation assays
The C. tropicalis flocculation assay was performed as previously described [66] with modifications. The wild type and PKA mutants were inoculated in SC liquid medium containing 10 mM GlcNAc and incubated at 30°C overnight (except the tpk1/tpk1 tpk2/tpk2 mutant, which was grown 2 days in YPD, washed twice with dH2O, transferred to SC medium and incubated at 30°C overnight). The cultures were vortexed, centrifuged for 1 min at 500 rpm, and photographed.
The biofilm formation assay was performed as previously described [48] with modifications. C. tropicalis strains were grown in YPD at 30°C overnight (except the tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed twice with dH2O, adjusted to 0.5 OD600 in 2 mL of SC medium, and inoculated into the non-tissue culture treated 12-well plate (Corning, Durham, NC, USA). The inoculated plate was incubated for 90 min at 37°C and 200 rpm, washed with PBS buffer twice, and re-filled with fresh SC medium. The plate was incubated for 24 h at 37°C and 200 rpm. Then, the culture medium was removed and the wells were washed twice with PBS buffer. Biofilm formation was visualized by applying 2 mL 0.05% crystal violet solution for 5 min. Excess stain was removed and the plates were washed twice with 2 mL of dH2O and photographed.
qRT-PCR
C. tropicalis strains were grown in YPD at 30°C overnight (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days) and washed twice with dH2O. Then, 0.2 OD600 cells were transferred to 10 mL of fresh liquid Spider medium and incubated for 4 h at 37°C. Total RNA was then extracted using TRIzol (Ambion, Carlsbad, CA, USA) and treated with TURBO DNA-free™ Kit (Thermo Fisher Scientific, Vilnius, Lithuania) to remove genomic DNA. cDNA synthesis was carried out using 200 ng of total RNA with the high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific, Vilnius, Lithuania) in a PCR machine (Biometra, Jena, Germany) following the manufacturer's manual. Transcript expression levels were analyzed by SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Vilnius, Lithuania) in a StepOnePlus machine (Applied Biosystems, Foster City, CA, USA) and normalized to the ACT1 gene using the method [67]. Accession numbers of the analyzed genes are ACT1 (CTRG_03626); ALS3 (CTRG_02293); HWP1 (CTRG_00477); EFG1 (KP314278); BCR1 (CTRG_00608); RIM101 (CTRG_03710); BRG1 (CTRG_04523); FLO8 (CTRG_02751); and GAL4 (CTRG_01928). The primers for qRT-PCR are shown in Table S1.
Murine model of systemic infection
Four- to five-week-old outbred male ICR mice were purchased from the bioLASCO Taiwan Co. (Taipei, Taiwan) (n = 10 for each group, except n = 9 for the wild type). The C. tropicalis wild-type strain (MYA3404), tpk1/tpk1 mutants (YC710 and YC757), and tpk2/tpk2 mutants (YC711 and YC681), were grown overnight in YPD broth at 30°C (except tpk1/tpk1 tpk2/tpk2 mutant, which was grown for two days), washed three times with PBS buffer (pH 7.4), resuspended in PBS buffer, and diluted to a concentration equal to 2.5 × 107 CFU/mL using a hemocytometer (Paul Marienfeld GmbH&Co. KG, Lauda-Königshofen, Germany). Then, 200 µL (5 × 106 cells) were used to infect mice by lateral tail vein injection. CFU and cell viability were verified by incubating cells on YPD agar plates 48 h at 30°C. Survival was monitored twice daily for 33 days and data were statistically analyzed by log-rank test using Prism 5.03.
To determine fungal burden, mice (n=5 for each group) were infected intravenously with 5 × 106 C. tropicalis wild type or mutant cells (in 200 µL) and were sacrificed three days post-infection. Kidneys and brains from the mice were dissected into two parts and weighed. One part of each organ was soaked in 10 mL PBS buffer and homogenized for 2 min at 1,750 rpm with 2010 Geno/Grinder (SPEX SamplePrep, Metuchen, NJ, USA). The tissue homogenates were serially diluted on YPD solid agar plates containing chloramphenicol (100 µg/mL) and incubated at 30°C for 24 to 48 h (96 h for the tpk1tpk1 tpk2/tpk2 mutant) to determine the CFU per gram of kidney or brain. The identities of the colonies recovered from the organs were further confirmed by PCR. The significance of differences in fungal burden was determined using the unpaired t-test compared to wild type. For histopathological analysis, the other sections of the organs were fixed in 10% formaldehyde solution (Sigma, St Louis, MO, USA), paraffin-embedded, and staining with hematoxylin-eosin (H&E) or periodic acid-Schiff (PAS) was performed by the School of Veterinary Medicine, National Taiwan University. After slide preparation, Candida colonization (PAS) and tissue necrosis (H&E) were examined by microscope.
Ethical statements
All experimental procedures were carried out according to NIH guidelines and were approved by the Institutional Animal Care and Use Committee at National Taiwan University (approval number NTU104-EL-00009).
Supplementary Material
Funding Statement
Ministry of Science & Technology (MOST), Taiwan; MOST 102-2320-B-002-041-MY2, 104-2320-B-002-063-MY3 and 106-2923-B-002-001-MY3
Acknowledgments
This work was financially supported by grants MOST 102-2320-B-002-041-MY2, 104-2320-B-002-063-MY3, and 106-2923-B-002-001-MY3 from the Ministry of Science & Technology (MOST), Taiwan. The authors thank Ms. Cecelia Wall for language editing.
References
- [1].Brown GD, Denning DW, Gow NA, et al.. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- [2].Silva S, Negri M, Henriques M, et al.. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012;36:288–305. doi: 10.1111/j.1574-6976.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- [3].Calderone RA, Clancy CJ. Candida and candidiasis. Washington: DC: ASM Press; 2012. [Google Scholar]
- [4].Negri M, Silva S, Henriques M, et al.. Insights into Candida tropicalis nosocomial infections and virulence factors. Eur J Clin Microbiol Infect Dis. 2012;31:1399–412. doi: 10.1007/s10096-011-1455-z. [DOI] [PubMed] [Google Scholar]
- [5].Nucci M, Colombo AL. Candidemia due to Candida tropicalis: clinical, epidemiologic, and microbiologic characteristics of 188 episodes occurring in tertiary care hospitals. Diagn Microbiol Infect Dis. 2007;58:77–82. doi: 10.1016/j.diagmicrobio.2006.11.009. [DOI] [PubMed] [Google Scholar]
- [6].Choi MJ, Won EJ, Shin JH, et al.. Resistance mechanisms and clinical features of fluconazole-fonsusceptible Candida tropicalis isolates compared with fluconazole-less-susceptible isolates. Antimicrob Agents Chemother. 2016;60:3653–61. doi: 10.1128/AAC.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Garcia-Effron G, Kontoyiannis DP, Lewis RE, et al.. Caspofungin-resistant Candida tropicalis strains causing breakthrough fungemia in patients at high risk for hematologic malignancies. Antimicrob Agents Chemother. 2008;52:4181–3. doi: 10.1128/AAC.00802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kothavade RJ, Kura MM, Valand AG, et al.. Candida tropicalis: its prevalence, pathogenicity and increasing resistance to fluconazole. J Med Microbiol. 2010;59:873–80. doi: 10.1099/jmm.0.013227-0. [DOI] [PubMed] [Google Scholar]
- [9].Yang YL, Ho YA, Cheng HH, et al.. Susceptibilities of Candida species to amphotericin B and fluconazole: the emergence of fluconazole resistance in Candida tropicalis. Infect Control Hosp Epidemiol. 2004;25:60–4. doi: 10.1086/502294. [DOI] [PubMed] [Google Scholar]
- [10].Yang YL, Lin CC, Chang TP, et al.. Comparison of human and soil Candida tropicalis isolates with reduced susceptibility to fluconazole. PLoS One. 2012;7:e34609. doi: 10.1371/journal.pone.0034609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yoo JI, Choi CW, Lee KM, et al.. National surveillance of antifungal susceptibility of Candida species in South Korean hospitals. Med Mycol. 2009;47:554–8. doi: 10.1080/13693780802354037. [DOI] [PubMed] [Google Scholar]
- [12].Gow NA, van de Veerdonk FL, Brown AJ, et al.. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 2012;10:112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Biswas S, Van Dijck P Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–76. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–48. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- [15].Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–28. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen YL, Yu SJ, Huang HY, et al.. Calcineurin controls hyphal growth, virulence, and drug tolerance of Candida tropicalis. Eukaryot Cell. 2014;13:844–54. doi: 10.1128/EC.00302-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fuller KK, Rhodes JC. Protein kinase A and fungal virulence: a sinister side to a conserved nutrient sensing pathway. Virulence. 2012;3:109–21. doi: 10.4161/viru.19396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hogan DA, Sundstrom P. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 2009;4:1263–70. doi: 10.2217/fmb.09.106. [DOI] [PubMed] [Google Scholar]
- [19].Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11:370–80. doi: 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol. 2012;10:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liebmann B, Müller M, Braun A, et al.. The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect Immun. 2004;72:5193–203. doi: 10.1128/IAI.72.9.5193-5203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brückner S, Mösch HU. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2012;36:25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- [23].Inglis DO, Sherlock G. Ras signaling gets fine-tuned: regulation of multiple pathogenic traits of Candida albicans. Eukaryot Cell. 2013;12:1316–25. doi: 10.1128/EC.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–48. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- [25].Souto G, Giacometti R, Silberstein S, et al.. Expression of TPK1 and TPK2 genes encoding PKA catalytic subunits during growth and morphogenesis in Candida albicans. Yeast. 2006;23:591–603. doi: 10.1002/yea.1377. [DOI] [PubMed] [Google Scholar]
- [26].Giacometti R, Kronberg F, Biondi RM, et al.. Catalytic isoforms Tpk1 and Tpk2 of Candida albicans PKA have non-redundant roles in stress response and glycogen storage. Yeast. 2009;26:273–85. doi: 10.1002/yea.1665. [DOI] [PubMed] [Google Scholar]
- [27].Giacometti R, Kronberg F, Biondi RM, et al.. Candida albicans Tpk1p and Tpk2p isoforms differentially regulate pseudohyphal development, biofilm structure, cell aggregation and adhesins expression. Yeast. 2011;28:293–308. doi: 10.1002/yea.1839. [DOI] [PubMed] [Google Scholar]
- [28].Bockmühl DP, Krishnamurthy S, Gerads M, et al.. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol Microbiol. 2001;42:1243–57. doi: 10.1046/j.1365-2958.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- [29].Sonneborn A, Bockmühl DP, Gerads M, et al.. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol Microbiol. 2000;35:386–96. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- [30].Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006;9:588–94. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- [31].Ramage G, Rajendran R, Sherry L, et al.. Fungal biofilm resistance. Int J Microbiol. 2012;2012:528521. doi: 10.1155/2012/528521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kuhn DM, George T, Chandra J, et al.. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob Agents Chemother. 2002;46:1773–80. doi: 10.1128/AAC.46.6.1773-1780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang Q, Tao L, Guan G, et al.. Regulation of filamentation in the human fungal pathogen Candida tropicalis. Mol Microbiol. 2016;99:528–45. doi: 10.1111/mmi.13247. [DOI] [PubMed] [Google Scholar]
- [34].Butler G, Rasmussen MD, Lin MF, et al.. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–62. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Toda T, Cameron S, Sass P, et al.. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–87. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- [36].Giacometti R, Kronberg F, Biondi RM, et al.. Cross regulation between Candida albicans catalytic and regulatory subunits of protein kinase A. Fungal Genet Biol. 2012;49:74–85. doi: 10.1016/j.fgb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- [37].Huang G, Yi S, Sahni N, et al.. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010;6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].D'Souza CA, Alspaugh JA, Yue C, et al.. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol. 2001;21:3179–91. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fuller KK, Richie DL, Feng X, et al.. Divergent Protein Kinase A isoforms co-ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus. Mol Microbiol. 2011;79:1045–62. doi: 10.1111/j.1365-2958.2010.07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cloutier M, Castilla R, Bolduc N, et al.. The two isoforms of the cAMP-dependent protein kinase catalytic subunit are involved in the control of dimorphism in the human fungal pathogen Candida albicans. Fungal Genet Biol. 2003;38:133–41. doi: 10.1016/S1087-1845(02)00520-0. [DOI] [PubMed] [Google Scholar]
- [41].Fichtner L, Schulze F, Braus GH. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288c. Mol Microbiol. 2007;66:1276–89. doi: 10.1111/j.1365-2958.2007.06014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gauthier GM, Keller NP. Crossover fungal pathogens: the biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet Biol. 2013;61:146–57. doi: 10.1016/j.fgb.2013.08.016. [DOI] [PubMed] [Google Scholar]
- [43].Silva S, Negri M, Henriques M, et al.. Adherence and biofilm formation of non-Candida albicans Candida species. Trends Microbiol. 2011;19:241–7. doi: 10.1016/j.tim.2011.02.003. [DOI] [PubMed] [Google Scholar]
- [44].Cullen PJ, Sprague GF Jr. The regulation of filamentous growth in yeast. Genetics. 2012;190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Finkel JS, Xu W, Huang D, et al.. Portrait of Candida albicans Adherence Regulators. PLoS Pathog. 2012;8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu J-Y, Li W-J, Shi C, et al.. Mutations in the Flo8 transcription factor contribute to virulence and phenotypic traits in Candida albicans strains. Microbiol Res. 2015;178:1–8. doi: 10.1016/j.micres.2015.05.007. [DOI] [PubMed] [Google Scholar]
- [47].Fox EP, Bui CK, Nett JE, et al.. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 2015;96:1226–39. doi: 10.1111/mmi.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nobile CJ, Fox EP, Nett JE, et al.. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–38. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–18. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- [50].Wang L, Renault G, Garreau H, et al.. Stress induces depletion of Cdc25p and decreases the cAMP producing capability in Saccharomyces cerevisiae. Microbiology. 2004;150:3383–91. doi: 10.1099/mic.0.27162-0. [DOI] [PubMed] [Google Scholar]
- [51].Cullen PJ, Sprague GF Jr. Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci U S A. 2000;97:13619–24. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–64. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cassola A, Parrot M, Silberstein S, et al.. Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot Cell. 2004;3:190–9. doi: 10.1128/EC.3.1.190-199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ding X, Cao C, Zheng Q, et al.. The regulatory subunit of Protein Kinase A (Bcy1) in Candida albicans plays critical roles in filamentation and white-opaque switching but is not essential for cell growth. Front Microbiol. 2016;7:2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Thevelein JM, Cauwenberg L, Colombo S, et al.. Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb Technol. 2000;26:819–25. doi: 10.1016/S0141-0229(00)00177-0. [DOI] [PubMed] [Google Scholar]
- [56].Mancera E, Porman AM, Cuomo CA, et al.. Finding a missing gene: EFG1 regulates morphogenesis in Candida tropicalis. G3 (Bethesda). 2015;5:849–56. doi: 10.1534/g3.115.017566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Araujo D, Henriques M, Silva S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017;25:62–75. doi: 10.1016/j.tim.2016.09.004. [DOI] [PubMed] [Google Scholar]
- [58].Moazeni M, Khorramizadeh MR, Teimoori-Toolabi L, et al.. Down-regulation of the ALS3 gene as a consequent effect of RNA-mediated silencing of the EFG1 gene in Candida albicans. Iranian Biomed J. 2012;16:172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bockmühl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157:1523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Park H, Myers CL, Sheppard DC, et al.. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell Microbiol. 2005;7:499–510. doi: 10.1111/j.1462-5822.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- [61].Lo HJ, Kohler JR, DiDomenico B, et al.. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–49. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- [62].Reuß O, Vik A, Kolter R, et al.. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–27. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- [63].De Backer MD, Maes D, Vandoninck S, et al.. Transformation of Candida albicans by electroporation. Yeast. 1999;15:1609–18. doi: 10.1002/(SICI)1097-0061(199911)15:15%3c1609::AID-YEA485%3e3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [64].Winter MB, Salcedo EC, Lohse MB, et al.. Global identification of biofilm-specific proteolysis in Candida albicans. MBio. 2016;7:e01514–16. doi: 10.1128/mBio.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- [66].Chin C, Lai WC, Lee TL, et al.. Dissection of the Candida albicans Cdc4 protein reveals the involvement of domains in morphogenesis and cell flocculation. J Biomed Sci. 2013;20:97. doi: 10.1186/1423-0127-20-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-DDC(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [68].Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


