Introduction
Despite three decades of successful, predominantly phenotype-driven, discovery of the genetic causes of monogenic disorders 1, up to half of children with severe developmental disorders (DDs) of likely genetic origin remain without a genetic diagnosis. Especially challenging are those disorders rare enough to have eluded recognition as a discrete clinical entity, those whose clinical manifestations are highly variable, and those that are difficult to distinguish from other, very similar, disorders. Here we demonstrate the power of embracing an unbiased genotype-driven approach 2 to identify subsets of patients with similar disorders. By studying 1,133 children with severe, undiagnosed DDs, and their parents, using a combination of exome sequencing 3–11 and array-based detection of chromosomal rearrangements, we discovered 12 novel genes causing DDs. These newly implicated genes increase by 10% (from 28% to 31%) the proportion of children that could be diagnosed. Clustering of missense mutations in six of these newly implicated genes suggest that normal development is being perturbed by an activating or dominant negative mechanism. Our findings demonstrate the value of adopting a comprehensive strategy, both genomewide and nationwide, to elucidating the underlying causes of rare genetic disorders.
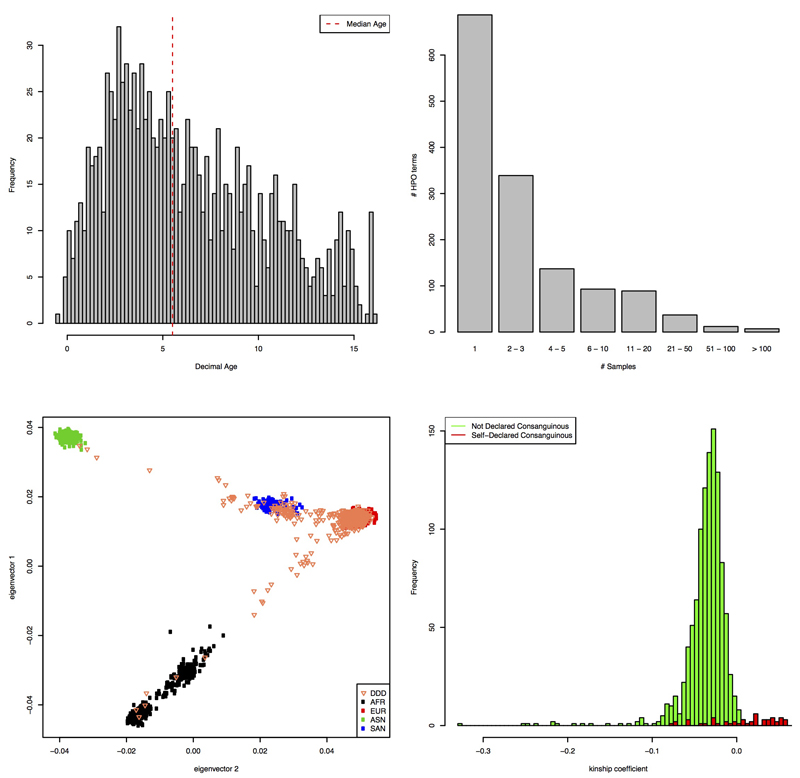
We established a network to recruit 1,133 children (median age 5.5, Extended Data Fig. 1A) with diverse, severe undiagnosed DDs, through all 24 regional genetics services of the UK National Health Service and Republic of Ireland. Among the most commonly observed phenotypes (Extended Data Fig. 1B, Supplementary Table 1) were intellectual disability or developmental delay (87% of children), abnormalities revealed by cranial MRI (30%), seizures (24%), and congenital heart defects (11%). These children are predominantly (~90%) of Northwest European ancestry (Extended Data Fig. 1C), with 47 pairs of parents (4.1%) exhibiting kinship equivalent to, or in excess of second cousins (Extended Data Fig. 1D, Supplementary Information). In most families (849/1,101), the child was the only affected family member, but 111 children had one or more parents with a similar DD, and 124 had a similarly affected sibling (Supplementary Information). Prior clinical genetic testing would have already diagnosed many children with easily recognized syndromes, or large pathogenic deletions and duplications, enriching this research cohort for less distinct syndromes, and novel genetic disorders.
We exome sequenced 1,133 affected children and their parents, from 1,101 families, representing 1,071 unrelated children and 30 sibships. We also performed exome-focused array comparative genomic hybridization (exome-aCGH) on the children (N=1,009) and UK controls (N=1,013) and genome-wide genotyping on the trios (N=1,006) to identify deletions, duplications, uniparental disomy (UPD) and mosaic large chromosome rearrangements. From our exome sequencing and exome-aCGH data, we detected an average of 19,811 coding or splicing single nucleotide variants (SNVs), 491 coding or splicing indels and 148 Copy Number Variants (CNVs) per child (Supplementary Information). From analyses of the genotyping array data 12 we identified 6 children with UPD and 5 children with mosaic large chromosomal rearrangements (Supplementary Information). The SNVs, indels and CNVs were analysed jointly in the following analyses, allowing, for example, the identification of compound heterozygous CNVs and SNVs affecting the same gene.
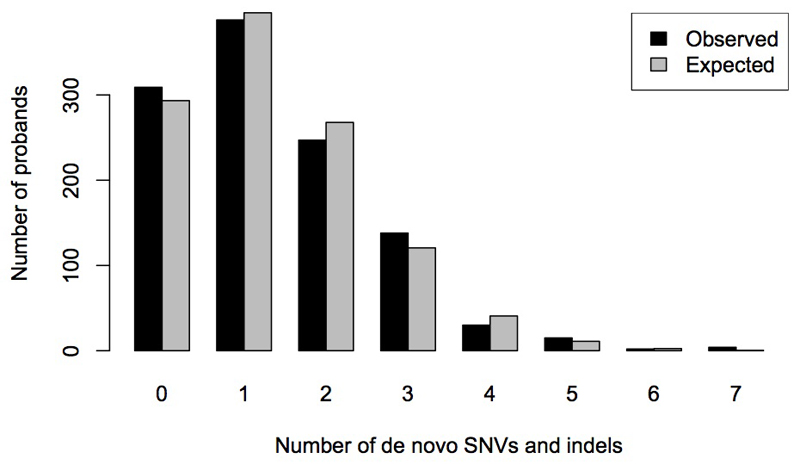
We discovered 1,618 de novo variants (1,417 SNVs, 114 indels and 87 CNVs) in coding and non-coding regions (Supplementary Tables 2 and 3), of which 1,596 (98.6%) were validated using a second, independent assay, and the remainder were validated clinically. This represents an average of 1.12 de novo SNVs and 0.09 de novo indels in coding or splicing regions per child, which is within the range of similar studies 3–11. The distribution of de novo SNVs and indels per child closely approximated the Poisson distribution expected for random mutational events (Extended Data Fig. 2).
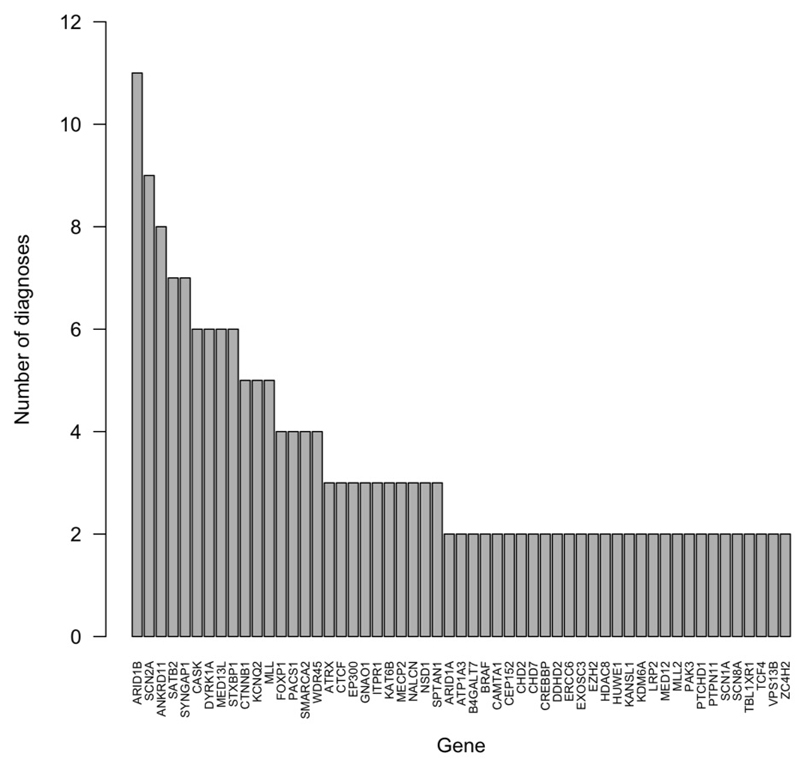
We classified 28% (N=317) of children with likely pathogenic variants (Supplementary Table 4 and 13) in 1,129 robustly implicated DD genes (published before Nov 2013), or with pathogenic deletions or duplications. The majority of these diagnoses involved de novo SNVs, indels or CNVs (Table 1). Females had a significantly higher diagnostic yield of autosomal de novo mutations than males (p=0.01, Fisher exact test). Among the single gene diagnoses, most DD genes (95/148) were only observed once, although eight (ARID1B, SATB2, SYNGAP1, ANKRD11, SCN1A, DYRK1A, STXBP1, MED13L) each accounted for 0.5-1% of children in our cohort (Extended Data Figure 3). For 17 of these children we identified two different genes with pathogenic variants, resulting in a composite clinical phenotype.
Table 1. Breakdown of diagnoses by mode and by sex.
| Female (%) | Male (%) | Total (%) | |
|---|---|---|---|
| Undiagnosed | 383 (69.6%) | 433 (74.3%) | 816 (72.0%) |
| Diagnosed | 167 (30.4%) | 150 (25.7%) | 317 (28.0%) |
| De novo mutation | 124 (22.5%) | 80 (13.7%) | 204 (18.0%) |
| chrX | 24 (4.4%) | 5 (0.9%) | 28 (2.6%) |
| autosomal | 100 (18.2%) | 75 (12.9%) | 176 (15.5%) |
| Autosomal Dominant* | 9 (1.6%) | 11 (1.9%) | 20 (1.8%) |
| Autosomal Recessive | 20 (3.6%) | 26 (4.5%) | 46 (4.1%) |
| X-linked Inherited | 1 (0.2%) | 19 (3.3%) | 20 (1.8%) |
| UPD/Mosaicism | 4 (0.7%) | 6 (1.0%) | 10 (0.9%) |
| Composite | 9 (1.6%) | 8 (1.4%) | 17 (1.5%) |
| Total | 550 | 583 | 1133 |
Inherited from an affected parent
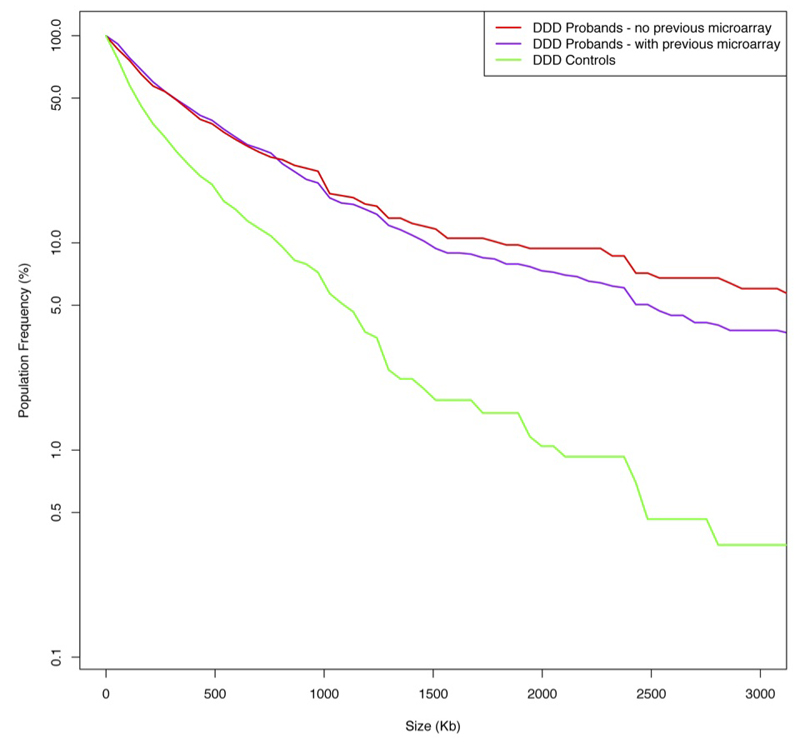
Analyses that assess the enrichment in patients of a particular class of variation, so-called ‘burden analyses’, both highlight classes of variants for detailed analysis, and enable estimation of the proportion of a particular class of variant that is likely to be pathogenic. We observed a significant (p=0.0004) burden of 87 de novo CNVs in the 1,133 DD children compared to 12 in 416 controls (Scottish Family Health Study14) despite most children (77%) having previously had clinical microarray testing (Extended Data Figure 4).
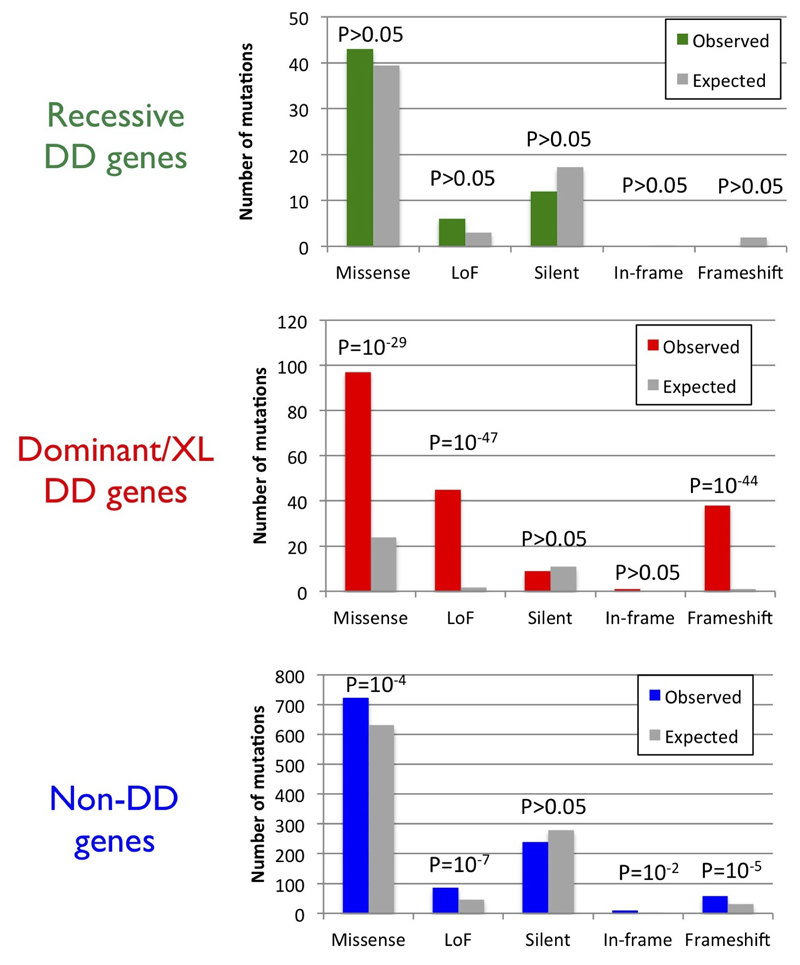
We used gene-specific mutation rates that account for gene length and sequence context 15 to assess the burden of different classes of de novo SNVs and indels (Supplementary Information). We observed no significant excess of any functional class of de novo SNVs or indels in autosomal recessive DD genes (Extended Data Figure 5), suggesting that few of these mutations are causally implicated. By contrast, we observed a highly significant excess of all ‘functional’ classes (coding and splice site variants excepting synonymous changes) of de novo SNVs and indels in the dominant and X-linked DD genes (Extended Data Figure 5) within which de novo mutations can be sufficient to cause disease. Not all protein-altering mutations in known dominant and X-linked DD genes will be pathogenic, and these burden analyses inform estimates of positive predictive values for different classes of mutations. The remaining, non-DD, genes in the genome also exhibit a more modest, but significant, excess of functional, but not silent, de novo SNVs and indels (Extended Data Figure 5).
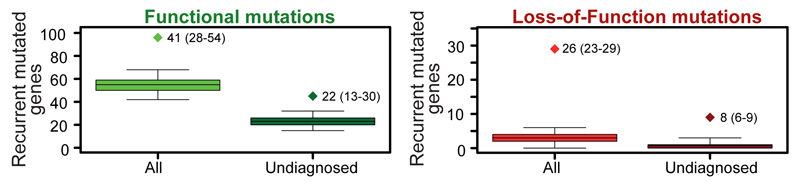
We observed 96 genes with recurrent, functional mutations (Figure 1A), a highly significant excess compared to the expected number derived from simulations (median=55, Supplementary Information). This enrichment is even more pronounced (observed:29, expected:3) for recurrent LoF mutations (Figure 1B). Among undiagnosed children, we observed an excess of 22 genes (observed: 45, expected: 23) with recurrent functional mutations (Figure 1A), and an excess of 8 genes (observed:9, expected:1) with recurrent LoF mutations (Figure 1B), implying that an appreciable fraction of these recurrently mutated genes are novel DD genes.
Figure 1. Excess of recurrently mutated genes.
Each panel shows the observed number of recurrently mutated genes (diamond) and the distribution of the number of recurrently mutated genes in 10,000 simulations (box indicates interquartile range, whiskers indicates 95% confidence interval) under a model of no gene-specific enrichment of mutations: a. all protein-altering mutations in all DDD children and undiagnosed DDD children, b. all LoF mutations in all DDD children and undiagnosed DDD children. Each diamond is annotated with the median excess of recurrently mutated genes, with 95% confidence intervals in brackets. P value of observed excess is <0.0001 for all four tests.
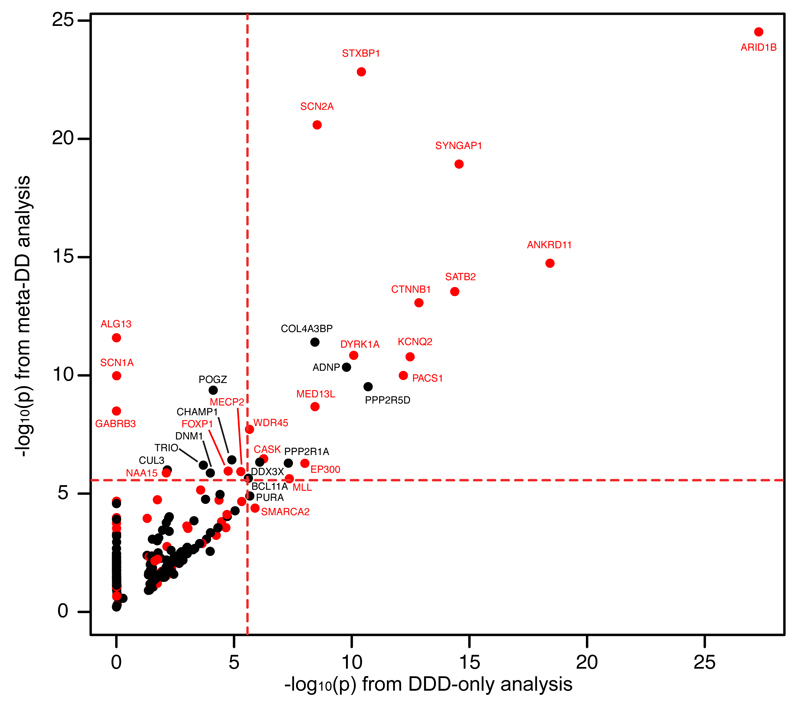
To identify individual genes enriched for damaging de novo mutations (Supplementary Information), we tested for a gene-specific overabundance of either de novo LoF mutations or clustered functional de novo mutations in 1,130 children (excluding one twin from each of 3 identical twin-pairs). To increase power to detect DD genes, we also meta-analysed our data with published de novo mutations from 2,347 DD trios with intellectual disability 4,9, epileptic encephalopathy 3, autism 6–8,10, schizophrenia 5, or congenital heart defects 11 (the ‘meta-DD’ dataset). These analyses (Figure 2) successfully re-discovered 20 known DD genes at genome-wide significance (p < 1.31 × 10-6, a Bonferroni p value of 0.05 corrected for 38,504 tests [Supplementary Information]). Thus, despite the broad phenotypic ascertainment in these datasets, we can robustly detect DD genes solely on statistical grounds.
Figure 2. Gene-specific significance of enrichment for DNMs.
The –log10(p) value of testing for mutation enrichment is plotted only for each gene with at least one mutation in DDD children. On the X-axis is the p value of the most significant test in the DDD dataset, and on the Y-axis is the minimal p value from the significance testing in the meta-analysis dataset. Red indicates genes already known to be associated with DDs (in DDG2P). Only genes with a p value of less than 0.05/18,272 (red lines) are labeled.
To increase our power to detect novel DD genes, we repeated the gene-specific analysis described above excluding the 317 individuals with a known cause of their DD. In this analysis the statistical genetics evidence was integrated with phenotypic similarity of patients, available data on model organisms and functional plausibility. We identified 12 novel disease genes with compelling evidence for pathogenicity (Table 2), nine of which exceeded the genome-wide significance threshold of 1.36 × 10-6 (Supplementary Information), with the remaining three genes (PCGF2, DNM1 and TRIO) just below this significance threshold. The two children with identical Pro65Leu mutations in PCGF2, which encodes a component of a Polycomb transcriptional repressor complex, share a strikingly similar facial appearance representing a novel and distinct dysmorphic syndrome. DNM1 was previously identified as a candidate gene for epileptic encephalopathy (EE) 3. Two of the three children we identified with DNM1 mutations also had seizures, and a heterozygous mouse mutant manifests seizures 16. In addition to two de novo missense SNVs in TRIO, we identified an intragenic de novo 82kb deletion of 16 exons. For several of these novel DD genes, the meta-DD analysis increased the significance of enrichment. For example, a total of five de novo LoF variants in POGZ were identified, two from our cohort, two from recent autism studies and one from a recent schizophrenia study. We also identified six genes with suggestive statistical evidence of being novel DD genes, defined as being a p value for mutation enrichment less than 1 × 10-4 and being plausible from a functional perspective (Extended Data Table 1). We anticipate that the majority of these genes will eventually accrue sufficient evidence to meet the stringent criteria we defined above for declaring a novel DD gene.
Table 2. Novel genes with compelling evidence for a role in DD.
| Evidence | Gene | de novos DDD (Missense, LoF) | de novos Meta (Missense, LoF) | P Value | Test | Mutation Clustering | Predicted Haploinsufficiency |
|---|---|---|---|---|---|---|---|
| De novo enrichment | COL4A3BP | 3 (3,0) | 5 (5,0) | 4.10E-12 | Meta | Yes | 14.7% |
| PPP2R5D | 4 (4,0) | 5 (5,0) | 6.01E-12 | DDD | Yes | 19.7% | |
| ADNP | 4 (0,4) | 5 (0,5) | 4.59E-11 | Meta | No | 9.8% | |
| POGZ | 2 (0,2) | 5 (0,5) | 4.31E-10 | Meta | No | 30.0% | |
| PPP2R1A | 3 (3,0) | 3 (3,0) | 2.03E-08 | DDD | Yes | 23.5% | |
| DDX3X | 4 (3,1) | 5 (3,2) | 2.26E-07 | DDD | No | 12.7% | |
| CHAMP1 | 2 (0,2) | 3 (0,3) | 4.58E-07 | Meta | No | 52.9% | |
| BCL11A | 3 (3,0) | 4 (3,1) | 1.03E-06 | DDD | Yes | 0.6% | |
| PURA | 3 (1,2) | 3 (1,2) | 1.14E-06 | DDD | No | 9.4% | |
| De novo enrichment + additional evidence | DNM1 | 3 (3,0) | 5 (5,0) | 1.43E-06 | Meta | No | 13.5% |
| TRIO | 2 (2,0) | 7 (7,0) | 5.16E-06 | Meta | Yes | 25.7% | |
| PCGF2 | 2 (2,0) | 2 (2,0) | 1.08E-05 | DDD | Yes | 37.7% | |
The table summarises the 12 genes with compelling evidence to be novel DD genes. The number of unrelated patients with independent functional or LoF mutations in the DDD cohort or the wider meta-analysis dataset including DDD patients is listed. The p value reported is the minimum p value from the testing of the DDD dataset and the meta-analysis dataset. The dataset that gave this minimal p value is also reported. Mutations are considered to be clustered if the p value of clustering of functional SNVs is less than 0.01. Predicted haploinsufficiency is reported as a percentile of all genes in the genome, with ~0% being highlight likely to be haploinsufficient and 100% very unlikely to be haploinsufficient, based on the prediction score described in Huang et al 26 updated to enable predictions for a higher fraction of genes in the genome. During submission, a paper was published online describing a novel DD caused by mutations in ADNP 27.
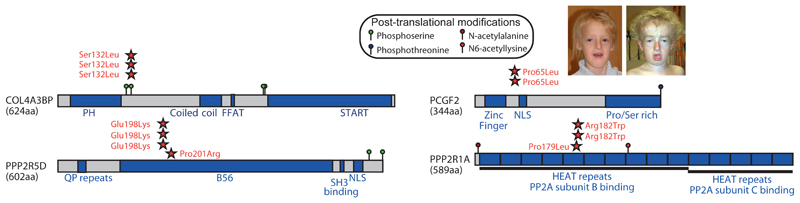
Strikingly, we observed identical missense mutations in unrelated, phenotypically similar, patients for four of these novel DD genes (PCGF2, COL4A3BP, PPP2R1A and PPP2R5D), and for a fifth gene, BCL11A, we identified highly significant clustering of non-identical missense mutations (Figure 3). We hypothesise that the mutations in some of these genes may be operating by either dominant negative or activating mechanisms. This hypothesis is supported by prior functional evidence for several of the mutated amino acids. The three identical Ser132Leu mutations in COL4A3BP, which encodes an intracellular transporter of ceramide, remove a serine that when phosphorylated down-regulates transporter activity from the ER to the golgi 17, presumably resulting in intra-cellular imbalances in ceramide and its downstream metabolic pathways. The two mutated amino acids (Arg182Trp and Pro179Leu) in PPP2R1A, which encodes the scaffolding A subunit of the Protein Phosphatase 2 complex, have been previously identified as sites of driver mutations in endometrial and ovarian cancer 18. It has previously been shown that mutating either of these two residues results in impaired binding of B subunits of the complex 18. Intriguingly, PPP2R5D encodes one of the possible B subunits of the same Protein Phosphatase 2 complex, suggesting that the clustered missense mutations (Pro201Arg and Glu198Lys) in this gene may similarly perturb interactions between subunits of this complex. Further functional studies will be required to confirm this hypothesis.
Figure 3. Five novel genes with clustered mutations.
The domains (blue), post-translational modifications, and mutation locations (red stars) are shown for five proteins with highly clustered de novo mutations in unrelated children with severe, undiagnosed DDs. For two proteins (COL4A3BP and PCGF2) where all observed mutations are identical, photos are shown to highlight the facial similarities of patients carrying the same mutation.
We assessed transmission biases of potentially pathogenic inherited SNVs in our probands (Supplementary Information) and observed a genome-wide trend (p=0.015) towards over-transmission to probands of very rare (MAF < 0.0005%) LoF variants, but not damaging missense variants. We also observed a 1.8-fold enrichment (p=0.04) of rare (MAF<5%) biallelic LoF variants (Supplementary Table 5) among probands without a likely dominant cause of their disorder, compared to those with either a diagnostic de novo mutation or an affected parent. Again we saw no enrichment in biallelic damaging missense variants (Extended Data Table 2), consistent with a similar observation in children with autism 19. These observations imply that although inherited LoF variants (both monoallelic and biallelic) are likely contributing to DD in our patients, much larger sample sizes will be required to pinpoint specific DD genes in this way.
To direct future, detailed functional experiments on the developmental role of a subset of candidate genes from this study we used two approaches. First, knockdown-induced phenotypes were recorded in early zebrafish development. Second we performed a systematic review of perturbed gene function in human, mouse, xenopus, zebrafish and drosophila. In both approaches the animal phenotypes were compared to those seen in individuals in our cohort
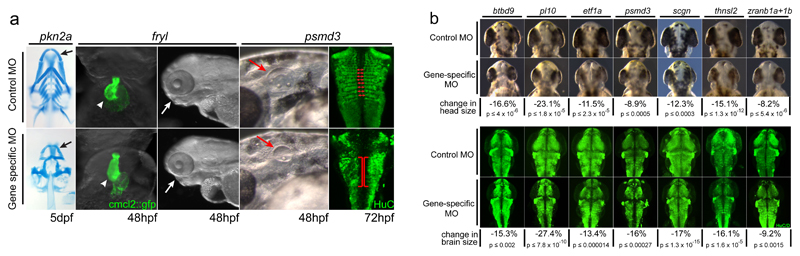
We undertook an antisense-based loss of function screen in zebrafish to assess 32 candidate DD genes with de novo LoF, de novo missense or biallelic LoF variants from exome sequencing (Supplementary Information and Supplementary Table 6). These candidate genes corresponded to 39 zebrafish orthologues. Knockdowns of these zebrafish genes were repeated at least twice and all morpholinos were co-injected with tp53 morpholino to eliminate off-target toxicity. Successful knockdown of the targeted mRNA could be confirmed using RT-PCR for 82.4% of genes (28/34) and 9/11 (82%) of genes that were tested gave an equivalent phenotype when knocked down by a second, independent morpholino. Knock-down of at least one or a pair of zebrafish orthologues of 65.6% of candidate DD genes (21 out of 32) resulted in perturbed embryonic and larval development (Figure 4, Extended Data Table 3, Supplementary Data and Supplementary Table 7). Large-scale mutagenesis 20 and morpholino 21 studies suggest knockout or knockdown of 6-12% genes give developmental phenotypes, suggesting at least a five-fold enrichment of developmentally non-redundant genes among the 32 selected for modelling. We then compared the phenotypes of the zebrafish morphants to those of the DDD individuals with de novo mutations or biallelic LoF variants in the orthologous genes (Extended Data Table 3). 11/21 (52.4%) of the genes were categorised as strong candidates based on phenotypic similarity (Figure 4A). 7/11 were potential microcephaly genes whose gene knockdown in zebrafish gives significant reductions in both head measurements, and neural tissue (Figure 4B, Supplementary Information). 6/21 (28.6%) genes resulted in severe morphant phenotypes which could not be meaningfully linked to patient phenotypes. As many of our candidate DD genes carried heterozygous LoF variants (de novo mutations), it is to be expected that the severity of LoF phenotypes in zebrafish may exceed that observed in our patient cohort. The genes with proven non-redundant developmental roles can reasonably be assigned higher priority for downstream functional investigations and genetic analyses.
Figure 4. Candidate gene Loss of Function modeling in zebrafish reveals enrichment for developmentally important proteins.
a, Examples of developmental phenotypes: Knockdown of pkn2a results in reduced cartilaginous jaw structures (black arrows), knockdown of fryl results in cardiac and craniofacial defects (white arrowheads and arrows, respectively), while knockdown of psmd3 results in smaller ear primordia (red arrows), and mis-patterned CNS neurons (compare red double arrows and brackets). b, Knockdown outcomes of 7 genes with variants present in microcephaly patients: Interocular measurements of brightfield images from control and LoF embryos reveal significant decreases in head size. A neuronal antibody stain (anti-HuC/D, green channel) labels the brains of control and morphant zebrafish. Measurements taken across the widest extent of the midbrain identify significant reductions in brain size, likely underlying the concomitant head size reductions seen in brightfield. In b, tables show average percentage reduction in head and brain width, and p-values of a t-test.
Our systematic review of gene perturbation in multiple species sought both confirmatory and contradictory (e.g. healthy homozygous knock-out) evidence from other animal models for these 21 apparently developmentally important genes. We identified 16 genes with solely confirmatory data, often from multiple different organisms, none with solely contradictory data, two with both confirmatory and contradictory evidence and three with no evidence either way (Supplementary Table 8).
In summary, our analyses validate a large-scale, genotype-driven strategy for novel DD gene discovery that is complementary to the traditional phenotype-driven strategy of studying patients with very similar presentations, and is particularly effective for discovering novel DDs with highly variable or indistinct clinical presentations. Our meta-analysis with previously published DD studies increased power to detect novel DD genes and highlights the shared genetic etiologies between diverse neurodevelopmental disorders such as intellectual disability, epilepsy, autism and schizophrenia 22. We identified significantly more pathogenic autosomal de novo mutations in females compared to males. An increased burden of monogenic disease among females with neurodevelopmental disorders has become more apparent 23,24, and our observations strengthen this proposition. Further investigations are required to assess whether males might be enriched for poly/oligogenic causation.
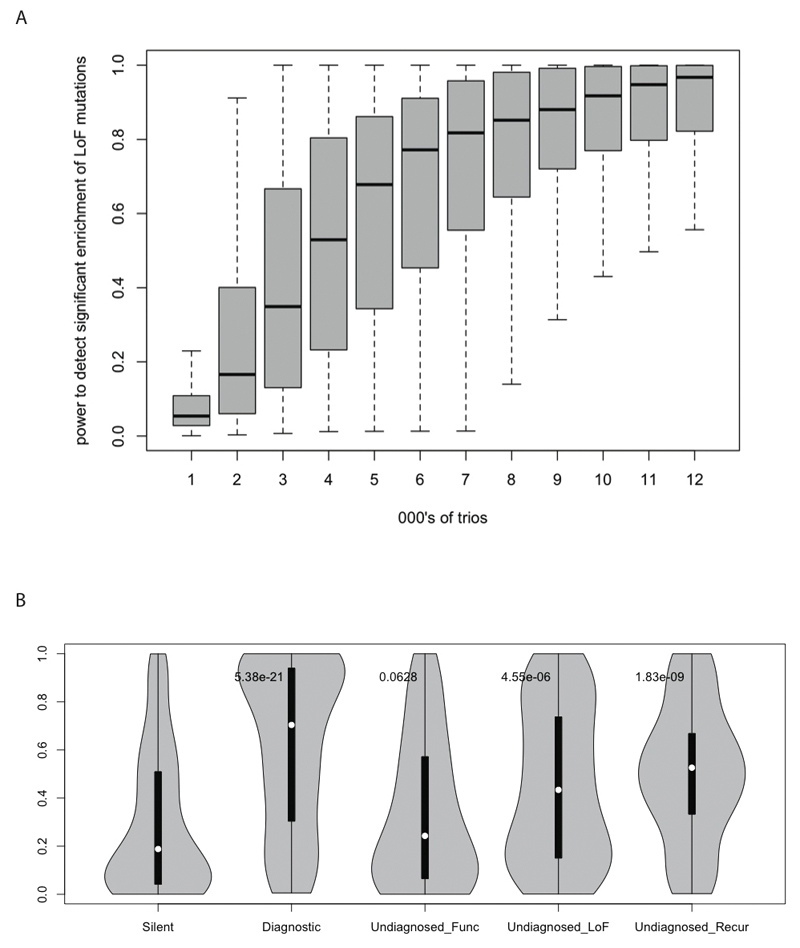
The 35 patients with pathogenic mutations in the 12 novel DD genes we discovered increased our diagnostic yield from 28% to 31%. What, then, are the causes of the DDs in the other 69% of patients? The undiagnosed patients are not obviously less severely affected than the diagnosed patients (e.g. fewer phenotype terms, older age of recruitment). We anticipate that there are many more pathogenic, monogenic, coding mutations in these undiagnosed patients that we have detected, but for which compelling evidence is currently lacking. This hypothesis is supported by four strands of evidence: (i) modeling statistical power suggests that studying ~1,000 trios has only 5-10% power to detect an averagely mutable haploinsufficient DD gene (Extended Data Figure 6A, Supplementary Information), (ii) the expectation that our power to detect novel DD genes that operate recessively or by gain-of-function mechanisms will be lower than for haplosufficient genes, (iii) the significant enrichment in undiagnosed patients of functional mutations in genes predicted to exhibit haploinsufficiency (Extended Data Figure 6B), and (iv) the strong enrichment for developmental phenotypes in the zebrafish knock-down screen.
Given our limited power to detect pathogenic mutations that act through dominant negative or activating mechanisms, it was notable that in four of our novel genes (COL4A3BP, PPP2R1A, PPP2R5D and PCGF2) we observed identical de novo mutations in unrelated trios. Two hypotheses might explain this observation: first, that there is a vast number of different gain-of-function mutations, of which we are just scratching the surface in this study, or second, that these particular variants are enriched in our cohort due to these mutations conferring a positive selective advantage in the germline 25. Analysis of larger datasets will be required to assess these hypotheses, although they are not necessarily mutually exclusive.
These considerations of the limited power of even nationwide studies such as ours motivate the international sharing of minimal genotypic and phenotypic data, for example through the DECIPHER web portal (http://decipher.sanger.ac.uk), to provide diagnoses for patients who would otherwise remain undiagnosed. Plausibly pathogenic variants observed in undiagnosed patients in our study (de novo SNVs, indels and CNVs, and biallelic LoF in genes not yet associated with disease) are shared through DECIPHER, and we encourage other, comparable studies to adopt a similar approach.
Extended Data
EDT1. Novel genes with suggestive evidence for a role in DD.
Six genes with suggestive evidence to be novel DD genes. The number of unrelated patients with independent functional or LoF mutations in the DDD cohort or the wider meta-analysis dataset including DDD patients is listed. The p value reported is the minimum p value from the testing of the DDD dataset and the meta-analysis dataset. The dataset that gave this minimal p value is also reported. Mutations are considered to be clustered if the p value of clustering of functional SNVs is less than 0.01. Predicted haploinsufficiency is reported as a percentile of all genes in the genome, with ~0% being highly likely to be haploinsufficient and 100% very unlikely to be haploinsufficient, based on the prediction score described in Huang et al 26 updated to enable predictions for a higher fraction of genes in the genome. NAA10 is already known to cause an X-linked recessive DD in males, but here we identified missense mutations in females, suggesting a different, X-linked dominant, disorder.
| Evidence | Gene | de novos DDD (Missense, LoF) | de novos Meta (Missense, LoF) | P Value | Test | Mutation Clustering | Predicted Haploinsufficiency |
|---|---|---|---|---|---|---|---|
| De novo enrichment + additional evidence | NAA15 | 1 (0,1) | 3 (0,3) | 1.64E-06 | Meta | No | 7.5% |
| ZBTB20 | 3 (1,2) | 3 (1,2) | 4.84E-06 | DDD | No | 0.2% | |
| NAA10 | 2 (2,0) | 3 (3,0) | 8.28E-06 | Meta | No | 34.1% | |
| TRIP12 | 3 (1,2) | 4(2,2) | 2.13E-05 | Meta | No | 3.8% | |
| USP9X | 3 (1,2) | 3 (1,2) | 5.14E-05 | DDD | No | 3.8% | |
| KAT6A | 2 (0,2) | 2 (0,2) | 7.91E-05 | DDD | No | 19.0% |
EDT2. Biallelic Loss of function and damaging functional variants.
Rare (MAF < 5%) biallelic loss-of-function and damaging functional variants in uninherited diplotypes and probands. ‘Likely dominant probands’ refers to probands with a reported de novo mutation or affected parents, and ‘other probands’ to all remaining probands. ‘DDG2P Biallelic’ refers to confirmed and probable DDG2P genes with a biallelic mode of inheritance. See Supplemental methods for details of variant processing.
| Biallelic Variant Types | Untransmitted Diplotypes (n=1080) | Likely Dominant Probands (n=270) | Other Probands (n=810) |
|---|---|---|---|
| LoF/LoF (Genome-wide) | 110 | 17 | 86 |
| LoF/Dam (Genome-wide) | 87 | 21 | 71 |
| Dam/Dam (Genome-wide) | 312 | 90 | 264 |
| LoF/LoF (DDG2P Biallelic) | 1 | 1 | 3 |
| LoF/Dam (DDG2P Biallelic) | 2 | 0 | 6 |
| Dam/Dam (DDG2P Biallelic) | 26 | 7 | 25 |
EDT3. Zebrafish modeling identifies 21 developmentally important candidate genes.
This table summarises the 21 genes whose knockdown results in developmental phenotypes in zebrafish. “# patients” column indicates how many patients were identified as carrying variants in these genes. Split numbers indicate the breakdown of variant types (eg. for BTBD9, 2/1 is two biallelic LoF and one de novo missense carrying patients). A summary of the patient phenotypes is listed, as well as the relevant phenotypes observed in zebrafish knockdown experiments. Phenotypic concordance categories indicate the degree of overlap between the zebrafish phenotyping and the patient phenotypes. Weak concordance typically is the result of severe, multisystem phenotypes in zebrafish. See Supplemental Materials for more detailed phenotype information.
| Gene | # patients | Variant | Patient phenotypes | Phenotypic concordance | Relevant knockdown phenotypes |
|---|---|---|---|---|---|
| BTBD9 | 2/1 | Biallelic LoF/De novo Missense | Seizures, microcephaly, hypertonia | Strong | Reduced head size, brain volume |
| CHD3 | 1/2 | De novo LoF/Missense | CNS and craniofacial defects | Strong | Abnormal head shape |
| DDX3X | 1/3 | De novo LoF/Missense | Moderately short stature, microcephaly, CNS defects | Strong | Reduced head size, brain volume |
| ETFl | 1 | De novo LoF | CNS and craniofacial defects, seizures, microcephaly, hypertelorism | Strong | Reduced head size, brain volume |
| FRYL | 1 | De novo LoF | Short stature, craniofacial and cardiac defects | Strong | Cardiac defects, reduced axis length |
| PKN2 | 1 | De novo Missense | CNS, cardiac, ear, and craniofacial defects, growth retardation | Strong | Cardiac, craniofacial cartilage, and growth defects |
| PSMD3 | 1 | De novo Missense | Microcephaly, muscular hypotonia, seizures, growth abnormality | Strong | Reduced head size and neural defects |
| SCGN | 1 | Biallelic LoF | Seizures, microcephaly, CNS defects | Strong | Reduced head size, brain volume |
| SETD5 | 1 | De novo LoF | Seizures, CNS and cardiac defects, poor motor coordination | Strong | Reduced head size, cardiac defects, abnormal locomotion |
| THNSL2 | 2 | Biallelic LoF | Microcephaly, CNS and ear defects | Strong | Reduced head size, brain volume, neural defects |
| ZRANB1 | 2 | De novo Missense | Microcephaly, muscle defects, seizures | Strong | Reduced heaa size and neural defects |
| DPEP2 | 1 | Biallelic LoF | CNS defects, growth retardation | Moderate | Growth reduction |
| PSD2 | 1 | De novo LoF | CNS defects, hypertonia, seizures | Moderate | Abnormal musculature, CNS and locomotion |
| SAP130 | 1 | De novo LoF | Short stature, hypotonia, hypotelorism | Moderate | Abnormal locomotion |
| CN0T1 | 1/1 | De novoLoF/Missense | Short stature, cardiac, CNS, ear and craniofacial defects | Weak | Multisystem |
| DTWD2 | 1 | De novo LoF | CNS defects, seizures | Weak | Multisystem |
| ILVBL | 1 | De novo LoF | CNS and craniofacial defects | Weak | Multisystem |
| NONO | 1 | De novo LoF | CNS and ear defects, hypotonia, growth retardation | Weak | Multisystem, with otic and growth defects |
| POGZ | 2 | De novoLoF | CNS and ear defects, hypotonia, seizures, coloboma | Weak | Multisystem |
| SMARCD1 | 1/1 | De novoLoF/Missense | CNS defects, hypotonia | Weak | Multisystem |
| WWC1 | 1 | De novo Missense | CNS defects, hypertelorism | None | None |
EDF1. Characteristics of the families.
A. Gestation Adjusted Decimal Age at Last Clinical Assessment. Histogram showing the distribution of the gestation adjusted decimal age at last clinical assessment across the 1133 probands. The dashed red line shows the median age. B. Frequency of HPO Term Usage. Bar plot showing, for each used HPO term, the number of times it was observed across the 1133 proband patient records. C. Projection PCA plot of the 1133 probands. PCA plot of 1133 DDD probands projected onto a PCA analysis using 4 different HapMap populations from the 1000 genomes project. Black: African, Red: European, Green: East Asian, Blue: South Asian and the 1133 DDD probands are represented by orange triangles. D. Self Declared and Genetically Defined Consanguinity. Overlaid histogram showing the distribution of kinship coefficients from KING comparing parental samples for each trio. Green: Trios where consanguinity was not entered in the patient record on DECIPHER. Red: Trios consanguinity was declared in the patient record on DECIPHER.
EDF2. Number of Validated de novo SNVs and indels per Proband.
Bar plot showing the distribution of the observed number of validated SNVs and indels per proband sample, and the expected distribution assuming a Poisson distribution with the same mean as the observed distribution.
EDF3. Number of Diagnoses per Gene.
Histogram showing the number of diagnoses per gene for genes with at least two diagnoses from different proband samples.
EDF4. Burden of Large CNVs in 1133 DDD Proband Samples.
Plot comparing the frequency of rare CNVs in three sample groups against CNV size. Y-axis is the on a log scale. Red: DDD probands who have not had previous microarray based genetic testing, Purple: DDD probands who have had negative previous microarray based genetic testing Green: DDD controls.
EDF5. Expected and observed numbers of de novo mutations.
The expected and observed numbers of mutations of different functional consequences in three mutually exclusive sets of genes are shown, along with the p value from an assessment of a statistical excess of observed mutations. The three classes of genes are described in the main text.
EDF6. Haploinsufficiency analyses.
A. Saturation analysis for detecting haploinsufficient DD genes. A boxplot showing the distribution of statistical power to detect a significant enrichment of LoF mutations across 18,272 genes in the genome, for different numbers of trios studied, from 1,000 trios to 12,000 trios. B. Distribution of haplinsufficiency scores in selected sets of de novo mutations. Violin plot of haploinsufficiency scores in five sets of de novo mutations: Silent - all synonymous mutations, Diagnostic - mutations in known DD genes in diagnosed individuals, Undiagnosed_Func - all functional mutations in undiagnosed individuals, Undiagnosed_LoF - All LoF mutations in undiagnosed individuals, Undiagnosed_recur - mutations in genes with recurrent functional mutations in undiagnosed individuals. P values for a Mann-Whitney test comparing each of the latter four distributions to that observed for the silent (synonymous) variants are plotted at the top of each violin.
Supplementary Information
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Acknowledgements
We dedicate this paper to John Tolmie and Louise Brueton for their unwavering and enthusiastic support of the DDD project, and in memory of deeply valued friends and colleagues. We are indebted to the families for their participation and patience. We thank Mark Daly and Kaitlin Samocha for access to unpublished mutation rate estimates. We are grateful to Stephan Saunders, Damian Smedley, Don Conrad, Avinash Ramu and Ni Huang for access to data and algorithms. We thank the UK National Blood Service and the Generation Scotland: Scottish Family Health Study for access to DNA from controls. Generation Scotland has received core funding from the Chief Scientist Office of the Scottish Government Health Directorates CZD/16/6 and the Scottish Funding Council HR03006. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.
Footnotes
Author information
Data can be accessed at the European Genome Phenome Archive under accession number EGAS00001000775.
References
- 1.OMIM. Online Mendelian Inheritance in Man, OMIM. 2014 <http://omim.org>
- 2.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 5.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/s0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 10.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King DA, et al. A novel method for detecting uniparental disomy from trio genotypes identifies a significant excess in children with developmental disorders. Genome Res. 2014;24:673–687. doi: 10.1101/gr.160465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright CF, et al. Deciphering Developmental Disorders: Clinical Genome Sequencing Implemented in a Large-Scale Rare Disease Study. Lancet. (in review) [Google Scholar]
- 14.Smith BH, et al. Cohort Profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. Int J Epidemiol. 2013;42:689–700. doi: 10.1093/ije/dys084. [DOI] [PubMed] [Google Scholar]
- 15.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014 doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boumil RM, et al. A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumagai K, Kawano M, Shinkai-Ouchi F, Nishijima M, Hanada K. Interorganelle trafficking of ceramide is regulated by phosphorylation-dependent cooperativity between the PH and START domains of CERT. J Biol Chem. 2007;282:17758–17766. doi: 10.1074/jbc.M702291200. [DOI] [PubMed] [Google Scholar]
- 18.Walter G, Ruediger R. Mouse model for probing tumor suppressor activity of protein phosphatase 2A in diverse signaling pathways. Cell Cycle. 2012;11:451–459. doi: 10.4161/cc.11.3.19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim ET, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–242. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kettleborough RN, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature. 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickart MA, et al. Genome-wide reverse genetics framework to identify novel functions of the vertebrate secretome. PLoS One. 2006;1:e104. doi: 10.1371/journal.pone.0000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Craddock N, Owen MJ. The Kraepelinian dichotomy - going, going… but still not gone. Br J Psychiatry. 2010;196:92–95. doi: 10.1192/bjp.bp.109.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacquemont S, et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Goriely A, Wilkie AO. Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease. Am J Hum Genet. 2012;90:175–200. doi: 10.1016/j.ajhg.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helsmoortel C, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.