Abstract
Long noncoding RNAs (lncRNAs) have been associated with hepatocellular carcinoma (HCC), but the underlying molecular mechanisms of their specific association with hepatocarcinogenesis have not been fully explored.
Methods: miR503HG was identified by microarray and validated by real-time PCR. Survival analysis was evaluated using the Kaplan-Meier method and assessed using the log-rank test. In vitro and in vivo assays were preformed to explore the biological effects of miR503HG in HCC cells. The interaction of miR503HG with HNRNPA2B1 was identified by RNA pull-down and RNA immunoprecipitation. Expression of HNRNPA2B1 was examined by western blotting, immunofluorescence and immunohistochemical analyses, while HNRNPA2B1 ubiquitination was detected by immunoprecipitation.
Results: We have identified 713 differentially expressed lncRNAs in 12 pairs of HCC tissues compared with corresponding noncancerous liver tissues. One of these lncRNAs, miR503HG, the host gene of miR503, is markedly decreased in HCC. Expression level of miR503HG is significantly associated with the time to recurrence and overall survival and is an independent risk factor for recurrence and survival. Enhanced expression of miR503HG could noticeably inhibit HCC invasion and metastasis in vitro and in vivo. Further investigation suggested that miR503HG could specifically interact with the heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1). miR503HG promoted HNRNPA2B1 degradation via the ubiquitin-proteasome pathway, which reduced the stability of p52 and p65 mRNA, and simultaneously suppressed the NF-κB signaling pathway in HCC cells. In addition, miR503HG can function synergistically with miR503 to inhibit HCC migration.
Conclusion: Our findings support a role for miR503HG in tumor recurrence risk and survival prediction in HCC patients. We demonstrate a novel mechanism by which miR503HG inhibits the NF-κB signaling pathway and exerts its metastatic tumor suppression function through modulating the ubiquitination status of HNRNPA2B1.
Keywords: lncRNA, miR503HG, prognosis, metastasis, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common solid tumors worldwide and the second leading cause of cancer-related death in China 1. Despite the use of innovative therapeutic strategies for HCC, the 5-year survival rate of HCC subjects remains poor. This is mainly due to the high rates of recurrence and metastasis after surgical resection 2. Previous studies identified multiple underlying molecular mechanisms regulated by protein-coding genes in HCC. Nevertheless, only approximately 1% of the human genome encodes protein, with another 4-9% transcribing many short or long RNAs with limited protein coding capacity. Recent studies have implicated small noncoding RNAs, i.e., microRNAs, in HCC development and progression 3-5. Several studies have uncovered the pathophysiological contributions of lncRNAs to the HCC tumorigenesis and metastasis; however, the functions and mechanisms of most lncRNAs remains unexplored, and the relationship between lncRNAs and miRNAs also remains largely unknown.
lncRNAs are a class of non-coding RNA transcripts that are longer than 200 nt and exhibit limited or no protein-coding potential 6. It has been reported that lncRNAs regulate gene expression in cis as enhancer-associated RNAs 7, through transcriptional regulation 8, transcription factor trapping 9, and chromatin looping 10. lncRNAs also regulate distant genes in trans through chromatin modification 11 and through serving as a scaffold for assembly of multiple regulatory molecules at a single locus 12. lncRNAs have been implicated in many cellular processes. In cancers, lncRNA-SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex 13. NEAT1 modulates the replication stress response and chemosensitivity 14. Currently, several lncRNAs, including RERT, lncRNA-LET, lncTCF7 and lncSOX4, are directly involved in the tumorigenesis of HCC 15-18. Given the fact that such large scales of lncRNAs are dysregulated in HCC, it is highly possible that lncRNAs are directly associated with the carcinogenesis of HCC, as the functions of the vast majority of lncRNAs remain to be identified.
Nuclear factor-κB (NF-κB) is a family of transcription factors that play critical roles in the regulation of immune function, inflammatory responses, and malignant transformation 19. Aberrant activation of the NF-κB signaling pathway is known to be associated with a wide range of cancers, including HCC 20-22. Recently, great progress has been made in the understanding of NF-κB activation. Many negative regulators, including proteins and miRNAs, that inhibit the activation of NF-κB signaling have been identified 23-25. The lncRNA NKILA, which is induced by NF-κB, mediates a negative feedback loop to suppress NF-κB signaling by binding the cytoplasmic NF-κB/IκB complex and preventing IκB phosphorylation in breast cancer 26. Nevertheless, whether lncRNA plays a role in the regulation of the NF-κB signaling pathway in HCC is not yet clear.
In the current study, we performed lncRNA expression profiling analysis in HCC tumor tissues and corresponding noncancerous liver tissues. We found that a downregulated lncRNA—the host gene of miR503, known as miR503HG—can inhibit HCC invasion and metastasis. Moreover, miR503HG can directly interact with heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) to inhibit metastasis and the NF-κB signaling pathway in HCC.
Methods
Patients and specimens
Human HCC tissues and adjacent noncancerous liver tissues for qPCR analysis of miR503HG (n=93) and HNRNPA2B1 (n=67) were acquired from the surgical specimen archives of Huashan Hospital and Zhongshan Hospital, respectively, Shanghai, China. Specimens from 80 patients with HCC and corresponding noncancerous liver tissues at Shanghai Outdo Biotech co., LTD. were used for tissue microarray construction and prognosis analysis. Tumor differentiation was graded using the Edmondson grading system. Clinical staging was performed according to the tumor-node-metastasis (TNM) staging system and the Barcelona Clinic Liver Cancer (BCLC) classification system. Overall survival (OS) and time to recurrence (TTR) were calculated from the date of surgery to the date of death and the first recurrence, respectively. The data were censored at the last follow-up visit or at the time of the patient's death without relapse. Institutional review board approval and written informed consent from each patient were obtained. The clinicopathological characteristics of the HCC patients are provided in Table S1 and Table S2.
Microarray analysis
Twelve HCC tissues and corresponding noncancerous liver tissues were used to synthesize double-stranded complementary DNA (cDNA). Double-stranded cDNA was labeled and hybridized to the 12x135K LncRNA Expression Microarray (Arraystar, Rockville, MD). After hybridization and washing, slides were scanned with an Axon GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA). Expression data were normalized using quantile normalization and the Robust Multichip Average (RMA) algorithm, which is included in the NimbleScan software (version 2.5; Roche NimbleGen, Inc., Madison, WI). P values were calculated using the paired t-test. The threshold we used to screen for the up- or down-regulation of lncRNAs was a fold change ≥ 1.5 with a P-value < 0.05. Hierarchical clustering was performed based on differentially expressed lncRNAs using the Cluster Treeview software from Stanford University (Palo Alto, CA).
Cell culture
The human HCC cell lines SMMC-7721 and Huh7, normal liver cell L02, as well as the human embryonic kidney cell line 293T, were cultured in Dulbecco's Modified Eagle's Medium (DMEM), supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin. All of the cell lines were cultured in a humidified chamber with 5% CO2 at 37 °C. The cells were tested regularly for mycoplasma (R&D Systems' new MycoProbe Mycoplasma Detection Kit).
Subcellular fractionation
Subcellular fractionation was performed with a PARIS™ Kit (Ambion, Austin, TX) according to the manufacturer's instructions. For nuclear fractionation, 107 SMMC-7721 and Huh7 cells were collected, resuspended in the cell fraction buffer and incubated on ice for 10 min. After centrifugation, the supernatant and nuclear pellet were preserved for RNA extraction using cell disruption buffer.
In situ hybridization
miR503HG and Pol II were probed in SMMC-7721 cells using an RNAscope Fluorescent Reagent Kit (Advanced Cell Diagnostics, Hayward, CA, USA). Cells were seeded in growth medium at 80% density, fixed in 10% neutral buffered formalin (NBF), and then dehydrated in an EtOH series. After dehydration, miR503HG and Pol II specific target probe hybridization were performed according to the manufacturer's instructions. The locations of miR503HG and Pol II in the cells were imaged using a LEICA TCS SP5 confocal microscope.
RNA pull-down assay
RNA pull-down assays were performed as described previously 27. Briefly, biotin-labeled RNAs were transcribed in vitro with the Biotin RNA labeling mix (Roche Diagnostics, Indianapolis, IN, USA) and SP6/T7 RNA polymerase (Roche), treated with RNase-free DNase I (Roche), and purified with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). One milligram of HCC cell extract was mixed with 50 pmol of biotinylated RNA. Sixty microliters of washed Streptavidin agarose beads (Invitrogen, Carlsbad, CA, USA) was added to each binding reaction, which was incubated at room temperature for one hour. The beads were washed briefly five times and boiled in SDS buffer, and the retrieved proteins were measured on SDS-PAGE gels for mass spectrometry (MS) or western blotting.
RNA immunoprecipitation
RNA immunoprecipitation (RIP) experiments were performed with a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer's instructions. The HNRNPA2B1 antibodies were used for RIP (Cell Signaling Technology, Beverly, MA, USA). The co-precipitated RNAs were detected by qPCR. Total RNAs (input controls) and isotype controls were assayed simultaneously to demonstrate that the detected signals were the result of RNAs specifically bound to HNRNPA2B1. The gene-specific primers used for detecting miR503HG, Cyp7a1 and Abca1 are presented in Table S5.
Immunoprecipitation
An immunoprecipitation (IP) assay was performed using the Thermo Scientific™ Pierce Classic IP Kit (Thermo Fisher Scientific, MA, USA) according to the manufacturer's instructions. SMMC-7721 with miR503HG overexpression and negative control cells were treated with MG132. Cells were lysed in IP Lysis/Wash Buffer containing protease inhibitors and RNase Inhibitor (Promega, Madison, WI., USA) and centrifuged at 13,000 × g for 10 min. The supernatants were incubated with Pierce Protein A/G Agarose overnight at 4 °C. After washing, the proteins were eluted with 2X Non-reducing Lane Marker Sample Buffer containing 20 mM DTT at 100 °C for 5-10 min. Immunocomplexes were analyzed by SDS/PAGE and immunoblotting with anti-HNRNPA2B1 antibody.
Immunofluorescence
Cells were cultured on glass cover slips, fixed for 10 min with 4% formaldehyde, and permeabilized with 0.5% Triton X-100 for 15 min at room temperature. Immunofluorescence analysis was performed using the following antibodies: HNRNPA2B1 (200 μg/mL) and Anti-mouse IgG Fab2 Alexa Fluor (R) 488 (1:2000, CST). Cell nuclei were stained with DAPI (4,6-diamidino-2-phenylindole). After immunostaining, the samples were observed using a LEICA TCS SP5 confocal microscope.
Immunohistochemistry (IHC)
TMAs were constructed by Shanghai Outdo Biotech co., LTD. (Shanghai, China) and immunohistochemistry was performed and integrated optical density (IOD) was measured as reported previously 28. Briefly, assessment for staining was based on the percentage of positively stained cells and staining intensity was analyzed by software Image-Pro Plus 6.0 (Media Cybernetics, Inc., Bethesda, MD). Immunohistochemical score was independently assessed by 2 pathologists without knowledge of patient characteristics.
Statistical analysis
The results are presented as mean ± SD. The data were analyzed using Student's t-test (two-tailed, with P < 0.05 considered significant) unless otherwise specified (paired t-test, χ2 test or Pearson's correlation). The cumulative survival and recurrence rates were evaluated using the Kaplan-Meier method (log-rank test). Cox multivariate regression analysis was used to determine the independent factors that influenced survival and recurrence. All of the statistical analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL).
Results
miR503HG is downregulated in HCC and is inversely associated with patient prognosis
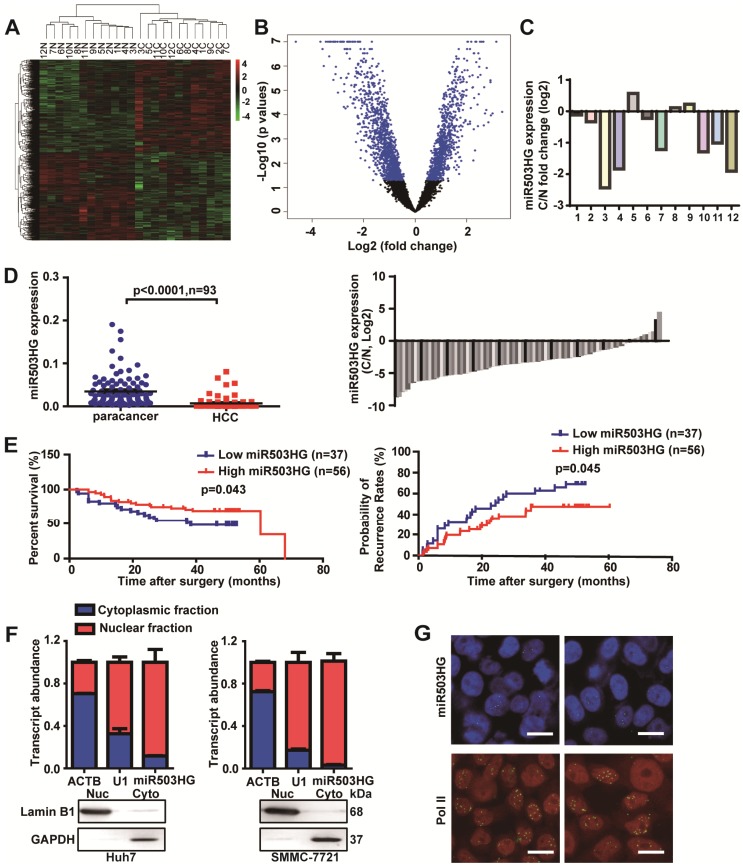
To identify lncRNAs involved in HCC, we analyzed total RNA from 12 pairs of HCC tissues and the corresponding noncancerous liver tissues with a noncoding RNA microarray (Arraystar, Rockville, MD). In total, we identified 713 dysregulated lncRNAs (fold change ≥ 1.5 and P<0.05), of which 51.3% were upregulated and 48.7% were downregulated in HCC tissues compared to the corresponding adjacent noncancerous liver tissues (Figure 1A-B and Table S3). Interestingly, we found five differentially expressed lncRNAs—BX648949, AK098707, CR612213, AK123264 and NR_024607—that are host genes for microRNAs (Figure S1A). We next assessed the expression of these lncRNAs in 36 pairs of HCC and corresponding noncancerous liver tissues from cohort 1. The results showed that only NR_024607 (the host gene of miR503, termed as miR503HG) was significantly decreased in HCC (P < 0.0001; Figure S1B), and it was expressed at a significantly lower level in most of the HCC tissues according to probe signal calculation of the microarray data (Figure 1C).
Figure 1.
miR503HG is downregulated in HCC and is inversely associated with the patient prognosis. (A) Heatmap and (B) volcano plot illustrating the 713 differentially expressed lncRNAs between 12 pairs of cancerous tissues and corresponding adjacent non-cancerous liver tissues of HCC patients (fold change ≥ 1.5, P-value < 0.05). (C) The fold change of miR503HG expression in 12 HCC tissues. (D) Left: The expression of miR503HG in 93 pairs of HCC and noncancerous liver tissues. Right: the fold change of miR503HG in 93 HCC tissues. (E) Kaplan-Meier analysis of the correlation between miR503HG expression and overall survival or recurrence in 93 patients with HCC. Log-rank tests were used to determine statistical significance. (F) Real-time PCR analysis of the subcellular location of miR503HG in HCC cells. U1 snRNA (nuclear retained) and ACTB mRNAs (exported to cytoplasm) were used as controls. The values represent the median of 3 technical replicates. Data are the mean ± SD. (G) In situ hybridization analysis of the subcellular location of miR503HG in SMMC-7721 cells; Pol II enriched in the nucleus was used as a control. Scale bar = 20 μm.
To further investigate the clinicopathological and prognostic significance of miR503HG in HCC patients, 93 pairs of HCC and corresponding noncancerous liver tissue samples from cohort 2 were examined. The results showed that the expression level of miR503HG was significantly lower in HCC tissues (P < 0.0001), and downregulated expression of miR503HG was observed in 80% (75/93) of HCC (Figure 1D). Kaplan-Meier analysis revealed that the expression level of miR503HG was significantly associated with overall survival (OS) (P = 0.045) and the time to tumor recurrence (TTR) (P = 0.043) (Figure 1E). Cox proportional hazards regression analysis further demonstrated that low miR503HG expression in HCC tissues was an independent prognostic factor for reduced overall survival (HR: 0.37; 95% CI: 0.16-0.84; P = 0.018) and TTR (HR: 0.48; 95% CI: 0.24-0.98; P = 0.045) (Table 1). These results suggested that miR503HG might be a potential tumor-suppressor lncRNA in HCC.
Table 1.
Univariate and Multivariate analyses of factors associated with OS and TTR of HCC (n=93).
| Variable | OS | TTR | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Univariate analysis | ||||||
| miR503HG (High vs. Low) | 0.51 | 0.26-0.99 | 0.048 | 0.57 | 0.33-0.99 | 0.049 |
| Sex (female vs. male) | 0.93 | 0.38-2.25 | 0.871 | 0.51 | 0.20-1.28 | 0.150 |
| Age, years (>55 vs. ≤50) | 0.89 | 0.46-1.71 | 0.723 | 0.85 | 048-1.48 | 0.558 |
| ALT (≥75 vs. <75U/L) | 1.35 | 0.48-3.84 | 0.572 | 1.94 | 0.82-4.58 | 0.131 |
| AFP, ng/mL (>20 vs. ≤20) | 2.02 | 1.02-4.00 | 0.044 | 1.65 | 0.93-2.91 | 0.086 |
| Liver cirrhosis (yes vs. no) | 1.31 | 0.51-3.38 | 0.575 | 1.10 | 052-2.34 | 0.808 |
| HBsAg (positive vs. negative) | 0.57 | 0.24-1.33 | 0.192 | 0.98 | 0.44-2.20 | 0.980 |
| Tumor size, cm (>5 vs. ≤5) | 1.94 | 1.00-3.76 | 0.051 | 1.51 | 0.87-2.64 | 0.144 |
| Tumor number (multiple vs. single) | 1.05 | 0.49-2.24 | 0.898 | 1.26 | 0.64-2.47 | 0.500 |
| Tumor capsule (complete vs. none) | 0.50 | 0.26-0.99 | 0.046 | 0.60 | 0.34-1.07 | 0.082 |
| Vascular invasion (yes vs. no) | 3.55 | 1.81-6.96 | <0.001 | 3.25 | 1.81-5.82 | <0.001 |
| Lymphatic and distant metastasis (yes vs. no) | 2.32 | 1.15-4.68 | 0.018 | 2.20 | 1.20-4.03 | 0.011 |
| BCLC stage (B and C vs. 0 and A) | 1.58 | 0.56-4.46 | 0.393 | 2.03 | 0.81-5.12 | 0.133 |
| TNM stage (III-IV vs. I-II) | 2.83 | 1.47-5.47 | 0.002 | 2.00 | 1.12-3.57 | 0.019 |
| Tumor differentiation (III-IV vs. I-II) | 2.75 | 1.36-5.56 | 0.005 | 2.23 | 1.20-4.15 | 0.011 |
| Multivariate analysis | ||||||
| miR503HG (High vs. Low) | 0.37 | 0.16-0.84 | 0.018 | 0.48 | 0.24-0.98 | 0.045 |
| AFP, ng/mL (>20 vs. ≤20) | 2.14 | 0.99-4.59 | 0.052 | 1.38 | 0.74-2.59 | 0.317 |
| Tumor capsule (complete vs. none) | 0.64 | 0.29-1.34 | 0.260 | 0.63 | 0.32-1.23 | 0.626 |
| Vascular invasion (yes vs. no) | 1.54 | 0.54-4.39 | 0.424 | 1.48 | 0.60-3.61 | 0.393 |
| Lymphatic and distant metastasis (yes vs. no) | 1.49 | 0.50-4.46 | 0.473 | 1.48 | 0.55-3.93 | 0.437 |
| TNM stage (III-IV vs. I-II) | 1.56 | 0.72-3.36 | 0.261 | 1.31 | 0.68-2.55 | 0.421 |
| Tumor differentiation (III-IV vs. I-II) | 1.97 | 0.85-4.55 | 0.114 | 1.68 | 0.81-3.49 | 0.167 |
The “low” or “high” expression of miR503HG level was defined according to their cutoff values, which were defined as the ROC curve analysis of the cohort of patients tested. Analysis was conducted using univariate analysis or multivariate Cox proportional hazards regression. In the univariate analysis, serum alpha-fetoprotein (AFP) level, tumor capsule, vascular invasion, lymphatic and distant metastasis, TNM stage and tumor differentiation were revealed to be significantly correlated with prognosis of HCC patients. miR503HG expression in HCC tissues were also significantly associated with both OS and TTR. Individual clinical characteristics that showed significance in the univariate analysis and miR503HG were adopted as covariates in a multivariate Cox proportional hazards regression model for further analysis.
The full-length sequence of miR503HG was obtained by RACE analyses. The results showed that miR503HG, located on human chromosome X, is composed of 786 nucleotides and consists of three exons and two introns (Figure S2A-B). We analyzed the coding potential of miR503HG using several prediction software, and the results showed that miR503HG did not have any coding potential (Figure S2C). In addition, ORFfinder predicted that the longest possible ORF of miR503HG only contains 45 amino acids (Figure S2D), which indicated miR503HG is unlikely to translate a protein. These results indicated that miR503HG is a non-coding RNA. Furthermore, subcellular fractionation and real-time PCR analyses showed that miR503HG is predominantly abundant in the nucleus of HCC cells (Figure 1F). In situ hybridization by RNAscope analyses also revealed that miR503HG is located mainly in the nucleus (Figure 1G).
miR503HG inhibits HCC cell invasion and metastasis in vitro and in vivo
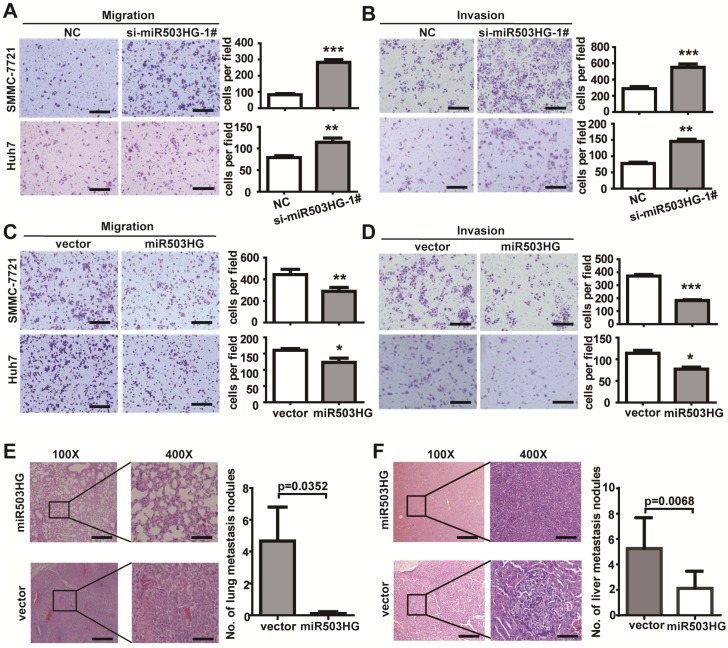
To elucidate the function of miR503HG on cell biological behavior, we first determined the expression levels of miR503HG in various HCC cell lines. The results showed that the expression level of miR503HG was relatively low in the HCC cell lines compared with normal liver cells L02 (Figure S3A). We knocked down miR503HG expression by transfection of two small-interfering RNAs (siRNAs) targeting miR503HG (Figure S3B). Transwell assays showed that knockdown of miR503HG significantly enhanced migration and invasion of SMMC-7721 and Huh7 cells compared with the negative control (Figure 2A-B and Figure S3C-D).
Figure 2.
miR503HG inhibits HCC cell invasion and metastasis in vitro and in vivo. (A-B) Migration assays of 5×104 HCC cells or invasion assays of 1×105 HCC cells with miR503HG knockdown by using siRNA-1#. Representative images are shown on the left, and the average number of cells per field at the indicated time points is shown on the right. Data are the mean ± SD. **P < 0.01 and ***P < 0.001. Scale bar = 100 μm. (C-D) Migration and invasion assays of SMCC-7721 and Huh7 cells that stably overexpress of miR503HG or control vector. Data are the mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001. Scale bar = 100 μm. (E) Hematoxylin and eosin (H&E) staining of metastatic nodules in the lungs of nude mice 8 weeks after tail vein injection of Huh7 cells infected with lentivirus expressing miR503HG or the control. The numbers of metastatic nodules in the lungs of each mouse were counted and analyzed using Student's t-test. Data are presented as the mean ± SD (n = 9 mice per group). Scale bar = 200 μm (100x) or 50 μm (400x). (F) SMMC-7721 cells infected with lentivirus expressing miR503HG or the control vector were orthotopically injected into the livers of nude mice. 8 weeks later, the mice were sacrificed, and the livers were subjected to H&E staining. The numbers of liver metastatic nodules of each mouse were counted and analyzed using Students' t-test. Data are presented as the mean ± SD (n = 8 mice per group). Scale bar = 200 μm (100x) or 50 μm (400x).
Next, we constructed a lentiviral vector to mediate the expression of miR503HG and established two stable cell lines (Figure S3B). Transwell assays showed that SMMC-7721 and Huh7 cells overexpressing miR503HG had dramatically reduced migratory and invasive capacity compared with the vector control (Figure 2C-D). To explore the effect of miR503HG on tumor metastasis in vivo, we implanted Huh7 cells with miR503HG stable overexpression or control cells into the tail vein of nude mice. There was a dramatic decrease in the number of metastatic lung nodules with the miR503HG-overexpressing cells compared with the control cells (P = 0.0352; Figure 2E). Furthermore, SMMC-7721 cells infected with the lentivirus expressing miR503HG or the control vector were orthotopically injected into the livers of nude mice. After 8 weeks, the mice were sacrificed and the intrahepatic metastatic nodules were examined. Results showed that miR503HG remarkably inhibited SMMC-7721 cells intrahepatic metastasis (Figure 2F). These observations are consistent with the in vitro results. Taken together, these findings indicate that miR503HG inhibits tumor invasion and metastasis of HCC cells in vitro and in vivo.
miR503HG physically interacts with HNRNPA2B1 in HCC cells
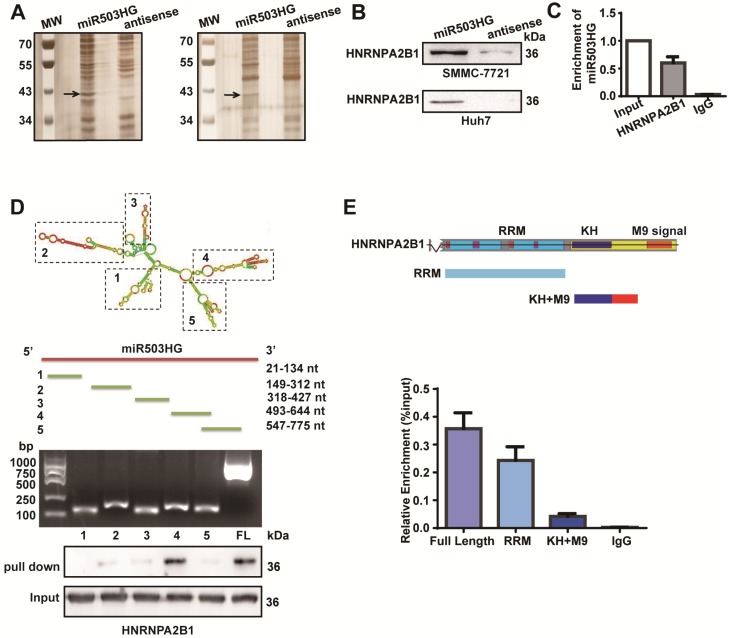
lncRNAs are often involved in molecular regulation pathways through their interactions with chromatin-modifying complexes or with heterogeneous nuclear ribonucleoproteins 29, 30. To identify binding partners for miR503HG, we incubated in vitro-transcribed biotinylated miR503HG as well as an antisense miR503HG control RNA with HCC cell extracts and subjected the RNA-binding proteins to mass spectrometry. The results in the two independent experiments repeatedly showed a specific protein band at approximately 40 kDa in the miR503HG pull-down samples, and 25 potential interacting proteins were obtained in the mass spectrometry assay (Figure 3A and Table S4). Notably, the heterogeneous nuclear ribonucleoprotein HNRNPA2B1 was confirmed as a specific binding partner for miR503HG by western blotting analysis in HCC cells (Figure 3B). We further validated this result using RNA immunoprecipitation (RIP) and found that miR503HG could be enriched in HNRNPA2B1 precipitates (Figure 3C). Moreover, deletion mapping analyses revealed that the truncated miR503HG fragment 493-644nt was responsible for the interaction of miR503HG with HNRNPA2B1 (Figure 3D). Next, RIP assays revealed that the RRM domain of HNRNPA2B1 was required for the interaction with miR503HG (Figure 3E). We also examined whether the interaction of miR503HG with HNRNPA2B1 regulated the binding between HNRNPA2B1 and other reported pre-mRNAs, such as Cyp7a1 and Abca1 31. RIP assays showed that overexpression of miR503HG caused a significant decreased enrichment of Abca1 mRNAs on HNRNPA2B1 compared with the negative control (Figure S4A). These results suggest that miR503HG physically interacts with HNRNPA2B1 in HCC cells.
Figure 3.
miR503HG physically interacts with HNRNPA2B1 in HCC cells. (A) RNA pull-down assays were performed in SMMC-7721 cells twice. Biotinylated miR503HG (left lane) or antisense RNA (right lane) was incubated with cell extracts and targeted with streptavidin beads; the associated proteins were resolved on a gel. The highlighted region was submitted for mass spectrometry. (B) Western blotting analysis of the specific association of HNRNPA2B1 with miR503HG in SMMC-7721 and Huh7 cells. (C) RIP enrichment was determined as RNA associated with the HNRNPA2B1 IP relative to an input control. The data represent the average and standard deviation of three independent experiments. (D) Deletion mapping of the HNRNPA2B1-binding domain in miR503HG. Top: graphic illustration of predicted miR503HG secondary structure (LNCipedia, http://www.lncipedia.org), and the truncation of miR503HG according to the stem-loop structure. Middle: the in vitro transcribed full-length and deletion fragments of miR503HG. Bottom: Western blotting analysis of HNRNPA2B1 in protein samples pulled down by the different miR503HG constructs. (E) Deletion mapping of the miR503HG-binding domain in HNRNPA2B1. Top: diagrams of full-length HNRNPA2B1 and the domain-truncated fragments RNA recognition motif (RRM), hnRNP K homology domain (KH), M9 signal (MN). Bottom: qPCR analysis of HNRNPA2B1 retrieved by full-length or domain-truncated HNRNPA2B1-HA using a HA antibody. RIP assays were performed using Huh7 cells transfected with the indicated vectors.
miR503HG mediates ubiquitination and degradation of HNRNPA2B1 in HCC cells
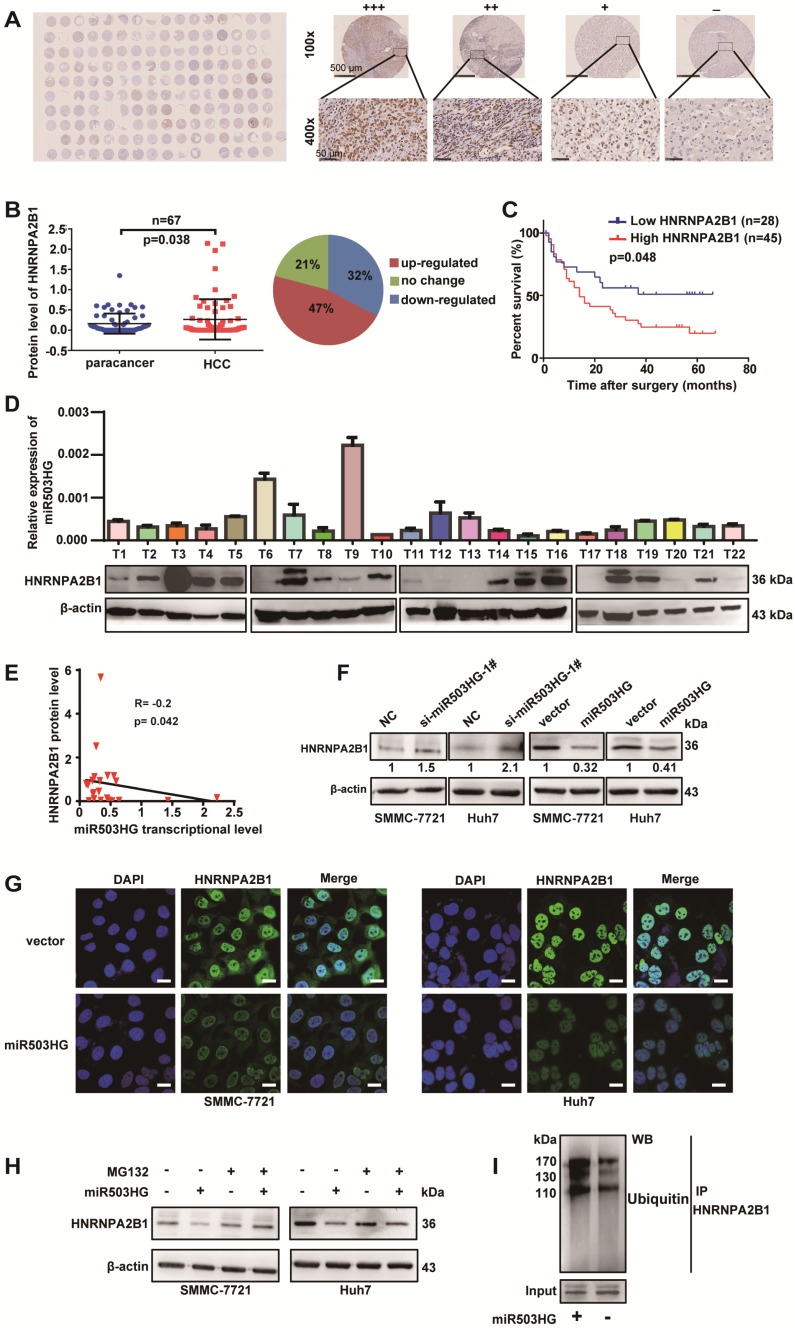
To explore whether miR503HG is involved in the regulation of HNRNPA2B1, we first examined the expression of HNRNA2B1 in 11 HCC cell lines. Our results showed that the protein expression level of HNRNPA2B1 was much upregulated in 10/11 of HCC cells compared to the human normal liver cell L02 (Figure S5A). We next detected the mRNA expression levels of HNRNPA2B1 and miR503HG in the same 67 pairs of HCC and adjacent noncancerous liver tissues from cohort 3. However, significantly dysregulated HNRNPA2B1 mRNA levels and correlation with miR503HG in HCC tissues were not observed (Figure S5B-C). Simultaneously, a tissue microarray with 80 pairs of HCC and corresponding noncancerous liver tissues was used to determine the protein level of HNRNPA2B1 (Figure 4A). Notably, we found that the protein level of HNRNPA2B1 was significantly increased in 47% of HCC tissues (Figure 4B). Kaplan-Meier survival analysis showed that HCC patients with high HNRNPA2B1 protein levels had worse OS than those with low HNRNPA2B1 protein levels (Figure 4C). In addition, we detected the transcriptional level of miR503HG and the protein level of HNRNPA2B1 in the same group of HCC tissues (Figure 4D). The results showed that the protein levels of HNRNPA2B1 were negatively correlated with miR503HG in 22 HCC tissues (Figure 4E). These results suggested that miR503HG might negatively regulate HNRNPA2B1 expression at the post-transcriptional level.
Figure 4.
miR503HG mediates ubiquitination and degradation of HNRNPA2B1 in HCC cells. (A) A tissue microarray with 80 pairs of HCC and corresponding noncancerous liver tissues was used to determine the protein expression level of HNRNPA2B1. (B) The protein level of HNRNPA2B1 was significantly increased in 47% of HCC tissues. (C) Kaplan-Meier analysis for OS was performed according to HNRNPA2B1 levels. (D) The transcriptional level of miR503HG and the protein expression level of HNRNPA2B1 in 22 HCC tissues. (E) Correlation analysis showed a negative relationship between miR503HG (x) and HNRNPA2B1 (y) in 22 HCC tissues (R = -0.2, P = 0.042). miR503HG and HNRNPA2B1 expression was normalized to β-actin. (F) HNRNPA2B1 protein expression in HCC cells with miR503HG knockdown or overexpression. (G) Immunofluorescence was used to compare the expression levels of HNRNPA2B1 between SMMC-7721 (or Huh7) cells with miR503HG overexpression and control cells. Scale bar = 20 μm. (H) SMMC-7721 and Huh7 cells expressing either miR503HG or control vector were cultured in the presence or absence of MG132 (20 μM) for 6 h. Cell lysates were then analyzed by western blotting. (I) Lysates from SMMC-7721 cells with miR503HG overexpression that were treated with MG132 were subjected to immunoprecipitation with anti-HNRNPA2B1 antibody followed by immunoblotting analysis with anti-ubiquitin or anti-HNRNPA2B1 antibody.
We next investigated the regulation mechanism of miR503HG in HNRNPA2B1 expression. We found that the transcriptional levels of HNRNPA2B1 were not affected by miR503HG in HCC cells (Figure S5D). However, knockdown of miR503HG resulted in increased expression of HNRNPA2B1 protein. In parallel, we observed a reduction in HNRNPA2B1 protein levels by overexpressing miR503HG (Figure 4F). These results were further confirmed by immunofluorescence in HCC cells after overexpression or knockdown of miR503HG (Figure 4G). To determine whether miR503HG reduced HNRNPA2B1 protein stability, we treated miR503HG-overexpressing or control HCC cells with the proteasome inhibitor MG-132. As shown in Figure 4H, MG-132 treatment abolished the downregulation of HNRNPA2B1 protein levels in HCC cells with overexpression of miR503HG (Figure 4H). Furthermore, we found that overexpression of miR503HG resulted in increased ubiquitination of HNRNPA2B1 (Figure 4I). These findings suggested that miR503HG downregulated HNRNPA2B1 protein abundance via the ubiquitin-proteasome pathway.
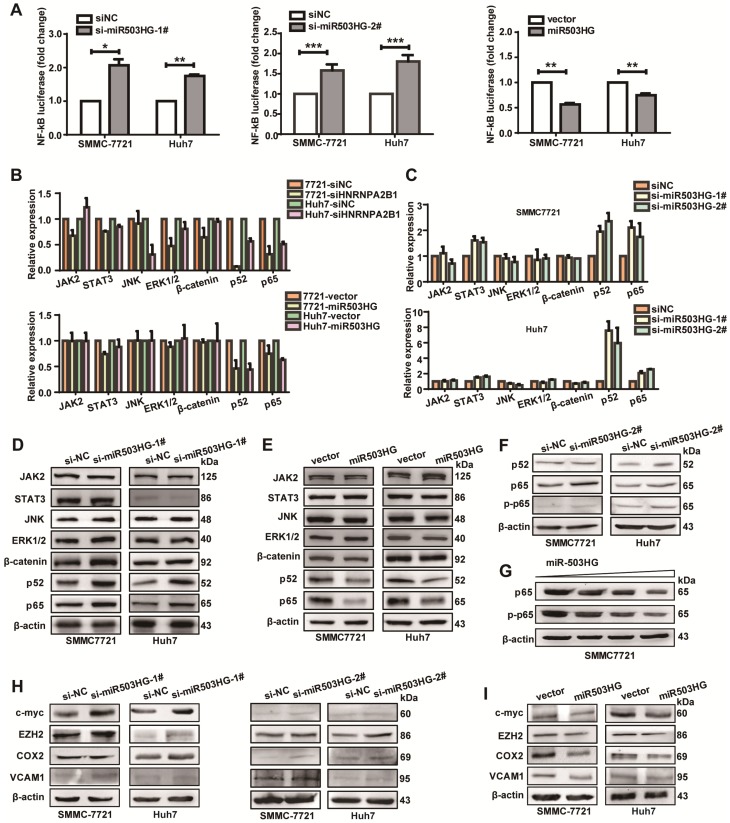
miR503HG inhibits the NF-κB signaling pathway in HCC cells
It has been known that HNRNPA2B1 is involved in several RNA-related biological processes of genes in some signal transduction pathways. Therefore, we hypothesized that miR503HG regulated some signaling pathways that are involved in cancer metastasis through HNRNPA2B1. To test this hypothesis, we first screened some signaling pathways that might be regulated by HNRNPA2B1. The signaling pathway reporter vectors were co-transfected with HNRNPA2B1 siRNA and luciferase activities were measured. Results showed that the NF-κB-dependent reporter activity was most significantly decreased compared to the negative control (Figure S6A). To further investigate the effect of miR503HG on NF-κB activities, we examined the luciferase activity of NF-κB in miR503HG-knockdown HCC cells. Results showed that inhibition of miR503HG lead to a more than 1.5-fold increase in NF-κB luciferase activity compared to that of the negative control. Nevertheless, overexpression of miR503HG dramatically inhibited NF-κB reporter activity compared with the negative control (Figure 5A). These results demonstrated that miR503HG was possibly involved in the regulation of the NF-κB pathway by modulating HNRNPA2B1.
Figure 5.
miR503HG inhibits the NF-κB signaling pathway in HCC cells. (A) Relative luciferase activity of NF-κB in miR503HG-downregulated or -upregulated SMMC-7721 and Huh7 cells. The values are presented as the mean ± standard error of the ratio of firefly luciferase activity to Renilla luciferase activity and are representative of three independent experiments. Data are the mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001. (B) Real-time PCR analysis of the expression of JAK2, STAT3, JNK, ERK1/2, β-catenin, p52 and p65 in HNRNPA2B1 silenced (top) and miR503HG-overexpressing HCC cells (bottom). (C) The mRNA levels of JAK2, STAT3, JNK, ERK1/2, β-catenin, p52, and p65 in HCC cells with miR503HG knockdown. (D-E) Western blotting analysis shows changes in JAK2, STAT3, JNK, ERK1/2, β-catenin, p52, and p65 in miR503HG-downregulated and miR503HG-upregulated HCC cells. β-actin was used as the loading control. (F) Knockdown of miR503HG could increase the phosphorylation levels of p65 in HCC cells. (G) Overexpression of miR503HG decreased endogenous p65 and phospho-p65 in a dose-dependent manner. (H-I) The protein levels of the NF-κB downstream effectors were determined by western blotting analyses in miR503HG-downregulated or -upregulated HCC cells.
Next, we examined the transcriptional levels of some of the genes in these pathways after HNRNPA2B1 knockdown. As expected, mRNA levels of p52 and p65, key mediators of the NF-κB pathway, were down-regulated. The same effects were also observed in miR503HG-overexpressing HCC cells (Figure 5B). Conversely, mRNA levels of p52 and p65 were increased in miR503HG-silenced HCC cells (Figure 5C). However, HNRNPA2B1 or miR503HG did not modulate the transcription levels of JAK2, STAT3, JNK, ERK1/2, and β-catenin significantly (Figure 5B-C). Additionally, we explored whether the stability of p52 and p65 mRNA were regulated by HNRNPA2B1. We treated HCC cells with actinomycin D to block new RNA synthesis and then measured p52 and p65 mRNA levels every 2 h. We found that knockdown of HNRNPA2B1 accelerated the degradation of p52 and p65 mRNA (Figure S6B). Consistent with this result, p52 and p65 mRNA degradation was also accelerated in miR503HG-overexpressing cells (Figure S6C). These results suggested that miR503HG reduced the stability of p52 and p65 mRNA, which might be through negative regulation of HNRNPA2B1.
We further detected the protein levels of signal transduction pathway genes in HCC cells with miR503HG knockdown or overexpression. The results revealed that knockdown of miR503HG increased the protein expression of p52 and p65 (Figure 5D). Conversely, overexpression of miR503HG resulted in decreased expression of p52 and p65 protein in HCC cells (Figure 5E). Furthermore, we showed that the loss of miR503HG could increase the phosphorylation levels of p65 in HCC cells (Figure 5F). Importantly, miR503HG decreased the protein levels of p65 and phospho-p65 in a dose-dependent manner in SMMC-7721 cells (Figure 5G). However, we didn't observe a significant change in the protein levels of JAK2, STAT3, JNK, ERK1/2, and β-catenin or the phosphorylation levels of JAK2, STAT3, JNK, and ERK1/2 with miR503HG knockdown or overexpression (Figure 5D-E and Figure S6D). Moreover, we observed that the inhibition of miR503HG increased the protein levels of a collection of NF-κB downstream effectors that are central coordinators of cancer cell metastasis, such as c-Myc, EZH2, COX2, and VCAM1, in SMMC-7721 and Huh7 cells (Figure 5H). Conversely, ectopic expression of miR503HG could decrease the protein levels of these genes (Figure 5I). Taken together, these results suggested that miR503HG could suppress the activity of the NF-κB pathway in HCC cells.
HNRNPA2B1 is required for miR503HG regulation of HCC cell migration and the NF-κB signaling pathway
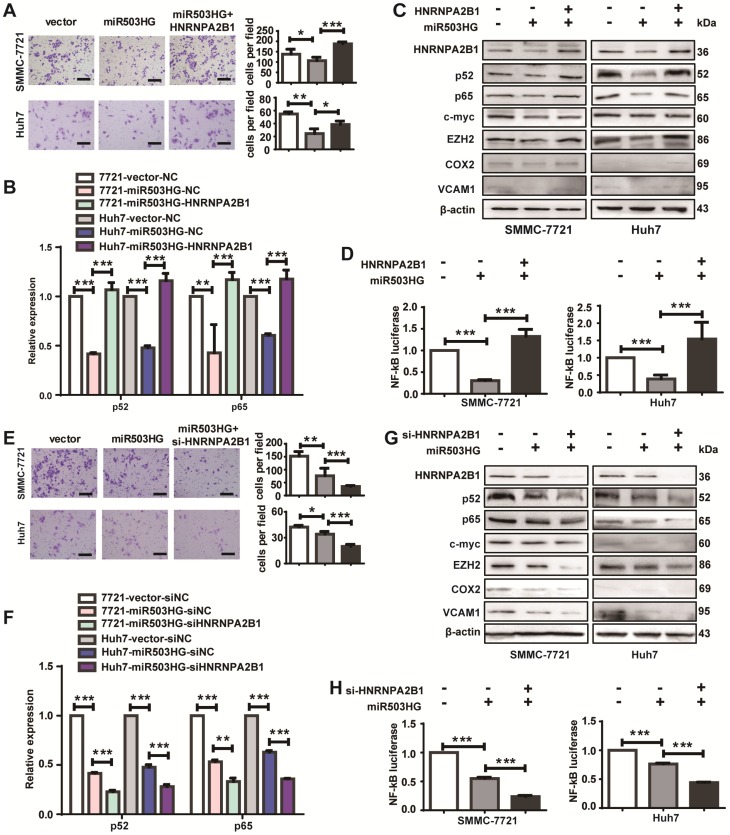
To determine whether HNRNPA2B1 is a critical mediator of miR503HG in HCC cell metastasis, cell migration after overexpression of HNRNPA2B1 in HCC cells with miR503HG overexpression was examined. We found that suppression of the migratory capacity by miR503HG was reversed when HNRNPA2B1 expression was simultaneously upregulated (Figure 6A). In addition, restoration of HNRNPA2B1 strongly increased the mRNA levels of p52 and p65 that are suppressed by miR503HG (Figure 6B). Concomitant overexpression of miR503HG and HNRNPA2B1 nullified the inhibitory effect of miR503HG on p52 and p65 protein and downstream genes of the NF-κB pathway and NF-κB-dependent reporter activity (Figure 6C-D). In contrast, HNRNPA2B1 knockdown in miR503HG-overexpressing cells resulted in a substantially decreased migratory capacity of HCC cells (Figure 6E). The inhibitory effects of the mRNA and protein expression levels of the NF-κB pathway by miR503HG were further reduced when the cells were co-transfected with siHNRNPA2B1 (Figure 6F-G), and a similar effect was observed for the NF-κB-dependent reporter activity (Figure 6H). Moreover, the increased migration of HCC cells by si-miR503HG was attenuated in cells co-transfected with si-HNRNPA2B1 and si-miR503HG (Figure S7A). The upregulation effects of the mRNA and protein expression levels of the NF-κB pathway or the NF-κB reporter activity regulated by miR503HG knockdown were diminished when HNRNPA2B1 was silenced (Figure S7B-D). These results indicated that HNRNPA2B1 is required for miR503HG regulation of HCC cell migration and the NF-κB signaling pathway.
Figure 6.
HNRNPA2B1 is required for miR503HG regulation of HCC cell migration and the NF-κB signaling pathway. (A) Migration assays of SMMC-7721 and Huh7 cells with overexpression of miR503HG and HNRNPA2B1. Scale bar = 100 μm. (B) The mRNA levels of p52 and p65 in SMMC-7721 and Huh7 cells that concomitantly overexpress miR503HG and HNRNPA2B1. (C) Western blotting analysis shows the changes of p52, p65 and the NF-κB downstream genes in HNRNPA2B1-restored HCC cells. (D) Relative luciferase activity of NF-κB in SMMC-7721 and Huh7 cells overexpressing HNRNPA2B1 with miR503HG. (E) Migration assays of HCC cells with miR503HG overexpression and HNRNPA2B1 knockdown. Scale bar = 100 μm. (F) The mRNA levels of p52 and p65 in miR503HG-overexpressing HCC cells with HNRNPA2B1 knockdown. (G) The protein levels of p52, p65 and the NF-κB downstream genes in miR503HG-overexpressing HCC cells with knockdown of HNRNPA2B1. (H) The NF-κB activity in HCC cells with miR503HG overexpression and HNRNPA2B1 knockdown. Data in (A-B, D-F, H) are the mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001.
miR503HG functions synergistically with miR503 to suppress HCC cell migration and invasion
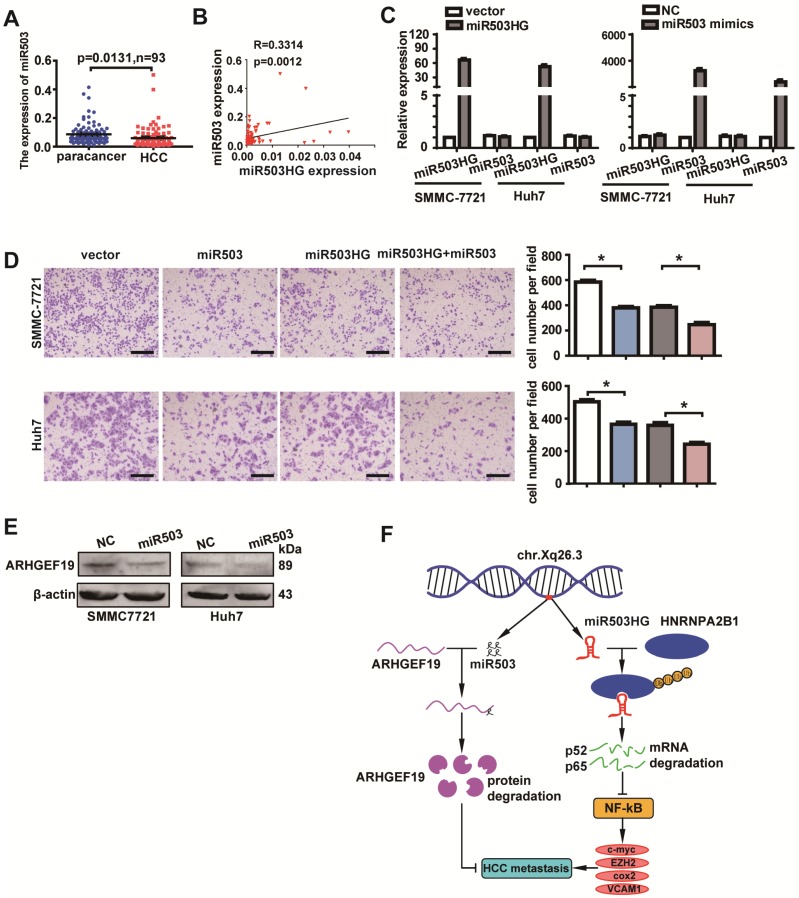
Because miR503HG is the host gene of miR503, we decided to investigate whether miR503HG and miR503 are transcribed together in HCC. We examined the expression of miR503 in 93 pairs of HCC and corresponding non-cancerous tissues. The results revealed that the expression level of miR503 was also significantly downregulated in HCC tissues compared with the corresponding normal tissues (Figure 7A). Importantly, the expression of miR503 was highly correlated with the expression of miR503HG (Figure 7B). These findings suggested that miR503 and its host gene miR503HG might be co-expressed in HCC. However, enhanced or decreased expression of miR503HG did not appreciably affect the level of miR503 (Figure 7C and Figure S8A). Similarly, the expression of miR503HG was not influenced by miR503 overexpression or knockdown in HCC cells (Figure 7C and Figure S8B). Intriguingly, transwell assays showed that increased miR503 expression significantly suppressed HCC cell migration (Figure 7D). In addition, western blotting revealed that ARHGEF19 was the potential target gene of miR503, as its protein level was reduced when miR503 was overexpressed (Figure 7E), which is consistent with a previous report 32. Importantly, we also found that elevation of miR503HG and miR503 simultaneously resulted in a further decrease in the cell migration of SMMC-7721 and Huh7 cells (Figure 7D). These results indicated that miR503HG functions synergistically with miR503 to suppress HCC migration.
Figure 7.
miR503HG functions synergistically with miR503 to suppress HCC cell migration and invasion. (A) Expression of miR503 in 93 pairs of HCC tissues and the corresponding normal liver tissues. U6B was used as the negative control. (B) The correlation of the expression levels of miR503 and miR503HG. Pearson's correlation was performed. (C) Expression of miR503 and miR503HG following overexpression of miR503HG and miR503 in SMMC-7721 and Huh7 cells. (D) Transwell migration assays of SMMC-7721 and Huh7 cells were performed after transient transfection of miR503 in miR503HG-upregulated or control HCC cells. Data are the mean ± SD. *P < 0.05. Scale bar = 100 μm. (E) Western blotting analysis shows changes in ARHGEF19 in miR503-upregulated SMMC-7721 and Huh7 cells. (F) A proposed model for illustrating the expression, function and mechanism of miR503HG in HCC metastasis.
Discussion
HCC is characterized by the dysregulation of numerous gene networks wherein both protein-coding genes and noncoding RNAs are involved. Of all the currently studied noncoding RNAs, lncRNAs are of increasing interest. Recently, we identified many lncRNAs that are aberrantly expressed in human HCC. Our results suggest that the deregulated lncRNAs may be biomarkers for distinguishing HCC tissues from noncancerous liver tissues. We further observed that miR503HG is significantly downregulated in HCC tumors. Given that low miR503HG expression levels were associated with significantly reduced overall survival and an increased risk of tumor recurrence, miR503HG may represent an independent prognostic biomarker in patients with HCC. Additionally, Kaplan-Meier analysis of the public database also demonstrated that low expression levels of miR503HG are associated with poor prognosis of patients with breast cancer, lung cancer and ovarian cancer (Figure S9) 33-35. Several of the described lncRNAs, for example, H19, MDIG, ATB, TSLNC8, and HOXD-AS1, may also be prognostic factors associated with recurrence in HCC 36-40, suggesting enormous potential for further development of lncRNA biomarkers for specific cancer histologies.
As miR503HG was commonly lowly expressed in HCC cells, to avoid its role in HCC progression would be somewhat artificial, Huh7 and SMMC-7721 with relatively higher miR503HG basal expression were selected to examine the effects of miR503HG. We identified that miR503HG can inhibit HCC cell metastasis in vitro and in vivo. To our knowledge, this is the first study to report that miR503HG regulates cellular metastasis in HCC. miR503HG inhibited migration and invasion in the JEG-3 choriocarcinoma cell line 41, which further supports the potential tumor suppressor role of miR503HG in cancers.
In this study, we found that miR503HG was associated with HNRNPA2B1 and promoted HNRNPA2B1 degradation via the ubiquitin-proteasome pathway in HCC cells. SELENBP1, which play a selenium-dependent role in ubiquitination-mediated protein degradation, was found in our mass spectrometry assay. However, we didn't observe an interaction between miR503HG and SELENBP1 by RNA pull-down or RIP experiments (data not shown). We have also performed RIP experiments with antibodies to the SKP2 and Pirh-2 proteins, which have been previously reported as E3 ligases in HCC 42, 43. Nevertheless, we also did not detect enrichment of miR503HG in the RIP deposition (data not shown). It is possible that miR503HG interacts with other E3 ligases. The interaction between miR503HG and HNRNPA2B1 changes the HNRNPA2B1 protein conformation and exposes the concealed ubiquitination sites, while other mechanisms are likely responsible for the ubiquitination of HNRNPA2B1.
HNRNPA2B1 is known to form complexes with lncRNAs and is emerging as an important mediator of lncRNA-induced transcriptional repression. A recent study has reported that lnc-HC regulates Cyp7a1 and Abca1 expression through forming a lnc-HC-HNRNPA2B1-mRNA complex 31. Here, we found that miR503HG decreased the stability of p52 and p65 and subsequently inhibited NF-κB transcriptional activity and the expression of multiple NF-κB signaling pathway downstream effectors that are central to the coordination of cell metastasis. Furthermore, we noted that ectopic overexpression of HNRNPA2B1 nullifies the inhibitory effect of miR503HG on the NF-κB signaling pathway and rescues the suppression of HCC migration. These results demonstrate that miR503HG may be involved in NF-κB signaling pathway regulation through binding to HNRNPA2B1, thus inhibiting HCC metastasis. Although our findings link miR503HG to the NF-κB pathway, the detailed molecular mechanism of miR503HG in NF-κB signaling is not currently understood in depth. It is likely that miR503HG regulates p52 and p65 expression by forming a miR503HG-HNRNPA2B1 complex and then the complex anchors to p52 and p65 to make the mRNAs fragile.
miR503HG is located on chromosome Xq26.3 and is the host gene of miR503. In the current study, we found that miR503 is also downregulated in HCC and that the expression level of miR503 is positively correlated with the level of miR503HG. Enhanced expression of miR503 did not appreciably affect the level of miR503HG, and overexpression of miR503HG did not affect the miR503 expression levels. These results indicate that miR503HG and miR503 are two different RNAs that are transcribed from the same transcript. The function of miR503HG is miR503-independent. More importantly, we identified that miR503HG plays a key role in tumor metastasis in that it can inhibit HCC metastasis in vitro and in vivo. Moreover, miR503HG can function synergistically with miR503 to inhibit HCC cell migration. These two different noncoding RNAs synergistically control the same biological process in different ways by regulating different downstream genes (Figure 7F).
In conclusion, miR503HG is often downregulated and co-expressed with miR503 in human HCC. It can suppress metastasis and acts synergistically with miR503 to inhibit HCC cell migration and metastasis. HNRNPA2B1 can physically interact with miR503HG and is required for miR503HG control of HCC metastasis and the NF-κB signaling pathway. These findings provide a new mechanism of HCC metastasis and a promising prognostic biomarker of HCC.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81201626) and the National 973 Key Basic Research Program (2013CB910504).
Abbreviations
- AFP
alpha-fetoprotein
- ALT
alanine aminotransferase
- BCLC
Barcelona Clinic Liver Cancer
- CI
confidence interval
- HCC
hepatocellular carcinoma
- HBsAg
hepatitis B surface antigen
- HR
hazard ratio
- lncRNA
long noncoding RNA
- OS
overall survival
- RACE
rapid amplification of cDNA ends
- RIP
RNA immunoprecipitation
- TNM
tumor-node-metastasis staging system
- TTR
time to recurrence.
References
- 1.Thomas MB, Jaffe D, Choti MM, Belghiti J, Curley S, Fong Y, Gores G. et al. Hepatocellular carcinoma: consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 2010;28:3994–4005. doi: 10.1200/JCO.2010.28.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrighetti L, Pulitano C, Catena M, Arru M, Guzzetti E, Halliday J, Ferla G. Liver resection with portal vein thrombectomy for hepatocellular carcinoma with vascular invasion. Ann Surg Oncol. 2009;16:1254. doi: 10.1245/s10434-009-0383-y. [DOI] [PubMed] [Google Scholar]
- 3.Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, Ge C. et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390–99. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 4.Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M. et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–56. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- 5.Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T, Chen Z. et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–44. doi: 10.1002/hep.27929. [DOI] [PubMed] [Google Scholar]
- 6.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ. et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–90. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M. et al. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–81. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–24. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA. et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–98. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B. et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–68. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun. 2016;7:12598. doi: 10.1038/ncomms12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X. et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–96. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Z, Gao X, He Y, Zhao H, Yu Q, Jiang D, Zhang P. et al. An insertion/deletion polymorphism within RERT-lncRNA modulates hepatocellular carcinoma risk. Cancer Res. 2012;72:6163–72. doi: 10.1158/0008-5472.CAN-12-0010. [DOI] [PubMed] [Google Scholar]
- 19.Mao X, Su Z, Mookhtiar AK. Long non-coding RNA: a versatile regulator of the nuclear factor-kappaB signalling circuit. Immunology. 2017;150:379–88. doi: 10.1111/imm.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, Zhang Q. et al. Hepatocyte nuclear factor 4alpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology. 2014;60:1607–19. doi: 10.1002/hep.27177. [DOI] [PubMed] [Google Scholar]
- 21.Song R, Song H, Liang Y, Yin D, Zhang H, Zheng T, Wang J. et al. Reciprocal activation between ATPase inhibitory factor 1 and NF-kappaB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology. 2014;60:1659–73. doi: 10.1002/hep.27312. [DOI] [PubMed] [Google Scholar]
- 22.Wilson CL, Jurk D, Fullard N, Banks P, Page A, Luli S, Elsharkawy AM. et al. NFkappaB1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat Commun. 2015;6:6818. doi: 10.1038/ncomms7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao C, Li Y, Huan L, Zhang Y, Zhao F, Wang Q, Liang L. et al. NF-kappaB signaling relieves negative regulation by miR-194 in hepatocellular carcinoma by suppressing the transcription factor HNF-1alpha. Sci Signal. 2015;8:ra75. doi: 10.1126/scisignal.aaa8441. [DOI] [PubMed] [Google Scholar]
- 24.Ding J, Huang S, Wang Y, Tian Q, Zha R, Shi H, Wang Q. et al. Genome-wide screening reveals that miR-195 targets the TNF-alpha/NF-kappaB pathway by down-regulating IkappaB kinase alpha and TAB3 in hepatocellular carcinoma. Hepatology. 2013;58:654–66. doi: 10.1002/hep.26378. [DOI] [PubMed] [Google Scholar]
- 25.Ruland J. Return to homeostasis: downregulation of NF-kappaB responses. Nat Immunol. 2011;12:709–14. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 26.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L. et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–81. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin H, Zhang Y, You H, Tao X, Wang C, Jin G, Wang N. et al. Prognostic significance of kynurenine 3-monooxygenase and effects on proliferation, migration, and invasion of human hepatocellular carcinoma. Sci Rep. 2015;5:10466. doi: 10.1038/srep10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–76. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Lan X, Yan J, Ren J, Zhong B, Li J, Li Y, Liu L. et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64:58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Tao Y, Peng C, Gu P, Wang W. miR-503 regulates metastatic function through Rho guanine nucleotide exchanger factor 19 in hepatocellular carcinoma. J Surg Res. 2014;188:129–36. doi: 10.1016/j.jss.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 34.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 35.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng C, Cheng S. et al. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol Lett. 2015;9:955–63. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata C, Otsuka M, Kishikawa T, Ohno M, Yoshikawa T, Takata A, Koike K. Diagnostic and therapeutic application of noncoding RNAs for hepatocellular carcinoma. World J Hepatol. 2015;7:1–6. doi: 10.4254/wjh.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Li Z, Liu L, Wang Q, Li S, Chen D, Hu Z. et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the IL-6/STAT3 signaling pathway. Hepatology. 2018;67:171–87. doi: 10.1002/hep.29405. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Huo X, Yang XR, He J, Cheng L, Wang N, Deng X. et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16:136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muys BR, Lorenzi JC, Zanette DL, Lima e Bueno Rde B, de Araujo LF, Dinarte-Santos AR, Alves CP. et al. Placenta-enriched lincRNAs miR503HG and LINC00629 decrease migration and invasion potential of JEG-3 cell line. PLoS One. 2016;11:e0151560. doi: 10.1371/journal.pone.0151560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calvisi DF, Ladu S, Pinna F, Frau M, Tomasi ML, Sini M, Simile MM. et al. SKP2 and CKS1 promote degradation of cell cycle regulators and are associated with hepatocellular carcinoma prognosis. Gastroenterology. 2009;137:1816–26. doi: 10.1053/j.gastro.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Wu G, Sun M, Zhang W, Huo K. AIG1 is a novel Pirh2-interacting protein that activates the NFAT signaling pathway. Front Biosci (Elite Ed) 2011;3:834–42. doi: 10.2741/E291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.