Abstract
Persistent viral infections result from evasion or avoidance of sterilizing immunity, extend the timeframe of virus transmission, and can trigger disease. Prior studies in mouse models of persistent infection have suggested that ineffective adaptive immune responses are necessary for persistent viral infection. However, recent work in the murine norovirus (MNV) model of persistent infection demonstrates that innate immunity can control both early and persistent viral replication independent of adaptive immune effector functions. Interferons (IFNs) are central to the innate control of persistent MNV, apart from a role in modulating adaptive immunity. Furthermore, subtypes of IFN play distinct tissue-specific roles in innate control of persistent MNV infection. Type I IFN (IFN-α/β) controls systemic replication and type III IFN (IFN-λ) controls MNV persistence in the intestinal epithelium. In this article, we will review recent findings in the MNV model, highlighting the role of IFNs and innate immunity in clearing persistent viral infection, and discussing the broader implications of these findings for control of persistent human infections.
Introduction to viral persistence and interferon
Upon recognition of a viral infection, the host ideally mounts an immune response that leads to viral clearance within a matter of days, and minimizes collateral damage to host tissues. However, many viruses persist weeks or months after an immune response has been initiated and some are never cleared by host immunity. These persistent viral infections can trigger disease, but they can also be neutral or even beneficial to the host [1–3]. From the host perspective, there is evolutionary pressure to avoid excessive tissue damage by tolerating infections that are not quickly resolved. From the virus perspective, persistence extends the timeframe for possible transmission to a new host and thereby provides a strong evolutionary advantage. Thus, evolutionary pressures have selected for a diverse array of persistent viruses with correspondingly diverse mechanisms of persistence. Persistent viruses can be broadly categorized into those involving periods of latency (e.g. lentiviruses and herpesviruses) and those that replicate continuously with no known mechanism of latency (e.g. flaviviruses and alphaviruses). Whereas latency is a built-in mechanism for avoiding the immune response, continuous replication represents a constant challenge to host immunity. This review will discuss the role of interferons (IFNs) during persistent infection with continuously-replicating viruses and emphasize recent insights resulting from studies in the murine norovirus (MNV) model system.
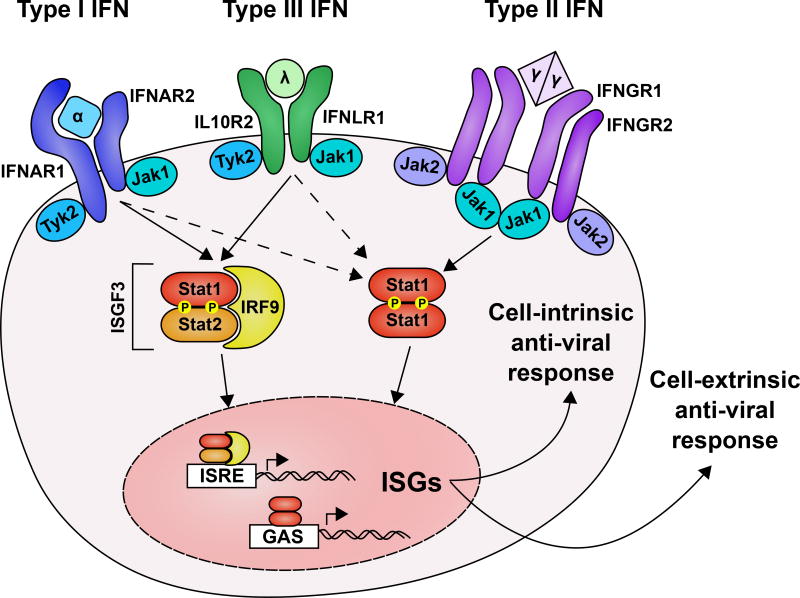
IFNs were discovered in 1957 as a soluble factor named for their ability to interfere with viral replication [4,5]. The importance of IFN is now abundantly clear and is reflected by the diverse viral mechanisms to avoid or evade their induction or action [6–12]. IFNs can be classified into three types based on their receptor usage (Figure 1). Type I IFNs (IFN-α’s, IFN-β, and others, subsequently referred to as IFN-α/β) signal through a receptor composed of IFNAR1 and IFNAR2 (IFNAR); type II IFN (IFN-γ) signals through a receptor composed of IFNGR1 and IFNGR2 (IFNGR); type III IFNs (IFN-λ’s) signal through a receptor composed of IFNLR1 and IL10R2 (IFNLR) [13,14]. IFN-α/β and IFN-λ genes have undergone gene duplication and evolutionary selection resulting in multi-gene families whereas IFN-γ is a single gene and is ostensibly under less evolutionary pressure [15]. In general, IFN receptor signaling induces a broad transcriptional response dependent on phosphorylation of signal transducer and activator of transcription (STAT) proteins by Janus kinase (JAK) family proteins [16,17]. IFNLR and IFNAR trigger an overlapping antiviral transcription program, but IFNLR expression is cell-type restricted relative to the more broadly expressed IFNAR [18–20]. Transcription downstream of IFNAR and IFNLR is driven predominantly by interferon stimulated gene factor 3 (ISGF3), which consists of STAT1, STAT2, and IRF9. Expression of interferon stimulated genes (ISGs) results in cell-intrinsic antiviral responses (e.g. block in translation, induction of apoptosis) and cell-extrinsic antiviral responses (e.g. recruitment of immune cells and stimulation of effector functions) [13,21–24]. Several recent reviews have focused on IFN-λ biology, including features that distinguish it from IFN-α/β [25–30]. Therefore, we will focus here on the role of IFN-λ and IFN-α/β pathways in the context of viral persistence.
Figure 1. Interferons and their receptors.
The three families of IFN are distinguished by receptor usage. Type I IFNs (IFN-α’s, IFN-β, and others) are recognized by a dimer of IFNAR1 and IFNAR2. Type III IFNs (IFN-λ’s) are recognized by a heterodimer of IL10R2 and IFNLR1. Type II IFN (IFN-γ) is recognized by a dimer of IFNGR1 and IFNGR2 heterodimers. JAK family kinases are pre-associated with the receptor cytoplasmic domains and are activated by ligand binding. STAT family transcription factors, predominantly STAT1 and STAT2 for IFN signaling, are phosphorylated and activated. STAT1 homodimers translocate to the nucleus and bind promoters with GAS elements. STAT1, STAT2 heterodimers associate with IRF9 to form ISGF3 which bind to promoters with ISRE elements and activate transcription. The solid and dashed arrows represent primary and secondary transactivation respectively of ISRE and GAS elements. Hundreds of ISGs are produced upon IFN signaling. Functions of ISGs can be broadly divided into cell-intrinsic antiviral immunity and cell- extrinsic antiviral immunity.
IFN as a regulator of adaptive immunity in the LCMV model of persistence
Insights from the extensively studied Lymphocytic choriomeningitis virus (LCMV) mouse model of persistence have led to mechanistic paradigms of persistent infection, and resulted in development of influential immunomodulatory therapies [31]. Therefore, prior to delving into the MNV model, we will provide some context by summarizing key findings regarding IFN in the LCMV model of persistence (see also Table 1 for additional comparison of LCMV and MNV mechanisms of persistence).
Table 1.
Comparison of LCMV persistence with systemic and enteric MNV persistence
| Virus | LCMV | MNV | |
|---|---|---|---|
| Location | systemic | systemic | enteric |
| Viral factors | Glycoprotein of LCMV clone 13 binds to α-dystroglycan and promotes infection of DCs and fibroblastic reticular cells [33,64]. This cellular tropism favors CTL down-regulation and viral persistence [36,65]. | Capsid. Polymorphisms in the protruding domain of the CW3 capsid (VP1) favor spread from the intestine to systemic organs[52,54,74]. This increased systemic spread correlates with enhanced IFN-α/β production and clearance of systemic CW3 [72]. | NS1. Polymorphisms in the NS1 gene of the persistent strain CR6 are necessary for early replication in the cecum and colon, the primary site of persistent replication [54]. NS1CR6 is necessary and sufficient for infection of IECs in vivo [68]. |
| Polymerase of LCMV clone 13 increases the rate of viral replication [34,122]. Increased replication results in increased antigen abundance which can favor CTL down-regulation and viral persistence [35,123]. | Capsid. Polymorphisms in the protruding domain of the CW3 capsid (VP1) correlate with increased IFN-λ production and favor clearance of CW3D94E following high dose inoculation [72]. | ||
| Others? Other viral determinants in CR6 are likely because this strain persists in the intestine regardless of inoculum dose, even with the VP1 gene from CW3 [54,72]. | |||
| Host factors | IFN-α/β controls LCMV replication [38], has both positive and negative impacts on sustained CTL function [38–40,43,44], and inhibits B cell responses [124–126]. | IFN-α/β is required for clearance of CW3. CW3 replicates uncontrollably and kills IFNAR1-deficient mice [77]. CR6 spreads from the intestine and persists systemically [72,89]. | IFN-λ. CR6 intestinal replication is increased in IFNLR1-deficient mice. Treatment with recombinant IFN-λ prevents persistence and clears established CR6 infection [72]. |
| CTLs kill LCMV infected cells and promote viral clearance. Loss of CTL function, through numerous mechanisms, correlates with LCMV persistence [32,35,37,38,127,128]. | Myeloid cell-intrinsic immunity. CW3 persists systemically when myeloid-lineage cells lack IFNAR1 [93]. Persistence results from defects in the myeloid cell-intrinsic response rather than downregulation of adaptive immunity [93]. | Epithelial cell-intrinsic immunity. Epithelial lineage specific deletion of IFNLR1 phenocopies full IFNLR1 deficiency [83]. Clearance of CR6 infection by IFN-λ treatment is independent of B and T cells [83]. IFNλ clears epithelial cell infection in vivo [68]. | |
| Antibody. B cells, CD4 T cell help, and LCMV neutralizing antibodies are required for sustained clearance of LCMV [129,130]. | B and T cells. B cells, CD4 T cells, and CD8 T cells all contribute to systemic clearance of CW3 [60,61]. CW3 persists systemically in mice with intact IFN-α/β response but no B or T cells (Rag1−/−) [49]. | Bacterial microbiome. Depletion of the bacterial microbiome with antibiotics inhibits establishment of persistent CR6 infection [89]. This promoting effect of intestinal bacteria is specific to intestinal tissue and is absent in IFNLR1-deficient mice [89]. | |
Infection with persistent strains of LCMV, such as clone 13, results in loss of LCMV-specific cytotoxic T lymphocyte (CTL) function [32–36], and restoration of CTL function promotes viral clearance [37]. Therefore, CTLs are a key determinant of persistence in the LCMV model and a major focus of study in the field. IFN-λ does not play a major role in control of LCMV persistence [18], which is consistent with low IFNLR expression on T cells and LCMV tropism for cells that do not express IFNLR. In contrast, IFN-α/β plays a crucial role in regulation of CTL function and LCMV clearance. Infection of Ifnar1−/− mice with the otherwise non-persistent LCMV Armstrong strain results in increased viral titers, dampened CTL function and delayed viral clearance [38]. Correspondingly, administration of recombinant IFN-α/β [39] or knocking out a negative regulator of IFN-α/β [40] enhances CTL function and promotes clearance of LCMV clone 13. Mechanistically, IFN-α/β promotes CTL function by signaling on T cells directly [41,42] and by regulating antigen abundance [35]. Recently, it has become clear that the beneficial roles of IFN-α/β are in a fragile balance with concurrently acting suppressive roles. In two separate studies, transient antibody blockade of IFNAR1 reduced production of the immunosuppressive cytokine, interleukin 10 (IL-10), increased CTL function, and enhanced viral clearance [43,44]. This suppressive role for IFN-α/β may represent negative feedback on the immune response to prevent immunopathology [45], and the balance of beneficial and detrimental roles for IFN-α/β on LCMV-specific CTL function ultimately depends on the timing and magnitude of IFN-α/β expression [39]. However, in all cases, the role of IFN-α/β in LCMV persistence has been clearly linked to regulation of a functional and sustained CTL response.
Murine norovirus and persistent infection in the intestine
Many persistent infection models, including LCMV, study infection of the circulatory system and internal organs. The intestinal mucosa, however, has evolved to tolerate abundant commensal microbes and thus poses a distinct set of challenges for clearance of viral infection. The MNV model of persistence in the intestine is ideal for study of antiviral immunity in the context of these challenges [46–49].
The founding strain of MNV (MNV-1) was discovered in 2003 by sequencing a lethal infectious agent isolated from brain tissue of Rag1−/−/Stat1−/− mice [49]. MNV belongs to the same genus as human noroviruses, which are a major cause of gastroenteritis spread by the fecal-oral route [50,51]. MNV-1 was initially passaged intra-cranially, but it is also infectious by the oral route [49]. Additionally, sequencing of feces from laboratory mice identified additional MNV strains that are 87–95% identical to MNV-1, indicating that MNV is a fecal-oral pathogen, like human noroviruses [46,47]. MNV-1 is cleared by wild-type (WT) mice, but it can be lethal in Stat1−/− mice [49,52], implicating IFN family cytokines in its clearance. Unlike MNV-1, fecal-isolated strains persist in intestine and draining mesenteric lymph nodes (MLNs) of WT mice and rarely kill Stat1−/− mice [46,53–55]. The initial discovery of MNV-1 and subsequent discovery of persistent strains that differ phenotypically formed the basis for a mouse model to study viral persistence in the intestine.
CTL responses differ between MNV strains but do not clear persistent infection
There are several persistent strains of MNV that have been studied [46,53,55–58]. For simplicity, we focus here on the persistent strain CR6 and the non-persistent strain CW3, a plaque purified descendent of MNV-1 [46,59]. CW3 persists systemically in Rag1−/− mice with B and T cells each contributing to clearance of this otherwise non-persistent strain [49,60,61]. These findings are consistent with a requirement for CTL effector functions in MNV clearance similar to the paradigm established in LCMV persistence. Furthermore, CW3 elicits a more robust expansion of MNV-specific CTLs in the intestine and spleen than CR6 [62]. However, mutation of a single amino acid difference in the CR6 CTL epitope (F525Y) results in robust expansion of MNV-specific CTLs, but does not prevent CR6 persistence [63]. Additionally, there is no evidence of clonal deletion or non-responsiveness of CR6-specific CTLs, as is seen in persistent LCMV infection [62,63]. Instead, CR6-specific CTLs remain functional but do not “see” continuous viral replication in the intestine, and the CTL transcriptome during CR6 persistence resembles the murine cytomegalovirus (MCMV) CTL transcriptome during MCMV latency [63]. Altogether, these data indicate that a robust CTL response does not clear persistent CR6 and remains largely ignorant of its continuous replication. Persistence of CR6, despite eliciting functional CTLs, indicates that the mechanism of MNV persistence in the intestine differs from that of systemic LCMV persistence
Viral genetic determinants of MNV persistence
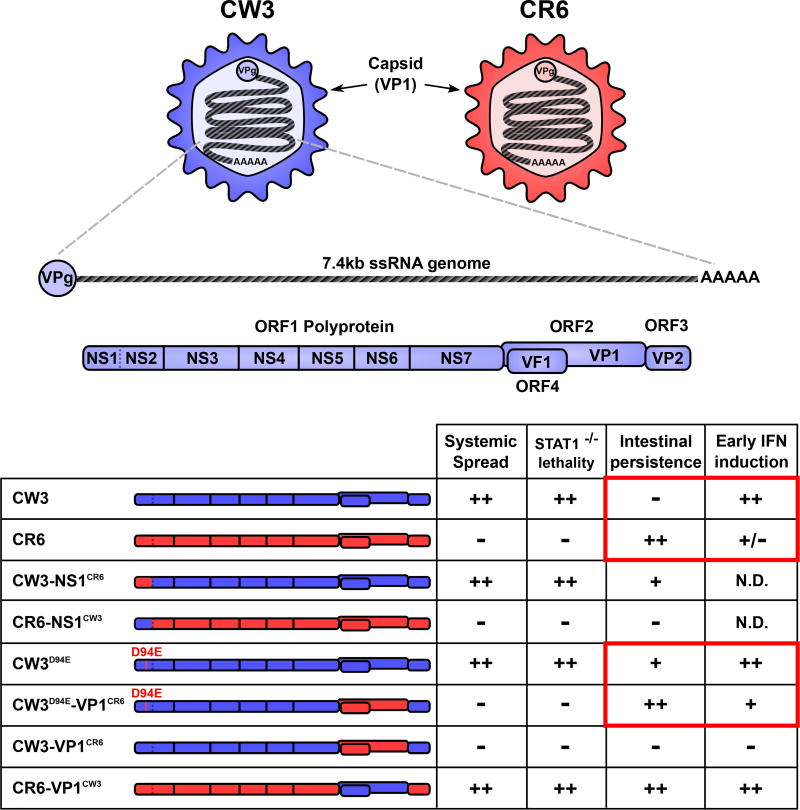
Genetic studies have been key to understanding mechanisms of persistence in the LCMV model [33,36,64,65]. Likewise, we used MNV reverse genetics to develop chimeric strains and identify genes required for persistence [52,59,66] (Figure 2). By swapping single genes reciprocally between CW3 and CR6, we found that the NS1 gene of CR6 (NS1CR6) is necessary and sufficient for persistence [54]. Furthermore, a single conservative amino acid change in NS1CW3 from aspartic acid to glutamic acid at position 94 (CW3D94E) is sufficient for persistence [54,67]. NS1CR6 promotes early growth in the cecum and proximal colon where MNV ultimately persists most abundantly [53,54,57]. Moreover, NS1CR6 is necessary and sufficient for intestinal epithelial cell (IEC) tropism in vivo, indicating that IECs of the cecum and colon are the cellular reservoir of persistence [68]. NS1 is an essential part of the viral replication complex, interacts with the membrane protein VAPA, and localizes with the replication complex on cytoplasmic membranes [69–71]. However, the molecular mechanism of NS1 in persistence remains a major gap in knowledge.
Figure 2. Viral genetics of MNV strain-dependent phenotypes.
MNV is a positive-sense single-stranded RNA virus with a ~7.4kb genome, covalently linked at the 5’ end to viral protein, genome-linked (VpG). Open reading frame (ORF) 1 encodes a polyprotein that is cleaved into six non-structural (NS) proteins by the viral protease (NS6). NS1–2 is further cleaved by a host protease (caspase 3) into NS1 and NS2 [118]. ORF2 encodes the major capsid protein (VP1), which self assembles to form the viral capsid and enclose the viral genome. Shown in the table are parental CW3 (blue) and CR6 (red) as well as several chimeric viruses. Selected phenotypic characteristics for each chimeric virus are shown. The ability to replicate in IECs and persist is determined by NS1CR6 or a D94E mutation in NS1CW3 [54,68]. The capacity to spread outside of the intestine correlates with lethality in Stat1−/− mice and is determined by the capsid [52,54,74]. The capsid also determines early IFN production in the peyer’s patches and MLN [72]. Red boxes indicate inverse correlations between early IFN production and intestinal persistence. N.D. = not determined.
In the LCMV model, increasing infectious dose can significantly promote viral persistence [38]. Conversely, we found that the frequency of mice persistently infected with CW3D94E is reduced when infectious dose is increased from 104 to 106 PFU/mouse [72]. CR6 persists at all infectious doses up to 107 PFU/mouse, suggesting that genetic differences between CW3 and CR6 other than D94E play a dose-dependent role in persistence. Combining CR6 capsid (VP1CR6) chimerism with the D94E mutation (CW3D94E-VP1CR6) enables this virus to persist at high dose [72]. Additionally, at high infectious dose, the CW3 capsid (VP1CW3) is necessary and sufficient for early stimulation of IFN-β and IFN-λ production in Peyer’s patches and MLNs one to two days after infection [72]. Therefore, the capsid gene is a second genetic determinant of MNV persistence, and early IFN production is implicated in blocking persistent infection.
The protruding domain of the capsid gene binds cell surface carbohydrates and the proteinaceous MNV receptor, CD300lf [55,73]. This domain is also the determinant of early IFN responses in MLNs [72], spread to extra-intestinal tissues (systemic spread) [54], and lethality in Stat1−/− mice [52,74]. These data suggest that differential VP1CW3 binding to cells could be the basis for systemic spread, early induction of IFN, and clearance from WT mice (Figure 2). However, CR6 with the capsid of CW3 (CR6-VP1CW3) spreads systemically and triggers early IFN production, but still persists in the intestine and MLN [54]. Therefore, additional gene(s) of CR6 may provide resistance to the effects of early IFN whereas their CW3 counterparts do not. The MNV ORF4 gene product, virulence factor 1 (VF1), is an attractive candidate because it inhibits ISG expression in vitro and is essential for replication in vivo [75]. Although further viral genetic studies are necessary, the results thus far demonstrate the importance of early infection of IECs and a diminished host IFN response in enabling MNV persistence in the intestine (Figure 2, Table 1).
IFN-λ is a key host determinant of epithelial tropism and persistence in the intestine
The crucial role of IFNs in control of CW3 replication was clear from the time of its discovery as a lethal virus in Stat1−/− mice [49]. In contrast, CR6 rarely kills Stat1−/− mice, but it does persist systemically and replicate to higher titers in the colon [52,72] (Figure 3B). Thus, STAT1-dependent immunity controls the extent of persistent replication in the intestine as well as systemic spread.
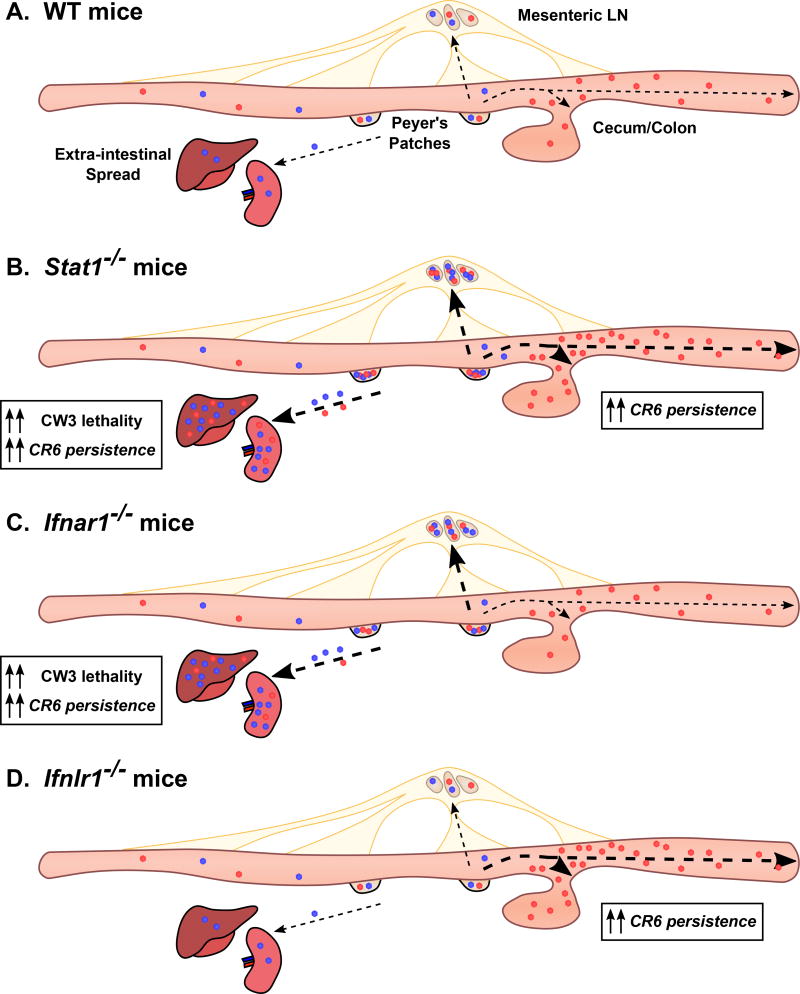
Figure 3. The cooperative roles of interferon in persistent MNV infection.
IFN-α/β and IFN-λ have compartmentalized roles in control of MNV. A. CW3 (blue) and CR6 (red) exhibit distinct tropism as depicted in Figure 2. MNV uses Peyer’s patches as an initial site of entry and subsequently spreads to the mesenteric lymph nodes (MLNs) [119–121]. CW3 is minimally shed into the feces and spreads to extra-intestinal tissues including the spleen and liver in an acute infection, whereas CR6 spreads to the cecum and colon and is robustly shed into the feces [52,54]. B. In the absence of STAT1, CW3 replicates to higher titers in all tissues and the mice die of uncontrolled viral replication. CR6 does not kill these mice but replicates to higher titer in the intestine and persists in extra-intestinal tissues [52,72]. C. As in Stat1−/− mice, CW3 replicates to higher titers and can kill Ifnar1−/− mice [77]. CR6 persists systemically in Ifnar1−/− mice, but intestinal persistence is unchanged [72,89]. D. CW3 replication is unchanged in Ifnlr1−/− mice. CR6 replicates to higher titers in the intestine of Ifnlr1−/− mice, but is undetected in systemic tissues [72,83,89].
Studies in Ifnar1−/− or Ifnlr1−/− mice reveal separate roles for IFN-λ and IFN-α/β in control of MNV replication (Figure 3). Strikingly, IFN-λ is uniquely responsible for control of early and persistent CR6 replication in the IECs of the colon [68,72]. Additionally, treatment with recombinant IFN-λ [72] or stimulation of IFN-λ production by intra-epithelial T cells [76] clears persistent MNV from the intestine. Conversely, IFN-α/β prevents systemic spread of CR6 and limits systemic replication of CW3 [72,77,78] (Figure 4A). This cooperative and compartmentalized role of IFN-λ and IFN-α/β was similarly shown in an acute model of reovirus infection: IFN-λ controls replication in the intestinal epithelium whereas IFN-α/β controls replication in other cell types and systemic dissemination [79]. The compartmentalized roles of IFN-λ and IFN-α/β are underpinned by compartmentalized expression of IFN receptors; IFNLR is highly expressed in IECs but not on most immune cells whereas IFNAR is broadly expressed on immune cell types but is expressed poorly by IECs of adult mice [79–82]. Indeed, IFNLR must be expressed specifically on IECs for IFN-λ to clear reovirus or MNV from the intestine [68,83]. From these studies, the specific role for IFN-λ in IECs is clear.
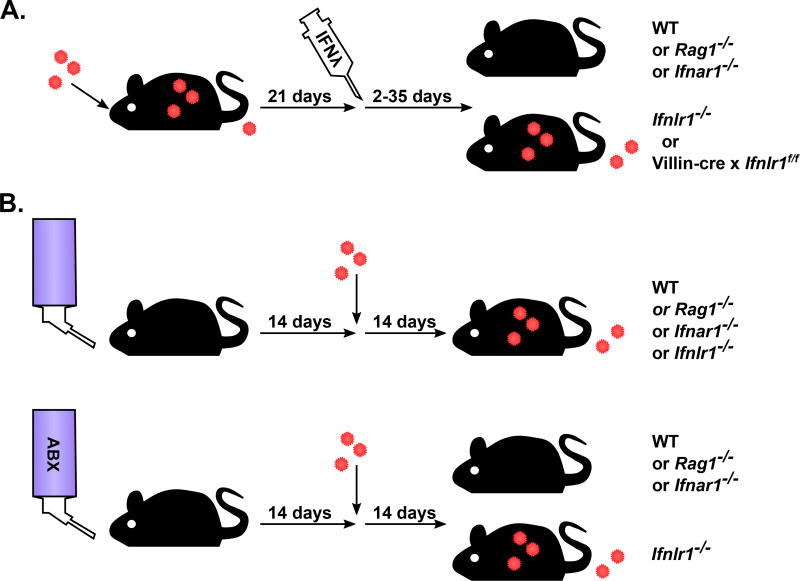
Figure 4. IFN-λ controls intestinal MNV persistence independent of adaptive immunity.
A. Therapeutic administration of IFN-λ to persistently infected mice leads to clearance. IFN-λ is effective in WT mice, Rag1−/− mice and Ifnar1−/− mice indicating that adaptive immunity and type I IFN are dispensable for the antiviral effects of IFN-λ [72]. IFN-λ treatment has no effect in mice without IFNLR1 or with IFNLR1 absent specifically in Villin-expressing IECs (Villin-cre × Ifnlr1f/f) indicating that epithelial cells must respond directly to IFN-λ [83]. B. Intestinal bacteria promote CR6 infection in an IFNLR1-dependent RAG1-independent manner. Depletion of commensal bacteria by administering a cocktail of antibiotics (ABX) in the drinking water results in resistance to persistent CR6 infection. Similar to therapeutic IFN-λ treatment, the bacteria-dependent phenotype is present in WT mice, Rag1−/− mice and Ifnar1−/− mice. Commensal bacteria are not required in Ifnlr1−/− mice indicating that the effect of bacteria depends on IFN-λ signaling. See Baldridge et al. [89] for additional host genetic analysis of this phenotype.
Recently, it has been demonstrated that the bacterial microbiome regulates intestinal immunity and can promote infection of multiple enteric viruses [84–88], including noroviruses [89–91]. Treating mice with a cocktail of antibiotics (ABX) to deplete intestinal bacteria diminishes early MNV-1 replication in the intestine [90,91] and establishment of CR6 persistence in the colon [89]. Bacteria promote CR6 persistence in WT or Rag1−/− mice but not Ifnlr1−/− mice, further emphasizing the crucial role of IFN-λ in the intestine and indicating that the role of bacteria is countered by IFN-λ signaling [89]. (Figure 4B). This pro-viral role for bacteria is unique to the intestine because acute splenic growth of CW3 or CR6-VP1CW3 is not impacted by ABX treatment whereas acute and persistent intestinal growth of CR6 or CR6-VP1CW3 is prevented [89]. Additionally, the role for bacteria appears to be in early establishment of persistence rather than maintenance because in rare cases where ABX-treated mice become infected they remain persistently infected [89]. Paradoxically, the bacterial microbiome stimulates increased ISG expression in intestinal tissue that correlates with its pro-viral role [89,92]. Further work is required to resolve this paradox by defining specific mechanisms by which bacteria impact IFN-λ signaling and antiviral immunity in the intestine.
Whereas there is a clear role for IFN-λ and IEC-intrinsic immunity in control of persistent intestinal replication, MNV-specific adaptive immunity plays a minor role in control of intestinal persistence. Indeed, persistent CR6 infection in Rag1−/− mice is cleared by treatment with recombinant IFN-λ and persistent shedding is increased in Rag1−/−/Ifnlr1−/− double knockout mice [72,83] (Figure 4A). Together with the central role of IECs in the IFN-λ response, these experiments demonstrate that IFN-λ controls the extent and duration of intestinal viral persistence without a requirement for B and T cells, in contrast to the central role that adaptive immune responses play in LCMV persistence (Table 1).
IFN-α/β and adaptive immunity play non-redundant roles in preventing systemic MNV persistence
IFN-α/β and adaptive immunity are both required for clearance of systemic MNV infection [49,60,61]. However, our recent work indicates that IFN-α/β plays a crucial role that is independent from B and T cell responses. CD11c-lineage-specific knockout of Ifnar1, which includes MNV infected myeloid cells, enables systemic CW3 persistence without compromising B and T cell responses [93]. In fact, increased early viral replication in these mice leads to enhanced MNV-specific CTL and antibody responses, but this adaptive response fails to clear infection for at least 35 days. Mixed bone marrow chimeras indicate that the increased replication is due to cell intrinsic defects in CD11c-expressing cells (Figure 5) [93]. Although the specific cell type that harbors persistent virus in these mice is unknown, these data are consistent with MNV replication in CD11c-positive myeloid cells [59]. So, IFN-α/β differs from IFN-λ in its co-operative, non-redundant relationship with adaptive immunity as well as the cellular reservoir it protects (myeloid vs. IEC). However, despite their contextual differences, IFN-α/β and IFN-λ are each required for clearance of MNV from a cellular reservoir of persistence independent of adaptive immunity (Table 1).
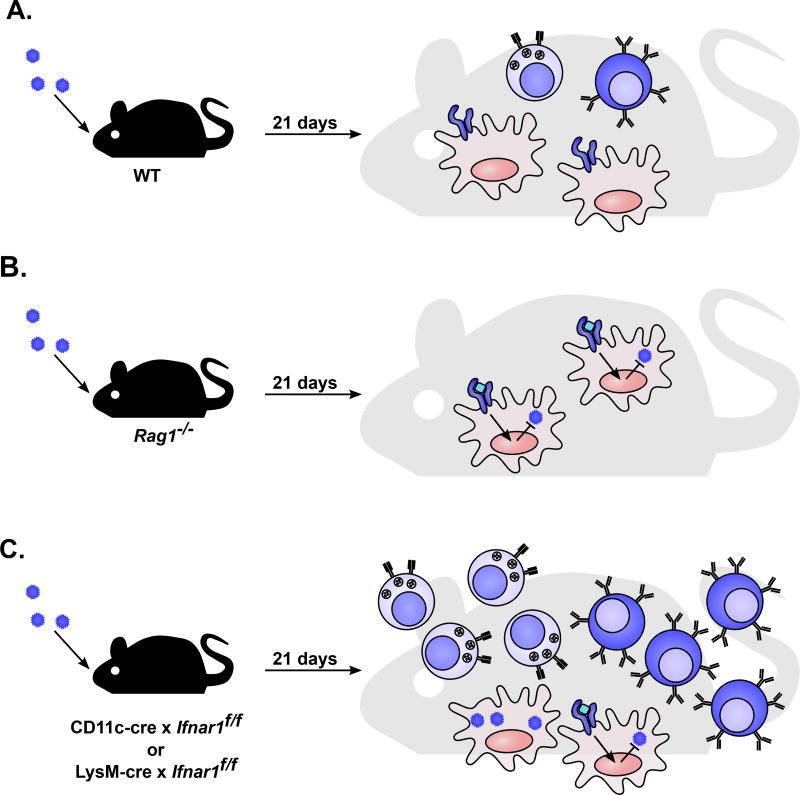
Figure 5. Separate roles for IFN-α/β and adaptive immunity in preventing systemic persistence.
Immune defects resulting in systemic persistence of CW3. A. WT mice mount a CW3-specific T cell (light blue cells) and B cell response (dark blue cells) [60–62]. IFNAR1-expressing myeloid cells (red cells) mount a cell-intrinsic antiviral response to facilitate clearance of viral infection [77]. B. Mice lacking B or T cells (Rag1−/−) do not have a CW3 specific adaptive immune response. IFNAR1-expressing myeloid cells mount a cell-intrinsic antiviral response that is insufficient for clearance of viral infection. C. Mice with a sub-population of myeloid cells deficient in IFNAR1 (LysM-cre × Ifnar1f/f or CD11c-cre Ifnar1f/f) mount an enhanced CW3-specific T cell and B cell response. These cells remain functional, but are insufficient for clearance of viral infection [93].
Open questions and future directions in MNV persistence
Identification of the molecular mechanism by which NS1CR6 enables tropism for IECs in vivo is a major open question. IECs do not commonly express the MNV receptor, CD300lf, whereas myeloid cells express it abundantly [73,94,95]. Additionally, work from our lab and others have not found MNV to grow in IECs in vitro unless they ectopically express CD300lf [68]. A receptor for human noroviruses is not known, but Estes and colleagues recently developed an in vitro method for human norovirus infection of primary human epithelial cells using bile acids to promote infection [96]. It will be important to determine whether bile acids or other co-factors also enable MNV infection of IECs and whether NS1 plays a role in this process. Additionally, characterization of the IECs in which MNV persists in vivo will provide clues to the mechanism by which NS1 enables infection of this niche.
In addition to the molecular mechanism of NS1, it is not clear why WT hosts do not produce enough endogenous IFN-λ to clear CR6 infection. Part of the answer to this question may come from defining the mechanism by which VP1CR6 results in reduced early production of IFN-λ [72]. However, other factors are also at play because CR6-VP1CW3 stimulates expression of IFN-λ, but persists [54,72]. Therefore, genetic differences between CW3 and CR6 other than the capsid may provide resistance to the anti-viral effects of endogenous IFN-λ (Figure 2). This may be part of NS1’s role, either alone or in co-operation with VF1 [68,75]. Identifying viral mechanisms of endogenous IFN-λ suppression and IFN-λ resistance will be important for understanding the immunological basis of intestinal persistence.
Another open question is how the bacterial microbiome stimulates ISG expression in the intestine [79,89], but paradoxically promotes CR6 persistence in an IFNLR1-dependent manner [89,92]. One explanation for this may be that ISGs stimulated by the bacterial microbiome are distinct from those stimulated by therapeutic IFN-λ administration. Another difference is that bacterially stimulated ISGs are of low magnitude and are tonic rather than acute - this may result in negative feedback de-sensitization to IFN-λ stimulation [97–101]. Interleukin-22 may also be involved because it is stimulated by bacteria, shares the IL-10R2 receptor subunit with IFN-λ, and promotes the IFN-λ response to rotavirus infection [102]. Understanding how bacterial stimulation differs transcriptionally from the therapeutic IFN-λ stimulation will provide insight into regulation of this pathway.
Finally, it is unclear why IFN-λ and IFN-α/β responses are compartmentalized. An attractive hypothesis is that constant bombardment of microbes at the epithelium necessitates separate regulatory mechanisms for clearing viruses without triggering immunopathology. The intestinal epithelium of neonatal mice and germ-free mice is responsive to IFN-α/β, suggesting that the loss of IFN-α/β responsiveness by IECs is, in fact, driven by the bacterial microbiome [58,82]. In addition to regulated receptor expression, differences in the magnitude and kinetics of downstream ISG expression my differentiate the outcome of IFN-α/β and IFN-λ signaling [103]. It will be important to continue uncovering the genetic and molecular basis of compartmentalized receptor expression and differential regulation of the ISG response.
Potential implications for human persistent viral infection
The pleiotropic effects of IFNs make it difficult to definitively determine the relative importance of human innate and adaptive immune responses to persistent viral infection. However, there is some recent evidence that clearance of multiple human viruses depends on IFN-stimulated innate immunity.
IFN-λ received increased notoriety following groundbreaking work that uncovered a correlation between a polymorphism (rs368234815) in the IFNL3/4 locus and clearance of hepatitis C virus (HCV) [104–107]. Unlike in mice, humans express IFNLR on hepatocytes, enabling a direct role for IFN-λ in HCV infection [18,19]. The protective IFNL3/4 SNP creates a premature stop codon in the IFNL4 gene and may promote HCV clearance through up-regulation of neighboring IFN-λ genes or up-regulation of ISG induction in response to IFN-α/β therapy [104,107–109]. HCV encodes inhibitors of IFN-α/β induction that are required for its replication, demonstrating the importance of ISG induction in cell-intrinsic HCV control [110]. Together, these studies suggest that IFN-λ, in conjunction with IFN-α/β, determines HCV persistence. Therefore, spontaneous and/or therapeutic clearance of HCV may be dependent on IFN-stimulated innate immunity.
IFN-λ may be crucial for controlling viral infection in humans when adaptive immunity is compromised. AIDS patients and transplant recipients each have reduced adaptive immune function and correspondingly have increasing dependence on the efficacy of innate immunity. The IFNL3/4 SNP correlates with human cytomegalovirus (HCMV) retinitis in AIDS patients [111] and HCMV pathology in transplant recipients [112]. These correlative findings suggest that the IFN-λ response controls persistent replication of HCMV, particularly when adaptive immunity is compromised. Pregnancy is another scenario correlated with reduced adaptive immunity and increased susceptibility to infections [113]. As in HCV patients, the IFNL3/4 polymorphism was recently shown to correlate with increased ISG expression in postpartum women and may provide enhanced innate immune clearance of viruses [114].
Although IECs are a major target of IFN-λ signaling in mice, more work is needed to understand the role of IFN-λ in human IECs. Studies so far indicate that intestinal cells can respond to IFN-λ [18,19,115,116], IFN-λ is preferentially produced by IECs [79,116,117], and IFN-λ signaling is uniquely dependent on p38 kinase activity [115]. It will be important to continue these and related studies to define IFN-λ production and response within the human intestine.
Concluding Remarks
In summary, the MNV model has revealed a key role for IFN-λ in clearance of persistent viral infection from IECs, independent of adaptive immunity. Additionally, IFN-α/β clears MNV from myeloid cells to prevent systemic viral persistence. Moreover, recent human studies have found correlation between IFN-λ polymorphisms and control of viral infection when adaptive immunity is compromised. Continued work to answer outstanding questions in the MNV model and in humans is essential if we are to fully appreciate the impact of IFNs in persistent viral infection.
References
- 1.Virgin HW, et al. Redefining Chronic Viral Infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 4.Isaacs a, Lindenmann J. Virus Interference. I. The Interferon. Proc. R. Soc. B Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 5.Isaacs A, et al. Virus Interference. II. Some Properties of Interferon. Proc. R. Soc. B Biol. Sci. 1957;147:268–273. [PubMed] [Google Scholar]
- 6.Diamond MS. Mechanisms of Evasion of the Type I Interferon Antiviral Response by Flaviviruses. J. Interf. Cytokine Res. 2009;29:521–530. doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- 7.Short JAL. Viral evasion of interferon stimulated genes. Bioscience Horizons. 2009;2:212–224. [Google Scholar]
- 8.Taylor KE, Mossman KL. Recent advances in understanding viral evasion of type I interferon. Immunology. 2013;138:190–197. doi: 10.1111/imm.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basler CF, Amarasinghe GK. Evasion of Interferon Responses by Ebola and Marburg Viruses. J. Interf. Cytokine Res. 2009;29:511–520. doi: 10.1089/jir.2009.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J. Biol. Chem. 2010;285:22741–7. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grandvaux N, et al. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 2002;15:259–267. doi: 10.1097/00001432-200206000-00008. [DOI] [PubMed] [Google Scholar]
- 12.Levy DE, García-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine and Growth Factor Reviews. 2001;12:143–156. doi: 10.1016/s1359-6101(00)00027-7. [DOI] [PubMed] [Google Scholar]
- 13.Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J. Interferon Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manry J, et al. Evolutionary genetic dissection of human interferons. J. Exp. Med. 2011;208:2747–59. doi: 10.1084/jem.20111680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aaronson DS. A Road Map for Those Who Don’t Know JAK-STAT. Science (80-.) 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 17.Schindler C, Plumlee C. Inteferons pen the JAK-STAT pathway. Seminars in Cell and Developmental Biology. 2008;19:311–318. doi: 10.1016/j.semcdb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheppard P, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 19.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 20.Sommereyns C, et al. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–25. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenschow DJ. Antiviral Properties of ISG15. Viruses. 2010;2:2154–68. doi: 10.3390/v2102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, Baldridge MT. Interferon-lambda: A potent regulator of intestinal viral infections. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syedbasha M, Egli A. Interferon Lambda: Modulating immunity in infectious diseases. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pott J, Stockinger S. Type I and III interferon in the gut: Tight balance between host protection and immunopathology. Front. Immunol. 2017;8:1–15. doi: 10.3389/fimmu.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotenko SV, Durbin JE. Contribution of type III interferons to antiviral immunity: Location, location, location. J. Biol. Chem. 2017;292:7295–7303. doi: 10.1074/jbc.R117.777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazear HM, et al. Interferon-λ: Immune Functions at Barrier Surfaces and Beyond. Immunity. 2015;43:15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wack A, et al. Guarding the frontiers: the biology of type III interferons. Nat. Immunol. 2015;16:802–809. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keir ME, et al. PD-1 and Its Ligands in Tolerance and Immunity. Annu. Rev. Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed R, et al. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matloubian M, et al. Genetic basis of viral persistence: single amino acid change in the viral glycoprotein affects ability of lymphocytic choriomeningitis virus to persist in adult mice. J. Exp. Med. 1990;172:1043–1048. doi: 10.1084/jem.172.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matloubian M, et al. Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J. Virol. 1993;67:7340–9. doi: 10.1128/jvi.67.12.7340-7349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wherry EJ, et al. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 2003;77:4911–27. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sevilla N, et al. Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J. Exp. Med. 2000;192:1249–1260. doi: 10.1084/jem.192.9.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 38.Ou R, et al. Critical role for alpha/beta and gamma interferons in persistence of lymphocytic choriomeningitis virus by clonal exhaustion of cytotoxic T cells. J. Virol. 2001;75:8407–23. doi: 10.1128/JVI.75.18.8407-8423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, et al. Timing and magnitude of type i interferon responses by distinct sensors impact CD8 T cell exhaustion and chronic viral infection. Cell Host Microbe. 2012;11:631–642. doi: 10.1016/j.chom.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee MS, et al. Negative Regulation of Type I IFN Expression by OASL1 Permits Chronic Viral Infection and CD8(+) T-Cell Exhaustion. PLoS Pathog. 2013;9:e1003478. doi: 10.1371/journal.ppat.1003478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolumam Ga, et al. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 2005;202:637–50. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava S, et al. Type I interferons directly inhibit regulatory T cells to allow optimal antiviral T cell responses during acute LCMV infection. J. Exp. Med. 2014;211:961–974. doi: 10.1084/jem.20131556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teijaro JR, et al. Persistent LCMV Infection Is Controlled by Blockade of Type I Interferon Signaling. Science (80-.) 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson EB, et al. Blockade of Chronic Type I Interferon Signaling to Control Persistent LCMV Infection. Science (80-.) 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trinchieri G. Type I interferon: friend or foe? J. Exp. Med. 2010;207:2053–63. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thackray LB, et al. Murine Noroviruses Comprising a Single Genogroup Exhibit Biological Diversity despite Limited Sequence Divergence. J. Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu CC, et al. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 2006;56:247–51. [PubMed] [Google Scholar]
- 48.Hsu CC, et al. Molecular characterization of three novel murine noroviruses. Virus Genes. 2007;34:147–155. doi: 10.1007/s11262-006-0060-1. [DOI] [PubMed] [Google Scholar]
- 49.Karst SM, et al. STAT1-dependent innate immunity to a Norwalk-like virus. Science (80-.) 2003;299:1575–8. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 50.Glass RI, et al. Norovirus gastroenteritis. N. Engl. J. Med. 2009;361:1776–85. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmed SM, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strong DW, et al. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J. Virol. 2012;86:2950–8. doi: 10.1128/JVI.07038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arias A, et al. Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon. Journal of General Virology. 2012;93:1432–1441. doi: 10.1099/vir.0.042176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nice TJ, et al. A Single-Amino-Acid Change in Murine Norovirus NS1/2 Is Sufficient for Colonic Tropism and Persistence. J. Virol. 2013;87:327–334. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taube S, et al. Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. J. Virol. 2012;86:5584–93. doi: 10.1128/JVI.06854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu S, et al. Identification of Immune and Viral Correlates of Norovirus Protective Immunity through Comparative Study of Intra-Cluster Norovirus Strains. PLoS Pathog. 2013;9:e1003592. doi: 10.1371/journal.ppat.1003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niendorf S, et al. Infection with the persistent murine norovirus strain MNV-S99 suppresses IFN-Beta release and activation of stat1 In vitro. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kernbauer E, et al. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–8. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wobus CE, et al. Replication of Norovirus in Cell Culture Reveals a Tropism for Dendritic Cells and Macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chachu KA, et al. Antibody is critical for the clearance of murine norovirus infection. J. Virol. 2008;82:6610–7. doi: 10.1128/JVI.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chachu KA, et al. Immune Mechanisms Responsible for Vaccination against and Clearance of Mucosal and Lymphatic Norovirus Infection. PLoS Pathog. 2008;4:e1000236. doi: 10.1371/journal.ppat.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomov VT, et al. Persistent Enteric Murine Norovirus Infection Is Associated with Functionally Suboptimal Virus-Specific CD8 T Cell Responses. J. Virol. 2013;87:7015–7031. doi: 10.1128/JVI.03389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomov VT, et al. Differentiation and Protective Capacity of Virus-Specific CD8+ T Cells Suggest Murine Norovirus Persistence in an Immune-Privileged Enteric Niche. Immunity. 2017;0:1–16. doi: 10.1016/j.immuni.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan BM, et al. Point mutation in the glycoprotein of lymphocytic choriomeningitis virus is necessary for receptor binding, dendritic cell infection, and long-term persistence. Proc. Natl. Acad. Sci. 2011;108:2969–2974. doi: 10.1073/pnas.1019304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller SN, et al. Viral targeting of fibroblastic reticular cells contributes to immunosuppression and persistence during chronic infection. Proc. Natl. Acad. Sci. 2007;104:15430–15435. doi: 10.1073/pnas.0702579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ward VK, et al. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. 2007;104:11050–11055. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borin BN, et al. Murine norovirus protein NS1/2 aspartate to glutamate mutation, sufficient for persistence, reorients side chain of surface exposed tryptophan within a novel structured domain. Proteins. 2013;82:1200–1209. doi: 10.1002/prot.24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee S, et al. Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine. Cell Host Microbe. 2017 doi: 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hyde JL, Mackenzie JM. Subcellular localization of the MNV-1 ORF1 proteins and their potential roles in the formation of the MNV-1 replication complex. Virology. 2010;406:138–148. doi: 10.1016/j.virol.2010.06.047. [DOI] [PubMed] [Google Scholar]
- 70.Ettayebi K, Hardy ME. Norwalk Virus Nonstructural Protein p48 Forms a Complex with the SNARE Regulator VAP-A and Prevents Cell Surface Expression of Vesicular Stomatitis Virus G Protein. J. Virol. 2003;77:11790–11797. doi: 10.1128/JVI.77.21.11790-11797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCune BT, et al. Noroviruses Co-opt the Function of Host Proteins VAPA and VAPB for Replication via a Phenylalanine-Phenylalanine-Acidic-Tract-Motif Mimic in Nonstructural Viral Protein NS1/2. MBio. 2017;8:1–17. doi: 10.1128/mBio.00668-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nice TJ, et al. Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science (80-.) 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orchard RC, et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science (80-.) 2016;353:933–936. doi: 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bailey D, et al. A Single Amino Acid Substitution in the Murine Norovirus Capsid Protein Is Sufficient for Attenuation In Vivo. J. Virol. 2008;82:7725–7728. doi: 10.1128/JVI.00237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McFadden N, et al. Norovirus Regulation of the Innate Immune Response and Apoptosis Occurs via the Product of the Alternative Open Reading Frame 4. PLoS Pathog. 2011;7:e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swamy M, et al. Intestinal intraepithelial lymphocyte activation promotes innate antiviral resistance. Nat. Commun. 2015;6:7090. doi: 10.1038/ncomms8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thackray LB, et al. Critical Role for Interferon Regulatory Factor 3 (IRF-3) and IRF-7 in Type I Interferon-Mediated Control of Murine Norovirus Replication. J. Virol. 2012;86:13515–13523. doi: 10.1128/JVI.01824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang S, et al. Nondegradative Role of Atg5–Atg12/ Atg16L1 Autophagy Protein Complex in Antiviral Activity of Interferon Gamma. Cell Host & Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahlakõiv T, et al. Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections. PLOS Pathog. 2015;11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mordstein M, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010;84:5670–7. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pott J, et al. IFN- determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin J, et al. Distinct Roles of Type I and Type III Interferons in Intestinal Immunity to Homologous and Heterologous Rotavirus Infections. PLoS Pathog. 2016;12:e1005600. doi: 10.1371/journal.ppat.1005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baldridge MT, et al. Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus. J. Virol. 2017;91:e02079–16. doi: 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kane M, et al. Successful Transmission of a Retrovirus Depends on the Commensal Microbiota. Science (80-.) 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuss SK, et al. Intestinal Microbiota Promote Enteric Virus Replication and Systemic Pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uchiyama R, et al. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 2014;210:171–82. doi: 10.1093/infdis/jiu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wilks J, et al. Mammalian Lipopolysaccharide Receptors Incorporated into the Retroviral Envelope Augment Virus Transmission. Cell Host Microbe. 2015;18:456–462. doi: 10.1016/j.chom.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson CMM, et al. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15:36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baldridge MT, et al. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science (80-.) 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones MK, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science (80-.) 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu S, et al. Norovirus antagonism of B-cell antigen presentation results in impaired control of acute infection. Mucosal Immunol. 2016;9:1559–1570. doi: 10.1038/mi.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stockinger S, et al. TRIF signaling drives homeostatic intestinal epithelial antimicrobial peptide expression. J. Immunol. 2014;193:4223–34. doi: 10.4049/jimmunol.1302708. [DOI] [PubMed] [Google Scholar]
- 93.Nice TJ, et al. Type I Interferon Receptor Deficiency in Dendritic Cells Facilitates Systemic Murine Norovirus Persistence Despite Enhanced Adaptive Immunity. PLOS Pathog. 2016;12:e1005684. doi: 10.1371/journal.ppat.1005684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood. 2013;121:1951–1960. doi: 10.1182/blood-2012-09-435057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haga K, et al. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc. Natl. Acad. Sci. 2016;113:E6248–E6255. doi: 10.1073/pnas.1605575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ettayebi K, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science (80-.) 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krebs DL, Hilton DJ. SOCS: physiological suppressors of cytokine signaling. J. Cell Sci. 2000;113(Pt 1):2813–9. doi: 10.1242/jcs.113.16.2813. [DOI] [PubMed] [Google Scholar]
- 98.Meuwissen MEC, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J. Exp. Med. 2016;213:1163–1174. doi: 10.1084/jem.20151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Malakhova OA, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X, et al. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee MS, et al. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat. Immunol. 2013;14:346–55. doi: 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- 102.Hernández PP, et al. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olagnier D, Hiscott J. Type I and type III interferon-induced immune response: It’s a matter of kinetics and magnitude. Hepatology. 2014;59:1225–1228. doi: 10.1002/hep.26959. [DOI] [PubMed] [Google Scholar]
- 104.Bibert S, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J. Exp. Med. 2013;210:1109–1116. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 106.Thomas DL, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prokunina-Olsson L, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sheahan T, et al. Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe. 2014;15:190–202. doi: 10.1016/j.chom.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hong M, et al. Interferon lambda 4 expression is suppressed by the host during viral infection. J. Exp. Med. 2016;213:2539–2552. doi: 10.1084/jem.20160437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li X-D, et al. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bibert S, et al. The IFNL3/4 ΔG variant increases susceptibility to cytomegalovirus retinitis among HIV-infected patients. AIDS. 2014;28:1885–9. doi: 10.1097/QAD.0000000000000379. [DOI] [PubMed] [Google Scholar]
- 112.Manuel O, et al. Influence of IFNL3/4 Polymorphisms on the Incidence of Cytomegalovirus Infection After Solid-Organ Transplantation. J. Infect. Dis. 2014;211:906–914. doi: 10.1093/infdis/jiu557. [DOI] [PubMed] [Google Scholar]
- 113.Kourtis AP, et al. Pregnancy and Infection. N. Engl. J. Med. 2014;370:2211–2218. doi: 10.1056/NEJMra1213566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Price AA, et al. Prolonged activation of innate antiviral gene signature after childbirth is determined by IFNL3 genotype. Proc. Natl. Acad. Sci. 2016;113:10678–10683. doi: 10.1073/pnas.1602319113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pervolaraki K, et al. Type I and Type III Interferons Display Different Dependency on Mitogen-Activated Protein Kinases to Mount an Antiviral State in the Human Gut. Front. Immunol. 2017;8:459. doi: 10.3389/fimmu.2017.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saxena K, et al. A paradox of transcriptional and functional innate interferon responses of human intestinal enteroids to enteric virus infection. Proc. Natl. Acad. Sci. 2017;114:E570–E579. doi: 10.1073/pnas.1615422114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Odendall C, et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–728. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sosnovtsev SV, et al. Cleavage Map and Proteolytic Processing of the Murine Norovirus Nonstructural Polyprotein in Infected Cells. J. Virol. 2006;80:7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gonzalez-Hernandez MB, et al. Efficient Norovirus and Reovirus Replication in the Mouse Intestine Requires Microfold (M) Cells. J. Virol. 2014;88:6934–6943. doi: 10.1128/JVI.00204-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gonzalez-Hernandez MB, et al. Murine norovirus transcytosis across an in vitro polarized murine intestinal epithelial monolayer is mediated by M-like cells. J. Virol. 2013;87:12685–93. doi: 10.1128/JVI.02378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Elftman MD, et al. Multiple effects of dendritic cell depletion on murine norovirus infection. J. Gen. Virol. 2013;94:1761–1768. doi: 10.1099/vir.0.052134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bergthaler A, et al. Viral replicative capacity is the primary determinant of lymphocytic choriomeningitis virus persistence and immunosuppression. Proc. Natl. Acad. Sci. 2010;107:21641–21646. doi: 10.1073/pnas.1011998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Richter K, et al. Antigen amount dictates CD8+T-cell exhaustion during chronic viral infection irrespective of the type of antigen presenting cell. Eur. J. Immunol. 2012;42:2290–2304. doi: 10.1002/eji.201142275. [DOI] [PubMed] [Google Scholar]
- 124.Fallet B, et al. Interferon-driven deletion of antiviral B cells at the onset of chronic infection. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aah6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sammicheli S, et al. Inflammatory monocytes hinder antiviral B cell responses. Sci. Immunol. 2016;1:1–11. doi: 10.1126/sciimmunol.aah6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moseman EA, et al. Type I interferon suppresses virus-specific B cell responses by modulating CD8(+) T cell differentiation. Sci. Immunol. 2016;1:2–13. doi: 10.1126/sciimmunol.aah3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zajac AJ, et al. Viral Immune Evasion Due to Persistence of Activated T Cells Without Effector Function. J. Exp. Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 2006;12:1301–9. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Planz O, et al. A critical role for neutralizing-antibody-producing B cells, CD4(+) T cells, and interferons in persistent and acute infections of mice with lymphocytic choriomeningitis virus: implications for adoptive immunotherapy of virus carriers. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6874–9. doi: 10.1073/pnas.94.13.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thomsen AR, et al. Exhaustion of CTL memory and recrudescence of viremia in lymphocytic choriomeningitis virus-infected MHC class II-deficient mice and B cell-deficient mice. J. Immunol. 1996;157:3074–80. [PubMed] [Google Scholar]