Abstract
The biogenesis of mitochondria, chloroplasts and Gram-negative bacteria requires insertion of β-barrel proteins into the outer membranes. Homologous Omp85 proteins are essential for membrane insertion of β-barrel precursors. It is unknown if precursors are threaded through the channel interior and exit laterally or if they are translocated into the membrane at the Omp85-lipid interface. We have mapped the interaction of a precursor in transit with the mitochondrial Omp85 channel Sam50 in the native membrane environment. The precursor is translocated into the channel interior, interacts with an internal loop and inserts into the lateral gate by β-signal exchange. Transport through the Omp85 channel interior followed by release through the lateral gate into the lipid phase may represent a basic mechanism for membrane insertion of β-barrel proteins.
β-Barrel proteins are of central importance in the outer membranes of mitochondria, chloroplasts and Gram-negative bacteria. In eukaryotic cells, β-barrel proteins are essential for the communication between the double membrane-bounded organelles and the rest of the cell. β-Barrel channels mediate the translocation of a large number of metabolites and the import of organellar precursor proteins that are synthesized in the cytosol. The machineries for the biogenesis of β-barrel proteins have been identified in mitochondria and bacteria, termed sorting and assembly machinery (SAM) and β-barrel assembly machinery (BAM), respectively (1–6). The core component of the β-barrel insertion machinery is a member of the Omp85 superfamily, conserved from bacteria (BamA) to humans (Sam50/Tob55), whereas accessory BAM and SAM subunits are not conserved (1, 2, 4, 5, 7–11). The most C-terminal β-strand of each precursor serves as signal recognized by the Omp85 machinery (12, 13) and the assembly of a β-barrel protein was shown to occur from the C-terminus (14). Upon closure of the barrel, the protein is released from the assembly machinery (15).
Members of the Omp85 superfamily form 16-stranded β-barrels, including BamA/Sam50, the filamentous haemagglutinin secretion protein FhaC, and the translocation and assembly module TamA (14, 16–19). In case of FhaC, a substrate protein was shown to be translocated across the bacterial outer membrane through the interior of the β-barrel channel (20). The substrates of BamA/Sam50/TamA, however, have to be inserted into the lipid phase to become integral outer membrane proteins. High resolution structures of BamA/TamA and disulfide scanning revealed a flexible interaction of the first and last β-strand, suggesting a lateral opening of a β-barrel gate toward the membrane and a distortion of the adjacent membrane lipids (16, 18, 21–27). Different models have been discussed for the BamA/Sam50/TamA-mediated insertion of β-barrel precursors into the outer membrane (5, 15, 16, 18, 21–38). In the BamA/Sam50-assisted model, the precursor is inserted at the protein-lipid interface; BamA/Sam50 creates a distortion and thinning of the membrane that favors spontaneous insertion of the precursor into the membrane. In the BamA/Sam50-budding model, the precursor is threaded through the β-barrel interior of BamA/Sam50 and laterally released via an opened lateral gate. The BamA structures, which were obtained in non-native environments and in the absence of precursor proteins (35), supported arguments for both models (16, 21–26) and thus the mechanism of β-barrel translocation via BAM/SAM is unknown.
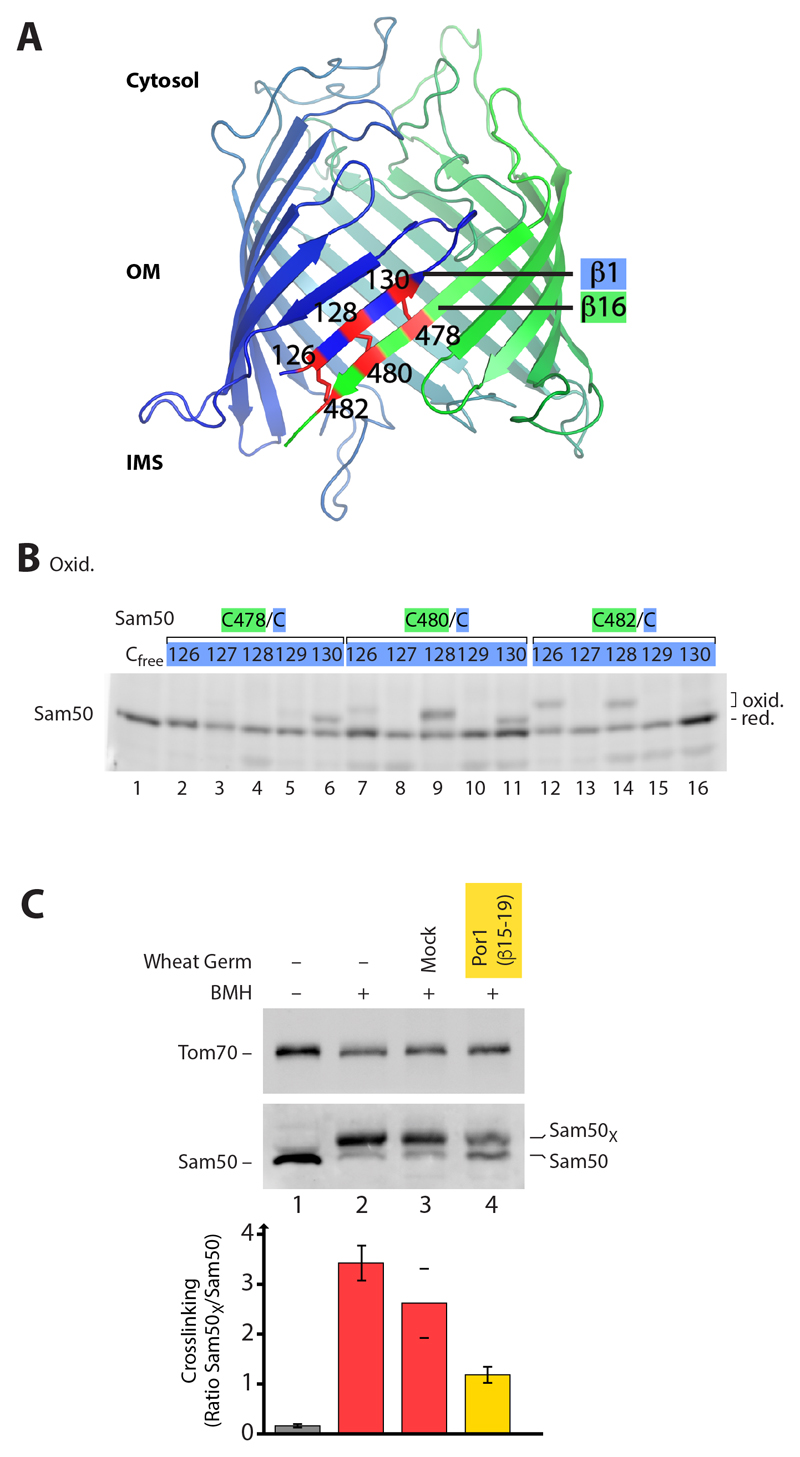
Lateral gate of the Sam50 β-barrel in the mitochondrial outer membrane
We developed a system to map the interaction of Sam50 with β-barrel precursors in transit in the native mitochondrial membrane environment. The β-barrel channel of Sam50 was modeled based on the BamA structures and cysteine/disulfide-scanning of β-strands 1 and 16 (Fig. 1, A and B, and fig. S1, A to C) (39, 40). In the absence of precursor proteins, β-strands 1 and 16 interacted, i.e. the putative lateral gate was closed (Fig. 1B and fig. S1C) (31). However, oxidation-induced disulfide formation between distinct cysteines also revealed a sliding of β-strands 1 and 16, i.e. a dynamic behavior of the gate (27). To probe for possible opening of the gate in the presence of substrate, we tested β-barrel precursors that contained the β-hairpin mitochondrial targeting signal (6) and imported them into isolated intact mitochondria, followed by position-specific SH-crosslinking of β-strands 1 and 16. The crosslinking reagent bismaleimidohexane (BMH) showed a high efficiency for stably linking strands 1 and 16 in the absence of substrate (Fig. 1C, lane 2, and fig. S1C). A C-terminal fragment of the major mitochondrial β-barrel protein Porin/VDAC (Por1), including the Por1 β-signal, considerably disturbed the interaction of Sam50 β-strands 1 and 16 (Fig. 1C, lane 4), indicating that the Por1 substrate interfered with gate closing.
Fig. 1. Intramolecular interaction between the first and last β-strand of Sam50.
(A) Model of the Sam50 β-barrel. Engineered disulfide bonds between the first and last β-strand of Sam50 are indicated in red. Additional disulfide bonds are possible due to the dynamic interaction of β-strands 1 and 16 (27). IMS, intermembrane space; OM, outer membrane. (B) Yeast strains expressing cysteine-free Sam50 (Cfree) and Sam50 cysteine variants (containing exactly two cysteine residues as indicated) were treated in vivo with the oxidant 4,4′-Dipyridyl disulfide (4-DPS), followed by non-reducing SDS-PAGE, Western blotting and immunodecoration. (C) Isolated Sam50C128/C480 mitochondria were incubated with a Por1 precursor construct (β-strands 15-19 corresponding to amino acid residues 210-283) and controls as indicated. Samples were treated with the crosslinker BMH and analyzed as in (B). Quantification of crosslinking efficiency (mean ± SEM, N = 3 for samples 1, 2 and 4; mean with range, N = 2 for sample 3). Sam50X, crosslinked Sam50.
β-Signal exchange in the lateral gate and release of the full-length β-barrel precursor
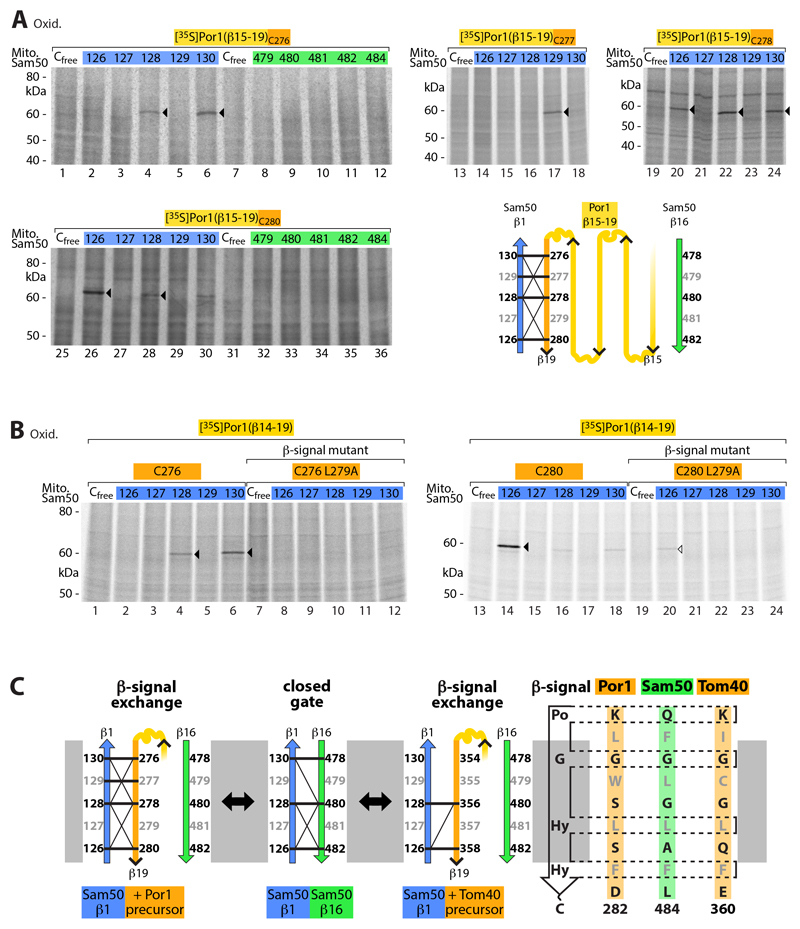
It has been speculated that the β-signal may be specifically recognized by BamA/Sam50 via exchange of the endogenous BamA/Sam50 β-signal (31, 33), yet experimental demonstration has been lacking (35). β-Strand 16 of BamA/Sam50 functions as β-signal and thus in the exchange model the β-signal of the precursor, corresponding to the C-terminal β-strand 19 of Por1, should interact with Sam50-β1. To test this hypothesis, we synthesized a 35S-labeled Por1 substrate carrying a single cysteine residue at distinct positions of the β-signal. After import into mitochondria containing Sam50 with a single cysteine residue at different positions in β-strands 1 or 16, we probed the proximity of the β-strands by disulfide formation. The Por1 β-signal indeed specifically aligned with Sam50-β1 such that residues predicted to point toward either the channel interior (black) or the lipid phase (gray) selectively interacted (Fig. 2A and fig. S2A).
Fig. 2. Interaction of Sam50 β-strand 1 with the C-terminal β-signal of precursor proteins.
(A) Radiolabeled Por1(β15-19) precursors containing one cysteine at the positions indicated were imported for 5 min into mitochondria isolated from yeast strains expressing Sam50 with the indicated cysteine residues, followed by oxidation with 4-DPS (lanes 1-12, 19-36) or CuSO4 (lanes 13-18). Samples were analyzed by non-reducing SDS-PAGE and autoradiography. Arrowheads, disulfide-bonded Sam50-Por1(β15-19) adducts. Schematic model, disulfide bond formation of Sam50 β-strand 1 with the β-signal (β19) of the Porin precursor β15-19; thick and thin lines indicate strong and weak formation of Sam50-Por1 adducts, respectively. (B) [35S]Por1(β14-19)C276, [35S]Por1(β14-19)C280 and the corresponding β-signal mutants (L279A) were incubated for 5 min with isolated mitochondria of Sam50 cysteine variants followed by oxidation with 4-DPS, non-reducing SDS-PAGE and autoradiography. Arrowheads, cysteine-specific Sam50-precursor adducts. (C) Schematic model illustrating the β-signal exchange observed in Fig. 2 and fig. S2.
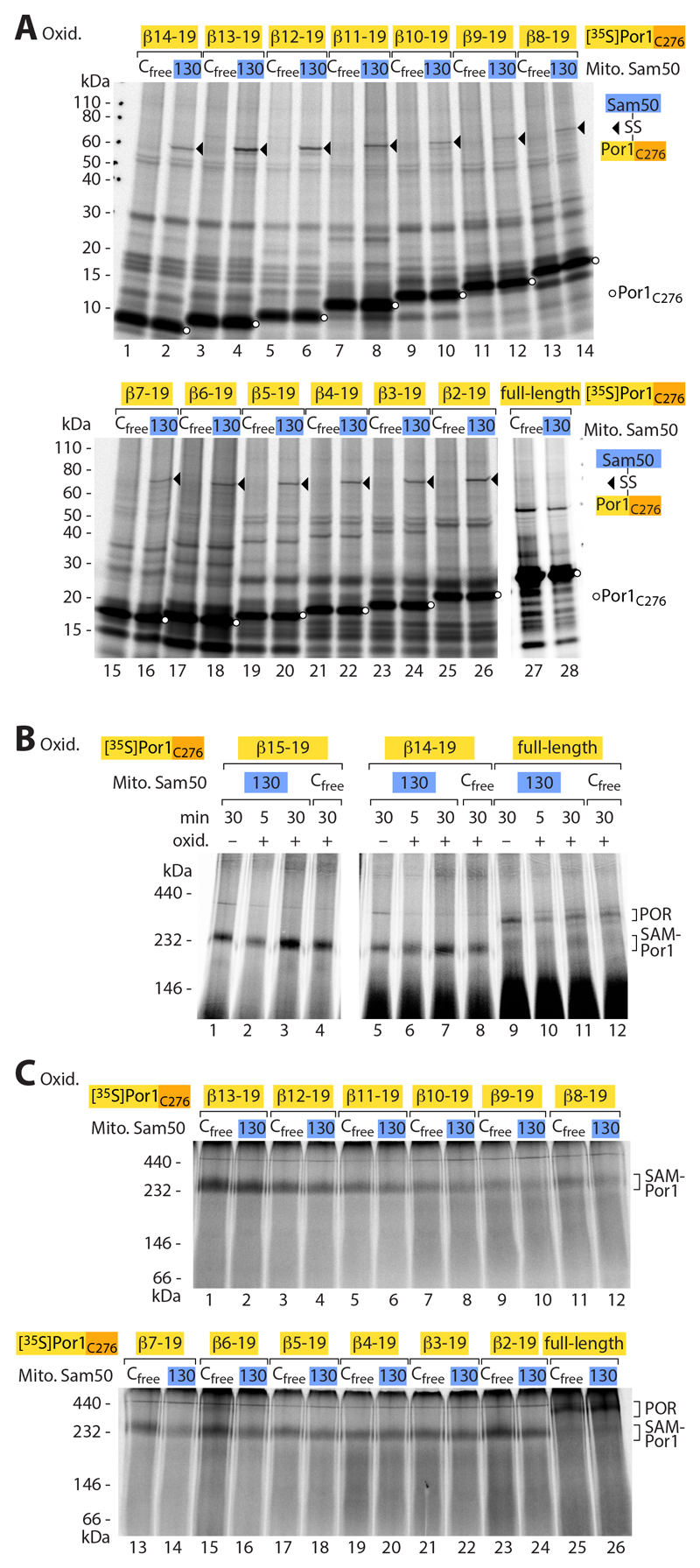
We performed several control experiments. (i) The Por1 β-signal selectively interacted with Sam50-β1, but not with Sam50-β16 (Fig. 2A and fig. S2A). (ii) To test a different β-signal, we imported a 35S-labeled C-terminal precursor of the mitochondrial import channel Tom40 and observed a comparable pairing with Sam50-β1 (fig. S2B). (iii) A precursor containing a mutant form of the Por1 β-signal (replacement of a conserved hydrophobic residue (13, 41) was strongly impaired in the interaction with Sam50-β1 (Fig. 2B). These results show that the β-signal of precursors specifically interacts with Sam50-β1 (Fig. 2C). (iv) We analyzed substrates of different size, covering the range from 5 to 18 β-strands, and observed disulfide formation between the Por1 β-signal and Sam50-β1 in each case (Fig. 2A, Fig. 3A and fig. S2A). (iv) Co-migration of the differently sized Por1 β-barrel precursors with the SAM complex observed by blue native gel analysis (1, 3, 8, 9, 13) showed that each substrate accumulated at the SAM complex (Fig. 3, B and C). (v) Only the full-length Por1 precursor, corresponding to 19 β-strands, was released from the SAM complex and assembled into the mature Porin complex (Fig. 3, B and C) (42–45).
Fig. 3. Release of full-length β-barrel precursor from the SAM complex.
(A) [35S]Por1C276 constructs of different length were incubated with isolated Sam50Cfree or Sam50C130 mitochondria for 30 min, followed by oxidation with 4-DPS, analysis by non-reducing SDS-PAGE and autoradiography. Lanes 27 and 28, shorter exposure to compensate for strong intensity of full-length Por1. Arrowheads, disulfide-bonded Sam50-precursor adducts; circles, Por1C276 precursor. (B and C) Samples as described for (A) were analyzed by blue native electrophoresis and autoradiography. Lanes 1–4 vs. lanes 5–12 of (B), linear adjustment of brightness and contrast to compensate for strong intensity of Por1(β15-19)C276. POR, assembled Porin complex; SAM-Por1, Porin1 precursor at SAM.
Taken together, we conclude that the β-signal of the precursor is bound by Sam50-β1 via exchange with the endogenous Sam50 β-signal (β16) (Fig. 2C). Porin precursors up to 18 β-strands accumulate at the SAM complex and only the full-size precursor is released into the lipid phase of the outer membrane.
β-Barrel precursors interact with both sides of the Sam50 gate
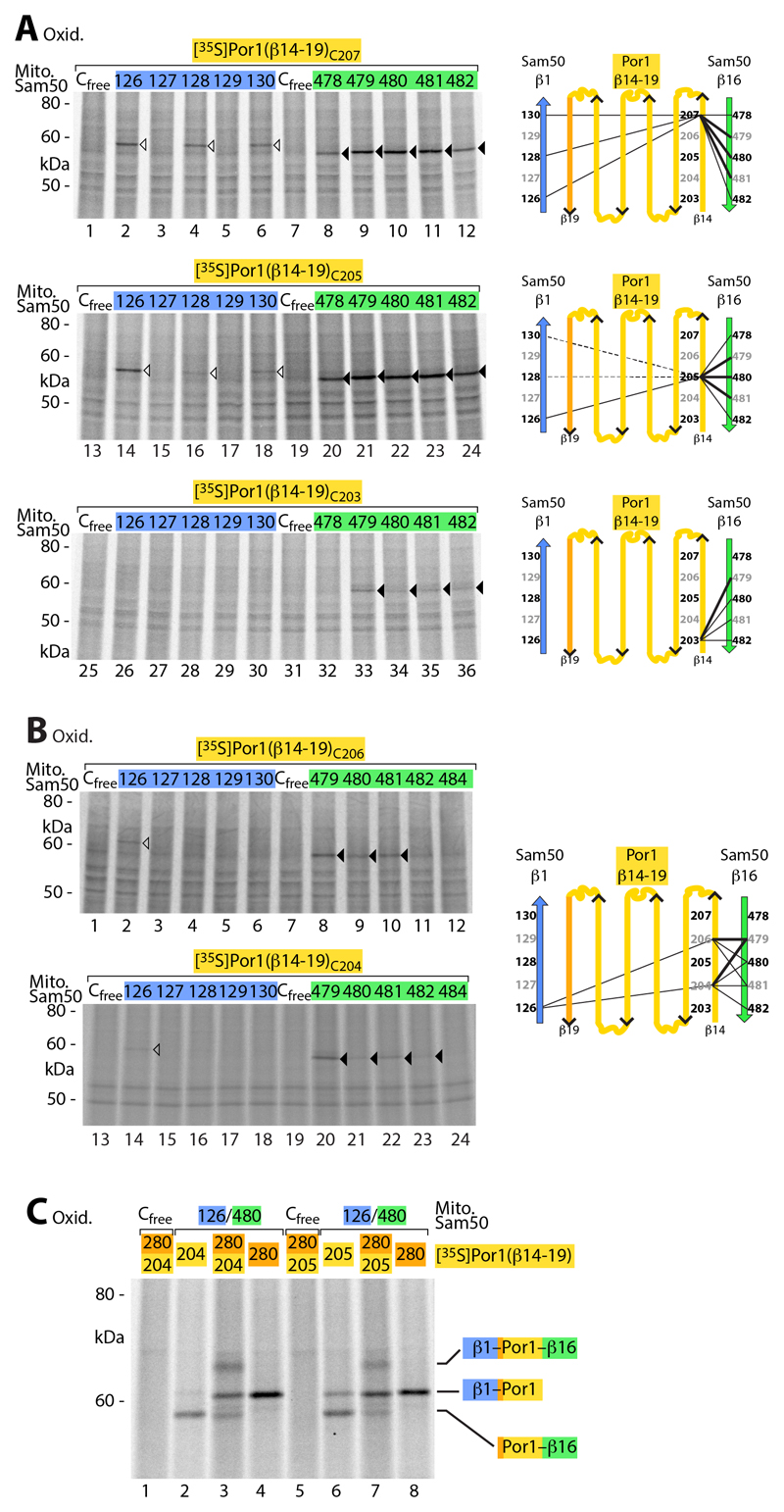
We asked if the substrate also interacted with β-strand 16 of Sam50 and performed disulfide scanning between this β-strand and the N-terminal region of the precursor, corresponding to β-strand 14 of mature Por1. We tested five distinct amino acid positions corresponding to Por1-β14 and observed disulfide formation with Sam50-β16 in each case (Fig. 4, A and B). However, the interaction showed a considerably higher flexibility than that of the β-signal of the precursor with Sam50-β1 (Fig. 2 and fig. S2). A Por1 precursor with a mutant β-signal strongly inhibited the interaction of the N-terminal precursor region with Sam50-β16 (fig. S3). Since the β-signal itself did not interact with Sam50-β16, this finding indicates that the specific binding of the β-signal to Sam50-β1 is a prerequisite for the accumulation of the N-terminal precursor region at Sam50-β16. To provide further evidence that the precursor was intercalated between β-strands 1 and 16 of Sam50, we studied if it interacted with both strands simultaneously. Por1 precursors containing two cysteine residues, one in the C-terminal β-signal and one in the N-terminal region, were accumulated at Sam50, carrying a cysteine residue in β1 as well as in β16, and subjected to oxidation. In addition to the single disulfides formed (like in Fig. 2, A and B, and Fig. 4, A and B), we observed the formation of two disulfides simultaneously (Fig. 4C, lanes 3 and 7).
Fig. 4. Interaction of Sam50 with the N-terminal β-strand of precursor proteins.
(A) [35S]Por1(β14-19) precursors containing a single cysteine residue as indicated were imported into mitochondria isolated from yeast strains expressing the indicated Sam50 cysteine variants, followed by oxidation with 4-DPS, non-reducing SDS-PAGE and autoradiography. Black and white arrowheads, cysteine-specific disulfide-bonded Por1(β14-19) adducts to the C-terminal and N-terminal β-strand of Sam50, respectively. Right, schematic models. (B) [35S]Por1(β14-19)C206 and [35S]Por1(β14-19)C204 were treated as described in (A). (C) [35S]Por1(β14-19) single and double cysteine variants were incubated with isolated mitochondria from yeast strains expressing Sam50Cfree or the double cysteine variant Sam50C126/C480, followed by oxidation with 4-DPS. Samples were analyzed as described in (A).
Our results indicate that β-barrel precursors are inserted into a Sam50 gate formed between β-strands 1 and 16. The C-terminal β-signal specifically exchanges with Sam50-β1, whereas the N-terminal region of the precursor undergoes a flexible interaction with Sam50-β16.
Translocation of β-barrel precursors into the Sam50 channel
The N-terminal region of the precursor (residues 204 to 207) was also found in close proximity to the first residue (126) of Sam50-β1 (Fig. 4, A and B). Sam50res126 is positioned at the intermembrane space opening of the Sam50 channel and predicted to point toward the channel interior (Fig. 1A). Por1res207, which is located toward the cytosolic side of mature Por1 (42–44), was not only found in proximity of Sam50res126 but also of further residues of Sam50-β1 predicted to face the channel interior (residues 128 and 130) (Fig. 4A and fig. S3). Disulfide formation between the N-terminal region of Por1 and Sam50-β1 was impaired when the Por1 β-signal was mutated (fig. S3). Thus, a functional C-terminal β-signal is a prerequisite for the observed proximity of the N-terminal precursor region with Sam50-β1 (pairing between Sam50-β1 and the β-signal involves hydrogen bonds of the polypeptide backbone and thus cysteine side chains are available for disulfide formation). These findings are compatible with a model that upon binding of the β-signal to Sam50-β1, the N-terminal region of the precursor is passing at the interior of Sam50-β1.
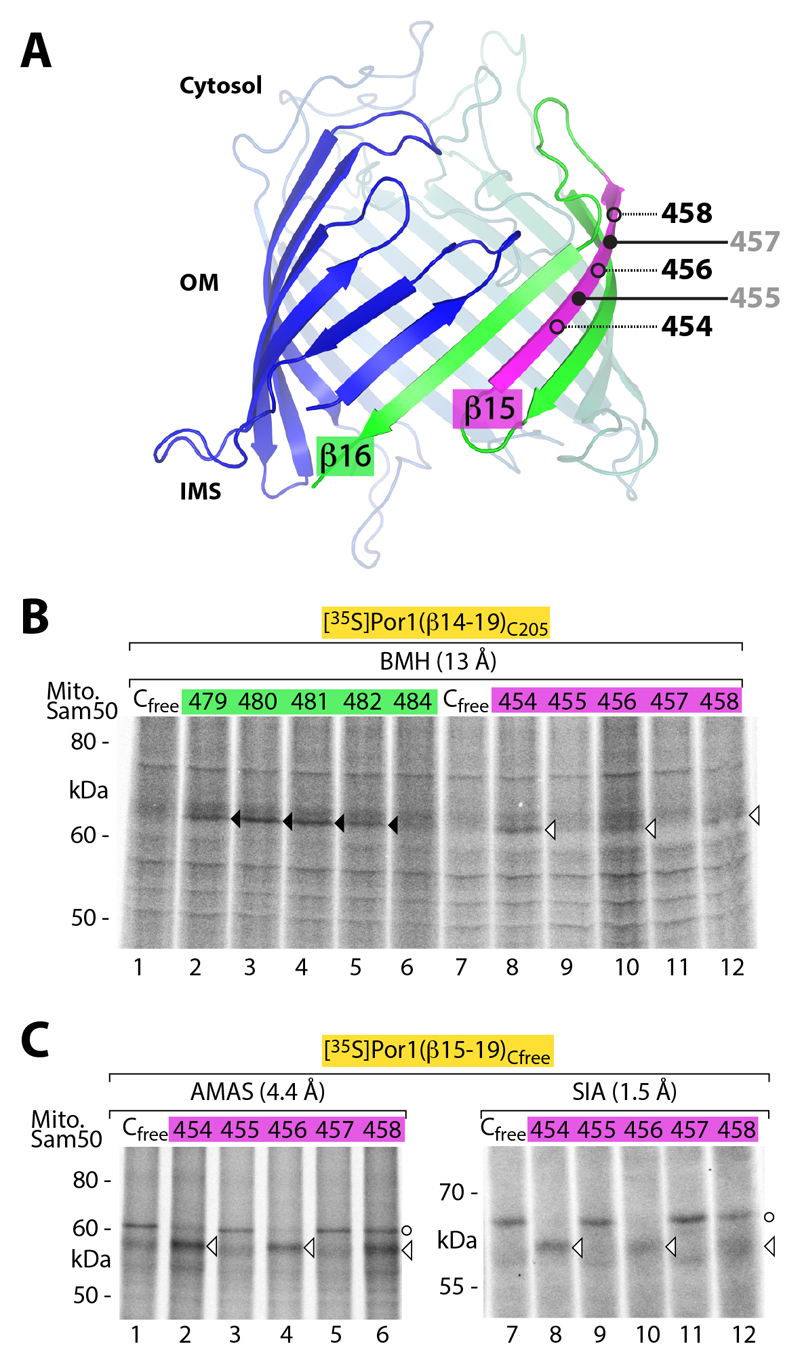
To obtain independent evidence that β-barrel precursors are using the interior of the Sam50 channel, we analyzed Sam50 β-strand 15 and compared residues predicted to face either the channel interior (black) or the lipid phase (gray) (Fig. 5A). A 35S-labeled Por1 precursor with a single cysteine residue in the N-terminal region (residue 205) was imported into Sam50 containing a single cysteine at different positions of either β-strand 15 or 16. In contrast to Sam50-β16, we did not observe disulfide formation between the precursor and Sam50-β15 upon oxidation (fig. S4), indicating that Por1res205 was not so close to Sam50-β15 to promote disulfide formation. Using SH-specific BMH, the precursor was crosslinked to Sam50-β15 and β16. Whereas the crosslinking occurred to various residues of Sam50-β16 (comparable to the oxidation assay), only residues of Sam50-β15 predicted to face the channel interior were crosslinked to the precursor (Fig. 5B). To probe further regions of the precursor, we used the short amine-to-sulfhydryl crosslinking reagents N-α-maleimidoacet-oxysuccinimide ester (AMAS) and succinimidyl iodoacetate (SIA) together with a cysteine-free Por1 precursor and Sam50 containing a single cysteine residue in β15. Cysteine-specific crosslinking occurred only to Sam50-β15 residues predicted to face the channel interior (Fig. 5C, arrowheads) (a larger non-specific band at 60 kDa was formed when no SH-group was available, i.e. also with cysteine-free Sam50). These results are fully compatible with the model that transfer of the Por1 precursor involves the interior of the Sam50 channel, but do not fit to a model in which the Por1 precursor is inserted at the protein-lipid interphase without getting access to the channel.
Fig. 5. Interaction of β-barrel precursor with Sam50 residues facing the channel interior.
(A) Model of the Sam50 β-barrel. β-Strand 15, purple; open and filled circles, residues facing the interior of the barrel (black) or the lipid phase (gray), respectively. (B and C) Radiolabeled Por1 precursor variants were imported into mitochondria of yeast strains expressing the indicated Sam50 variants. Samples were crosslinked with BMH, AMAS or SIA and analyzed by non-reducing SDS-PAGE and autoradiography. Black and white arrowheads, cysteine-specific precursor adducts to Sam50-β16 or Sam50-β15, respectively; circles, unspecific/cysteine-independent adducts (in the absence of free SH-groups, AMAS and SIA can react with other amino acids).
Sam50 loop 6 is required for β-signal binding
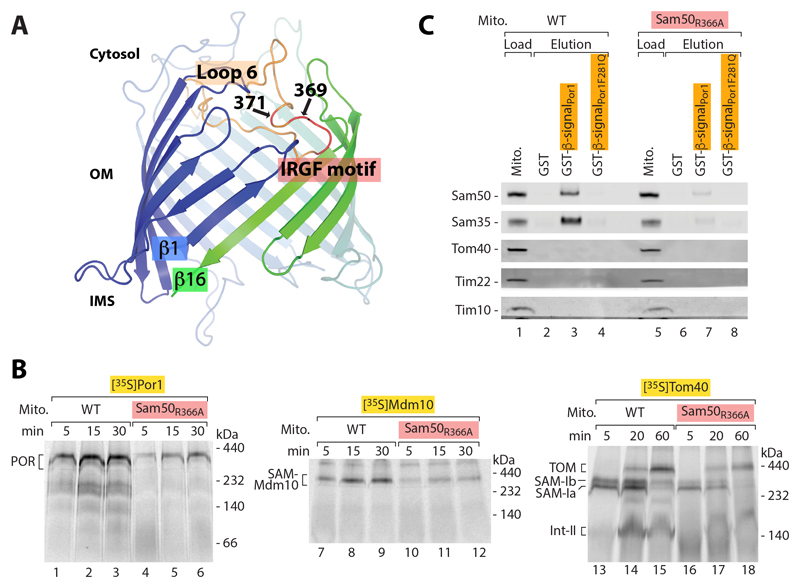
In addition to the β-barrel channel, Sam50 possesses two major characteristic elements, an N-terminal polypeptide transport associated (POTRA) domain exposed to the intermembrane space and a highly conserved loop 6 that extends from the cytosolic side of the β-barrel. (i) Whereas bacterial BamA proteins contain several POTRA domains that interact with β-barrel precursors and are crucial for precursor transfer from the periplasm into the outer membrane (17, 46–49), Sam50 contains a single POTRA domain that is not essential for cell viability (13, 50, 51). Disulfide formation between the Por1 precursor and Sam50 β-strands 1 and 16 was not blocked in mitochondria lacking the entire POTRA domain (fig. S5). Together with blue native gel analysis (13, 45), this result indicates that the single POTRA domain is not crucial for precursor transfer to Sam50. (ii) Loop 6 extends from the outside/cytosolic side into the channel interior in all Omp85 high resolution structures analyzed (Fig. 6A) (16, 18, 21–25, 52). Deletion of Sam50 loop 6 was lethal to yeast cells. When wild-type Sam50 was depleted, expression of a Sam50 mutant form lacking the conserved segment of loop 6 did not rescue growth and led to strong defects of the import of 35S-labeled β-barrel precursors such as Por1 and Tom40 into mitochondria (fig. S6, A and B). The steady-state levels of β-barrel proteins and various Tom proteins were decreased (fig. S6C). As the TOM complex imports a large number of precursor proteins, this mutant did not permit a selective analysis of the function of loop 6. We thus generated point mutants of the conserved IRGF motif of loop 6 (53, 54). Sam50R366A yeast exhibited a temperature-sensitive growth phenotype on non-fermentable medium (fig. S7A). Mitochondria isolated upon growth of the mutant cells on permissive temperature showed normal steady-state levels of SAM, TOM and further control proteins (fig. S7, B and C). The import of 35S-labeled β-barrel precursors such as Por1, Mdm10 and Tom40 was strongly inhibited (Fig. 6B), whereas the import of matrix-targeted and intermembrane-space-targeted precursors, which depend on the TOM complex but not on SAM, was not or only mildly affected (fig. S7D). The import of [35S]Tom40 can be dissected into distinct stages by blue native gel analysis (1, 3, 8, 9). Sam50R366A mitochondria were impaired in the formation of SAM-bound intermediates (Fig. 6B). We conclude that loop 6 of Sam50 is required for a stable interaction of the precursor with SAM. It has been reported that both Sam50 and Sam35 are needed for binding of a β-barrel precursor to the SAM complex (13). To directly test the contribution of loop 6, we performed affinity purification from lysed mitochondria using a purified β-signal-fusion protein, leading to the co-purification of Sam50 and Sam35 from wild-type mitochondria; a mutant β-signal did not pull down Sam50-Sam35 (Fig. 6C) (13). The interaction of Sam50-Sam35 with the β-signal was strongly disturbed in Sam50R366A mitochondria (Fig. 6C), demonstrating that loop 6 is required for stable precursor binding to Sam50-Sam35.
Fig. 6. Loop 6 of Sam50 is essential for β-barrel biogenesis.
(A) Model of the Sam50 β-barrel indicating loop 6 in peach and the conserved IRGF motif at the tip of loop 6 in red. The positions of residues 369 and 371 used for crosslinking in Fig. 7 are indicated. (B) Assembly of full-length β-barrel precursor proteins [35S]Por1, [35S]Mdm10 and [35S]Tom40 in wild-type (WT) and Sam50R366A mitochondria was analyzed by blue native electrophoresis and autoradiography. POR, Porin complex; SAM-Mdm10, SAM-Mdm10 complex; SAM-Ia, SAM-Ib and Int-II, assembly intermediates of Tom40. (C) Immobilized glutathione-S-transferase (GST)-fusion proteins carrying the Por1 β-signal were incubated with digitonin-lysed WT and Sam50R366A mitochondria. The β-signal was released by thrombin protease cleavage and eluates were analyzed by SDS-PAGE and immunodecoration. Load 12.5%; elution 100%.
β-Hairpin-like transport of precursor proteins by Sam50
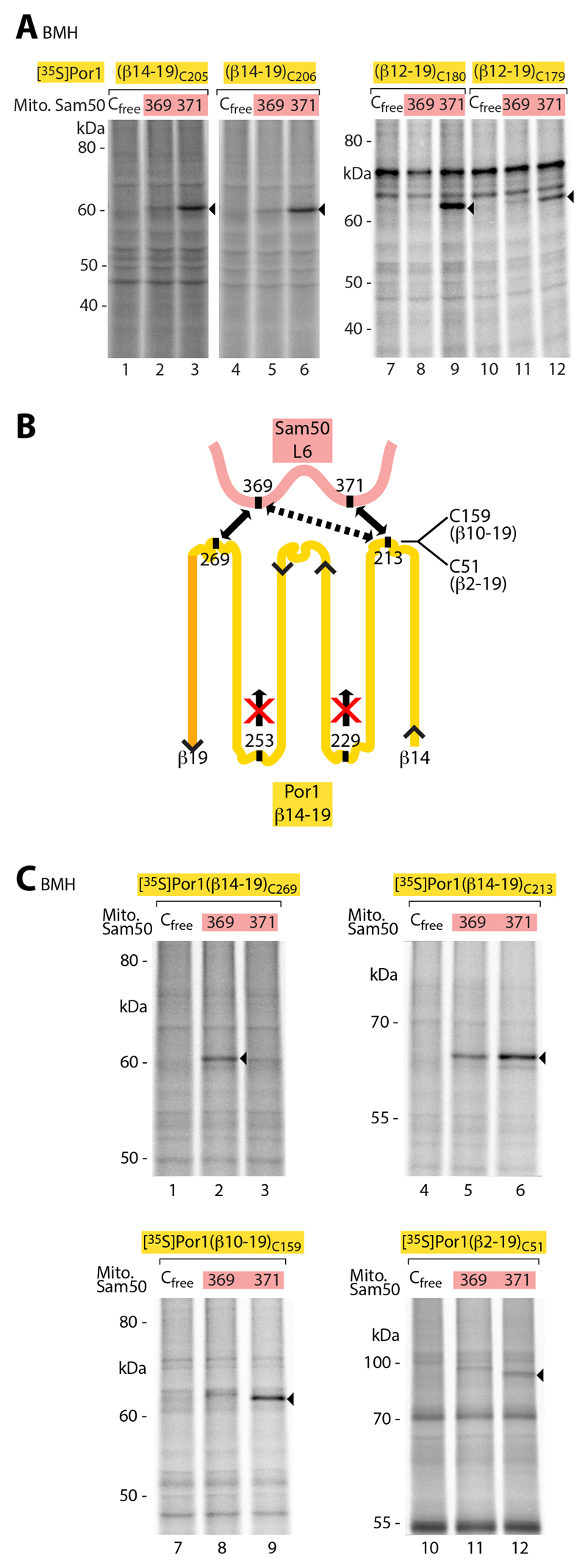
To determine if a precursor in transit was in proximity to loop 6, 35S-labeled Por1 precursors with a single cysteine residue in the N-terminal region were imported into mitochondria containing Sam50 with a single cysteine residue in loop 6. By SH-specific crosslinking, the precursors were linked to residue 371 of loop 6 (Fig. 7A). A mutant β-signal prevented crosslinking of the N-terminal precursor region to loop 6 (fig. S8A), whereas the β-signal itself was not found in proximity of loop 6 (fig. S8B, lanes 1-6), supporting our conclusion that a functional β-signal is a prerequisite for further translocation steps of the precursor. It has been suggested that β-barrel precursors transported by SAM/BAM may be partially folded such that β-hairpins consisting of two adjacent β-strands are formed (35, 55). We used distinct approaches to assess this view. (i) Using precursors of different length, covering 5, 6, 7 or 8 β-strands of mature Por1, only precursors corresponding to an even number of β-strands were crosslinked to loop 6 (Fig. 7A and fig. S8B, lanes 7-30). (ii) We analyzed an internal precursor region that corresponds to a β-hairpin in mature Por1 by inserting a pair of cysteine residues at the putative adjacent β-strands and a tobacco etch virus (TEV) protease cleavage site at the predicted loop between the β-strands. Upon import of the [35S]precursor into mitochondria and lysis, TEV protease cleaved the precursor into two fragments (fig. S9A). When SH-specific crosslinking was performed before lysis, the fragments were not separated, demonstrating that the corresponding cysteines of the predicted adjacent β-strands were indeed in close, hairpin-like proximity. (iii) We inserted single cysteine residues into precursor regions that correspond to cytosolic loops or intermembrane space-exposed turns of mature Por1 and imported them into mitochondria containing a single cysteine in Sam50-loop 6 (summarized in Fig. 7B). The predicted most C-terminal precursor loop was crosslinked to residue 369 of Sam50-loop 6, whereas the predicted most N-terminal precursor loop was preferentially crosslinked to residue 371 (Fig. 7C and fig. S9B; precursors of different length and SH-specific crosslinkers with different spacer length yielded a comparable pattern). Cysteines inserted into the predicted precursor turns were not crosslinked to Sam50 loop 6 (Fig. 7B and fig. S9C). (iv) The specific pairing of the C-terminal β-signal of the precursor with Sam50-β1 (Fig. 2 and fig. S2) indicates that the β-signal is likely in a β-strand conformation. These results suggest that β-precursors interacting with Sam50 are not in a random conformation, but are partially folded and contain β-hairpin-like elements.
Fig. 7. β-Barrel precursors in transit are in close proximity to Sam50 loop 6.
(A) [35S]Por1(β14-19) precursors carrying a cysteine in β-strand 14 at position 205 or 206 and [35S]Por1(β12-19) precursors carrying a cysteine in β-strand 12 at position 180 or 179 were imported for 5 min into isolated mitochondria containing Sam50 variants, followed by crosslinking with BMH. Samples were analyzed by non-reducing SDS-PAGE and autoradiography. Arrowheads, cysteine-specific Sam50-Por1 precursor crosslinking products. (B) Schematic model summarizing the crosslinking results of Fig 7C and fig. S9, B and C. (C) Radiolabeled Por1 constructs were imported into mitochondria of yeast strains expressing the indicated Sam50 variants for 5 min. Samples were treated with BMH and analyzed as described in (A).
Taken together, loop 6 of Sam50 is in proximity of the precursor in transit and plays a crucial role in β-barrel biogenesis. Thus, in contrast to the POTRA domain, the functional importance of loop 6 in precursor transfer has been conserved from the bacterial Omp85 proteins FhaC and BamA (53, 54, 56) to Sam50. The analysis of precursor interaction with Sam50 supports the view that precursor insertion involves β-hairpin-like conformations.
Discussion
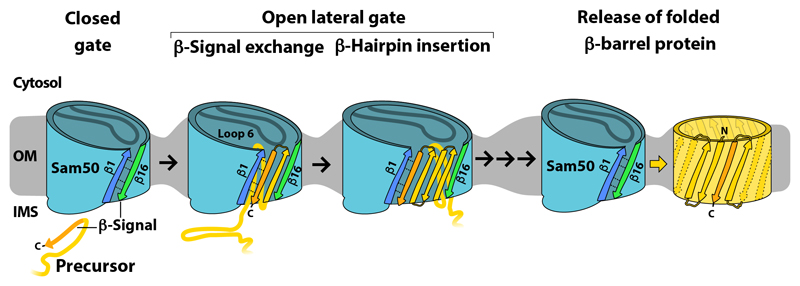
We conclude that the biogenesis of mitochondrial β-barrel precursors involves the gate formed by the first and last β-strands of Sam50. The analysis in the native mitochondrial system provides strong evidence for both the exchange model of β-signal recognition and the lateral release model of precursor exit through the Sam50 β-barrel gate (31, 33, 35, 36). Our findings suggest the following translocation path of a mitochondrial β-barrel precursor through SAM (Fig. 8). The precursor enters the interior of the Sam50 channel from the intermembrane space side in close proximity to Sam50 β-strand 1. The C-terminal β-signal of the precursor is specifically bound to Sam50-β1 by exchange with the endogenous Sam50 β-signal (Sam50-β16), leading to an opening of the lateral gate. The conserved loop 6 of Sam50 is involved in precursor transfer to the lateral gate. More and more N-terminal portions of the precursor are threaded through the gate in close proximity to Sam50-β16. Upon translocation of the entire precursor polypeptide chain by Sam50, the full-length β-barrel can be formed and released from the SAM complex (13).
Fig. 8. Putative model for sorting of β-barrel precursors through the lateral gate of Sam50.
β-Barrel precursors are translocated from the intermembrane space (IMS) side into the lumen of Sam50. The C-terminal β-signal of the precursor specifically binds to β-strand 1 of Sam50 by replacing the endogenous β-signal of Sam50 (β-strand 16). This induces an opening of the lateral gate of Sam50 between β-strands 1 and 16. Further strands of the precursor are inserted into the lateral gate in β-hairpin-like structures. Loop 6 of Sam50 promotes transfer of the precursor into the lateral gate. The full-length precursor is released from the lateral gate into the lipid phase of the outer membrane (OM). Thinning of the membrane in proximity to the lateral gate facilitates membrane insertion of the β-barrel protein.
When comparing mitochondrial and bacterial β-barrel biogenesis, the pathways start in different locations (eukaryotic vs. bacterial cytosol) and converge at the central Sam50/BamA β-barrel. Three main stages can be distinguished. (i) Initial translocation into the intermembrane space/periplasm is mediated by non-related translocases: the TOM complex of the mitochondrial outer membrane and the Sec complex of the bacterial plasma membrane (5, 6). (ii) Subsequent precursor transfer to the outer membrane is performed by in part related machineries, including intermembrane space/periplasmic chaperones and POTRA domains (46–49, 57–59). The bacterial transfer machinery is considerably more complex than that of mitochondria, likely reflecting the large number of bacterial β-barrel substrates (60). Bacteria use multiple POTRA domains and several periplasm-exposed Bam proteins (5, 15), whereas mitochondria contain a single non-essential POTRA domain and no accessory intermembrane space-exposed proteins (13, 50). The two cytosol-exposed peripheral Sam proteins are involved in formation of a TOM-SAM supercomplex (Sam37) and stabilization of the SAM-bound form of the precursor (Sam35) (9–11, 13, 39, 41). (iii) Finally, the membrane insertion process occurs via the highly conserved membrane-integral part of Sam50/BamA. The β-signal has been well conserved and several examples were reported that the β-signal is exchangeable between bacteria, mitochondria and chloroplasts (12, 13, 61), underscoring the conservation of basic mechanisms of β-barrel biogenesis. β-Barrel proteins are anchored in the lipid phase by a hydrophobic belt; the diminished hydrophobic area near the Sam50/BamA lateral gate is thought to cause a membrane thinning (16, 21). In vitro studies on β-barrel membrane protein insertion demonstrate that membrane defects and BamA mediated membrane distortion support membrane insertion (62–64). Sam50/BamA induced membrane thinning may contribute to β-barrel membrane protein biogenesis in vivo by facilitating protein membrane insertion upon release from the SAM/BAM lateral gate. We propose that elements of both controversially discussed mechanisms, budding model and assisted model, will be employed in the lateral gate sorting mechanism shown here.
The large diversity of bacterial β-barrel proteins and the involvement of multiple POTRA domains and accessory Bam proteins (5, 15, 51, 60) raise the possibility that additional precursor-specific folding pathways may complement the central mechanism of β-signal exchange and sorting via the lateral gate elucidated here. For example assembly of oligomeric β-barrels in bacteria might be stalled at the BAM complex until all subunits are assembled (65), similar to the arrest of shortened precursor constructs of monomeric β-barrels (Fig. 3). We envision that precursor insertion through the β-barrel channel and lateral gate demonstrated with mitochondrial Sam50 represents a basic mechanism that can also be employed by β-barrel assembly machineries of bacteria and chloroplasts.
Materials and methods
Site-directed mutagenesis
Mutagenesis was performed using the centromeric plasmid pFL39 (66) containing the wild-type open reading frame of Saccharomyces cerevisiae SAM50, TOM40 or POR1 and their corresponding native promoter and terminator sequences (Table S1). Primers listed in Table S2, containing the specific mutational changes, were used for PCR with the high fidelity polymerases KOD (Sigma-Aldrich) or Q5 (NEB). After DpnI (NEB) template digestion (3 h at 37°C), PCR products were transformed into competent XL-1 Blue Escherichia coli cells (Stratagene). Plasmids were isolated by using the QIAprep Spin Miniprep Kit (Qiagene). Successful mutagenesis was confirmed by sequencing.
Yeast strains and growth conditions
Since SAM50 is an essential gene, the plasmid shuffling method was used to exchange SAM50 wild-type with mutated versions of sam50 in a YPH499 background (67). The shuffling strain sam50Δ contains a chromosomal deletion of SAM50 and expresses a wild-type copy of SAM50 on a YEp352 plasmid with a URA3 marker (7). After transformation of the centromeric TRP1 plasmid pFL39 containing a mutated sam50 allele, positive clones were selected on medium lacking tryptophan. By growth on plates containing 5-fluoroorotic acid (5-FOA) (Melford), cells that lost the URA3 plasmid expressing wild-type SAM50 were selected. Subsequently, yeast cells were grown on non-fermentable medium containing glycerol to rule out the loss of mitochondrial DNA. At each step, plates were incubated at 23°C to minimize possible temperature sensitive growth defects.
Yeast cells were cultured in liquid YPG medium (1% [w/v] yeast extract (Becton Dickinson), 2% [w/v] bacto peptone (Becton Dickinson), 3% [w/v] glycerol (Sigma), pH 5 - HCl (Roth)) at 23°C and shaking with 130 rpm. For growth tests, single yeast cells were picked and incubated overnight in 5 ml YPG. Cells corresponding to an OD600 of 1 were taken from yeast strains indicated and resuspended in 1 ml autoclaved and distilled H2O. The suspension was further diluted by factors of 1:10, 1:100, 1:1,000 and 1:10,000. 3 or 5 µl were dropped on solid YPG (1% [w/v] yeast extract, 2% [w/v] bacto peptone, 3% [w/v] glycerol, 2.5% [w/v] agar (Becton Dickinson)) and YPD (1% [w/v] yeast extract, 2% [w/v] bacto peptone, 2% [w/v] glucose (Roth), 2.5% [w/v] agar). Plates were incubated at indicated temperatures.
Yeast cells expressing Sam50 lacking loop 6 (sam50Δloop6) did not yield colonies after plasmid shuffling. Therefore, the plasmid encoding Sam50Δloop6 was transformed into a YPH499 strain expressing SAM50 under the control of a galactose promoter. After selection on galactose (Sigma-Aldrich) containing medium lacking tryptophan, the shutdown of SAM50 wild-type was performed by growth in liquid SL-medium (0.3% [w/v] yeast nitrogen base w/o amino acids (Becton Dickinson), 0.077% [w/v] complete supplement mix (-TRP) (MP biomedicals), 0.05% [w/v] NaCl (Roth), 0.05% [w/v] CaCl2 (Roth), 0.06% [w/v] MgCl2 (Roth), 0.1% [w/v] NH4Cl (Roth), 0.1% [w/v] KH2PO4 (Roth), 0.6% [w/v] NaOH (Roth), 2.2% [v/v] lactic acid (Roth), 0.05% [w/v] glucose) (11, 13, 68). Yeast cells were diluted approximately every 20 h with fresh medium. Yeast strains are listed in Table S3.
Isolation of mitochondria
Yeast cells were cultivated in YPG medium for 2 days as a preculture. The main culture was inoculated with the preculture and incubated for at least 15 h with shaking at 130 rpm and 30°C. Yeast expressing Sam50Δloop6 were grown in SL-Medium at 30°C for 42.5 h to ensure proper shutdown of SAM50 wild-type.
Yeast cells were harvested during log-phase by centrifugation at 1,700 × g (maximal relative centrifugal force; 4,000 rpm, H-12000 Thermo-Fisher Scientific) for 10 min at room temperature. Yeast cells were washed twice with distilled H2O, and incubated with 2 ml/g wet weight DTT buffer (100 mM Tris(hydrosymethyl)aminomethane (Tris)/H2SO4 (MP Biomedicals and Roth), pH 9.4, 10 mM dithiothreitol (DTT, Roth)) for 20 min with shaking at 130 rpm and 30°C. Yeast cells were reisolated by centrifugation for 5 min at 2,700 × g (4,000 rpm, SLA-3000 Sorvall) and incubated for 30-45 min in 6.5 ml/g wet weight Zymolyase buffer (16 mM K2HPO4 (Roth), 4 mM KH2PO4, pH 7.4, 1.2 M sorbitol (Roth), 3 mg/ml Zymolyase 20T (Seikagaku Kaygyo Co.)) with shaking at 130 rpm and 30°C. The resulting spheroplasts were pelleted by centrifugation for 5 min at 1,500 × g (3,000 rpm, SLA-3000 Sorvall) and washed with buffer (16 mM K2HPO4, 4 mM KH2PO4, pH 7.4, 1.2 M sorbitol). The pellet was resuspended in homogenization buffer (0.6 M sorbitol, 10 mM Tris/HCl, pH 7.4, 1 mM ethylenediaminetetraacetic acid (EDTA, Calbiochem), 0.4% [w/v] bovine serum albumin (Sigma), 1 mM phenylmethyl sulfonyl fluoride (PMSF, Sigma)). The spheroplasts were mechanically opened using a glass-Teflon potter by homogenizing the solution 17 times on ice. Mitochondria were isolated using a four-centrifugation step procedure. To remove cell debris, the solution was spun for 15 min, 1,450 × g (3,500 rpm, SS-34, Sorvall) at 4°C, followed by a high speed spinning step to pellet mitochondria at 18,500 × g (12,500 rpm, SS-34, Sorvall) for 15 min at 4°C. The mitochondrial pellet was resuspended in ice cold SEM buffer (250 mM sucrose (MP Biomedicals), 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS, Sigma), pH 7.2, 1 mM EDTA) containing 1 mM PMSF and both centrifugation steps were repeated. The mitochondrial pellet was resuspended in ice cold SEM and the protein concentration was determined using the Bradford protein assay (69). The concentration was adjusted to 10 mg mitochondrial protein per 1 ml SEM. Mitochondria were aliquoted, snap-frozen in liquid nitrogen and stored at -80°C (70).
Oxidation assays with whole yeast cells
Yeast cells were grown overnight in YPG medium at 30°C. Cells corresponding to an OD600 of 1 were taken and harvested by centrifugation for 10 min at 1,500 × g (4,000 rpm, FA-45-24-11, Eppendorf). Cells were resuspended in 100 µl buffer A (2 mM PMSF, 2× protease inhibitor w/o EDTA (Roche), 1 mM EDTA) and oxidized by adding 0.2 mM 4,4’-Dipyridyl disulfide (4-DPS, Sigma-Aldrich) (39). Cells were incubated on ice for 30 min followed by addition of 50 mM iodoacetamide (IA, Sigma) and further incubated for 15 min on ice. After addition of 60 mM NaOH, cells were centrifuged for 10 min at 1,700 × g (4,000 rpm, FA 45-30-11, Eppendorf) and 4°C, resuspended in Laemmli buffer and heated to 65°C for 10 min shaking vigorously.
Sam50 modeling
Potential templates were identified with the HHPRED server restricting the search sequence to the Sam50 β-barrel domain (residues 125-484) (71). The hidden-Markov model based homology search identified templates in the PDB with good p- and E-values. This included structures from FhaC [PDB code: 4QL0 (51)] and TamA [PDB code: 4C00 (18)] as well as several structures from BamA [PDB codes: 4K3B and 4K3C (16); 4C4V (21); 4N75 (22) and 5EKQ (23)], which exhibit considerable variations in the interaction between the β1 and β16-strands. A Sam50 model was calculated from each template using Modeller (72). The model obtained from the BamA structure with PDB code 4N75 (22) with optimized alignment fit best to the experimental results of disulfide bonds and crosslink formation (Model S1).
Despite low sequence identity of 14%, the β-barrel model of Sam50 shows a very good agreement with the structure of BamA (PDB code: 4n75) with a core RMSD of 1.6 Å (Cα-atoms of 310 out of 360 residues). Ramachandran analysis using RAMPAGE (73) showed similar geometrical quality of the model compared to the template (favored/allowed/outlier residues, model: 90.2% / 7.3% / 2.5% and template: 94.7% / 4.5% / 0.8%). Also, the distribution of charged and aromatic residues in respect to barrel inward and outward facing side chains agrees well between model and structure. In order to evaluate the position of loop 6, we superimposed the model with five BamA structures (PDB codes: 4K3B, 4K3C, 4C4V, 4N75 and 5EKQ) as well as the TamA structure (PDB code: 4C00). They all show a highly similar overall structure for loop 6, with identical positions for the conserved IRGF motif including side chain orientations. IRGF faces the inside wall of the barrel (strands 13-16). Noteworthy is for instance the interaction between the guanidino group of the motif’s arginine residue with an aromatic side chain of β-barrel strand 13. The Sam50 model agrees overall with the structures of the loop and the position of IRGF side chains, for instance R366 is interacting with the aromatic ring of F413. Also, positions and orientations of residues 369-371 in the Sam50 model agree with those of the aforementioned structures. In addition, the side chain orientations of the Sam50 β-signal (strand 16) toward either the β-barrel lumen or the lipid phase agree with the structure of the conserved β-signal of mitochondrial VDAC/Porin (42–44).
For graphical presentations, cysteine residues were included in silico at relevant positions and disulfide bonds formed using coot (74) before figures were generated with Pymol (The PyMOL Molecular Graphics System, Version 1.6 Schrödinger, LLC.). The Sam50 β-barrel models were oriented according to the localization of the N-terminal POTRA domain in the mitochondrial intermembrane space (13, 50).
In vitro transcription/translation
Plasmids containing the coding region of the gene of interest and carrying an upstream SP6 promoter binding region were incubated with TNT SP6 quick coupled kit (Promega), an in vitro eukaryotic translation system based on rabbit reticulocytes, in the presence of [35S]methionine (PerkinElmer). The reaction was incubated for at least 90 min at 25°C with shaking at 300 rpm. Reactions were stopped upon addition of 20 mM unlabeled methionine (Roth). A clarifying step was performed at 125,000 × g (45,000 rpm, TLA-55, Beckman) for 30 min at 4°C. 0.3 M sucrose was added to the supernatant and the lysate was snap-frozen and stored at -80°C. Successful transcription/translation was checked by SDS-PAGE and autoradiography.
Template DNA of cysteine mutants of Por1 and Tom40 constructs was generated by PCR using 2× REDTaq ReadyMix (Genaxxon). Forward primers contained a RTS™ wheat germ kit (5prime) specific 5’-CTTTAAGAAGGAGATATACC-3’ sequence upstream of the start codon. The corresponding reverse primers contained downstream of the stop codon a 5’-TGATGATGAGAACCCCCCCC-3’ wheat germ sequence. Cysteine mutagenesis was performed using a primer encoding the desired mutation. Successful mutations were confirmed by sequencing. In case, the methionine radiolabeling of the protein fragment was not sufficient, the methionine encoding sequence 5’-ATGATGATG-3’ was added instead of the start codon and before the stop codon. PCR products were analyzed by inspection of the DNA bands on 2% agarose (Biozym) gels. Products were purified using the QIAquick PCR Purification Kit (Qiagen). A consecutive PCR was performed according to the RTS™ wheat germ LinTempGenSet His6-tag (5prime) manual using 2× REDTaq ReadyMix. PCR products were purified and concentrated using the MinElute PCR purification kit (Qiagen). Wheat germ lysate, an eukaryotic cell-free protein expression system based on wheat germ, was used as described in the RTS™ 100 wheat germ CECF Kit (5prime) with modification for radiolabeled lysates, including [35S]methionine in the reaction solution and supplementation of unlabeled methionine with [35S]methionine in the feeding solution. After incubation for 24 h, lysates were clarified by centrifugation at 125,000 × g (45,000 rpm, TLA-55, Beckman) for 1 h at 4°C. The supernatant was transferred to a fresh tube, snap-frozen in liquid nitrogen and stored at -80°C. Successful translation was checked by SDS-PAGE and autoradiography.
In vitro import into mitochondria and crosslinking/oxidation
Mitochondria were thawed on ice. For one import reaction, 50 µg mitochondria (protein amount) were resuspended in 100 µl import buffer (3% [w/v] bovine serum albumin, 250 mM sucrose, 80 mM KCl (Roth), 5 mM MgCl2, 2 mM KH2PO4, 5 mM methionine (Sigma), 10 mM MOPS-KOH (Roth), pH 7.2) with 4 mM ATP (Roche), 4 mM NADH (Roth), 5 mM creatine phosphate (Roche) and 0.1 mM creatine kinase (Roche). To deplete the membrane potential (-Δψ), AVO (8 µM antimycin A (Sigma), 1 µM valinomycin (Sigma), 20 µM oligomycin (Sigma), final concentrations) was added and NADH was omitted (75). When Tim9 was imported, bovine serum albumin was omitted. 4-10% [v/v] of rabbit reticulocyte lysate or wheat germ lysate containing the precursor proteins were incubated with mitochondria at 25°C with shaking at 300 rpm. Membrane potential dependent import reactions were stopped by addition of AVO, before the import reactions were transferred on ice. The mitochondria were pelleted for 10 min at 4°C and 18,500 × g (13,200 rpm, FA 45-30-11, Eppendorf), the supernatant was discarded and the pellet was washed with 100 µl SEM. Mitochondria were resuspended in Laemmli buffer (0.2 M Tris, pH 6.8, 2% [w/v] dodecylsulfate-Na-salt (SDS, Serva), 4% [v/v] glycerol, 12,5% [w/v] bromphenol blue (Sigma), 1 mM PMSF, 50 mM iodoacetamide) and incubated for 10 min at 65°C shaking vigorously.
In case of experiments combining protein import and crosslinking or oxidation, mitochondria were incubated in 100 µl SEM including energy mix (4 mM ATP, 4 mM NADH, 5 mM creatine phosphate, 0.1 mM creatine kinase) and 2-6% [v/v] precursor-containing lysate was added. Import was conducted at 25°C for 5 to 30 min, shaking at 300 rpm, and the reactions were transferred on ice. To oxidize proteins, 0.36 mM 4-DPS or 2.5 mM CuSO4 (Roth) was added to the reaction. For crosslinking experiments, 1 mM crosslinking reagent 1,6-bismaleimidohexane (BMH, Thermo-Fisher Scientific), bismaleimidoethane (BMOE, Thermo-Fisher Scientific), 1,3-propanediylbismethanethiosulfonat (M3M, Interchim), 1,1-methanediylbismethanethiosulfonat, (M1M, Interchim), N-α-maleimidoacet-oxysuccinimide ester (AMAS, Thermo-Fisher Scientific) or succinimidyl iodoacetate (SIA, Thermo-Fisher Scientific) were added from a 10 mM stock solution prepared in dimethyl sulfoxide (DMSO, Roth). Samples were gently mixed and incubated on ice for 30 min. Oxidation/crosslinking reactions were stopped by addition of 50 mM iodoacetamide and incubated on ice for 15 min. Reactions were laid on top of 500 µl S500EM (500 mM sucrose, 10 mM MOPS, pH 7.2, 1 mM EDTA) and centrifuged for 15 min at 4°C and 20,800 × g (14,000 rpm, FA 45-30-11, Eppendorf) for purification. The pellet was resuspended in 100 µl SEM and processed as described above. Samples analyzed on blue native PAGE were resuspended in digitonin buffer (0.1 mM EDTA, 10% [v/v] glycerol, 50 mM NaCl, 1 mM PMSF, 20 mM Tris/HCl, pH 7.4, 1% [w/v] digitonin (Calbiochem)) and incubated on ice for 15 min before addition of blue native loading dye (0.5% [w/v] Coomassie blue G (Serva), 50 mM 6 aminocaproic acid, 10 mM Bis Tris/HCl, pH 7).
β-Signal affinity assay
The method was performed as described in Kutik et al. (13). Briefly, E. coli cells expressing glutathione-S-transferase (GST), GST-β-signalPor1 and GST-β-signalPor1F281Q were lysed and GST constructs were bound to glutathione sepharose 4B beads (GE Healthcare). Mitochondria were solubilized in GST buffer L (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, Roth), 100 mM potassium acetate (KOAc, Roth), 10 mM magnesium acetate (Mg(OAc)2, Roth), 10% [v/v] glycerol, 1% [w/v] digitonin (Calbiochem), 1 mM PMSF). After centrifugation for 10 min at 4°C and 20,800 × g (14,000 rpm, FA 45-30-11, Eppendorf), the supernatant was transferred to GST bound sepharose beads and incubated for 30 min at 4°C shaking end over end. Samples were centrifuged for 1 min at 4°C and 500 × g and washed at least seven times with GST buffer W (20 mM HEPES, 100 mM KOAc, 10 mM Mg(OAc)2, 10% [v/v] glycerol, 0.5% [w/v] digitonin). To cleave bound proteins from GST, samples were incubated overnight at 4°C, shaking at 800 rpm, in 200 µl GST buffer T (20 mM HEPES, 100 mM KOAc, 10 mM Mg(OAc)2, 10% [v/v] glycerol, 0.5% [w/v] digitonin, 2.5 mM CaCl2, 80 units thrombin protease (Calbiochem)). Columns were centrifuged for 1 min at 4°C and 500 × g. The flow-through was mixed with Laemmli buffer including 1% [v/v] β-mercaptoethanol (Roth) and heated to 95°C for 5 min. Samples were analyzed by SDS-PAGE.
TEV protease cleavage assay
In vitro import into mitochondria followed by crosslinking using BMOE was conducted as described above. After purification, samples were resuspended in solubilization buffer (20 mM Tris/HCl, pH 7.4, 0.1 mM EDTA, 50 mM NaCl) containing 6 M guanidinium hydrochloride (Roth). Samples were heated to 95°C and diluted 1:4 in solubilization buffer. TEV protease (Thermo-Fisher Scientific) was added and incubated for 30 min on ice. Samples were precipitated using 14% [w/v] trichloracetic acid (TCA, Roth) and 0.0125% [w/v] sodium deoxycholate (Sigma). Samples were resuspended in Laemmli buffer containing 1 mM PMSF and 10 mM IA and heated to 65°C for 10 min shaking vigorously. Samples were separated by 4-12% NuPAGE gels (Thermo-Fisher Scientific) according to the manufacturer’s protocol.
Swelling assay
To test the integrity of the mitochondrial outer membrane, 100 µg mitochondria (protein amount) were thawed on ice and resuspended in either 100 µl hypotonic swelling buffer (1 mM EDTA, 10 mM MOPS/KOH, pH 7.2) or isotonic SEM buffer. Mitochondria/mitoplasts were incubated on ice for 15 min before the indicated amount of proteinase K (Roche) was added. The samples were further incubated for 15 min on ice. Proteinase K was inactivated by addition of 2 mM PMSF and further incubated on ice for 10 min. Mitochondria/mitoplasts were pelleted and washed with SEM buffer including 1 mM PMSF. Samples were resuspended in Laemmli buffer, including 1% [v/v] β-mercaptoethanol and 1 mM PMSF, and separated by SDS-PAGE.
SDS-PAGE, NuPAGE, Tris-Tricine PAGE, Blue Native PAGE and Western Blotting
SDS-PAGE was performed using 10% polyacrylamide gels and SDS running buffer (25 mM Tris/HCl, pH 8.8, 191 mM glycine (MP Biomedicals), 1% [w/v] SDS). Gels were run at 30-35 mA for 3-5 h. NuPAGE bis-tris pre-cast gels (10%, Thermo-Fisher Scientific) were used according to the manufacturer’s instructions. Tris-Tricine PAGE gels consistent of a 4-16% polyacrylamide gradient (48% [w/v] acrylamide (Roth), 1.5% [w/v] bisacrylamide (Serva)). Gels were run using anode buffer (0.2 M Tris/HCl, pH 8.9) and cathode buffer (0.1 M Tris, 0.1 M Tricine (Roth), 0.1% [w/v] SDS, pH 8.25) at 70 mA for 3-5 h. For all above mentioned gel electrophoresis procedures, samples were resuspended in Laemmli buffer containing 1 mM PMSF, heated to 65°C for 10 min shaking vigorously. When samples were crosslinked or oxidized, no DTT or β-mercaptoethanol was added but 50 mM iodoacetamide.
Native protein complexes were analyzed using blue native PAGE (76). After import of radiolabeled proteins, mitochondria were resuspended in cold digitonin buffer (0.1 mM EDTA, 10% [v/v] glycerol, 50 mM NaCl, 1 mM PMSF, 20 mM Tris/HCl, pH 7.4, 0.35-1% [w/v] digitonin) and incubated on ice for 15 min. Blue native loading dye (0.5% [w/v] Coomassie blue G (Serva), 50 mM 6-aminocaproic acid (Sigma), 10 mM Bis/Tris (Roth), pH 7) was added. Samples were centrifuged at 4°C for 15 min at 20,800 × g (14,000 rpm, FA 45-30-11, Eppendorf) and the supernatant was loaded on a 6-16.5% discontinuous gradient gel. 8.5 cm gels were run in a cooled Hoefer SE600 vertical electrophoresis chamber using anode buffer (50 mM Bis/Tris/HCl, pH 7) and cathode buffer (50 mM tricine, pH 7, 15 mM Bis/Tris, 0.02% [w/v] Coomassie G) at 90 mA and 600 V for 90 min.
With the exception of blue native gels, gels containing radiolabeled samples were stained and fixed using staining buffer (30% [v/v] ethanol, 10% [v/v] acetic acid (Roth), 0.2% [w/v] Coomassie R250 (Roth)) followed by destaining with destain buffer (50% [v/v] methanol (Roth), 20% [v/v] acetic acid) until protein bands were clearly visible. Gels were dried onto Whatman paper (Macherey-Nagel) and exposed using PhosphorImager screens (GE Healtcare and Fuji), followed by autoradiographic detection (Storm PhosphorImager, GE Healthcare; FLA9000, Fujifilm).
When immunoblotting was performed, gels were incubated for 5 min in SDS running buffer after gel electrophoresis. Gel contents were transferred onto PVDF membranes (Immobilon-P, Millipore) using standard semi dry western blotting (77) at 250 mA for 2 h using blotting buffer (20 mM Tris, 150 mM glycine, 0.02% [w/v] SDS, 20% [v/v] methanol). PVDF membranes were stained with staining buffer, destained using destain buffer until visible bands confirmed equal loading, and completely destained using 100% methanol. Blocking was performed for 1 h using 5% [w/v] fat-free dried milk powder (Frema Reform) in TBST (200 mM Tris/HCl, pH 7.5, 1.25 M CaCl2, 0.1% [v/v] Tween20 (Sigma)) at room temperature. After washing in TBST, membranes were incubated with the designated primary antibodies listed in Table S4, overnight at 4°C or for at least 1 h at room temperature. After a second washing step in TBST, membranes were decorated with secondary anti-rabbit IgG antibody (Sigma), diluted 1:5,000, that was coupled to horse radish peroxidase in 5% [w/v] fat-free dried milk powder in TBST for 1 h. After washing a third time in TBST, membranes were incubated in ECL solution (GE Healthcare) and the chemiluminescence signal was detected by the LAS-4000 system (Fujifilm).
Supplementary Material
One Sentence Summary.
Membrane protein insertion by a β-barrel channel involves precursor translocation through the channel interior and exit through the lateral gate.
Acknowledgments
We thank Dr. Chris Meisinger for discussion. This work was supported by the European Research Council (ERC) Consolidator Grant No. 648235, the Deutsche Forschungsgemeinschaft (PF 202/8-1; BE 4679/2-1), the Sonderforschungsbereiche 746 and 1140, and the Excellence Initiative of the German federal and state governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School). Work included in this study has also been performed in partial fulfillment of the requirements for the doctoral theses of A.I.C.H. and C.L. and the diploma thesis of A.I.C.H. at the University of Freiburg. The data presented in this paper are tabulated in the main paper and the supplementary materials.
References
- 1.Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 2.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 3.Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 4.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Khane D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Hagan CL, Silhavy TJ, Kahne D. β-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 6.Jores T, Klinger A, Groß LE, Kawano S, Flinner N, Duchardt-Ferner E, Wöhnert J, Kalbacher H, Endo T, Schleiff E, Rapaport D. Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat Commun. 2016;7:12036. doi: 10.1038/ncomms12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- 8.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, Pfanner N, Meisinger C. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem. 2004;279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- 11.Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, Rapaport D. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Stubenrauch C, Belousoff MJ, Hay ID, Shen H-H, Lillington J, Tuck KL, Peters KM, Phan M-D, Lo AW, Schembri MA, Strugnell RA, et al. Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nat Microbiol. 2016;1:16064. doi: 10.1038/nmicrobiol.2016.64. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, Kahne DE. Characterization of a stalled complex on the β-barrel assembly machine. Proc Natl Acad Sci USA. 2016;113:8717–8722. doi: 10.1073/pnas.1604100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. Structural insight into the biogenesis of β-barrel membrane proteins. Nature. 2013;501:385–390. doi: 10.1038/nature12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clantin B, Delattre A-S, Rucktooa P, Saint N, Méli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 18.Gruss F, Zähringer F, Jakob RP, Burmann BM, Hiller S, Maier T. The structural basis of autotransporter translocation by TamA. Nat Struct Mol Biol. 2013;20:1318–1320. doi: 10.1038/nsmb.2689. [DOI] [PubMed] [Google Scholar]
- 19.Selkrig J, Mosbahi K, Webb CT, Belousoff MJ, Perry AJ, Wells TJ, Morris F, Leyton DL, Totsika M, Phan M-D, Celik N, et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol. 2012;19:506–510. doi: 10.1038/nsmb.2261. [DOI] [PubMed] [Google Scholar]
- 20.Baud C, Guérin J, Petit E, Lesne E, Dupré E, Locht C, Jacob-Dubuisson F. Translocation path of a substrate protein through its Omp85 transporter. Nat Commun. 2014;5:5271. doi: 10.1038/ncomms6271. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht R, Schütz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, Zeth K. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D Biol Crystallogr. 2014;70:1779–1789. doi: 10.1107/S1399004714007482. [DOI] [PubMed] [Google Scholar]
- 22.Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, Huang Y. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J. 2014;28:2677–2685. doi: 10.1096/fj.13-248450. [DOI] [PubMed] [Google Scholar]
- 23.Bakelar J, Buchanan SK, Noinaj N. The structure of the β-barrel assembly machinery complex. Science. 2016;351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 25.Han L, Zheng J, Wang Y, Yang X, Liu Y, Sun C, Cao B, Zhou H, Ni D, Lou J, Zhao Y, et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol. 2016;23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 26.Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, Ranson NA. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat Commun. 2016;7:12865. doi: 10.1038/ncomms12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doerner PA, Sousa MS. Extreme dynamics in the BamA β-barrel seam. Biochemistry. 2017;56:3142–3149. doi: 10.1021/acs.biochem.7b00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein A, Israel L, Lackey SWK, Nargang FE, Imhof A, Baumeister W, Neupert W, Thomas DR. Characterization of the insertase for β-barrel proteins of the outer mitochondrial membrane. J Cell Biol. 2012;199:599–611. doi: 10.1083/jcb.201207161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Berg B. Lateral gates: β-barrels get in on the act. Nat Struct Mol Biol. 2013;20:1237–1239. doi: 10.1038/nsmb.2709. [DOI] [PubMed] [Google Scholar]
- 30.Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci USA. 2014;111:5878–5883. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. Lateral opening and exit pore formation are required for BamA function. Structure. 2014;22:1055–1062. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinschmidt JH. Folding of β-barrel membrane proteins in lipid bilayers — Unassisted and assisted folding and insertion. Biochim Biophys Acta. 2015;1848:1927–1943. doi: 10.1016/j.bbamem.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Noinaj N, Rollauer SE, Buchanan SK. The β-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr Opin Struct Biol. 2015;31:35–42. doi: 10.1016/j.sbi.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang XC, Han L. How does a β-barrel integral membrane protein insert into the membrane? Protein Cell. 2016;7:471–477. doi: 10.1007/s13238-016-0273-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noinaj N, Gumbart JC, Buchanan SK. The β-barrel assembly machinery in motion. Nat Rev Microbiol. 2017;15:197–204. doi: 10.1038/nrmicro.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bamert RS, Lundquist K, Hwang H, Webb CT, Shiota T, Stubenrauch CJ, Belousoff MJ, Goode RJA, Schittenhelm RB, Zimmerman R, Jung M, et al. Structural basis for substrate selection by the translocation and assembly module of the β-barrel assembly machinery. Mol Microbiol. 2017;106:142–156. doi: 10.1111/mmi.13757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estrada Mallarino L, Fan E, Odermatt M, Müller M, Lin M, Liang J, Heinzelmann M, Fritsche F, Apell H-J, Welte W. TtOmp85 a β-barrel assembly protein, functions by barrel augmentation. Biochemistry. 2015;54:844–852. doi: 10.1021/bi5011305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Y, Zeng Y, Wang Z, Dong C. BamA β16C strand and periplasmic turns are critical for outer membrane protein insertion and assembly. Biochem J. 2017;474:3951–3961. doi: 10.1042/BCJ20170636. [DOI] [PubMed] [Google Scholar]
- 39.Qiu J, Wenz L-S, Zerbes RM, Oeljeklaus S, Bohnert M, Stroud DA, Wirth C, Ellenrieder L, Thornton N, Kutik S, Wiese S, et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell. 2013;154:596–608. doi: 10.1016/j.cell.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 40.Shiota T, Imai K, Qiu J, Hewitt VL, Tan K, Shen H-H, Sakiyama N, Fukasawa Y, Hayat S, Kamiya M, Elofsson A, et al. Molecular architecture of the active mitochondrial protein gate. Science. 2015;349:1544–1548. doi: 10.1126/science.aac6428. [DOI] [PubMed] [Google Scholar]
- 41.Wenz L-S, Ellenrieder L, Qiu J, Bohnert M, Zufall N, van der Laan M, Pfanner N, Wiedemann N, Becker T. Sam37 is crucial for formation of the mitochondrial TOM–SAM supercomplex, thereby promoting β-barrel biogenesis. J Cell Biol. 2015;210:1047–1054. doi: 10.1083/jcb.201504119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ujwal R, Cascio D, Colletier J-P, Faham S, Zhang J, Toro L, Ping P, Abramson J. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stroud DA, Becker T, Qiu J, Stojanovski D, Pfannschmidt S, Wirth C, Hunte C, Guiard B, Meisinger C, Pfanner N, Wiedemann N. Biogenesis of mitochondrial β-barrel proteins: the POTRA domain is involved in precursor release from the SAM complex. Mol Biol Cell. 2011;22:2823–2833. doi: 10.1091/mbc.E11-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Khane D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 47.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 49.Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007;12:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habib SJ, Waizenegger T, Niewienda A, Paschen SA, Neupert W, Rapaport D. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J Cell Biol. 2007;176:77–88. doi: 10.1083/jcb.200602050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfitzner A-K, Steblau N, Ulrich T, Oberhettinger P, Autenrieth IB, Schütz M, Rapaport D. Mitochondrial-bacterial hybrids of BamA/Tob55 suggest variable requirements for the membrane integration of β-barrel proteins. Sci Rep. 2016;6:39053. doi: 10.1038/srep39053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maier T, Clantin B, Gruss F, Dewitte F, Delattre A-S, Jacob-Dubuisson F, Hiller S, Villeret V. Conserved Omp85 lid-lock structure and substrate recognition in FhaC. Nat Commun. 2015;6:7452. doi: 10.1038/ncomms8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delattre A-S, Clantin B, Saint N, Locht C, Villeret V, Jacob-Dubuisson F. Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily. FEBS J. 2010;277:4755–4765. doi: 10.1111/j.1742-4658.2010.07881.x. [DOI] [PubMed] [Google Scholar]
- 54.Leonard-Rivera M, Misra R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J Bacteriol. 2012;194:4662–4668. doi: 10.1128/JB.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Höhr AIC, Straub SP, Warscheid B, Becker T, Wiedemann N. Assembly of β-barrel proteins in the mitochondrial outer membrane. Biochim Biophys Acta. 2015;1853:74–88. doi: 10.1016/j.bbamcr.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Rigel NW, Ricci DP, Silhavy TJ. Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for β-barrel assembly in Escherichia coli. Proc Natl Acad Sci USA. 2013;110:5151–5156. doi: 10.1073/pnas.1302662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
- 58.Hoppins SC, Nargang FE. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem. 2004;279:12396–12405. doi: 10.1074/jbc.M313037200. [DOI] [PubMed] [Google Scholar]
- 59.Sklar JG, Wu T, Khane D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fairman JW, Noinaj N, Buchanan SK. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol. 2011;21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ulrich T, Rapaport D. Biogenesis of beta-barrel proteins in evolutionary context. Int J Med Microbiol. 2015;305:259–264. doi: 10.1016/j.ijmm.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 62.Patel GJ, Kleinschmidt JH. The lipid bilayer-inserted membrane protein BamA of Escherichia coli facilitates insertion and folding of outer membrane Protein A from its complex with Skp. Biochemistry. 2013;52:3974–3986. doi: 10.1021/bi400103t. [DOI] [PubMed] [Google Scholar]
- 63.Danoff EJ, Fleming KG. Membrane defects accelerate outer membrane β-barrel protein folding. Biochemistry. 2015;54:97–99. doi: 10.1021/bi501443p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schiffrin B, Calabrese AN, Higgins AJ, Humes JR, Ashcroft AE, Kalli AC, Brockwell DJ, Radford SE. Effects of periplasmic chaperones and membrane thickness on BamA-catalysed outer membrane protein folding. J Mol Biol. 2017;23:3776–3792. doi: 10.1016/j.jmb.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sikdar R, Peterson JH, Anderson DE, Bernstein HD. Folding of a bacterial integral outer membrane protein is initiated in the periplasm. Nat Commun. 2017;8:1309. doi: 10.1038/s41467-017-01246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 67.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 69.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 70.Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- 71.Hildebrand A, Remmert M, Biegert A, Söding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77:128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 72.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2014;47:1–32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 73.Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 74.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- 76.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 77.Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.