Here, Lee et al. investigated the molecular mechanisms by which acetyl-CoA production impacts gene expression and how acetyl-CoA promotes malignant phenotypes. Their findings show that acetyl-CoA can enhance H3K27ac in a locus-specific manner and that expression of cell adhesion genes is driven by acetyl-CoA in part through activation of Ca2+–NFAT signaling.
Keywords: acetyl-CoA, histone acetylation, metabolism, calcium, NFAT1, glioblastoma
Abstract
The metabolite acetyl-coenzyme A (acetyl-CoA) is the required acetyl donor for lysine acetylation and thereby links metabolism, signaling, and epigenetics. Nutrient availability alters acetyl-CoA levels in cancer cells, correlating with changes in global histone acetylation and gene expression. However, the specific molecular mechanisms through which acetyl-CoA production impacts gene expression and its functional roles in promoting malignant phenotypes are poorly understood. Here, using histone H3 Lys27 acetylation (H3K27ac) ChIP-seq (chromatin immunoprecipitation [ChIP] coupled with next-generation sequencing) with normalization to an exogenous reference genome (ChIP-Rx), we found that changes in acetyl-CoA abundance trigger site-specific regulation of H3K27ac, correlating with gene expression as opposed to uniformly modulating this mark at all genes. Genes involved in integrin signaling and cell adhesion were identified as acetyl-CoA-responsive in glioblastoma cells, and we demonstrate that ATP citrate lyase (ACLY)-dependent acetyl-CoA production promotes cell migration and adhesion to the extracellular matrix. Mechanistically, the transcription factor NFAT1 (nuclear factor of activated T cells 1) was found to mediate acetyl-CoA-dependent gene regulation and cell adhesion. This occurs through modulation of Ca2+ signals, triggering NFAT1 nuclear translocation when acetyl-CoA is abundant. The findings of this study thus establish that acetyl-CoA impacts H3K27ac at specific loci, correlating with gene expression, and that expression of cell adhesion genes are driven by acetyl-CoA in part through activation of Ca2+–NFAT signaling.
Metabolic reprogramming in cancer cells facilitates the acquisition and utilization of nutrients necessary for proliferation and survival within the tumor microenvironment. In addition to direct roles in metabolic processes, metabolites can be substrates of enzymes that carry out post-translational modifications and thus serve parallel functions as signaling molecules that regulate gene expression, impacting cell differentiation and function (Metallo and Vander Heiden 2010; Wellen and Thompson 2012; Pavlova and Thompson 2016).
One metabolic intermediate that links metabolism, signaling, and the epigenome is acetyl-coenzyme A (acetyl-CoA), the acetyl donor for acetylation reactions (Lee et al. 2015; Pietrocola et al. 2015; Kinnaird et al. 2016). Histone acetylation levels correlate with acetyl-CoA abundance (Cai et al. 2011; Lee et al. 2014; Cluntun et al. 2015). In diverse types of mammalian cells, the acetyl-CoA-producing enzyme ATP-citrate lyase (ACLY) regulates histone acetylation levels in a nutrient-dependent manner (Wellen et al. 2009; Donohoe et al. 2012; Covarrubias et al. 2016; Zhao et al. 2016; Wong et al. 2017; Sivanand et al. 2018). ACLY is localized to both the cytosol and nucleus and generates acetyl-CoA from citrate, thus directly linking mitochondrial metabolism to nuclear processes, such as histone acetylation, and cytosolic processes, such as lipid biosynthesis. Under glucose- or oxygen-depleted conditions or when ACLY function is impaired, high concentrations of acetate can compensate to maintain global histone acetylation (Wellen et al. 2009; Gao et al. 2016; Zhao et al. 2016).
We established previously that nutrient-dependent fluctuations in acetyl-CoA abundance impact global histone acetylation levels and gene expression profiles in glioblastoma multiforme (GBM) cells (Lee et al. 2014). Under low-glucose conditions, acetate supplementation rescues acetyl-CoA production and the expression of a subset of glucose-regulated genes, which we termed “acetyl-CoA-up-regulated” genes. Among the acetyl-CoA-up-regulated genes that we identified in GBM cells are those involved in cell adhesion, extracellular matrix (ECM) interaction, cytoskeletal dynamics, and integrin signaling (Lee et al. 2014). GBM, the most common brain tumor in adults, is highly diffuse and invasive, limiting the efficacy of surgical resection (Furnari et al. 2007; Jhanwar-Uniyal et al. 2015). Whether metabolic alterations contribute to the invasive phenotype in GBM is unclear. In this study, we aimed to define the mechanisms through which acetyl-CoA abundance regulates expression of cell adhesion and migration genes in GBM cells, since such insights could guide therapeutic strategies to limit the invasive characteristics of this lethal cancer.
Building on our previous findings that global histone acetylation levels are responsive to the availability of glucose and acetate (Lee et al. 2014), we analyzed the impact of nutrient availability on histone acetylation throughout the genome using an exogenous reference genome for normalization to account for changes in total acetylation between conditions (Orlando et al. 2014). Remarkably, manipulation of glucose and acetate in the culture medium had only a modest impact on histone H3 Lys27 acetylation (H3K27ac) peaks near the transcription start site (TSS) of most genes but potently impacted H3K27ac at specific loci, correlating with gene expression. Thus, changes in acetyl-CoA abundance resulted in site-specific changes in H3K27ac, raising the question of how this specificity is mediated by a metabolite. We hypothesized that particular transcription factors might be regulated in an acetyl-CoA-dependent manner. We identified nuclear factor of activated T cells 1 (NFAT1) as a key acetyl-CoA- and ACLY-responsive transcription factor that promotes GBM cell migration and adhesion to the ECM. Mechanistically, we discovered that acetyl-CoA abundance impacts NFAT1 nuclear localization through modulation of intracellular calcium (Ca2+) levels. These data thus demonstrate that changes in acetyl-CoA abundance result in site-specific modulation of histone acetylation and identify the regulation of Ca2+–NFAT1 signaling by acetyl-CoA as a key link between cellular metabolism and gene regulation that promotes GBM cell interaction with the ECM and migration.
Results
Acetyl-CoA availability regulates expression of cell migration and adhesion genes in GBM cells
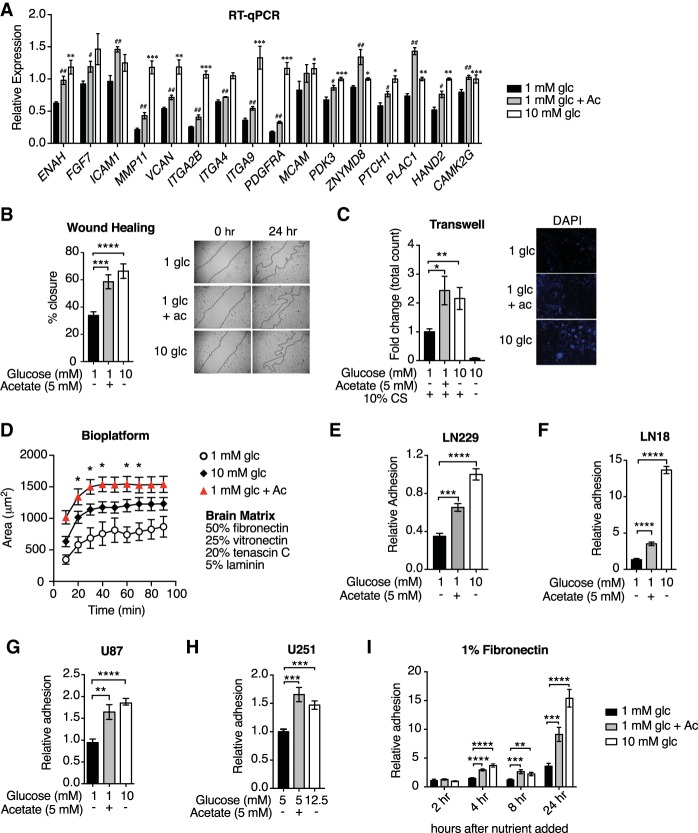
Global histone acetylation levels decline when glucose is limited, correlating with reduced acetyl-CoA abundance, and both acetyl-CoA and global histone acetylation levels can be restored upon supplementation of acetate (Wellen et al. 2009; Lee et al. 2014), which enters the cell and is directly converted to acetyl-CoA by acetyl-CoA synthetase enzymes (ACSS1 in mitochondria and ACSS2 in the cytosol) (Schug et al. 2016). We previously identified several gene sets enriched specifically when acetyl-CoA abundance is high in GBM cells (Lee et al. 2014). Among these were annotated pathways involved in cell adhesion, migration, and cytoskeletal dynamics (PANTHER: integrin signaling pathway; KEGG [Kyoto Encyclopedia of Genes and Genomes]: regulation of actin cytoskeleton; KEGG: cell adhesion molecules; Reactome: integrin cell surface interactions; and KEGG: ECM–receptor interaction) (Lee et al. 2014). To confirm these findings, mRNA expression patterns for select genes within these sets were validated by RT-qPCR in LN229 GBM cells, comparing low-glucose (1 mM glucose), high-glucose (10 mM glucose), and low-glucose plus acetate (1 mM glucose + 5 mM acetate) conditions (Fig. 1A). Given this regulation of cell adhesion and migration genes by glucose and acetate, we next investigated whether acetyl-CoA abundance impacts the ability of cells to migrate using wound healing and transwell migration assays. In both assays, cell migration was impaired in low-glucose conditions and rescued by acetate supplementation (Fig. 1B,C).
Figure 1.
Acetyl-CoA promotes cell adhesion and migration in GBM cells. (A) Relative mRNA levels of acetyl-CoA-up-regulated genes as determined by RT-qPCR in LN229 cells. (#) P < 0.05; (##) P < 0.01, significance of acetate treated over 1 mM glucose. (**) P < 0.01; (***) P < 0.001, significance of 10 mM glucose treated over 1 mM glucose. (B) Wound healing assay in LN229 cells. (***) P < 0.001; (****) P < 0.0001. (Right panel) Lines indicate the boundary of the scratch. Photos were captured at 0 h and after 24 h. (C) Transwell migration of LN229 across an 8.0-µm polycarbonate membrane. (*) P < 0.05; (**) P < 0.01. (Right panel) Cells on the membrane were stained with Hoechst, and photos were captured 24 h after seeding. (D) Adhesion quantified on biomaterial platform with ECM components of the brain. Cells were pretreated with the indicated conditions for 24 h, and the area covered by cells was measured over time. (*) P < 0.05, significance of acetate conditions over 1 mM glucose conditions determined by Tukey's post hoc test. (E–H) Relative adhesion to 1% fibronectin after 24 h of the indicated treatments in LN229 (E), LN18 (F), U87 (G), and U251 (H) cells. (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. (I) LN229 cells were incubated in 1 mM glucose overnight, and then the medium was changed to the indicated conditions. Quantification of adhesion to 1% fibronectin after the indicated hours of treatment. (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. All panels show mean ± SEM of triplicates.
Integrin-mediated adhesion to the ECM is a crucial component of cancer cell migration and invasion (Pickup et al. 2014). To test whether acetyl-CoA abundance promotes GBM cell adhesion to the ECM, we used a brain-inspired (i.e., modeled on the ECM composition of the brain) biomaterial platform comprised of 50% fibronectin, 25% vitronectin, 20% tenascin C, and 5% laminin (Barney et al. 2015). After incubating cells in high or low glucose with or without acetate supplementation, cells were seeded onto the ECM, and their adhesion kinetics were quantified. Both glucose and acetate enhanced LN229 cell adhesion to the brain-inspired ECM (Fig. 1D) as well as to fibronectin alone (Fig. 1E). Similar observations were also made with three other GBM cell lines (Fig. 1F–H). Importantly, acetate rescued adhesion and migration without impacting cell doubling time (Lee et al. 2014), AMPK activation (Supplemental Fig. S1A), or markers of the unfolded protein response (Supplemental Fig. S1B), suggesting that the adhesion and migration phenotypes are not secondary to effects on proliferation, bioenergetics, or endoplasmic reticulum (ER) stress. On the other hand, acetate restored histone acetylation levels in low-glucose conditions, and inhibition of the lysine acetyltransferase (KAT) p300 suppressed glucose- and acetate-dependent increases in global H3K27ac and cell adhesion to the ECM (Supplemental Fig. S1C,D), consistent with a potential role for histone acetylation in promoting these phenotypes. These data suggest that acetyl-CoA promotes GBM cell adhesion to ECM in a p300-dependent manner.
Next, to assess whether acetyl-CoA-dependent cell adhesion is likely to require transcription, we analyzed the time required for cells to adhere following glucose and acetate supplementation. Cells were incubated overnight in 1 mM glucose, then glucose or acetate was added, and adhesion was subsequently measured over 24 h. Increased fibronectin adhesion was observed beginning 4 h after glucose or acetate addition and further increased after 24 h (Fig. 1I). These data are consistent with a mechanism whereby gene transcription rather than a more acute signaling mechanism mediates glucose- and acetate-induced cell adhesion to the ECM.
Glucose-regulated cell adhesion and migration require ACLY
We next sought to distinguish whether acetate is used exclusively for acetyl-CoA production in the cytosol or whether its use in mitochondria is also relevant to the adhesion phenotype. Acetate can be converted to acetyl-CoA by ACSS2 in the cytosol and by ACSS1 in mitochondria (Comerford et al. 2014; Schug et al. 2015, 2016), and it has been shown to feed into the TCA cycle in GBM (Mashimo et al. 2014). Consistent with this, acetate supplementation significantly rescued citrate levels under low-glucose conditions (Supplemental Fig. S2A), and 13C-acetate tracing confirmed that acetate carbon contributes significantly to the synthesis of citrate, ACLY's substrate, in both high- and low-glucose conditions (Supplemental Fig. S2B). Moreover, supplementation of either pyruvate or fatty acids, each of which can be metabolized to acetyl-CoA in mitochondria, resulted in significant rescue of cell adhesion to fibronectin, similar to that observed with acetate supplementation (Supplemental Fig. S2C,D). These data indicate that acetyl-CoA production in mitochondria can promote cell adhesion.
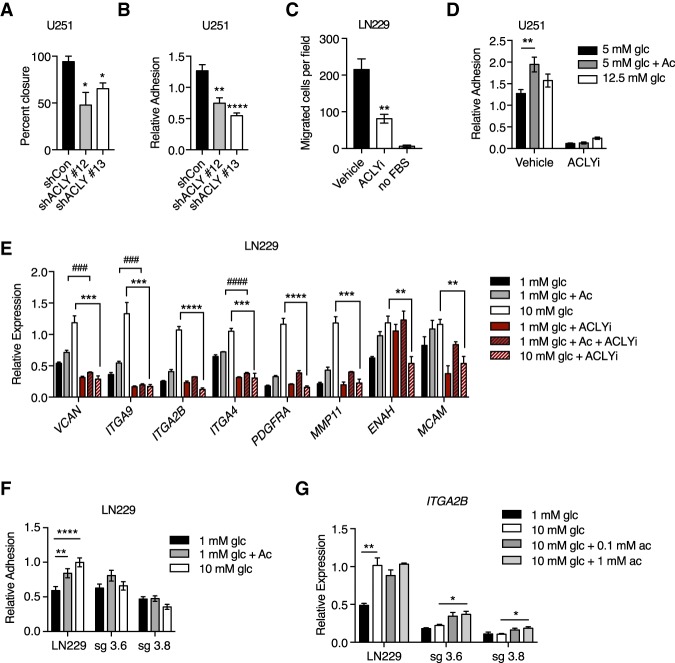
Since regulation of histone acetylation by acetyl-CoA requires its production outside of mitochondria and since acetyl-CoA transfer from mitochondria to cytosol and the nucleus is mediated by citrate export and cleavage by ACLY, these findings prompted us to investigate whether ACLY promotes GBM cell adhesion and migration. Indeed, both wound healing and adhesion to fibronectin were impaired upon ACLY silencing (Fig. 2A,B). Biochemical inhibition of ACLY also potently suppressed transwell migration (Fig. 2C), cell adhesion (Fig. 2D), and glucose-dependent expression of adhesion- and migration-related genes (Fig. 2E; Supplemental Fig. S2E). ACLY inhibition also suppressed acetate-dependent induction of some of these genes (Fig. 2E), indicating that acetate's effects on gene expression are partially dependent on ACLY. To further validate these findings, we deleted ACLY from LN229 cells using CRISPR/Cas9 gene editing (Zhao et al. 2016). Consistent with RNAi and inhibitor data, nutrient-dependent adhesion to ECM (Fig. 2F) and expression of the integrin ITGA2B (Fig. 2G) were suppressed upon genetic deletion of ACLY in LN229 cells. Moreover, xenograft tumor growth was markedly impaired in the absence of ACLY (Supplemental Fig. S2F,G), and several adhesion- and migration-related genes exhibited lower expression in tumors generated from ACLY knockout cells (Supplemental Fig. S2H), supporting the relevance of ACLY in modulating expression of these genes in vivo. Collectively, these data establish that ACLY-dependent acetyl-CoA production promotes GBM cell adhesion and migration.
Figure 2.
Glucose-dependent cell migration requires ACLY. (A) Wound healing assay in U251 with stable knockdown of ACLY. (*) P < 0.05. (B) Relative adhesion in U251 cells with knockdown of ACLY. (**) P < 0.01; (****) P < 0.0001. (C) Transwell migration of LN229 cells after treatment with 50 µM ACLY inhibitor (ACLYi; BMS303141) or vehicle in 10 mM glucose. No FBS is a negative control. (**) P < 0.01. (D) Adhesion assay in with U251 cells after treatment with 50 µM ACLYi or vehicle for 24 h. (E) Relative mRNA expression of acetyl-CoA-regulated genes in LN229 cells treated with 50 µM ACLYi. (###) P < 0.001; (####) P < 0.0001; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. (F) Relative adhesion in two different ACLY knockout clones (sg3.6 and sg3.8) on 1% fibronectin. Parental cells were infected with vector expressing Cas9 but without guide RNA. (**) P < 0.01; (****) P < 0.0001. (G) Relative expression of ITGA2B in cells described in F. (*) P < 0.05; (**) P < 0.01. All panels show mean ± SEM.
Nutrient availability alters histone acetylation in a site-specific manner
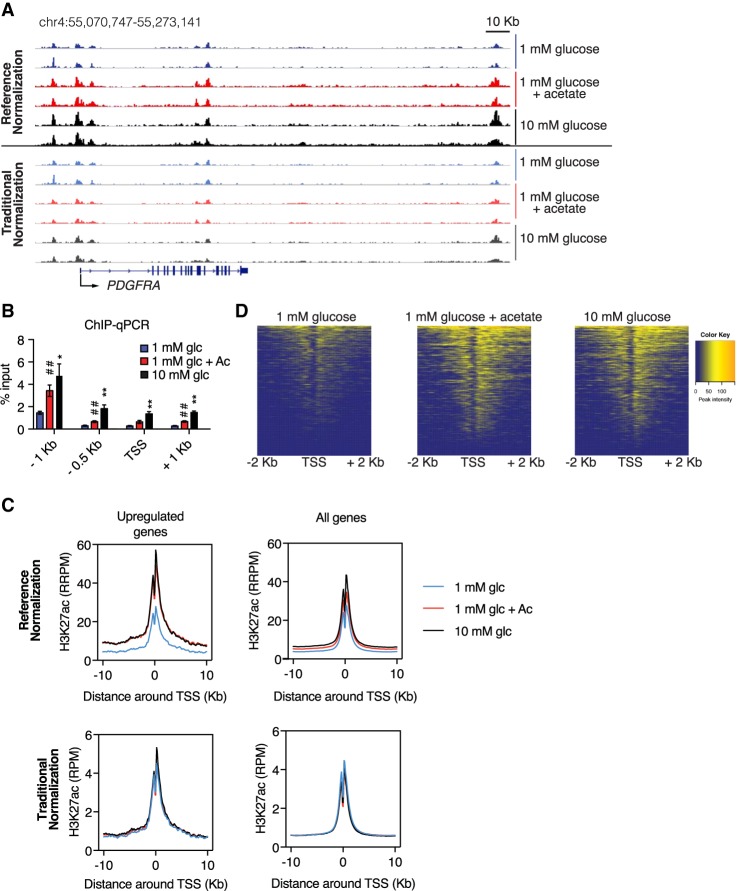
Since acetyl-CoA abundance was found to impact global histone acetylation levels and expression of distinct sets of genes associated with cell adhesion and migration, we next aimed to define the specific loci at which histone acetylation was regulated in response to glucose and acetate availability. To investigate this, we performed H3K27ac chromatin immunoprecipitation (ChIP) coupled with next-generation sequencing (ChIP-seq). We focused on H3K27ac, since it has important roles in gene regulation and is mediated by CBP/p300 (Feller et al. 2015), which is also required for glucose- and acetate-dependent adhesion (Supplemental Fig. S1D). The overall distribution of H3K27ac peaks throughout the genome was similar between conditions (Supplemental Fig. S3A). Since traditional ChIP-seq data normalization to percentage of reads fails to account for differences in the abundance of histone modification between conditions, we used an exogenous reference genome (Drosophila melanogastor chromatin spike-in) for normalization (ChIP-Rx) (Orlando et al. 2014). As expected, the ratio of total H3K27ac reads aligning to the Homo sapiens genome versus the D. melanogaster genome decreased markedly in low glucose and increased with acetate supplementation (Supplemental Fig. S3B). These data are consistent with the pronounced changes in global H3K27ac observed in these conditions by Western blot (Supplemental Fig. S1C). We next aimed to broadly evaluate the genome-wide loci at which H3K27ac was gained in high-glucose or acetate-supplemented conditions compared with the 1 mM glucose condition. Using a stringent computational tool, DiffBind, we identified 1091 regions that gained peaks. When the nearest genes were analyzed, only a small fraction overlapped with the acetyl-CoA-up-regulated genes defined previously by RNA sequencing (RNA-seq) (Supplemental Fig. S3C). These differential regions were primarily within introns and intergenic regions (Supplemental Figs. S3D, S4A); thus, we reasoned that some of these peaks are likely to be putative enhancers, and it is also possible that some peaks are gained in regions not directly involved in transcriptional control.
We next specifically interrogated the relationship between H3K27ac and gene expression in response to acetyl-CoA levels. Inspection of candidate loci encoding acetyl-CoA-up-regulated genes (genes from which mRNA expression is reduced in low glucose and increased with high glucose or acetate) (Lee et al. 2014) such as PDGFRA and MMP11 revealed markedly reduced signal in low-glucose conditions relative to high-glucose or acetate-supplemented conditions, in agreement with ChIP-qPCR (ChIP coupled with quantitative PCR) data (Fig. 3A,B; Supplemental Fig. S4B). Notably, these differences were obscured when using a traditional normalization to the percentage of reads, highlighting the importance of the reference genome normalization (Fig. 3A; Supplemental Fig. S4B). For genes at which mRNA expression was not altered by glucose or acetate, such as RPL19, ChIP-Rx reported no distinct change in H3K27ac (Supplemental Fig. S4C). Metagene analysis of all acetyl-CoA-up-regulated genes demonstrated that H3K27ac was potently suppressed near the TSS under low- versus high-glucose conditions and rescued upon acetate supplementation (Fig. 3C,D). Normalization to percentage of reads concealed this regulation (Fig. 3C). In contrast to the acetyl-CoA-up-regulated genes, metagene analysis of all genes revealed only a modest suppression of signal in low-glucose conditions (Fig. 3C). Together, these data demonstrate that acetyl-CoA abundance results in a widespread but modest tuning of H3K27ac at promoter TSS loci across the genome but also that H3K27ac at specific genes is highly responsive to acetyl-CoA, correlating with expression of those genes. These findings suggest that acetyl-CoA abundance might impact expression of distinct sets of genes as well as H3K27ac at these genes through specific upstream mechanisms.
Figure 3.
H3K27ac near TSSs is sensitive to acetyl-CoA availability. (A) PDGFRA ChIP-seq tracks with reference normalization (top) and traditional normalization (bottom). (B) H3K27ac ChIP-qPCR analyzing the PDGFRA promoter region. (C) Metagene analysis of H3K27ac in the region −10 to +10 kb around the TSS of acetyl-CoA-up-regulated genes identified in Lee et al. (2014) and for all genes. (Top) Reference genome-normalized data. (Bottom) Traditional normalization. (D) Heat map of H3K27ac at acetyl-CoA-up-regulated genes (reference-normalized).
Cellular adhesion and migration genes are regulated by NFAT family transcription factors
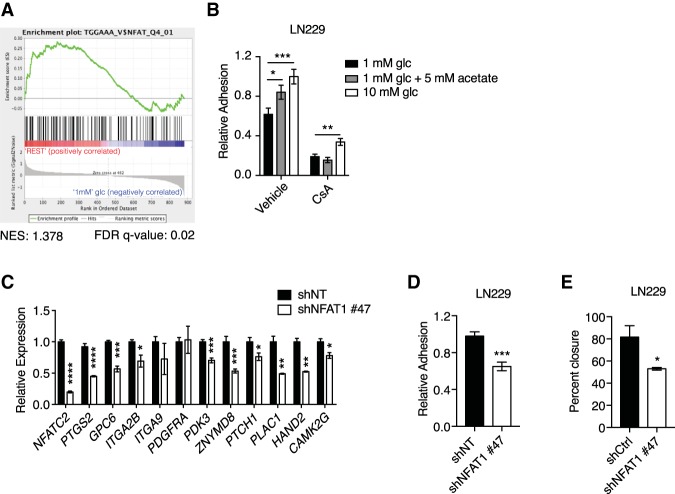
We thus postulated that acetyl-CoA levels elicit distinct patterns of gene expression by activating specific acetyl-CoA-responsive transcription factors. A query of the Broad Institute Molecular Signatures Database (MSigDB) revealed that the NFAT-binding consensus motif TGGAAA (Badran et al. 2002) was the most enriched transcription factor motif within 2 kb upstream of and downstream from the TSSs of acetyl-CoA-up-regulated genes (Supplemental Fig. S5A; Mootha et al. 2003; Subramanian et al. 2005). Gene set enrichment analysis (GSEA) specifically interrogating an NFAT signature confirmed this enrichment (Fig. 4A; Supplemental Fig. S5B).
Figure 4.
The transcription factor NFAT1 mediates acetyl-CoA-dependent cell adhesion and migration. (A) GSEA of acetyl-CoA-up-regulated genes, comparing 1 mM glucose with “rest” (10 mM glucose and 1 mM glucose + acetate conditions). (B) LN229 cell adhesion onto 1% fibronectin after 24 h of treatment with 10 µM cyclosporin A (CsA) or vehicle control. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (C) Relative mRNA expression of acetyl-CoA-up-regulated genes after knockdown of NFAT1 in LN229 cells. Representative genes from the list generated from GSEA in A are shown (see also Supplemental Fig. S5B). (D) Relative adhesion onto 1% fibronectin after shRNA-mediated knockdown of NFAT1 in LN229 cells. (***) P < 0.001. (E) Wound healing assay in LN229 cells after shRNA-mediated knockdown of NFAT1. (*) P < 0.05. All panels show mean ± SEM.
NFAT comprises a family of four Ca2+-regulated transcription factors, which control diverse cellular processes in a wide range of cell types (Mancini and Toker 2009; Muller and Rao 2010). NFAT isoforms also interact with the coactivator p300 (Garcia-Rodriguez and Rao 1998), a KAT that facilitates nutrient-dependent adhesion (Supplemental Fig. S1D). Notably, NFAT has a well-established role in not only regulating migration of normal cells but also tissue invasion by cancer cells (Qin et al. 2014) and has been implicated in the pathogenesis of GBM (Tie et al. 2013; Wang et al. 2015). However, few NFAT-regulated genes have been identified in gliomas, and NFAT sensitivity to acetyl-CoA availability has not been explored.
To investigate the role of NFAT, we examined the impact on cell adhesion of inhibiting NFAT activation with the calcineurin (CaN) inhibitor cyclosporin A (CsA) (Schreiber and Crabtree 1992). CsA potently suppressed LN229 and U251 cell adhesion to fibronectin in the presence of either glucose or acetate (Fig. 4B; Supplemental Fig. S6A). qPCR analysis revealed that NFAT1 (NFATC2) is the most highly expressed NFAT isoform in LN229 cells (Supplemental Fig. S6B). Silencing of NFAT1 suppressed expression of known NFAT target genes such as PTGS2 as well as putative NFAT target genes identified by our analysis of acetyl-CoA-responsive genes (Fig. 4C; Supplemental Fig. S6C,D). In addition, NFAT1 silencing reduced cell migration and adhesion to fibronectin (Fig. 4D,E), consistent with a mechanism in which these acetyl-CoA-dependent responses are mediated by NFAT1.
NFAT1 nuclear localization is regulated by acetyl-CoA availability and mediates acetyl-CoA-dependent cell adhesion
NFAT is retained in an inactive hyperphosphorylated form in the cytoplasm, and its activation is triggered by Ca2+-dependent CaN-mediated dephosphorylation. Dephosphorylation exposes a nuclear localization signal that promotes NFAT nuclear translocation, allowing it to bind to its canonical DNA motif (Okamura et al. 2000; Mancini and Toker 2009; Muller and Rao 2010).
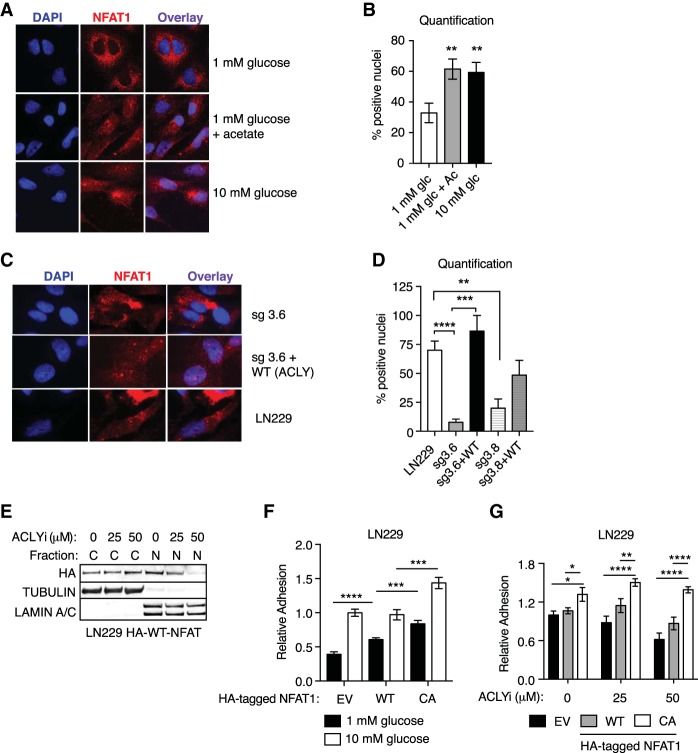
Since the data suggested that acetyl-CoA abundance might regulate cell adhesion and migration through NFAT1 and since NFAT1 transcriptional activity depends on its nuclear localization, we asked whether acetyl-CoA alters NFAT1 localization. We indeed observed markedly fewer cells containing nuclear NFAT1 under low-glucose conditions than in high-glucose or acetate-supplemented conditions (Fig. 5A,B). Furthermore, NFAT1 was largely excluded from nuclei of ACLY-deficient cells, and nuclear localization was rescued upon reconstituting these cells with murine ACLY (Fig. 5C,D). ACLY inhibition similarly suppressed NFAT nuclear localization (Fig. 5E). These data indicate that acetyl-CoA production promotes NFAT1 nuclear localization. To further test the role for NFAT1 in acetyl-CoA-dependent cell adhesion, we next asked whether NFAT1 activation is sufficient to rescue cell adhesion defects when acetyl-CoA abundance is limited. Indeed, we found that constitutively active NFAT1 (CA) (Okamura et al. 2000) rescued defects in cell adhesion produced by low-glucose conditions or ACLY inhibition (Fig. 5F,G). Thus, acetyl-CoA regulates GBM cell adhesion to ECM by controlling NFAT1 nuclear localization.
Figure 5.
NFAT1 nuclear localization is acetyl-CoA-regulated and required for cell adhesion. (A) Immunofluorescent detection of endogenous NFAT1 localization in LN229 cells. Representative images are shown. (B) Quantification of NFAT1 detected in the nucleus, as shown in A, as percentage of cells in each field. At least 170 cells were scored per condition. (**) P < 0.01. (C) Immunofluorescent detection of endogenous NFAT1 localization in ACLY knockout sg3.6 cells with or without murine wild-type ACLY cDNA expression. Representative images are shown. (D) Quantification of endogenous NFAT1 detected in the nucleus, as shown in C, as percentage of cells in each field. (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. (E) Murine HA-tagged NFAT1 was transiently expressed in LN229 cells, cells were treated with the indicated doses of ACLYi, and nuclear and cytosolic fractions were prepared and analyzed by Western blot. (F) Glucose regulation of cell adhesion on 1% fibronectin in LN229 cells expressing empty vector, murine wild-type NFAT1, or constitutively active (CA) NFAT1 (CA-NFAT1). (G) ACLYi regulation of cell adhesion on 1% fibronectin in cells expressing empty vector (plx303), wild-type NFAT1, or CA-NFAT1. (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001.
Acetyl-CoA-triggered cytoplasmic Ca2+ signals control NFAT1 activation and cell adhesion
Since NFAT1 nuclear localization is controlled by its Ca2+-dependent dephosphorylation, we asked whether NFAT1 phosphorylation is regulated in response to changes in acetyl-CoA abundance. Both low-glucose conditions and ACLY inhibition reduced NFAT gel mobility relative to high-glucose or acetate-treated conditions, consistent with hyperphosphorylation. Treatment with the Ca2+ ionophore ionomycin induces NFAT dephosphorylation, and ionomycin treatment reversed the low-glucose-induced or ACLY inhibition-induced shift in NFAT mobility (Supplemental Fig. S7A,B), suggesting that acetyl-CoA promotes Ca2+-dependent dephosphorylation of NFAT.
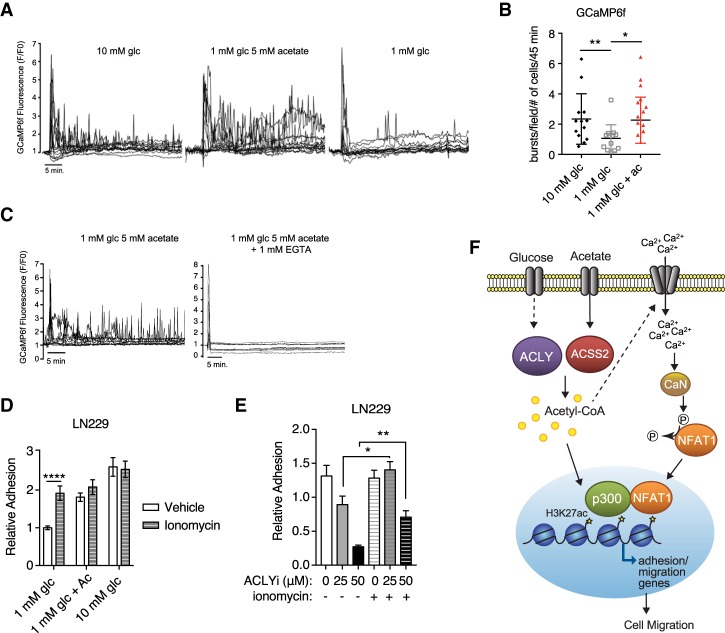
To directly test whether acetyl-CoA mobilizes Ca2+, we performed single-cell measurements of cytosolic Ca2+ in LN229 cells that express the genetically encoded calcium reporter GCaMP6f (Chen et al. 2013). GCaMP6f-expressing cells were cultured overnight in low glucose (1 mM), and, 18–20 h later, Ca2+ measurements were initiated following replacement with fresh medium containing either 1 mM glucose, 10 mM glucose, or 1 mM glucose + 5 mM acetate. Under all conditions, a large Ca2+ spike was observed immediately following addition of fresh serum-containing medium (Fig. 6A). However, after this initial spike, subsequent Ca2+ bursts occurred with higher frequency in the presence of either high-glucose or acetate medium than in low-glucose medium (Fig. 6A,B). Addition of the calcium chelating agent EGTA to the medium inhibited these persistent Ca2+ bursts (Fig. 6C), indicating that acetate-dependent Ca2+ oscillations require an influx of extracellular ions. Collectively, these data suggest that acetyl-CoA production promotes NFAT dephosphorylation and nuclear localization through regulation of Ca2+ influx. To further validate this role for acetyl-CoA-dependent Ca2+ signaling, we tested cells’ ability to adhere in low-acetyl-CoA conditions if treated with ionomycin. Ionomycin treatment rescued defective cell adhesion under low-glucose conditions or upon ACLY inhibition (Fig. 6D,E), indicating that Ca2+ signals are sufficient to mediate adhesion when acetyl-CoA is low. Together, these data establish that acetyl-CoA promotes cell adhesion at least in part by mobilizing Ca2+, which promotes NFAT1 nuclear localization and NFAT1-dependent gene expression (Fig. 6F).
Figure 6.
Ca2+ signaling is acetyl-CoA-regulated. (A) Representative Ca2+ traces from the GCaMP6f reporter in the indicated conditions. Each trace represents one cell, and at least 15 cells were analyzed from two different fields in each condition. (B) Quantification of Ca2+ spikes after nutrient add-back. At least 116 cells were scored in each condition in a blinded manner. (*) P < 0.05; (**) P < 0.01. (C) Representative Ca2+ traces in the presence or absence of EGTA are shown. (D) LN229 cell adhesion onto 1% fibronectin after 20 h of incubation in the indicated concentrations of glucose and acetate followed by 4 h of treatment with 0.5 µM ionomycin or vehicle control. (****) P < 0.0001. (E) Relative adhesion after 20 h of treatment with ACLYi followed by 4 h of treatment with 0.5 µM ionomycin or vehicle control in LN229 cells expressing murine wild-type NFAT1. (*) P < 0.05; (**) P < 0.01. (F) Model showing how acetyl-CoA promotes GBM cell adhesion and migration through Ca2+–NFAT signaling.
Discussion
Metabolites are now recognized to exert potent signaling effects that can impact cancer phenotypes. Using genome-wide approaches, we found that in response to nutrient-dependent fluctuations in acetyl-CoA abundance, H3K27ac is potently regulated near the TSSs of specific genes (correlating with gene expression) rather than uniformly at all genes. Acetyl-CoA-responsive genes in GBM cells include many associated with cell migration and adhesion to the ECM. Mechanistically, we identify NFAT1 as a key mediator of acetyl-CoA-dependent regulation of adhesion- and migration-related genes. We further establish that acetyl-CoA abundance regulates NFAT1 through control of Ca2+ homeostasis, triggering NFAT1 dephosphorylation and nuclear translocation when acetyl-CoA is abundant (Fig. 6F). Further work, including NFAT1 ChIP studies, will be needed to confirm the genes that NFAT1 directly regulates in GBM.
Our findings expand on prior studies that revealed that there is substantial cross-talk between Ca2+–NFAT signaling and glucose metabolism (Lawrence et al. 2002; Koenig et al. 2010; Ho et al. 2015; Vaeth et al. 2017). Specifically, the glycolytic metabolite phosphoenolpyruvate (PEP) was shown in T cells to inhibit SERCA, resulting in sustained increases in cytosolic Ca2+- and NFAT1-dependent transcription (Ho et al. 2015). While PEP abundance decreases under low-glucose conditions in GBM cells, it is not rescued by acetate supplementation (Supplemental Fig. S8A), indicating that PEP levels do not account for the activation of Ca2+–NFAT signaling observed in response to acetate. Instead, we provide evidence that acetyl-CoA is a key product of glucose metabolism that promotes elevated intracellular Ca2+ in a manner dependent on Ca2+ entry into the cell (Fig. 6C). While we did not determine the mechanism by which acetyl-CoA controls Ca2+ dynamics, there are a limited number of candidate calcium channels expressed in GBM cells, including voltage-operated calcium channels, ligand-gated calcium channels, and store-operated calcium channels (Leclerc et al. 2016). Store-operated calcium entry (SOCE) occurs following IP3 receptor-mediated Ca2+ release from ER into the cytosol (Putney 1986). The subsequent decrease in ER Ca2+ levels activates STIM proteins (Liou et al. 2005), which oligomerize and relocalize in the ER membrane to regions juxtaposed to the plasma membrane, where they activate ORAI1–3 channels responsible for the sustained influx of extracellular Ca2+ (Roos et al. 2005; Feske et al. 2006; Vig et al. 2006; Prakriya and Lewis 2015). Delineating the precise mechanisms that link acetyl-CoA to Ca2+ signaling will be an important focus of future studies.
These data contribute to a growing body of evidence that the deregulation of Ca2+ signaling in the cytoplasm can contribute to tumorigenesis by regulating proliferation, apoptosis, migration, and tissue invasion of transformed cells (Stewart et al. 2015; Monteith et al. 2017). Substantial prior data also tie NFAT transcription factors to tumor progression (Mancini and Toker 2009; Qin et al. 2014). In GBM, Ca2+ dynamics and STIM1/Orai-mediated SOCE have been implicated in tumor growth and invasion (Chigurupati et al. 2010; Motiani et al. 2013; Zhang et al. 2017), and both NFAT1 and NFAT2 have been shown to promote invasive phenotypes of GBM cells (Tie et al. 2013; Wang et al. 2015). The findings of this study now link acetyl-CoA abundance to Ca2+–NFAT signaling and GBM cell adhesion and migration. Additional investigation using autochthonous tumor models is needed to more thoroughly evaluate the potential for targeting acetyl-CoA metabolism and Ca2+–NFAT signaling in GBM.
While we focused on GBM cells in this study, NFAT plays important roles in numerous cell types, including T cells. Prior work has shown that mitochondrial function is required for efficient NFAT activation in T cells, and this involves SOCE and reactive oxygen species production (Hawkins et al. 2010; Sena et al. 2013). Similarly, we noted that mitochondrial complex I inhibition suppressed NFAT1 nuclear localization in GBM cells (Supplemental Fig. S8B), suggesting that common mechanisms between cell types might be involved. Acetyl-CoA metabolism has also been implicated in gene regulation in T cells. Ifng expression, for example, is regulated in part by glycolysis-dependent histone acetylation (Peng et al. 2016), and ACLY has been shown to regulate cytokine-induced gene expression (Osinalde et al. 2016). Thus, understanding whether acetyl-CoA also impacts Ca2+–NFAT signaling cell types in addition to GBM cells, such as T cells, is of substantial interest.
To date, the functions of metabolic control of chromatin modification and its roles in the regulation of specific genes have largely remained mysterious. This study addresses both the genome-wide regulation of H3K27ac in response to nutrient availability and the mechanisms through which specific genes are regulated by acetyl-CoA. The marked reduction in H. sapiens versus spiked-in D. melanogaster reads observed by H3K27ac ChIP-Rx under low-glucose conditions (Supplemental Fig. S3B) is consistent with the striking suppression in global H3K27ac seen by Western blot (Supplemental Fig. S1C) as well as with glucose-dependent alterations in multiple histone acetyl marks recorded by mass spectrometry (Lee et al. 2014). While potent regulation of H3K27ac at the promoter TSSs of acetyl-CoA-up-regulated genes was observed (Fig. 3C,D), these changes occur at a relatively small number of genes and cannot account for the global changes in acetylation observed. Notably, many of the other differentially regulated loci were in intronic and intergenic regions of the genome (Supplemental Fig. S3D), and, while many of these peaks could be in enhancers, understanding the significance of these sites for gene regulation requires further investigation. It is possible that the broader modest tuning of H3K27ac peak height observed across all genes accounts for a portion of the global changes in histone acetylation (Fig. 3C), without impacting gene expression. Although histone acetylation and gene expression are closely linked, precedence does exist for regulation of histone acetylation independent of gene regulation. For example, acetate mobilization from chromatin has been proposed to be a mechanism of intracellular pH regulation, since acetate export from the cell is proton-coupled (McBrian et al. 2013). It has also been suggested that histones might represent a source of acetate that could be mobilized for energetic purposes (Martinez-Pastor et al. 2013), although little direct experimental evidence for this possibility has yet emerged. Relatedly, histones have also been proposed as a methyl sink, and SAM-dependent changes in histone methylation were found to be uncoupled from transcriptional regulation in yeast (Ye et al. 2017). Thus, our data are consistent with a model in which (1) global histone acetylation changes may be largely separate from gene regulation, and (2) acetyl-CoA coordinates more specific upstream mechanisms of gene regulation (such as regulation of transcription factors that function together with KATs) to control locus-specific H3K27ac and gene expression. Consistent with the notion that acetyl-CoA-dependent regulation of transcription factors may be a major mechanism dictating the effects of this metabolite on gene expression, a recent study found that breast cancer metastasis increases upon acetyl-CoA carboxylase inhibition due to acetyl-CoA accumulation and increased Smad2 acetylation (Rios Garcia et al. 2017). Thus, it is possible that acetyl-CoA abundance may regulate multiple transcription factors in different contexts through both acetylation-dependent and acetylation-independent mechanisms. We anticipate that further investigation into the distinct transcriptional programs that are regulated by acetyl-CoA abundance will provide crucial insights into mechanisms through which oncogenic metabolic reprogramming and microenvironmental nutrient availability control cell functions and fates.
Materials and methods
Cells
All cell lines used in this study were regularly checked for mycoplasma and authenticated using STR profiling. LN229 and LN18 cells were normally cultured in RPMI with 10% CS (Hyclone) and supplemented with 1% L-glutamine (Gibco). U87 and U251 cells were normally cultured in DMEM (Gibco) with 10% heat-inactivated FBS (Gemini Bio-products) and supplemented with 1% L-glutamine (Gibco). For experiments, cells were grown in glucose-free medium supplemented with 10% dialyzed FBS (dFBS) (Gibco) and the indicated amounts of glucose and acetate for 24 h unless otherwise indicated. Cells were cultured at 37°C and 5% CO2. Cells expressing NFAT constructs were used within 96 h of transduction.
Reagents (plasmids, antibodies, drugs, and metabolites)
The antibodies used were Tubulin (Sigma), NFAT1 (Cell Signaling for Western; Abcam for immunofluorescence), HA (Thermo-Scientific), H3K27ac (Abcam), H4K12ac (Millipore), AMPKα (Cell Signaling Technology), p-phospho-AMPKα T172 (Cell Signaling Technology), IRDye 800CW goat anti-rabbit IgG secondary (Licor), and IRDye 680RD goat anti-mouse IgG secondary (Licor). NFAT1 shRNA were cloned into a pLKO.1 backbone (sense sequence #47: GTGAACTTCTACGTCATCAAT). ACLY shRNA #12 and #13 were published previously (Lee et al. 2014; Sivanand et al. 2017). siRNA against NFAT1 was a SMARTpool from Dharmacon. CA-NFAT1 (Addgene, plasmid 11792) and wild-type NFAT1 (Addgene, plasmid 11791) were cloned into pLX303. ACLY inhibitor (ACLYi: BMS303141), CsA (TEVA), and ionomycin (Tocris) were dissolved in ethanol. C646 (p300 inhibitor) was dissolved in DMSO. Glucose and sodium acetate stock solutions were purchased from Sigma. GlcNAc (Sigma) was dissolved in water. Fatty acids (palmitate: oleate mix) were conjugated to BSA as described previously (Shah et al. 2016). Pyruvate was purchased from Gibco, and dimethyl-α-ketoglutarate was purchased from Sigma.
Transwell migration assay
Transwell migration assays were completed on 8-µm pore membranes in six-well format (Corning). Cells were pretreated with 1 mM glucose, 25 × 104 cells were seeded with glucose and/or acetate in the upper and lower chambers, and dFBS was added only to the lower chamber as a chemoattractant. After 24 h, cells that did not migrate were cleaned off the top of the membrane with a cotton swab, and migrated cells were fixed, stained with DAPI, and mounted onto slides.
Scratch assay
Cells were grown in a monolayer in a six-well plate format and scratched with a pipet tip. Cell positions were recorded at pretreatment and 24 h after treatment with the indicated conditions. Cell positions were recorded on a Leica staged microscope to allow for accurate pretreatment and post-treatment imaging.
Brain ECM adhesion assay
LN229 GBM cells were pretreated as described above. Cells were then trypsinized and reseeded for adhesion experiments on coverslips prepared as described previously (Barney et al. 2015) with brain-inspired ECM proteins: 50% fibronectin (EMD Millipore), 25% vitronectin (R&D Systems), 20% tenascin C (R&D Systems), and 5% laminin (Life Technologies) (all weight percentages). Briefly, glass coverslips were UV ozone-treated for 10 min (Bioforce Nanoscience) and functionalized through sequential deposition of (3-aminopropyl)triethoxysilane (Sigma), N,N-disuccinimidyl carbonate (Sigma), diisopropylethylamine (Sigma), and then ECM proteins. For adhesion experiments, cells were seeded at 4000 cells per square centimeter in low-glucose medium and imaged 10 min after seeding at 5-min intervals for 2 h using a Zeiss Axio Observer Z1 microscope (Carl Zeiss). ImageJ (National Institutes of Health) was used to manually trace cell areas.
Fibronectin adhesion assay
Before use, 96-well plates were coated with 1% human plasma fibronectin-purified protein (Millipore). Cells were treated with nutrients and/or inhibitors for the indicated amount of time, typically 24 h, in a six-well format. After treatment, media were collected, and cells were washed once with PBS before trypsin was added. Trypsin was quenched with the collected media to prevent changes in nutrient availability during the adhesion assay. For LN229 and LN18 cells, 6 × 104 cells were seeded per well and allowed to adhere for 15 min. For U251 and U87 cells, 4 × 104 cells were seeded per well and allowed to adhere for 15 min. Wells were washed three times with PBS and stained with crystal violet solution for 30 min. Crystal violet was washed with tap water, and the plate was air-dried overnight. Crystal violet was solubilized in 10% acetic acid solution, and the absorbance was read at 590 nm.
Calcium imaging
LN229 cells stably expressing (G418-selected) pGP-CMV-GCaMP6f (Chen et al. 2013) were seeded onto fibronectin-coated coverslips and maintained in low-glucose culture medium (RPMI, 10% dFBS, 1 mM glucose, supplemented with L-glutamine) for 16 h prior to the recordings. Cells were maintained in a temperature- and CO2-controlled chamber mounted on the stage of a fluorescence microscope for the experiment, and GCaMP6f images of individual cells were captured every 15 sec for 1 h following addition of medium containing 1 mM glucose, 1 mM glucose with 5 mM acetate, 10 mM glucose, and 1 mM glucose with 5 mM acetate plus 1 mM EGTA. For visualization of the calcium traces, GCaMP6f fluorescence intensity was normalized to the cells' resting GCaMP6f intensity at the beginning of the recording (F/F0), and each trace contained representative samples from two separate fields. For quantification of calcium signals, only fields that underwent medium-dependent changes were included. Calcium spikes were analyzed by calculating the number of spikes per number of cells in each field.
Metabolomics: PEP and citrate
Cells were grown on six-well dish format. After treatment with 1 mM glucose overnight, glucose and/or acetate was added to the medium for the indicated amount of time. At harvest, the medium was aspirated off the cells, and the cells were scraped directly into 1 mL of supercold 80:20 MetOH:water. Samples were transferred to a 1.5-mL tube. Standards for PEP were added to 1 mL of MetOH:water. Internal standard (20 µL of 0.05 µg/µL U-13C-citrate) was added to every sample and standard. Samples were centrifuged at 17,000 rcf for 10 min at 4°C, and the supernatant was transferred to glass tubes for drying under nitrogen. Pellets were resuspended in 100 µL of water, and mass analysis was determined on a TSQ instrument.
Immunofluorescence
Cells were grown on 22-mm glass coverslips in a six-well dish. Cells were fixed with 3% formaldehyde in 1× PBS for 10 min at room temperature. Each well was washed twice with 1× PBS to remove traces of formaldehyde and then permeablized for 5 min on ice with 0.1% Triton in PBS (PBS-T). The following steps were all performed at room temperature. Coverslips were blocked with 3% nonfat milk in PBS-T (filtered) for 15 min followed by incubation with antibody at 1:100 in 3% milk in PBS-T for 30 min. After incubation, coverslips were washed three times with PBS-T for 1 min and incubated with secondary antibody diluted in 3% milk in PBS-T at 1:1250 for 30 min, protected from light. Coverslips were washed three times with PBS-T for 1 min, stained with Hoechst, washed twice with PBS-T, and mounted. Photos were taken on an Olympus inverted microscope. Merged images were generated by ImageJ. Quantification of the percent nuclear was completed in a blinded manner.
Animals
Xenograft tumor experiments were approved by the Animal Care and Use Committee at the University of Pennsylvania. Briefly, seven female 7-wk-old athymic nude mice (Charles River) were injected subcutaneously with 1.5 million vector control LN229 cells or sg3.8 cells in the left or right flank, respectively. Before each injection, cells were resuspended in 200 µL of DMEM mixed with an equal volume of Matrigel (BD Bioscience). Once palpable tumors were established, tumor size was measured twice a week with a digital caliper. After 6 wk, mice were sacrificed by CO2 euthanasia, and xenograft tumors were dissected, weighed, and snap-frozen with liquid nitrogen until further analysis.
Transcription factor identification
The up-regulated subset of genes from the 881 acetyl-CoA-regulated genes (Lee et al. 2014) was entered into MSigDB version 4.0 with a false discovery rate (FDR) q-value set to <0.05. The top 10 transcription factor targets were queried. The FDR q-value was calculated by MSigDB. The 881 genes were then tested for enrichment of TGGAAA_V$NFAT_Q4_01 using GSEA version 3.0 with 1000 permutations and gene set permutation type as recommended on the GSEA user guide.
Cell lysis for Western blot
Nuclear and cytoplasmic fractionation protocols were followed as published previously (Sivanand et al. 2017). Whole-cell lysate was prepared with RIPA buffer or direct lysis with 2× LDS (Invitrogen). All buffers were supplemented with protease inhibitor cocktail (Sigma) and phosphatase inhibitor tablets (Roche). Samples were quantified by BCA (Thermo Fisher) prepared for Western blot analysis and resolved through a 4%–12% Bis-Tris NuPAGE and Bolt gel systems (Invitrogen). Usually, 20 µg of protein was loaded per lane.
Acid extraction of histones for Western blot
Cells grown on a six-well plate format were pretreated in the indicated conditions for 24 h, and histones were harvested by acid extraction as described (Lee et al. 2014). Usually, 2 µg of protein was loaded for per lane.
RT-qPCR
RNA isolated with Trizol was quantified, and 1–2 µg was used for cDNA synthesis using a kit (Invitrogen). qPCR was completed with Power SYBR (Invitrogen) on ViiA 7 (Applied Biosystems) for a total of 40 cycles. Results were analyzed using ΔΔCt. Primers for cDNA are listed in Supplemental Table S1. Gene expression was normalized to ribosomal protein RPL19.
ChIP library preparation
LN229 cells grown on a 15-cm plate format were pretreated in the indicated conditions for 24 h. Drosophila S2 cells were not exposed to the glucose or acetate treatments, and one additional flask was also used to count the number of cells. Additional plates per condition were used for cell counts. All cells were cross-linked in 1% methanol-free formaldehyde in PBS for 15 min at room temperature. Cells were removed with a cell lifter and transferred to 50-mL conical tubes. Cell pellets were resuspended in lysis buffer 1 (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1× inhibitors [protease inhibitor, sodium butyrate, and phosphatase inhibitors] added fresh). Cells were spun at 3000 rpm at 4°C, and the pellet was resuspended in lysis buffer 2 (10 mM Tris-HCl at pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× inhibitors [protease inhibitor, sodium butyrate, and phosphatase inhibitors] added fresh). Samples were pelleted at 3000 rpm at 4°C and resuspended in 1 mL of lysis buffer 2. Drosophila S2 and LN229 cells were combined at a ratio of two Drosophila cells for every one human cell. This ratio allowed us to check our normalization by qPCR prior to preparation for sequencing. After combining LN229 and Drosophila S2 cells, traditional ChIP protocol as described previously (Shah et al. 2013) was followed with slight changes. Briefly, chromatin was suspended in lysis buffer 3 (10 mM Tris-HCl at pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1× inhibitors [protease inhibitor, sodium butyrate, and phosphatase inhibitors] added fresh) and sheared with a Covaris Sonicator (S220). Equal aliquots of sonicated chromatin were used per immunoprecipitation reaction—H3K27ac (Abcam, ab4729), IgG (Cell Signaling Technology, 2729S), and histone H3 (Abcam, ab1791)—overnight at 4°C. The beads were then washed, and DNA was reverse-cross-linked and purified. Following ChIP, immunoprecipitated DNA was quantified by qPCR using Power SYBR (Life Technologies) on ViiA 7 real-time PCR (Life Technologies). Libraries were prepared using the New England Biolabs Next Ultra II DNA library kit with 25 nM starting material. Samples were multiplexed and sequenced on an Illumina Next-Seq platform.
ChIP-Rx analysis
Sequenced reads were aligned to the human genome (hg19) and Drosophila genome (dm3) following methods described previously (Orlando et al. 2014). All samples were trimmed for low-quality ends of reads and adaptors by Trimmomatic (Bolger et al. 2014) with the parameters ILLUMINACLIP:TruSeq3-SE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:3:3 MINLEN:30 followed by alignment to the human genome (hg19) and Drosophila genome (dm3) using Bowtie2 (Langmead and Salzberg 2012) with the parameters -N 1 --phred33. H3 and H3K27ac samples were peak-called against the paired input samples using MACS (Zhang et al. 2008) after removing duplicate reads and mitochondria reads by SAMtools (Li 2011). Next, all peaks were annotated by HOMER annotatePeaks.pl (Heinz et al. 2010). As described previously (Orlando et al. 2014), read counts (depth) were adjusted with traditional normalization (per base pair per peak [TSS] on top of the standard total mapped tag normalization of 10 million tags) or reference-adjusted normalization (per base pair per peak [TSS] on top of the total mapped tag normalization of Drosophila reference). To create the browser tracks shown in Figure 3 and Supplemental Figure S3, we created bedGraph and bigwig files from each human SAM file using the HOMER-made University of California at Santa Cruz (UCSC) file (Heinz et al. 2010). The bigwig files were then visualized in the Integrated Genomics Viewer (IGV) (Thorvaldsdottir et al. 2013) browser to produce the gene tracks. Processed ChIP-seq files and raw FASTQ files are available on the Gene Expression Omnibus repository under study GSE109340.
For metagene analysis of TSS regions, two sets of genes (all protein-coding genes and acetyl-CoA-up-regulated genes [as defined in Lee et al. 2014], shown as purple and orange clusters) were used for comparison between traditional normalization and reference-adjusted normalization. Each TSS was separated into 100 equally sized bins, and the number of ChIP-seq reads mapping into each bin was calculated. These counts were then scaled by either traditional or reference-adjusted normalization, and, finally, the column-wise average was visualized. For the acetyl-CoA-regulated genes, genes were identified from the RNA-seq analysis with significantly differential expression.
Heat maps were created by gplots heatmap.2 (https://CRAN.R-project.org/package=gplots) after HOMER annotatePeaks.pl separated the peak signals of each gene into 10 equally sized bins between −2 kb and +2 kb from the center of the TSS. To identify differentially regulated regions, DiffBind (http://bioconductor.org/packages/3.7/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf) and DESeq2 (Love et al. 2014) with two replicates of each condition were used. The cutoff threshold was set at FDR-adjusted P-value <0.01, and peak width was set at 2500 base pairs × 2 from summit.
Statistics
All results reported in the figures and Supplemental Material are presented as mean ± SEM unless specified otherwise. Data were reported as average of biological replicates. P-values were calculated based on two-tailed unpaired Student's t-tests.
Supplementary Material
Acknowledgments
This work was supported by R01CA174761 and a Pew Biomedical Scholar Award to K.E.W. J.V.L. was supported by predoctoral fellowship 1F31CA189744 and an Advancing Graduate Studies (AGS) Patel Family Scholar Award. S.Z. is supported by F99CA222741. S.F. is supported by Penn-PORT (Penn-Post-doctoral Opportunities in Research and Teaching) Institutional Research and Academic Career Development Award post-doctoral fellowship K12 GM081259. S.T. is supported by American Diabetes Association post-doctoral fellowship 1-18-PDF-144. K.K. received Jumpstart for Juniors undergraduate summer funding from the University of Pennsylvania. In addition, this work was funded by a grant from the National Institutes of Health (1DP2CA186573-01) awarded to S.R.P. S.R.P. is a Pew Biomedical Scholar supported by the Pew Charitable Trusts. L.E.B. was partially supported by National Research Service Award T32 GM008515 from the National Institutes of Health. A.D.S. was supported by a National Science Foundation Graduate Research Fellowship (1451512). N.W.S. was supported by K22ES026235 and a Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program (CURE) grant.
Author contributions: J.V.L. and K.E.W. conceptualized the study, designed the research, and wrote the manuscript. J.V.L. performed the majority of experiments. C.T.B. and B.D.F. measured Ca2+ signals and edited the manuscript. S.T. and N.W.S. performed metabolite measurements. L.E.B., A.D.S., and S.R.P. measured cell adhesion to the brain-inspired matrix. K.K., A.C., S.Z., and S.F. performed experiments. P.S. and S.L.B. assisted in ChIP-Rx optimization. T.K. analyzed ChIP-Rx data. All authors reviewed data and provided feedback on the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.311027.117.
References
- Badran BM, Wolinsky SM, Burny A, Willard-Gallo KE. 2002. Identification of three NFAT binding motifs in the 5′-upstream region of the human CD3γ gene that differentially bind NFATc1, NFATc2, and NF-κB p50. J Biol Chem 277: 47136–47148. [DOI] [PubMed] [Google Scholar]
- Barney LE, Dandley EC, Jansen LE, Reich NG, Mercurio AM, Peyton SR. 2015. A cell-ECM screening method to predict breast cancer metastasis. Integr Biol 7: 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. 2011. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42: 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigurupati S, Venkataraman R, Barrera D, Naganathan A, Madan M, Paul L, Pattisapu JV, Kyriazis GA, Sugaya K, Bushnev S, et al. 2010. Receptor channel TRPC6 is a key mediator of Notch-driven glioblastoma growth and invasiveness. Cancer Res 70: 418–427. [DOI] [PubMed] [Google Scholar]
- Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW. 2015. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, Walters H, Tantawy MN, Fu A, Manning HC, et al. 2014. Acetate dependence of tumors. Cell 159: 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias AJ, Aksoylar HI, Yu J, Snyder NW, Worth AJ, Iyer SS, Wang J, Ben-Sahra I, Byles V, Polynne-Stapornkul T, et al. 2016. Akt–mTORC1 signaling regulates Acly to integrate metabolic input to control of macrophage activation. Elife 5: e11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. 2012. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell 48: 612–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller C, Forne I, Imhof A, Becker PB. 2015. Global and specific responses of the histone acetylome to systematic perturbation. Mol Cell 57: 559–571. [DOI] [PubMed] [Google Scholar]
- Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441: 179–185. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. 2007. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710. [DOI] [PubMed] [Google Scholar]
- Gao X, Lin SH, Ren F, Li JT, Chen JJ, Yao CB, Yang HB, Jiang SX, Yan GQ, Wang D, et al. 2016. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat Commun 7: 11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez C, Rao A. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J Exp Med 187: 2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins BJ, Irrinki KM, Mallilankaraman K, Lien YC, Wang Y, Bhanumathy CD, Subbiah R, Ritchie MF, Soboloff J, Baba Y, et al. 2010. S-glutathionylation activates STIM1 and alters mitochondrial homeostasis. J Cell Biol 190: 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, Tsui YC, Cui G, Micevic G, Perales JC, et al. 2015. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162: 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhanwar-Uniyal M, Labagnara M, Friedman M, Kwasnicki A, Murali R. 2015. Glioblastoma: molecular pathways, stem cells and therapeutic targets. Cancers (Basel) 7: 538–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird A, Zhao S, Wellen KE, Michelakis ED. 2016. Metabolic control of epigenetics in cancer. Nat Rev Cancer 16: 694–707. [DOI] [PubMed] [Google Scholar]
- Koenig A, Linhart T, Schlengemann K, Reutlinger K, Wegele J, Adler G, Singh G, Hofmann L, Kunsch S, Buch T, et al. 2010. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology 138: 1189–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Bhatt HS, Easom RA. 2002. NFAT regulates insulin gene promoter activity in response to synergistic pathways induced by glucose and glucagon-like peptide-1. Diabetes 51: 691–698. [DOI] [PubMed] [Google Scholar]
- Leclerc C, Haeich J, Aulestia FJ, Kilhoffer MC, Miller AL, Neant I, Webb SE, Schaeffer E, Junier MP, Chneiweiss H, et al. 2016. Calcium signaling orchestrates glioblastoma development: facts and conjunctures. Biochim Biophys Acta 1863: 1447–1459. [DOI] [PubMed] [Google Scholar]
- Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan ZF, Lim HW, Liu S, et al. 2014. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab 20: 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JV, Shah S, Carrer A, Wellen KE. 2015. A cancerous web: signaling, metabolism, and the epigenome. Mol Cell Oncol 2: e965620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr, Meyer T. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol 15: 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M, Toker A. 2009. NFAT proteins: emerging roles in cancer progression. Nat Rev Cancer 9: 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor B, Cosentino C, Mostoslavsky R. 2013. A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer Discov 3: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, et al. 2014. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159: 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB, et al. 2013. Histone acetylation regulates intracellular pH. Mol Cell 49: 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Vander Heiden MG. 2010. Metabolism strikes back: metabolic flux regulates cell signaling. Genes Dev 24: 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteith GR, Prevarskaya N, Roberts-Thomson SJ. 2017. The calcium–cancer signalling nexus. Nat Rev Cancer 17: 367–380. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. [DOI] [PubMed] [Google Scholar]
- Motiani RK, Hyzinski-Garcia MC, Zhang X, Henkel MM, Abdullaev IF, Kuo YH, Matrougui K, Mongin AA, Trebak M. 2013. STIM1 and Orai1 mediate CRAC channel activity and are essential for human glioblastoma invasion. Pflugers Arch 465: 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MR, Rao A. 2010. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol 10: 645–656. [DOI] [PubMed] [Google Scholar]
- Okamura H, Aramburu J, Garcia-Rodriguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell 6: 539–550. [DOI] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. 2014. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9: 1163–1170. [DOI] [PubMed] [Google Scholar]
- Osinalde N, Mitxelena J, Sanchez-Quiles V, Akimov V, Aloria K, Arizmendi JM, Zubiaga AM, Blagoev B, Kratchmarova I. 2016. Nuclear phosphoproteomic screen uncovers ACLY as mediator of IL-2-induced proliferation of CD4+ T lymphocytes. Mol Cell Proteomics 15: 2076–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. 2016. The emerging hallmarks of cancer metabolism. Cell Metab 23: 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO. 2016. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354: 481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup MW, Mouw JK, Weaver VM. 2014. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep 15: 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. 2015. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab 21: 805–821. [DOI] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS. 2015. Store-operated calcium channels. Physiol Rev 95: 1383–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney JW Jr. 1986. A model for receptor-regulated calcium entry. Cell Calcium 7: 1–12. [DOI] [PubMed] [Google Scholar]
- Qin JJ, Nag S, Wang W, Zhou J, Zhang WD, Wang H, Zhang R. 2014. NFAT as cancer target: mission possible? Biochim Biophys Acta 1846: 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios Garcia M, Steinbauer B, Srivastava K, Singhal M, Mattijssen F, Maida A, Christian S, Hess-Stumpp H, Augustin HG, Muller-Decker K, et al. 2017. Acetyl-CoA carboxylase 1-dependent protein acetylation controls breast cancer metastasis and recurrence. Cell Metab 26: 842–855.e5. [DOI] [PubMed] [Google Scholar]
- Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, et al. 2005. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol 169: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL, Crabtree GR. 1992. The mechanism of action of cyclosporin A and FK506. Immunol Today 13: 136–142. [DOI] [PubMed] [Google Scholar]
- Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, et al. 2015. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27: 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug ZT, Vande Voorde J, Gottlieb E. 2016. The metabolic fate of acetate in cancer. Nat Rev Cancer 16: 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. 2013. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Carriveau WJ, Li J, Campbell SL, Kopinski PK, Lim HW, Daurio N, Trefely S, Won KJ, Wallace DC, et al. 2016. Targeting ACLY sensitizes castration-resistant prostate cancer cells to AR antagonism by impinging on an ACLY–AMPK–AR feedback mechanism. Oncotarget 7: 43713–43730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanand S, Rhoades S, Jiang Q, Lee JV, Benci J, Zhang J, Yuan S, Viney I, Zhao S, Carrer A, et al. 2017. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol Cell 67: 252–265 e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanand S, Viney I, Wellen KE. 2018. Spatiotemporal control of acetyl-CoA metabolism in chromatin regulation. Trends Biochem Sci 43: 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart TA, Yapa KT, Monteith GR. 2015. Altered calcium signaling in cancer cells. Biochim Biophys Acta 1848: 2502–2511. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie X, Han S, Meng L, Wang Y, Wu A. 2013. NFAT1 is highly expressed in, and regulates the invasion of, glioblastoma multiforme cells. PLoS One 8: e66008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaeth M, Maus M, Klein-Hessling S, Freinkman E, Yang J, Eckstein M, Cameron S, Turvey SE, Serfling E, Berberich-Siebelt F, et al. 2017. Store-operated Ca2+ entry controls clonal expansion of T cells through metabolic reprogramming. Immunity 47: 664–679 e666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, et al. 2006. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 312: 1220–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Z, Li J, Zhang W, Ren F, Yue W. 2015. NFATc1 activation promotes the invasion of U251 human glioblastoma multiforme cells through COX-2. Int J Mol Med 35: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB. 2012. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol 13: 270–276. [DOI] [PubMed] [Google Scholar]
- Wong BW, Wang X, Zecchin A, Thienpont B, Cornelissen I, Kalucka J, Garcia-Caballero M, Missiaen R, Huang H, Bruning U, et al. 2017. The role of fatty acid β-oxidation in lymphangiogenesis. Nature 542: 49–54. [DOI] [PubMed] [Google Scholar]
- Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. 2017. A metabolic function for phospholipid and histone methylation. Mol Cell 66: 180–193 e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cruickshanks N, Yuan F, Wang B, Pahuski M, Wulfkuhle J, Gallagher I, Koeppel AF, Hatef S, Papanicolas C, et al. 2017. Targetable T-type calcium channels drive glioblastoma. Cancer Res 77: 3479–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Torres A, Henry RA, Trefely S, Wallace M, Lee JV, Carrer A, Sengupta A, Campbell SL, Kuo YM, et al. 2016. ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep 17: 1037–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.