Abstract
The emergence of microRNA as regulators of organogenesis and tissue differentiation has stimulated interest in the ablation of microRNA expression and function during discrete periods of development. To this end, inducible, conditional modulation of microRNA expression with doxycycline-based tetracycline-controlled transactivator and tamoxifen-based estrogen receptor systems has found widespread use. However, the induction agents and components of genome recombination systems negatively impact pregnancy, parturition, and postnatal development; thereby limiting the use of these technologies between late gestation and the early postnatal period. MicroRNA inhibitor (antimiR) administration also represents a means of neutralizing microRNA function in vitro and in vivo. To date, these studies have used direct (parenteral) administration of antimiRs to experimental animals. As an extension of this approach, an alternative means of regulating microRNA expression and function is described here: the maternal–placental–fetal transmission of antimiRs. When administered to pregnant dams, antimiRs were detected in offspring and resulted in a pronounced and persistent reduction in detectable steady-state free microRNA levels in the heart, kidney, liver, lungs, and brain. This effect was comparable to direct injection of newborn mouse pups with antimiRs, although maternal delivery resulted in fewer off-target effects. Furthermore, depletion of steady-state microRNA levels via the maternal route resulted in concomitant increases in steady-state levels of selected microRNA targets. This novel methodology permits the temporal regulation of microRNA function during late gestation and in neonates, without recourse to conventional approaches that rely on doxycycline and tamoxifen, which may confound studies on developmental processes.
Keywords: microRNA, antimiR, development, organogenesis, maternal transmission
INTRODUCTION
MicroRNA are emerging as pivotal regulators of cell differentiation and organogenesis during embryonic and postnatal development (Ivey and Srivastava 2015). Powerful technologies exist that facilitate studies on microRNA function, including inducible, conditional deletion-ready mouse strains, where the induction or ablation of gene expression can be achieved by combining the appropriate “floxed” mouse strain with a suitable conditional (cell-type or tissue-specific) mouse Cre-recombinase driver-line (Le and Sauer 2001). The inclusion of inducible systems, such as those based on the tetracycline-controlled transactivator (tTA) (Gossen and Bujard 1992) or the ligand-binding domain of the estrogen receptor (ER) (Metzger et al. 1995) or ER variants (ERT and ERT2) (Indra et al. 1999) introduce the possibility of modulating gene expression in a temporal fashion, where activation (or repression) of microRNA expression with the tetracycline analog doxycycline, or the estradiol analog 4-hydroxytamoxifen, at a specific, predefined time-point is possible.
However, powerful technologies such as these are not without drawbacks, which fall into two categories. The components of inducible systems, and the induction agent itself, may have toxic or off-target effects. Expression of the reverse tetracycline-transactivator (rtTA) alone in pulmonary epithelial cells causes emphysema in mice, even in the absence of the induction agent doxycycline (Sisson et al. 2006); while in humans, tetracycline is contraindicated in pregnancy, and is not used in neonates or adolescents due to disturbances to bone and tooth development (Mylonas 2011). Similarly, doxycycline alone has direct effects, as a pan-matrix metalloproteinase inhibitor (Golub et al. 1987), and anti-angiogenic agent (Gilbertson-Beadling et al. 1995). Off-target effects have also been noted for tamoxifen, the induction agent in the ER-coupled systems (Flynn et al. 2017). Also, while tamoxifen is well-tolerated in newborn mice, appreciable toxicity has been noted during concomitant use of tamoxifen with injurious stimuli used to drive pathological transformation in disease models (Ruiz-Camp et al. 2017).
Apart from off-target effects and toxicity, the timing of induction agent administration is complicated during pregnancy, when genome recombination during late gestation or in the early postnatal stages might be desired; particularly when the estrogen analog, tamoxifen is used (Lizen et al. 2015). Tamoxifen is teratogenic (Cunha et al. 1987), is associated with congenital abnormalities (Braems et al. 2011), and delays labor in mice (Fang et al. 1996). Placental anomalies and fetal loss were also noted in mice fed doxycycline (Moutier et al. 2003), and a pronounced arrest of postnatal lung development was noted in newborn rats receiving oral doxycycline (Hosford et al. 2004). These reports highlight a need for an alternative means of modulating microRNA expression during late gestation and the early postnatal period, for studies on organogenesis, and on organ maturation in neonates.
Such an alternative means have emerged with the advent of synthetic microRNA inhibitors (antimiRs), exemplified by the original pioneering work of Jan Krützfeldt and coworkers, who documented the utility of 2′-O-methoxyethyl-modified oligonucleotides (antagomiRs) for the first time in vivo (Krützfeldt et al. 2005, 2007). Strategies to modulate microRNA function using synthetic oligonucleotides continue to evolve (Stenvang et al. 2012). Much emphasis is currently focused on the in vivo application of a spectrum of antimiRs that are chemically stabilized against degradation. For example, locked-nucleic acid (LNA) antimiRs are a class of bicyclic RNA analogs that contain a furanose ring in the sugar–phosphate backbone that is chemically “locked” in a RNA mimicking N-type (C3′-endo) conformation by the introduction of a 2′-O,4′-C methylene bridge (Obika et al. 1997; Koshkin et al. 1998). To this end, the present study explored the possibility of directly modulating microRNA function in newborn mouse pups, and during the early postnatal phase, by maternal transmission of LNA antimiRs in utero.
RESULTS
AntimiR-29a-3p reduced steady-state miR-29a-3p levels in multiple organs
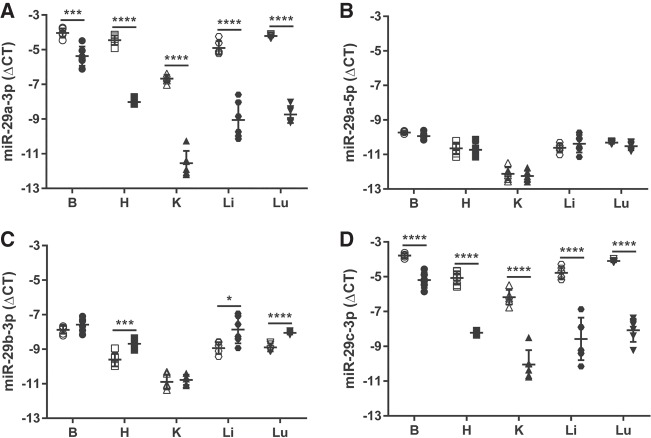
Either scrambled LNA antimiR or LNA antimiR-29a-3p was administered by single i.p. injection to newborn mouse pups on the day of birth (Fig. 1A), and 7 d post-administration, the brain, heart, kidney, liver, and lungs were harvested from mouse pups, and screened for steady-state free miR-29a-3p levels by real-time RT-PCR analysis of total RNA pools. Steady-state free miR-29a-3p levels were reduced in all five organs in mouse pups that received antimiR-29a-3p, compared with mouse pups that received scrambled antimiR (Fig. 2A). The impact of antimiR-29a-3p on free miR-29a-3p steady-state levels was strongest and approximately equivalent for the kidney (ΔΔCT ≈ 5, equivalent to a ±32-fold change) and the lungs (ΔΔCT ≈ 5, equivalent to a ±32-fold change), followed by the liver (ΔΔCT ≈ 4, equivalent to a ±16-fold change) and heart (ΔΔCT ≈ 3.5, equivalent to a ±11-fold change). The brain was least impacted (ΔΔCT ≈ 1.5, equivalent to a ±threefold change). No impact of antimiR-29a-3p on steady-state free levels of the complementary strand, miR-29a-5p, was noted, in any organ (Fig. 2B).
FIGURE 1.
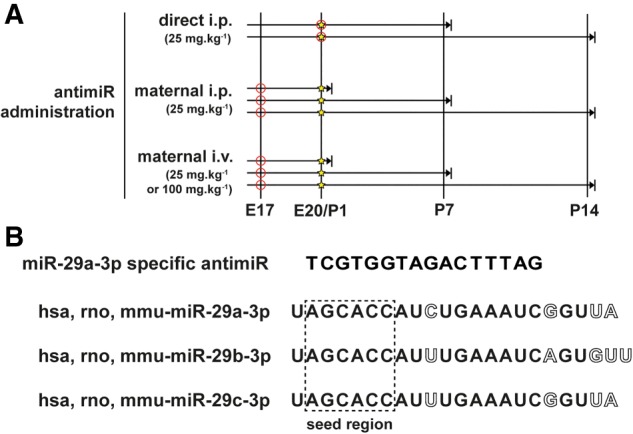
Schematic illustration of the antimiR administration protocol. (A) An antimiR directed against miR-29a-3p (or a scrambled antimiR) was administered by injection (○) via the intraperitoneal (i.p.) route (25 mg kg−1) on the day of birth [postnatal day (P)1; ⋆] to newborn pups; or via either the i.p. (25 mg kg−1) or intravenous (i.v.; 25 or 100 mg kg−1) routes to pregnant dams on the 17th day of gestation, here denoted embryonic day (E)17. Organs were harvested (→|) on P1, P7, or P14. (B) The antimiR-29a-3p sequence corresponded to part of the seed sequence (contained within the dashed-line box) of the miR-29 family of microRNA, and as such, targeted all three miR-29 family members in mice: miR-29a-3p, miR-29b-3p, and miR-29c-3p. Nucleotide base mismatches comparing the three miR-29a family members are indicated with hollow letters. (hsa) Homo sapiens; (mmu) Mus musculus; (rno) Rattus norvegicus.
FIGURE 2.
The miR-29 family members can be targeted in mouse pups directly injected with an antimiR. Either a scrambled antimiR (open symbols) or an antimiR directed against miR-29a-3p (closed symbols) was administered by intraperitoneal injection (25 mg kg−1) to newborn mouse pups on the day of birth. Organs were harvested from mouse pups on the seventh day of life: (B) brain, (H) heart, (K) kidney, (Li) liver, (Lu) lung. Total RNA pools were screened by real-time RT-PCR for (A) miR-29a-3p, (B) miR-29a-5p, (C) miR-29b-3p, and (D) miR-29c-3p. The miR steady-state expression levels are described by the mean ΔCT ± SD. (n = 5–6 animals, per group; each symbol represents an individual animal), where ΔCT = CT(Rnu6) − CT(miR of interest). Mean values were compared between the scrambled versus miR-29a-3p-specific antimiR-treated mouse pups by unpaired Student's t-test. (*) P < 0.05; (***) P < 0.001; (****) P < 0.0001.
The impact of antimiR-29a-3p on steady-state levels of free miR-29b-3p and miR-29c-3p, the remaining two members of the miR-29 family, was also assessed, since the antimiR spanned the seed region shared by all three miRs (Fig. 1B). There was an impact of antimiR-29a-3p on steady-state levels of free miR-29b-3p noted in the heart, liver, and lung, albeit to a lesser extent than that observed for miR-29a-3p; where an approximately twofold increase in steady-state levels of free miR-29b-3p was noted in these three organs (Fig. 2C). In contrast, the impact of antimiR-29a-3p on steady-state free miR-29c-3p levels effectively paralleled the trends, both in magnitude and in the organs studies, that were noted for miR-29a-3p (Fig. 2D).
The in vivo impact of antimiR-29a-3p on free miR-29a-3p steady-state levels persists over time
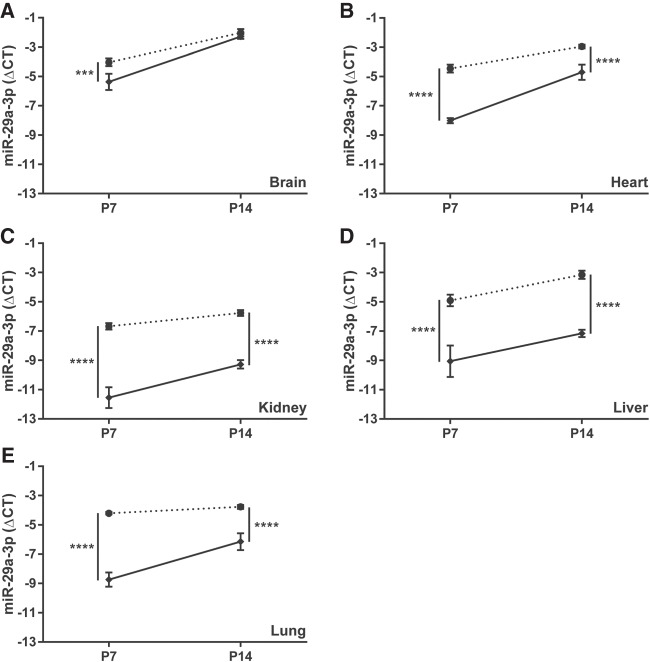
After direct injection of mouse newborns, the impact of antimiR-29a-3p on steady-state free miR-29a-3p levels persisted over time, in all organs studied except the brain (Fig. 3). The comparatively moderate (threefold) difference in free miR-29a-3p steady-state levels noted in the brain 7 d after antimiR administration was lost by the fourteenth day of postnatal life, where steady-state free miR-29a-3p levels in the brains of scrambled antimiR-treated versus antimiR-29a-3p-treated mice were equivalent (Fig. 3A). While the magnitude of the impact of antimiR-29a-3p on steady-state free miR-29a-3p levels was reduced comparing P7 with P14 in the heart (Fig. 3B), kidney (Fig. 3C), and the lungs (Fig. 3E), the steady-state miR-29a-3p levels always remained at least fourfold different versus scrambled antimiR-treated controls. For the liver, the magnitude of the effect of antimiR-29a-3p on steady-state miR-29a-3p levels was the same at P7 and P14 (Fig. 3D).
FIGURE 3.
The impact of a directly injected antimiR directed against miR-29a-3p persists, but is slowly lost over the first fourteen days of postnatal life in mouse pups. Either a scrambled antimiR (closed circle, dotted line) or an antimiR directed against miR-29a-3p (closed diamond, solid line) was administered by intraperitoneal injection (25 mg kg−1) to newborn mouse pups on the day of birth. The (A) brain, (B) heart, (C) kidney, (D) liver, and (E) lung were harvested en bloc from mouse pups on postnatal day (P)7 or P14. The impact of antimiR-29a-3p administered at P1, on steady-state miR-29a-3p levels in these five organs was assessed at P7 and P14. The miR steady-state expression levels are described by the mean ΔCT ± SD. (n = 5–6 animals, per group; data points for each individual animal are omitted, for clarity), where ΔCT = CT(Rnu6) − CT(miR-29a-3p). Mean values were compared between the scrambled versus miR-29a-3p-specific antimiR-treated mouse pups by unpaired Student's t-test. (***) P < 0.001; (****) P < 0.0001.
AntimiR-29a-3p administration to pregnant dams impacts free miR-29a-3p steady-state levels in dams and offspring
Administration of a high (100 mg kg−1) dose of antimiR-29a-3p to pregnant dams via the i.v. route on the 17th day of gestation resulted in a pronounced impact on steady-state free miR-29a-3p levels in the brain, heart, kidney, liver, and lung at PP1 (Fig. 4), with an approximately 512-fold difference in detectable free miR-29a-3p steady-state levels noted in the kidney (the most affected organ), comparing antimiR-29a-3p-treated dams with scrambled antimiR-treated dams. The effects of antimiR-29a-3p administration remained pronounced also at PP14 (Fig. 4).
FIGURE 4.
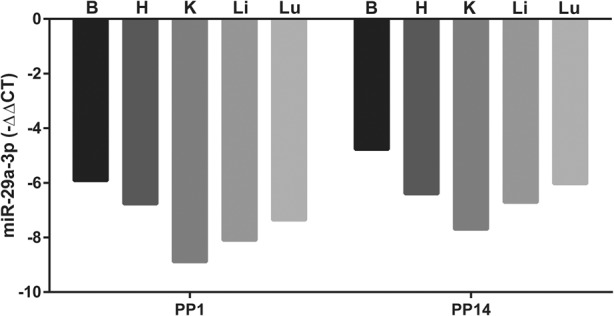
Reduced steady-state free miR-29a-3p levels are detected in the organs of antimiR-29a-3p-injected dams. The steady-state free miR-29a-3p levels were assessed by real-time RT-PCR in the brain (B), heart (H), kidney (K), liver (Li), and lungs (Lu) of dams at postpartum day (PP)1 and PP14, after intravenous injection of either a scrambled antimiR (100 mg kg−1) or antimiR-29a-3p (100 mg kg−1) at gestational day 17. Data are presented as mean ΔΔCT, where ΔΔCT reflects ΔCT(antimiR-29a-3p treatment)–ΔCT(scrambled antimiR treatment), the negative value reflecting reduced detectable free miR-29a-3p in antimiR-29a-3p-injected dams. The original ΔCT values were obtained by ΔCT = CT(Rnu6) − CT(miR-29a-3p).
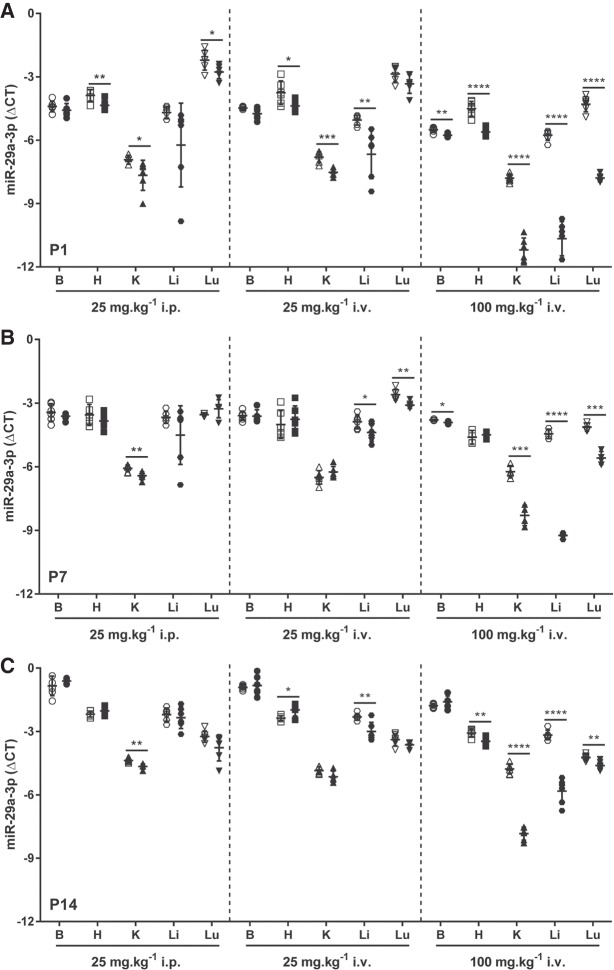
Administration of antimiR-29a-3p to pregnant dams via the i.p. route on the 17th day of gestation at a dose of 25 mg kg−1 was without impact on the body mass of pups at P1 (the day of birth) compared to pups delivered by dams that received scrambled antimiR (Fig. 5A). However, antimiR-29a-3p administration did impact the steady-state free miR-29a-3p levels in newborn pups, in the heart, kidney, and lungs (Fig. 6A, left-hand panel). This impact did not persist in the heart and lungs at P7 (Fig. 6B, left-hand panel) or P14 (Fig. 6C, left-hand panel), where steady-state free miR-29a-3p levels remained affected only in the kidney. Interestingly, miR-29a-3p administration to dams did impact the body mass of pups at P7 (Fig. 5B) and P14 (Fig. 5C), compared with pups from scrambled antimiR-treated dams.
FIGURE 5.
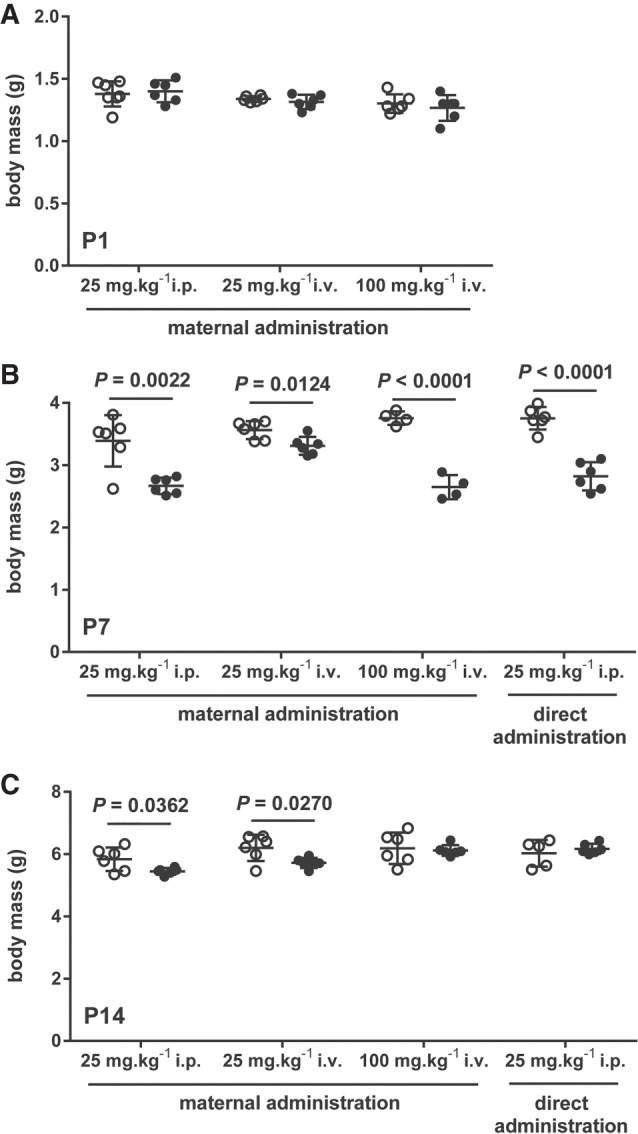
Maternal and direct administration of antimiR-29a-3p did impact postnatal growth of mouse pups. The body mass of mouse pups delivered by dams treated with intraperitoneal (i.p.) or intravenous (i.v.) scrambled antimiR (open circles) or antimiR-29a-3p (closed circles) was assessed on (A) the day of birth, postnatal day (P)1; as well as on (B) P7 and (C) P14. Additionally, the body mass of mouse pups directly injected at P1 (via the i.p. route) with scrambled antimiR (open circles) or antimiR-29a-3p (closed circles) was also assessed at (B) P7 and (C) P14. Data reflect mean body mass ± SD. (n = 5–6, per group). Mean values were compared between the scrambled versus miR-29a-specific antimiR-treated mouse pups by unpaired Student's t-test. All significant differences (P < 0.05) are illustrated, and the P-value is provided above the pair of data sets.
FIGURE 6.
Steady-state levels of miR-29a-3p in mouse pups are influenced by administration of antimiR-29a-3p to pregnant dams. Either a scrambled antimiR (open symbols) or an antimiR directed against miR-29a-3p (closed symbols) was administered by intraperitoneal (25 mg kg−1) or intravenous (i.v.; 25 or 100 mg kg−1) injection to pregnant dams on the 17th day of gestation. Organs were harvested from mouse pups, either on the day of birth, postnatal day (P)1, or at P7 or P14: (B) brain, (H) heart, (K) kidney, (Li) liver, (Lu) lung. Total RNA pools were screened by real-time RT-PCR for steady-state miR-29a-3p levels in organs from pups at (A) P1, (B) P7, and (C) P14. The miR steady-state expression levels are described by the mean ΔCT ± SD. (n = 4–6 animals, per group; each symbol represents an individual animal), where ΔCT = CT(Rnu6) − CT(miR-29a-3p). Mean values were compared between the scrambled versus miR-29a-specific antimiR-treated mouse pups by unpaired Student's t-test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001.
When the route of administration of antimiR-29a-3p was changed from i.p. to i.v. on the 17th day of gestation—also at a dose of 25 mg kg−1—a similar pattern was obtained to that obtained using the i.p. administration protocol, both in terms of an impact on offspring body mass (Fig. 5A–C) as well as on free microRNA-29a-3p steady-state levels (Fig. 6A–C, middle panels), where no impact on the miR-29a-3p levels was noted in the brain at any time-point investigated. In contrast, a persistent impact of antimiR-29a-3p on free miR-29a-3p steady-state levels in the liver was noted, at all time-points investigated: P1, P7, and P14.
Remaining with the i.v. route on the 17th day of gestation, when the antimiR dose was increased from 25 mg kg−1 to 100 mg kg−1, paradoxically, no impact on body mas was noted at P1 or P14, although a reduction in body mass (compared to pups from scrambled antimiR-treated dams) was noted at P7 (Fig. 5A–C). The steady-state levels of free miR-29a-3p were impacted in all organs investigated, including the brain, at P1 (Fig. 6A, right-hand panel). The effect persisted in the brain, kidney, liver, and the lungs at P7 (Fig. 6B, right-hand panel). By P14, the effect of antimiR-29a-3p was lost in the brain, but was now noted in the heart, and persisted in the kidney, liver, and the lungs (Fig. 6C, right-hand panel). By way of comparison, body masses of directly injected pups followed a similar trajectory to body-masses of pups from dams that received 100 mg kg−1 i.v. antimiR-29a-3p (Fig. 5B,C). In sum, the 100 mg kg−1 dose applied via the i.v. route to pregnant dams yielded the largest magnitude of effect on free miR-29a-3p steady-state levels in offspring.
Antimir-29a-3p detection in mouse organs
AntimiR-29a-3p was detected in the lung, liver, and kidney of mouse pups at P7 that were delivered by pregnant dams injected via the i.v. route with antimiR-29a-3p (dose, 100 mg kg−1) using a stem–loop primer-based real-time RT-PCR approach (Fig. 7). This approach delineated the signal attributable to antimiR-29a-3p from that of the signal from antimiR sequences present in pri- and pre-miR-29 family members, which were present in the background. The magnitude of the fold-change was smaller in lung tissue (≈75-fold) in comparison to liver (≈1750-fold) and kidney (≈1600-fold) (Fig. 7).
FIGURE 7.
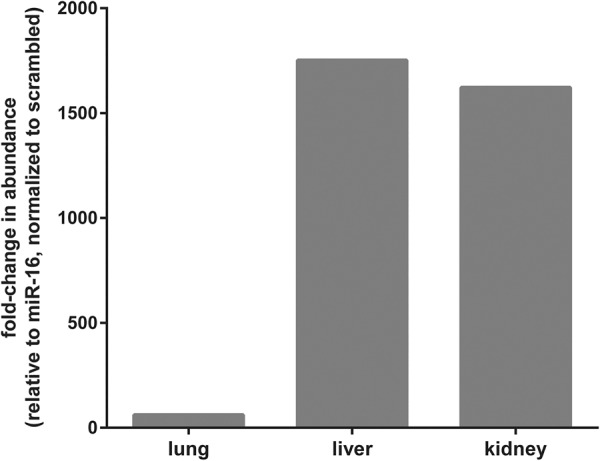
AntimiR-29a-3p was detected in selected organs of pups born to antimiR-29a-3p-treated dams by stem–loop real-time RT-PCR. Scrambled antimiR or antimiR-29a-3p was administered (100 mg kg−1, via the intravenous route) to pregnant dams on gestational day 17. The antimiR-29a-3p was detected by stem–loop real-time RT-PCR in total RNA pools from the lungs, liver, and kidneys of offspring at postnatal day 7. The data reflect the mean fold-change in the abundance of PCR product detected by stem–loop real-time RT-PCR, relative to miR-16, and normalized to the scrambled antimiR-treated group, as described in Materials and Methods.
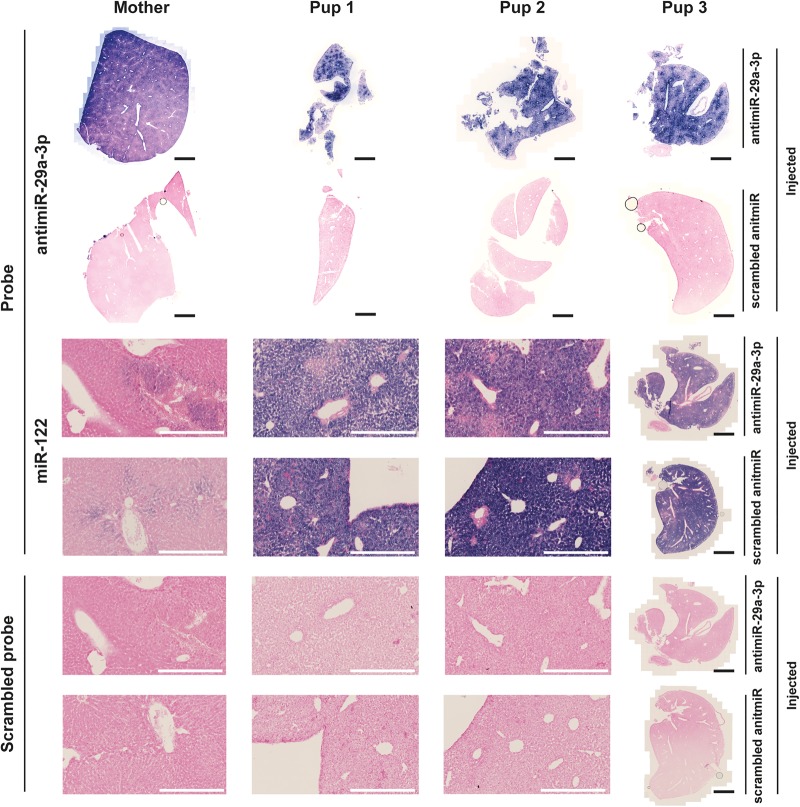
As conformation of these data, antimiR-29a-3p was also detected in the livers of mouse pups at P7, that were delivered by dams injected via the i.v. route with antimiR-29a-3p (dose, 100 mg kg−1), as well as in the liver of a nursing dam at PP7, by in situ hybridization. The antimiR-29a-3p in situ hybridization signal was prominent in the livers of both pups and the dam. As a positive control, an in situ hybridization signal for miR-122 was also used. It is noteworthy that miR-122 expression is developmentally regulated in the liver (Hsu et al. 2016), with a pronounced increase in expression in the second week of postnatal life (over P7-P14), which then dramatically drops, and is substantially reduced in adulthood: hence, the increased miR-122 in situ hybridization signal in the lungs of mouse pups at P7 (“Pup1,” “Pup2,” and “Pup3,” in Fig. 8), compared with the nursing dam (“Mother” in Fig. 8).
FIGURE 8.
AntimiR-29a-3p was detected in selected organs of pups born to antimiR-29a-3p-treated dams by in situ hybridization. AntimiR-29a-3p was administered (100 mg kg−1, via the intravenous route) to pregnant dams on gestational day 17. The antimiR-29a-3p was detected by in situ hybridization in the livers of offspring at postnatal day 7. Illustrated are liver sections from a dam (Mother) and from three different pups born to dams treated with either scrambled antimiR or with antimiR-29a-3p. AntimiR-29a-3p was detected with an in situ probe directed against antimiR-29a-3p, while a probe directed against the developmentally regulated miR-122 served as a positive control for the in situ hybridization reaction, and a scrambled in situ hybridization probe served as a negative control for the in situ hybridization reaction. A low-magnification view is illustrated for all samples from Pup 3, while higher-magnification views are illustrated for the dam (Mother), as well as Pup 1 and Pup 2. Black scale bars represent 2 mm, while white scale bars 400 µm.
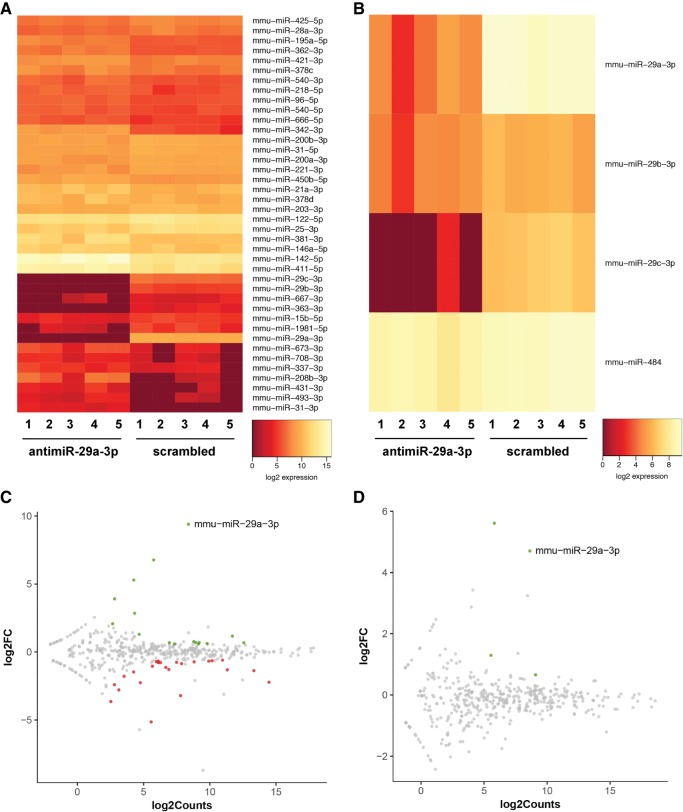
Off-target effects on other microRNA species in the liver were noted after application of the antimiR-29a-3p. In directly injected mouse pups (25 mg kg−1, i.p.), changes in the free, steady-state levels of 40 microRNA species were noted (Fig. 9A,C) at P7. Noteworthy is that the change of the greatest magnitude was observed in miR-29a-3p, where free miR-29b and free miR-29c were also changed in abundance. Also interesting was the observation that 24 of the 40 changed microRNA species in the livers of directly injected pups exhibited increased levels (Fig. 9C). This might indicate a secondary impact of the reduced levels of microRNA that were apparently targeted by the antimiR-29a-3p, on the expression of these 24 microRNA.
FIGURE 9.
Administration of antimiR-29a-3p had multiple off-target effects on free microRNA steady-state levels in directly injected pups, but limited off-target effects after maternal delivery. Either scrambled antimiR or antmiR-29a-3p was administered to pregnant mice at gestational day 17 (dose, 100 mg kg−1, intravenous) or directly to newborn mouse pups at postnatal day (P)1 (the day of birth; dose, 25 mg kg−1, intraperitoneal). Livers were harvested from offspring from injected pregnant mice and from directly injected pups, at P7. An RNA-seq screen was undertaken on microRNA pools from harvested livers. Complete RNA-seq data are available at the NCBI Gene Expression Omnibus under GEO Series accession number GSE110888. Heatmaps reflecting RNA-seq data are provided for (A) livers from directly injected pups, and (B) livers from offspring of injected pregnant dams. All microRNA species are illustrated that exhibited a false-discovery rate of <0.1, a minimum of five reads, and a fold-change of at least 1.5. In addition, M/A plots are provided for (C) livers from directly injected pups, and (D) livers from offspring of injected pregnant dams, to relate log2(fold-change) to log2(RNA-seq counts), where green spots indicate decreased transcript abundance in antimiR-29a-3p-treated groups, gray spots exhibit no change in transcript abundance comparing the antimiR-29a-3p-treated versus scrambled antimiR-treated groups, and red spots indicate increased transcript abundance in antimiR-29a-3p-treated groups.
Off-target effects of antimiR-29a-3p on the miRNome in mouse pup livers
Fewer (four) changes in free steady-state levels of microRNA species were noted in the livers of mouse pups at P7 that were delivered by pregnant dams injected via the i.v. route with antimiR-29a-3p (dose, 100 mg kg−1) (Fig. 9B,D). Apart from an impact on miR-484, these changes were confined to the miR-29 family.
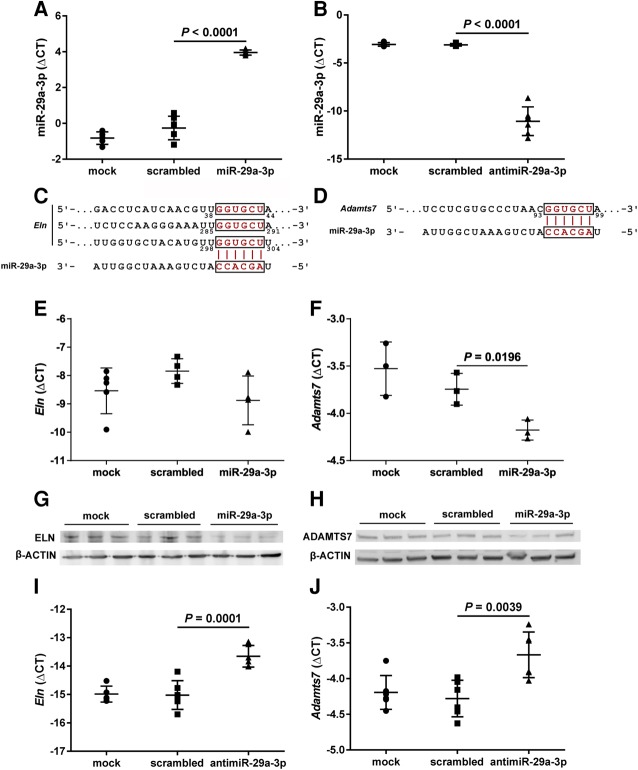
miR-29a-3p targets the Eln and Adamts7 mRNA in vitro
A miR-29a-3p mimic was successfully used to increase steady-state miR-29a-3p levels in NIH/3T3 cells in vitro (Fig. 10A), while conversely, an antimiR-29a-3p reduced free miR-29a-3p steady-state levels in NIH/3T3 cells in vitro (Fig. 10B). An in silico analysis revealed the Eln mRNA (encoding elastin, ELN) and the Adamts7 mRNA (encoding ADAM metallopeptidase with thrombospondin type 1 motif 7; ADAMTS7) to be candidate targets of miR-29a-3p, with three (two 8mer and one 7mer-m8) predicted miR-29a-3p binding-sites in the 3′-UTR of the Eln mRNA (Fig. 10C) and one 7mer-A1 predicted miR-29a-3p binding-site in the 3′-UTR of the Adamts7 mRNA (Fig. 10D). Both candidate miR-29a-3p targets could be validated in vitro. While a miR-29a-3p mimic applied to NIH/3T3 cells had no appreciable impact on steady-state Eln mRNA levels assessed by real-time RT-PCR (Fig. 10E), reduced steady-state tropoelastin protein levels were noted in the background of miR-29a-3p mimic application, assessed by immunoblot (Fig. 10G). In contrast, miR-29a-3p mimic application to NIH/3T3 cells caused a reduction in both the steady-state Adamts7 mRNA levels assessed by real-time RT-PCR (Fig. 10F), and the steady-state ADAMTS7 protein levels, assessed by immunoblot (Fig. 10H). Consistent with the decreased free miR-29a-3p steady-state levels noted in NIH3T3 cells after antimiR-29a-3p application, an increased steady-state mRNA abundance of both Eln mRNA (Fig. 10I) as well as Adamts7 mRNA (Fig. 10J) was noted.
FIGURE 10.
Validation of Eln and Adamts7 mRNA as targets of miR-29a-3p in NIH/3T3 cells in vitro. The NIH/3T3 cells were mock-transfected, or were transfected with either a scrambled microRNA mimic, a miR-29a-3p mimic, or an antimiR-29a-3p for 48 h (all at 80 nM). Total RNA pools from NIH/3T3 cells were screened by real-time RT-PCR for steady-state levels of free miR-29a-3p, after (A) transfection of a miR-29a-3p mimic, or (B) an antimiR-29a-3p. In silico analyses revealed both the (C) Eln mRNA and (D) the Adamts7 mRNA to be candidate targets of miR-29a-3p; where the Eln mRNA 3′-untranslated region (UTR) contained three (8mer34–44, 8mer285–291, and 7mer-m8298–304) predicted miR-29a-3p binding-sites, and the Adamts7 mRNA 3′-UTR contained a single (7mer-A193–99) predicted miR-29a-3p binding-site. Therefore, total RNA pools from miR-29a-3p mimic-treated NIH/3T3 cells were also screened by real-time RT-PCR for steady-state expression levels of (E) Eln and (F) Adamts7. Total protein extracts from NIH/3T3 cells were screened by immunoblot for steady-state protein expression levels of (G) elastin (ELN) and (H) ADAM metallopeptidase with thrombospondin type 1 motif 7 (ADAMTS7), where β-ACTIN served as a loading control. Total RNA pools from antimiR-29a-3p-treated NIH/3T3 cells were also screened by real-time RT-PCR for steady-state expression levels of (I) Eln and (J) Adamts7. For RT-PCR studies, steady-state free miR or mRNA expression levels are described by the mean ΔCT ± SD. (n = 3–6 separate transfection experiments, per group; each symbol represents an independent transfection experiment, where some symbols may coalesce). For microRNA analyses, ΔCT = CT(Rnu6) – CT(miR-29a-3p), while for mRNA analyses, ΔCT = CT(Polr2a) − CT(gene of interest). Mean values were compared between treatment conditions by one-way ANOVA with Tukey's post hoc modification, and P-values are provided for scrambled versus miR/antimiR treatments.
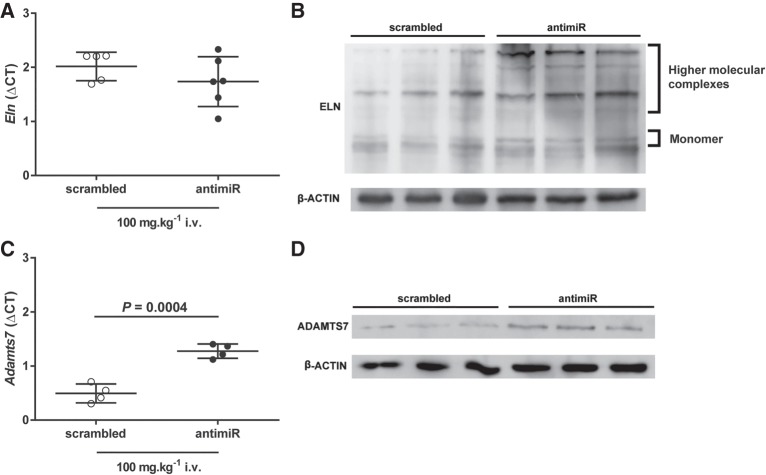
Maternal administration of antimiR-29a-3p protects steady-state levels of miR-29a-3p targets in offspring
Administration of antimiR-29a-3p via the i.v. route to pregnant dams did not impact steady-state mRNA levels of the miR-29a-3p target Eln mRNA in livers harvested from mouse pups at P7 (Fig. 11A). However, consistent with the in vitro studies reported above, a pronounced increase in steady-state protein levels of both tropoelastin (the elastin monomer), as well as higher-order elastin complexes, was noted in livers harvested from mouse pups at P7 that were born to dams that were treated with antimiR-29a-3p (Fig. 11B). Also consistent with the in vitro data provided above, administration of antimiR-29a-3p via the i.v. route to pregnant dams increased steady-state mRNA levels of the miR-29a-3p target Adamts7 mRNA (Fig. 11C) in livers harvested from mouse pups at P7. Similarly, steady-state levels of ADAMTS7 protein were also increased in the same livers that had been harvested from P7 mouse pups derived from antimiR-29a-3p-treated pregnant dams (Fig. 11D).
FIGURE 11.
Maternal transmission of antimiR-29a-3p to mouse pups increases steady-state expression levels of miR-29a-3p target-genes. Either a scrambled antimiR (open symbols) or an antimiR directed against miR-29a-3p (closed symbols) was administered by intravenous (i.v.) injection at a dose of 100 mg kg−1 to pregnant dams on the 17th day of gestation. The liver was harvested from mouse pups on postnatal day (P)7. Steady-state mRNA expression levels were assessed by real-time RT-PCR for the miR-29a-3p targets (A) Eln, and (C) Adamts7, described by the mean ΔCT ± SD. (n = 4–6 animals per group; each symbol represents an animal, where some symbols may coalesce), where ΔCT = CT(Polr2a) − CT(gene of interest). Mean values were compared between treatment conditions by unpaired Student's t-test. P-values are provided when P < 0.05. Total liver protein extracts from pups at P7 were screened by immunoblot for steady-state protein expression levels of (B) elastin (ELN) and (D) ADAM metallopeptidase with thrombospondin type 1 motif 7 (ADAMTS7), where β-ACTIN served as a loading control. For immunoblots, protein extracts from three scrambled antimiR-treated mice and three antimiR-29a-3p-treated mice were run side-by-side, on the same gel.
DISCUSSION
Many microRNA species have been—and continue to be—implicated as key mediators of normal and pathological pre- and postnatal development (Ivey and Srivastava 2015) of a spectrum of organs: Aberrant expression of the miR-23b polycistron (containing miR-23b, miR-27b, and miR-24) deregulates bile duct development in newborns (Rogler et al. 2017); the miR-379/miR-410 cluster in the liver controls metabolic adaptation at birth (Labialle et al. 2014); miR-124a is required for retinal development, and for proper axonal development of hippocampal neurons in newborns (Sanuki et al. 2011); and miR-489 (Olave et al. 2016), the miR-17/miR-92 cluster (Rogers et al. 2015; Robbins et al. 2016), and the miR-29 family (Durrani-Kolarik et al. 2017) are all associated with altered lung development in newborns (Nardiello and Morty 2016; Surate Solaligue et al. 2017). These (and other) reports underscore the importance of being able to modulate microRNA function at birth and in the early postnatal period, without recourse to conventional genome recombination approaches that rely on induction agents such as doxycycline and tamoxifen, which disturb developmental processes.
Currently, the widely used genome recombination technologies that rely on the induction of recombination with doxycycline (Gossen and Bujard 1992) or tamoxifen (Metzger et al. 1995; Indra et al. 1999) can be unsuitable for use during late gestation, parturition, and the neonatal period; due to confounding effects of these induction agents, and components of the recombination systems, on pregnancy, the developing fetus, labor and childbirth, and immediate postnatal organ development (Cunha et al. 1987; Golub et al. 1987; Gilbertson-Beadling et al. 1995; Fang et al. 1996; Moutier et al. 2003; Hosford et al. 2004; Sisson et al. 2006; Braems et al. 2011; Mylonas 2011; Lizen et al. 2015; Flynn et al. 2017; Ruiz-Camp et al. 2017). These concerns have driven the present study, which explored the possibility of administering microRNA antimiRs to pregnant dams, with the aim of assessing the impact of the antimiR on the expression and function of the target microRNA in offspring.
To address this idea, miR-29a-3p was selected as a target to study maternal transmission of an antimiR directed against miR-29a-3p. The miR-29a-3p was selected as a target because mice deficient in miR-29a, through targeted deletion of the miR-29a/miR-29b-1 locus are healthy, viable, and fecund (Papadopoulou et al. 2011), despite the association of the miR-29 family with disordered lung development, described above. As such, the risk to the health or the reproductive capacity of mice in which miR-29a-3p was targeted with an antimiR was considered low. Furthermore, an LNA (Obika et al. 1997; Koshkin et al. 1998) structure was used to enhance stability of the antimiR in vivo. The data reported here indicate that microRNA antimiRs can be delivered to offspring via the maternal–placental–fetal unit. The antimiR itself was detected in offspring by two independent detection approaches: a stem–loop real-time RT-PCR analysis protocol that was designed to negate the influence of target antimiR sequences in the endogenous pri- and pre-miR present in the tissues, which would generate a background signal. The antimiR was also detected directly, by in situ hybridization. Additionally, changes in the target microRNA levels (in this case, miR-29a-3p) were noted in the nursing dams, as well as in offspring. It is important to note that antimiRs are thought to bind to and block microRNA targets, and not necessarily promote degradation of the microRNA targets. As such, a loss of RT-PCR signal was interpreted as a reduction in the steady-state “free” levels of a microRNA species (where the blocked microRNA target can no longer function as a PCR template). Changes in the mRNA and protein expression of two miR-29a-3p target genes, Eln and Adamts7, were also noted in offspring, indicating that the maternally delivered antimiR was functional in offspring. Our proof-of-principle study has focused on a single antimiR species. It is important to note that the optimal route, dosing, and duration of effect of other antimiRs will have to be individually determined for delivery of a specific antimiR to a specific organ at a specific time-point of embryonic or postnatal life, since other antimiRs may exhibit different organ transit kinetics.
Direct injection of antimiR-29a-3p into newborn mouse pups resulted in a broad organ distribution of the effects of the antimiR: the brain, heart, kidney, liver, and the lungs. The brain was the least impacted organ, where a comparatively moderate change in miR-29a-3p free steady-state levels was noted. This is a most interesting observation, since pioneering studies addressing the in vivo targeting of microRNA with antagomiRs reported that antagomiRs could not cross the blood brain barrier, which is notoriously impermeable to exogenous agents (Krützfeldt et al. 2005). However, a close examination of the northern blots presented in that study do suggest a very mild impact on steady-state miR-16 levels (where an antagomiR-16 was applied via the i.v. route 24 h prior to organ harvest).
The effect of antimiR-29a-3p directly injected into newborn pups on the day of birth generally persisted over the first two weeks of life. This was not the case for the brain, where the comparatively moderate decrease in free miR-29a-3p steady-state levels noted at P7 was lost by P14. However, in the heart, kidney, liver, and the lungs, the impact of antimiR-29a-3p on free miR-29a-3p steady-state levels persisted up to P14. It is noteworthy that a mouse increases in mass from ≈1 g at birth, to ≈6 g at P14 (Fig. 5). This most likely accounts for the gradual “recovery” of free miR-29a-3p steady-state levels, concomitant with pup growth, and consequent dilution of the antimiR in the body. However, the effect of the antimiR remained pronounced, given that the solid and dashed lines in the graphs in Figure 3B–E are almost parallel. Concerning changes in mouse pup growth trajectory, it is noteworthy that administration of antimiR-29a-3p did impact mouse pups growth relative to that of scrambled antimiR-treated mice, both for directly injected pups, as well as maternally delivered antimiR (Fig. 5). Although the antimiR-29a-3p was selected as it was thought to be relatively inert in terms of mouse health, there is clearly an impact of the antimiR on mouse pup growth trajectory over the first two weeks of life.
No impact of antimiR-29a-3p was noted on the complementary strand, miR-29a-5p. Concerning the 3p strands of the other two miR-29 family members, parallel changes in steady-state levels of miR-29c-3p were noted (compared with miR-29a-3p), while miR-29b-3p was comparatively less affected. These discordant effects of antimiR-29a-3p on miR-29b-3p and miR-29c-3p may be explained in terms of sequence homology (Fig. 1B), where miR-29a-3p and miR-29c-3p differ by a single nucleotide base substitution, while miR-29a-3p and miR-29b-3p differ by four nucleotide base substitutions, and also, one nucleotide base addition in the miR-29b-3p sequence.
In any approach to modulate microRNA function, it would be important to document an effect not only on the microRNA of interest, but also on mRNA targets of the microRNA of interest. To this end, an in silico analysis of predicted miR-29a-3p targets was undertaken, utilizing multiple online microRNA-target detection algorithms such as TargetScan, Diana microT, and miRBase (Cristiano and Veltri 2016), which revealed over 1000 predicted targets. Among these were two miR-29a-3p validated targets that are relevant to organogenesis and cardiopulmonary physiology: Eln mRNA (Ott et al. 2011; Zhang et al. 2012) and Adamts7 mRNA (Du et al. 2012). Historically, microRNA were thought to function in mammals by repressing protein target translation, without impacting steady-state mRNA levels; while in plants, microRNA were thought to regulate gene expression by destabilizing mRNA (Huntzinger and Izaurralde 2011), although the latter mechanisms are also known to be operative in animal cell cultures (Jonas and Izaurralde 2015). For these reasons, target mRNA were selected to include microRNA targets that were impacted by both mechanisms of microRNA-mediated regulation. Using miR-29a-3p mimics in vitro, the Eln mRNA was validated as a miR-29a-3p target, where, although no impact of a miR-29a-3p mimic was noted on steady-state Eln mRNA levels, ELN protein levels were reduced. This is consistent with prevailing thinking that microRNA act primarily by repressing translation, without mRNA destabilization, although mRNA destabilization may also be noted (Baek et al. 2008). The latter situation was indeed the case with Adamts7, where miR-29a-3p mimics reduced both steady-state Adamts7 mRNA and ADAMTS7 protein levels in NIH/3T3 cells.
To evaluate whether maternal delivery of antimiRs to offspring was accompanied by modulation of validated mRNA targets, the steady-state mRNA and corresponding protein levels of Eln and Adamts7 were assessed in the livers of offspring of antimiR-29a-3p-treated pregnant dams, at P7. The liver was selected to demonstrate proof-of-principle as this highly vascularized organ that receives ≈30% of the resting cardiac output, exhibited the most persistent effects of antimiR-29a-3p administration in both directly injected and maternally “treated” mouse pups. Consistent with the decreased steady-state miR-29a-3p levels noted in the livers of P7 offspring of antimiR-29a-3p-treated pregnant dams; concomitantly increased steady-state Adamts7 mRNA levels, and concomitantly increased ELN and ADAMTS7 protein levels were noted. These data conclusively demonstrate that it is possible to modulate steady-state microRNA levels in offspring via the maternal–placental–fetal unit, by treating pregnant dams with antimiRs. Furthermore, these interventions not only target steady-state microRNA levels, but also the steady-state levels of cognate microRNA targets.
This new methodology described in the present report introduces an alternative to the more classical genome recombination-based approaches to the modulation of gene expression, which may not be suitable for studies on developmental biology or parturition, due to concerns about toxicity, off-target effects, and an impact on infant delivery of induction agents and components of the recombination systems. Clearly, there are limitations and caveats to the new methodology introduced here. The first of these concerns off-target effects where antimiRs may exhibit a broader spectrum of targets than would genetic ablation of a discrete locus. For example, the miR-29a-3p used here, also impacted other miR-29 family members: miR-29b-3p and miR-29c-3p. Genetic ablation of the miR-29a/miR-29b-1 locus would have a more restricted effect. This concern about off-target effects is highlighted by the RNA-seq data (Fig. 9), where direct injection of mouse pups with antimiR29a-3p impacted the free steady-state levels of 37 microRNA species, in addition to the three miR-29 family members. This concern is nullified in genetic ablation approaches. A second caveat concerns the cell- or tissue-restriction of antimiR function. Genetic ablation approaches, in combination with conditional (tissue- or cell-type-specific) systems would restrict modulation of microRNA expression to a particular cell- or tissue-type. AntimiR action cannot (yet) be restricted to a particular cell or tissue type.
However, antimiR approaches also present several advantages over the gene ablation approaches. One such advantage of antimiRs over gene ablation is temporal control. AntimiRs can be applied at selected time-points during development, while in the absence of an inducible system (which comes with the toxicity and off-target concerns raised above), constitutive genetic ablation would exist from conception, and as such, there is no temporal control of gene expression. A second clear advantage of antimiRs is that the genetic ablation approaches cannot target the 5p strand without targeting the 3p strand (or vice versa), whereas an antimiR approach permits the discrete targeting of one or the other microRNA strand. Maternal administration of antimiRs has the additional benefit of avoiding repeat injections in newborn mice, which could cause considerable local trauma, and present difficulties with accurate dosing.
Both antimiR and gene ablation approaches have the drawbacks of costs and concerns about the survival and health of affected experimental animals. Studies involving preexisting transgenic conditional and/or inducible deletion-ready (“floxed”) mouse strains are cheaper than antimiR administration, where antimiRs that are chemically stabilized against degradation in vivo remain expensive reagents. The cost of the LNA antimiRs for the in vivo experiments for this study was ≈€5400. However, this is substantially cheaper than the de novo generation of conditional deletion-ready mouse strains. Concerning the potential impact of antimiRs and gene ablation studies on experimental animals, as highlighted in the present study (Fig. 5), antimiRs are not necessarily inert in terms of pup health, where a change in pup growth trajectory was noted during antimiR administration. However, this is also true for transgenic animals, and depends entirely on the gene being targeted.
In sum, a novel experimental methodology is presented here allowing for the temporal control of microRNA expression and function in newborns over the critical parturition and neonatal period that is exquisitely sensitive to more conventional approaches to the ablation of gene expression. This new methodology may be useful in developmental biology studies and other investigations that address very late embryonic development, parturition, and the early neonatal period.
MATERIALS AND METHODS
Administration of microRNA antimiRs to pregnant dams and newborn mice
All animal studies were approved by the local authorities, the Regierungspräsidium Darmstadt, housing the German equivalent of an Institutional Animal Care and Use Committee (approval numbers B2/1051 and B2/1108). C57BL/6J mice were maintained on a 12 h:12 h dark:light cycle, and provided with food and water ad libitum. Newborn mouse pups received a single antimiR injection on the day of birth, either with an LNA-stabilized scrambled antimiR (5′-ACGTCTATACGCCCA-3′; Exiqon) or an LNA-stabilized antimiR directed against miR-29a-3p (antimiR-29a-3p; 5′-GATTTCAGATGGTGCT-3′; Exiqon), at a dose of 25 mg kg−1. AntimiRs were dissolved in nuclease-free water, for injection, and were injected via the intraperitoneal (i.p.) route in a volume of 15 µL.g−1. Directly injected newborn mouse pups were euthanized on postnatal day (P)7 or P14, and the brain, heart, kidney, liver, and lungs were harvested for total RNA and total protein isolation. Pregnant dams received a single injection of LNA-stabilized scrambled antimiR or antimiR-29a-3p on the 17th day of gestation, either via the i.p. route (25 mg kg−1), or via the tail vein for intravenous (i.v.) application (at a dose of 25 mg kg−1 or 100 mg kg−1). Injections in pregnant dams were made in a delivery volume of 2 µL g−1, for both i.p. and i.v. injections. After delivery, mouse pups and dams were euthanized on the day of birth [postnatal day (P)1], or at P7, or P14, and the brain, heart, kidney, liver, and lungs were harvested for total RNA and total protein isolation. The antimiR administration protocol is depicted schematically in Figure 1A.
Total RNA and total protein isolation
Total RNA was isolated from mouse organs and NIH/3T3 cells using a miRNeasy Mini Kit (Qiagen, 217004). For solid organs, tissue was homogenized in a Precellys 24-Dual homogenizer (PEQLAB). For RNA isolation from NIH/3T3 cells, cells were homogenized by scraping in RNA lysis buffer. For RNA-seq analyses, small RNA pools were isolated from mouse livers using a PureLink miRNA Isolation Kit (Invitrogen, K157001). For protein isolation, NIH/3T3 cells were homogenized by scraping in 20 mM Tris-Cl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% (v/v) NP-40, 1 mM sodium ortho-vanadate and 1× Complete protease inhibitor cocktail (Roche, 11836145001). Protein concentration was determined by Bradford assay.
Real-time RT-PCR analysis of detectable free microRNA and mRNA steady-state levels
For microRNA analysis, cDNA was prepared using a miScript II RT Kit (Qiagen, 218161). For mRNA analysis, cDNA was prepared as described previously (Alejandre-Alcázar et al. 2007). For real-time RT-PCR analysis, ΔCT values were calculated as mean CT(reference gene) − mean CT(gene of interest), where the Rnu6 gene (Qiagen, MS00033740) or the Polr2a gene (forward, 5′-CTAAGGGGCAGCCAAAGAAAC-3′; reverse, 5′-CCATTCAGCATACAACTCTAGGC-3′) served as reference genes for microRNA and mRNA, respectively. For steady-state expression levels of free miR-29 family members, primers were obtained from Qiagen: miR-29a-3p (MS00001372), miR-29a-5p (MS00032641), miR-29b-3p (MS00005936), and miR-29c-3p (MS00001379). For steady-state expression levels of miR-29a target mRNA, the following primer pairs were used: Eln (forward, 5′-TGTCCCACTGGGTTATCCCAT-3′; reverse, 5′-CAGCTACTCCATAGGGCAATTTC-3′) and Adamts7 (forward, 5′-GCAGGCTTCGTCTGCTTTCTA-3′; reverse, 5′-GCCATCAGATAAGGGTTGGTGG-3′).
AntimiR-29a-3p and miR-16 detection by stem–loop real-time RT-PCR
The LNA antimiR-29a-3p was detected in microRNA pools (including other small RNA) isolated from the livers, lungs, and kidneys of offspring (P7) from antimiR-29a-3p-treated pregnant dams, using a stem–loop quantitative real-time RT-PCR assay (Chen et al. 2005). Small RNA (including LNA antimiRs) were isolated from mouse organs using a mirVana miRNA Isolation Kit (Thermo Fisher, AM1560). Taqman analysis was undertaken with stem–loop primers targeting LNA-antimiR-29a-3p (primers custom synthesized by Thermo Fisher, custom assay CTXGPY7) and miR-16 (Thermo Fisher, 4427975; custom assay 000391). The abundance of the antimiR-29a-3p signal was first related to the abundance of miR-16, this relative abundance of antimiR-29a-3p signal in the antimiR-29a-3p-treated samples was normalized to the relative abundance of the antimiR-29a-3p signal in the scrambled antimiR-treated samples. The advantage of this approach is that endogenous background levels of pri- and pre-miR-29a in the sample (which also contain the antimiR-29a-3p sequence) are irrelevant. Indeed, fold-changes in the abundance of the exogenous antimiR-29a-3p over background levels of the pri- and pre-miR were noted. The stem–loop real-time RT-PCR was undertaken as a commercial service by Axolabs GmbH.
AntimiR-29a-3p and miR-122 detection by in situ hybridization
The LNA antimiR-29a-3p was also detected in the livers of P7 offspring from scrambled antimiR-29a-3p-treated pregnant dams, as well as in the pregnant dams at postpartum day (PP)7, by in situ hybridization, essentially as described in Nielsen et al. (2011) and modified in Gallo Cantafio et al. (2016). Mouse livers were perfused via the hepatic artery with PBS, and fixed in 10% (m/v) formalin for 48 h. Livers were embedded in paraffin, and sectioned at 5 µm. Hybridizations were performed using a double-digoxigenin-conjugated probe (5′-AGCACCATCTGAAATC-3′; 5 nM, hybridized at 56°C) directed against antimiR-29a-3p. In parallel, the miR-122 served as a positive control for the in situ hybridization reaction, and was detected with a double-digoxigenin-conjugated 5′-CAAACACCATTGTCACACTCCA-3′ probe (10 nM, hybridized at 58°C). Similarly, a double-digoxigenin-conjugated scrambled probe was prepared using the sequence 5′-TCTAACACGTCTATACGCCCA-3′ (10 nM, hybridized at 58°C). Probe binding was detected with an alkaline phosphatase-conjugated anti-digoxigenin antibody. Images were acquired with a Zeiss Axioscan slide scanner. The in situ hybridization was undertaken as a commercial service by Bioneer A/S.
RNA-seq analyses
RNA pools were screened for miRNA content and integrity on a BioAnalyzer 2100 (Agilent), and 300 ng of the small RNA fraction (<200 nt) was used as input in a TruSeq Small RNA Library Kit (Illumina) reaction. The library preparation proceeded according to routinely established protocols, with the exception that a PAGE-based size selection was omitted prior to PCR; and rather, a complete cDNA pool was amplified and libraries were pooled based on the amount of fragments within the range of 145–160 bp that correspond to insert sizes from 15–30 bp. The size selection of this pool was performed in a PippinHT system (Sage Science) on 3% agarose cassettes; with a target size set to 154 bp. Sequencing was performed on a NextSeq500 sequencer (Illumina) using v2 chemistry, resulting in an average of 9 million reads per sample with a 1 × 75 bp single-end setup. The resulting raw reads were assessed for quality, adapter content, and duplication rates with FastQC software. Sequence data were analyzed using the MIRPIPE application (Kuenne et al. 2014), a free, publicly available application developed at the Max Planck Institute for Heart and Lung Research in Bad Nauheim, Germany (https://bioinformatics.mpi-bn.mpg.de/). The MIRPIPE application enables the rapid and simple browser-based miRNA homology detection and quantification, and features automatic trimming of raw RNA-seq reads originating from various sequencing instruments, and facilitates the processing of isomiRs, as well as the quantification of miRNAs from RNA-seq studies, that can be compared to annotated public, as well as user-developed reference databases. The raw read data were processed to remove an adapter sequence and trim according to a quality threshold (default Q ≥ 20) by cutadapt. Only reads of the desired size range were selected to limit the pool to likely mature miRNAs (default: 18–28 nt). Duplicate reads were collapsed to decrease the number of necessary homology searches. Only those sequences present at a minimum number (default = 5) were retained for further analyses. Read counts from isomiRs of the same miRNA were combined. The resulting read sequences were compared against annotated sequences held in the murine mirBase database of miRNAs. Differentially expressed miRNAs were identified using DESeq2 version 1.62 (Love et al. 2014). Only genes with a maximum Benjamini–Hochberg corrected P-value of 0.1, and a minimum combined mean of five counts were deemed to be significantly differentially expressed. To control for false-discovery rates, corrected P-values were calculated, using the method of Benjamini and Hochberg (Benjamini and Hochberg 1995). The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE110888.
Immunoblot analyses
Antibodies used for immunoblot analysis were: rabbit anti-mouse ADAMTS7 (abcam, ab203027, dilution 1:100) and rabbit anti-mouse elastin (GeneTex, GTX37428, dilution 1:100). Loading equivalence was determined using a rabbit anti-mouse β-ACTIN antibody (Cell Signaling Technology, Inc.; #4967, dilution 1:1000).
Culture and transfection of NIH/3T3 cells
NIH/3T3 cells were obtained from the American Type Culture Collection (CRL-1658) and maintained in DMEM (GIBCO, 21885–025) supplemented with 10% (v/v) FBS (HI FBS; GIBCO, 10082), and 1% (v/v) penicillin-streptomycin (GE Healthcare, P11-010), in incubator at 37°C and humidified in 5% (v/v) CO2 atmosphere. NIH/3T3 cells were either mock transfected with Lipofectamine 2000/Opti-MEM (Life Technologies, 11668-019 and 31985-047), or transfected with scrambled oligonucleotide (Qiagen, 1027280), a miR-29a-3p mimic (Qiagen, MSY0000535), or miR-29a-3p inhibitor (Qiagen, MIN0000535). Cells were transfected at a concentration of 80 nM mimic or inhibitor for 48 h.
In silico analyses
Predicted miR-29a-3p targets were identified using the target prediction algorithms TargetScan version 7.1. (Agarwal et al. 2015) using the online portal at http://www.targetscan.org; Diana microT version 5.4 (Maragkakis et al. 2009) using the online portal at http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index; and miRBase version 21 (Griffiths-Jones et al. 2006) using the online portal at http://www.mirbase.org/search.shtml. Validated targets were then identified by literature screen at PubMed.
Statistical analyses
Data are presented as mean ΔCT ± SD. Differences between two-member groups were evaluated by a two-tailed unpaired Student's t-test. For multiple comparisons, a one-way ANOVA with Tukey's post hoc modification was applied. P-values <0.05 were regarded as significant. All statistical analyses were performed with GraphPad Prism 6.0. The presence of statistical outliers was tested by Grubbs’ test; when outliers were found, the sample was removed from all analyses. Statistical approaches for the analysis of RNA-seq data are described in the RNA-seq subsection.
ACKNOWLEDGMENTS
The authors thank Alan R. Kimmel (National Institutes of Health, Bethesda, MD) for constructive comments on our manuscript; Arthur Krieg (Checkmate Pharmaceuticals, Cambridge, MA) for helpful advice; Jochen Deckert (Axolabs GmbH, Kulmbach, Germany) for performing the stem–loop real-time RT-PCR analyses; Boye Schnack Nielsen (Bioneer A/S, Hørsholm, Denmark) for performing the in situ hybridization; and Roman Kurek (Qiagen) for expert advice. The authors acknowledge Tanja Enders and Kevin Wilson for performing the tail-vein and intraperitoneal injections to pregnant dams. This study was financially supported by the Max Planck Society; Rhön Klinikum AG grant FI_66; University Hospital Giessen and Marburg grant UKGM62589135; the Federal Ministry of Higher Education, Research and the Arts of the State of Hessen “LOEWE Programme,” the German Center for Lung Research (Deutsches Zentrum für Lungenforschung), and the German Research Foundation (Deutsche Forschungsgemeinschaft) through Excellence Cluster EXC147, Collaborative Research Center SFB1213/1, Clinical Research Unit KFO309/1 (projects P6 and P8), and individual research grant Mo 1789/1.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063206.117.
REFERENCES
- Agarwal V, Bell GW, Nam JW, Bartel DP. 2015. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4: e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejandre-Alcázar MA, Kwapiszewska G, Reiss I, Amarie OV, Marsh LM, Sevilla-Pérez J, Wygrecka M, Eul B, Köbrich S, Hesse M, et al. 2007. Hyperoxia modulates TGF-β/BMP signaling in a mouse model of bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 292: L537–L549. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. 2008. The impact of microRNAs on protein output. Nature 455: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. [Google Scholar]
- Braems G, Denys H, De Wever O, Cocquyt V, Van den Broecke R. 2011. Use of tamoxifen before and during pregnancy. Oncologist 16: 1547–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano F, Veltri P. 2016. Methods and techniques for miRNA data analysis. Methods Mol Biol 1375: 11–23. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Taguchi O, Namikawa R, Nishizuka Y, Robboy SJ. 1987. Teratogenic effects of clomiphene, tamoxifen, and diethylstilbestrol on the developing human female genital tract. Hum Pathol 18: 1132–1143. [DOI] [PubMed] [Google Scholar]
- Du Y, Gao C, Liu Z, Wang L, Liu B, He F, Zhang T, Wang Y, Wang X, Xu M, et al. 2012. Upregulation of a disintegrin and metalloproteinase with thrombospondin motifs-7 by miR-29 repression mediates vascular smooth muscle calcification. Arterioscler Thromb Vasc Biol 32: 2580–2588. [DOI] [PubMed] [Google Scholar]
- Durrani-Kolarik S, Pool CA, Gray A, Heyob KM, Cismowski MJ, Pryhuber GS, Lee LJ, Yang Z, Tipple TE, Rogers LK. 2017. Mir-29b supplementation decreases expression of matrix proteins and improves alveolarization in mice exposed to maternal inflammation and neonatal hyperoxia. Am J Physiol Lung Cell Mol Physiol 313: L339–L349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Wong S, Mitchell BF. 1996. Relationships among sex steroids, oxytocin, and their receptors in the rat uterus during late gestation and at parturition. Endocrinology 137: 3213–3219. [DOI] [PubMed] [Google Scholar]
- Flynn M, Heale KA, Alisaraie L. 2017. Mechanism of off-target interactions and toxicity of tamoxifen and its metabolites. Chem Res Toxicol 30: 1492–1507. [DOI] [PubMed] [Google Scholar]
- Gallo Cantafio ME, Nielsen BS, Mignogna C, Arbitrio M, Botta C, Frandsen NM, Rolfo C, Tagliaferri P, Tassone P, Di Martino MT. 2016. Pharmacokinetics and pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in mice and non-human primates. Mol Ther Nucleic Acids 5: e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson-Beadling S, Powers EA, Stamp-Cole M, Scott PS, Wallace TL, Copeland J, Petzold G, Mitchell M, Ledbetter S, Poorman R. 1995. The tetracycline analogs minocycline and doxycycline inhibit angiogenesis in vitro by a non-metalloproteinase-dependent mechanism. Cancer Chemother Pharmacol 36: 418–424. [DOI] [PubMed] [Google Scholar]
- Golub LM, McNamara TF, D'Angelo G, Greenwald RA, Ramamurthy NS. 1987. A non-antibacterial chemically-modified tetracycline inhibits mammalian collagenase activity. J Dent Res 66: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci 89: 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosford GE, Fang X, Olson DM. 2004. Hyperoxia decreases matrix metalloproteinase-9 and increases tissue inhibitor of matrix metalloproteinase-1 protein in the newborn rat lung: association with arrested alveolarization. Pediatr Res 56: 26–34. [DOI] [PubMed] [Google Scholar]
- Hsu SH, Delgado ER, Otero PA, Teng KY, Kutay H, Meehan KM, Moroney JB, Monga JK, Hand NJ, Friedman JR, et al. 2016. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology 64: 599–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110. [DOI] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. 1999. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ERT and Cre-ERT2 recombinases. Nucleic Acids Res 27: 4324–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D. 2015. microRNAs as developmental regulators. Cold Spring Harb Perspect Biol 7: a008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16: 421–433. [DOI] [PubMed] [Google Scholar]
- Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. 1998. LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 4: 3607–3630. [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438: 685–689. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. 2007. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res 35: 2885–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenne C, Preussner J, Herzog M, Braun T, Looso M. 2014. MIRPIPE: quantification of microRNAs in niche model organisms. Bioinformatics 30: 3412–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labialle S, Marty V, Bortolin-Cavaille ML, Hoareau-Osman M, Pradere JP, Valet P, Martin PG, Cavaille J. 2014. The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J 33: 2216–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Y, Sauer B. 2001. Conditional gene knockout using Cre recombinase. Mol Biotechnol 17: 269–275. [DOI] [PubMed] [Google Scholar]
- Lizen B, Claus M, Jeannotte L, Rijli FM, Gofflot F. 2015. Perinatal induction of Cre recombination with tamoxifen. Transgenic Res 24: 1065–1077. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. 2009. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 37: W273–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. 1995. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci 92: 6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutier R, Tchang F, Caucheteux SM, Kanellopoulos-Langevin C. 2003. Placental anomalies and fetal loss in mice, after administration of doxycycline in food for tet-system activation. Transgenic Res 12: 369–373. [DOI] [PubMed] [Google Scholar]
- Mylonas I. 2011. Antibiotic chemotherapy during pregnancy and lactation period: aspects for consideration. Arch Gynecol Obstet 283: 7–18. [DOI] [PubMed] [Google Scholar]
- Nardiello C, Morty RE. 2016. MicroRNA in late lung development and bronchopulmonary dysplasia: the need to demonstrate causality. Mol Cell Pediatr 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen BS, Jørgensen S, Fog JU, SøkildeIb R, Christensen J, Hansen U, Brünner N, Baker A, Møller S, Nielsen HS. 2011. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis 28: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obika S, Nanbu D, Hari Y, Morio KI, In Y, Ishida T, Imanishi T. 1997. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3, -endo sugar puckering. Tetrahedron Lett 38: 8735–8738. [Google Scholar]
- Olave N, Lal CV, Halloran B, Pandit K, Cuna AC, Faye-Petersen OM, Kelly DR, Nicola T, Benos P, Kaminski N, et al. 2016. Regulation of alveolar septation by microRNA-489. Am J Physiol Lung Cell Mol Physiol 310: L476–L487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott CE, Grunhagen J, Jager M, Horbelt D, Schwill S, Kallenbach K, Guo G, Manke T, Knaus P, Mundlos S, et al. 2011. MicroRNAs differentially expressed in postnatal aortic development downregulate elastin via 3′ UTR and coding-sequence binding sites. PLoS One 6: e16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, Zuklys S, Hollander GA, Matthys P, Gray DH, et al. 2011. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat Immunol 13: 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins ME, Dakhlallah D, Marsh CB, Rogers LK, Tipple TE. 2016. Of mice and men: correlations between microRNA-17 approximately 92 cluster expression and promoter methylation in severe bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 311: L981–L984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LK, Robbins M, Dakhlallah D, Yang Z, Lee LJ, Mikhail M, Nuovo G, Pryhuber GS, McGwin G, Marsh CB, et al. 2015. Attenuation of miR-17/92 cluster in bronchopulmonary dysplasia. Ann Am Thorac Soc 12: 1506–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler CE, Matarlo JS, Kosmyna B, Fulop D, Rogler LE. 2017. Knockdown of miR-23, miR-27, and miR-24 alters fetal liver development and blocks fibrosis in mice. Gene Expr 17: 99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Camp J, Rodríguez-Castillo JA, Herold S, Mayer K, Vadász I, Tallquist MD, Seeger W, Ahlbrecht K, Morty RE. 2017. Tamoxifen dosing for Cre-mediated recombination in experimental bronchopulmonary dysplasia. Transgenic Res 26: 165–170. [DOI] [PubMed] [Google Scholar]
- Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, et al. 2011. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci 14: 1125–1134. [DOI] [PubMed] [Google Scholar]
- Sisson TH, Hansen JM, Shah M, Hanson KE, Du M, Ling T, Simon RH, Christensen PJ. 2006. Expression of the reverse tetracycline-transactivator gene causes emphysema-like changes in mice. Am J Respir Cell Mol Biol 34: 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. 2012. Inhibition of microRNA function by antimiR oligonucleotides. Silence 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surate Solaligue DE, Rodríguez-Castillo JA, Ahlbrecht K, Morty RE. 2017. Recent advances in our understanding of the mechanisms of late lung development and bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 313: L1101–L1153. [DOI] [PubMed] [Google Scholar]
- Zhang P, Huang A, Ferruzzi J, Mecham RP, Starcher BC, Tellides G, Humphrey JD, Giordano FJ, Niklason LE, Sessa WC. 2012. Inhibition of microRNA-29 enhances elastin levels in cells haploinsufficient for elastin and in bioengineered vessels—brief report. Arterioscler Thromb Vasc Biol 32: 756–759. [DOI] [PMC free article] [PubMed] [Google Scholar]