Abstract
Metal oxide nanoparticles (MO-NPs)are known to effectively inhibit the growth of a wide range of Gram-positive and Gram-negative bacteria. They have emerged as promising candidates to challenge the rising global issue of antimicrobial resistance. However, a comprehensive understanding of their mechanism of action and identifying the most promising NP materials for future clinical translation remain a major challenge due to variations in NP preparation and testing methods. With various types of MO-NPs being rapidly developed, a robust, standardized, in vitro assessment protocol for evaluating the antibacterial potency and efficiency of these NPs is needed. Calculating the number of NPs that actively interact with each bacterial cell is critical for assessing the dose response for toxicity. Here we discuss methods to evaluate MO-NPs antibacterial efficiency with focus on issues related to NPs in these assays. We also highlight sources of experimental variability including NP preparation, initial bacterial concentration, bacterial strains tested, culture microenvironment, and reported dose.
Keywords: Metal Oxides, bacteria, nanoparticles, protein corona, colony forming units, assembly/agglomeration
Antibacterial resistance is rapidly increasing among many species of bacteria, becoming a major clinical and public health issue across the globe. Each year, 2 million cases are identified in United States alone, where antibiotic resistant bacteria have added substantial health burden to patients and financial burden to the healthcare system(1). Nanoscale engineering of surfaces and nanoparticles (NPs) provides new paths for antibacterial drug design that augment traditional tools of organic chemistry(2, 3). Metal oxide NPs (MO-NPs) have shown especially promising results in inhibiting bacterial growth and challenging antibacterial resistance(4–6). MO-NPs are further desirable as antimicrobial pharmaceuticals due to their durability, high stability, and low mammalian cell toxicity when compared to organic NPs(7). Their nanoscale and variable surface chemistry allow MO-NPs to induce bacterial toxicity through several modes of actions such as, lipid peroxidation, oxidative stress, cell membrane lysis, enzyme inhibition, proteolysis, etc(4, 8, 9). Literature suggests that due to these NPs’ multiple mechanisms of attack, bacteria would be challenged to develop strong resistance pathways(10), while mammalian cells and tissues have better protection toward similar chemical pathways(11). Despite this promise, the explosion of novel formulations of MO-NPs has not yet translated to clinically relevant antimicrobial treatments.
This delay in translation is a reflection of two conceptual problems. First, while MO-NPs appear effective against medically relevant pathogens, their exposure to the environment through either waste disposal(12, 13)or usage in food packaging industry(14–16) poses a great threat to many ‘friendly’ species of bacteria. Likewise, these materials have broad antimicrobial spectrum and therefore the potential for significant disruption of the healthy human microbiome which can lead to devastating diseases such as Clostridium difficile colitis and peptic ulcer disease. Second, and the primary focus of this article, is the high degree of variability and inconsistency in characterizing, evaluating, and reporting of experimental results for MO-NPs. To be clear, most of the studies on NP formulations are performed with rigor and diligence. However, the NP synthesis and characterization methods have many differences with traditional organic synthesis protocols. The methods of NP standardization and metrics, especially for biomedical purposes are still being established(17).
The proper protocols for antibacterial efficacy and mammalian cell toxicity evaluations for NPs are still being developed. Hence one can observe dramatic variations in the various experimental protocols and outcomes between publications. This makes developing a comprehensive, fundamental description of MO-NP antibiotics, and comparisons of specific formulations for potential clinical translation based on the current set of data quiet challenging.
Here we will focus on the specific issues that challenge interpretation of the antimicrobial MO-NP literature. By highlighting variability across studies, we will demonstrate the significant potential to tune MO-NPs for a variety of purposes and address the need to selectively inhibit pathogens while sparing or even stimulating ‘friendly’ members of the microbiome. While the potential is great, the challenge of translation is equally great. The intent of this work is to generate awareness of the variability within the literature and provide guidance for designing future experiments and reporting of results that will allow rigor and reproducibility not just within research groups, but across the antimicrobial NP field. First, we describe how the mechanism of action of a NP defines the appropriate dosing unit and how NP synthesis technique influences their biochemical mechanism. Then, different methods for evaluating antimicrobial efficacy are discussed and specific sources of variability in results are identified. Finally, we describe how the surrounding growth media or matrix provides additional variability in experimental results and provide a rational framework to describe the dose-response relationship between bacteria and MO-NPs.
Mechanism of action influences dosing unit
The efficacy of antibiotics against a specific microorganism is typically measured by the minimum inhibitory concentration (MIC). The MIC is the lowest concentration that will inhibit the visible growth of a microorganism after 24-hour culture. For traditional pharmaceutical antibiotics this is expressed in mass concentration (e.g., μg/ml). This makes sense and offers little controversy for small molecule drugs like penicillin that typically work by inhibiting a specific enzyme. There is a predictable, linear relationship between the mass and the number of active molecules. For NPs, however, the water becomes quite muddy. As a simple example, take the seminal work by Raghupathi et al. on zinc oxide (ZnO) NPs against Staphylococcus aureus(18). They showed that decreasing particle size increases antibacterial potency with the exact same molar concentration (i.e., 6 mM). With NPs, there is no longer a simple or even predictable relationship between mass and molecule – or particle – number. Further, it is unclear from that paper whether 6 mM represents the molar concentration of ZnO atoms or ZnO particles. The potential difference is astounding. That is, the mass of ZnO required for 6 mmoles of 30 nm NPs is orders of magnitude less than 6 mmoles of 300 nm NPs. Although their work is an exceptional illustration of the potential of nanotechnology to have dramatic impact on biological functions, it shows that our traditional conceptualization of drug dosing as applied to NPs, is not sufficient to describe the phenomena and will require a better conceptual framework.
One possible explanation for the inverse relationship between NP size and antibacterial activity described above and by others is that decreasing NP size leads to increasing effective surface area. We are fully aware of its limitations in the view of the importance of shape on antibacterial activity of NPs(19), and their biomimetic properties(20), however, effective surface area is an essential physico-chemical parameter involved in several potential mechanisms for antibacterial activity. Under that paradigm, surface area concentration maybe a more appropriate unit of dose, suggesting whereby that the mechanism of action of the NP against bacteria is based on the NP’s surface chemistry. Therefore, we can infer that knowledge of the mechanism of action of a NP against bacteria will provide insight into the appropriate dosing unit. Furthermore, this will inform tuning of the NP synthesis for maximum activity. As such, understanding the mechanism of action is crucial to interpreting the results of MO-NP experiments and in particular comparing various synthesis techniques.
Some of the most noted and researched MO-NPs include, silver oxide (Ag2O), copper oxide (CuO), iron oxide (Fe2O3), magnesium oxide (MgO), titanium oxide (Ti2O), and ZnO. Certain NPs made out of copper and silver have been shown to kill bacteria most likely via the release of Cu 2+ and Ag + ions(21–26). It should be noted that resistance to these particles has been shown to develop over time, although its mechanisms are not understood. The mechanism of antibacterial activity for NPs of Ti2O, MgO, and ZnO is even more controversial(27, 28). Proposed mechanisms have been comprehensively reviewed in the past and include release of toxic ions as well as oxidative stress, lipid peroxidation, cell membrane damage, enzyme inhibition, and proteolysis(4).
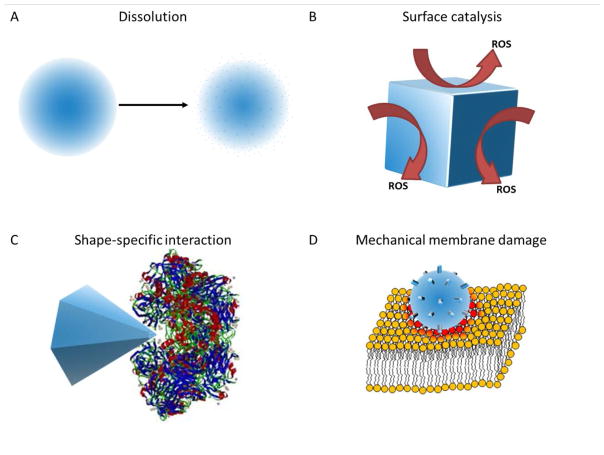
However, we will provide a conceptual framework for how a particular mechanism of action leads to consideration of a specific dosing unit, depicted in Figure 1. Let us first consider the already mentioned release of Ag+ ions. Xiu et al. provided a definitive study demonstrating the requirement of Ag ion release for Ag-NP antibacterial activity(24). Therefore, the effective dose can be related to the total mass and/or moles of Ag+ ions released by a particular NP. The primary dosing unit would then be mass concentration. Since many NPs will not completely dissolve, the effective concentration may be some fraction of the total concentration within the NPs. In addition, the rate of release may be dependent on the solvated surface area of the particle. Small particles with high surface area to weight ratio may generate a greater effective mass concentration than larger particles. Ultimately, the effective dose unit is the amount of ions in solution. For this scenario we recommend measuring and reporting the ion concentration using techniques such as inductively coupled plasma mass spectroscopy. In addition, results should be compared to a highly soluble salt with same ion concentration as a positive control.
Figure 1.
Mechanism of action conceptualization. (A) NPs dissolve in to constitutive toxic ions which diffuse into the media. (B) NP catalyze the production of toxic substances (e.g., ROS) on their surface. (C) NP vertices or edges interact with biomolecules such as proteins or DNA in a shape-specific, biomimetic manner. (D) Rough NPs come into contact with bacterial membranes causing mechanical damage.
Alternatively, for ZnO-NPs, the catalytic production of reactive oxygen species (ROS) is a commonly proposed mechanism of action. While this remains a matter of significant debate, it creates a very different scenario in terms of dose. This mechanism is based on the catalytic properties of ZnO-NP surface. Specifically, ZnO is a wide-band gap semiconductor which can generate ROS on its surface(29–31). While not required, this phenomenon is enhanced by excitation with ultraviolet light(25, 32). Under this mechanism the total mass of the NP is less important. Rather, the total available surface area available for catalysis is the most important unit to consider. This scenario is also true for NPs that have been coated with specific bio-organic substances such as enzymes, peptides, or individual amino acids. Again the mass of the NP is much less important than the bioactive surface area. Here detailed knowledge of NP morphology is required to determine the effective surface area dose. However, it should be noted that this will be complicated by NP propensity toward agglomeration which reduces the effective surface area.
Another unique facet of MO-NPs is the wide variety of shapes and morphologies that can be generated including spheres, plates, pyramids, rods, wires, stars, and flowers(2). Complex morphologies that have multiple reactive features can be generated. For instance, Cha et al demonstrated that ZnO nanopyramids could effectively inhibit enzyme activity through binding to the enzyme without protein denaturation(19). They suggest that specific edges or vertices of the pyramid shape allowed binding and conformational frustration of the enzyme. Interestingly, this specific shape was much more effective in inhibiting bacterial growth than other ZnO shapes. This suggests that protein binding and enzyme inhibition may play a role in the mechanism of action. Pyramidal and star-shaped NPs therefore may have multiple edges or vertices that are “active sites” on a single particle. In this case, neither the surface area nor the mass are the most important factors in the dose; it is the number of effective “active sites” that are most important. Ideally, a detailed understanding of the molecular mechanism and identity of potential interacting biomolecules is required to report the effective ‘stoichiometry’ and therefore appropriate dose.
Finally, many suggest that MO-NPs interact with bacterial cell membrane and provide a mechanical disruption of the membrane(4). This results in loss of membrane integrity, dissipation of ion gradients, arrest of cellular metabolism, and ultimately cell death via lysis(4). This mechanism shifts focus from the chemical properties of the NPs to their mechanical properties. Here, one can imagine a hard, rough NP interacting with a relatively soft and fluid cell membrane, essentially abrading the cell surface causing local damage. There are ample electron microscopic evidences of cell surface damage caused by MO-NPs in the literature(3, 4). Although it remains unclear if membrane damage is a direct action of the NPs or a downstream event in the cell dying process, this proposed mechanism is plausible. Therefore, appropriate NP dose in this case would likely be a combination of NP surface roughness as well as available surface area. Again, detailed reporting of NP morphology is critical for proper comparison with other data.
This is certainly not an exhaustive list of mechanisms of action for MO-NPs. However, there is evidence in the literature that all of these phenomena can occur individually or simultaneously. Therefore, when making comparisons with regards to the potency of MO-NPs, it is important to understand the mechanism of activity of the NPs on the bacteria. This will inform the dosing unit to be used for comparison purposes.
NP synthesis and surface modifications influence antibacterial mechanism of action
There are a variety of techniques for preparing MO-NPs, including sonochemical, reflux, biosynthesis, solvothermal, etc(33–35). These synthesis methods result in vastly different surface chemistry, size, and shapes of the MO-NPs. These differences are further magnified by the use of various surfactants, capping agents, and stabilizers(36). The additives function to modulate shape, surface chemistry, and stability, as well as potentiate interaction with specific cells(37). Ultimately, these variables define the activity of MO-NPs against bacteria. That is, positively charged NPs may interact with the surface of negatively charged bacteria (e.g, Escherichia coli and Staphylococcus aureus)(38) causing cell wall or membrane disruption. Alternatively, hydrophobic NPs may dissolve through the membrane to interact with and inhibit cytosolic contents such as proteins or nucleic acids. Or, as noted above, specific NP surface chemistry may release or catalyze the generation of toxic substances. Most of the MO-NP publications are motivated by the discovery and development of novel NP preparations followed by subsequent testing, however, what is lacking is a comprehensive understanding of how specific NP features (surface chemistry, shape, and size) enhance antibacterial function. Recently, strong indications that chirality of the individual NPs(39–42) and their assemblies(43) needs to be considered as a part of the parameter set(44). On the contrary, some features may lead to particle instability, degradation, or agglomeration which may significantly reduce the NP efficacy or longevity. Being able to assess which properties lead to increased potency, longevity, or selectivity would then inform more intelligent design of next generation NP preparations. Currently, there are computer aided designs to simulate these possible interactions between NPs and target biomolecules. The compositions of coronas which form due to biomolecule - NP interactions vary over time with continuous protein association and dissociation events. Literature suggests that computer simulations that are calibrated by experimental protein - NP binding affinities serve as strong predictors of NP stability and interaction with cells(45–48). As a matter of fact, scientists have used machine learning and developed Quantitative Nanostructure-Activity Relationship (QNAR) modeling, which seems to reproducibly predict biological activity profiles of novel nanomaterials, and prioritize design and manufacturing of nanomaterials towards better and safer products(48). Improvement in such software modeling technologies will greatly expand the understanding of NP antibacterial mechanism.
Evaluating antimicrobial efficacy
MO-NP studies report antibacterial effect using a variety of terms, such as percent of cells (directly visualized by microscopy), zone of inhibition, log reduction in colony forming units (CFU), change in bacterial suspension turbidity (i.e., optical density), live/dead fluorescence ratio, MIC, etc. While all of these techniques are validated in the literature, they lead to varied interpretations. For instance, how do you compare 104 live cells to an optical density of 0.1 to a live/dead fluorescence ratio of 10%? It is important to understand the differences in the interpretation of these results and to ensure that appropriate comparisons are being made between different studies. In addition, the details of the specific experimental protocol including, initial starting concentration of bacteria, bacterial species and strain, dose of NPs, and time of exposure are important considerations. In this section, we will discuss pitfalls of interpreting various techniques and the sources of experimental variability with specific focus on unique issues related NPs.
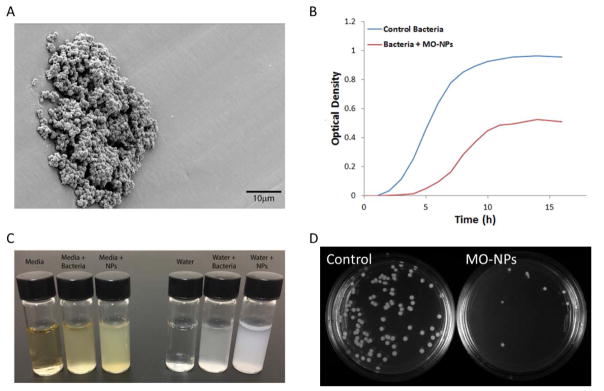
Direct visualization and counting of bacterial cells using microscopy is a common method to evaluate antimicrobial effect. This can be done using a hemocytometer and conventional light microscopy to generate a cell per volume concentration for test and control samples. Likewise, electron microscopy (EM) can be used to assess number of cells. However, many cells can appear intact but may very well be dead or at least uncultureable(49). The exquisite detail provided by EM is enticing but is subject to significant sample processing artifact(4). This process requires extensive sample preparation where NP exposed cells are fixed with an aldehyde fixative and serially dehydrated with ethanol for electron imaging. Sample dehydration requires multiple centrifugations at high speeds to precipitate bacteria and resuspend in higher concentrations of alcohol. This introduces significant artifacts where NPs attached to cell surfaces are removed, or free floating NPs in solution falsely attach to cell surfaces. This sample processing provides great difficulty in interpreting the physical interaction between cell surface and NPs under EM. In addition, the small number of cells visualized in a single field of views opens the door for significant sampling bias. NPs may further confound sampling bias by causing agglomerations of cells and NPs leading to heterogeneous distribution (Figure 2A).
Figure 2.
(A) Scanning electron micrograph of S. aureus grown on a MO-NP coated surface demonstrating a heterogeneous distribution. (B) Example growth curves, measured by optical density, of bacteria with and without MO-NPs. (C) Demonstration of how cells and NPs can have similar turbidity. (D) Bacterial culture plates demonstrating enumeration of colonies with and without MO-NPs.
Zone of inhibition assays are commonly used as a clinical test of antibiotic susceptibility. Antibiotic-impregnated test disks are placed on agar plates inoculated with bacteria. Effective antimicrobials will inhibit growth or kill bacterial leaving an area around the disk where the bacteria are not visible. While they are common and well standardized they provide only qualitative assessment. Furthermore, they depend on the diffusion of the antibiotic material from the test disk. This is straightforward for small molecule antibiotics ( e.g., penicillin) but less so for larger NPs or NP conjugates which have decreased diffusivity or constituents with varied diffusivity from the test disk.
The colloidal nature of bacteria allows indirect detection of their growth by monitoring turbidity or optical density of aqueous suspensions. Typically a dilute suspension of cells is exposed to a certain concentration of NPs. Spectrophotometry measurements are taken as a function of time generating a typical sinusoidal growth curve (Figure 2B). This provides excellent relative measurements between samples being run simultaneously and is a standard in microbiology laboratories. However, many NPs are optically active or at least provide their own turbidity. While in some instances this can be corrected for, particle agglomeration or interaction with cells can lead to colloidal instability and unpredictable changes in optical density over time. Also, the increased turbidity of the base NP suspension can drive the optical density out of the linear range once bacteria are added (Figure 2C).
One of the best methods for reporting antibacterial activity is reduction in CFUs. In this case a sample exposed to a test NP suspension is then serially diluted and spread on agar plates. This spatial separation of cells allows enumeration of individual cells as colonies (Figure 2D). It is highly quantitative and because the cells have to generate colonies they have to be alive. However, it is time consuming and labor intensive. It also neglects a phenotype of cells known to be live but unculturable(50). The significance of these cells remains a question. They may be permanently “wounded” or a persister cell that is metabolically quiescent but able to switch back to a rapidly dividing cell at a later date(51). There is another issue with enumeration of CFU that is unique to NPs. That is, it may confound the exposure time. If a NP remains in contact with or internalized within a cell it will not be serial diluted from the suspension. Therefore, it will continue to provide exposure during the plating process. This is a problem when comparing to a NP that remains in the solution and is serially diluted to sub lethal concentrations during the plating process.
Fluorescent staining of live and dead cells using a combination of cell membrane permeant and impermeant dyes is another means to evaluate antimicrobial efficacy(25, 52–54). This can be semi-quantitative by evaluating general fluorescent intensity via fluorescence spectroscopy of the two stains or full quantitative using flow cytometry, fluorescent microscopy, or confocal laser scanning microscopy. The use of fluorescent dyes in evaluating NPs can be a source of significant error. There is a high tendency for the NPs to interact with dyes causing systemic bias(4). Also, NP agglomerations may be miscounted as cells on a flow cytometer therefore, specific considerations for gating are necessary.
If there is a gold standard for evaluating antibacterial efficacy it is the MIC and/or the minimum bactericidal concentration (MBC). The MIC by definition is the lowest concentration that prevents visible growth of a bacterium overnight. Similarly the MBC is the lowest concentration causing bacterial cell death. MIC is normally performed by making serial dilutions of bacterial culture into various concentration of antibacterial substance, and observing turbidity after 24-hour incubation at 37°C(55). MBC is observed by taking the least grown culture after NP exposure and plating it on agar for colony enumeration. This gives a value for the number of cells that died during NP exposure. Measuring the MIC and MBC for NPs is challenging for two main reasons. First, as mentioned above, NP suspensions may be turbid at baseline due to facile assembly of NPs into supraparticles(56, 57) with and without biological components(58). Second, NPs have varied dissolution rates and colloidal stability in bacterial culture media. Some NPs stay in suspension easily in growth media, either due to capping reagents or ligands, while others become unstable over time. Instability results in NP precipitation and sequestration at the bottom of the test vessel. Cells significantly separated from NPs in the supernatant are free to grow. Changes in NP surface chemistry due to cellular interaction or binding of organic substance in the media over time can lead to NP agglomeration.
Sources of experimental variability
Clearly there is no perfect method for evaluating antimicrobial efficacy. A thorough evaluation will required a combination of methodologies. Regardless of the technique, there are a number of experimental conditions that must be considered, controlled, and reported in detail. First, the results of almost all assays described above are highly dependent on the initial concentration of cells, as large number of cells can outgrow a chosen antibacterial concentration, while few cells are more susceptible at that same concentration. MIC testing of small molecule antibiotics is typically standardized by an initial optical density, instead of the cell number(55). Given the issues of turbidity already discussed, we agree with others in the literature suggest that using ~106 bacterial cells per antibacterial concentration. Of note, currently used initial concentrations varying anywhere between 104–108 cells(55). The difference affected by initial bacterial concentration was highlighted by Leung et al for ZnO and TiO2. They varied the initial concentration from 106 to 108 and observed changes in cell growth inhibition as well as protein expression(59). If a specific number is followed by all testing the efficiency of NPs against bacteria, many of the MBC and mechanistic assays can be standardized and reported back in terms of colony enumeration over a period of time.
Second, antibacterial efficacy is a function of both dose and exposure time. In other words, NP antibacterial effect depends on whether a single dose is strong enough (acute dose) or whether low doses are administered over an extended period of time (chronic dose). In the case of bacteria, where many species are observed to divide every 20–40 minutes in a nutrient rich environment at ambient temperatures, providing a consistent exposure dose of NPs with respect to the growing number of bacteria is challenging.
Third, as with all antimicrobials there is significant variation in the antibacterial spectrum. That is, some species are more or less susceptible than others. Worse still, there is significant variability between strains within the same species. Of particular concern in the NP literature is the use of bacterial strains which are not typically considered pathogenic or of industrial significance. The most common are K12 strains of E. coli which are commonly used in molecular biology laboratories as expression or cloning vectors(60, 61). Although these strains are convenient, they are engineered strains and typically lack many of the highly evolved and redundant defense mechanism of true pathogens. Studies limited to these strains while scientifically interesting provide little motivation for translation to clinical application. Table 1 uses ZnO-NPs as an example and provides a brief summary of the challenges associated with NP efficacy assessment by providing examples from literature where various bacterial strains, cell numbers, and NP preparations, are used and evaluated by different methods. Based on these results, it is difficult to assess which types of ZnO-NPs are most effective in killing the listed bacteria and whether ZnO-NPs are more or less efficient when compared to Fe2O3 or MgO as an example. Therefore a more standardized and organized method is necessary when reporting MO-NP antibacterial data to best evaluate the effectiveness of the rapidly developing NP materials.
Table 1.
Summary of NP antimicrobial efficacy
| Metal oxide |
Synthesis | Bacterial strains tested |
Nanoparticle size (nm) |
Shape | Antimicrobial efficacy | Exposure time |
Initial bacterial number |
Highest NP concentration |
Ref |
|---|---|---|---|---|---|---|---|---|---|
| ZnO | Sol gel | Staphylococcus aureus and Escherichia coli strains unknown | 3.9±0.5 | sphere |
S. aureus - 6 log E. coli - 5 log |
24hr | 106 | 0.3% w/v | (68) |
| ZnO | Combustion |
Escherichia coli, strain BL21 DE3 Bacillus subtilis, strain 168 |
Length: 150–200 Diameter: 10 |
nanorod, tetrapod | OD = 0.0 | 24hr | OD 0.1 | 1mg/mL | (69) |
| ZnO | Reflux |
Klebsiella pneumoniae, LM2, Escherichia coli, UTI89 and MG1655 Staphylococcus aureus SH1000, Staphylococcus epidermidis RP62A |
20–25 | hexagonal pyramids |
K. pneumoniae - OD=1.6 E. coli - OD=1.0 S. aureus - OD=0.25 S. epidermidis - OD=0.25 |
10hrs | OD 0.15 | 667ug/mL | (65) |
| ZnO | Reflux |
Klebsiella pneumoniae, LM2, Escherichia coli, UTI89 and MG1655 Staphylococcus aureus SH1000, Staphylococcus epidermidis RP62A |
Diameter: 20 Thickness: 3.5 |
plates |
K. pneumoniae - OD=1.6 E. coli - OD=1.0 S. aureus - OD=0.7 S. epidermidis - OD=0.4 |
10hrs | OD 0.15 | 667ug/mL | (65) |
| ZnO | Reflux |
Klebsiella pneumoniae, LM2, Escherichia coli, UTI89 and MG1655 Staphylococcus aureus SH1000, Staphylococcus epidermidis RP62A |
4.5 | sphere |
K. pneumoniae - OD=1.6 E. coli - OD=1.0 S. aureus - OD=0.28 S. epidermidis - OD=0.25 |
10hrs | OD 0.15 | 667ug/mL | (65) |
| ZnO | Sonochemical, photo reduction |
Staphylococcus aureus, ATCC 29213 Escherichia coli, ATCC 25922 |
30 | irregular | S. aureus - 90% live, E. coli -75% live | 10min | Unknown | 0.1mg/mL | (32) |
| ZnO + light | Sonochemical, photo reduction |
Staphylococcus aureu, ATCC 29213 Escherichia coli, ATCC 25922 |
30 | irregular |
S. aureus - 25% live E. coli - 10% live |
10min | Unknown | 0.1mg/mL | (32) |
| Fe2O3 | Sol gel | Staphylococcus aureus and Escherichia coli strains unknown | 17.3 ± 4.6 | sphere-like |
S. aureus - 0 log E. coli - 0 log |
24hr | 1.00E+06 | 0.3% w/v | (68) |
| MgO | Combustion |
Escherichia coli, strain BL21 DE3 Bacillus subtilis, strain 168 |
50 | cubic |
E. coli - OD=1.2 B. subtilis - OD=0.3 |
24hr | OD 0.1 | 1mg/mL | (69) |
Role of surrounding media/microenvironment on NP efficacy
Lastly, the type of surrounding media (e.g., nutrient rich or minimal media) can have a profound effect on results. Not only are certain types of media species and/or stain specific but there are significant NP considerations. Most investigations of NP toxicity on bacterial cells is tested in growth medium such as tryptic soy broth (TSB) or lysogeny broth (LB), both of which contain high concentrations of amino acids, small peptides, and salts as well as trace minerals and vitamins. Many are also supplemented with glucose as well. These media constituents can greatly affect the surface chemistry of MO-NPs by changing zeta potential, dissolving the particle, or blocking interactions. Media contents can make NPs unstable through ion/molecule/protein adsorption or loss of surface functionality, resulting in the formation of aggregates(36, 62). The role of complex NP corona ( i.e., biological components that surround NPs in dispersion(63, 64)) may stabilize the NPs but also affect the outcome of their interactions with bacteria.
The colloidal stability and/or agglomeration rate of NPs in specific media and shaking (used to aerate media) conditions is not typically described or discussed in most papers. These processes further influence the NP behavior and significantly alter their diffusivity. However, at the same time, the growth media is the best condition to test NP toxicity, as bacteria have regulated their gene expression for maximal replication rate. Therefore, it is imperative to test the effects of the specific media on key NP features before drawing conclusions on their toxicity and efficiency in killing bacteria. Specifically, NPs exposed to media (without bacteria) can be tested for changes in zeta potential, addition of functional groups through fourier transform infrared spectroscopy, optic properties via spectroscopy, and size and shape via EM and spectroscopic techniques. However, it should be noted that once NP antimicrobial activity is confirmed or optimized and the role of the surrounding media elucidated for a given NP preparation repeat testing in the solvent conditions of the given application (blood, urine, food preparations, etc.) will be required as new solvent constituents may alter the NP in different and unpredictable ways.
In addition to the static constituents in cell culture media, there are dynamics changes to the surrounding microenvironment. At times, increased cell proliferation changes the pH and chemical composition of the media. The production of acid metabolites and waste products begin to take the place of nutrients and raw materials. This effect is much more challenging to test. However, if cells and NPs can be centrifuged out separately, where cells precipitate faster than NPs due to their mass, the remaining NPs can be removed and tested using techniques noted above.
A framework for the dose-response relationship
The argument thus far is that the current state of the art fails to provide a comprehensive description of how the cell culture microenvironment changes the surface chemistry, agglomeration rate, and decomposition of MO-NPs. More importantly, it fails to elucidate how these factors are dictated by the specific NP dose of interest. Therefore, it is difficult to develop a NP efficacy assessment in a straightforward manner. We contend that the most interesting factor is the number of NPs that could be actively interacting with a single bacterial cell in a given period of time. Here we offer a framework for assessing NP and bacterial cell interaction by suggesting calculations of NP particle numbers from a given mass concentration and the assessment of individual and agglomerate NPs to cell ratio as a means to more objectively and rigorously describe the antibacterial efficiency of a candidate NP preparation.
Based on the previous discussion, evaluating the difference between a toxic but nonlethal dose and a lethal dose for NPs is extremely difficult since their mechanism of action against bacteria is not clearly understood and they have varied behavior in different growth media solutions. Presently, there are three methods to measure MO-NPs for dosing: (1) mass/weight of a dissolved substance per volume (gram per liter), (2) molar concentration of a dissolved amount of substance (number of atoms, to be calculated from the molecular weight) per volume (mol per liter) or (3) particle density or particle concentration per volume (particle counts per volume).
Given the frequent crystallinity of MO-NPs, many of them form well-defined geometric shapes, where structures such as spheres, rods, and cubes are seen commonly. Therefore, once individual particle dimensions are determined via transmission electron microscopy (TEM), the values can be used for accurate calculations of volume and conversion from mass concentration to particle number concentration. Most NP dispersions are made by weighing dry NP powder and then sonicating or suspending into the experimental solvent, followed by exposure to cells. We suggest not only reporting the mass concentration of experimental conditions, but also the number of particles or agglomerates potentially interacting with certain number of cells in that sample. In essence, this generates a ratio of NPs to cells and provides an upper limit to the number of NPs that can be interacting with a single cell. One way of calculating the number of particle in concentrations is using the volume (V) of the NP shape and the density (ρ) of the material. This method was used by McGuffie et. al(65);, essentially V was calculated from TEM measurements and multiplied by the ρ to give the mass of a single NP. Then, dividing the experimental mass concentration by the individual particle mass (ρV) yields the number of particles in that particular concentration. As a result, they showed how changing the dosing unit altered the conclusions regarding the role of shape on NP efficacy.
These calculations can be applied to particles of various shapes and sizes, as long as their shape is geometric and the density of the nanomaterial is known. There are however, major challenges involved with strongly predicting how many NPs are directly interacting with cells. This is because NP assembly is a common phenomenon due to van-der-Waals and other forces(66), substances being secreted by the bacteria, the growth media contents, or other sources. Therefore, we also suggest reporting the average NP agglomerate size/shape in respective media over a period of time as well. This can be evaluated through light scattering measurements(67). If NP antibacterial activity is reported in terms of initial cell numbers used, number of NPs present, and the number of CFU after NP exposure, NP potency can be assessed better.
Conclusion
Assessing MO-NP toxicity and dosage for actively dividing bacteria is highly challenging due to complex effects of NP synthesis techniques, NP agglomeration, NP confounding of standard microbiology assays, and interaction of NPs with cells and media constituents. Considering a fundamental dosing unit of the number of particles internalized or interacting per cell will provide a mechanism for comparative studies. We propose that using density of NP material and volume of the NP shape, individual NP mass can be calculated. By reporting the number of initial cells used, number of potential particles and NP aggregates exposed to these cells, exposure time, and the log reduction of CFU, a standard model of NP toxicity towards bacterial cells could be potentially prepared. There are challenges in reporting the exact number of NPs that are in contact with bacterial cells, as many MO-NPs may assemble/agglomerate and are likely to develop the protein coronas in the growth media. The solubility of these NPs is time dependent and plays a major role in the NP toxicity as well. Additionally, several factors should be considered when assessing dose response and antibacterial mechanism of MO-NPs. Some of these include the NP mechanism of action, synthesis, surface textures or topography, zeta-potential, particle shape, stability in growth media, and the band gap energy levels as oxygen vacancies on the MO-NPs surface could induce cellular oxidative stress. Although the literature is demonstrating that MO-NPs are effective antibacterial agents, this data can be challenged by discrepancies across studies in the amount of cell death/inhibition, the types of bacterial species used, initial number of cells exposed to NP, and methods for preparing NPs and their suspensions. Continual testing is necessary to assess the efficacy of MO-NP antibacterial toxicity according to a standard toxicity assessment. Further studies should be performed to better understand the MO-NP mechanism of action against bacteria to bring better alternatives for antibiotics and disinfectants in biomedical applications.
Footnotes
Conflict of Interest
The authors have no competing conflicts of interest related to this work.
References
- 1.Ventola CL. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharmacy and Therapeutics. 2015;40(4):277–83. [PMC free article] [PubMed] [Google Scholar]
- 2.Bozon-Verduraz F, Fievet F, Piquemal J-Y, Brayner R, Kabouss K, Soumare Y, et al. Nanoparticles of metal and metal oxides: some peculiar synthesis methods, size and shape control, application to catalysts preparation. Braz J Phys. 2009:39. [Google Scholar]
- 3.Jiang W, Mashayekhi H, Xing B. Bacterial toxicity comparison between nano- and micro-scaled oxide particles. Environmental pollution. 2009;157(5):1619–25. doi: 10.1016/j.envpol.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Djurisic AB, Leung YH, Ng AM, Xu XY, Lee PK, Degger N, et al. Toxicity of metal oxide nanoparticles: mechanisms, characterization, and avoiding experimental artefacts. Small. 2015;11(1):26–44. doi: 10.1002/smll.201303947. [DOI] [PubMed] [Google Scholar]
- 5.Dizaj SM, Lotfipour F, Barzegar-Jalali M, Zarrintan MH, Adibkia K. Antimicrobial activity of the metals and metal oxide nanoparticles. Materials Science and Engineering. 2014;44:278–84. doi: 10.1016/j.msec.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Horie M, Fujita K, Kato H, Endoh S, Nishio K, Komaba LK, et al. Association of the physical and chemical properties and the cytotoxicity of metal oxide nanoparticles: metal ion release, adsorption ability and specific surface area. Metallomics: integrated biometal science. 2012;4(4):350–60. doi: 10.1039/c2mt20016c. [DOI] [PubMed] [Google Scholar]
- 7.Deravi LF, Swartz JD, Wright DW. The biomimetic synthesis of metal oxide nanomaterials. Wiley; 2010. [Google Scholar]
- 8.Corr SA. Nanoscience: Volume 1: Nanostructures through Chemistry. 1. The Royal Society of Chemistry; 2013. Metal oxide nanoparticles; pp. 180–207. [Google Scholar]
- 9.Allahverdiyev AM, Kon KV, Abamor ES, Bagirova M, Rafailovich M. Coping with antibiotic resistance: combining nanoparticles with antibiotics and other antimicrobial agents. Expert Review of Anti-infective Therapy. 2011;9(11):1035–52. doi: 10.1586/eri.11.121. [DOI] [PubMed] [Google Scholar]
- 10.Stankic S, Suman S, Haque F, Vidic J. Pure and multi metal oxide nanoparticles: synthesis, antibacterial and cytotoxic properties. Journal of Nanobiotechnology. 2016;14(1):73. doi: 10.1186/s12951-016-0225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liong M, Lu J, Kovochich M, Xia T, Ruehm SG, Nel AE, et al. Multifunctional Inorganic Nanoparticles for Imaging, Targeting, and Drug Delivery. ACS nano. 2008;2(5):889–96. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Microbiology: Nanoparticles kill resistant bacteria. Nature. 2016;537(7620):282–3. doi: 10.1038/537282d. [DOI] [PubMed] [Google Scholar]
- 13.Miller RJ, Lenihan HS, Muller EB, Tseng N, Hanna SK, Keller AA. Impacts of Metal Oxide Nanoparticles on Marine Phytoplankton. Environmental Science & Technology. 2010;44(19):7329–34. doi: 10.1021/es100247x. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Zhang C, Hu Z. Impact of metallic and metal oxide nanoparticles on wastewater treatment and anaerobic digestion. Environmental Science: Processes & Impacts. 2013;15(1):39–48. doi: 10.1039/c2em30655g. [DOI] [PubMed] [Google Scholar]
- 15.Freyre-Fonseca V, Delgado-Buenrostro NL, Chirino YI, Gutiérrez-López GF. Safety Studies of Metal Oxide Nanoparticles Used in Food Industry. In: Hernández-Sánchez H, Gutiérrez-López GF, editors. Food Nanoscience and Nanotechnology. Cham: Springer International Publishing; 2015. pp. 243–65. [Google Scholar]
- 16.Martirosyan A, Schneider Y-J. Engineered Nanomaterials in Food: Implications for Food Safety and Consumer Health. International Journal of Environmental Research and Public Health. 2014;11(6):5720–50. doi: 10.3390/ijerph110605720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong B, Seog JH, Graham LM, Lee SB. Experimental considerations on the cytotoxicity of nanoparticles. Nanomedicine (London, England) 2011;6(5):929–41. doi: 10.2217/nnm.11.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekhon BS. Nanotechnology in agri-food production: an overview. Nanotechnology, Science and Applications. 2014;7:31–53. doi: 10.2147/NSA.S39406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cha S-H, Hong J, McGuffie M, Yeom B, VanEpps JS, Kotov NA. Shape-Dependent Biomimetic Inhibition of Enzyme by Nanoparticles and Their Antibacterial Activity. ACS Nano. 2015;9(9):9097–105. doi: 10.1021/acsnano.5b03247. [DOI] [PubMed] [Google Scholar]
- 20.Kotov NA. Inorganic Nanoparticles as Protein Mimics. Science. 2010;330(6001):188–9. doi: 10.1126/science.1190094. [DOI] [PubMed] [Google Scholar]
- 21.Hans M, Erbe A, Mathews S, Chen Y, Solioz M, Mücklich F. Role of Copper Oxides in Contact Killing of Bacteria. Langmuir. 2013;29(52):16160–6. doi: 10.1021/la404091z. [DOI] [PubMed] [Google Scholar]
- 22.Bao H, Yu X, Xu C, Li X, Li Z, Wei D, et al. New Toxicity Mechanism of Silver Nanoparticles: Promoting Apoptosis and Inhibiting Proliferation. PLOS ONE. 2015;10(3):e0122535. doi: 10.1371/journal.pone.0122535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung WK, Koo HC, Kim KW, Shin S, Kim SH, Park YH. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Applied and Environmental Microbiology. 2008;74(7):2171–8. doi: 10.1128/AEM.02001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiu Z-m, Zhang Q-b, Puppala HL, Colvin VL, Alvarez PJJ. Negligible Particle-Specific Antibacterial Activity of Silver Nanoparticles. Nanoletters. 2012;12(8):4271–5. doi: 10.1021/nl301934w. [DOI] [PubMed] [Google Scholar]
- 25.Raghupathi KR, Koodali RT, Manna AC. Size-dependent bacterial growth inhibition and mechanism of antibacterial activity of zinc oxide nanoparticles. Langmuir. 2011;27(7):4020–8. doi: 10.1021/la104825u. [DOI] [PubMed] [Google Scholar]
- 26.Prabhu S, Poulose EK. Silver nanoparticles: mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. International Nano Letters. 2012;2(1):32. [Google Scholar]
- 27.Arijit Kumar C, Ruchira C, Tarakdas B. Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology. 2014;25(13):135101. doi: 10.1088/0957-4484/25/13/135101. [DOI] [PubMed] [Google Scholar]
- 28.Amna T, Hassan MS, Barakat NA, Pandeya DR, Hong ST, Khil M-S, et al. Antibacterial activity and interaction mechanism of electrospun zinc-doped titania nanofibers. Appl Microbiol Biotecech. 2012:93. doi: 10.1007/s00253-011-3459-0. [DOI] [PubMed] [Google Scholar]
- 29.Dutta RK, Nenavathu BP, Gangishetty MK, Reddy AV. Studies on antibacterial activity of ZnO nanoparticles by ROS induced lipid peroxidation. Colloids and surfaces B, Biointerfaces. 2012;94:143–50. doi: 10.1016/j.colsurfb.2012.01.046. [DOI] [PubMed] [Google Scholar]
- 30.Yang M, Sun K, Kotov NA. Formation and Assembly–Disassembly Processes of ZnO Hexagonal Pyramids Driven by Dipolar and Excluded Volume Interactions. Journal of the American Chemical Society. 2010;132(6):1860–72. doi: 10.1021/ja906868h. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Ji Z, Xia T, Meng H, Low-Kam C, Liu R, et al. Use of Metal Oxide Nanoparticle Band Gap To Develop a Predictive Paradigm for Oxidative Stress and Acute Pulmonary Inflammation. ACS Nano. 2012;6(5):4349–68. doi: 10.1021/nn3010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He W, Kim HK, Wamer WG, Melka D, Callahan JH, Yin JJ. Photogenerated charge carriers and reactive oxygen species in ZnO/Au hybrid nanostructures with enhanced photocatalytic and antibacterial activity. J Am Chem Soc. 2014:136. doi: 10.1021/ja410800y. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Xu H, Chen Z-S, Chen G. Biosynthesis of nanoparticles by microorganisms and their applications. J Nanomater. 2011:2011. [Google Scholar]
- 34.Raliya R, Tarafdar J. Biosynthesis and characterization of zinc, magnesium and titanium nanoparticles: an eco-friendly approach. Int Nano Letters. 2014:4. [Google Scholar]
- 35.Das VL, Thomas R, Varghese RT, Soniya EV, Mathew J, Radhakrishnan EK. Extracellular synthesis of silver nanoparticles by the Bacillus strain CS 11 isolated from industrialized area. 3. Biotech. 2014;4(2):121–6. doi: 10.1007/s13205-013-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore TL, Rodriguez-Lorenzo L, Hirsch V, Balog S, Urban D, Jud C, et al. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chemical Society Reviews. 2015;44(17):6287–305. doi: 10.1039/c4cs00487f. [DOI] [PubMed] [Google Scholar]
- 37.Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. International Journal of Nanomedicine. 2012;7:5577–91. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halder S, Yadav KK, Sarkar R, Mukherjee S, Saha P, Haldar S, et al. Alteration of Zeta potential and membrane permeability in bacteria: a study with cationic agents. SpringerPlus. 2015;4:672. doi: 10.1186/s40064-015-1476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaaff TG, Whetten RL. Giant Gold–Glutathione Cluster Compounds: Intense Optical Activity in Metal-Based Transitions. The Journal of Physical Chemistry B. 2000;104(12):2630–41. [Google Scholar]
- 40.Noguez C, Garzon IL. Optically active metal nanoparticles. Chemical Society Reviews. 2009;38(3):757–71. doi: 10.1039/b800404h. [DOI] [PubMed] [Google Scholar]
- 41.Gautier C, Bürgi T. Chiral Gold Nanoparticles. Chem Phys Chem. 2009;10(3):483–92. doi: 10.1002/cphc.200800709. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y, Yang M, Sun K, Tang Z, Kotov NA. Similar Topological Origin of Chiral Centers in Organic and Nanoscale Inorganic Structures: Effect of Stabilizer Chirality on Optical Isomerism and Growth of CdTe Nanocrystals. Journal of the American Chemical Society. 2010;132(17):6006–13. doi: 10.1021/ja906894r. [DOI] [PubMed] [Google Scholar]
- 43.Chen W, Bian A, Agarwal A, Liu L, Shen H, Wang L, et al. Nanoparticle Superstructures Made by Polymerase Chain Reaction: Collective Interactions of Nanoparticles and a New Principle for Chiral Materials. Nano letters. 2009;9(5):2153–9. doi: 10.1021/nl900726s. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki N, Wang Y, Elvati P, Qu Z-B, Kim K, Jiang S, et al. Chiral Graphene Quantum Dots. ACS Nano. 2016;10(2):1744–55. doi: 10.1021/acsnano.5b06369. [DOI] [PubMed] [Google Scholar]
- 45.Vilanova O, Mittag JJ, Kelly PM, Milani S, Dawson KA, Rädler JO, et al. Understanding the Kinetics of Protein–Nanoparticle Corona Formation. ACS Nano. 2016;10(12):10842–50. doi: 10.1021/acsnano.6b04858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dell’Orco D, Lundqvist M, Oslakovic C, Cedervall T, Linse S. Modeling the Time Evolution of the Nanoparticle-Protein Corona in a Body Fluid. PLOS ONE. 2010;5(6):e10949. doi: 10.1371/journal.pone.0010949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epa VC, Burden FR, Tassa C, Weissleder R, Shaw S, Winkler DA. Modeling Biological Activities of Nanoparticles. Nano letters. 2012;12(11):5808–12. doi: 10.1021/nl303144k. [DOI] [PubMed] [Google Scholar]
- 48.Fourches D, Pu D, Tassa C, Weissleder R, Shaw SY, Mumper RJ, et al. Quantitative Nanostructure-Activity Relationship (QNAR) Modeling. ACS nano. 2010;4(10):5703–12. doi: 10.1021/nn1013484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu JL, Luo Z, Bashir S. A progressive approach on inactivation of bacteria using silver-titania nanoparticles. Biomaterials Science. 2013;1(2):194–201. doi: 10.1039/c2bm00010e. [DOI] [PubMed] [Google Scholar]
- 50.Stewart EJ. Growing Unculturable Bacteria. Journal of Bacteriology. 2012;194(16):4151–60. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y. Persisters, persistent infections and the Yin-Yang model. Emerg Microbes Infect. 2014;3:e3. doi: 10.1038/emi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Y, Strømme M, Welch K. Bacteria viability assessment after photocatalytic treatment. 3. Biotech. 2014;4(2):149–57. doi: 10.1007/s13205-013-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gopinath V, Priyadarshini S, Loke MF, Arunkumar J, Marsili E, MubarakAli D, et al. Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity. Arabian Journal of Chemistry [Google Scholar]
- 54.Boda SK, Broda J, Schiefer F, Weber-Heynemann J, Hoss M, Simon U, et al. Cytotoxicity of Ultrasmall Gold Nanoparticles on Planktonic and Biofilm Encapsulated Gram-Positive Staphylococci. Small. 2015;11(26):3183–93. doi: 10.1002/smll.201403014. [DOI] [PubMed] [Google Scholar]
- 55.European Committee for Antimicrobial Susceptibility Testing of the European Society of Clinical M, Infectious D. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clinical Microbiology and Infection. 2003;9(8):ix–xv. [Google Scholar]
- 56.Xia Y, Nguyen TD, Yang M, Lee B, Santos A, Podsiadlo P, et al. Self-assembly of self-limiting monodisperse supraparticles from polydisperse nanoparticles. Nat Nano. 2011;6(9):580–7. doi: 10.1038/nnano.2011.121. [DOI] [PubMed] [Google Scholar]
- 57.Allen AJ, Hackley VA, Jemian PR, Ilavsky J, Raitano JM, Chan S-W. In situ ultra-small-angle X-ray scattering study of the solution-mediated formation and growth of nanocrystalline ceria. Journal of Applied Crystallography. 2008;41(5):918–29. [Google Scholar]
- 58.Park JI, Nguyen TD, de Queirós Silveira G, Bahng JH, Srivastava S, Zhao G, et al. Terminal supraparticle assemblies from similarly charged protein molecules and nanoparticles. Nature Communications. 2014;5:3593. doi: 10.1038/ncomms4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leung YH, Xu X, Ma APY, Liu F, Ng AMC, Shen Z, et al. Toxicity of ZnO and TiO2 to Escherichia coli cells. Scientific Reports. 2016;6:35243. doi: 10.1038/srep35243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhukova LV. Evidence for Compression of Escherichia coli K12 Cells under the Effect of TiO2 Nanoparticles. ACS Applied Materials & Interfaces. 2015;7(49):27197–205. doi: 10.1021/acsami.5b08042. [DOI] [PubMed] [Google Scholar]
- 61.Rosales-Colunga LM, Martínez-Antonio A. Engineering Escherichia coli K12 MG1655 to use starch. Microbial Cell Factories. 2014;13:74. doi: 10.1186/1475-2859-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kendall M, Hodges NJ, Whitwell H, Tyrrell J, Cangul H. Nanoparticle growth and surface chemistry changes in cell-conditioned culture medium. Philosophical Transactions of the Royal Society B: Biological Sciences. 2015;370(1661) doi: 10.1098/rstb.2014.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cedervall T, Lynch I, Lindman S, Berggård T, Thulin E, Nilsson H, et al. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proceedings of the National Academy of Sciences. 2007;104(7):2050–5. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pino Pd, Pelaz B, Zhang Q, Maffre P, Nienhaus GU, Parak WJ. Protein corona formation around nanoparticles - from the past to the future. Materials Horizons. 2014;1(3):301–13. [Google Scholar]
- 65.McGuffie MJ, Hong J, Bahng JH, Glynos E, Green PF, Kotov NA, et al. Zinc oxide nanoparticle suspensions and layer-by-layer coatings inhibit staphylococcal growth. Nanomedicine: Nanotechnology, Biology and Medicine. 12(1):33–42. doi: 10.1016/j.nano.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang M, Chan H, Zhao G, Bahng JH, Zhang P, Král P, et al. Self-assembly of nanoparticles into biomimetic capsid-like nanoshells. Nat Chem. 2017;9(3):287–94. doi: 10.1038/nchem.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hackley VA, Clogston JD. Measuring the Hydrodynamic Size of Nanoparticles in Aqueous Media Using Batch-Mode Dynamic Light Scattering. In: McNeil SE, editor. Characterization of Nanoparticles Intended for Drug Delivery. Totowa, NJ: Humana Press; 2011. pp. 35–52. [DOI] [PubMed] [Google Scholar]
- 68.Gordon T, Perlstein B, Houbara O, Felner I, Banin E, Margel S. Synthesis and characterization of zinc/iron oxide composite nanoparticles and their antibacterial properties. Colloid Surf A. 2011:374. [Google Scholar]
- 69.Vidic J, Stankic S, Haque F, Ciric D, Goffic R, Vidy A, et al. Selective antibacterial effects of mixed ZnMgO nanoparticles. J Nanoparticle Res. 2013:15. doi: 10.1007/s11051-013-1595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]