Abstract
Filamentous fungi are prolific producers of secondary metabolites with drug-like properties, and their genome sequences have revealed an untapped wealth of potential therapeutic leads. To better access these secondary metabolites and characterize their biosynthetic gene clusters, we applied a new platform for screening and heterologous expression of intact gene clusters that uses fungal artificial chromosomes and metabolomic scoring (FAC-MS). We leverage FAC-MS technology to identify the biosynthetic machinery responsible for production of acu-dioxomorpholine, a metabolite produced by the fungus, Aspergilllus aculeatus. The acu-dioxomorpholine nonribosomal peptide synthetase features a new type of condensation domain (designated CR) proposed to use a noncanonical arginine active site for ester bond formation. Using stable isotope labeling and MS, we determine that a phenyllactate monomer deriving from phenylalanine is incorporated into the diketomorpholine scaffold. Acu-dioxomorpholine is highly related to orphan inhibitors of P-glycoprotein targets in multidrug-resistant cancers, and identification of the biosynthetic pathway for this compound class enables genome mining for additional derivatives.
Graphical abstract
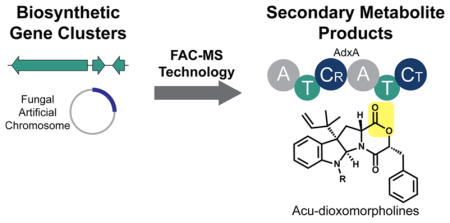
Fungal secondary metabolites have been a valuable source of therapeutics, including drugs such as penicillin, lovastatin, and cyclosporine.1 Over the past decade, it has become apparent that fungal genomes represent an untapped wealth of novel secondary metabolites, often containing >50 biosynthetic gene clusters (BGCs) per species.2,3 Associating these BGCs to their secondary metabolites is a low-throughput and challenging task, requiring labor-intensive heterologous expression approaches or genetic manipulations for fungal species which often lack such tools.4
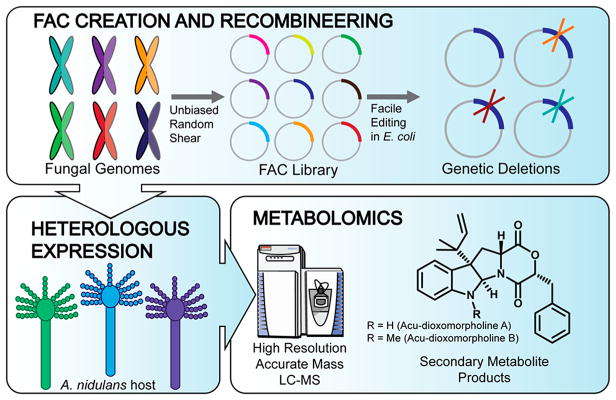
To address this challenge of associating BGCs with their secondary metabolites on a large scale, we recently reported the development of a platform designed to systematically access these fungal BGCs via heterologous expression in Aspergillus nidulans with detection and scoring of data produced by mass spectrometry-based metabolomics (Figure 1).5,6 Here we utilize FAC-MS technology for further dissection of one of these previously described FACs, thus identifying the biosynthetic pathway for the known metabolite acu-dioxomorpholine and a desmethyl intermediate, here designated acu-dioxomorpholine B and A, respectively.7,8 Acu-dioxomorpholine is highly related to other indole alkaloids like javanicunine, mollenine, and shornephine/PF1233 (Figure S1).9–12 Several of these metabolites inhibit P-glycoprotein transporters, key mediators of chemotherapeutic drug efflux in cancer subtypes that are resistant to first line chemotherapeutics.11,12 While therapeutic interest in these compounds is growing, the biosynthesis of diketomorpholines is currently unknown.
Figure 1.
Platform for discovery of fungal secondary metabolites and their biosynthetic pathways using fungal artificial chromosomes and mass spectrometry-based metabolomic scoring (FAC-MS). Fungal genomes are randomly sheared, and ~100 kb fragments with BGCs are inserted into FACs (top), which are A. nidulans/E. coli shuttle vectors. This enables facile deletion of biosynthetic genes (top, right). FACs are transformed into A. nidulans, a model fungus which serves as a heterologous host (bottom, at left). MS-based metabolomics is utilized to detect secondary metabolites associated with each FAC and to determine the effect of BGC mutants (bottom, at right).
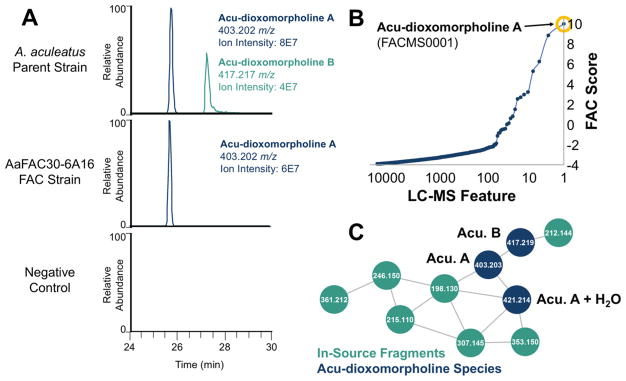
In using the FAC-MS platform, we detected an unknown metabolite from heterologous expression of one particular FAC (AaFAC30-6A16) with an m/z value of 403.2020 and a high FAC-Score of 10 (Figures 2A and 2B). Note that FAC-Scores for putative hits range from 0 to 27.6 The 403.2020 m/z compound was validated as shown in the bottom panel of Figure 2A, was consistent with a molecular formula of C25H27N2O3 (+1.0 ppm error), and was designated as compound FACMS0001. Comparison of metabolite extracts from A. nidulans harboring AaFAC30-6A16 and the A. aculeatus parental strain revealed that the same 403 m/z compound was present in both (Figure 2A). To visualize the relatedness of metabolites in this two-strain data set, we turned to spectral networking (Figure 2C), which clusters structurally and biosynthetically related metabolites using their mass spectrometric fragmentation patterns.13 We observed that the MS/MS fragmentation pattern of the 403 m/z compound was highly similar to that of a known A. aculeatus metabolite, acu-dioxomorpholine, dereplicated by accurate mass (417.2181 m/z, C26H29N2O3, +2.1 ppm error) and by comparison to previously reported ion fragmentation data for acu-dioxomorpholine.8 Manual analysis of MS/MS spectra confirmed that these metabolites are structurally related by a methyl group, with both the intact masses and the three major fragment ions differing in mass consistent with a CH2 moiety (Figure S2). On the basis of high resolution MS and MS/MS data analysis, we have designated these compounds as acu-dioxomorpholine A (C25H27N2O3) and B (C26H29N2O3), an update from previously used acu-dioxomorpholine nomenclature.7,8
Figure 2.
FAC-MS data enabling the identification of the acu-dioxomorpholine biosynthetic gene cluster. (A) Both acu-dioxomorpholine A and B were detected in the A. aculeatus parent strain; however, only acu-dioxomorpholine A was detected in AaFAC30-6A16. Neither metabolites were detected in a negative control FAC (no insert). (B) A metabolite feature corresponding to acu-dioxomorpholine A was detected in the FAC strain AaFAC30-6A16. This feature was the highest scoring ion for this strain using a FAC Score which ranks features based on their uniqueness within the entire FAC library. (C) Mass spectral networking of A. aculeatus metabolomics data reveals structurally related features corresponding to the reported structure of acu-dioxomorpholine B, a desmethyl variant, acu-dioxomorpholine A, a hydrolyzed version of acu-dioxomorpholine A, and several fragment ions produced in the electrospray source of the mass spectrometer. Acu-dioxomorpholine A and B are abbreviated as Acu. A and Acu. B, respectively.
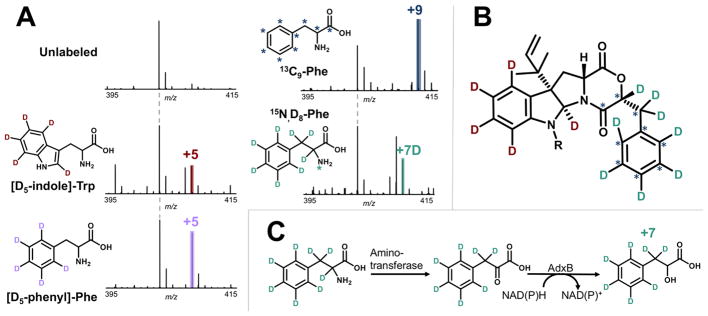
To confirm identification of acu-dioxomorpholine A and B and to probe their biosynthesis, we utilized stable isotope feeding of biosynthetic precursors. Their structures contain an indoline moiety expected to be derived from tryptophan and a benzyl group expected to be derived from phenylalanine. Labeling with [D5-indole]-Trp and [D5-phenyl]-Phe resulted in a shift of +5 Da in each case (Figure 3A and Figure S3), indicating that the acu-dioxomorpholine scaffold results from condensation of tryptophan with a phenylalanine-derived precursor. Additionally, full retention of all indole deuterons is consistent with prenylation at the C3 position as previously reported for acu-dioxomorpholine B, as prenylation at a different indole position would result in retention of only four deuterons. MS/MS analysis of the D5-indole-labeled acu-dioxomorpholine A and B species confirmed that the difference between these two metabolites is N-methylation (Figure S4).
Figure 3.
Atom tracking in acu-dioxomorpholine A biosynthesis using stable isotope labeling and MS. (A) Feeding with (D5-indole)-Trp resulted in a shift of +5 Da (408.2325 m/z, −1.2 ppm error), confirming Trp is a precursor. A + 4 labeled species was also detected, likely corresponding to loss of the C2 deuteron during prenylation and ring closure. Feeding with (D5-phenyl)-Phe resulted in a shift of +5 Da (408.2331 m/z, 0.2 ppm error), consistent with Phe being the source of the phenyl ring in acu-dioxomorpholine. Feeding with 13C9-Phe resulted in a shift of +9 Da (412.2319 m/z, 0.3 ppm error), providing further support for phenylalanine incorporation. Feeding with D8, 15N-Phe resulted in a shift of +7 Da (410.2465 m/z, 2.3 ppm error), indicating that phenylalanine is incorporated into acu-dioxomorpholine with loss of the nitrogen and a single deuteron. A +6 Da species was also detected from D8, 15N-Phe labeling, likely resulting from phenylpyruvate tautomerization. (B) Structure of acu-dioxomorpholine A and B showing the positions labeled with stable isotopes. (C) Summary of the phenylalanine to phenyllactate transformation supported by stable isotope labeling. Phenylalanine is converted to phenylpyruvate as a step in phenylalanine catabolism. AdxB reduces phenylpyruvate to form phenyllactate.
To further investigate the mechanism for phenylalanine incorporation into acu-dioxomorpholine A and B, we also used 13C9-Phe and D8,15N-Phe labeling. Feeding with 13C9-Phe resulted in a shift of +9 Da (Figure 3A), providing further evidence that this portion of the diketomorpholine scaffold is phenylalanine-derived. Feeding with D8,15N-Phe resulted in a shift corresponding to +7 deuterons, consistent with loss of a nitrogen and deuteron prior to incorporation into acu-dioxomorpholine A and B. A major peak with a mass shift of +6 Da was also observed for D8, 15N-Phe labeling (Figure 3A, fifth panel), consistent with partial loss of deuterons at the β position of phenylpyruvate due to ketoenol tautomerization. These data suggest that phenylalanine is transaminated to form phenylpyruvate (a known reaction in phenylalanine catabolism resulting in loss of the nitrogen and α-proton), and then phenylpyruvate is reduced to form the α-hydroxy acid, phenyllactate (Figure 3C). This same pathway is known to occur in lactic acid bacteria and the yeast Wickerhamia fluorescens but has not been observed in other fungi to our knowledge.14,15
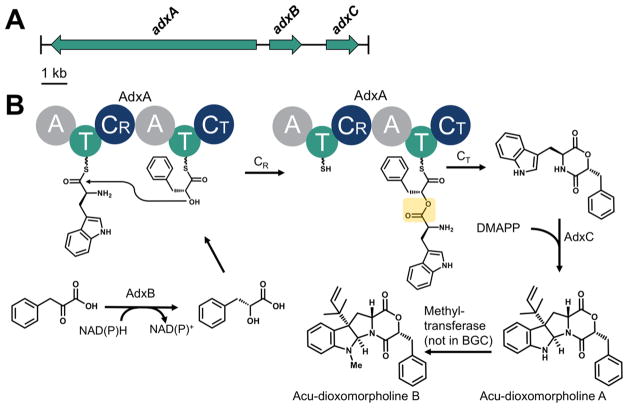
The gene cluster captured by AaFAC30-6A16 was predicted to contain eight genes by the Secondary Metabolite Unique Regions Finder (SMURF) (Table S1).2 To determine which of these genes are essential for acu-dioxomorpholine biosynthesis, we deleted seven predicted genes individually using FAC recombineering in E. coli. After transformation, growth in A. nidulans, and MS-based metabolomics of their extracts, acu-dioxomorpholine A production was found to be completely abolished in ΔadxA, ΔadxB, and ΔadxC deletants, corresponding to a predicted nonribosomal peptide synthetase (NRPS), NAD(P)H-dependent reductase, and aromatic prenyltransferase, respectively (Figure S5). The other four deletants had no significant effect on acu-dioxomorpholine A production.
Based on heterologous expression, high resolution mass spectrometry, stable isotope metabolic feeding, and genetic deletion, we propose the biosynthesis of acu-dioxomorpholine A and B (Figure 4). In this scheme, phenyllactate is formed via reduction of phenylpyruvate catalyzed by AdxB, which we designate a phenylpyruvate reductase. AdxB is predicted to bind NAD(P)H and catalyze the carbonyl to alcohol reduction to afford phenyllactate (Figure S6). It has 47% sequence identity to a predicted NADPH-dependent aldehyde reductase (GenBank XP_001276060) and 30% sequence identity to an experimentally characterized aldehyde reductase (UniProt Q9UUN9) which catalyzes NADPH-dependent reduction of alkyl and aryl aldehydes to their corresponding alcohols.16 Therefore, these findings are consistent with the phenylpyruvate reductase activity asserted for AdxB in acu-dioxomorpholine biosynthesis.
Figure 4.
Proposed biosynthetic pathway for acu-dioxomorpholine A and B production. (A) The acu-dioxomorpholine gene cluster contains a nonribosomal peptide synthetase gene (adxA), an NAD(P)-dependent reductase gene (adxB), and a prenyltransferase gene (adxC). (B) AdxB catalyzes reduction of phenylpyruvate to form phenyllactate. AdxA dimerizes tryptophan and phenyllactate via an ester bond-forming condensation domain containing an unusual arginine active site (CR). The depsipeptide intermediate is released by cyclization catalyzed by AdxA-CT, resulting in the diketomorpholine. Reverse prenylation catalyzed by AdxC results in acu-dioxomorpholine A. An unidentified methyltransferase (proposed to not be in the BGC) affords acu-dioxomorpholine B.
AdxA is a two-module NRPS proposed to assemble the diketomorpholine core from tryptophan and phenyllactate (Figure 4B). The first adenylation domain of AdxA (AdxA-A1) is predicted to recognize a hydrophobic amino acid such as tryptophan (Figure S7A). However, the second adenylation domain (AdxA-A2) has an unusual glycine in place of the typically conserved aspartate. This aspartate is usually responsible for helping to coordinate the positively charged amino group of the loaded amino acid (Asp235 for the classic example GrsA).17 Thus, AdxA-A2 is unlikely to load an amino acid; however, it would be expected to be able to load phenyllactate. Consistent with this, the substrate binding residues of AdxA-A2 bear similarity to those of other fungal adenylation domains which recognize α-hydroxy acids, including PF1022 synthetase which recognizes phenyllactate (Figure S7B).18 Collectively, this analysis of the adxA gene suggests that AdxA-A1 and AdxA-A2 are responsible for activating tryptophan and phenyllactate, respectively.
Loading of Trp followed by phenyllactate on AdxA suggests that the normal peptide condensation reaction catalyzed by NRPSs cannot occur. This is entirely consistent with the structure of acu-dioxomorpholine, which contains an ester bond within the diketomorpholine ring. Notably, the first condensation domain (designated CR) in the center of AdxA has a noncanonical arginine (R197) in place of the normal and highly conserved His, which ordinarily functions as the catalytic base for peptide condensation (Figure S8A). This suggests that R197 is involved in ester formation in one of two ways, as diagramed in Figure S9. One possibility is that the dielectric constant of the local protein microenvironment suppresses the pKa of the R197 guanidino group, so that it can function as a general base. While arginine is not typically considered a good candidate for general base catalysis, a handful of non-NRPS enzymes is known to utilize arginine in this role.19 Alternatively, it has been proposed elsewhere that the canonical catalytic histidine of NRPS condensation domains may serve to stabilize a tetrahedral oxyanion intermediate rather than direct nucleophile activation for attack of the upstream thioester, since pKa estimations of TycC His3256 suggest that it is protonated and thus unlikely to act as a catalytic base.20 Since arginine is expected to be protonated at physiological pH, R197 of AdxA would be a good candidate to serve this role.
To determine the prevalence of such atypical condensation domains containing an Arg vs a His among fungi, we analyzed 240 condensation domains from experimentally validated BGCs deposited in Minimum Information about a Biosynthetic Gene Cluster (MIBiG) (Figure S8B).21 We found that only 8/240 condensation domains also contained a proposed catalytic arginine in place of the expected His3256, corresponding to condensation domains within the lovastatin and D-lysergic acid peptide pathways with no apparent relation to ester formation (Figure S8C). This data thus shows that the activity proposed for the AdxA CR domain is unique. While the replacement of the canonical histidine with an arginine in the condensation domains suggests a catalytic role for arginine, further experimental evidence will be needed as validation.
The final steps in production of the acu-dioxomorpholine core invoke AdxC for reverse prenylation of the diketomorpholine intermediate at the C3 position as well as ring closure between C2 and the diketomorpholine nitrogen (Figure 4B). Such prenylation reactions are commonly observed in fungal indole alkaloids and result from addition of a dimethylallyl diphosphate (DMAPP) cofactor from the allylic position (termed “normal” prenylation) or the vinylic position (termed “reverse” prenylation). AdxC has 53% sequence identity to the roquefortine prenyltransferase RoqD (UniProt B6HJU1), which also catalyzes indole reverse prenylation at the C3 position.22 Interestingly, both AdxC and RoqD lack a catalytic lysine that has been proposed to abstract a proton from the C4 position, suggesting this lysine may have future value in predicting the regiospecificity of aromatic prenyltransferases (Figure S10).23 Alternatively, prenylation may occur at an earlier step in the pathway.
The final step of the biosynthetic pathway involves methylation to form acu-dioxomorpholine B. Interestingly, no predicted methyltransferase was found on AaFAC30-6A16 nor in the genomic vicinity in the A. aculeatus genome, suggesting that a background, promiscuous methyltransferase activity is involved. Consistent with this observation, only the desmethyl acu-dioxomorpholine A was detected in AaFAC30-6A16 extracts. This suggests that the major biosynthetic product of this BGC is acu-dioxomorpholine A not B.
In addition to diketomorpholines, depsipeptides containing α-hydroxy acids have a wide variety of biological activities, including the enniatin family.24 The identification of the acu-dioxomorpholine BGC enables the targeted discovery of additional diketomorpholine and depsipeptide metabolites with therapeutic potential. To test this possibility, we searched for NRPS BGCs containing AdxB homologues and identified putative diketomorpholine clusters in A. udagawae and A. clavatus (Figure S11). Notably, each of these clusters contains an NRPS adenylation domain with substrate binding residues similar to those of AdxA-A2, suggesting that these NRPSs activate phenyllactate or another α-hydroxy acid.
In conclusion, we have utilized the FAC-MS platform to identify the first BGC for diketomorpholine alkaloids, a scaffold with therapeutic potential as P-glycoprotein inhibitors for multidrug-resistant cancers. This work also identified an unusual ester bond formation on an NRPS assembly line and biosynthetic signatures related to the biogenesis of this class of compounds. Assignment of this BGC will advance their translational development by enabling the search for related compounds and facilitating their larger-scale production. Though fungal genomes represent an untapped wealth of novel secondary metabolites, the ability to prioritize BGCs based on potential for bioactivity is lacking. By providing a systematic and large-scale way of accessing and prioritizing these fungal BGCs, FAC-MS technology can reinvigorate early discovery pipelines.
METHODS
Gene Cluster Editing via FAC Recombineering
Predicted biosynthetic genes on AaFAC30-6A16 were deleted using Red/ET tools as mentioned.7 FAC transformation into A. nidulans and culture conditions were performed as mentioned.7
A. aculeatus Growth and Isotope Labeling
A. aculeatus was inoculated from a concentrated spore stock in solid GMM supplemented with 1 mM 13C9-Phe; D8,15N-Phe; [D5-indole]-Trp; or [D5-phenyl]-Phe. Cultures were incubated at 30 °C for 3 days and extracted with 20 mL hexanes. Extracts were dried in vacuo, reconstituted at 2 mg mL−1 in 50% acetonitrile, and analyzed by LC-MS.
LC-MS Analysis
Metabolite extracts were analyzed by LC-MS using a Thermo Q Exactive mass spectrometer in line with an Agilent 1200 HPLC as described.7 Mass spectral networking was performed using in-house software as mentioned.25
Bioinformatics Analyses
Minimum Information about a Biosynthetic Gene Cluster (MIBiG) sequences were filtered for ascomycete NRPS gene clusters.21 Excised condensation domain sequences (Pfam 00668, n = 240) were aligned using MUSCLE.26 Profile Hidden Markov Models were created using HMMER3.27 Other sequence alignments were also performed using MUSCLE.
Supplementary Material
Acknowledgments
The authors thank T. Toby at Northwestern University for assistance with the artwork in Figure 1. This work was funded by the US National Institutes of Health SBIR grant R44AI118086 to C.C.W., J.W.B., and N.L.K.; grant R01AI065728 to N.P.K.; grant R01AT009143 to N.L.K.; grant T32GM008449 to M.T.R.; and grant F32GM120869 to K.D.C. M.T.R was supported in part by the Northwestern University Graduate School Cluster in Biotechnology, Systems, and Synthetic Biology.
Footnotes
Notes
The authors declare the following competing financial interest(s): C.C.W., R.Y., M.N.I., C.C., M.S., and E.W. are employees of Intact Genomics which sells unbiased Random Shear BAC and FAC libraries and services for genome discovery and DNA research, as well as BAC cloning, DNA end repairing, E. coli competent cells, and other enzyme kits for DNA and protein research.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.8b00024.
Supplemental figures, spectra, chromatograms, bioinformatics analyses, and proposed mechanisms (PDF)
References
- 1.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism – from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 2.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger, and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anyaogu DC, Mortensen UH. Heterologous production of fungal secondary metabolites in Aspergilli. Front Microbiol. 2015 doi: 10.3389/fmicb.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bok JW, Ye R, Clevenger KD, Mead D, Wagner M, Krerowicz A, Albright JC, Goering AW, Thomas PM, Kelleher NL, Keller NP, Wu CC. Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics. 2015 doi: 10.1186/s12864-015-1561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, Chen C, Yang K, Robey MT, Gao P, Lamprecht M, Thomas PM, Islam MN, Palmer JM, Wu CC, Keller NP, Kelleher NL. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat Chem Biol. 2017;13:895–901. doi: 10.1038/nchembio.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen LM, Hoeck C, Frisvad JC, Gotfredsen CH, Larsen TO. Dereplication guided discovery of secondary metabolites of mixed biosynthetic origin from Aspergillus aculeatus. Molecules. 2014;19:10898–10921. doi: 10.3390/molecules190810898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overy D, Correa H, Roullier C, Chi WC, Pang KL, Rateb M, Ebel R, Shang Z, Capon R, Bills G, Kerr R. Does osmotic stress affect natural product expression in fungi? Mar Drugs. 2017;15:254. doi: 10.3390/md15080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HJ, Gloer JB, Wicklow DT, Dowd PF. Mollenines A and B: new dioxomorpholines from the ascostromata of Eupenicillium molle. J Nat Prod. 1998;61:804–807. doi: 10.1021/np9704056. [DOI] [PubMed] [Google Scholar]
- 10.Nakadate S, Nozawa K, Horie H, Fujii Y, Nagai M, Komai S, Hosoe T, Kawai K, Takashi Y, Fukushima K. New dioxomorpholine derivatives, Javanicunine A and B, from Eupenicillium javanicum. Heterocycles. 2006;68:1969–1972. [Google Scholar]
- 11.Khalil ZG, Huang XC, Raju R, Piggott AM, Capon RJ. Shornephine A: structure, chemical stability, and P-glycoprotein inhibitory properties of a rare diketomorpholine from an Australian marine-derived Aspergillus sp. J Org Chem. 2014;79:8700–8705. doi: 10.1021/jo501501z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aparicio-Cuevas MA, Rivero-Cruz I, Sanchez-Castellanos M, Menendez D, Raja HA, Joseph-Nathan P, Gonzalez MDC, Figueroa M. Dioxomorpholines and derivatives from a marine-facultative Aspergillus species. J Nat Prod. 2017;80:2311–2318. doi: 10.1021/acs.jnatprod.7b00331. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen DD, Wu CH, Moree WJ, Lamsa A, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C, Ballesteros J, Sanchez J, Watrous JD, Phelan VV, van de Wiel C, Kersten RD, Mehnaz S, De Mot R, Shank EA, Charusanti P, Nagarajan H, Duggan BM, Moore BS, Bandeira N, Palsson BO, Pogliano K, Gutierrez M, Dorrestein PC. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci U S A. 2013;110:E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu W, Yu S, Zhu L, Zhang T, Jiang B. Recent research on 3-phenyllactic acid, a broad-spectrum antimicrobial compound. Appl Microbiol Biotechnol. 2012;95:1155–1163. doi: 10.1007/s00253-012-4269-8. [DOI] [PubMed] [Google Scholar]
- 15.Fujii T, Shimizu M, Doi Y, Fujita T, Ito T, Miura D, Wariishi H, Takaya N. Novel fungal phenylpyruvate reductase belongs to d-isomer-specific 2-hydroxyacid dehydrogenase family. Biochim Biophys Acta, Proteins Proteomics. 2011;1814:1669–1676. doi: 10.1016/j.bbapap.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Kita K, Fukura T, Nakase K, Okamoto K, Yanase H, Kataoka M, Shimizu S. Cloning, Overexpression, and mutagenesis of the Sporobolomyces salmonicolor AKU4429 gene encoding a new aldehyde reductase, which catalyzes the stereoselective reduction of ethyl 4-chloro-3-oxobutanoate to ethyl (S)-4-chloro-3-hydroxybutanoate. Appl Environ Microbiol. 1999;65:5207–5211. doi: 10.1128/aem.65.12.5207-5211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 18.Weckwerth W, Miyamoto K, Iinuma K, Krause M, Glinski M, Storm T, Bonse G, Kleinkauf H, Zocher R. Biosynthesis of PF1022A and related cyclooctadepsipeptides. J Biol Chem. 2000;275:17909–17915. doi: 10.1074/jbc.M001084200. [DOI] [PubMed] [Google Scholar]
- 19.Guillen Schlippe YV, Hedstrom L. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch Biochem Biophys. 2005;433:266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Dusterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJ, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kotter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nutzmann HW, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gomez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage AA, Caffrey P, Cheng YQ, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, van der Donk WA, Dorrestein PC, Doyle S, Driessen AJ, Ehling-Schulz M, Entian KD, Fischbach MA, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Hofte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon HJ, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Mendez C, Metsa-Ketela M, Micklefield J, Mitchell DA, Moore BS, Moreira LM, Muller R, Neilan BA, Nett M, Nielsen J, O’Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet JL, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas JA, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Sussmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, Glockner FO. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Estrada C, Ullan RV, Albillos SM, Fernandez-Bodega MA, Durek P, von Dohren H, Martin JF. A single cluster of coregulated genes encodes the biosynthesis of the mycotoxins roquefortine C and meleagrin in Penicillium chrysogenum. Chem Biol. 2011;18:1499–1512. doi: 10.1016/j.chembiol.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Metzger U, Schall C, Zocher G, Unsold I, Stec E, Li SM, Heide L, Stehle T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci U S A. 2009;106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prosperini A, Berrada H, Ruiz MJ, Caloni F, Coccini T, Spicer LJ, Perego MC, Lafranconi A. A review of the mycotoxin enniatin B. Front Public Health. 2017;5 doi: 10.3389/fpubh.2017.00304. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henke MT, Soukup AA, Goering AW, McClure RA, Thomson RJ, Keller NP, Kelleher NL. New aspercryptins, lipopeptide natural products, revealed by HDAC inhibition in Aspergillus nidulans. ACS Chem Biol. 2016;11:2117–2123. doi: 10.1021/acschembio.6b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.