Significance
Understanding how microbes adapt to novel environments is essential to understanding acute bacterial infection and long-term disease, as genetic architecture underlying the production and maintenance of genetic variation influences a population’s potential for adaptation. In this in-depth analysis of a highly replicated Escherichia coli long-term evolution experiment, we observe rapid diversification into stable subpopulations in response to several environmental variables. This niche separation creates novel genetic backgrounds upon which new traits, such as differential nutrient utilization or antimicrobial resistance, can arise. The observed genetic changes, in a simple and tractable experimental system, mimic events known to occur during bacterial infections.
Keywords: biofilm, niche specialization, complex environment
Abstract
How genetic variation is generated and maintained remains a central question in evolutionary biology. When presented with a complex environment, microbes can take advantage of genetic variation to exploit new niches. Here we present a massively parallel experiment where WT and repair-deficient (∆mutL) Escherichia coli populations have evolved over 3 y in a spatially heterogeneous and nutritionally complex environment. Metagenomic sequencing revealed that these initially isogenic populations evolved and maintained stable subpopulation structure in just 10 mL of medium for up to 10,000 generations, consisting of up to five major haplotypes with many minor haplotypes. We characterized the genomic, transcriptomic, exometabolomic, and phenotypic differences between clonal isolates, revealing subpopulation structure driven primarily by spatial segregation followed by differential utilization of nutrients. In addition to genes regulating the import and catabolism of nutrients, major polymorphisms of note included insertion elements transposing into fimE (regulator of the type I fimbriae) and upstream of hns (global regulator of environmental-change and stress-response genes), both known to regulate biofilm formation. Interestingly, these genes have also been identified as critical to colonization in uropathogenic E. coli infections. Our findings illustrate the complexity that can arise and persist even in small cultures, raising the possibility that infections may often be promoted by an evolving and complex pathogen population.
Comprising most of the tree of life and constituting a major determinant of human health and disease, microbes are the most diverse organisms on earth, but how this diversity and genetic variation is generated and maintained remains a central question in evolutionary and population biology. For over 30 y, long-term evolution experiments have examined how prokaryotes and single-celled eukaryotes adapt to novel environments ranging from nutrient-poor environments where Escherichia coli are presented with simple carbohydrates as carbon sources (1–3); spatially structured environments where bacteria and yeast evolve strategies to survive oxygen limitation, migration, and cell crowding (4–6); and engineered communities, where microbes evolve under cooperative scenarios (7, 8). Through these experiments microbes have been shown to rapidly adapt to laboratory settings (4, 5, 9–11) by evolving key innovations, such as the ability to utilize a previously unusable carbon source (12), development of cross-feeding behaviors (13), changes in cellular morphology (4), and extreme levels of polyploidization (14, 15).
In nature, microbes are more often faced with ecologically complex environments, and the combination of nutritional and spatial heterogeneity can introduce many novel niches in which microbes may adapt. Nutritional heterogeneity can allow for the evolution of generalist/specialist strategies (16–19), and nutrient-rich environments offer many potential, evolutionary trajectories (20). Further, in spatially structured environments, the two segregated populations may independently evolve and create additional genetic diversity (4, 21, 22).
Here, we report the results of a long-term evolution experiment consisting of 100 E. coli populations. These populations have been propagated from either WT K-12 or mismatch-repair deficient genetic backgrounds, at five different transfer densities, for over 3 y (up to 10,000 generations), in growth conditions allowing for both spatial and nutritional specialization. We applied a systems approach for assessing both the evolving populations and derived clones, using genomic, transcriptomic, and exometabolomic data, and growth phenotypes associated with lag time, maximum growth rate, competitive growth (relative fitness when cocultured with the progenitor strain), auxotrophy (inability to create essential nutrients such as amino acids), and spatial variation (location of haplotypes within the culture tube). Long-term population substructure rapidly and stably evolves in these populations, with subpopulation haplotypes being characterized by spatial affinity, differential regulation of metabolism genes, and differential utilization of amino acids.
Results
Long-Term Populations Harbor Multiple Dominant Haplotypes.
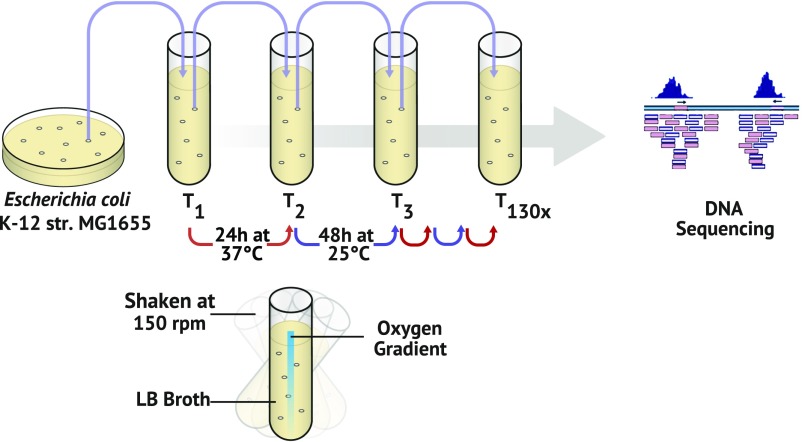
For over 3 y we have maintained 100 lines of WT (MMR+) and repair-deficient (MMR−) E. coli populations by serial transfer in 16- × 100-mm culture tubes incubated in alternating conditions of 37 °C for 24 h and 25 °C for 48 h while shaking at 175 rpm (Fig. 1). Due to the dimensions of the culture tubes, shaking is inefficient at environmental homogenization, resulting in an oxygen gradient that creates the opportunity for niche-heterogeneity development. In an attempt to manipulate effective population size, five different daily transfer volumes (1 mL, 100 μL, 10 μL, 1 μL, and 10 nL) were used to inoculate 10 mL of LB each transfer. These differences in transfer volume also affected the number of generations experienced by the populations before carrying capacity is reached after each transfer. As such, an approximately fivefold difference in cell division number per transfer exists between the largest and smallest transfer densities. Early in the experiment, we observed widespread development of biofilm at the liquid–air interface. Although cultures are thoroughly vortexed before each transfer, the biofilm rapidly and consistently reforms within 24 h.
Fig. 1.
Illustration of culture methods for long-term evolution. Fifty WT and 50 mutL− (mismatch-repair deficient) E. coli K-12 MG1655 populations were established from a single isolated colony and grown in glass culture tubes containing 10 mL of LB broth. Cultures were propagated by alternating 37 °C incubation for 24 h with 25 °C incubation for 48 h. Further, five different evolution treatments were established by varying the volume of the transfer bottleneck. Populations were thoroughly vortexed before each transfer, and whole communities were subjected to whole-genome sequencing every 6 mo.
Every 6 mo, a fully homogenized sample of each population was collected and sequenced at 100× coverage to identify and track new and segregating polymorphisms. Whole-community sequencing of 100 populations after 1 y of evolution (800–3,400 generations) identified polymorphisms expected to encourage [fimE: a repressor of the type 1 fimbriae; hns: histone-like global regulator which regulates fimB and fimE (23, 24)] and inhibit [acrB: a subunit of the MDR family efflux pump AcrAB/TolC (25)] biofilm formation, as well as many other polymorphisms that accounted for significant portions of the populations.
To gain more insight into the potential linkage of these polymorphisms within distinct haplotypes, we focused on 10 populations shown from population sequencing to contain a combination of polymorphisms in fimE, hns, and/or acrB (Table 1 and Datasets S1 and S2). At the 2.5-y collection, we isolated eight randomly selected clones from each of these 10 populations for whole-genome sequencing and phenotypic assessment. Initial plating of the clones on Congo red agar confirms that all clones are capable of forming biofilms, whereas the WT progenitor is not (26, 27) (SI Appendix, Fig. S1). Further, when grown as monocultures, all of these clones formed visible biofilms at the liquid–air interface.
Table 1.
Summary of genetic background, daily transfer population size, and number of cumulative generations for each population analyzed in this study
| Population* | Initial genotype | Biofilm-related polymorphisms† | Transfer volume | Generations‡ |
| 113 | ∆mutL | acrB; fimE | 1 mL | 2,000 |
| 125 | WT | hns; fimE | 1 mL | 2,000 |
| 126 | ∆mutL | acrB; fimE | 1 mL | 2,000 |
| 129 | WT | acrB; fimE; hns | 1 mL | 2,000 |
| 210 | ∆mutL | acrB; fimE; hns | 100 μL | 4,150 |
| 221 | WT | hns | 100 μL | 4,150 |
| 233 | ∆mutL | acrB; fimE; hns | 100 μL | 4,150 |
| 326 | ∆mutL | hns | 10 μL | 6,330 |
| 410 | ∆mutL | acrB; fimE | 1 μL | 8,500 |
| 417 | ∆mutL | acrB; fimE | 1 μL | 8,500 |
The first digit of each population encodes the transfer volume treatment.
After 1 y.
After 2.5 y.
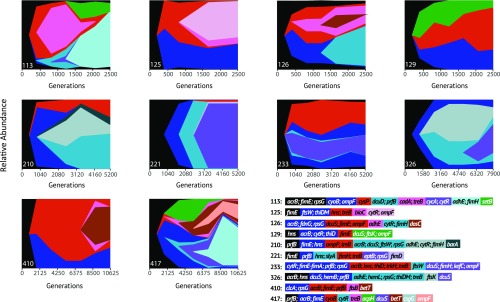
Genomic sequence of the clones revealed multiple haplotypes coexisting in all assayed populations, from two major clades in the WT populations to five clades in repair-deficient populations (SI Appendix, Fig. S2). Further, these haplotypes contribute to a stable population structure. Specifically populations 410 and 417, our populations evolving with the smallest transfer sizes, have been cultivated for at least 10,000 generations and have yet to experience a population-wide fixation event (Fig. 2; red: haplotype A, blue: haplotype B, green: haplotype C). Sequenced clones were additionally surveyed for cooccurrence of biofilm-promoting (fimE and hns) and biofilm-inhibiting (acrB) mutations and only one population, 417 (106 transfer density, MMR−), harbored clones containing null mutations (frameshift, IS-element insertion, or large deletion) in both fimE and acrB.
Fig. 2.
Muller plots representing relative abundances of haplotypes over 3 y of evolution. Shades of green, red, and blue denote different haplotypes, with variation in shade both within the figure and the mutation legend representing new variants arising within these haplotypes. Mutations in black occurred and fixed within populations before divergence of subpopulations. Relative abundances of haplotypes were inferred via whole-population sequencing performed every 6 mo, combining with whole-genome sequencing of eight individual clones from each population revealed haplotype associations. Numbers in the legend correspond to a particular population, while mutations included in the legend are present for at least two sampling points at a frequency of <0.05 in the population. Datasets S1 and S2 contain a complete list of mutations identified in each isolated clone.
Parallel Mutation Is Extensive Across Populations.
Across all 10 focus populations, extensive parallel mutation, beyond biofilm-associated genes, is observed. We identified 66 genes that acquired either an SNP or indel mutation in at least 5 of 10 populations (expectation based on 10,000 random permutations = 33, P value <10−4) (SI Appendix, Table S1). Interestingly, the order in which several shared mutations arose and spread is similar across populations. For example, a nonsynonymous change in trkH (potassium transporter) commonly occurred early and increased to high frequencies, followed by reversion of a known laboratory-associated mutation in prfB (peptide chain release factor RF-2) to its WT form (28) and nonsynonymous changes in dcuS (sensor kinase for dicarboxylates), ompF (outer membrane porin), and treB (trehalose importer), with mutations in adhE (acetaldehyde-CoA dehydrogenase) and fimH (minor component of type 1 fimbriae) reliably occurring later in evolutionary time (SI Appendix, Table S2). We additionally identified 23 loci that acquired an IS-element insertion or a long deletion in at least two populations (SI Appendix, Table S3). As in a recent study on human gut colonization (29, 30), we also observed a high incidence of parallel deletion of the gat operon (6 out of 10 populations).
To ensure that the parallel mutations observed in these populations were due to selection and not an artifact of target size, we conducted a log-likelihood ratio test (G-test) for both the WT and repair-deficient populations (31). We then compared the resulting G-scores to an expectation calculated by bootstrapping 1,000 random G-score distributions based on the total number of nonsynonymous mutations observed in each genetic background (55 mutations in WT; 5,371 mutations in repair-deficient) and significance was determined with a z-test (31). For both genetic backgrounds, calculated G-scores were significantly different from expected (WT: Gobs = 472, Gexp = 522.53, P = 0.00147; repair-deficient: Gobs = 5,736.32, Gexp = 7,756.81, P = 1.39 × 10−35) (Dataset S3).
We additionally compared the parallel mutations identified in our populations to the mutations identified across two different publicly available datasets (SI Appendix, Table S4): (i) Lenski’s most-characterized population, Ara-1 (9), contained only one gene, yghJ (since renamed sslE), a putative secreted and surface-associated lipoprotein mucinase, that also exhibited mutations in our 10 focus populations (Jaccard index = 0.011, P = 0.396) and (ii) Helling’s population characterized by Kinnersley et al. (32) shared 11 of the 66 genes containing parallel mutations within our populations (Jaccard index = 0.016, P = 0.26). Although the Lenski and Helling experiments also focus on long-term evolution of E. coli strains, differences exist in both culture conditions and E. coli genetic background with respect to our study. Specifically, Lenski’s populations involve E. coli B and are batch-cultured in a minimal glucose medium, and the Helling lines were evolved in a chemostat containing a minimal glucose medium, despite the shared usage of K-12–derived E. coli. Given the different experimental conditions, the difference in the evolved molecular differences between these sets of lines is not surprising.
Evolved Isolates and Populations Show Increased Fitness in Heterogeneous Environments but Not in Homogeneous Environments.
To associate the evolved genotypes within our populations to phenotypes, we focused further on eight clones isolated from population 125, which experienced the fewest SNPs throughout the experiment (26 SNPs) and has a WT (MMR+) background. Through whole-genome sequencing of the individual clones, we identified two major haplotype groups from population 125, with clones 125-1, 125-3, and 125-6 (haplotype A) characterized by an early IS-element insertion upstream of hns and clones 125-2, 125-4, 125-5, 125-7, and 125–8 (haplotype B) characterized by an IS-mediated deletion of thiDM, rendering the latter clones auxotrophic for the B vitamin thiamine. Individual haplotypes are defined as members of a clade and are not identical to each other, although they share defining and presumably early mutations.
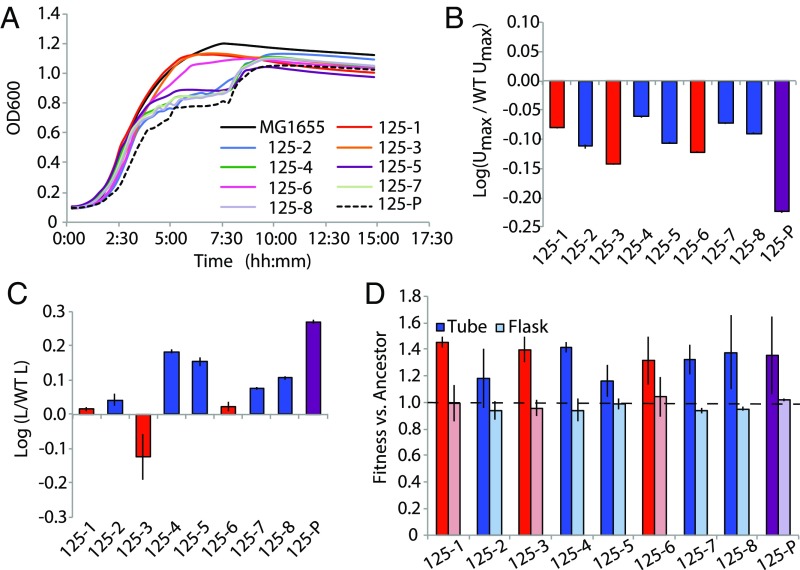
All individuals isolated from population 125 had significantly slower maximum growth rates than the WT progenitor strain when assayed in a 96-well plate (Welch’s t = −14.236, P = 2.11 × 10−9) (Fig. 3 A and B). Additionally, a revived sample of population 125 collected at the same time the clones were isolated (after 2.5 y of evolution or 2,000 generations) had an even slower maximum growth rate than each of the individual clones (Welch’s t = 10.792, P = 8.52 × 10−7). Further, when we examined lag time, all clones except 125-3 experienced longer lag times than the WT progenitor strain when grown in isolation (Fig. 3C).
Fig. 3.
Differences in growth phenotypes of clones isolated from population 125. (A) Growth curves (measured by optical density at a wavelength of 600 nm) for clones isolated from population 125 (MMR+) and revived population 125 (125-P) cultured in LB broth for 24 h at 37 °C. Warmer colors (red, orange, and pink) represent clones belonging to haplotype A and cooler colors (blue, green, and purple) represent clones belonging to haplotype B. MG1655 is the progenitor from which the evolved populations originated. (B) Relative maximum growth rates (Umax) for clones isolated from population 125 determined by fitting a modified Gompertz equation to optical density data. For all bar graphs, red bars represent clones belonging to haplotype A, blue bars represent clones belonging to haplotype B, and purple bars (125-P) represent a revived sample of population 125 after 2.5 y. (C) Relative lag time (L) for clones isolated from population 125 determined by fitting a modified Gompertz equation to optical-density data. (D) Competitive fitness of clones and revived sample of population 125 when cocultured with a rifampicin-resistant MG1655 strain and counted through replica plating determined by the equation , where Ei and Ef are the initial and final counts of the evolved clones (population) and Ai and Af are the initial and final counts of the MG1655 progenitor within the coculture. Rifampicin resistance was used as a selective marker as this phenotype can be conferred without a measurable fitness effect. Dark bars represent fitness in a culture tube (heterogeneous environment) and light bars represent fitness in a flask (homogeneous environment).
Surprisingly, these slower growth rates did not negatively impact the fitness of the individual clones in competition with the progenitor, as they all out-performed the progenitor when cocultured in growth tubes (with paired t test significance ranging from P = 0.078 for clone 125-2 to P = 0.006 for clone 125-4) (Fig. 3D, dark bars) under conditions similar to the evolution experiment. However, when the individual isolates were cocultured with the progenitor in a well-mixed, homogeneous environment, all performed either the same or worse than the progenitor strain (with paired t test significance ranging from P = 0.792 for clone 125-1 to P = 0.006 for clone 125-2) (Fig. 3D, light bars).
In light of the extreme fitness differences between the individual derived clones and the WT progenitor, we asked if the presence of haplotype A and B within a population is truly stable. To this end, we reconstructed the population by coculturing each of the haplotype A clones (125-1, 125-3, and 125–6) with each of the haplotype B clones (125-2, 125-4, 125-5, 125-7, and 125-8) and transferring 1 mL of culture to a new culture tube containing fresh media every day for 7 d (∼22 generations). Comparison of the frequencies of the two haplotypes after 1 and 7 d revealed no difference in the abundance of each haplotype between these two time points (paired t test: t = 1.6787; P = 0.0961), implying mutual stability (SI Appendix, Fig. S3). Further, we calculated the minimum expected change between the two haplotypes in the absence of population stability using the fitness data shown in Fig. 3D. For example, assuming fitness is transitive, the fitness of haplotype A’s clone 125-1 relative to haplotype B’s clone 125-4 is ∼1.45/1.42, or ∼1.02, ignoring sampling error. If we let the starting proportion of clone 125-1 (p0) equal 0.402, as it is in SI Appendix, Fig. S3A, and the strength of selection (s) equal 0.02, as determined by the relative fitness of clone 125-1, then the expected proportion of clone 125-1 after 22 generations (pt) can be solved for with the equation (33), or ∼0.510. However, the proportion of clone 125-1 found in the experimental data is 0.410, or essentially unchanged, suggesting that the presence of the two subpopulations is, in fact, stable.
Individual Clones Show Differential Utilization of Key Nutrients.
Stable subpopulation structure is often engineered or evolved on the basis of syntrophy—cross-feeding of nutrients (34, 35) or specialization on a specific novel nutrient (36). As LB is a complex growth medium with many potential nutrients, specifically amino acids that clones could potentially specialize on, we looked for evidence for syntrophy in the study population. First, we plated all characterized individual clones from population 125 on M9 minimal media to determine if any auxotrophy had evolved within the population. If a particular clone is auxotrophic, then growth is observed on LB media but not on M9, as the latter forces the bacteria to biosynthesize their own amino acids and vitamins. While no amino acid auxotrophy is observed, we observed auxotrophy to the B vitamin thiamin in haplotype B. This auxotrophy was confirmed as haplotype B clones have an observed deletion of the thiamin biosynthesis genes (thiDM) and are able to grow on M9 media supplemented with thiamin.
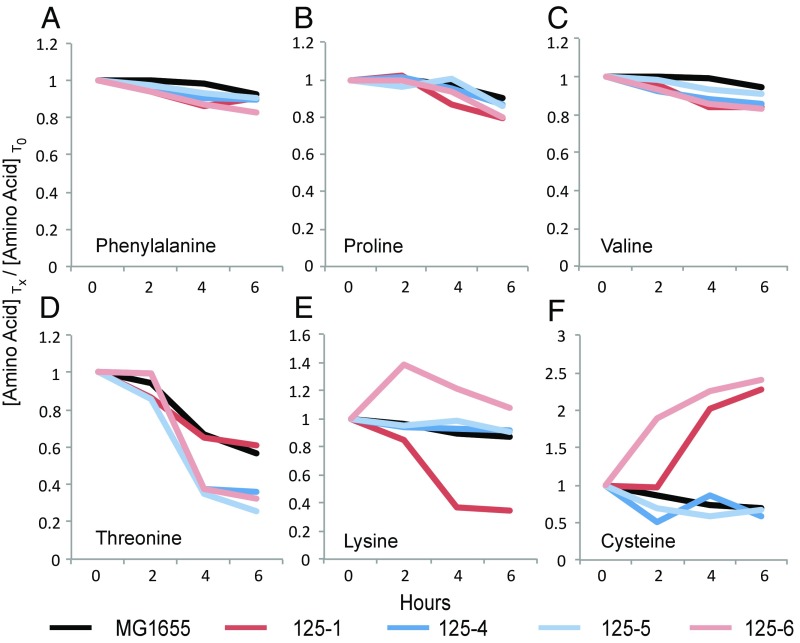
Next, to further investigate if differential amino acid usage could be a source of syntrophy, we used an exometabolomic approach to survey the spent media of four individual clones representing each haplotype in population 125 as well as the WT progenitor (SI Appendix, Table S5). For 14 of the 17 amino acids surveyed, no distinguishable difference was detected in utilization compared with the WT progenitor (Fig. 4 A–C). However, there are extreme differences within and among haplotypes in the utilization of threonine, lysine, and cysteine (Fig. 4 D–F and Dataset S4).
Fig. 4.
Uptake/release of free amino acids from single clones in LB broth through an initial 6 h of growth. Amino acid availability is demonstrated as a ratio of the concentration of a particular amino acid at a particular time point (Tx) and that for the same amino acid at 0 h (T0) as detected by GC-MS. (A) phenylalanine, (B) proline, and (C) valine show no significant difference in net uptake across samples, while (D) threonine, (E) lysine, and (F) cysteine show significant differences in net uptake and net excretion between the evolved clonal isolates and the WT progenitor.
Individual Clones Showed Major Transcriptional Changes Suggestive of Their Potential Niche.
While parallel mutation can highlight candidate genes that may be important in establishing subpopulation structure, genomic data alone are unable to confirm if nonsynonymous mutations result in gain or loss of function, or if there are downstream effects on metabolic or regulatory pathways. Additionally, any epigenetic changes that occur, such as adenine methylation (which can be associated with phenotypes regulated by phase variation), would go undetected by genomic analysis alone. Thus, we performed transcriptome analysis of mRNA extracted at midlog phase from clones isolated from population 125 and the WT progenitor.
Analysis of the transcriptomes revealed that the similarity of clones based on differential gene expression resembles their phylogenetic relationship (SI Appendix, Fig. S4 and Dataset S5). Major differences between the two haplotypes include up-regulation of the nupG and the deo operon (both associated with nucleotide salvage) in haplotype A and down-regulation of two cold-shock proteins, cspG (haplotype A) and cspC (haplotype B). Within haplotype A, there appears to be further distinction between clone 125-1 and clones 125-3 and -4 with an additional 38 differentially regulated genes, suggesting further structure within this particular haplotype. Specifically, up-regulation of the cad operon (involved in lysine import and catabolism) is detected in clone 125-1, a possible cause for the increased utilization of lysine in this clone as observed in the exometabolomic analysis. Up-regulation of hydrogenase-2 (hybACO) and spermidine acetyltransferase (speG) and down-regulation of cytochrome bd-II (cbdAB) are also observed in clone 125-1 but not in the other clones belonging to haplotype A.
In addition to haplotype-specific regulation, some loci experienced parallel differential expression in both haplotypes A and B. Clones 125-3 and -6 (haplotype A) and clones 125-2, -4, -5, -7, and -8 (haplotype B) have increased expression of the tdc operon (threonine import and catabolism) consistent with the increase in threonine uptake, while clones 125-3 and -6 (haplotype A) and clone 125-2 (haplotype B) have up-regulation in flu, an epigenetically regulated phase-variable gene encoding a biofilm-promoting autotransporter. In addition to influencing biofilming, flu also appears to play a role in flocculation, potentially lending insight into the spatial segregation of haplotypes within the evolving population (37, 38).
Haplotypes Show Affinity for Different Spatial Niches.
Finally, we evaluated whether the different haplotypes display an affinity for different spatial niches within the growth environment. As previously mentioned, haplotype B clones are auxotrophic for thiamine, which allows simple quantification of the two haplotypes by replica plating from solid LB media to M9. Thus, we aspirated 500 μL of cells from liquid-culture replicates of population 125 in two assays, one from the top half of the liquid culture and one from the bottom. In both assays, despite being the less-common haplotype (accounting for only ∼30% of the entire population according to whole-population sequencing), haplotype B comprised 75% of the cells isolated from the planktonic, more anaerobic, niche, while haplotype A accounted for the other 25%. Further, when the biofilm itself was surveyed, only individuals belonging to haplotype B were present, contrary to expectations based on the genomically surveyed genotypes. Two possibilities for the underrepresentation of haplotype A in our sampling are that (i) haplotype A is concentrated in flocs, making aspiration of those individuals more difficult, or (ii) some members of haplotype A have become so sensitive to traditional plating methods that we are unable to recover them.
To determine whether aerobic growth could impede our ability to sample clones without bias, we again aspirated 500 μL of cells from both the top half and bottom half of six liquid-culture replicates of population 125. From each aspirate, 100 μL of cells were spread on two LB plates before incubating one replicate aerobically and the other replicate in an anaerobic jar. While the difference in cell counts between the two treatments was not statistically significant (paired t testtop = 0.828, P = 0.454; paired t testbottom = 1.80, P = 0.146), there was a trend of higher cell counts on the anaerobic plates, suggesting the existence of a subset of individuals not captured with aerobic plating (SI Appendix, Fig. S5).
Discussion
Parallel Mutation Provides Evidence for Possible Adaptive Targets.
Within this study, we have identified a substantial number of parallel mutations among replicate populations. While some mutations, such as IS-element insertions upstream of hns, have such broad downstream effects that it is difficult to elucidate the specific adaptive effects, others such as deletion of the gat operon [which is notably highly expressed in ancestral K-12 (39)] have the adaptive potential to reduce bioenergetic costs by reducing expression. This is further supported through our transcriptional analysis, where gatB and gatY were observed to be within the top 1.5% of all expressed genes in the progenitor line. A previous study also observed the gat operon as a deletion hotspot during long-term evolution of E. coli K-12 in chemostats and concluded that the benefits of this deletion could be attributed to conservation of resources otherwise wasted by constitutive expression of gat (40).
In addition to this shared evolutionary behavior of the gat operon across studies, we also identified similarities between mutations in our population and another earlier long-term evolution experiment (13, 32, 41), even though the structural and nutritional environments differed: Helling’s population as characterized by Kinnersley et al. (2) originated from a clone (JA122) harboring the same IS-element insertion in fimE acquired across our experimental populations. It is possible that impairing the function of fimE creates an adaptive constraint, as 4 of the 11 genes experiencing parallel mutation between our populations and the Helling populations (cytR, dcuS, fimH, and rpsG) are mutationally overrepresented as indicated by their high G-scores (SI Appendix, Table S4).
Potential Drivers of Subpopulation Structure.
The stable coexistence of multiple dominant haplotypes over thousands of generations may have a number of causes. However, our analyses suggest the evolution of subpopulation structure in population 125 was a two-step process: spatial differentiation, followed by metabolic differentiation. Early mutations affecting biofilm-related genes along with the absence of haplotype A from the biofilm at the liquid–air interface support this hypothesis. While both haplotypes within population 125 experience some sympatric interaction as evidenced by their cooccurrence within the planktonic niche, it is possible that the two haplotypes hardly compete at all within the experimental culture environment, with the majority of haplotype A existing within flocs in the bottom of the culture tube. As such, the coexistence of the two major haplotypes may not involve frequency-dependent selection (42, 43) or clonal reinforcement (32). If a frequency-dependent relationship is operating, then it is most likely occurring within haplotype A, where clone 125-1 appears quite different both genotypically and transcriptionally from the other haplotype A clones, 125-3 and 125-6. We observe that haplotype A clone 125-6 overproduces lysine, whereas haplotype A clone 125-1 overutilizes lysine; the overproduction of lysine has been previously shown to support cooperation in synthetic communities (34).
Earlier studies have suggested that inhabiting the liquid–air interface is advantageous, owing to the availability of oxygen within this niche (4). However, few explanations, if any, have been offered for the potential advantage of occupying the oxygen-poor bottom-dwelling niche. Is haplotype B simply the fitter haplotype and rewarded with the oxygen-rich niche, or does haplotype A have a distinct advantage in the oxygen-poor niche?
One potential benefit of the bottom-dwelling niche that is unavailable at the surface–air interface involves nutrients scavenged from dead cells. Transcriptome data revealed that the major regulatory difference between haplotypes A and B is the overexpression of nupG and the deo operon, both responsible for nucleotide salvage in haplotype A. Free nucleotides are not an abundant component of LB media (44), which is composed mainly of amino acids. However, the main byproduct of amino acid metabolism is ammonia, which causes a rapid increase in pH within the culture environment. While E. coli is well-equipped to handle acid stress, the common result of growth in glucose (44), E. coli cells do quite poorly in basic environments, with growth generally arrested at pH levels exceeding 9. In our experimental cultures, the pH increases to 8.7 within 8 h after transfer. The effect of this increase was observed in our initial exometabolomic assays, where results become complicated after 6 h, as cell lysis caused a substantial release of amino acids into the culture medium. This resulting cellular debris would introduce a source of nutrients primarily accessible to clones specializing in the bottom-dwelling niche.
Another example of a transcriptional difference affecting niche specialization is the differential expression of speG (spermidine acetyltransferase). Haplotype A clone 125-1 overexpresses speG while clones 125-2, -3, -4, -5, -7, and -8 (belonging to both haplotypes A and B) all have speG activity knocked down due to an IS-element insertion in ynfB, which is located upstream in the same operon as speG. Overexpression of speG may be beneficial in a basic environment as a means of increasing catabolism of free spermidine, as higher pH exacerbates polyamine stress (45). This stress is less of an issue for clones located in flocs (125-3 and -6; haplotype A) or in the surface biofilm (125-2, -4, -5, -7, and -8; haplotype B), as these complex structures insulate cells from environmental stressors (46, 47). The presence of these seven clones within complex structures (flocs and biofilms) is further supported by their increased utilization of threonine (Fig. 4). Recently, threonine has been shown to be an important nutrient in biofilms and other hypoxic, amino acid-rich environments, as threonine can be fermented into 1-propanol (48). This fermentation via the tdc operon, which is also overexpressed in these seven clones, preserves cellular redox balance and is expected to have a fitness advantage during anaerobic and biofilm growth.
Clinical Relevance
The long-term evolution experiment described here provides an excellent model for examining adaptation, specialization, and diversification within microbial populations experiencing environmental spatial heterogeneity. The usefulness of this model extends from questions of microbial speciation to those with clinical implications. For example, the products of both fimE and hns regulation are genes known to be important to uropathogenic E. coli for adherence to the epithelial wall, allowing colonization of the urinary tract (49–51). Further, as in LB medium, adaptation to a high environmental pH is advantageous for long-term colonization of the urinary tract, as nitrogen-based compounds (which metabolize to ammonia) are also the major nutrient source in urine (52).
Particularly relevant results to the diagnosis and treatment of infection are (i) the rapidity with which subpopulation structure develops, with many of the characteristic mutations appearing before our first sampling point, and (ii) the possibility that traditional plating methods may exclude some subpopulations of cells. These observations suggest that the diagnostic practice of characterizing a single infective isolate may be misleading in understanding and combating chronic urinary tract infections. With the power of whole-population sequencing, it is now possible to investigate clinical samples to determine if similar population structure arises during infection as in our experimental system, potentially changing the way we think about infection from an isolate view to a population view.
Materials and Methods
Strain Information and Culture of Populations.
The WT progenitor strain PFM2 is a prototrophic derivative of the E. coli K-12 reference strain MG1655, while the repair-deficient strain PFM5 is a derivative of PFM2 with the mutL gene replaced by an in-frame scar sequence (53). Populations were established by inoculating 10 mL of LB-Miller broth (BD Difco) with a single-isolated progenitor colony cultivated overnight at 37 °C on LB agar plates. Resulting cultures were maintained in 16- × 100-mm glass culture tubes containing 10 mL LB broth shaking at 175 rpm, alternating transfers after 24 h of growth at 37 °C and 48 h of growth at 25 °C (Fig. 1). Cultures were thoroughly vortexed immediately before transfer to ensure equal likelihood that planktonic and biofiliming individuals would be transferred to the next culture tube. Populations were also subjected to one of five different evolutionary treatments aimed at manipulating effective population size through different bottleneck sizes at each transfer. Bottlenecks were created by either varying the transfer volume (109 cells: 1 mL; 108: 100 µL; 107: 10 µL; 106: 1 µL) or by diluting 1 µL of culture in 100 µL of PBS and inoculated with 1 µL of the resulting diluted cell culture (104 cells). Although populations were incubated at both 25 °C and 37 °C, populations were only characterized at 37 °C.
Nucleic Acid Isolation from Populations.
DNA was isolated from the remaining fully vortexed populations following transfer at each 6-mo time point with the Wizard Genomic DNA Purification Kit (Promega) following the standard protocol. The rest of the remaining culture was frozen and stored in 10% glycerol at −80 °C as backup. Paired-end sequencing libraries were then constructed using the Nextera DNA Library Preparation Kit (Illumina) following an augmented protocol for optimization of reagent use (54) and submitted to the Hubbard Center for Genome Studies at the University of New Hampshire for sequencing on an Illumina HiSeq2500 2 × 150 rapid run.
Isolation of Clones.
Thoroughly vortexed samples from E. coli populations actively evolving for 2.5 y were streaked onto LB agar plates and cultured overnight at 37 °C. From each population plate, eight random isolated colonies (80 total) were selected and restreaked on LB agar plates. Each individual isolate was then collected for DNA extraction, frozen for long-term storage in 10% glycerol at −80 °C, or stored on LB agar at 4 °C for phenotypic assessment.
Nucleic Acid Isolation from Clones.
DNA was isolated from midlog-phase LB broth cultures using the Wizard Genomic DNA Purification Kit (Promega) and submitted to the Center for Genomics and Bioinformatics at Indiana University for library preparation and sequencing. Paired-end sequencing libraries were constructed using the Nextera DNA Library Preparation Kit (Illumina) following an augmented protocol for optimization of reagent use (54) and sequenced on an Illumina NextSeq500 300-cycle, midoutput run. RNA was isolated from midlog-phase LB broth cultures using the FastRNA Pro Blue Kit (MP Biomedicals) and purified with the RNeasy Kit (Qiagen) to remove DNases before submission to the Center for Genomics and Bioinformatics at Indiana University for library preparation and sequencing on an Illumina NextSeq500 75-cycle, high-output run. Sequencing data are available on the SRA database at NCBI, BioProject PRJNA448766. For more information on RNA library preparation see SI Appendix, Extended Materials and Methods.
Assembly and Analysis of Genomic Sequence.
To identify mutation variation within clonal isolates, paired-end sequencing reads were quality-controlled using the cutadapt software (55) and mapped to the E. coli K-12 MG1655 reference sequence (NC_000913.3) (56) with bwa-mem version 0.7.10 (57) (The entire workflow can be found at https://github.com/behrimg/MuriSam.) Further, we applied GATK version 3.4–0 (58) for base quality score recalibration, indel realignment, and duplicate removal and performed SNP and INDEL discovery across all eight clones per population simultaneously using standard hard filtering parameters according to GATK Best Practices (59, 60). Variants identified with GATK were then annotated with SnpEff version 4.1 (61). To detect structural variants in our clonal sequences, we used Trimmomatic version 0.32 (62) to crop sequencing reads to 90 bp to infer structural variation with GRASPER (63, 64). To identify mutational variation within populations across 6-mo intervals, paired-end sequencing reads were mapped to the E. coli K-12 MG1655 reference sequence and mutational variants identified with breseq version 0.27.1 (65, 66). For information on visualization of genomic data see SI Appendix, Extended Materials and Methods.
Assembly and Analysis of Transcriptomic Sequence.
To identify gene expression differences between individual clones, single-end RNA sequencing reads were mapped to the E. coli K-12 MG1655 reference sequence (NC_000913.3) using the Tuxedo protocol for RNA expression analysis (67). Following differential expression analysis, downstream statistical analysis and visualization were conducted using the cummRbund package for R.
Determination of Auxotrophy.
To assess if auxotrophy had evolved in individual clones during the course of long-term evolution, isolates were plated on solid M9 minimal media and cultured for 96 h at 37 °C, checking every 24 h for growth. To screen for the nutrient requirement for which the clones were auxotrophic, supplementation tests were performed with crossed pools of amino acids, nucleotides, and vitamins (68).
Determination of Relative Growth Phenotypes.
To conduct the growth assays, individual clones were grown overnight in culture tubes containing 10 mL of LB broth at 37 °C. We then inoculated 100 µL of fresh liquid LB-Miller with 1 µL of the overnight culture in octuplicate in 96-well culture plates. On every growth assay culture plate we included eight wells of inoculum-free LB media, followed by alternating rows of eight wells inoculated with evolved clones and eight wells inoculated with the MG1655 progenitor strain. Culture plates were loaded into a SynergyMx Microplate Reader (BioTek) and incubated at 37 °C with shaking for 24 h, recording OD readings at 600 nm, every 15 min. Maximum growth rate was calculated by fitting a modified Gompertz equation to OD readings (https://github.com/LennonLab/Growth_Curves).
Determination of Competitive Growth Phenotypes.
A selectable competition strain was created by culturing 108 cells from the MG1655 progenitor strain on 5 LB agar + 0.01% rifampicin plates at 37 °C for 24 h. Eight random colonies were then selected from the five plates and prepared for long-term storage. To determine which isolate acquired resistance to rifampicin while maintaining the fitness phenotype of the WT progenitor, the eight rifampicin-resistant isolates and the MG1655 progenitor were each grown overnight in culture tubes containing 10 mL of LB broth at 37 °C. A fresh culture tube of 10 mL LB broth was then inoculated with 100 µL of a single rifampicin-resistant isolate and 100 µL of the MG1655 progenitor. Immediately after inoculation 100 µL of well-vortexed culture was serial diluted in PBS and spread onto LB agar for initial colony counting. Following an overnight incubation at 37 °C, 100 µL of well-vortexed culture was serially diluted in PBS and spread onto LB agar plates and incubated at 37 °C overnight for final plate counting. To separate the rifampicin-resistant colonies from the MG1655 progenitor colonies, a replica plating technique was used. Initial and final countable LB agar plates were stamped onto velveteen felts and transferred to LB agar plates + 0.01% rifampicin. The LB–rifampicin plates were also incubated at 37 °C overnight for plate counting. The following equation was then used to determine fitness, where ω represents relative fitness, A represents the number of MG1655 CFUs after subtracting E, the number of rifampicin-resistant CFUs counted on the LB–rifampicin plates from the total number of CFUs on the LB agar:
We then selected the isolate with the ω value closest to 1 to represent the ancestral phenotype in further competitive growth assays. To determine the relative fitness of population 125, the coculture fitness assessment experiment was repeated with triplicate cultures of population 125 as well as clonal isolates from 125 competed against the rifampicin-resistant progenitor strain in both glass culture tubes as well as 10 mL of culture in 100-mL flasks.
Determination of Growth Behavior.
To confirm the potential for biofilm-forming behavior, an isolated colony of the evolved clones and the WT progenitor were each streaked on Congo red BHI agar and incubated overnight at 37 °C. Presence of bright-red colonies, opposed to pale-pink, confirm the ability to form biofilms (27). We then aimed to identify the localization of the different haplotypes of population 125 within the long-term culture environment. Using the evolved thiamine auxotrophy of haplotype B in population 125 we are able to separate and count colonies of haplotype A and haplotype B using a replica plating technique. Triplicate cultures of population 125 were grown overnight in 16- × 100-mm polyurethane culture tubes containing LB broth. We concluded that the substitution of polyurethane for glass did not disrupt the subpopulation structure to the persistence of the biofilm at the surface–air interface. By inserting a 20-gauge needle through the side of the tube, we aspirated 0.50 mL of culture from both the bottom quarter and the upper quarter of the culture tube. After aspiration, cells belonging to the biofilm were then collected from the side of the tube where the air–liquid interface once resided. Once collected, cells were cultured at 10−6 dilutions, 37 °C overnight on LB to obtain accurate colony counts. To identify colonies of auxotrophic clones aspirated from the different regions, colonies were replica-plated on M9 minimal media using velveteen felts and incubated at 37 °C for 72 h. To confirm the presence of oxygen-sensitive clones this experiment was repeated by culturing six replicates of population 125 at 10−6 dilutions on LB and incubating both aerobically and in anaerobic jars (Alcore International) containing an anaerobic gas pack (AnaeroGen; Thermo Scientific) at 37 °C overnight.
Finally, we used replica plating to investigate the stability of the subpopulation structure by reconstructing the population through coculture of a single clone from each haplotype. Cocultures were established by inoculating culture tubes containing 10 mL of LB with 100 µL of an overnight culture containing a clone from haplotype A and 100 µL of an overnight culture containing a clone from haplotype B in triplicate. Cocultures were then shaker-incubated at 37 °C for 24 h before fully vortexing and transferring 1 mL of coculture into a fresh culture tube containing 10 mL of LB broth. After 24 h, 72 h, and 168 h, fully vortexed samples were plated in triplicate on LB and incubated at 37 °C before colony counting and replica plating onto M9.
Determination of Amino Acid Uptake.
A single colony from the WT progenitor and clones 125-1, 125-2, 125-3, and 125-5 were used to inoculate culture tubes containing 10 mL of LB-Miller and incubated overnight at 37 °C. We transferred 100 µL from each culture tube into four 100-mL flasks containing fresh LB broth. Each flask thus represented a time point in which the spent media was to be surveyed (0, 2, 4, and 6 h). The sets of 2-, 4-, and 6-h treatments were incubated at 37 °C, shaking at 200 rpm, while the 0-h treatments were collected immediately. Spent media from all treatments was collected via filter centrifugation and immediately refrigerated before submission for GC-MS analysis (for more information about GC-MS sample preparation and analysis see SI Appendix, Extended Materials and Methods).
Supplementary Material
Acknowledgments
We thank D. A. Drummond, P. Foster, W.-C. Ho, J.-F. Gout, J. Lennon, H. Long, J. McKinlay, and the reviewers for their helpful suggestions. Library construction and sequencing was conducted by Center for Genomics and Bioinformatics at Indiana University. The High Performance Computing Cluster was maintained by the National Center for Genome Analysis Support at Indiana University. This work was supported by Army Research Office Grants ARO65308-LS-MU and W911NF-14-1-0411 and National Institutes of Health Grant F32GM123703.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequences for all clones and populations analyzed in this paper have been deposited at the NCBI SRA database as BioProject PRJNA448766.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708371115/-/DCSupplemental.
References
- 1.Friesen ML, Saxer G, Travisano M, Doebeli M. Experimental evidence for sympatric ecological diversification due to frequency-dependent competition in Escherichia coli. Evolution. 2004;58:245–260. [PubMed] [Google Scholar]
- 2.Kinnersley MA, Holben WE, Rosenzweig F. E unibus plurum: Genomic analysis of an experimentally evolved polymorphism in Escherichia coli. PLoS Genet. 2009;5:e1000713. doi: 10.1371/journal.pgen.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rozen DE, Philippe N, Arjan de Visser J, Lenski RE, Schneider D. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol Lett. 2009;12:34–44. doi: 10.1111/j.1461-0248.2008.01257.x. [DOI] [PubMed] [Google Scholar]
- 4.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 5.Frenkel EM, et al. Crowded growth leads to the spontaneous evolution of semistable coexistence in laboratory yeast populations. Proc Natl Acad Sci USA. 2015;112:11306–11311. doi: 10.1073/pnas.1506184112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kryazhimskiy S, Rice DP, Desai MM. Population subdivision and adaptation in asexual populations of Saccharomyces cerevisiae. Evolution. 2012;66:1931–1941. doi: 10.1111/j.1558-5646.2011.01569.x. [DOI] [PubMed] [Google Scholar]
- 7.Harcombe W. Novel cooperation experimentally evolved between species. Evolution. 2010;64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 8.Shou W, Ram S, Vilar JM. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA. 2007;104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol. 2009;74:119–129. doi: 10.1101/sqb.2009.74.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc Natl Acad Sci USA. 2013;110:E250–E259. doi: 10.1073/pnas.1207025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy SF, et al. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature. 2015;519:181–186. doi: 10.1038/nature14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helling RB, Vargas CN, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataram S, et al. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell. 2016;166:1585–1596.e22. doi: 10.1016/j.cell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerstein AC, Chun H-JE, Grant A, Otto SP. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxer G, Doebeli M, Travisano M. The repeatability of adaptive radiation during long-term experimental evolution of Escherichia coli in a multiple nutrient environment. PLoS One. 2010;5:e14184. doi: 10.1371/journal.pone.0014184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandberg TE, Lloyd CJ, Palsson BO, Feist AM. Laboratory evolution to alternating substrate environments yields distinct phenotypic and genetic adaptive strategies. Appl Environ Microbiol. 2017;83:e00410–e00417. doi: 10.1128/AEM.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puentes-Téllez PE, Hansen MA, Sørensen SJ, van Elsas JD. Adaptation and heterogeneity of Escherichia coli MC1000 growing in complex environments. Appl Environ Microbiol. 2013;79:1008–1017. doi: 10.1128/AEM.02920-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper TF, Lenski RE. Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evol Biol. 2010;10:11. doi: 10.1186/1471-2148-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davlieva M, et al. A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin. Nucleic Acids Res. 2015;43:4758–4773. doi: 10.1093/nar/gkv321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habets MG, Rozen DE, Hoekstra RF, de Visser JA. The effect of population structure on the adaptive radiation of microbial populations evolving in spatially structured environments. Ecol Lett. 2006;9:1041–1048. doi: 10.1111/j.1461-0248.2006.00955.x. [DOI] [PubMed] [Google Scholar]
- 22.Lind PA, Farr AD, Rainey PB. Evolutionary convergence in experimental Pseudomonas populations. ISME J. 2017;11:589–600. doi: 10.1038/ismej.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 24.Olsen PB, Klemm P. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol Lett. 1994;116:95–100. doi: 10.1111/j.1574-6968.1994.tb06681.x. [DOI] [PubMed] [Google Scholar]
- 25.Kvist M, Hancock V, Klemm P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol. 2008;74:7376–7382. doi: 10.1128/AEM.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Römling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bak G, et al. Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci Rep. 2015;5:15287. doi: 10.1038/srep15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uno M, Ito K, Nakamura Y. Functional specificity of amino acid at position 246 in the tRNA mimicry domain of bacterial release factor 2. Biochimie. 1996;78:935–943. doi: 10.1016/s0300-9084(97)86715-6. [DOI] [PubMed] [Google Scholar]
- 29.Lourenço M, et al. A mutational hotspot and strong selection contribute to the order of mutations selected for during Escherichia coli adaptation to the gut. PLoS Genet. 2016;12:e1006420. doi: 10.1371/journal.pgen.1006420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barroso-Batista J, et al. The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 2014;10:e1004182. doi: 10.1371/journal.pgen.1004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenaillon O, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinnersley M, et al. Ex uno plures: Clonal reinforcement drives evolution of a simple microbial community. PLoS Genet. 2014;10:e1004430. doi: 10.1371/journal.pgen.1004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crow JF, Kimura M. An Introduction to Population Genetics Theory. Harper & Row; New York: 1970. pp. 192–193. [Google Scholar]
- 34.Mee MT, Collins JJ, Church GM, Wang HH. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA. 2014;111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doebeli M, Knowlton N. The evolution of interspecific mutualisms. Proc Natl Acad Sci USA. 1998;95:8676–8680. doi: 10.1073/pnas.95.15.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasman H, Chakraborty T, Klemm P. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol. 1999;181:4834–4841. doi: 10.1128/jb.181.16.4834-4841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beloin C, Roux A, Ghigo J-M. Escherichia coli biofilms. Curr Top Microbiol Immunol. 2008;322:249–289. doi: 10.1007/978-3-540-75418-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon SH, et al. Comparative multi-omics systems analysis of Escherichia coli strains B and K-12. Genome Biol. 2012;13:R37. doi: 10.1186/gb-2012-13-5-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong S, Khodursky A, Dykhuizen DE, Dean AM. Evolutionary genomics of ecological specialization. Proc Natl Acad Sci USA. 2004;101:11719–11724. doi: 10.1073/pnas.0404397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: Genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang GI, Botstein D, Desai MM. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics. 2011;188:647–661. doi: 10.1534/genetics.111.128942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maddamsetti R, Lenski RE, Barrick JE. Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics. 2015;200:619–631. doi: 10.1534/genetics.115.176677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yohannes E, Thurber AE, Wilks JC, Tate DP, Slonczewski JL. Polyamine stress at high pH in Escherichia coli K-12. BMC Microbiol. 2005;5:59. doi: 10.1186/1471-2180-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charles CJ, et al. Floc formation reduces the pH stress experienced by microorganisms living in alkaline environments. Appl Environ Microbiol. 2017;83:e02985-16. doi: 10.1128/AEM.02985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davey ME, O’toole GA. Microbial biofilms: From ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Létoffé S, et al. Biofilm microenvironment induces a widespread adaptive amino-acid fermentation pathway conferring strong fitness advantage in Escherichia coli. PLoS Genet. 2017;13:e1006800. doi: 10.1371/journal.pgen.1006800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim JK, et al. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect Immun. 1998;66:3303–3310. doi: 10.1128/iai.66.7.3303-3310.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Struve C, Krogfelt KA. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology. 1999;145:2683–2690. doi: 10.1099/00221287-145-10-2683. [DOI] [PubMed] [Google Scholar]
- 51.Connell I, et al. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franz M, Hörl WH. Common errors in diagnosis and management of urinary tract infection. I: Pathophysiology and diagnostic techniques. Nephrol Dial Transplant. 1999;14:2746–2753. doi: 10.1093/ndt/14.11.2746. [DOI] [PubMed] [Google Scholar]
- 53.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci USA. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baym M, et al. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One. 2015;10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 56.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKenna A, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H, Popodi E, Foster PL, Tang H. Detection of structural variants involving repetitive regions in the reference genome. J Comput Biol. 2014;21:219–233. doi: 10.1089/cmb.2013.0129. [DOI] [PubMed] [Google Scholar]
- 64.Lee H, Doak TG, Popodi E, Foster PL, Tang H. Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli. Nucleic Acids Res. 2016;44:7109–7119. doi: 10.1093/nar/gkw647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrick JE, et al. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics. 2014;15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis RW, Roth JR, Botstein D. Advanced Bacterial Genetics: A Manual for Genetic Engineering. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1980. pp. 15–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.