Significance
High-density lipoprotein (HDL), popularly known as “good cholesterol,” is an assembly of lipids and proteins that is protective against cardiovascular diseases. HDL begins to develop when two copies of the protein apolipoprotein A-I (APOA1) mediate the removal of excess lipids from peripheral cells and form a nanodisc. How the proteins adjust their structures as the nanodisc increases in diameter, mediates the conversion of cholesterol to cholesteryl esters, and becomes spherical is crucial to understanding HDL maturation and relevant pathologies. This study combines multimicrosecond atomistic molecular dynamics simulations, molecular modeling, and cross-linking experiments to reveal APOA1 structure in two nascent HDLs. The structures provide a mechanistic understanding for HDL maturation and revise previous models of nascent HDL.
Keywords: APOA1, HDL, molecular dynamics simulation, Rosetta, chemical cross-linking
Abstract
Understanding the function of high-density lipoprotein (HDL) requires detailed knowledge of the structure of its primary protein, apolipoprotein A-I (APOA1). However, APOA1 flexibility and HDL heterogeneity have confounded decades of efforts to determine high-resolution structures and consistent models. Here, molecular dynamics simulations totaling 30 μs on two nascent HDLs, each with 2 APOA1 and either 160 phospholipids and 24 cholesterols or 200 phospholipids and 20 cholesterols, show that residues 1–21 of the N-terminal domains of APOA1 interact via strong salt bridges. Residues 26–43 of one APOA1 in the smaller particle form a hinge on the disc edge, which displaces the C-terminal domain of the other APOA1 to the phospholipid surface. The proposed structures are supported by chemical cross-linking, Rosetta modeling of the N-terminal domain, and analysis of the lipid-free ∆185APOA1 crystal structure. These structures provide a framework for understanding HDL maturation and revise all previous models of nascent HDL.
HDL is an assembly of lipids and proteins. The primary lipids are phospholipid and unesterified cholesterol (UC), and the primary protein is APOA1 (1), encoded by the APOA1 gene in humans. Studies have established the association of low plasma HDL cholesterol concentration with an elevated risk of cardiovascular diseases (2), and more recently, the importance of HDL particle size in the plasma (3, 4). The atheroprotective property of HDL is attributed to its role in a multistep process called reverse cholesterol transport (RCT) (5). RCT starts when APOA1 interacts with the ATP-binding cassette subfamily A member 1 (ABCA1). This yields nascent HDL, a discoidal bilayer stabilized by two molecules of APOA1. Nascent HDL then interacts with lecithin: cholesterol acyltransferase (LCAT) (6), which converts cholesterol to cholesteryl ester, resulting in spherical HDL (α-HDL). In the direct RCT, scavenger receptor class B member 1 (SR-BI) mediates the absorption of α-HDL to the liver (7). Atomistic-level characterization of RCT and other biological functions of HDL (8) depend on high-resolution structures of lipid-bound APOA1 and other proteins implicated in HDL metabolism, such as ABCA1, LCAT, and SR-BI. However, despite decades of efforts, HDL particles have not lent themselves to high-resolution structural techniques, such as NMR or X-ray crystallography, and low-resolution techniques have been unable to describe local APOA1 conformations that may be potential docking sites for other proteins implicated in HDL functions, such as LCAT and ABCA1.
Although nascent HDL is short-lived in plasma because it readily interacts with LCAT and transforms to spherical HDL, it can be reconstituted in vitro and exhibits many traits of native HDL. When palmitoyl-oleoyl-phosphatidylcholine (POPC) and APOA1 are reconstituted to form discoidal HDL (rHDL), four relatively monodisperse nanoparticles are generated. They contain ∼50:2, 100:2, 160:2, and 200:2 POPC:APOA1, and their diameters are close to 80 Å, 90 Å, 100 Å, and 110 Å, respectively, with some variation depending on the experiment (gel electrophoresis, size exclusion chromatography, and EM) (9–11). Herein, these particles are denoted rHDL-2-80, rHDL-2-90, rHDL-2-100, and rHDL-2-110, based on the number of APOA1 molecules and the approximate disc diameter. As the number of POPC further increases, the numbers of APOA1 per HDL particle can increase to three or four.
A fundamental question concerning the maturation process of HDL is how APOA1 adjusts its conformation to produce HDL size quantization. A comprehensive nascent HDL model should relate APOA1 conformations in different-size discs to disc quantization. This paper concerns rHDL-2-100 and rHDL-2-110, whose sizes are close to the predominant plasma nascent HDLs (1).
There are two crystal structures of truncated lipid-free APOA1 (12, 13), and one structure of truncated lipid-bound APOA1 determined from NMR, EPR, and transmission EM data (14). Based on theoretical considerations and experimental studies, models have been proposed for the structure of full-length APOA1 in nascent HDL. In the detailed structural model proposed by Segrest et al. (15) in 1999, known as the “double belt,” α-helical pairs of APOA1 form a smooth and essentially planar double ring around the lipid disc. Strong intermolecular salt bridges cause the two proteins to stack in an antiparallel fashion with pairwise overlapping of residues 121–142 (Helix 5), producing a registry called LL5/5 (15) (SI Appendix, Fig. S1); this registry leads to a slight overlap (or partial registry) of the N-terminal domains (NTDs), as shown in SI Appendix, Fig. S2A, for rHDL-2-110.
As is evident from SI Appendix, Fig. S2B, a top-down view of rHDL-2-110, a smooth double belt is too large to fit tightly around the nanodisc. Since the protein/lipid circumference mismatch increases as the disc size decreases, the original double belt model (herein denoted “planar double belt”) requires adjustments to allow APOA1 to conform to the disc size. Jones et al. (16) refined the planar double belt in rHDL-2-100 (160:24 POPC:UC) using molecular dynamics simulated annealing (MDSA), and showed a strong association of the NTDs and mobility of C-terminal domains (CTDs). Additionally, as commonly found from relatively short MD simulations, APOA1 adapted a “zig-zag” arrangement to optimize contact of the protein and the disc edge. Three other models are noteworthy: (i) “solar flare” [Wu et al. (9)], from hydrogen-deuterium exchange experiments for a disc of 200 POPC (rHDL-2-110): residues 159–180 of each APOA1 form a protruding solvent-exposed loop (akin to a solar flare) that was proposed to activate LCAT; (ii) “belt buckle” [Bhat et al. (17)], from chemical cross-linking/MS data on a disc of 145 POPC: both NTDs and CTDs fold back onto the belt similar to hairpins; and (iii) “looped belt” [Martin et al. (18)], from EPR and FRET for rHDL-2-100: there exists a central loop-like structure from residues 133–146. Although all these models share the LL5/5 registry, they differ significantly, especially in their description of terminal domains, and have different implications for the maturation process of nascent HDL.
Computational methods have complemented experiments to characterize structural features, such as the LL5/5 registry (15), but they have had their own limitations. For example, previous MD studies on HDL have been either short atomistic simulations (16, 19), which suffer from limited sampling, or long coarse-grained simulations (19, 20), which lack high resolution and accuracy and do not allow for secondary structure dynamics in the protein. Recently, a special-purpose supercomputer, Anton-2 (21), was developed, which enables multimicrosecond MD simulations of systems of HDL size. In this study, Anton-2 was employed to characterize the structure of nanodiscs consisting of 160:24:2 (rHDL-2-100) and 200:20:2 (rHDL-2-110) POPC:UC:APOA1 in atomistic detail. The smaller and larger particles were simulated for 20 μs and 10 μs, respectively, and showed reasonable convergence within these simulation times. These long time scales allowed for significant conformational changes of proteins to yield structures that are substantially different from previous models. The structures are consistent with results of chemical cross-linking experiments and Rosetta modeling carried out for this study, and lead to a model for APOA1 stabilization of HDL.
Results
For ease of describing the simulation results, the two (identical) proteins are denoted “1” and “2” and rendered in blue and red, respectively, in molecular graphics images.
Simulation 1 (20 µs): 160:24:2 POPC:UC:APOA1 (rHDL-2-100).
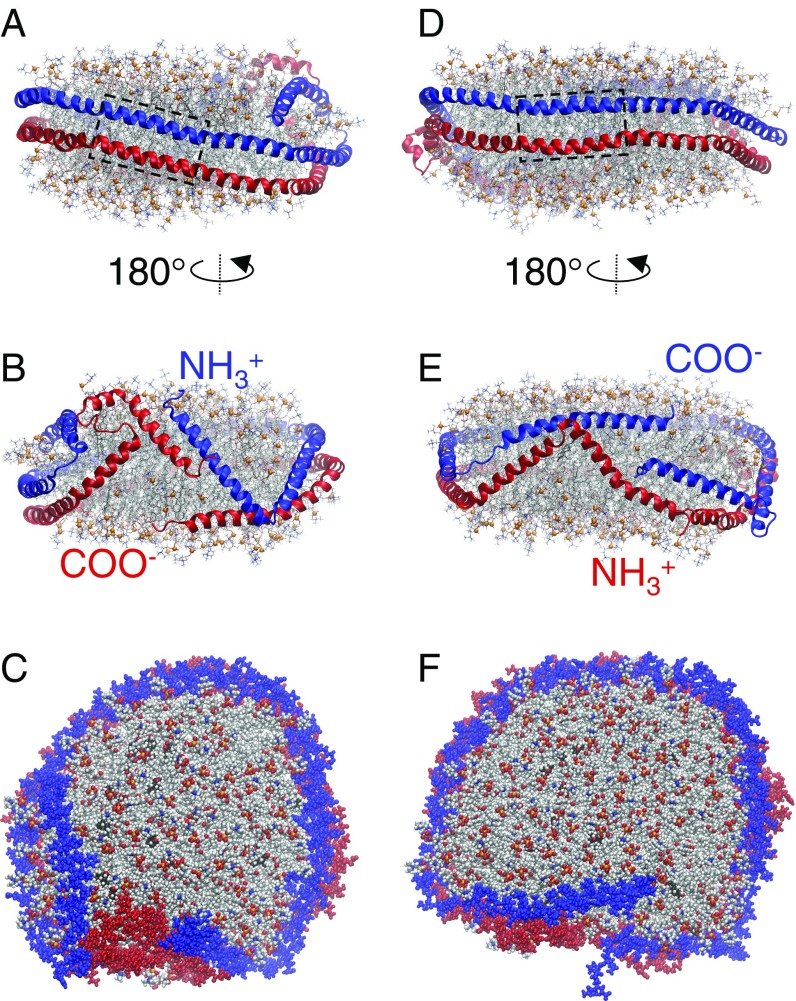
The simulation was initialized from the planar double belt similar to the conformation shown in SI Appendix, Fig. S2, and evolved over the course of 20 μs to the structure shown in Fig. 1 A–C. The disc is flat, and H5 and nearby helices maintained their registries throughout the simulation (Fig. 1A). The helices near the NTDs and CTDs at the other side of the disc showed substantial rearrangement (Fig. 1B). The top-down view (Fig. 1C) indicates that the disc is nearly circular and compacted compared with the initial condition.
Fig. 1.
Final structures of two simulated discoidal HDLs. (A–C) Simulation 1: 160:24:2 POPC:UC:APOA1, t = 20 μs. (D–F) Simulation 2: 200:20:2 POPC:UC:APOA1, t = 10 μs. Boxes highlight LL5/5 registry in A and D. Waters are omitted for clarity. Panels A, B, D, and E show side views (proteins, ribbon; phosphorus of POPC, ball; other lipid atoms, stick). Panels C and F show top-down views (all atoms in space-filling). Color codes: proteins, blue or red; POPC (phosphorus, orange; oxygen, red; nitrogen, blue; carbon, gray); UC carbon, black.
The disc started to bend within 3 ns of the simulation to decrease the protein–lipid circumference mismatch. A number of residues, including K195–A196 of Protein 1 and S36 of Protein 2, lost their α-helical structures and formed helical bends. The gradual increase of the bending angle of the NTD of Protein 2 induced bending of the CTD of Protein 1 at T197–T202 (1-μs snapshot in SI Appendix, Fig. S3A). The structure did not change significantly until ∼10 μs when the NTD of Protein 2 formed turns at two more regions, G26 and N43–K45 (time series of the secondary structure in SI Appendix, Fig. S4A). This created a small condensed domain that deflected the CTD of Protein 1, causing it to fold back onto itself between 10–16 μs (SI Appendix, Fig. S3B). The NTD of Protein 1 did not form a condensed region similar to that of Protein 2; it only bent at residues L38–G39. However, residues F33–L44 of this protein showed transient changes of α-helicity to coil, turn, and β structures (SI Appendix, Figs. S3C and S4A). These structural changes were not sufficient to deflect the CTD, and Protein 2 remained on the disc edge throughout the simulation. Residues 1–21 of the two proteins were loosely associated for the first 16 µs and formed two stable intermolecular salt bridges (R10–D13) for the remainder of the trajectory, tightening the proteins around the disc and making the assembly flatter.
The time evolution of the rmsd of the conformation of proteins with respect to the planar double belt as the reference structure shows the major folding events (SI Appendix, Fig. S5A). The rmsd spiked shortly after the start of the simulation, showing a transitory zig-zag adaptation of proteins to the disc size. The increase of rmsd of Protein 1 until 10 µs, followed by a plateau (black line in SI Appendix, Fig. S5A), is due to the migration of its CTD to the disc surface. The slight decrease after 16 µs is due to flattening of the disc caused by increased overlap of NTDs. The rmsd change in the last 4 μs of the simulation is due to the fluctuation of the disc shape; the NTD condensation, CTD bending, and the overlap between residues 1–21 do not change within this time interval. The rmsd with respect to the last configuration (20 μs) averaged over the two proteins decreases from ∼22 Å in the beginning of the simulation to 6 Å within 16–20 µs (SI Appendix, Fig. S5C). The disc diameter averaged over 16–20 μs is 97.0 ± 0.2 Å (SI Appendix, Fig. S6A) and is consistent with the experimentally determined diameter of 100 Å.
Simulation 2 (10 µs): 200:20:2 POPC:UC:APOA1 (rHDL-2-110).
Simulation 2 was initialized from the planar double belt (SI Appendix, Fig. S2), similar to Simulation 1 but with a larger lipid disc, and yielded the structure shown in Fig. 1 D–F. As SI Appendix, Fig. S4B shows, Proteins 1 and 2 bend at residues A196–T202 and L38–G39, respectively (the same residues as in Simulation 1), and the NTD of Protein 2 deflects the CTD of Protein 1 from the edge to the edge-surface boundary. However, in contrast to Simulation 1, the NTD bent at L38–G39 only and was unable to induce a significant displacement of the CTD. Thus, the CTD remained on the edge-surface boundary throughout the simulation. Residues 1–21 of the two proteins increased their overlap starting from 4 μs, and residues R10 and D13 formed two intermolecular salt bridges, similar to Simulation 1, for the remainder of the trajectory.
The rmsd with respect to the initial configuration (planar double belt) spiked at the start of simulation but to a lesser degree than in Simulation 1 (SI Appendix, Fig. S5B). It took 6 µs for the proteins to relax. The rmsd with respect to the last configuration (10 μs) averaged over the two proteins decreases from 13 Å in the beginning of the simulation to 7 Å within 6–10 μs (SI Appendix, Fig. S5D). The disc diameter averaged over 6–10 μs is 105.3 ± 0.1 Å (SI Appendix, Fig. S6B), in keeping with the experimental diameter of 110 Å.
Additional Modeling of the N Terminus.
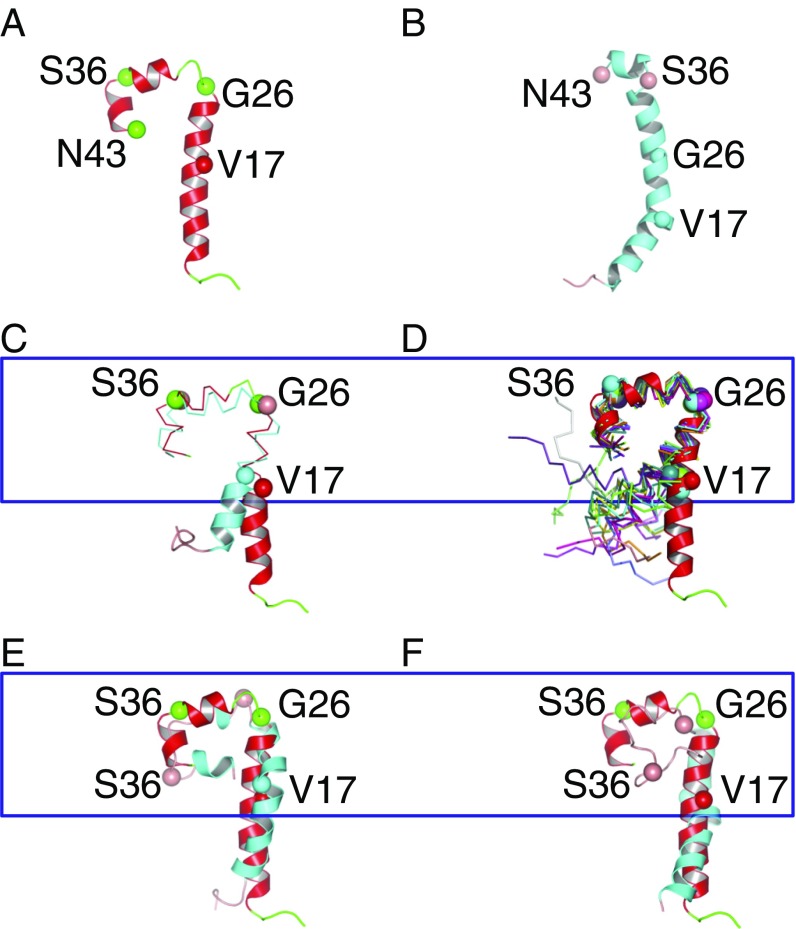
Fig. 2A shows the structure of residues 1–43 of Protein 2 at 20 μs of Simulation 1. This segment contains residues 26–43—henceforth referred to as the Hinge—that bent in Simulations 1 and 2 to modulate the adaptation of APOA1 around the two differently sized discs. Comparison with residues 3–43 from the C-terminally truncated crystal structure of lipid-free APOA1 (13) (Fig. 2B) shows that residue S36 creates a helical bend in both structures.
Fig. 2.
Structures of APOA1 NTDs obtained from different methods. (A) Twenty-microsecond MD simulation. (B) Lipid-free ∆185 APOA1 X-ray structure (13). (C) Representative structure of the most populated cluster of Rosetta models. (D) Thirteen structures extracted from Rosetta fold-and-dock simulation having the smallest rmsd from the structure in A. (E and F) Fifteen-nanosecond in vacuo MD simulation. Twenty-microsecond MD simulation structure (red; A) was used as a reference for alignments of C–F. Cαs of Val17, Gly26, Ser36, and Asn43 are shown in space-filling representation; and colors represent secondary structures (coil, green; coil, pink; helix, red; helix, cyan). Blue boxes highlight aligned residues 17–43.
Rosetta modeling was performed to further elucidate the folding tendencies of the NTD of APOA1. Specifically, the pair of APOA1 residues 1–43 of the planar double belt (SI Appendix, Fig. S2) was modeled by Rosetta fold-and-dock in two separate runs. The first run generated 10,000 models. The rmsd clustering analysis determined that the most populated cluster contained 22% of the models. Residues 17–43 of a representative structure of this cluster aligned with the same residues in the 20-μs MD structure, with an rmsd of 3.18 Å (Fig. 2C). The second run generated 500 models. Residues 17–43 were aligned with the same residues in the MD structure and 13 structures had an rmsd <3 Å (Fig. 2D). These results show that Rosetta, whose energy functions are different from those used in MD, also yields similar bending of NTDs.
Finally, a 15-ns in vacuo MD simulation of the pair of APOA1 residues 1–43 of the planar double belt was carried out to examine the robustness of the Hinge motif. Both segments formed condensed structures between residues G26 and N43 (Fig. 2 E and F), similar to the Hinge structure in Simulation 1, hence indicating the intrinsic tendency of the NTD to bend at the Hinge. However, the relative arrangement of secondary structural elements (SI Appendix, Fig. S7) differs from that obtained in Simulations 1 and 2, as expected from such a simplified model.
Chemical Cross-Linking and MS/MS Analysis of rHDL-2-100 Particles.
Close inter- and intramolecular contacts between the side chains of Lys and Glu/Asp were determined in rHDL-2-100 particles (160:8:2 POPC:UC:APOA1) using the zero-length cross-linker EDC (1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide hydrochloride) (22), followed by MS. EDC reacts with Lys and acidic residues (Asp and Glu), resulting in the formation of an amide bond (22). The maximum distance between the backbone Cα atoms is the sum of the length of two side chains plus the length of the amide bond (10.5 Å for K–D linkage and 12.1 Å for K–E linkage). 15N-labeled APOA1 [heavy (H)] and 14N-labeled APOA1 [light (L)] were used to distinguish intermolecular and intramolecular cross-links in rHDL discs. Under these conditions, rHDL particles containing LL, LH, and HH in the ratio of 1:2:1 should be generated (23).
Inter- and intramolecular distances between Cα atoms were calculated for Simulation 1 (averaged over 16–20 μs) to compare with cross-linking data. The cutoff radius was 15.1 Å (12.1 Å plus 3 Å motion-averaging factor). Fig. 3 presents the distance maps; SI Appendix, Tables S1 and S2 list simulated distances (average and range) for the 16 experimentally observed cross-links. The simulations resulted in different structures of the two proteins, hence the asymmetry of distance maps and different distances for each protein in the supporting tables. In contrast, cross-links from experiments are symmetric because a residue involved in a cross-link cannot be attributed to a particular APOA1; only the inter- and intramolecular cross-links can be distinguished.
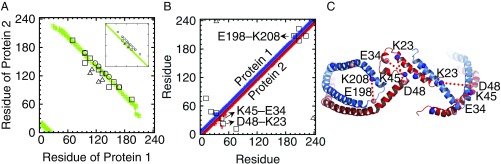
Fig. 3.
Cross-links of rHDL-2-100. Intermolecular (A) and intramolecular (B) distance maps of Cα from simulation (colored points, 15.1 Å cutoff) compared with experimental cross-linking data with three qualitative signal strengths (circle, strong; square, medium; triangle, weak). (C) Twenty-microsecond snapshot from Simulation 1 highlighting critical intramolecular cross-links; Cα atoms of residues involved in experimentally observed cross-links are shown as balls, and cross-links that are consistent with the experiment are shown by red dotted lines. Inset of A shows an intermolecular distance map of Cα from an ideal belt model with LL5/4 registry (see SI Appendix, Fig. S8, for LL4/4 and LL5/2 registries).
Ten intermolecular cross-links of light and heavy peptides were detected experimentally (Fig. 3A and SI Appendix, Table S1), all of which exhibited the anticipated ratio of LL, LH, and HH peptides. Seven out of the ten cross-links were consistent with the distance map obtained from Simulation 1. These are all between helices 2 and 8 and are consistent with the LL5/5 registry used in the simulation. The distances of the remaining three experimentally observed cross-links are all >25 Å in the simulation (SI Appendix, Table S1). However, they are all <15.1 Å when the ideal belt structure is generated with LL5/4 registry (Fig. 3A, Inset). These results indicate the coexistence of LL5/5 and LL5/4 registries in the experimental sample. As SI Appendix, Fig. S8 shows, no other registries are consistent with any of the cross-links. Three of the cross-links had strong MS/MS intensities, five were medium, and two were weak. The three strong signals corresponded to cross-links whose residues formed stable salt bridges in the simulation, and the two weak signals corresponded to cross-links consistent with LL5/4 registry. Note, however, that signal strength is not well correlated to the abundance of cross-linked peptides, so comparison of peaks yields only qualitative trends.
Six intramolecular cross-links were detected (Fig. 3B and SI Appendix, Table S2), three of which were consistent with the simulation distance map: K23–D48, K45–E34, and K208–E198. The average Cα–Cα distance between K23 and D48 was larger than the cutoff (15.1 Å). However, they became as close as 13.6 Å in both Proteins 1 and 2. There is a steric hindrance between K23 and D48 in Protein 2, but not in Protein 1. Therefore, the simulation implies that this cross-link may be due to the bending of one protein, but not necessarily both. Cα atoms of K45 and E34 became as close as 7.6 Å and 11.7 Å, with average values of 12.4 Å and 15.5 Å in Proteins 1 and 2, respectively. Therefore, while this cross-link is consistent with both structures of Proteins 1 and 2 in the simulation, it is more consistent with that of Protein 1. Likewise, the average and closest distances of Cα atoms of K208 and E198 indicate that this cross-link is more consistent with the bending of Protein 1 in the CTD. Note that an intramolecular distance map for a protein in the planar double belt exhibits a straight line of contacts along the diagonal. However, the special folds of NTDs and CTDs in Simulation 1 lead to off-diagonal contacts that match with the three cross-links. Of the remaining three, residues involved in K12–E76 and K238–E34 are separated by >40 Å in the simulation. K208 and E223 are closer (22.8 ± 0.1 Å and 23.0 ± 0.1 Å in Proteins 1 and 2, respectively) but are still ∼8 Å outside the cutoff. Fig. 3C shows the cross-links in the final structure of Simulation 1.
Discussion
It is notable that the simulations, although initialized from structures developed using reliable experimental constraints (high α-helical content and LL5/5 registry of the two APOA1 and particle stoichiometry), still required trajectories of 10–20 μs to yield converged results. Shorter conventional MD simulations are not adequate for probing major rearrangements of APOA1 helices.
The flexibility of APOA1 is an essential feature of its function, enabling it to stabilize both discoidal and spherical assemblies with varying lipid stoichiometries. This dynamic behavior makes the classification and orientation determination of structures challenging when using techniques such as cryo-electron tomography. Despite the low resolution, many recent EM images of HDL show density on the disc surface, indicating a surface-associated segment (see figure 2A of ref. 11 and figure 9B of ref. 24). The solar flare, belt buckle, and looped belt are in clear discrepancy with this important observation. In contrast, the current multimicrosecond simulation of rHDL-2-100 yielded a surface-associated segment. This segment is the CTD (H9, H10, and parts of H8) that is deflected by the Hinge on the disc edge. As opposed to terminal domains, H2 to H7 remained smooth and their registries did not change from that of the planar double belt. Of note, the simulations reported here were started from the simplest model, the planar double belt, to minimize the bias toward a certain configuration, and sampled a huge array of protein, lipid, and water conformations.
The simulation structures provide a mechanistic understanding of how nascent HDL matures from 100 Å to 110 Å particles via the interplay between NTDs and CTDs. In rHDL-2-100, the Hinge of one protein condenses and ejects the CTD of the other protein to the surface. In rHDL-2-110, the Hinge is only slightly bent and is unable to eject the CTD of the other. The adaptation of APOA1 to different-size discs is due to three essential molecular characteristics of APOA1: (i) strong intermolecular salt bridges that are crucial to the stability of the complex (15, 25), (ii) flexibility that enables the double belt to bend depending on the particle size, and (iii) the ability of the NTD to form a hinge. The third characteristic provides the driving force for the deflection of the CTDs to the disc surface.
A key feature of the present study is the experimental validation of critical distances in simulated rHDL-2-100 using chemical cross-linking and MS/MS analysis with a zero-order cross-linker. An advantage of this approach is the strong distance constraints that a zero-order cross-linker, which mimics salt bridges in proteins, places on the conformation of proteins. This leads to identification of Lys and Glu/Asp side chains that are in close proximity, thus allowing for rigorous testing of a structure. Three intramolecular cross-links are consistent with the simulation structure. K208–E198 supports the CTD loop, the hallmark of the structure. K45–E34 and K23–D48 indicate a condensed region, which can arise from either or both proteins. It is noteworthy that the intermolecular cross-links that did not fit the simulation structure were consistent with an ideal belt model with LL5/4 registry. The coexistence of particles with LL5/5 and LL5/4 registries was postulated based on theoretical considerations (15, 26) and confirmed by FRET (27). The current study provides another piece of evidence for that using the zero-order cross-linker. Particles with LL5/5 and LL5/2 registries were also hypothesized to coexist (25, 27–29), but neither the cross-links nor the simulations in this study indicated their existence. Heterogeneity of registries provides a plausible explanation for the three intramolecular cross-links that did not fit the distance maps. The K12–E76 cross-link might also be explained by the presence of lipid-free APOA1 resulting from rHDL preparation by the cholate dialysis method (24). The Cαs of K12 and E76 are separated by 14.2 Å (within the cutoff for the observed cross-link) in the crystal structure of the C-terminally truncated lipid-free APOA1 (13), and this might also hold in solution.
There are lines of evidence, in addition to intramolecular cross-links (Fig. 3B), that support the finding of a small, condensed NTD between residues G26 and N43 determined by the 20-μs MD simulation (the Hinge; Fig. 1B, Protein 2). The lipid-free Δ185APOA1 crystal structure (13) (Fig. 2B) contains a helical turn at S36; this residue in Protein 2 initiates the deflection of the CTD of Protein 1. The APOA1 model for lipid-poor nascent HDL developed by Sorci-Thomas et al. (30) also exhibits a helical turn in the Hinge. Moreover, Rosetta fold-and-dock modeling and in vacuo MD simulation of the NTD each resulted in structural condensation between residues G26 and N43 that aligned well with the structure of the Hinge (Fig. 2 C–F). These results strongly support the conclusion that the structure of the Hinge represents a stable low-energy conformation. Finally, Lagerstedt et al. (31) used EPR spectroscopy and hypothesized a hinge in the same region as residues 26–43. The mobility and solvent-accessibility of spin-labeled residues 14, 19, 34, 37, 41, and 58 in rHDLs with different sizes were analyzed. Residues 34, 37, and 41 showed spectral changes relative to rHDL particle size, whereas residues 14, 19, and 58 were not affected by particle-size changes. These findings are in excellent agreement with Simulations 1 and 2, in which residues 34, 37, and 41 are contained in the Hinge (which condenses or expands depending on the particle size); residues 14 and 19 belong to the first 21 residues that are similarly interlocked in both particles (SI Appendix, Fig. S9); and residue 58 is further away from the Hinge and thus unaffected.
In both rHDL-2-100 and rHDL-2-110, two salt bridges between R10 and D13 (SI Appendix, Fig. S9) contribute the most to the strong interaction between NTDs, inhibiting their migration to the disc surface. These salt bridges are absent in the planar double belt (initial condition), and their formation tightened the proteins around the nanodiscs. R10 and D13 are well conserved among 31 species of animals (32) and, as a hypothesis, their mutation should impede HDL reconstitution.
The current study also lends insight to three components of HDL heterogeneity that pose challenges for model derivation based on experimental data:
-
i)
The first stems from the coexistence of subpopulations of particles of the same size but different registries of APOA1. The large number of data points (one set of cross-links from each registry in this study) complicates analysis and can lead to incorrect models. The simulation structure obtained here for rHDL-2-100 is not consistent with all of the observed cross-links, nor should it be.
-
ii)
The second component stems from differences in tertiary structures of the two proteins in an HDL particle. The simulations here showed how the asymmetry of the two APOA1 modulates particle size. Experimental techniques, in contrast, have not distinguished the two proteins in HDL particles and obscure potentially crucial differences in their tertiary structures. For example, the two APOA1 in belt buckle, looped belt, and solar flare models are incorrectly assumed to be identical. The structural asymmetry, however, does not hold for all rHDL particles. Bibow et al. (14) deduced a symmetrical structure of a truncated APOA1 dimer, lacking residues 1–54 and Helix 5, based on a single peak per residue in NMR data.
-
iii)
The third component arises from different-sized particles in the experimental mixture. The simulation of rHDL-2-110 (whose cross-links were not experimentally determined here) provides insight into this size effect. SI Appendix, Table S3 lists the distances calculated for rHDL-2-110 for three critical cross-links observed experimentally in rHDL-2-100. SI Appendix, Tables S2 (the simulated distances for rHDL-2-100) and S3, reveal that only K45–E34 is common to both particles. D48–K23 and E198–K208 are present in rHDL-2-100 but not rHDL-2-110.
APOA1 flexibility and HDL heterogeneity inform the discussion of previous models. Simulation 2 did not support the solar flare model (9), which concerns rHDL-2-110. A simulation study that used the solar flare model as the initial condition showed the instability of the solvent-exposed residues (33). Hence, extending Simulation 2 beyond 10 μs is unlikely to yield such a model. Comparing the structures with the belt buckle model (17), the arrangements of both CTDs and NTDs are substantially different from the structures in the present study, although H5 and nearby helices are similarly arranged. This model was derived to satisfy all cross-linking data, which may correspond to a mixture of particles with different registries. Indeed, one of the critical cross-links in the belt buckle model (K12–K182) is also consistent with an LL5/2 registry (28). Comparing the structures with the looped belt model (18), which concerns rHDL-2-100 or smaller particles, the pairwise H5 helices are differently arranged and no bending tendency was observed between residues K133 and E146 to indicate a loop formation.
The structure of rHDL-2-100 in this study also differs from the MDSA-generated structure of Jones et al. (16), although there are similarities, such as N-terminal contact. However, the short MDSA resulted in a structure with a prominent zig-zag arrangement of the APOA1 around the disc edge. In the present study, it took 16 μs for rHDL-2-100 and 6 μs for rHDL-2-110 to gradually flatten, almost three orders of magnitude longer than the simulation time of Jones et al. (16). Valid determination of molecular docking sites of discoidal HDL to LCAT or ABCA1 will be greatly aided by starting with well-equilibrated structures, such as those in the present study.
In conclusion, multimicrosecond MD simulations yield structures of two nascent HDL particles of 160:24:2 and 200:20:2 POPC:UC:APOA1 stoichiometry that are supported by cross-linking data and differ in important details from previous models. Two distinct features emerged. First, NTDs strongly interact and tighten the proteins around the nanodisc. Second, APOA1 modulates disc size by the formation of a condensed domain in the NTD—the Hinge—which, in turn, controls the deflection of the CTD of the other protein to the disc surface. These features provide a framework for understanding the ability of APOA1 to regulate the size and shapes of HDL, and to form binding sites for proteins involved with RCT. Such binding sites likely include the loop in the CTD and/or the Hinge.
Materials and Methods
MD Simulations.
The planer double belt (15) was used as the initial configuration of proteins. CHARMM 36 (34, 35) was used for lipid and protein parameters. The systems were solvated with water and 0.15 mM NaCl and contained ∼325,000 atoms. Simulations were prepared using the CHARMM program (36) on an in-house supercomputer (LoBoS) and run on the Anton-2 supercomputer (21). More details are presented in SI Appendix.
Rosetta Modeling.
The symmetric fold-and-dock protocol (37) in Rosetta was used for de novo structure predictions of the pair of residues 1–43. Trimer and ninemer fragment libraries were generated using the amino acid sequences on the Robetta server (robetta.bakerlab.org). Two monomers were anchored via a virtual residue, and the moves were replicated to preserve the symmetry of monomers. A total of 10,000 models were generated by alternation of fragment insertion and rigid body perturbation, followed by all-atom refinement. The models were subjected to cluster analysis (details in SI Appendix). Subsequently, 500 structures from Rosetta fold-and-dock were aligned with residues 1–43 and the rmsd for Cα of residues 17–43 was calculated. Thirteen models with an rmsd <3 Å were extracted.
Cross-Linking Reaction and Proteolytic Digestion.
rHDL particles were made with light (14N) and heavy labeled (15N) APOA1 in a 1:1 ratio (10). The ratio between lipids and APOA1 was POPC:UC:APOA1 160:8:2. Samples were then purified by high-resolution size-exclusion chromatography (Supradex 200) and confirmed by calibrated ion mobility analysis (38). rHDL particles were exchanged into PBS (pH 6.5) before EDC cross-linking. Reactions were carried out at a total protein concentration of 0.56 mg/mL and an EDC concentration of 20 mM for 12 h at 4 °C. The reaction mixture was again separated by high-resolution size-exclusion chromatography (Supradex 200, 350 μL flow/min) to remove any inter–cross-linked particles. Collected rHDLs were then incubated overnight at 37 °C with sequencing grade modified trypsin (Promega) at a ratio of 20:1 (wt:wt) protein:trypsin in 100 mm NH4HCO3, pH 8. Digestion was halted by acidification (pH 2–3) with TFA.
Capillary LC-ESI-MS/MS and Data Analysis.
Capillary LC-ESI-MS/MS was performed using an IntegraFrit capillary trapping column packed with 1.5 cm of C18 (150 μm × 11 cm, New Objective; Magic C18, 5 μm, 200 Å; Michrom BioResources), a capillary analytical column packed with 15 cm of C18 (75 μm × 15 cm, Magic C18, 5 μm, 100 Å; Michrom BioResources), an LTQ Orbitrap XL mass spectrometer (Thermo Electron), and a nanoACQUITY UPLC system (Waters). In each experiment, 1 μg of tryptic digest was injected onto a trapping column, and an 80-min gradient between 2% and 40% mobile-phase B (0.1% TFA in acetonitrile) was implemented. Charge-state rejection was enabled for 1+ and 2+ charge states. MS/MS data contained in mzXML files were subsequently searched against the database for cross-links using xQuest (version 2.1.1) (39, 40). For our searches, xQuest was used with default settings, except that mass shifts for cross-linking products were manually set to –18.01056 for the “xlink mass-shift.” The modified residues were set to Lys, Asp, and Glu. Cross-link matches were required to have an FDR <0.05. All matches were manually inspected to ensure that both peptides in the cross-linked product were sequenced and that the majority of abundant fragment ions could be assigned in the MS/MS spectra.
Supplementary Material
Acknowledgments
We thank Jens Meiler and Jonathan H. Sheehan for helpful discussions on the use and interpretation of Rosetta results, Martin K. Jones for advice on creating the contact maps, and Sean Davidson for helpful discussions. This research was supported in part by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute (NHLBI; M.P. and R.W.P.), and by the NHLBI Grant P01HL128203 (J.P.S., H.D.S., Y.H., and J.W.H.), and utilized the high-performance computational capabilities at NIH, Bethesda, MD (LoBoS cluster), and the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. Anton computer time was provided by the Pittsburgh Supercomputing Center (PSC) through Grant R01GM116961 from NIH and PSC. The Anton machine at PSC was generously made available by D. E. Shaw Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Trajectories and final configurations of Simulations 1 and 2 are available at https://zenodo.org/record/897027#.Wt4oSX__rcs.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721181115/-/DCSupplemental.
References
- 1.Duong PT, et al. Characterization of nascent HDL particles and microparticles formed by ABCA1-mediated efflux of cellular lipids to apoA-I. J Lipid Res. 2006;47:832–843. doi: 10.1194/jlr.M500531-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Assmann G, Schulte H, von Eckardstein A, Huang Y. High-density lipoprotein cholesterol as a predictor of coronary heart disease risk: The PROCAM experience and pathophysiological implications for reverse cholesterol transport. Atherosclerosis. 1996;124:S11–S20. doi: 10.1016/0021-9150(96)05852-2. [DOI] [PubMed] [Google Scholar]
- 3.Mackey RH, et al. High-density lipoprotein cholesterol and particle concentrations, carotid atherosclerosis, and coronary events: MESA (Multi-Ethnic Study of Atherosclerosis) J Am Coll Cardiol. 2012;60:508–516. doi: 10.1016/j.jacc.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson WS. HDL-C vs HDL-P: How changing one letter could make a difference in understanding the role of high-density lipoprotein in disease. Clin Chem. 2014;60:e1–e3. doi: 10.1373/clinchem.2014.232769. [DOI] [PubMed] [Google Scholar]
- 5.Remaley AT, Norata GD, Catapano AL. Novel concepts in HDL pharmacology. Cardiovasc Res. 2014;103:423–428. doi: 10.1093/cvr/cvu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu X, et al. A systematic investigation of structure/function requirements for the apolipoprotein A-I/lecithin cholesterol acyltransferase interaction loop of high-density lipoprotein. J Biol Chem. 2016;291:6386–6395. doi: 10.1074/jbc.M115.696088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu S, et al. Apolipoproteins of HDL can directly mediate binding to the scavenger receptor SR-BI, an HDL receptor that mediates selective lipid uptake. J Lipid Res. 1997;38:1289–1298. [PubMed] [Google Scholar]
- 8.Gordon SM, Hofmann S, Askew DS, Davidson WS. High density lipoprotein: It’s not just about lipid transport anymore. Trends Endocrinol Metab. 2011;22:9–15. doi: 10.1016/j.tem.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, et al. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. [DOI] [PubMed] [Google Scholar]
- 10.Cavigiolio G, et al. The interplay between size, morphology, stability, and functionality of high-density lipoprotein subclasses. Biochemistry. 2008;47:4770–4779. doi: 10.1021/bi7023354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, et al. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. J Lipid Res. 2011;52:175–184. doi: 10.1194/jlr.D010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci USA. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mei X, Atkinson D. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J Biol Chem. 2011;286:38570–38582. doi: 10.1074/jbc.M111.260422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibow S, et al. Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nat Struct Mol Biol. 2017;24:187–193. doi: 10.1038/nsmb.3345. [DOI] [PubMed] [Google Scholar]
- 15.Segrest JP, et al. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 16.Jones MK, Gu F, Catte A, Li L, Segrest JP. “Sticky” and “promiscuous”, the yin and yang of apolipoprotein A-I termini in discoidal high-density lipoproteins: A combined computational-experimental approach. Biochemistry. 2011;50:2249–2263. doi: 10.1021/bi101301g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin DDO, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J Biol Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 19.Shih AY, Sligar SG, Schulten K. Maturation of high-density lipoproteins. J R Soc Interface. 2009;6:863–871. doi: 10.1098/rsif.2009.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones MK, et al. Assessment of the validity of the double superhelix model for reconstituted high density lipoproteins: A combined computational-experimental approach. J Biol Chem. 2010;285:41161–41171. doi: 10.1074/jbc.M110.187799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw DE, et al. Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. IEEE Press; Piscataway, NJ: 2014. Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer; pp. 41–53. [Google Scholar]
- 22.Hermanson GT. Bioconjugate Techniques. Academic; San Diego: 1996. [Google Scholar]
- 23.Lima DB, et al. Characterization of homodimer interfaces with cross-linking mass spectrometry and isotopically labeled proteins. Nat Protoc. 2018;13:431–458. doi: 10.1038/nprot.2017.113. [DOI] [PubMed] [Google Scholar]
- 24.Chen B, et al. Apolipoprotein AI tertiary structures determine stability and phospholipid-binding activity of discoidal high-density lipoprotein particles of different sizes. Protein Sci. 2009;18:921–935. doi: 10.1002/pro.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silva RAGD, Hilliard GM, Li L, Segrest JP, Davidson WS. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Li S, Jones MK, Segrest JP. Rotational and hinge dynamics of discoidal high density lipoproteins probed by interchain disulfide bond formation. Biochim Biophys Acta. 2012;1821:481–489. doi: 10.1016/j.bbalip.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li HH, et al. ApoA-I structure on discs and spheres: Variable helix registry and conformational states. J Biol Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 28.Davidson WS, Thompson TB. The structure of apolipoprotein A-I in high density lipoproteins. J Biol Chem. 2007;282:22249–22253. doi: 10.1074/jbc.R700014200. [DOI] [PubMed] [Google Scholar]
- 29.Cuellar LÁ, Prieto ED, Cabaleiro LV, Garda HA. Apolipoprotein A-I configuration and cell cholesterol efflux activity of discoidal lipoproteins depend on the reconstitution process. Biochim Biophys Acta. 2014;1841:180–189. doi: 10.1016/j.bbalip.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Sorci-Thomas MG, et al. Nascent high density lipoproteins formed by ABCA1 resemble lipid rafts and are structurally organized by three apoA-I monomers. J Lipid Res. 2012;53:1890–1909. doi: 10.1194/jlr.M026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagerstedt JO, et al. Structure of apolipoprotein A-I N terminus on nascent high density lipoproteins. J Biol Chem. 2011;286:2966–2975. doi: 10.1074/jbc.M110.163097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bashtovyy D, Jones MK, Anantharamaiah GM, Segrest JP. Sequence conservation of apolipoprotein A-I affords novel insights into HDL structure-function. J Lipid Res. 2011;52:435–450. doi: 10.1194/jlr.R012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih AY, Sligar SG, Schulten K. Molecular models need to be tested: The case of a solar flares discoidal HDL model. Biophys J. 2008;94:L87–L89. doi: 10.1529/biophysj.108.131581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das R, et al. Simultaneous prediction of protein folding and docking at high resolution. Proc Natl Acad Sci USA. 2009;106:18978–18983. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouillette CG, et al. Structural studies of apolipoprotein A-I/phosphatidylcholine recombinants by high-field proton NMR, nondenaturing gradient gel electrophoresis, and electron microscopy. Biochemistry. 1984;23:359–367. doi: 10.1021/bi00297a027. [DOI] [PubMed] [Google Scholar]
- 39.Rinner O, et al. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walzthoeni T, et al. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat Methods. 2012;9:901–903. doi: 10.1038/nmeth.2103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.