Significance
Fixed nitrogen is essential for plant growth. Some plants, such as legumes, can host nitrogen-fixing bacteria within cells in root organs called nodules. Nodules are considered to have evolved in parallel in different lineages, but the genetic changes underlying this evolution remain unknown. Based on gene expression in the nitrogen-fixing nonlegume Parasponia andersonii and the legume Medicago truncatula, we find that nodules in these different lineages may share a single origin. Comparison of the genomes of Parasponia with those of related nonnodulating plants reveals evidence of parallel loss of genes that, in legumes, are essential for nodulation. Taken together, this raises the possibility that nodulation originated only once and was subsequently lost in many descendant lineages.
Keywords: symbiosis, biological nitrogen fixation, evolution, comparative genomics, copy number variation
Abstract
Nodules harboring nitrogen-fixing rhizobia are a well-known trait of legumes, but nodules also occur in other plant lineages, with rhizobia or the actinomycete Frankia as microsymbiont. It is generally assumed that nodulation evolved independently multiple times. However, molecular-genetic support for this hypothesis is lacking, as the genetic changes underlying nodule evolution remain elusive. We conducted genetic and comparative genomics studies by using Parasponia species (Cannabaceae), the only nonlegumes that can establish nitrogen-fixing nodules with rhizobium. Intergeneric crosses between Parasponia andersonii and its nonnodulating relative Trema tomentosa demonstrated that nodule organogenesis, but not intracellular infection, is a dominant genetic trait. Comparative transcriptomics of P. andersonii and the legume Medicago truncatula revealed utilization of at least 290 orthologous symbiosis genes in nodules. Among these are key genes that, in legumes, are essential for nodulation, including NODULE INCEPTION (NIN) and RHIZOBIUM-DIRECTED POLAR GROWTH (RPG). Comparative analysis of genomes from three Parasponia species and related nonnodulating plant species show evidence of parallel loss in nonnodulating species of putative orthologs of NIN, RPG, and NOD FACTOR PERCEPTION. Parallel loss of these symbiosis genes indicates that these nonnodulating lineages lost the potential to nodulate. Taken together, our results challenge the view that nodulation evolved in parallel and raises the possibility that nodulation originated ∼100 Mya in a common ancestor of all nodulating plant species, but was subsequently lost in many descendant lineages. This will have profound implications for translational approaches aimed at engineering nitrogen-fixing nodules in crop plants.
Nitrogen sources such as nitrate or ammonia are key nutrients for plant growth, but their availability is frequently limited. Some plant species in the related orders Fabales, Fagales, Rosales, and Cucurbitales—collectively known as the nitrogen-fixing clade—can overcome this limitation by establishing a nitrogen-fixing endosymbiosis with Frankia or rhizobium bacteria (1). These symbioses require specialized root organs, known as nodules, that provide optimal physiological conditions for nitrogen fixation (2). For example, nodules of legumes (Fabaceae, order Fabales) contain a high concentration of hemoglobin that is essential to control oxygen homeostasis and protect the rhizobial nitrogenase enzyme complex from oxidation (2, 3). Legumes, such as soybean (Glycine max), common bean (Phaseolus vulgaris), and peanut (Arachis hypogaea), represent the only crops that possess nitrogen-fixing nodules, and engineering this trait in other crop plants is a long-term vision in sustainable agriculture (4, 5).
Nodulating plants represent ∼10 related clades that diverged >100 Mya, supporting a shared evolutionary origin of the underlying capacity for this trait (1). Nevertheless, these nodulating clades are interspersed with many nonnodulating lineages. This has led to two hypotheses explaining the evolution of nodulation (1). The first is that nodulation has a single origin in the root of the nitrogen-fixation clade, followed by multiple independent losses. The second is that nodulation originated independently multiple times, preceded by a single hypothetical predisposition event in a common ancestor of the nitrogen-fixing fixation clade. The latter of these hypotheses is more widely accepted (6–12).
Genetic dissection of rhizobium symbiosis in two legume models—Medicago truncatula (medicago) and Lotus japonicus (lotus)—has uncovered symbiosis genes that are essential for nodule organogenesis, bacterial infection, and nitrogen fixation (Dataset S1). These include genes encoding LysM-type receptors that perceive rhizobial lipochitooligosaccharides (LCOs; also known as Nod factors) and transcriptionally activate the NODULE INCEPTION (NIN) transcription factor (13–18). Expression of NIN is essential and sufficient to set in motion nodule organogenesis (17, 19–21). Some symbiosis genes have been coopted from the more ancient and widespread arbuscular mycorrhizal symbiosis (22, 23). However, causal genetic differences between nodulating and nonnodulating species have not been identified (24).
To obtain insight into the molecular-genetic changes underlying evolution of nitrogen-fixing root nodules, we conducted comparative studies by using Parasponia (Cannabaceae, order Rosales). The genus Parasponia is the only lineage outside the legume family establishing a nodule symbiosis with rhizobium (25–28). Similarly as shown for legumes, nodule formation in Parasponia is initiated by rhizobium-secreted LCOs (29–31). This suggests that Parasponia and legumes use a similar set of genes to control nodulation, but the extent of common gene use between distantly related nodulating species remains unknown. The genus Parasponia represents a clade of five species that is phylogenetically embedded in the closely related Trema genus (32). Like Parasponia and most other land plants, Trema species can establish an arbuscular mycorrhizal symbiosis (SI Appendix, Fig. S1). However, they are nonresponsive to rhizobium LCOs and do not form nodules (28, 31). Taken together, Parasponia is an excellent system for comparative studies with legumes and nonnodulating Trema species to provide insights into the molecular-genetic changes underlying evolution of nitrogen-fixing root nodules.
Results
Nodule Organogenesis Is a Genetically Dominant Trait.
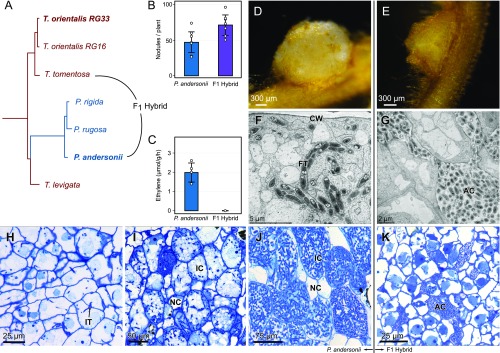
First, we took a genetics approach to understanding the rhizobium symbiosis trait of Parasponia by making intergeneric crosses (SI Appendix, Table S1). Viable F1 hybrid plants were obtained only from the cross Parasponia andersonii (2n = 20) × Trema tomentosa (2n = 4x = 40; Fig. 1A and SI Appendix, Fig. S2). These triploid hybrids (2n = 3x = 30) were infertile, but could be propagated clonally. We noted that F1 hybrid plants formed root nodules when grown in potting soil, similar to earlier observations for P. andersonii (33). To further investigate the nodulation phenotype of these hybrid plants, clonally propagated plants were inoculated with two different strains, Bradyrhizobium elkanii strain WUR3 (33) or Mesorhizobium plurifarium strain BOR2. The latter strain was isolated from the rhizosphere of Trema orientalis in Malaysian Borneo and showed to be an effective nodulator of P. andersonii (SI Appendix, Fig. S3). Both strains induced nodules on F1 hybrid plants (Fig. 1 B, D, and E and SI Appendix, Fig. S4) but, as expected, not on T. tomentosa, nor on any other Trema species investigated. By using an acetylene reduction assay, we noted that, in contrast to P. andersonii nodules, in F1 hybrid nodules of plant H9 infected with M. plurifarium BOR2 there is no nitrogenase activity (Fig. 1C). To further examine this discrepancy, we studied the cytoarchitecture of these nodules. In P. andersonii nodules, apoplastic M. plurifarium BOR2 colonies infect cells to form so-called fixation threads (Fig. 1 F and H–J), whereas, in F1 hybrid nodules, these colonies remain apoplastic and fail to establish intracellular infections (Fig. 1 G and K). To exclude the possibility that the lack of intracellular infection is caused by heterozygosity of P. andersonii whereby only a nonfunctional allele was transmitted to the F1 hybrid genotype, or by the particular rhizobium strain used for this experiment, we examined five independent F1 hybrid plants inoculated with M. plurifarium BOR2 or B. elkanii WUR3. This revealed a lack of intracellular infection structures in nodules of all F1 hybrid plants tested, irrespective which of the two rhizobium strains was used (Fig. 1 G and K and SI Appendix, Fig. S4), confirming that heterozygosity of P. andersonii does not play a role in the F1 hybrid infection phenotype. These results suggest, at least partly, independent genetic control of nodule organogenesis and rhizobium infection. Because F1 hybrids are nodulated with similar efficiency as P. andersonii (Fig. 1B), we conclude that the network controlling nodule organogenesis is genetically dominant.
Fig. 1.
Nodulation phenotype of P. andersonii and interspecific P. andersonii × T. tomentosa F1 hybrid plants. (A) Phylogenetic reconstruction based on whole chloroplast of Parasponia and Trema. The Parasponia lineage (blue) is embedded in the Trema genus (red). Species selected for interspecific crosses are indicated and species used for reference genome assembly are in bold. All nodes had a posterior probability of 1. (B) Mean number of nodules on roots of P. andersonii and F1 hybrid plants (n = 7). (C) Mean nitrogenase activity in acetylene reductase assay of P. andersonii and F1 hybrid nodules (n = 4). Bar-plot error bars indicate SDs; dots represent individual measurements. (D) P. andersonii nodule. (E) F1 hybrid nodule. (F and G) Ultrastructure of nodule tissue of P. andersoni (F) and F1 hybrid (G). Note the intracellular fixation thread (FT) in the cell of P. andersonii in comparison with the extracellular, apoplastic colonies of rhizobia (AC) in the F1 hybrid nodule. (H–J) Light-microscopy images of P. andersonii nodules in three subsequent developmental stages. (H) Stage 1: initial infection threads (IT) enter the host cells. (I) Stage 2: progression of rhizobium infection in nodule host cell. (J) Stage 3: nodule cells completely filled with fixation threads. Note difference in size between the infected (IC) and noninfected cells (NC). (K) Light-microscopy image of F1 hybrid nodule cells. Note rhizobium colonies in apoplast, surrounding the host cells (AC). Nodules have been analyzed 6 wk post inoculation with M. plurifarium BOR2. CW, cell wall.
Parasponia and Trema Genomes Are Highly Similar.
Based on preliminary genome size estimates made by using FACS measurements, three Parasponia and five Trema species were selected for comparative genome analysis (SI Appendix, Table S2). K-mer analysis of medium-coverage genome sequence data (∼30×) revealed that all genomes had low levels of heterozygosity, except those of Trema levigata and T. orientalis accession RG16 (SI Appendix, Fig. S5). Based on these k-mer data, we also generated more accurate estimates of genome sizes. Additionally, we used these data to assemble chloroplast genomes, based on which we obtained additional phylogenetic evidence that T. levigata is sister to Parasponia (Fig. 1A and SI Appendix, Figs. S6–S8). Graph-based clustering of repetitive elements in the genomes (calibrated with the genome size estimates based on k-mers) revealed that all selected species contain approximately 300 Mb of nonrepetitive sequence and a variable repeat content that correlates with the estimated genome size that ranges from 375 to 625 Mb (SI Appendix, Fig. S9 and Table S3). Notably, we found a Parasponia-specific expansion of ogre/tat LTR retrotransposons comprising 65–85 Mb (SI Appendix, Fig. S9B). We then generated annotated reference genomes by using high-coverage (∼125×) sequencing of P. andersonii accession WU1 (30) and T. orientalis accession RG33 (SI Appendix, Tables S4 and S5). These species were selected based on their low heterozygosity levels in combination with relatively small genomes. T. tomentosa was not used for a high-quality genome assembly because it is an allotetraploid (SI Appendix, Fig. S5 and Tables S2 and S3).
We generated orthogroups for P. andersonii and T. orientalis genes and six other Eurosid species, including arabidopsis (Arabidopsis thaliana) and the legumes medicago and soybean. From both P. andersonii and T. orientalis, ∼35,000 genes could be clustered into >20,000 orthogroups (SI Appendix, Table S6 and Dataset S2; note that there can be multiple orthologous gene pairs per orthogroup). Within these orthogroups, we identified 25,605 P. andersonii–T. orientalis orthologous gene pairs based on phylogenetic analysis as well as whole-genome alignments (SI Appendix, Table S6). These orthologous gene pairs had a median percentage nucleotide identity of 97% for coding regions (SI Appendix, Figs. S10 and S11). This further supports the recent divergence of the two species and facilitates their genomic comparison.
Common Utilization of Symbiosis Genes in Parasponia and Medicago.
To assess commonalities in the utilization of symbiosis genes in Parasponia species and legumes, we employed two strategies. First, we performed phylogenetic analyses of close homologs of genes that were characterized to function in legume–rhizobium symbiosis. This revealed that P. andersonii contains putative orthologs of the vast majority of these legume symbiosis genes (96 of 126; Datasets S1 and S3). Second, we compared the sets of genes with enhanced expression in nodules of P. andersonii and medicago. RNA sequencing of P. andersonii nodules revealed 1,719 genes that are functionally annotated and have a significantly enhanced expression level (fold change >2, P < 0.05, DESeq2 Wald test) in any of three nodule developmental stages compared with uninoculated roots (SI Appendix, Fig. S12 and Dataset S4). For medicago, we generated a comparable data set of 2,753 nodule-enhanced genes based on published RNA sequencing data (34). We then determined the overlap of these two gene sets based on orthogroup membership and found that 382 orthogroups comprise both P. andersonii and medicago nodule-enhanced genes. This number is significantly greater than is to be expected by chance (permutation test, P < 0.00001; SI Appendix, Fig. S13 and Dataset S5). Based on phylogenetic analysis of these orthogroups, we found that in 290 cases putative orthologs have been utilized in P. andersonii and medicago root nodules (Datasets S5 and S6). Among these 290 commonly utilized genes are 26 putative orthologs of legume symbiosis genes, e.g., the LCO-responsive transcription factor NIN and its downstream target NUCLEAR TRANSCRIPTION FACTOR-YA1 (NFYA1) that are essential for nodule organogenesis (19, 20, 35, 36) and RHIZOBIUM DIRECTED POLAR GROWTH (RPG) involved in intracellular infection (37). Of these 26, five are known to function also in arbuscular mycorrhizal symbiosis (namely VAPYRIN, SYMBIOTIC REMORIN, the transcription factors CYCLOPS and SAT1, and a cysteine proteinase gene) (38–45). To further assess whether commonly utilized genes may be coopted from the ancient and widespread arbuscular mycorrhizal symbiosis, we determined which fraction is also induced upon mycorrhization in medicago based on published RNA sequencing data (46). This revealed that only 8% of the commonly utilized genes have such induction in both symbioses (Dataset S5).
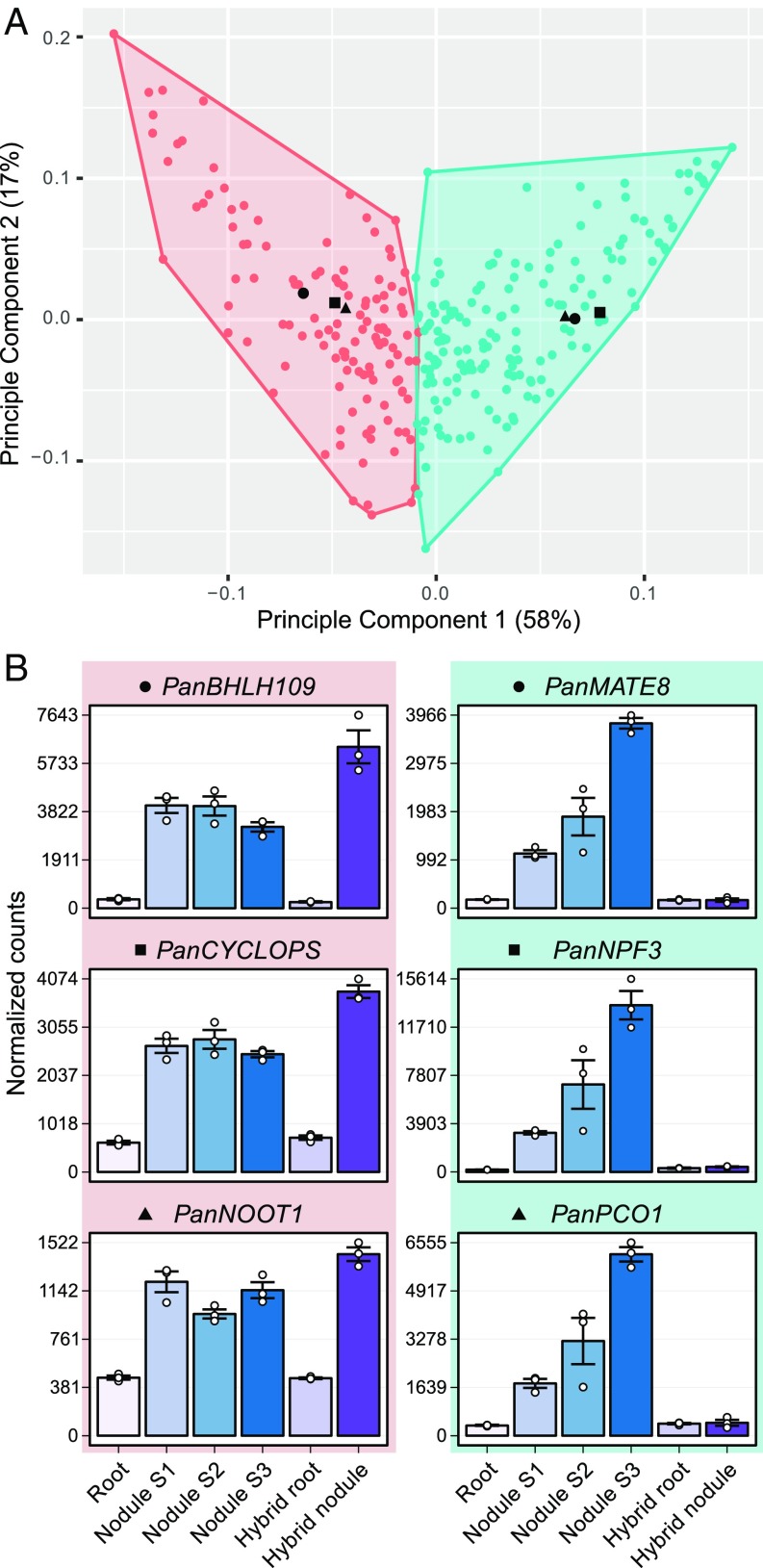
By exploiting the insight that nodule organogenesis and rhizobial infection can be genetically dissected using hybrid plants, we classified these commonly utilized genes into two categories based on their expression profiles in roots and nodules of both P. andersonii and F1 hybrids (Fig. 2). The first category comprises 126 genes that are up-regulated in both P. andersonii and hybrid nodules and that we associate with nodule organogenesis. The second category comprises 164 genes that are up-regulated in only the P. andersonii nodule and that we therefore associate with infection and/or fixation (Dataset S5). Based on these results, we conclude that Parasponia and medicago utilize orthologous genes that commit various functions in at least two different developmental stages of the root nodule.
Fig. 2.
Clustering of commonly utilized symbiosis genes based on expression profile in P. andersonii. (A) Principal component analysis plot of the expression profile of 290 commonly utilized symbiosis genes in 18 transcriptome samples: P. andersonii roots and nodules (stage 1–3) and hybrid roots and nodules (line H9). All samples have three biological replicates. The first two components are shown, representing 75% of the variation in all samples. Colors indicate clusters (k-means clustering using Pearson correlation as distance measure, k = 2) of genes with similar expression patterns. The three genes with the highest Pearson correlation to the cluster centroids are indicated as black dots, triangles, and squares, and their expression profiles are given in B. Cluster 1 (pink) represents genes related to nodule organogenesis: these genes are up-regulated in P. andersonii and hybrid nodules. Cluster 2 (green) represents genes related to infection and fixation: these genes are highly up-regulated in P. andersonii nodules but do not respond in the hybrid nodule. PanBHLH109, BASIC HELIX–LOOP–HELIX DOMAIN CONTAINING PROTEIN 109; PanMATE8, MULTI ANTIMICROBIAL EXTRUSION PROTEIN 8; PanNOOT1, NODULE ROOT 1; PanNPF3, NITRATE/PEPTIDE TRANSPORTER FAMILY 3; PanPCO1, PLANT CYSTEINE OXIDASE 1.
Lineage-Specific Adaptation in Parasponia HEMOGLOBIN 1.
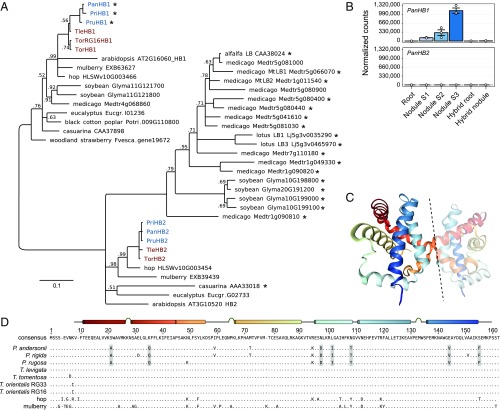
Notable exceptions to the pattern of common utilization in root nodules are the oxygen-binding hemoglobins. Earlier studies showed that Parasponia and legumes have recruited different hemoglobin genes (47). Whereas legumes use class II LEGHEMOGLOBIN to control oxygen homeostasis, Parasponia recruited the paralogous class I HEMOGLOBIN 1 (HB1) for this function (Fig. 3 A and B). Biochemical studies have revealed that P. andersonii PanHB1 has oxygen affinities and kinetics that are adapted to their symbiotic function, whereas this is not the case for T. tomentosa TtoHB1 (47, 48). We therefore examined HB1 from Parasponia species, Trema species, and other nonsymbiotic Rosales species to see if these differences are caused by a gain of function in Parasponia or a loss of function in the nonsymbiotic species. Based on protein alignment, we identified Parasponia-specific adaptations in 7 amino acids (Fig. 3 C and D). Among these is Ile(101), for which it is speculated to be causal for a functional change in P. andersonii HB1 (48). Hemoglobin-controlled oxygen homeostasis is crucial to protect the rhizobial nitrogen-fixing enzyme complex Nitrogenase in legume rhizobium-infected nodule cells (2, 3). Therefore, Parasponia-specific gain of function adaptations in HB1 may have comprised an essential evolutionary step toward functional nitrogen-fixing root nodules with rhizobium endosymbionts.
Fig. 3.
Parasponia-specific adaptations in class 1 hemoglobin protein HB1. (A) Phylogenetic reconstruction of class 1 (OG0010523) and class 2 hemoglobins (OG0002188). Symbiotic hemoglobins are marked with an asterisk; legumes and the actinorhizal plant casuarina have recruited class 2 hemoglobins for balancing oxygen levels in their nodules. Conversely, Parasponia has recruited a class 1 hemoglobin PanHB1, confirming parallel evolution of symbiotic oxygen transport in this lineage: M. truncatula (Medtr), G. max (Glyma), P. trichocarpa (Potri), F. vesca (Fvesca), E. grandis (Eugr), A. thaliana (AT). Node values indicate posterior probabilities below 1; scale bar represents substitutions per site. Parasponia marked in blue, Trema in red. (B) Expression profile of PanHB1 and PanHB2 in P. andersonii roots, stage 1–3 nodules, and P. andersonii × T. tomentosa F1 hybrid roots and nodules (line H9). Expression is given in DESeq2-normalized read counts; error bars represent SE of three biological replicates and dots represent individual expression levels. (C) Crystal structure of the asymmetric dimer of PanHB1 as deduced by Kakar et al. (48). Dashed line separates the two units. (D) Protein sequence alignment of class 1 hemoglobins from Parasponia spp.,Trema spp., hop (H. lupulus), and mulberry (M. notabilis). Only amino acids that differ from the consensus are drawn. A linear model of the crystal structure showing α-helices and turns is depicted above the consensus sequence. There are seven amino acids (marked gray) that consistently differ between all Parasponia and all other sampled species: Ala(21), Gln(35), Asp(97), Ile(101), Thr(108), Val(144), and Phe(155). These differences therefore correlate with the functional divergence between P. andersonii PanHB1 and T. tomentosa TtoHB1 (47, 48).
Parallel Loss of Symbiosis Genes in Trema and Other Relatives of Parasponia.
Evolution of complex genetic traits is often associated with gene copy number variations (CNVs) (49). To test if CNVs were associated with the generally assumed independent evolution of nodulation in Parasponia, we focused on two gene sets: (i) close homologs and putative orthologs of the genes that were characterized to function in legume-rhizobium symbiosis and (ii) genes with a nodule-enhanced expression and functional annotation in P. andersonii (these sets partially overlap and together comprise 1,813 genes; SI Appendix, Fig. S14). We discarded Trema-specific duplications as we considered them irrelevant for the nodulation phenotype. To ensure that our findings are consistent between the Parasponia and Trema genera and not the result of species-specific events, we analyzed the additional draft genome assemblies of two Parasponia and two Trema species (SI Appendix, Table S5). As these additional draft genomes were relatively fragmented, we sought additional support for presence and absence of genes by mapping sequence reads to the P. andersonii and T. orientalis reference genomes and by genomic alignments. This procedure revealed only 11 consistent CNVs in the 1,813 symbiosis genes examined, further supporting the recent divergence between Parasponia and Trema (SI Appendix, Fig. S15). Because of the dominant inheritance of nodule organogenesis in F1 hybrid plants, we anticipated finding Parasponia-specific gene duplications that could be uniquely associated with nodulation. Surprisingly, we found only one consistent Parasponia-specific duplication in symbiosis genes, namely, for a HYDROXYCINNAMOYL-COA SHIKIMATE TRANSFERASE (HCT; SI Appendix, Figs. S16 and S17). This gene has been investigated in the legume forage crop alfalfa (Medicago sativa), in which it was shown that HCT expression correlates negatively with nodule organogenesis (50, 51). Therefore, we do not consider this duplication relevant for the nodulation capacity of Parasponia. Additionally, we identified three consistent gene losses in Parasponia, among which is the ortholog of EXOPOLYSACCHARIDE RECEPTOR 3 that, in lotus, inhibits infection of rhizobia with incompatible exopolysaccharides (52, 53) (SI Appendix, Figs. S18–S20 and Table S7). Such gene losses may have contributed to effective rhizobium infection in Parasponia, and their presence in T. tomentosa could explain the lack of intracellular infection in the F1 hybrid nodules. However, they cannot explain the dominance of nodule organogenesis in the F1 hybrid.
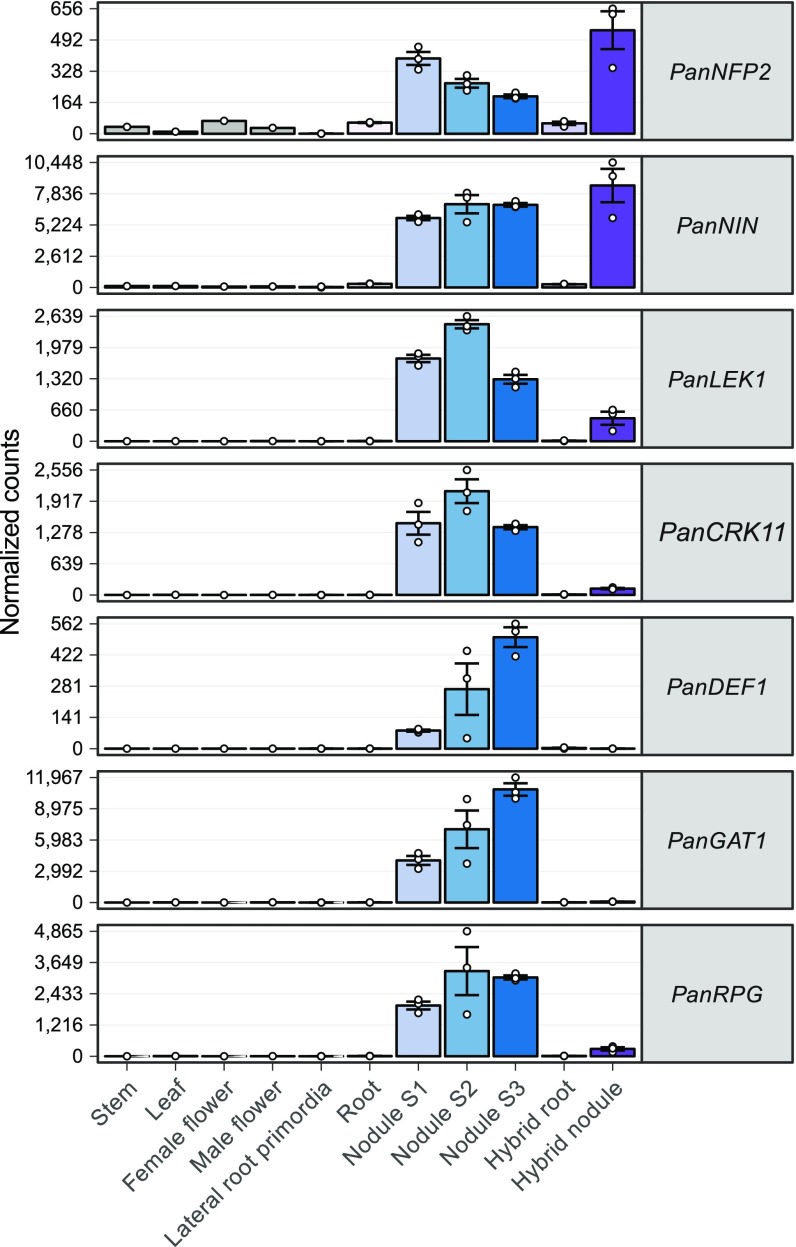
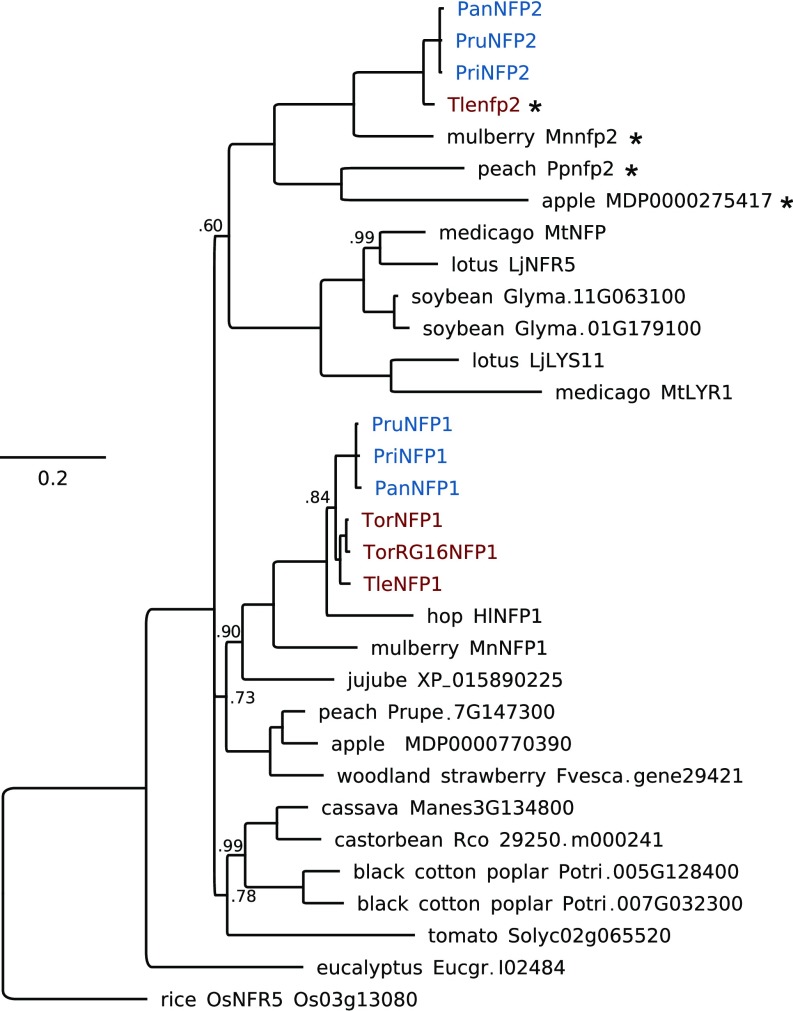
Contrary to our initial expectations, we discovered consistent loss or pseudogenization of seven symbiosis genes in Trema (SI Appendix, Figs. S21–S23 and Table S7). Based on our current sampling, these genes have a nodule-specific expression profile in P. andersonii, suggesting that they function exclusively in symbiosis (Fig. 4). Three of these are orthologs of genes that are essential for establishment of nitrogen-fixing nodules in legumes: NIN, RPG, and the LysM-type LCO receptor NFP/NFR5. In the case of NFP/NFR5, we found two close homologs of this gene, NFP1 and NFP2, a duplication that predates the divergence of legumes and Parasponia (Fig. 5). In contrast to NFP1, NFP2 is consistently pseudogenized in Trema species (Fig. 5 and SI Appendix, Figs. S22 and S23). In an earlier study, we used RNAi to target PanNFP1 (previously named PaNFP), which led to reduced nodule numbers and a block of intracellular infection by rhizobia as well as arbuscular mycorrhiza (30). However, we cannot rule out that the RNAi construct unintentionally also targeted PanNFP2, as both genes are ∼70% identical in the 422-bp RNAi target region. Therefore, the precise functioning of both receptors in rhizobium and mycorrhizal symbiosis remains to be elucidated. Based on phylogenetic analysis, the newly discovered PanNFP2 is the ortholog of the legume MtNFP/LjNFR5 genes encoding rhizobium LCO receptors required for nodulation, whereas PanNFP1 is most likely a paralog (Fig. 5). Also, PanNFP2 is significantly more highly expressed in nodules than PanNFP1 (SI Appendix, Fig. S25). Taken together, this indicates that PanNFP2 may represent a key LCO receptor required for nodulation in Parasponia.
Fig. 4.
Expression profile of P. andersonii symbiosis genes that are lost in Trema species. Expression of symbiosis genes in P. andersonii stem, leaf, female and male flowers, lateral root primordia, roots, and three nodule stages (S1–S3), and in F1 hybrid roots and nodules (line H9). Expression is given in DESeq2-normalized read counts; error bars represent SE of three biological replicates for lateral root primordia, root, and nodule samples. Dots represent individual expression levels. PanCRK11, CYSTEINE-RICH RECEPTOR KINASE 11; PanDEF1, DEFENSIN 1; PanLEK1, LECTIN RECEPTOR KINASE 1; PanNFP2, NOD FACTOR PERCEPTION 2; PanNIN, NODULE INCEPTION; PanRPG, RHIZOBIUM DIRECTED POLAR GROWTH.
Fig. 5.
Parasponia NFP2 are putative orthologs of legume LCO receptors MtNFP/LjNFR5. Phylogenetic reconstruction of the NFP/NFR5 orthogroup based on kinase domain. Protein sequences deduced from pseudogenes are marked with an asterisk. Included species are P. andersonii (Pan), Parasponia rigida (Pri), Parasponia rugosa (Pru), T. orientalis RG33 (Tor), T. orientalis RG16 (TorRG16), T. levigata (Tle), medicago (M. truncatula, Mt), lotus (L. japonicus, Lj), soybean (G. max, Glyma), peach (P. persica, Ppe), woodland strawberry (F. vesca, Fvesca), black cotton poplar (P. trichocarpa, Potri), eucalyptus (E. grandis, Eugr), jujube (Z. jujuba), apple (M. × domestica), mulberry (M. notabilis), hops (H. lupulus), cassava (Manihot esculenta), rice (O. sativa), tomato (S. lycopersicum), and castor bean (Ricinus communis). Node numbers indicate posterior probabilities below 1; scale bar represents substitutions per site. Parasponia proteins are marked in blue, Trema in red.
Based on expression profiles and phylogenetic relationships, we also postulate that Parasponia NIN and RPG commit essential symbiotic functions similarly as in other nodulating species (Fig. 3 and SI Appendix, Figs. S25–S28) (17, 19, 37, 54, 55). Compared with uninoculated roots, expression of PanRPG is >300-fold higher in P. andersonii nodules that become intracellularly infected (nodule stage 2), whereas, in F1 hybrid nodules, which are devoid of intracellular rhizobium infection, this difference is less than 20-fold (Fig. 4). This suggests that PanRPG commits a function in rhizobium infection, similarly as found in medicago (37). The transcription factor NIN has been studied in several legume species as well as in the actinorhizal plant casuarina (Casuarina glauca) and, in all cases, shown to be essential for nodule organogenesis (17, 19, 54, 55). Loss of NIN and possibly NFP2 in Trema species can explain the genetic dominance of nodule organogenesis in the Parasponia × Trema F1 hybrid plants.
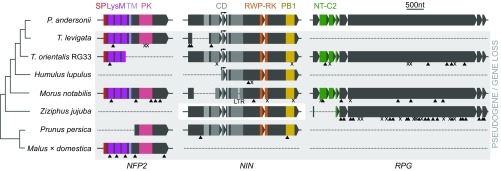
Next, we assessed whether loss of these symbiosis genes also occurred in more distant relatives of Parasponia. We analyzed nonnodulating species representing six additional lineages of the Rosales clade, namely hop (Humulus lupulus, Cannabaceae) (56), mulberry (Morus notabilis, Moraceae) (57), jujube (Ziziphus jujuba, Rhamnaceae) (58), peach (Prunus persica, Rosaceae) (59), woodland strawberry (Fragaria vesca, Rosaceae) (60), and apple (Malus × domestica, Rosaceae) (61). This revealed a consistent pattern of pseudogenization or loss of NFP2, NIN, and RPG orthologs, the intact jujube ZjNIN being the only exception (Fig. 6). We note that, for peach, NIN was previously annotated as a protein-coding gene (59). However, based on comparative analysis of conserved exon structures, we found two out-of-frame mutations (SI Appendix, Fig. S28). We therefore conclude that the NIN gene is also pseudogenized in peach. Because the pseudogenized symbiosis genes are largely intact in most of these species and differ in their deleterious mutations, the loss of function of these essential symbiosis genes should have occurred relatively recently and in parallel in at least seven Rosales lineages.
Fig. 6.
Parallel loss of symbiosis genes in nonnodulating Rosales species. Pseudogenization or loss of NFP2, NIN, and RPG in two phylogenetically independent Trema lineages, H. lupulus (hop), M. notabilis (mulberry), Z. jujuba (jujube), P. persica (peach), and M. × domestica (apple). In H. lupulus, NIN is pseudogenized, whereas NFP2 and RPG were not found (this may be because of the low N50 of the publicly available assembly). In Z. jujuba, NFP2 is lost and RPG is pseudogenized, but NIN is intact. In F. vesca, all three genes are lost (not shown). Introns are indicated but not scaled. Triangles indicate frame shifts; “X” indicates premature stop codons; “LTR” indicates LTR retrotransposon insertion (not scaled); arrows indicate alternative transcriptional start site in NIN. CD, 4 conserved domains (gray); LysM, 3 Lysin motif domains (magenta); NT-C2, N-terminal C2 domain (green); PB1, Phox and Bem1 domain (yellow); PK, protein kinase (pink); RWP-RK, conserved amino acid domain (orange); SP, signal peptide (red); TM, transmembrane domain (lilac).
Discussion
Here we present the nodulating nonlegume Parasponia as a comparative system to obtain insights in molecular genetic changes underlying evolution of nitrogen-fixing root nodules. We show that nodulation is a genetically dominant trait and that P. andersonii and the legume medicago share a set of 290 genes that have a nodule-enhanced expression profile. Among these are NIN and RPG, two genes that, in legumes, are essential for nitrogen-fixing root nodulation (17, 19, 37, 54). Both of these genes, as well as a putative ortholog of the NFP/NFR5-type LysM receptor for rhizobium LCO signal molecules—named NFP2 in Parasponia—are consistently pseudogenized or lost in Trema and other nonnodulating species of the Rosales order. This challenges the current view on the evolution of nitrogen-fixing plant–microbe symbioses.
Evolution of nodulation is generally viewed as a two-step process: first an unspecified predisposition event in the ancestor of all nodulating species, bringing species in the nitrogen-fixing clade to a precursor state for nodulation; and subsequently, nodulation originated in parallel: eight times with Frankia and twice with rhizobium (1, 6–12). This hypothesis is most parsimonious and suggests a minimum number of independent gains and losses of symbiosis. Based on this hypothesis, it is currently assumed that nonhost relatives of nodulating species are generally in a precursor state for nodulation (9).
Our results are difficult to explain under the hypothesis of parallel origins of nodulation. The functions of NFP2, NIN, and RPG currently cannot be linked to any nonsymbiotic processes. Therefore, it remains obscure why these symbiosis genes were maintained over an extended period of time in nonnodulating plant species and were subsequently independently lost. Additionally, the hypothesis of parallel origins of nodulation would imply convergent recruitment of at least 290 genes to commit symbiotic functions in Parasponia and legumes. Because these 290 genes encode proteins with various predicted functions (e.g., from extracellular signaling receptors to sugar transporters; Dataset S5), as well as comprise at least two different developmental expression patterns (nodule organogenesis and intracellular infection and/or fixation; Fig. 2 and Dataset S5), this would imply parallel evolution of a genetically complex trait.
Alternatively, the parallel loss of symbiosis genes in nonnodulating plants can be interpreted as parallel loss of nodulation (1). Under this hypothesis, nodulation possibly evolved only once in an ancestor of the nitrogen-fixing clade. Subsequently, nodulation was lost in most descendant lineages. This single-gain/massive-loss hypothesis fits our data better in two ways. First, a single gain explains the origin of the conserved set of at least 290 symbiosis genes utilized by Parasponia and medicago because they then result from the same ancestral recruitment event. Second, it more convincingly explains the parallel loss of symbiosis genes in nonnodulating plants because then gene loss correlates directly with loss of nodulation. Additionally, the single-gain/massive-loss model eliminates the predisposition event, a theoretical concept that currently cannot be addressed experimentally. We therefore favor this alternative hypothesis over the currently most widely held assumption of parallel origins of nodulation.
Loss of nodulation is not controversial, as it is generally considered to have occurred at least 20 times in the legume family (9, 10). Nevertheless, the single-gain/massive-loss hypothesis implies many more evolutionary events than the current hypothesis of parallel gains. On the contrary, it is conceptually easier to lose a complex trait, such as nodulation, than to gain it (11). Genetic studies in legumes demonstrated that nitrogen-fixing symbioses can be abolished by a single KO mutation in tens of different genes, among which are NFP/NFR5, NIN, and RPG (Dataset S1). Because parsimony implies equal weights for gains and losses, it therefore may not be the best way to model the evolution of nodulation.
Preliminary support for the single-gain/massive-loss hypothesis can be found in fossil records. Putative root nodule fossils have been discovered from the late Cretaceous (∼84 Mya), which corroborates our hypothesis that nodulation is much older than is generally assumed (62). Legumes are the oldest and most diverse nodulating lineage, but the earliest fossils that can be definitively assigned to the legume family appeared in the late Paleocene (∼65 Mya) (63). Notably, the age of the nodule fossils coincides with the early diversification of the nitrogen-fixing clade that has given rise to the four orders Fabales, Rosales, Cucurbitales, and Fagales (10). As it is generally agreed that individual fossil ages provide minimum bounds for dates of origins, it is therefore not unlikely that the last common ancestor of the nitrogen-fixing clade was a nodulator.
Clearly, the single-gain/massive-loss hypothesis that is supported by our comparative studies with Parasponia requires further substantiation. First, the hypothesis implies that many ancestral species in the nitrogen-fixing clade were able to nodulate. This should be further supported by fossil evidence. Second, the hypothesis implies that actinorhizal plant species maintained NIN, RPG, and possibly NFP2 (the latter only in case LCOs are used as symbiotic signal) (64). Third, these genes should be essential for nodulation in these actinorhizal plants as well as in Parasponia. This can be shown experimentally, as was done for NIN in casuarina (55).
Loss of symbiosis genes in nonnodulating plant species is not absolute, as we observed a functional copy of NIN in jujube. This pattern is similar to the pattern of gene loss in species that lost endomycorrhizal symbiosis in which, occasionally, endomycorrhizal symbiosis genes have been maintained in nonmycorrhizal plants (65, 66). Conservation of NIN in jujube suggests that this gene has a nonsymbiotic function. Contrary to NFP2, which is the result of a gene duplication near the origin of the nitrogen-fixing clade, functional copies of NIN are also present in species outside the nitrogen-fixing clade (SI Appendix, Fig. S26). This suggests that these genes may have retained—at least in part—an unknown ancestral nonsymbiotic function in some lineages within the nitrogen-fixing clade. Alternatively, NIN may have acquired a new nonsymbiotic function within some lineages in the nitrogen-fixing clade.
As hemoglobin is crucial for rhizobium symbiosis in legumes (3), it is striking that Parasponia and legumes do not use orthologous copies of hemoglobin genes in their nodules (47). Superficially, this seems inconsistent with a single gain of nodulation. However, hemoglobin is not crucial for all nitrogen-fixing nodule symbioses because several Frankia microsymbionts possess intrinsic physical characteristics to protect the Nitrogenase enzyme for oxidation (67–70). In line with this, Ceanothus spp. (Rhamnaceae, Rosales)—which represent actinorhizal nodulating relatives of Parasponia—do not express a hemoglobin gene in their Frankia-infected nodules (68–70). Consequently, hemoglobins may have been recruited in parallel after the initial gain of nodulation as parallel adaptations to rhizobium microsymbionts. Based on the fact that Parasponia acquired lineage-specific adaptations in HB1 that are considered to be essential for controlling oxygen homeostasis in rhizobium root nodules (47, 48), a symbiont switch from Frankia to rhizobium may have occurred recently in an ancestor of the Parasponia lineage.
Our study provides leads for attempts to engineer nitrogen-fixing root nodules in agricultural crop plants. Such a translational approach is anticipated to be challenging (71), and the only published attempt so far, describing transfer of eight LCO signaling genes, was unsuccessful (72). Our results suggest that transfer of symbiosis genes may not be sufficient to obtain functional nodules. Even though F1 hybrid plants contain a full haploid genome complement of P. andersonii, they lack intracellular infection. This may be the result of haploinsufficiency of P. andersonii genes in the F1 hybrid or because of an inhibitory factor in T. tomentosa. For example, inhibition of intracellular infection may be the result of a dominant-negative factor or the result of heterozygosity negatively affecting the formation of, e.g., LysM receptor complexes required for appropriate perception of microsymbionts. Such factors may also be present in other nonhost species. Consequently, engineering nitrogen-fixing nodules may require gene KOs in nonnodulating plants to overcome inhibition of intracellular infection. Trema may be the best candidate species for such a (re)engineering approach because of its high genetic similarity with Parasponia and the availability of transformation protocols (73). Therefore, the Parasponia–Trema comparative system may not only be suited for evolutionary studies, but also can form an experimental platform to obtain essential insights for engineering nitrogen-fixing root nodules.
Materials and Methods
Parasponia–Trema Intergeneric Crossing and Hybrid Genotyping.
Parasponia and Trema are wind-pollinated species. A female-flowering P. andersonii individual WU1.14 was placed in a plastic shed together with a flowering T. tomentosa WU10 plant. Putative F1 hybrid seeds were germinated (SI Appendix, Supplementary Methods) and transferred to potting soil. To confirm the hybrid genotype, a PCR marker was used that visualizes a length difference in the promoter region of LIKE-AUXIN 1 (LAX1; primers, LAX1-forward, ACATGATAATTTGGGCATGCAACA; LAX1-reverse, TCCCGAATTTTCTACGAATTGAAA; amplicon size, P. andersonii, 974 bp; T. tomentosa, 483 bp). Hybrid plant H9 was propagated in vitro (30, 74). The karyotype of the selected plants was determined according to Geurts and de Jong (75).
Assembly of Reference Genomes.
Cleaned DNA sequencing reads were de novo assembled by using ALLPATHS-LG (release 48961) (76). After filtering of any remaining adapters and contamination, contigs were scaffolded with two rounds of SSPACE-standard (v3.0) (77) with the mate-pair libraries using default settings. We used the output of the second run of SSPACE scaffolding as the final assembly (full details and parameter choices are provided in SI Appendix, Supplementary Methods). Validation of the final assemblies showed that 90–100% of the genomic reads mapped back to the assemblies (SI Appendix, Table S4), and 94–98% of CEGMA (78) and BUSCO (79) genes were detected (SI Appendix, Table S5).
Annotation of Reference Genomes.
Repetitive elements were identified following the standard Maker-P recipe (weatherby.genetics.utah.edu/MAKER/wiki/index.php/Repeat_Library_Construction-Advanced, accessed October 2015) as described on the GMOD site: (i) RepeatModeler with Repeatscout v1.0.5, Recon v1.08, RepeatMasker version open4.0.5, using RepBase version 20140131 (80) and TandemRepeatFinder; (ii) GenomeTools LTRharvest and LTRdigest (81); (iii) MITEhunter with default parameters (82). We generated species-specific repeat libraries for P. andersonii and T. orientalis separately and combined these into a single repeat library, filtering out sequences that are >98% similar. We masked both genomes by using RepeatMasker with this shared repeat library.
To aid the structural annotation, we used 11 P. andersonii and 6 T. orientalis RNA-sequencing (RNA-seq) datasets (SI Appendix, Table S8). All RNA-seq samples were assembled de novo by using genome-guided Trinity (83), resulting in one combined transcriptome assembly per species. In addition, all samples were mapped to their respective reference genomes by using BWA-MEM and processed into putative transcripts by using cufflinks (84) and transdecoder (85). As protein homology evidence, only UniProt (86) entries filtered for plant proteins were used. This way we included only manually verified protein sequences and prevented the incorporation of erroneous predictions. Finally, four gene-predictor tracks were used: (i) SNAP (87) trained on P. andersonii transdecoder transcript annotations; (ii) SNAP trained on T. orientalis transdecoder transcript annotations; (iii) Augustus (88), as used in the BRAKER pipeline, trained on RNA-seq alignments (89); and (iv) GeneMark-ET, as used in the BRAKER pipeline, trained on RNA-seq alignments (90).
First, all evidence tracks were processed by Maker-P (91). The results were refined with EVidenceModeler (EVM) (92), which was used with all of the same tracks as Maker-P, except for the Maker-P blast tracks and with the addition of the Maker-P consensus track as additional evidence. Ultimately, EVM gene models were preferred over Maker-P gene models except when there was no overlapping EVM gene model. Where possible, evidence of both species was used to annotate each genome (i.e., de novo RNA-seq assemblies of both species were aligned to both genomes).
To take maximum advantage of annotating two highly similar genomes simultaneously, we developed a custom reconciliation procedure involving whole-genome alignments. The consensus annotations from merging the EVM and Maker-P annotations were transferred to their respective partner genome by using nucmer (93) and RATT revision 18 (94) (i.e., the P. andersonii annotation was transferred to T. orientalis and vice versa) based on nucmer whole-genome alignments (SI Appendix, Fig. S10). Through this reciprocal transfer, both genomes had two candidate annotation tracks. This allowed for validation of annotation differences between P. andersonii and T. orientalis, reduced technical variation, and consequently improved all downstream analyses. After automatic annotation and reconciliation, 1,693 P. andersonii genes and 1,788 T. orientalis genes were manually curated. These were mainly homologs of legume symbiosis genes and genes that were selected based on initial data exploration.
To assign putative product names to the predicted genes, we combined BLAST results against UniProt, TrEMBL, and nr with InterProScan results (custom script). To annotate Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) enzyme codes we used Blast2GO based on the nr BLAST results and InterProScan results. Finally, we filtered all gene models with hits to InterPro domains that are specific to repetitive elements.
Orthogroup Inference.
To determine relationships between P. andersonii and T. orientalis genes, as well as with other plant species, we inferred orthogroups with OrthoFinder version 0.4.0 (95). As orthogroups are defined as the set of genes that are descended from a single gene in the last common ancestor of all of the species being considered, they can comprise orthologous as well as paralogous genes. Our analysis included proteomes of selected species from the Eurosid clade: A. thaliana TAIR10 (Brassicaceae, Brassicales) (96) and Eucalyptus grandis v2.0 (Myrtaceae, Myrtales) from the Malvid clade (97); Populus trichocarpa v3.0 (Salicaeae, Malpighiales) (98), legumes M. truncatula Mt4.0v1 (99) and G. max Wm82.a2.v1 (Fabaceae, Fabales) (100), F. vesca v1.1 (Rosaceae, Rosales) (60), and P. andersonii and T. orientalis (Cannabaceae, Rosales) from the Fabid clade (Dataset S2). Sequences were retrieved from phytozome (www.phytozome.net).
Gene CNV Detection.
To assess orthologous and paralogous relationships between Parasponia and Trema genes, we inferred phylogenetic gene trees for all 21,959 orthogroups comprising Parasponia and/or Trema genes by using the neighbor-joining clustering algorithm (101). Based on these gene trees, for each Parasponia gene, its relationship to other Parasponia and Trema genes was defined as follows: (i) orthologous pair indicates that the sister lineage is a single gene from the Trema genome, suggesting that they are the result of a speciation event; (ii) inparalog indicates that the sister lineage is a gene from the Parasponia genome, suggesting that they are the result of a gene duplication event; (iii) singleton indicates that the sister lineage is a gene from a species other than Trema, suggesting that the Trema gene was lost; and (iv) multiortholog indicates that the sister lineage comprises multiple genes from the Trema genome, suggesting that the latter are inparalogs. For each Trema gene, the relationship was defined in the same way but with respect to the Parasponia genome (SI Appendix, Table S6). Because phylogenetic analysis relies on homology, we assessed the level of conservation in the multiple-sequence alignments by calculating the trident score using MstatX (https://github.com/gcollet/MstatX) (102). Orthogroups with a score below 0.1 were excluded from the analysis. Examination of orthogroups comprising >20 inparalogs revealed that some represented repetitive elements; these were also excluded. Finally, orthologous pairs were validated based on the whole-genome alignments used in the annotation reconciliation.
Nodule-Enhanced Genes.
To assess gene expression in Parasponia nodules, RNA was sequenced from the three nodule stages described earlier as well as uninoculated roots (SI Appendix, Table S8). RNA-seq reads were mapped to the Parasponia reference genome with HISAT2 version 2.02 (103) using an index that includes exon and splice site information in the RNA-seq alignments. Mapped reads were assigned to transcripts with featureCounts version 1.5.0 (104). Normalization and differential gene expression were performed with DESeq2. Nodule enhanced genes were selected based on >2.0-fold change and P ≤ 0.05 in any nodule stage compared with uninoculated root controls. Genes without functional annotation or orthogroup membership or from orthogroups with low alignment scores (<0.1 trident score, as detailed earlier) or representing repetitive elements were excluded from further analysis. To assess expression of Parasponia genes in the hybrid nodules, RNA was sequenced from nodules and uninoculated roots. Here, RNA-seq reads were mapped to a combined reference comprising two parent genomes from P. andersonii and T. tomentosa. To assess which genes are nodule-enhanced in medicago, we reanalyzed published RNA-seq read data from Roux et al. (34) [archived at the National Center for Biotechnology Information (NCBI) under sequence read archive (SRA) study ID code SRP028599]. To assess which of these genes may be coopted from the ancient and widespread arbuscular mycorrhizal symbiosis, we generated a set of 575 medicago genes induced upon mycorrhization in medicago by reanalyzing published RNA-seq read data from Afkhami and Stinchcombe (archived at the NCBI under SRA study ID code SRP078249) (46) Both medicago data sets were analyzed as described earlier for Parasponia but by using the medicago genome and annotation version 4.0v2 as reference (99).
To assess common recruitment of genes in nodules from Parasponia and medicago, we counted orthogroups comprising P. andersonii and medicago nodule-enhanced genes. To assess whether this number is higher than expected by chance, we performed the hypergeometric test as well as three different permutation tests in which we randomized the Parasponia gene set, the medicago gene set, or both sets with 10,000 permutations. We then determined putative orthology between the Parasponia and medicago genes within the common orthogroups based on phylogenetic analysis. Parasponia and medicago genes were considered putative orthogroups if they occurred in the same subclade with more than 50% bootstrap support; otherwise, they were considered close homologs.
Availability of Data and Materials.
The data reported in this study are tabulated in Datasets S1–S7 and SI Appendix; sequence data are archived at NCBI (https://www.ncbi.nlm.nih.gov) under BioProject numbers PRJNA272473 and PRJNA272482; draft genome assemblies, phylogenetic datasets, and orthogroup data are archived at the Dryad Digital Repository (https://doi.org/10.5061/dryad.fq7gv88). Analyzed data can also be browsed or downloaded through a Web portal at www.parasponia.org. All custom scripts and code are available online at https://github.com/holmrenser/parasponia_code.
Supplementary Material
Acknowledgments
We thank Shelley James, Thomas Marler, Giles Oldroyd, and Johan van Valkenburg for providing germplasm and Ries deVisser (IsoLife) for supporting acetylene reduction assays.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. PRJNA272473 and PRJNA272482). Draft genome assemblies, phylogenetic datasets, and orthogroup data are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.fq7gv88). All custom scripts and code are available online at https://github.com/holmrenser/parasponia_code.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721395115/-/DCSupplemental.
References
- 1.Soltis DE, et al. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci USA. 1995;92:2647–2651. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 3.Ott T, et al. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol. 2005;15:531–535. doi: 10.1016/j.cub.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 4.Burrill TJ, Hansen R. Is symbiosis possible between legume bacteria and non-legume plants? Bull Univ Ill Agric Expt Stn. 1917;202:115–181. [Google Scholar]
- 5.Stokstad E. The nitrogen fix. Science. 2016;353:1225–1227. doi: 10.1126/science.353.6305.1225. [DOI] [PubMed] [Google Scholar]
- 6.Swensen SM. The evolution of actinorhizal symbioses: Evidence for multiple origins of the symbiotic association. Am J Bot. 1996;83:1503–1512. [Google Scholar]
- 7.Doyle JJ. Phylogenetic perspectives on nodulation: Evolving views of plants and symbiotic bacteria. Trends Plant Sci. 1998;3:473–478. [Google Scholar]
- 8.Doyle JJ. Phylogenetic perspectives on the origins of nodulation. Mol Plant Microbe Interact. 2011;24:1289–1295. doi: 10.1094/MPMI-05-11-0114. [DOI] [PubMed] [Google Scholar]
- 9.Werner GDA, Cornwell WK, Sprent JI, Kattge J, Kiers ET. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat Commun. 2014;5:4087. doi: 10.1038/ncomms5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H-L, et al. Large-scale phylogenetic analyses reveal multiple gains of actinorhizal nitrogen-fixing symbioses in angiosperms associated with climate change. Sci Rep. 2015;5:14023. doi: 10.1038/srep14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle JJ. Chasing unicorns: Nodulation origins and the paradox of novelty. Am J Bot. 2016;103:1865–1868. doi: 10.3732/ajb.1600260. [DOI] [PubMed] [Google Scholar]
- 12.Martin FM, Uroz S, Barker DG. Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science. 2017;356:eaad4501. doi: 10.1126/science.aad4501. [DOI] [PubMed] [Google Scholar]
- 13.Limpens E, et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 14.Madsen EB, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 15.Radutoiu S, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 16.Arrighi JF, et al. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279, and erratum (2007) 143:1078. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh JF, et al. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broghammer A, et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 20.Soyano T, Kouchi H, Hirota A, Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9:e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernié T, et al. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell. 2015;27:3410–3424. doi: 10.1105/tpc.15.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 23.Oldroyd GED. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 24.Geurts R, Xiao TT, Reinhold-Hurek B. What does it take to evolve a nitrogen-fixing endosymbiosis? Trends Plant Sci. 2016;21:199–208. doi: 10.1016/j.tplants.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Clason EW. 1936. The vegetation of the upper-Badak region of mount Kelut (East Java). Bull Jard Bot Buitenzorg Ser 3 13:509–518.
- 26.Trinick MJ. Symbiosis between Rhizobium and the non-legume, Trema aspera. Nature. 1973;244:459–460. [Google Scholar]
- 27.Akkermans ADL, Abdulkadir S, Trinick MJ. Nitrogen-fixing root nodules in Ulmaceae. Nature. 1978;274:190. [Google Scholar]
- 28.Becking JH. The Rhizobium symbiosis of the nonlegume Parasponia. In: Stacey G, Burris RH, Evans HJ, editors. Biological Nitrogen Fixation. Routledge, Chapman and Hall; New York: 1992. pp. 497–559. [Google Scholar]
- 29.Marvel DJ, Torrey JG, Ausubel FM. Rhizobium symbiotic genes required for nodulation of legume and nonlegume hosts. Proc Natl Acad Sci USA. 1987;84:1319–1323. doi: 10.1073/pnas.84.5.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Op den Camp R, et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331:909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- 31.Granqvist E, et al. Bacterial-induced calcium oscillations are common to nitrogen-fixing associations of nodulating legumes and nonlegumes. New Phytol. 2015;207:551–558. doi: 10.1111/nph.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang M-Q, et al. Molecular phylogenetics and character evolution of Cannabaceae. Taxon. 2013;62:473–485. [Google Scholar]
- 33.Op den Camp RHM, et al. Nonlegume Parasponia andersonii deploys a broad rhizobium host range strategy resulting in largely variable symbiotic effectiveness. Mol Plant Microbe Interact. 2012;25:954–963. doi: 10.1094/MPMI-11-11-0304. [DOI] [PubMed] [Google Scholar]
- 34.Roux B, et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014;77:817–837. doi: 10.1111/tpj.12442. [DOI] [PubMed] [Google Scholar]
- 35.Combier J-PP, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baudin M, et al. A phylogenetically conserved group of nuclear factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 2015;169:2761–2773. doi: 10.1104/pp.15.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arrighi J-F, et al. The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc Natl Acad Sci USA. 2008;105:9817–9822. doi: 10.1073/pnas.0710273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kistner C, et al. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell. 2005;17:2217–2229. doi: 10.1105/tpc.105.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deguchi Y, et al. Transcriptome profiling of Lotus japonicus roots during arbuscular mycorrhiza development and comparison with that of nodulation. DNA Res. 2007;14:117–133. doi: 10.1093/dnares/dsm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yano K, et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pumplin N, et al. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 2010;61:482–494. doi: 10.1111/j.1365-313X.2009.04072.x. [DOI] [PubMed] [Google Scholar]
- 42.Horváth B, et al. Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant Microbe Interact. 2011;24:1345–1358. doi: 10.1094/MPMI-01-11-0015. [DOI] [PubMed] [Google Scholar]
- 43.Murray JD, et al. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 2011;65:244–252. doi: 10.1111/j.1365-313X.2010.04415.x. [DOI] [PubMed] [Google Scholar]
- 44.Tóth K, et al. Functional domain analysis of the remorin protein LjSYMREM1 in Lotus japonicus. PLoS One. 2012;7:e30817. doi: 10.1371/journal.pone.0030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiasson DM, et al. Soybean SAT1 (symbiotic ammonium transporter 1) encodes a bHLH transcription factor involved in nodule growth and NH4+ transport. Proc Natl Acad Sci USA. 2014;111:4814–4819. doi: 10.1073/pnas.1312801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afkhami ME, Stinchcombe JR. Multiple mutualist effects on genomewide expression in the tripartite association between Medicago truncatula, nitrogen-fixing bacteria and mycorrhizal fungi. Mol Ecol. 2016;25:4946–4962. doi: 10.1111/mec.13809. [DOI] [PubMed] [Google Scholar]
- 47.Sturms R, Kakar S, Trent J, 3rd, Hargrove MS. Trema and parasponia hemoglobins reveal convergent evolution of oxygen transport in plants. Biochemistry. 2010;49:4085–4093. doi: 10.1021/bi1002844. [DOI] [PubMed] [Google Scholar]
- 48.Kakar S, et al. Crystal structures of Parasponia and Trema hemoglobins: Differential heme coordination is linked to quaternary structure. Biochemistry. 2011;50:4273–4280. doi: 10.1021/bi2002423. [DOI] [PubMed] [Google Scholar]
- 49.Żmieńko A, Samelak A, Kozłowski P, Figlerowicz M. Copy number polymorphism in plant genomes. Theor Appl Genet. 2014;127:1–18. doi: 10.1007/s00122-013-2177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shadle G, et al. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry. 2007;68:1521–1529. doi: 10.1016/j.phytochem.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 51.Gallego-Giraldo L, et al. Lignin modification leads to increased nodule numbers in alfalfa. Plant Physiol. 2014;164:1139–1150. doi: 10.1104/pp.113.232421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawaharada Y, et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature. 2015;523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 53.Kawaharada Y, et al. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun. 2017;8:14534. doi: 10.1038/ncomms14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borisov AY, et al. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 2003;131:1009–1017. doi: 10.1104/pp.102.016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clavijo F, et al. The Casuarina NIN gene is transcriptionally activated throughout Frankia root infection as well as in response to bacterial diffusible signals. New Phytol. 2015;208:887–903. doi: 10.1111/nph.13506. [DOI] [PubMed] [Google Scholar]
- 56.Natsume S, et al. The draft genome of hop (Humulus lupulus), an essence for brewing. Plant Cell Physiol. 2015;56:428–441. doi: 10.1093/pcp/pcu169. [DOI] [PubMed] [Google Scholar]
- 57.He N, et al. Draft genome sequence of the mulberry tree Morus notabilis. Nat Commun. 2013;4:2445. doi: 10.1038/ncomms3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J, et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016;12:e1006433. doi: 10.1371/journal.pgen.1006433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verde I, et al. International Peach Genome Initiative The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 2013;45:487–494. doi: 10.1038/ng.2586. [DOI] [PubMed] [Google Scholar]
- 60.Shulaev V, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velasco R, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 62.Herendeen PS, Magallon-Puebla S, Lupia R, Crane PR, Kobylinska J. A preliminary conspectus of the Allon flora from the Late Cretaceous (late Santonian) of central Georgia, USA. Ann Mo Bot Gard. 1999;86:407–471. [Google Scholar]
- 63.Bruneau A, Mercure M, Lewis GP, Herendeen PS. Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany. 2008;86:697–718. [Google Scholar]
- 64.Nguyen TV, et al. An assemblage of Frankiacluster II strains from California contains the canonical nod genes and also the sulfotransferase gene nodH. BMC Genomics. 2016;17:796. doi: 10.1186/s12864-016-3140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delaux P-M, et al. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci USA. 2015;112:13390–13395. doi: 10.1073/pnas.1515426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamel L, Keller-Pearson M, Roux C, Ané J-M. Biology and evolution of arbuscular mycorrhizal symbiosis in the light of genomics. New Phytol. 2017;213:531–536. doi: 10.1111/nph.14263. [DOI] [PubMed] [Google Scholar]
- 67.Winship LJ, Martin KJ, Sellstedt A. The acetylene reduction assay inactivates root nodule uptake hydrogenase in some actinorhizal plants. Physiol Plant. 1987;70:361–366. [Google Scholar]
- 68.Silvester WB, Winship LJ. Transient responses of nitrogenase to acetylene and oxygen in actinorhizal nodules and cultured frankia. Plant Physiol. 1990;92:480–486. doi: 10.1104/pp.92.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silvester WB, Berg RH, Schwintzer CR, Tjepkema JD. Oxygen responses, hemoglobin, and the structure and function of vesicles. In: Pawlowski K, Newton WE, editors. Nitrogen-Fixing Actinorhizal Symbioses, Nitrogen Fixation: Origins, Applications, and Research Progress. Springer; Dordrecht, The Netherlands: 2007. pp. 105–146. [Google Scholar]
- 70.Silvester WB, Harris SL, Tjepkema JD. Oxygen regulation and hemoglobin. In: Schwintzer CR, Tjepkema JD, editors. The Biology of Frankia and Actinorhizal Plants. Academic; New York: 1990. pp. 157–176. [Google Scholar]
- 71.Rogers C, Oldroyd GED. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J Exp Bot. 2014;65:1939–1946. doi: 10.1093/jxb/eru098. [DOI] [PubMed] [Google Scholar]
- 72.Untergasser A, et al. One-step Agrobacterium mediated transformation of eight genes essential for rhizobium symbiotic signaling using the novel binary vector system pHUGE. PLoS One. 2012;7:e47885. doi: 10.1371/journal.pone.0047885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao Q, Op den Camp R, Kalhor MS, Bisseling T, Geurts R. Efficiency of Agrobacterium rhizogenes–mediated root transformation of Parasponia and Trema is temperature dependent. Plant Growth Regul. 2012;68:459–465. [Google Scholar]
- 74.Davey MR, et al. Effective nodulation of micro-propagated shoots of the non-legume Parasponia andersonii by Bradyrhizobium. J Exp Bot. 1993;44:863–867. [Google Scholar]
- 75.Geurts R, de Jong H. Fluorescent in situhybridization (FISH) on pachytene chromosomes as a tool for genome characterization. Methods Mol Biol. 2013;1069:15–24. doi: 10.1007/978-1-62703-613-9_2. [DOI] [PubMed] [Google Scholar]
- 76.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 78.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 79.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 80.Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gremme G, Steinbiss S, Kurtz S. GenomeTools: A comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinformatics. 2013;10:645–656. doi: 10.1109/TCBB.2013.68. [DOI] [PubMed] [Google Scholar]
- 82.Han Y, Wessler SR. MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 89.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: Unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2016;32:767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lomsadze A, Burns PD, Borodovsky M. Integration of mapped RNA-seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 2014;42:e119. doi: 10.1093/nar/gku557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Campbell MS, et al. MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 2014;164:513–524. doi: 10.1104/pp.113.230144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Otto TD, Dillon GP, Degrave WS, Berriman M. RATT: Rapid annotation transfer tool. Nucleic Acids Res. 2011;39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Myburg AA, et al. The genome of Eucalyptus grandis. Nature. 2014;510:356–362. doi: 10.1038/nature13308. [DOI] [PubMed] [Google Scholar]
- 98.Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 99.Young ND, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 101.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 102.Valdar WSJ. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 103.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this study are tabulated in Datasets S1–S7 and SI Appendix; sequence data are archived at NCBI (https://www.ncbi.nlm.nih.gov) under BioProject numbers PRJNA272473 and PRJNA272482; draft genome assemblies, phylogenetic datasets, and orthogroup data are archived at the Dryad Digital Repository (https://doi.org/10.5061/dryad.fq7gv88). Analyzed data can also be browsed or downloaded through a Web portal at www.parasponia.org. All custom scripts and code are available online at https://github.com/holmrenser/parasponia_code.