Significance
Microsite evolution involving ecological divergence due to geological, edaphic, or climatic conditions requires adaptive complexes to environmental stresses. The higher drought tolerance of wild barley populations inhabiting Terra Rossa soil at the Tabigha Evolution Slope has been described, but the underlying genetic mechanisms remain unknown. Using genome resequencing and RNA-sequencing technologies of wild barley genotypes from contrasting Terra Rossa and basalt soil types, we identified genes in selection sweep regions on chromosomes 6H and 7H, showing divergence in the barley populations from Terra Rossa and basalt soils with significant roles in plant drought tolerance. Our results set a solid foundation for future work on gene discovery and on drought adaptation mechanisms in barley related to the rhizosphere environment.
Keywords: drought adaptation, evolution models, genome resequencing, Hordeum spontaneum, microsites
Abstract
Ecological divergence at a microsite suggests adaptive evolution, and this study examined two abutting wild barley populations, each 100 m across, differentially adapted to drought tolerance on two contrasting soil types, Terra Rossa and basalt at the Tabigha Evolution Slope, Israel. We resequenced the genomes of seven and six wild barley genotypes inhabiting the Terra Rossa and basalt soils, respectively, and identified a total of 69,192,653 single-nucleotide variants (SNVs) and insertions/deletions in comparison with a reference barley genome. Comparative genomic analysis between these abutting wild barley populations involved 19,615,087 high-quality SNVs. The results revealed dramatically different selection sweep regions relevant to drought tolerance driven by edaphic natural selection within 2,577 selected genes in these regions, including key drought-responsive genes associated with ABA synthesis and degradation (such as Cytochrome P450 protein) and ABA receptor complex (such as PYL2, SNF1-related kinase). The genetic diversity of the wild barley population inhabiting Terra Rossa soil is much higher than that from the basalt soil. Additionally, we identified different sets of genes for drought adaptation in the wild barley populations from Terra Rossa soil and from wild barley populations from Evolution Canyon I at Mount Carmel. These genes are associated with abscisic acid signaling, signaling and metabolism of reactive oxygen species, detoxification and antioxidative systems, rapid osmotic adjustment, and deep root morphology. The unique mechanisms for drought adaptation of the wild barley from the Tabigha Evolution Slope may be useful for crop improvement, particularly for breeding of barley cultivars with high drought tolerance.
Barley is the fourth largest cereal crop worldwide and is also an excellent model species for genetic and physiological studies (1, 2). Its unique genetic adaptation and tolerance to abiotic stresses are providing insights relevant to the improvement in other cereal crops. Wild barley (Hordeum spontaneum L.) is the progenitor of cultivated barley (Hordeum vulgare L.) and is a hardy plant species, harboring a myriad of distinctive genes, alleles, and regulators with potential for increasing the resistance of cultivated barley to abiotic stresses such as drought, salinity, and temperature (3–5). Genome sequences, both coding and noncoding, provide a solid basis for understanding biological traits and their regulation, and the recently released, high-quality reference barley genome sequences (6, 7) are providing important resources for comprehensive genetic and genomic studies of cereals. The reference genome provides a basis for positional cloning and functional analysis of key genes, optimizing population genomics and comparative genomic analysis as well as facilitating genome resequencing to investigate relationships between genomic variation and phenotypic differences.
Drought hampers crop production and the global food supply (5, 8). Crops often experience periods of atmospheric or soil water deficit, which are often accompanied by high temperatures, poor nutrient uptake, and aggravated soil salinity stress (9). Thus, typical features enabling drought tolerance of plants include, but are not limited to, deep and large root systems, thick and complex cuticular waxes, efficient stomatal regulation, and regulation of drought-responsive genes and metabolites (5, 8, 10, 11). As a relatively drought-tolerant crop for dryland agriculture in many countries, several studies aiming to elucidate the mechanisms of drought tolerance have been conducted on barley through genomic approaches (12–15). Using transcriptome analysis, ecotype-specific transcripts have been found in two wild barley ecotypes that differentially adapt them to drought stress (12) and genes related to differential reproduction under drought stress (15). Thus, investigating drought tolerance in wild barley may facilitate a better understanding of the genetic basis of this trait and the identification of effective genetic or genomic approaches toward barley improvement (14).
The complexity of drought stress requires the investigation of tolerance mechanisms in different targeted environments (5, 16). Wild barley is likely to suffer from different types of drought due to its wide geographic range, i.e., growth under different climate conditions or in different soil types. It was proposed that wild barley has evolved distinctive mechanisms to cope with different types of drought such as those experienced at the Tabigha Evolution Slope located north of the Lake of Galilee in Israel; our study complements those at the Evolution Canyon microsite (see E.N. publications of Evolution Canyons at evolution.haifa.ac.il.). At the Tabigha Evolution Slope, Middle Eocene hard limestones weather into Terra Rossa soils and abut Pleistocene volcanic basalt flows that weather into basaltic soils; this results in soils that display dramatic chemical and physical differences (17, 18). The basaltic soil possesses a greater water-holding capacity compared with Terra Rossa soil. A pioneering study of edaphic differentiation in wild barley has been made at the Tabigha Evolution Slope (17), taking advantage of the sharp microscale ecological divergence of plants growing on these two soil types. Allozyme polymorphisms were found, which were suggested to be at least partly adaptive and differentiated due to natural selection mediated by edaphic factors rather than by stochastic processes and/or neutrality of allozyme variants (17). A study in Aegilops peregrina L. at the same site also demonstrated allozymic divergence in two esterase loci between plants from Terra Rossa and basalt soils (19).
Plants thriving on the calcareous Terra Rossa naturally experience more intense drought than plants inhabiting the moist siliceous clay of the basalt soil, and a study (18) of wild barley populations from these soil types showed divergent phenotypic responses to water stress. A high degree of phenotypic variation was found in the wild barley populations from the Terra Rossa and basalt soils when drought treatments were imposed, resulting in significant genotype × treatment and soil type × treatment interactions. Terra Rossa genotypes exhibited significantly better drought adaptation and were more stable under drought stress, important traits for improving adaption to drought in cultivated barley (18). This contrasting drought tolerance in the wild barley populations at the Tabigha Evolution Slope appears to be related to adaptation to different soil conditions. However, the genomic basis of this difference remains unknown. These studies (17–19) provided the rationale for the present genome-wide comparison of wild barley populations at the Tabigha Evolution Slope, and we extend the limitation of allozymic markers to examine the diversity of the entire wild barley genome in two contrasting populations. We provide strong evidence for edaphic adaptations to drought across the whole genome in wild barley.
Results
Whole-Genome Resequencing and Genomic Diversity of Wild-Barley Genotypes from the Tabigha Evolution Slope.
A total of 1,300 Gb of clean data were obtained from 13 wild barley genotypes at the Tabigha Evolution Slope, with an average of 100 Gb for each genotype at 20 × genome coverage. There were, on average, 88.46% clean reads mapped to the reference genome of barley (cv. Zangqing320, www.ibgs.zju.edu.cn/ZJU_barleygenome.htm), with the mapped ratios ranging from 85.96 to 91.01% (SI Appendix, Table S1).
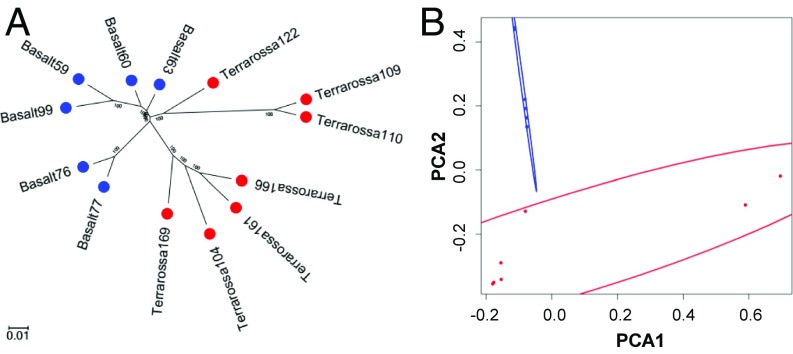
Based on the reads uniquely mapped to the reference genome, we identified a total of 69,192,653 single-nucleotide variants (SNVs) and insertions/deletions, ranging from 16,053,119 to 27,020,003 for each genotype (SI Appendix, Table S1). The large variations have enabled further analysis for population structure and genetic divergence of wild barley. We filtered the raw SNVs and obtained 19,615,087 high-quality SNVs for the construction of a phylogenetic tree (Fig. 1A) and for principal component analysis (PCA) (Fig. 1B), which demonstrated a significant difference between the two wild barley populations inhabiting the two soil types. The PCA clearly divided these barley genotypes into two groups that corresponded with the soil types (Fig. 1B). The phylogenetic tree may involve some mixing due to ongoing gene flow especially near the interface.
Fig. 1.
Phylogenetic tree (A) and PCA (B) of wild barley populations inhabiting Terra Rossa and basalt soils at the Tabigha Evolution Slope. (A) Phylogenetic tree was constructed using the neighbor-joining method, and the percentage of trees from 1,000 bootstrap replications in which the associated taxa clustered together are shown next to the branches. Circles in both A and B indicate barley genotypes from Terra Rossa (red) and basalt (blue) soil types. PCA 1, the first principal component; PCA 2, the second principal component.
Genetic Divergence of Wild Barley Inhabiting the Terra Rossa and Basalt Soils.
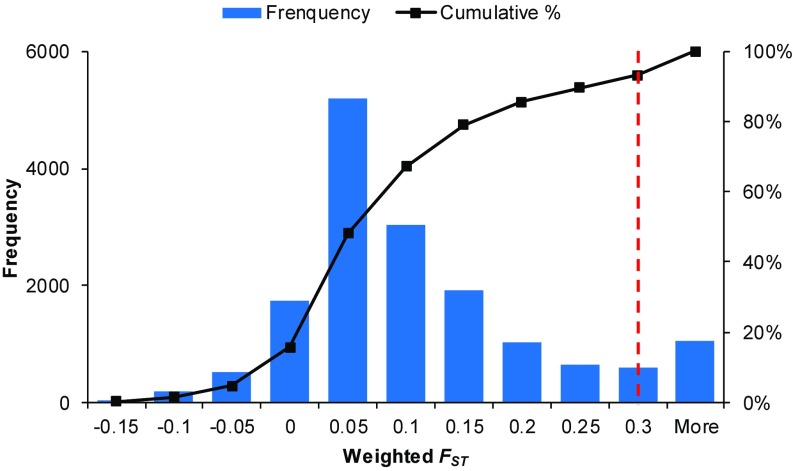
To identify potentially unique genomic regions modulated by environmental selections, Wright’s F-statistic (FST) was calculated based on the 19,615,087 high-quality SNVs. The results show that genetic differences between wild barley populations in Terra Rossa and basalt soils follow a normal distribution, resulting in a cumulative 93.39% with a weighted FST value of ≤0.3 (Fig. 2). This value (0.3) was used as the threshold to conduct a selective sweep analysis.
Fig. 2.
Distribution of FST values across the whole genome between wild barley populations inhabiting Terra Rossa and basalt soils at the Tabigha Evolution Slope. The FST value was calculated in each 1-Mb region with steps of 250 kb. The x axis indicates the value of FST, and the y axis shows the FST value frequency (left) and cumulative percentage (right). The red dashed line indicates the threshold value chosen based on the distribution of all windowed FST.
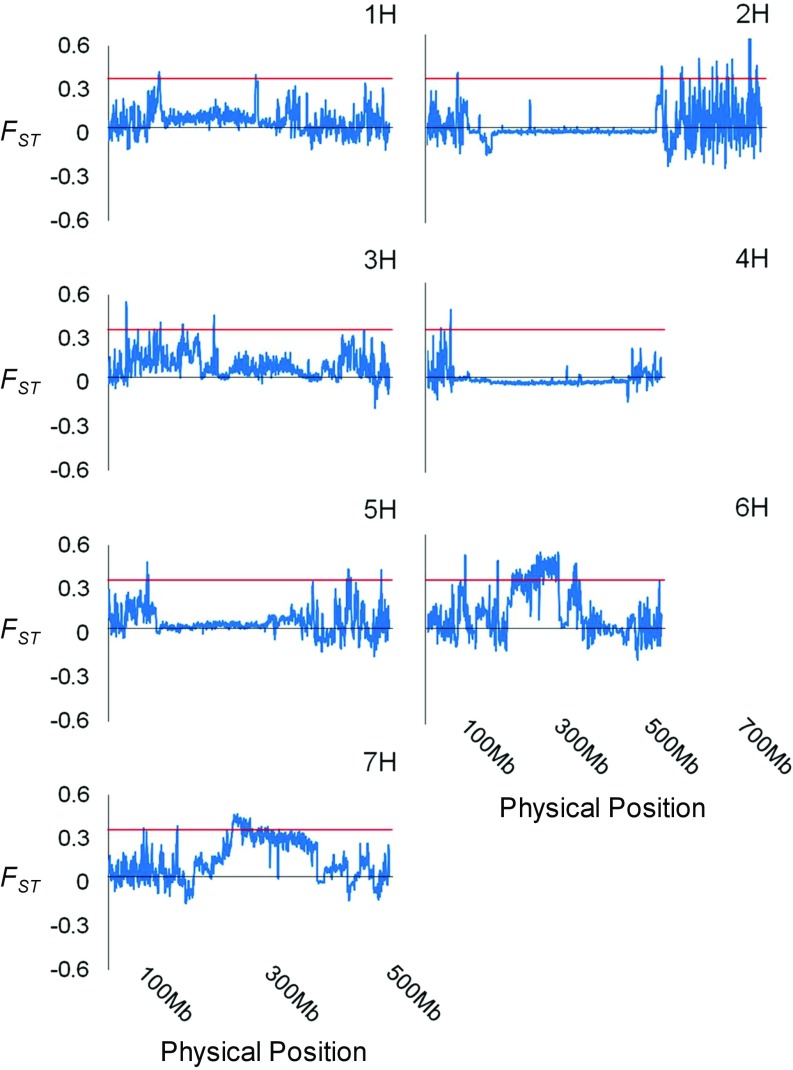
There were distinct patterns in the distribution of windowed FST values for each chromosome between the barley populations collected from the two soils (Fig. 3). Higher drought tolerance of the wild barley population inhabiting Terra Rossa soil than that in basalt soil (18) was measured using the fixation index FST. As a result, 7% of the FST windows were under strong selective sweeps (Fig. 2). The total length of genomic regions with FST values above 0.3 was 0.36 Gb and contained 2,577 high-confidence genes (e.g., those related to plant hormones, antioxidants, and osmoprotectants); these selected regions were assumed to be subject to diversifying selection (SI Appendix, Table S2 and Dataset S1). We found evidence of strong environmental selection in two genomic regions, each about 250 Mb in length located around the centromeres of chromosome 6H (between 194,500,001 and 302,750,000 bp) and chromosome 7H (between 219,250,001 and 382,750,000 bp) (Fig. 3).
Fig. 3.
Distribution of windowed FST values along each chromosome between wild barley populations from the Terra Rossa and basalt soils at the Tabigha Evolution Slope. The x axis indicates the physical position, and the y axis shows the value of FST for each 1-Mb genomic region with steps of 250 kb. The red solid lines indicate the threshold FST value (0.3) delimiting regions considered to be under strong selective sweeps.
To clarify the genetic differences between wild barley populations inhabiting Terra Rossa and basalt soils, we conducted a genetic diversity analysis using the SNVs of the 2,577 selected genes (Dataset S1). There were 881 genes carrying 6,926 SNVs with no missing sites for polymorphism analysis in each of the 13 wild barley genotypes, including 3,396 SNVs showing genetic diversity. Among these, we further selected the genes with all SNVs having no missing sites and obtained 77 genes showing genetic diversity between the two barley populations (Dataset S2). These genes included those encoding protein phosphatase 2C (PP2C, MLOC_15036), MIZU-KUSSEI 1 (MIZ1, MLOC_54802), alkaline neutral invertase CINV2 (MLOC_7525), and ECERIFERUM 1 (CER1, MLOC_11693). PP2C is an important negative regulator in ABA signaling and drought tolerance (20), MIZ1 plays a role in lateral root development by maintaining auxin levels (21), CINV2 regulates sugar-mediated root development by controlling sucrose catabolism in root cells (22), and CER1 participates in epicuticular wax biosynthesis (23). However, sequences of these genes in genotypes from basalt soil showed no genetic diversity (SI Appendix, Table S2 and Dataset S2).
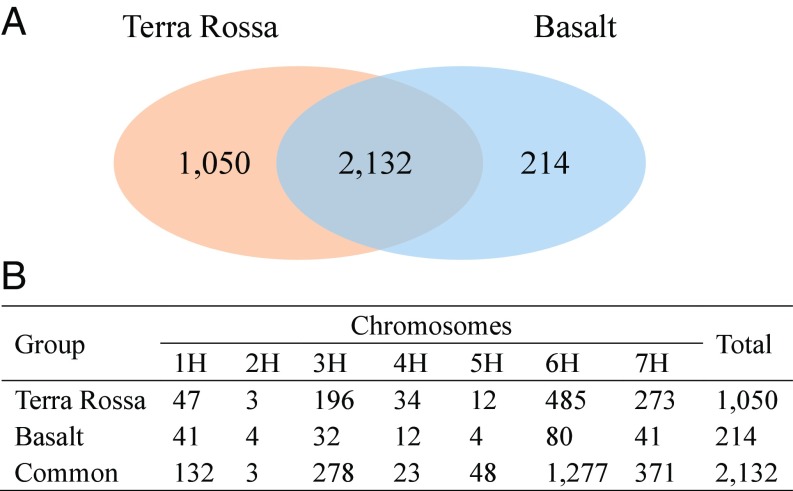
The average π value, an indicator of genetic diversity, for the wild barley population inhabiting the Terra Rossa soil (π = 0.208) was much higher than that of the population from the basalt soil (π = 0.147). In addition, there were 1,050 and 214 unique SNVs in the wild barley populations inhabiting Terra Rossa and basalt soils, respectively (Fig. 4A). Moreover, the Terra Rossa population showed a large number of unique SNVs in chromosomes 6H and 7H (Fig. 4B). We randomly chose 50-Mb regions on chromosome 5H (without selection signature) and chromosomes 6H and 7H (with genetic signature of selective sweeps) to conduct genetic diversity analysis. The average π value of SNVs in 50-Mb regions on chromosome 5H was 0.214 for the population inhabiting the Terra Rossa soil and 0.130 for the population on basalt. The results indicated that the genetic diversity of the populations inhabiting Terra Rossa soil is larger than that from the basalt soil, a phenomenon reported in other higher-stress environments (24).
Fig. 4.
Genetic diversity of two wild barley populations inhabiting Terra Rossa and basalt soils at the Tabigha Evolution Slope. (A) Venn diagram shows unique and common SNVs. (B) Distribution of SNVs with genetic diversity along chromosomes for each population.
Transcriptome Analysis of Wild-Barley Populations from Evolution Canyon I at Mount Carmel Under Drought Treatment.
Drought or soil water availability is one of the key climatic stresses that may distinguish the environment of the tropical hot and dry African Slope (AS) from the abutting temperate cool and humid forested European Slope (ES) of the Evolution Canyon I (ECI) at Mount Carmel, Israel, and 70 km west from the Tabigha Evolution Slope (25–28). Therefore, we conducted RNA sequencing to explore differential gene expression profiles of five wild barley genotypes each from the AS and the ES from ECI under drought treatment (SI Appendix, Fig. S1). In total, 3.47 billion raw reads (519.96 Gb) for 60 samples of the 10 wild barley genotypes (with two treatments and three biological replicates) were obtained by transcriptome sequencing. On average, 97.82% were clean reads, with the mean clean reads of 57,424,477 for each genotype, and 93.20% of the clean reads on average were mapped to the barley reference genome (1) (SI Appendix, Table S3).
Phylogenetic analysis (SI Appendix, Fig. S1), Venn diagram (SI Appendix, Fig. S2), and volcano plots of differentially expressed genes (DEGs) (SI Appendix, Fig. S3) at ECI showed clear divisions between the two populations. We identified 276 and 285 DEGs in five genotypes from both the AS and the ES; 59 DEGs were common to all 10 genotypes (SI Appendix, Fig. S2A). About 75.58% of the DEGs specific to the AS genotypes were up-regulated, while nearly 74.34% of the DEGs specific to the ES genotypes were down-regulated (SI Appendix, Fig. S2A), suggesting contrasting modes of response to drought stress between the barley genotypes from these tropical hot and dry AS and the temperate cool and humid ES. Furthermore, phenotypic analysis of these genotypes from the contrasting AS and ES showed that AS genotypes have significantly lower stomatal conductance and higher intrinsic leaf water-use efficiency at a population level (SI Appendix, Fig. S4).
Mechanisms Underlying the Genetic Difference of Drought Tolerance in Wild Barley at the Tabigha Evolution Slope.
To help understand the mechanisms of drought tolerance in wild barley from the Tabigha Evolution Slope, we analyzed the 2,577 genes in the selective genomic regions of genotypes from Terra Rossa and basalt soil types (Dataset S1), and the 502 DEGs of the genotypes from the AS and ES at ECI (Dataset S3), resulting in 22 common genes (SI Appendix, Fig. S2B), including one related to proline metabolism (MLOC_52074) (Datasets S1 and S3). Interestingly, we identified specific genes for drought adaptation of wild barley from the Terra Rossa soil.
Overall, many drought-responsive and ABA-signaling genes were identified in the selected genomic regions of wild barley from the Terra Rossa soil. For example, there were genes associated with stomatal development [mitogen-activated kinase kinase kinase, YODA (MLOC_51546), transmembrane leucine-repeat receptor (LRR)-like protein, TMM (MLOC_17359)], ABA synthesis and degradation [Cytochrome P450 protein, CYP (MLOC_7007), ABA receptor, PYL2 (MLOC_49654)], protein kinases [SNF1-related kinase, SnRK (ZLOC_8934), CBL-interacting protein kinase, CIPK9 (ZLOC_12175), serine threonine kinases (e.g., BLUS1: MLOC_57740), G-type lectin S-receptor–like serine threonine-kinases (MLOC_67098), lectin-domain–containing receptor kinases (MLOC_21948)], and protein phosphatases [serine threonine phosphatase, PP1 (MLOC_64374), phosphoinositide phosphatase, SAC, (MLOC_44139)] (SI Appendix, Table S2 and Dataset S1). These genes also encoded transcription factors [MYBs (MLOC_6171), NACs (MLOC_39910)], reactive oxygen species (ROS) and nitric oxide (NO) signaling [superoxide dismutase, SOD (MLOC_17760), peroxiredoxins, PRX (ZLOC_22743), nitrate reductase, NIA (MLOC_3293)], and Ca2+ binding and signaling [calmodulin-like (ZLOC_1443)] (SI Appendix, Table S2 and Dataset S1). Moreover, major ion channels [slow anion channels SLACs (MLOC_67347), SLAH1s (ZLOC_30254), SLAH2s (ZLOC_21671)], K+ transporters (MLOC_11780), cotransporters [ATP-binding cassette, ABC (MLOC_56261), nitrate transporter, NRT (MLOC_60308), high-affinity K+ transporter, HKT (MLOC_55066), cation/H+ exchanger, CHX (ZLOC_25185), cation-chloride cotransporter (MLOC_64607)], pumps [vacuolar H+-ATPase, VHAs (MLOC_72577), Ca2+-ATPase, ACAs (MLOC_34557)] were also found in the selective genomic regions of wild barley populations from the Tabigha Evolution Slope (SI Appendix, Table S2 and Dataset S1).
Some of these genes in the populations from the Tabigha Evolution Slope were also differentially in the populations from the AS and ES sites (Dataset S3). For example, there were nine genes and five DEGs encoding CYPs in wild barley of the Tabigha Evolution Slope and ECI; two (MLOC_23018, MLOC_64838) were significantly up-regulated in the wild barley from the AS and three (MLOC_51434, MLOC_7476, MLOC_65039) were down-regulated in genotypes from the ES (Datasets S1 and S3). BTB/POZ/MATH proteins function as a negative regulator, affecting stomatal behavior and responses to ABA in Arabidopsis (29). There were seven selected genes and three DEGs encoding BTB-POZ-MATH proteins in wild barley of the Tabigha Evolution Slope and ECI; MLOC_38371 was significantly up-regulated in wild barley from the AS, and two (MLOC_63926, MLOC_51464) were down-regulated in accessions from the ES (Datasets S1 and S3). H2O2 and NO are two key secondary messengers mediating signal transduction in response to many biotic and abiotic stresses (30–32). We identified 7 genes encoding peroxidases and 2 encoding glutaredoxins, 10 encoding glutathione synthesis and 3 genes encoding SOD, respectively, under selection in wild barley from the Tabigha Evolution Slope (SI Appendix, Table S2 and Dataset S1), but none was found in wild barley materials from ECI. LRR receptor-like serine threonine-kinases are key regulators of ABA signaling (RPKs), plant innate immunity (FLSs), and coping with adverse soil conditions (GSOs) (33–35). There were 12 LRR receptor-like serine threonine-kinases including the major RPKs, FLSs, and GSOs in the unique genomic regions of the wild barley from the Tabigha Evolution Slope, and two LRRs (including GSO1) were found in the wild barley from ECI (SI Appendix, Table S2 and Dataset S1).
In addition, there were differences in genes from the selective genomic regions encoding osmoprotectants. Seven of the 10 genes associated with proline accumulation were located on 6H and 7H of the genotypes from the Tabigha Evolution Slope (SI Appendix, Table S2 and Dataset S1), providing evidence of strong environmental selection in the two genomic regions. In contrast, none of the three DEGs associated with proline in the genotypes from ECI were located on these two chromosomes (Dataset S3). We also identified three genes related to root development [e.g., Root Hairless 1 (RTH1)] and four genes associated with auxin signaling from the selective genomic regions of the genotypes from the Tabigha Evolution Slope (SI Appendix, Table S2 and Dataset S1). In contrast, very few DEGs in ECI were related to these genes, which might contribute to the drought tolerance mechanisms in the wild barley inhabiting Terra Rossa soil (Dataset S3).
Discussion
Adaptive Edaphic Selection of Wild Barley at the Tabigha Evolution Slope.
Wild barley has adapted to various ecological and environmental conditions due to long-term adaptation driven by natural selection, including differences in water availability, soil type, temperature, and altitude (3, 17, 36–38). The average dry weight of wild barley from Terra Rossa was reduced by only 22.7% after a water stress treatment was imposed whereas a 78.0% decrease was detected for accessions from basalt soil, showing a significant genotypic difference between these populations (18). In this study, we performed genome resequencing of two wild barley populations adapted to these two contrasting soil types at the Tabigha Evolution Slope. Significantly, we identified two large genomic regions, 18.54% of chromosome 6H and 24.85% of chromosome 7H, that have been strongly subjected to environmental selection (Fig. 3). The results indicated a large genomic diversity between the two wild barley populations. This interslope genetic diversity at Tabigha between genotypes from calcareous Terra Rossa and siliceous basalt, a large part of which may involve adaptive pathways rich in drought resistance, is driven by edaphic differences, particularly in the drier Terra Rossa, highlighting the importance of edaphic factors in adaptive evolution.
In contrast, the long, nondiverse regions of chromosomes 4H and 5H show a close genetic relationship between these two wild barley populations (Fig. 3), which is consistent with the minor branches of the neighbor-joining tree (Fig. 1A). This is to be expected as gene flow is likely to occur in both directions primarily in the vicinity of the interface between the two soils at the Tabigha Evolution Slope (17). Thus, our results revealed that the wild barley genotypes from the two soils have a close genetic relationship at the whole-genome level but, interestingly, have distinct selection sweep regions due to the adaptation to different soil types, which, as discussed below, may be primarily mediated by drought, which is higher on the Terra Rossa than the basalt soil (17–19).
Drought Tolerance Mechanisms of Wild Barley at the Tabigha Evolution Slope.
Plants have evolved many strategies to maintain growth and development when water availability is restricted or unpredictable (5, 8). We have detected a set of genes relevant to drought adaptation in the wild barley population inhabiting the Terra Rossa soil in the Tabigha Evolution Slope in the selection sweep genomic regions (SI Appendix, Table S2 and Dataset S1). Coincidentally, several quantitative trait loci (QTL) associated with drought tolerance (39–42) have also been identified in the same chromosomes in field-grown barley, such as four QTLs on chromosome 6H and two QTLs on chromosome 7H controlling relative water content (42).
To obtain more supporting evidence for drought mediating the selection sweep, we also analyzed the drought-responsive DEGs of wild barley from ECI, another classic evolution microsite in Israel (28), for comparative analysis of the specific drought-tolerance mechanisms in materials from the Tabigha Evolution Slope. ECI provides a microcosmic ecological model of life with contrasting biodiversity, where the AS is characterized by higher solar radiation, higher temperature, less water, and wider spatiotemporal heterogeneity and fluctuation than the ES (28). Therefore, drought-adaptive mechanisms in wild barley genotypes at ECI may result mainly from climate conditions (28) and may be different from those found at the Tabigha Evolution Slope. More than 75% of the DEGs in the genotypes from the AS were up-regulated, while around 75% of those in genotypes from the ES were down-regulated (SI Appendix, Fig. S2A). Therefore, the up-regulation of these key drought-adaptive genes in wild barley genotypes from the AS may be responsible for overcoming drought.
Comparison of these 502 DEGs from wild barley from ECI with the 2,577 genes from wild barley from the Tabigha Evolution Slope demonstrated that most are found in the selective genomic regions of the wild barley from the Tabigha Evolution Slope, but also show distinct expression in the wild barley genotypes from ECI (Datasets S1 and S3). We summarize here a few highlights of the novel mechanisms of drought tolerance in the wild barley from the Tabigha Evolution Slope. It was reported that drought tolerance in barley may be attributed to ABA accumulation, osmotic adjustment, dehydrin expression, stomatal regulation, and root elongation of postgermination seedlings (9, 10, 13, 43). We suggest that the wild barley population inhabiting Terra Rossa soil from the Tabigha Evolution Slope has adapted to drought conditions by fine-tuning the ABA-modulated stomatal aperture opening to balance water use efficiency and CO2 assimilation. The ABA-signaling pathway is one of the major pathways regulating drought tolerance and seed germination in plants (9, 11, 44). ABA can bind to the PYR/PYL/RCAR ABA receptors to inhibit clade A type 2C protein phosphatases (PP2Cs), thus releasing SnRK2 protein kinases from inhibition by PP2Cs and activating downstream signaling components for stomatal closure (20, 45). Activated SnRK2s can phosphorylate downstream effectors, such as SLAC1, and then trigger stomatal closure (46). Genes within the selection sweep regions may participate in this pathway.
Moreover, ABA-mediated stress responses also involve Ca2+-dependent and -independent signaling and ROS and NO signaling, regulating anion channels and other transporters for stomatal closure under drought stress (9, 30, 31). H2O2 is the primary ROS (32, 47) and can cause oxidative cell damage such as lipid peroxidation and membrane damage (48). To protect cellular systems from cytotoxic ROS, plants express peroxidase and superoxide dismutase and antioxidants, such as glutathione (47, 49). There were seven genes related to ROS and NO signaling under selection in the wild barley from the Tabigha Evolution Slope (SI Appendix, Table S2 and Dataset S1). Nitrate-reductase–generated NO production, mediated by genes such as NIA1 and NIA2, regulates stress-responsive genes, such as TFs and enzymes, either through modification of cysteine residues of proteins (S-nitrosylation) or by direct or indirect interaction with biomolecules like fatty acids or hormones (30, 31). Significantly, drought-induced down-regulation of NIAs in the wild barley of the ES suggests that these plants may not have efficient regulation of NO production during drought stress (Dataset S3). In summary, many genes at key nodes of the ABA-signaling pathway were found in the selected regions of wild barley from Terra Rossa soil, suggesting that they have important roles in drought-tolerance mechanisms.
Soil conditions have a strong influence on plant root morphology. To adapt to water limitation, wild barley can sacrifice root number for deeper-growing roots to obtain water from lower soil levels (6). Drought tolerance in wild barley seedlings has been attributed to the root length of postgermination seedlings and the formation of root hairs (13, 50). In the current study, we identified a gene encoding RTH1 and two genes encoding Root Primordium Defective 1 (RPD1) under selection at the Tabigha Evolution Slope. Auxin also plays an important role in root formation (51). We detected four genes related to auxin under selection at the Tabigha Evolution Slope, and all of them were located on chromosome 7H. To adapt to dry and poor soils, plants form a Casparian strip as a hydrophobic barrier on endodermal cells that restricts lateral diffusion of ions and water between the root vascular bundles and the soil. Sulfated peptides, Casparian strip integrity factors (CIFs), are required for contiguous Casparian strip formation in Arabidopsis roots (33). These CIFs specifically bind to the endodermis-expressed leucine-rich repeat receptor kinases GSO1 and GSO2 for Casparian strip regulation and serve as active facilitators to cope with adverse soil conditions (33). Both GSO1 and GSO2 were found in the selective sweep genomic regions of the wild barley from the Tabigha Evolution Slope (SI Appendix, Table S2 and Dataset S1), and GSO1 was up-regulated by drought in the wild barley of the AS (Dataset S3), implying an active strategy for roots to control water and nutrient uptake under drought. Our results suggest that root morphology adjustment is another crucial adaptive mechanism for the wild barley genotypes inhabiting Terra Rossa at the Tabigha Evolution Slope that allows them to cope with drought stress.
Conclusions and Prospects.
In conclusion, wild barley populations inhabiting Terra Rossa soil at the Tabigha Evolution Slope have evolved a suite of drought-tolerance mechanisms under the long-term natural selection mediated by soil conditions. These mechanisms include the expression and regulation of genes related to ABA signaling, antioxidative defense systems, and root morphology for a genomic adaptation of wild barley to drought resistance at the Tabigha Evolution Slope. The comparison with the transcriptome of wild barley in ECI suggests unique genomic mechanisms for drought tolerance in the wild barley at the Tabigha Evolution Slope. Future studies could examine in depth the unique functional drought resistance found at the Tabigha Evolution Slope at genetic, genomic, and epigenomic levels.
Materials and Methods
Plant Materials.
We used 13 wild barley (H. spontaneum) genotypes from the Tabigha Evolution Slope (SI Appendix, Table S1) to conduct whole-genome resequencing, including 7 genotypes from Terra Rossa and 6 genotypes from basalt soils. All of the materials were collected and previously characterized by Nevo and coworkers (17, 18, 52).
Selective Sweep Analysis and Genetic Diversity Analysis.
Selective sweep analysis is one of the major methods used to detect selection signatures at the genomic level in many organisms where the value of FST is used to detect the signals of strong recent selection with reduced pooled heterozygosity (53). Selective sweep analysis was conducted by measuring the patterns of allele frequencies in each 1-Mb fragment with a step of 250 kb along all chromosomes, using the 19,615,087 high-quality SNVs randomly distributed on chromosomes. Genomic regions under selective sweeps were measured by the FST using VCFtools v0.1.13 (54) with parameters of “–fst-window-size 1000000–fst-window-step 250000.” Genomic regions with FST values >0.3 were considered under strong selective sweeps. Genetic diversity (π) was calculated using VCFtools v0.1.13 (54).
Population Structure Analysis.
A phylogenetic tree was constructed using FastTree (55) with 1,000 replicates for bootstrap confidence analysis. MEGA v5.05 (56) was applied to draw the constructed tree. PCA was performed by SNPRelate v1.10.2 (57) and Car v2.1.5 (58) packages of R (version 3.4.0).
Additional experimental details of drought treatment and transcriptome sequencing analysis of wild barley genotypes from ECI, DNA library preparation, deep sequencing, reads mapping, and SNVs and insertions/deletions calling can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ms. Zheying Wang (Hangzhou Guhe Information and Technology), Ms. Sanling Wu (Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University), and Mr. Yunkun Lu (Department of Cell Biology and Program in Molecular Cell Biology, Zhejiang University) for their assistance in sequencing and bioinformatics analysis. This study was supported by the Natural Science Foundation of China (Grants 31471480, 31620103912, and 31571578); the Natural Science Foundation of Zhejiang Province (Grant LR15C130001); Fundamental Research Funds for the Central Universities; the Jiangsu Collaborative Innovation Center for Modern Crop Production; and the Ancell-Teicher Research Foundation of Genetics and Molecular Evolution.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences of raw data have been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/, and the data accession numbers are listed in SI Appendix, Tables S1 and S3.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721749115/-/DCSupplemental.
References
- 1.Dawson IK, et al. Barley: A translational model for adaptation to climate change. New Phytol. 2015;206:913–931. doi: 10.1111/nph.13266. [DOI] [PubMed] [Google Scholar]
- 2.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 3.Nevo E. Evolution of wild barley and barley improvement. In: Zhang GP, Li CD, Liu X, editors. Advance in Barley Sciences. Springer; Berlin: 2012. pp. 1–16. [Google Scholar]
- 4.Zohary D, Hopf M, Weiss E. Domestication of Plants in the Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and the Mediterranean Basin. Oxford Univ Press; Oxford: 2012. [Google Scholar]
- 5.Nevo E, Chen G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ. 2010;33:670–685. doi: 10.1111/j.1365-3040.2009.02107.x. [DOI] [PubMed] [Google Scholar]
- 6.Dai F, et al. Assembly and analysis of a qingke reference genome demonstrate its close genetic relation to modern cultivated barley. Plant Biotechnol J. 2018;16:760–770. doi: 10.1111/pbi.12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mascher M, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544:427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 8.Pennisi E. Plant genetics. The blue revolution, drop by drop, gene by gene. Science. 2008;320:171–173. doi: 10.1126/science.320.5873.171. [DOI] [PubMed] [Google Scholar]
- 9.Chen ZH, et al. Molecular evolution of grass stomata. Trends Plant Sci. 2017;22:124–139. doi: 10.1016/j.tplants.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Cai S, Papanatsiou M, Blatt MR, Chen ZH. Speedy grass stomata: Emerging molecular and evolutionary features. Mol Plant. 2017;10:912–914. doi: 10.1016/j.molp.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedada G, et al. Transcriptome sequencing of two wild barley (Hordeum spontaneum L.) ecotypes differentially adapted to drought stress reveals ecotype-specific transcripts. BMC Genomics. 2014;15:995. doi: 10.1186/1471-2164-15-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G. 2005. Drought resistance in wild barley, Hordeum spontaneum, from Israel: Physiology, gene identification, and QTL mapping. PhD dissertation (Institute of Evolution, University of Haifa, Haifa, Israel)
- 14.Guo P, et al. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot. 2009;60:3531–3544. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hübner S, Korol AB, Schmid KJ. RNA-seq analysis identifies genes associated with differential reproductive success under drought-stress in accessions of wild barley Hordeum spontaneum. BMC Plant Biol. 2015;15:134. doi: 10.1186/s12870-015-0528-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Korff M, et al. Quantitative trait loci associated with adaptation to Mediterranean dryland conditions in barley. Theor Appl Genet. 2008;117:653–669. doi: 10.1007/s00122-008-0787-2. [DOI] [PubMed] [Google Scholar]
- 17.Nevo E, Brown AHD, Zohary D, Storch N, Beiles A. Microgeographic edaphic differentiation in allozyme polymorphisms of wild barley (Hordeum spontaneum, Poaceae) Plant Syst Evol. 1981;138:287–292. [Google Scholar]
- 18.Ivandic V, et al. Phenotypic responses of wild barley to experimentally imposed water stress. J Exp Bot. 2000;51:2021–2029. doi: 10.1093/jexbot/51.353.2021. [DOI] [PubMed] [Google Scholar]
- 19.Nevo E, Krugman T, Storch N. Edaphic natural selection of allozyme polymorphisms in Aegiops peregrina at a Galilee microsite in Israel. Heredity. 1994;72:109–112. [Google Scholar]
- 20.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moriwaki T, et al. Hormonal regulation of lateral root development in Arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiol. 2011;157:1209–1220. doi: 10.1104/pp.111.186270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barratt DH, et al. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA. 2009;106:13124–13129. doi: 10.1073/pnas.0900689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aarts MG, Keijzer CJ, Stiekema WJ, Pereira A. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell. 1995;7:2115–2127. doi: 10.1105/tpc.7.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nevo E. Molecular evolution and ecological stress at global, regional and local scales: The Israeli perspective. J Exp Zool. 1998;282:95–119. [Google Scholar]
- 25.Nevo E. Asian, African and European biota meet at “Evolution Canyon,” Israel: Local tests of global biodiversity and genetic diversity patterns. Proc Biol Sci. 1995;262:149–155. [Google Scholar]
- 26.Nevo E. “Evolution Canyon”: A microcosm of life’s evolution focusing on adaptation and speciation. Isr J Ecol Evol. 2006;52:485–506. [Google Scholar]
- 27.Nevo E. Evolution in action across life at “Evolution Canyon,” Israel. Trends Evol Biol. 2009;1:e3. [Google Scholar]
- 28.Nevo E. “Evolution Canyon,” a potential microscale monitor of global warming across life. Proc Natl Acad Sci USA. 2012;109:2960–2965. doi: 10.1073/pnas.1120633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechner E, et al. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev Cell. 2011;21:1116–1128. doi: 10.1016/j.devcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Chen ZH, et al. Nitrate reductase mutation alters potassium nutrition as well as nitric oxide-mediated control of guard cell ion channels in Arabidopsis. New Phytol. 2016;209:1456–1469. doi: 10.1111/nph.13714. [DOI] [PubMed] [Google Scholar]
- 31.Gupta KJ, Fernie AR, Kaiser WM, van Dongen JT. On the origins of nitric oxide. Trends Plant Sci. 2011;16:160–168. doi: 10.1016/j.tplants.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Møller IM, Sweetlove LJ. ROS signalling: Specificity is required. Trends Plant Sci. 2010;15:370–374. doi: 10.1016/j.tplants.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama T, et al. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science. 2017;355:284–286. doi: 10.1126/science.aai9057. [DOI] [PubMed] [Google Scholar]
- 34.Osakabe Y, et al. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005;17:1105–1119. doi: 10.1105/tpc.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stegmann M, et al. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science. 2017;355:287–289. doi: 10.1126/science.aal2541. [DOI] [PubMed] [Google Scholar]
- 36.Dai F, et al. Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA. 2012;109:16969–16973. doi: 10.1073/pnas.1215265109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai F, et al. Transcriptome profiling reveals mosaic genomic origins of modern cultivated barley. Proc Natl Acad Sci USA. 2014;111:13403–13408. doi: 10.1073/pnas.1414335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nevo E, Beiles A, Storch N, Doll H, Andersen B. Microgeographic edaphic differentiation in hordein polymorphisms of wild barley. Theor Appl Genet. 1983;64:123–132. doi: 10.1007/BF00272719. [DOI] [PubMed] [Google Scholar]
- 39.Diab AA, et al. Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor Appl Genet. 2004;109:1417–1425. doi: 10.1007/s00122-004-1755-0. [DOI] [PubMed] [Google Scholar]
- 40.Guo P, et al. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica. 2008;163:203–214. [Google Scholar]
- 41.Teulat B, et al. Several QTLs involved in osmotic adjustment trait variation in barley (Hordeum vulgare L.) Theor Appl Genet. 1998;96:688–698. [Google Scholar]
- 42.Teulat B, et al. QTL for relative water content in field-grown barley and their stability across Mediterranean environments. Theor Appl Genet. 2003;108:181–188. doi: 10.1007/s00122-003-1417-7. [DOI] [PubMed] [Google Scholar]
- 43.Grossi M, Cattivelli L, Terzi V, Stanca AM. Modification of gene-expression induced by ABA, in relation to drought and cold stress in barley shoots. Plant Physiol Biochem. 1992;30:97–103. [Google Scholar]
- 44.Pornsiriwong W, et al. A chloroplast retrograde signal, 3′-phosphoadenosine 5′-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife. 2017;6:e23361. doi: 10.7554/eLife.23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geiger D, et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol. 2016;67:25–53. doi: 10.1146/annurev-arplant-043015-111854. [DOI] [PubMed] [Google Scholar]
- 48.Liebler DC, Kling DS, Reed DJ. Antioxidant protection of phospholipid bilayers by alpha-tocopherol. Control of alpha-tocopherol status and lipid peroxidation by ascorbic acid and glutathione. J Biol Chem. 1986;261:12114–12119. [PubMed] [Google Scholar]
- 49.Larson RA. The antioxidants of higher plants. Phytochemistry. 1988;27:969–978. [Google Scholar]
- 50.Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 51.Ahkami AH, et al. Distribution of indole-3-acetic acid in Petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta. 2013;238:499–517. doi: 10.1007/s00425-013-1907-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nevo E, et al. Genomic microsatellite adaptive divergence of wild barley microclimatic stress in “Evolution Canyon,” Israel. Biol J Linn Soc Lond. 2005;84:205–224. [Google Scholar]
- 53.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 54.Danecek P, et al. 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.John F, Sanford W. An R Companion to Applied Regression. 2nd Ed Sage; Thousand Oaks, CA: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.