Abstract
CD1 tetramers loaded with lipid antigens facilitate the identification of rare lipid-antigen specific T cells present in human blood and tissue. Because CD1 proteins are structurally non-polymorphic, these tetramers can be applied to genetically diverse human populations, unlike MHC-I and MHC-II tetramers. However, there are no standardized assays to quantify and characterize lipid antigen-specific T cells present within clinical samples. We incorporated CD1b tetramers loaded with the mycobacterial lipid glucose monomycolate (GMM) into a multi-parameter flow cytometry assay. Using a GMM-specific T-cell line, we demonstrate that the assay is linear, reproducible, repeatable, precise, accurate, and has a limit of detection of approximately 0.007%. Having formally validated this assay, we performed a cross-sectional study of healthy U.S. controls and South African adolescents with and without latent tuberculosis infection (LTBI). We show that GMM-specific T cells are specifically detected in South African subjects with LTBI and not in U.S. healthy controls. This assay can be expanded to include additional tetramers or phenotypic markers to characterize GMM-specific T cells in studies of mycobacterial infection, disease, or vaccination.
Keywords: Human, T cells, CD1b, Tetramer, Mycobacteria, Assay validation
2. INTRODUCTION
T cells respond to mycobacterial cell wall lipids presented by functionally non-polymorphic CD1 molecules (Beckman et al., 1994; Gilleron et al., 2004; Layre et al., 2009; Moody et al., 2004; Moody et al., 2000; Moody et al., 1997). The human CD1 locus contains five genes (CD1A, CD1B, CD1C, CD1D, and CD1E) encoding five proteins (CD1a, CD1b, CD1c, CD1d, and CD1e) capable of processing and presenting lipid antigens to T cells. These proteins vary in the configuration of their binding grooves, patterns of cellular expression, and subcellular trafficking (Van Rhijn et al., 2013b). Canonically, CD1a binds lipopeptides, CD1b binds glycolipids, and CD1c binds phospholipids. T-cell responses to these lipids are detectable in the blood of Mycobacterium tuberculosis (M.tb)-infected humans (Gilleron et al., 2004; Layre et al., 2009; Montamat-Sicotte et al., 2011; Moody et al., 2000; Seshadri et al., 2015). However, it is currently unknown whether lipid-specific T-cells become activated or expand as a result of mycobacterial vaccination. A major barrier to progress has been the lack of formally validated assays to quantify and characterize T-cell responses to lipid antigens.
Tetramers take advantage of multimerization to generate high avidity reagents that can bind to and track rare antigen-specific T cells within a larger mixed population of T cells. Tetramers based on the major histocompatibility complex (MHC) Class I and Class II proteins have significantly advanced our understanding of T cell responses to peptide antigens but are limited by the highly polymorphic nature of MHC (Altman et al., 1996). On the other hand, CD1 genes are structurally non-polymorphic, so a CD1 tetramer can in principle be used on everyone, thus permitting a truly global analysis of antigen-specific T cell responses for the first time. The development of soluble lipid-loaded CD1 tetramers changed the landscape for ex-vivo investigation of T-cell phenotypes and functions (Benlagha et al., 2000; Karadimitris et al., 2001; Matsuda et al., 2000). These tetramers allowed engagement of more than one copy of the T cell receptor (TCR) on the surface of a T cell, resulting in increased avidity of the interaction and allowing identification of antigen-specific T cells by flow cytometry, even those present at low frequencies. Initially developed for CD1d, tetramers have now been extended to CD1a, CD1b, and CD1c, including those loaded with mycobacterial lipid antigens to facilitate studies in patients with latent and active tuberculosis (James et al., 2018; Kasmar et al., 2013, 2011; Ly et al., 2013). However, these reagents have not yet found their way into validated end point assays that could be employed in clinical settings.
Here, we present the formal validation of an assay using CD1b tetramers loaded with glucose monomycolate (GMM), a major component of the mycobacterial cell wall (Brennan et al., 1970). GMM comprises up to 2% of total extractable lipid and is produced by many mycobacterial species, including Mycobacterium bovis BCG, M. tuberculosis, M. smegmatis, M. leprae, and M. phlei (Brennan et al., 1970; Moody et al., 2000; Moody, 1997; Silva, 1985). Because the glucose moiety is host-derived, GMM in infected tissues signals the presence of pathogenic mycobacteria and provides an antigenic target for T cells (Moody et al., 2000). Thus, GMM has been observed to be an immunodominant antigen in experimental infection of cattle and studies of humans with latent tuberculosis (Nguyen et al., 2009; Seshadri et al., 2015).
We used GMM-specific T-cell lines to establish the operating characteristics in a flow cytometry assay and to optimize and validate the tetramer assay according to the following parameters: linearity, range, limit of detection, repeatability, reproducibility, intermediate precision, and accuracy. We used this assay to study a cohort of healthy subjects and find that GMM-CD1b tetramer positive cells are specifically detected in South African adolescents at high risk for M.tb exposure but not in U.S. subjects at low risk for exposure. We expect that this assay will find utility in natural history studies of M.tb exposure and disease as well as investigations into the immunogenicity of novel whole cell mycobacterial vaccines.
2. METHODS
2.1. Culture Media
Media (R10) for washing peripheral blood mononuclear cells (PBMC) consisted of RPMI 1640 (Gibco, Waltham, MA) supplemented with 10% fetal calf serum (Hyclone, Logan, UT). Our base T cell media (TCM) consisted of RPMI 1640 supplemented with 10% fetal calf serum, 100 U/mL Penicillin, 100 mg/mL Streptomycin, 55 mM 2-mercaptoethanol, 0.3X Essential Amino Acids, 60 mM Non-essential Amino Acids, 11 mM HEPES, and 800 mM L-Glutamine (Gibco, Waltham, MA) sterile-filtered. Our TCM containing human serum (TCM/HS) consisted of 10% human serum (derived from healthy donors), 100 U/mL Penicillin, 100 mg/mL Streptomycin, and 400 mM L-Glutamine (Gibco, Waltham, MA).
2.2. Preparation and storage of GMM lipids
Glucose monomycolate (C32-GMM) isolated from Rhodococcus equi was generously provided by the laboratory of D. Branch Moody. Stock GMM was solvated in chloroform:methanol (2:1, v:v) at a concentration of 1 mg/mL and sonicated for three minutes in a 37°C water bath to resuspend the lipid. The resulting suspension was divided into single-use glass vial aliquots (21 ug/vial) as repeated sonication may lead to solvent evaporation and increased concentration of the reagent. The solvent from each aliquot was then evaporated under a stream of nitrogen, and the vials were stored at −20°C.
2.3. Generation of GMM-loaded CD1b tetramers
Soluble biotinylated CD1b monomers were provided by the National Institutes of Health Tetramer Core Facility (Emory University, Atlanta, GA). The CD1b monomer loading protocol for GMM was derived from a previously published protocol (Kasmar et al., 2011). Briefly, one aliquot of 21 ug of GMM was sonicated into 41 uL of a 50 mM sodium citrate buffer at pH 4, containing 0.25% with 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (Sigma, St. Louis, MO) for two minutes at 37°C. In parallel, mock, unloaded tetramer was generated similarly to the loaded tetramer but in the absence of lipid. Subsequently, 9 uL of CD1b monomer (2 mg/mL) was added, and the resulting suspension contained GMM at 100-fold molar excess of CD1b monomer. The acidic buffer facilitates monomer loading by allowing the lipid to access the antigen-binding groove. This mixture of lipid and monomer was incubated in a 37°C water bath for two hours with vortexing every 30 minutes. At the end of the incubation, the solution was neutralized to pH 7.4 using 6 uL of 1 M Tris pH 9. Returning the solution to physiological pH prevents dissociation of the lipid-monomer complex. Equal amounts of a streptavidin conjugate were then added every 10 minutes for 100 minutes until a final volume of 15.43 uL streptavidin allophycocyanin (Life Technologies, Carlsbad, CA) or 28.83 uL of streptavidin phycoerythrin (Life Technologies, Carlsbad, CA) had been added. The stepwise addition of streptavidin ensured that each molecule bound to the maximum number of monomers and optimized for tetramerization. The final product was filtered through a SpinX column (Sigma, St. Louis, MO) to remove aggregates. The reagent was then stored at 4°C until use.
Biotinylated hCD1b monomer has a molecular weight of 57,106.2 daltons (Da), and 9 uL of a 2 mg/mL stock solution is used, which is equal to 3.15 * 10−10 moles (mol). C32 GMM is used at 100-fold molar (M) excess and has a molecular weight of 659 Da (Moody et al., 1997). Thus, 3.15 * 10−8 mol of C32 GMM is needed.
Our GMM stock is stored at 1 mg/mL, so 20.76 uL is needed for this reaction. Streptavidin is a complex made up of four subunits, and each has one binding site for biotin. For every four molecules of hCD1b monomer, one molecule of streptavidin is needed, and streptavidin is used at a 25% excess to ensure each hCD1b monomer is bound.
Streptavidin conjugates are provided at 1 mg/mL (Life Technologies, Carlsbad, CA), and the weight of the fluorophore is included. Streptavidin-APC has a molecular weight of 156,800 Da (6.378 * 10−6 M), and streptavidin-PE has a molecular weight of 292,800 Da (3.415 * 10−6 M).
2.4. Derivation of T cell lines
PBMC were isolated from healthy South African adults. The cells were depleted of CD14-expressing monocytes for a separate study and cryopreserved. For this study, the monocyte-depleted PBMC were thawed, washed in warm, sterile-filtered R10 supplemented with Benzonase (Millipore, Billerica, MA) at 10 uL/mL, and enumerated using Trypan blue exclusion. The cells were rested overnight in a 37°C humidified incubator supplemented with 5% CO2 at a density of three million cells per well in a 24-well plate in the presence of TCM. The following day, PBMC were washed and blocked with human serum (Valley Biomedical, Winchester, VA) in FACS buffer (1x phosphate-buffered saline (PBS) (Gibco, Waltham, MA) supplemented with 0.2% bovine serum albumin (BSA) (Sigma, St. Louis, MO)) mixed 1:1 for 10 min at 4°C. The samples were washed twice with PBS and stained with Aqua Live/Dead stain (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Following two additional PBS washes, cells were resuspended in 50 mL FACS buffer and 1 uL of either unloaded CD1b tetramer or GMM-loaded CD1b tetramer and incubated at room temperature for 40 minutes in the dark. Finally, cells were stained with anti-CD3-Phycoerythrin-Texas Red (ECD) (UCHT1; Beckman Coulter, Brea, CA) washed twice in TCM, and screened through a cell strainer tube (Falcon, Tewksbury, MA) to ensure a single cell suspension prior to sorting. Tetramer-positive T cells were sorted at the UW Department of Immunology Flow Cytometry Core using a FACS Aria II (BD Biosciences, San Jose, CA) cell sorter equipped with blue (488 nm), red (641 nm), and violet (407 nm), lasers.
Sorted T cells were washed and resuspended in TCM/HS. Eight wells of a 96-well U-bottom tissue culture plate were seeded with 150,000 cells per well with irradiated PBMC obtained from a local healthy blood donor to act as feeder cells. The sorted T cells were divided among the eight wells, and phytohaemagglutinin (PHA) (Remel, San Diego, CA) was added at a final concentration of 1.6 mg/mL. After two days in culture at 37°C/5% CO2, natural human IL-2 (Hemagen, Columbia, MD) was added and maintained in the culture media at a 1:20 dilution. Half the media was replaced every two days with TCM/HS and natural IL-2. When the cell clusters were large and round (approximately after eight days of growth), they were pooled into a 24-well plate. After 10 days in culture, cell lines were screened by GMM-CD1b tetramer staining (data not shown). Further expansion of the T cell line was achieved using a modified version of a previously published rapid expansion protocol (Riddell et al., 1992). Briefly, 200,000 T cells were mixed with 5 million irradiated EBV-transformed B cells and 25 million irradiated PBMC to act as feeder cells in T25 tissue culture flasks (Costar, St.Louis, MO) with 25 mL TCM. Anti-CD3 (clone OKT3) was added at a final concentration of 30 ng/mL and the mixture was incubated overnight at 37°C/5% CO2. The following day, recombinant human IL-2 (rIL-2) (Prometheus Pharmaceuticals through UWMC Clinical Pharmacy) was added to a final concentration of 50 U/mL. On day 4, the cells were washed twice in TCM to remove the anti-CD3 antibody and resuspended in fresh media supplemented with rIL-2 at 50 U/mL. Half the media was replaced every three days or split into new T25 flasks as determined by cell confluency. After 13 days in culture, GMM antigen-specific T cell expansion was confirmed through GMM tetramer staining (data not shown) and IFN-γ ELISPOT. The resulting T-cell lines were then cryopreserved on day 14 until use.
2.5. IFN-γ ELISPOT
We used an IFN-γ ELISPOT to test the functional response of T cell lines. Multiscreen-IP filter plates (Millipore, Billerica, MA) were coated with 1D1K antibody (Mabtech, Sweden) diluted 1:400 in PBS and incubated overnight at 4 C. The following day, two thousand T cells were plated 1:25 with human myelogenous leukemia cells (K562) stably transfected with CD1b or with a mock vector (EV) (de Jong et al., 2010). GMM lipid antigen was dried using a nitrogen stream and was sonicated into media to obtain a 4 μg/mL suspension and was plated with the cells at a final concentration of 1 μg/mL. The cells were incubated at 37°C/5% CO2 for approximately 16 hours. The following day, the cells were washed twice with sterile water to lyse the cells, and the plates were incubated with 7-B6-1 detection antibody (Mabtech, Sweden) diluted 1:3000 in 0.5% FBS/PBS and incubated for two hours at room temperature. The samples were then washed five times with PBS and incubated in ExtrAvidin-Alkaline Phosphatase (Sigma, St. Louis, MO) diluted 1:1000 in PBS and incubated for one hour at room temperature. Finally, the cells were washed with PBS again and incubated with BCIP/NBT substrate (Sigma, St. Louis, MO) for five minutes in the dark at room temperature to develop the membrane. Detection of IFN-γ spots was measured using an ImmunoSpot S6 Core Analyzer (Cellular Technology Limited, Cleveland, OH).
2.6. Flow Cytometry
For validation experiments, PBMC or T-cell lines were plated at one million cells per well in a 96-well U-bottom plate. Cells were washed and blocked with human serum (Valley Biomedical, Winchester, VA) prepared in FACS buffer (1x PBS (Gibco, Waltham, MA) supplemented with 0.2% BSA (Sigma, St. Louis, MO)) mixed 1:1 for 10 minutes at 4°C. Cells were centrifuged at 1800 rpm for 3 minutes, resuspended in 50 uL of FACS buffer containing 2 uL of GMM-loaded CD1b and 2 uL of unloaded control CD1b tetramers, then incubated at room temperature for 60 minutes. The tetramer titers were determined prior to use in the present study (data not shown). For this step and all subsequent steps, the cells were kept in the dark. At the end of the incubation period, the cells were washed twice with PBS and stained with Live/Dead Fixable Aqua or with Live/Dead Fixable Green Dead Cell Stain Kit (Life Technologies, Carlsbad, CA) per the manufacturer’s instructions. Following a 15 minute incubation at room temperature, the cells were washed twice in PBS and then labelled with anti-CD3 ECD (Beckman Coulter, Brea, CA) and anti-TCR Vα7.2 (TRAV1-2) Brilliant Violet 510 (clone 3C10; Biolegend, San Diego, CA) antibodies for 30 minutes at 4°C. The optimal titers of all antibodies were determined prior to use. After two final washes in FACS buffer, the cells were fixed in 1% Paraformaldehyde (PFA) (Electron Microscopy Sciences, Hatfield, PA) and acquired on a BD LSRFortessa (BD Biosciences, San Jose, CA) equipped with blue (488 nm), green (532 nm), red (628 nm), violet (405 nm), and ultraviolet (355 nm) lasers using standardized good clinical laboratory practice (GCLP) procedures to minimize the variability of data generated. Additional flow cytometers used in the intermediate precision and reproducibility assays included a LSRII (BD Biosciences, San Jose, CA) located at the University of Washington South Lake Union campus in Seattle and a LSRII (BD Biosciences, San Jose, CA) located at the HIV Vaccines Trials Network in Seattle, both equipped with blue (488 nm), green (532 nm), red (628 nm), violet (405 nm), and ultraviolet (355 nm) lasers.
For ex vivo analysis in U.S. and South African human cohorts, we expanded the GMM-CD1b tetramer assay to a 13-color multi-parameter flow cytometry panel. This panel contains the five-color panel validated in the present study as well as tetramers for other donor-unrestricted T cell populations and antibodies targeting T cell lineage and memory markers (Supplemental Table 1).
2.7. Ethics
The study protocols were approved by the IRBs of the University of Washington, the Fred Hutchinson Cancer Research Center, or the University of Cape Town. Written informed consent was obtained from all adult participants as well as from the parents and/or legal guardians of the adolescents who participated. In addition, written informed assent was obtained from the adolescents.
2.8. Clinical Cohorts
For creating T cell lines above, we used cryopreserved PBMC from four healthy South African adults. For ex vivo analysis of GMM-CD1b tetramer positive cells, we studied two cohorts of healthy subjects. First, U.S. healthy controls were recruited and enrolled at the Seattle HIV Vaccine Trials Unit as part of a cohort of healthy adults used to provide blood samples for developing and testing new assays. PBMC collected by leukapheresis from 20 HIV-seronegative individuals with a known T-cell response to CMV were used here. Second, we studied a subset of 6363 South African adolescents that were enrolled into a study that aimed to determine the incidence and prevalence of tuberculosis infection and disease (Mahomed et al., 2011). 12- to 18-year-old adolescents were enrolled at eleven high schools in the Worcester region of the Western Cape of South Africa. Subjects were screened for the presence of latent tuberculosis by a tuberculin skin test and IFN-γ release assay (IGRA) QuantiFERON-TB GOLD In-Tube (QFTG) (Cellestis Inc.) at study entry. PBMC were isolated from freshly collected heparinized blood via density centrifugation and cryopreserved. For this work, a sample of 10 M.tb-infected (QFTG+) and 8 M.tb-uninfected (QFTG-) adolescents were selected based on availability of PBMC.
2.9. Statistical Analysis
Initial compensation, gating, and quality assessment of flow cytometry data was performed using FlowJo version 9 (FlowJo, TreeStar Inc, Ashland OR). Unless otherwise-stated, the gating strategy for the qualification assays began with gating of singlets and viable cells. Lymphocytes were then identified by size gating, and T cells were identified using anti-CD3. The T cells were further examined for antigen-specificity using GMM-CD1b tetramers. Statistical analysis was performed using the OpenCyto framework in the R programming environment (Finak et al., 2014; R Core Team, 2016). To calculate the limit of detection, the N-1 Chi Squared test was used to determine the lowest dilution at which the signal was detectable above the background. The limit of the blank and limit of detection were then calculated (Armbruster and Pry, 2008), using the frequency of events when no GMM-specific cells were added and the 0.005% dilution. GMM frequencies were compared between subject groups using a Kruskal-Wallis test in R. If the p-value was below 0.05, then Dunn’s post-hoc test was performed with Benjamini-Hochberg multiple comparisons corrections.
3. RESULTS
3.1. Derivation of T cell lines and tetramer batch validation
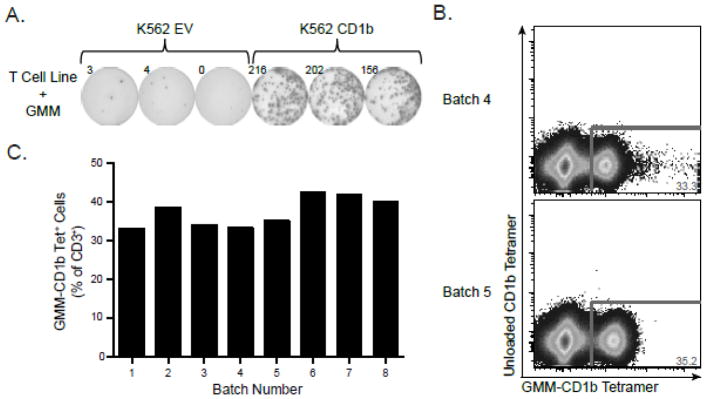
MHC tetramers require peptide ligand to ensure stable folding and expression. By contrast, CD1b tetramers are stable in the absence of ligand, so we required a method to validate each batch of GMM-CD1b tetramer produced in the lab. To accomplish this, we first generated GMM-specific T cell lines as described in Materials and Materials. GMM-specific T cell lines were incubated with K562 cells that were either mock transfected with empty vector (EV) or stably transfected with CD1b. We only observed IFN-γ production in the presence of both GMM and CD1b (Figure 1A). These data confirmed the antigen-specificity and functional reactivity of GMM-specific T cell lines and established a reagent that could be used to “benchmark” each batch of GMM-CD1b tetramer. We observed that two batches of tetramers created several months apart detected GMM-specific T cells at similar frequencies (Figure 1B). Examining eight batches of GMM-CD1b tetramer produced over 18 months, we found that the frequency of tetramer positive cells detected was remarkably consistent, with a mean of 37.36% of CD3+ T cells and coefficient of variation of 10.39% (Figure 1C). Thus, we were able to generate CD1b tetramers that were consistently and maximally loaded with GMM for validation studies.
Figure 1. Validation of GMM-CD1b tetramer using GMM-specific T cell lines.
(A) GMM-specific T cell line was tested for antigen-specificity using IFN-γ ELISPOT. T cells were incubated with K562 cells that were stably transfected with CD1b (K562 CD1b) or mock transfected with an empty vector (K562 EV) in the presence of 1 g/mL GMM followed by quantification of the number of IFN-γ spots present. (B) GMM-specific T cell line was stained with two batches of GMM-CD1b tetramer generated three months apart to assess batch-to-batch variation of the lipid-loaded tetramer. (C) Summary of GMM-CD1b tetramer staining from eight batches of tetramer prepared over 18 months. Data are depicted as a proportion of total CD3+ cells. Results in panels A and B are representative of at least eight independent experiments.
3.2. Assay validation vs. qualification
The terms validation and qualification have specific meanings in the context of Good Clinical Laboratory Practices. For full validation of an assay, eight parameters need to be assessed. These are described in the International Conference on Harmonization (ICH) Q2 (R1) document (“Validation of Analytical Procedures: Text and Methodology Q2(R1),” 2005). They are also listed in the FDA Guidance for Industry (“Bioanalytical Method Validation,” 2013). These parameters are accuracy, repeatability, intermediate precision, limit of detection, limit of quantitation, linearity, range, and specificity. To determine these parameters for the GMM CD1b tetramer, we used antigen-specific T-cell lines described above. This reagent allowed us to vary the detectable signal in a biological sample and determine assay performance over a range of expected precursor frequencies. We determined specificity by examining populations at low and high risk for M.tb exposure.
3.2.1. Assessing the repeatability, intermediate precision, and reproducibility of the CD1 tetramer assay
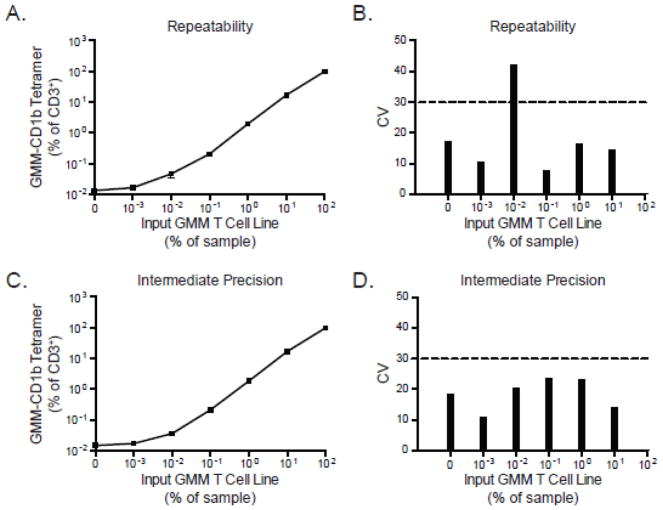
To determine the repeatability of the CD1b-GMM tetramer assay, the GMM-specific T cell line was first serially diluted in PBMC derived from a local area healthy donor and stained with a viability dye, anti-CD3 antibody, and CD1b-GMM tetramer. The frequency of tetramer positive T cells was quantified and compared among three replicate experiments performed by a single operator using the same reagents, T-cell lines, and flow cytometer. As the precursor frequency of GMM-specific T cells increased, the frequency of GMM-specific T cells detected by tetramer increased concordantly and consistently among the three replicate experiments (Figure 2A). The pre-determined acceptable threshold for the coefficient of variation (CV) was chosen as 30% based on prior experience validating a flow cytometric assay for detection of antigen-specific T cells (Horton et al., 2007). The CV for most dilutions, except one, fell below 30%, establishing the repeatability of the assay (Figure 2B). Notably, we were able to detect tetramer-positive cells at a frequency of 0.01% even in the absence of added CD1b-GMM tetramer. We interpreted this result as evidence of GMM-specific T cells in the blood of the healthy donor PBMC used as diluent for the experiments.
Figure 2. Assessing the repeatability, intermediate precision, and reproducibility of the CD1 tetramer assay.
The GMM-specific T-cell line was serially diluted in PBMC from a healthy donor and stained with GMM-CD1b tetramer. (A) Summary data from three independent experiments performed by a single operator in which the mean and standard error of the mean are displayed for each dilution tested. (B) Coefficient of variation (CV) across the three experiments, where the CV value of 30% is indicated (dashed line). To assess intermediate precision, three different operators performed the CD1 tetramer assay on three different flow cytometers using the same batch of T-cell lines and GMM-CD1b tetramer. Shown are the (C) mean and standard error of the mean at each dilution as well as the (D) CV of the experiments acquired between the three operators. Data for intermediate precision are representative of one experiment.
To assess intermediate precision and reproducibility, we examined results obtained by different operators using the same samples on the same day. Three operators shared the same aliquots of diluent PBMC and GMM-specific T cell line, and each operator stained the samples and collected the data on three different flow cytometers. The results of this experiment show that each operator detected the same frequency of tetramer-positive T cells at each dilution tested (Figure 2C). The CV of all dilutions tested was below 30% showing that this assay is reproducible despite varying the operator and instrument.
3.2.2. Measuring the limit of detection of the CD1 tetramer assay
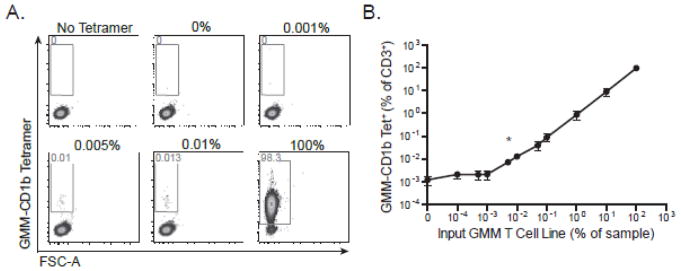
The limit of detection is difficult to quantify as there is no definitive assay to confirm the presence of antigen-specific cells in any particular sample. However, if we can determine the limit of quantitation and the limit of the blank (e.g. background), we can then assume that the limit of detection lies between these two values (Armbruster and Pry, 2008). Average background staining using diluent PBMCs was ~0.017% of total CD3+ events (Figure 2A and data not shown), which likely represented detection of real GMM-specific T cells and was therefore not a true measurement of the background of the assay. Therefore, we chose to use a different cell line, which is specific for the canonical glycosphingolipid, α-galactosylceramide (α-GalCer), as our diluent instead of PBMC. Because this T-cell line is specific for a CD1d-presented antigen, we expected it would not stain with a CD1b-specific reagent and so facilitate measurement of the true background of our assay. Indeed, in the CD1d-specific T-cell line, we observed positive events at a frequency of 0.001% in the absence and presence of tetramer, fully one order of magnitude lower than what we observed using PBMC (data not shown). We then serially diluted the GMM-specific T cell line with the α-GalCer T-cell line and stained the resulting mixture with GMM-CD1b tetramer (Figure 3A). The number of tetramer positive events in each sample was compared to the number of events at the zero dilution using chi-square test of proportions. Across three replicate experiments, the 0.005% dilution was the lowest condition observed to be statistically significant, and the calculated limit of detection was determined to be 0.007% (Figure 3B).
Figure 3. Quantitative validation of the limit of detection of the CD1 tetramer assay.
The GMM-specific T-cell line was serially diluted with CD1d-restricted T-cells specific for α-GalCer and then stained with GMM-CD1b tetramer. (A) Representative staining from one experiment in which GMM T cells were diluted with CD1d-restricted T ranging from no GMM-specific T-cells (0%) up to only GMM-specific T-cells (100%). ‘No Tetramer’ refects the condition in which no tetramer was added to a 1:1 mixture of GMM T cells and α-GalCer T cells). (B) Summary analysis of three independent experiments. Data are depicted as a percent of total CD3+ cells. Shown are the mean and standard error of the mean at each dilution. Chi-square analysis demonstrates that 0.007% is the limit of detection of the assay (marked with an asterisk).
3.2.3. Determining the accuracy of the CD1 tetramer assay
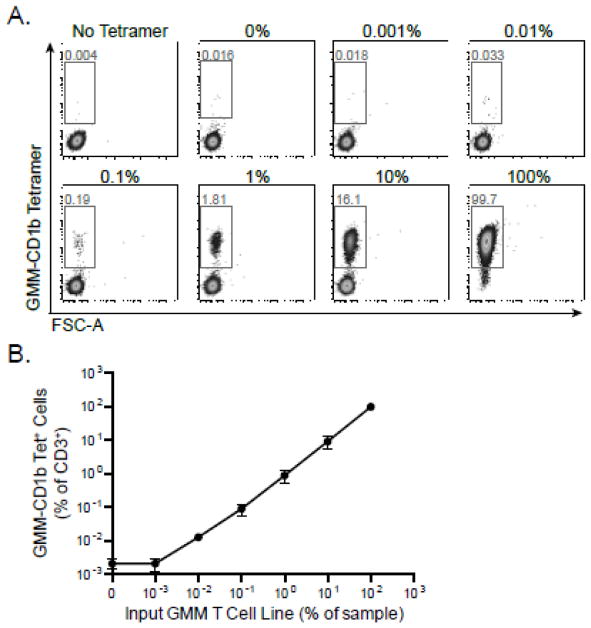
Accuracy refers to the ability of an assay to quantify the true value of a parameter that has already been determined by a “gold standard” reference assay. However, the true value of the number of antigen-specific T cells in a sample cannot be determined, since there is no standard reference assay to measure it. To circumvent this challenge, we took advantage of the fact that nearly 82% of the T-cells present in our GMM-specific T-cell line express T cell receptor alpha (TCR-α) variable segment, TRAV1-2 (data not shown). This provided a parallel method to quantify GMM-specific T cells and assess accuracy of the GMM-CD1b tetramer. TRAV1-2 is contained within a subset of GMM-specific T cells as well as mucosal associated invariant T (MAIT) cells (Porcelli et al., 1993; Van Rhijn et al., 2013a). Because MAIT cells are abundantly present in human blood, we wished to avoid using healthy donor PBMC as a diluent for these experiments. Instead, we used our α-GalCer-specific T cell line, which is negative for both GMM-CD1b tetramer and TRAV1-2 (data not shown). We serially diluted the GMM-specific cell line with the α-GalCer-specific cell line and measured the frequency of both GMM-specific T cells and TRAV1-2 expressing T cells (Figure 4A). These two quantities were highly correlated across all dilutions tested (Figure 4B). The CD1d-specific T-cell line is 98.5% positive for its antigen, and stains with TRAV1-2 at a background frequency of 0.017%. Whether the TRAV1-2 positive events are due to nonspecific staining or from TRAV1-2 positive cells that were inadvertently sorted and underwent in vitro expansion is unknown. Thus, our data reveal that the tetramer assay is accurate at detecting GMM-specific T cells at a frequency of 0.01%, which is slightly higher than the limit of detection.
Figure 4. Determining the accuracy of the CD1 Tetramer Assay.
A GMM-specific T cell line in which the majority of cells are known to express the T cell receptor alpha (TCR-α) variable segment, TRAV1-2, was serially diluted with an α-GalCer-specific CD1d-restricted cell line, which does not express TRAV1-2. The cells were co-stained with GMM-CD1b tetramer and anti-TRAV1-2. (A) Representative staining from three dilutions (0.01%, 0.1%, and 1%) in which the fraction of CD3+ cells that stain with GMM-CD1b tetramer (top) or with TRAV1-2 (bottom) is shown. (B) Correlation between GMM-CD1b and TRAV1-2 staining. TRAV1-2 staining in the absence of GMM-specific T cells is likely reflective of low frequency TRAV1-2+ cells present in the α-GalCer T cell line. Data are representative of three independent experiments.
3.2.4. Evaluating the linearity and range of the CD1 tetramer assay
To assess the linearity of the assay, GMM-specific T cells were serially diluted with PBMC from a healthy donor with samples ranging from 100% down to 0% GMM-specific T cells (Figure 5A). We performed linear regression using results from three independent experiments (Figure 5B). We noted a slope of 0.99 and adjusted R2 of 0.99, consistent with a linear fit to the data.
Figure 5. Evaluating the linearity and range of the CD1 Tetramer Assay.
GMM-specific T cells were serially diluted with PBMC from a healthy donor, and samples ranged from 100% to 0% GMM-specific T cells. The cells were stained with GMM-CD1b tetramer, and the frequency of GMM-specific T cells was measured. (A) Representative data in which the fraction of CD3+ T cells that stain with the tetramer over 10-fold dilutions of the GMM-specific T cell line. (B) Summary analysis of three independent experiments. Shown are the mean and standard error of the mean at each dilution. GMM tetramer staining is linear down to a precursor frequency of 0.001% input GMM-specific T cells.
We defined the range based on the linearity and the quantification limit as the set of dilutions in which the assay has acceptable precision, accuracy, and linearity. From the data presented in Figures 2, 3 and 4, we were able to demonstrate that this assay is repeatable with CV less than 20% and showed accurate identification of GMM-antigen specific T cells in a linear manner. We additionally demonstrated that the lower bounds of this assay has a limit of detection of 0.007% and is capable of detecting 100% of GMM-specific T cells in a given sample.
3.3. Frequencies of GMM-specific T cells in a South African cohort
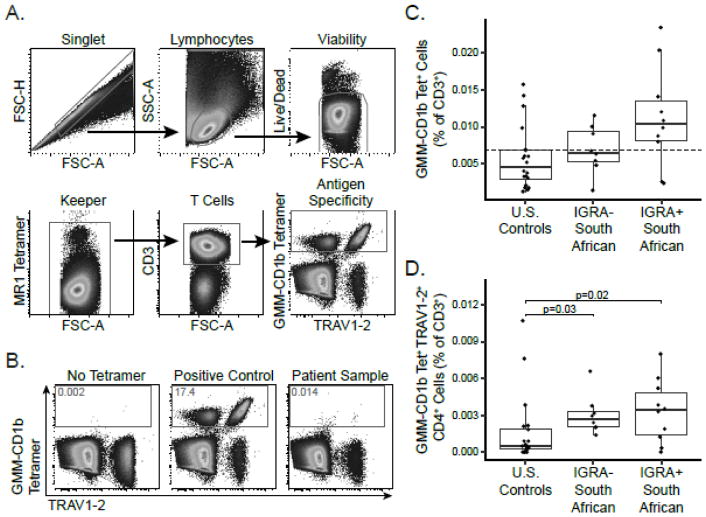
The data presented thus far show how we were able to address most of the validation parameters for our assay using GMM-specific T cell lines. The final parameter, specificity, required us to examine the performance of our assay in clinical samples where GMM-specific T cells are absent. Since there are no reference assays for GMM-specific T cells, we cannot formally assess specificity and thus consider our assay validated based on all the parameters that could be tested. However, we attempted to assess the specificity of our assay by examining the frequency of GMM-specific T cells in three groups of healthy subjects: U.S. adults at low risk of M.tb exposure (n=20) as well as IGRA-positive (n=10) and IGRA-negative (n=8) South African adolescents. To accomplish this, we incorporated GMM-CD1b tetramers into a multi-parameter flow cytometry panel that included additional CD1 tetramers and an MR1 tetramer (Supplementary Table 1). The data were collected in two batches in which individuals from each group were evenly distributed. The gating strategy proceeded from single cell events to lymphocytes by size gating and then to viable cells. A keeper gate using MR1 tetramers was then used to remove artifactual staining, and T cells were identified using anti-CD3 (Figure 6A). Donor PBMC mixed with GMM-specific T cell lines were used as a positive control. GMM-specific T cells in a test sample were identified on the basis of staining with GMM-CD1b tetramer and anti-TRAV1-2 with the inclusion gate set with respect to a No Tetramer control (Figure 6B, left) or positive control (Figure 6B, center).
Figure 6. Cross-sectional analysis of healthy controls in the United States and South Africa.
GMM-CD1b tetramers were incorporated into a multi-parameter flow cytometry assay to measure the frequencies and phenotypes of a variety of donor-unrestricted T cell populations. (A) The gating strategy proceeded from single cell events to lymphocytes by size gating and then to viable cells. A keeper gate using MR1 tetramers was then used to remove staining artifact, and T cells were identified using anti-CD3. Co-staining with GMM-CD1b tetramer and TRAV1-2 allowed the quantification of germline encoded mycolyl-reactive (GEM) T cells. (B) The tetramer positive gate was defined by a ‘No Tetramer’ negative control and a positive control using GMM-specific T cell lines diluted in donor PBMC (left and center). Representative staining from a South African adolescent (right). The frequency of GMM-CD1b tetramer positive T cells in the blood was quantified using cryopreserved PBMC obtained from three groups of healthy subjects: U.S. controls at low risk for M.tb exposure (n=20), South African adolescents with latent tuberculosis (IGRA-positive, n=10), and South African adolescents without latent tuberculosis (IGRA-negative, n=8). (C) Boxplots depict the median and interquartile range of tetramer-positive events as a fraction of CD3+ events. The limit of detection (0.007%) is noted as a dashed line. Frequency of tetramer-positive cells did not differ among the three groups (Kruskal-Wallis p=0.06). (D) Boxplots depict the median and interquartile range of the frequency of GEM T cells in the three groups defined as GMM-CD1b tetramer positive, TRAV1-2 positive, and CD4 positive. The frequency of GEM T cells is higher in the IGRA-negative (Dunn post-test p=0.03) and IGRA-positive adolescents (Dunn post-test p=0.02) compared to U.S. controls (Kruskal-Wallace p=0.02).
The GMM-specific T cells in a representative IGRA-positive adolescent were identified at a frequency of 0.014% of total CD3+ T cells, which was well-above the limit of detection (Figure 6B, right). Among 20 U.S. healthy donors, tetramer-positive T cells were present above the limit of detection in only 5 (25%) subjects. By contrast, tetramer-positive cells were present above the limit of detection in 8 (80%) of 10 IGRA-positive South African adolescents (Figure 6C). Although the frequency of GMM-specific T cells was not statistically different between U.S. healthy donors and South African adolescents (Kruskal-Wallace p=0.06), these data suggest that our validated assay detects GMM-specific T cells in a larger proportion of healthy subjects with latent TB infection. Germline-encoded mycolyl lipid-reactive (GEM) T cells are a subset of GMM-specific T cells that express the CD4 co-receptor and TRAV1-2 gene segment, and may be more specifically associated M.tb exposure (Van Rhijn et al., 2013a). Indeed, we found a significant difference in the frequency of GEM T cells among the three groups (Kruskal-Wallace p=0.02), which was driven by the difference observed between U.S. healthy donors and IGRA-negative South Africans (Dunn post-test p=0.03) and between U.S. healthy donors and IGRA-positive South Africans (Dunn post-test p=0.02) (Figure 6D). Notably, the frequency of GEM T cells did not differ between IGRA-positive and IGRA-negative adolescents (Dunn post-test p=0.40), perhaps a result of the strong M.tb infection pressure in South Africa. Taken together, these data establish specificity of the GMM-CD1b tetramer assay and demonstrate its application in human cohort studies.
4. DISCUSSION
In summary, we have qualitatively and quantitatively validated a flow cytometry assay to detect T cells specific for the mycobacterial glycolipid, glucose monomycolate (GMM). This assay is linear, reproducible, repeatable, precise, and accurate. Our experiments using human blood samples further reveal that CD1b-GMM tetramers have low background rates of detection in subjects at low risk for M.tb infection but convincingly reveal tetramer positive cells in subjects with known M.tb infection. Thus, this reagent also fulfills the criteria for specificity and could be used to detect changes in the frequency of GMM-specific T cells after infection or vaccination with mycobacteria.
We were able to detect GMM-specific T cells above the limit of detection in the blood of some healthy controls from Seattle who are at low risk for M.tb exposure or infection. Because GMM is known to be expressed by mycobacteria other than M.tb, it is possible that exposure to non-tuberculous mycobacteria that are ubiquitous in the environment can prime T-cell responses to GMM (Gotoh et al., 1991). Future studies will aim to develop tetramers using lipids that are uniquely expressed by M.tb. One candidate is sulfoglycolipids, which is dependent on the phoP transcription factor and for which we recently published a tetramer (Chesne-Seck et al., 2008; James et al., 2018). We also observed similar frequencies of GMM-CD1b tetramer positive and GEM T cells in IGRA-positive and IGRA-negative South African adolescents. These data are consistent with results of a CD1b functional assay that we published and confirm that T-cell responses to mycobacterial lipids and proteins are not necessarily concordant (Seshadri et al., 2015). GMM-CD1b T cells in IGRA-negative adolescents may represent exposure to non-tuberculous mycobacteria as above or even M.tb in an endemic setting.
T-cells specific for mycobacterial glycolipids have been shown to facilitate the lysis of M.tb infected cells through direct cell lysis or via production of IFN-γ, which activate antibacterial mechanisms within infected macrophages (Stenger et al., 1997). Pulmonary granulomas have recently been shown to express CD1b, so lipid-specific T cells might home to the lung and contribute to protective immunity at the site of infection (Chancellor et al., 2017). Lipid-specific T cells might also serve as a correlate of protective immunity in natural history and vaccine studies. Investigation of these myriad possibilities requires an assay that has been formally validated, as we have done here.
We expect that the main application of this assay in the near-term will be in evaluating the immunogenicity of novel whole cell mycobacterial vaccines. A number of these are currently in development and would be expected to boost T-cell responses to multiple peptide and non-peptide antigens (Scriba et al., 2016). SRL-172 is a heat killed non-tuberculous mycobacterial vaccine that was recently shown to prevent tuberculosis in a large study of HIV-infected adults in Tanzania (Von Reyn et al., 2010). A reformulated version (DAR-901) has recently entered Phase I studies. VPM1002 is a recombinant M. bovis strain that has been engineered to express bacterial listeriolysin and showed enhanced immunogenicity in a Phase I study (Grode et al., 2013). Finally, MTBVAC is an attenuated strain of M.tb that is currently in Phase I trials (Spertini et al., 2015). All of these whole cell vaccines have entered Phase I testing, so our assay could be used to determine the immunogenicity of T-cells specific for mycobacterial lipids. Our formally validated end point assay will allow regulatory agencies to determine whether lipid-specific T cell responses are induced or increased in frequency by these new vaccines.
We plan to further develop this assay to facilitate the use of limited samples from clinical trials, for example among South African infants and adults vaccinated with BCG, and are expanding this assay to allow identification and interrogation of other donor-unrestricted T cells, including iNKT cells and MAIT cells. A multi-tetramer assay would permit broad assessment of lipid-specific T-cell immunity during mycobacterial infection and vaccination in a manner similar to such panels for viral peptide antigens (Newell et al., 2009). If any whole cell vaccine is shown to be clinically efficacious, our assay can be used to evaluate lipid-specific T-cell responses as an immunologic correlate of protective immunity.
Supplementary Material
Acknowledgments
The work was supported by the the U.S. National Institutes of health (R01-AI125189, UM1 AI068618, P30 AI027757) and Doris Duke Charitable Foundation (CS). We thank Dr. Thomas Hawn, University of Washington, for providing PBMC for the limit of detection experiments and Charlotte James for assistance with intermediate precision experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altman JD, Moss Pa, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael aJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–6. doi: 10.1126/science.274.5284.94. https://doi.org/10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29(Suppl 1):S49–52. https://doi.org/citeulike-article-id:3416410. [PMC free article] [PubMed] [Google Scholar]
- Beckman EM, Porcelli Sa, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994 doi: 10.1038/372691a0. https://doi.org/10.1038/372691a0. [DOI] [PubMed]
- Benlagha K, Weiss a, Beavis a, Teyton L, Bendelac a. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. https://doi.org/10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [accessed 2.1.18];Bioanalytical Method Validation. 2013 URL https://www.fda.gov/downloads/drugs/guidances/ucm368107.pdf.
- Brennan PJ, Lehane DP, Thomas DW. Acylglucoses of the Corynebacteria and Mycobacteria. Eur J Biochem. 1970;13:117–123. doi: 10.1111/j.1432-1033.1970.tb00906.x. https://doi.org/10.1111/j.1432-1033.1970.tb00906.x. [DOI] [PubMed] [Google Scholar]
- Chancellor A, Tocheva AS, Cave-Ayland C, Tezera L, White A, Al Dulayymi JR, Bridgeman JS, Tews I, Wilson S, Lissin NM, Tebruegge M, Marshall B, Sharpe S, Elliott T, Skylaris C-K, Essex JW, Baird MS, Gadola S, Elkington P, Mansour S. CD1b-restricted GEM T cell responses are modulated by Mycobacterium tuberculosis mycolic acid meromycolate chains. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1708252114. 201708252. https://doi.org/10.1073/pnas.1708252114. [DOI] [PMC free article] [PubMed]
- Chesne-Seck ML, Barilone N, Boudou F, Asensio JG, Kolattukudy PE, Martín C, Cole ST, Gicquel B, Gopaul DN, Jackson M. A point mutation in the two-component regulator PhoP-PhoR accounts for the absence of polyketide-derived acyltrehaloses but not that of phthiocerol dimycocerosates in Mycobacterium tuberculosis H37Ra. J Bacteriol. 2008;190:1329–1334. doi: 10.1128/JB.01465-07. https://doi.org/10.1128/JB.01465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong A, Peña-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat Immunol. 2010;11:1102–1109. doi: 10.1038/ni.1956. https://doi.org/10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, Frelinger J, Jiang W, Newell EW, Ramey J, Davis MM, Kalams SA, De Rosa SC, Gottardo R. OpenCyto: An Open Source Infrastructure for Scalable, Robust, Reproducible, and Automated, End-to-End Flow Cytometry Data Analysis. PLoS Comput Biol. 2014:10. doi: 10.1371/journal.pcbi.1003806. https://doi.org/10.1371/journal.pcbi.1003806. [DOI] [PMC free article] [PubMed]
- Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Böhmer G, Prandi J, Mori L, Puzo G, De Libero G. Diacylated Sulfoglycolipids Are Novel Mycobacterial Antigens Stimulating CD1-restricted T Cells during Infection with Mycobacterium tuberculosis. J Exp Med J Exp Med. 2004;36491100:649–659. doi: 10.1084/jem.20031097. https://doi.org/10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh K, Mitsuyama M, Imaizumi S, Kawamura I, Yano I. Mycolic Acid-Containing Glycolipid as a Possible Virulence Factor of Rhodococcus equi for Mice. Microbiol Immunol. 1991;35:175–185. doi: 10.1111/j.1348-0421.1991.tb01546.x. https://doi.org/10.1111/j.1348-0421.1991.tb01546.x. [DOI] [PubMed] [Google Scholar]
- Grode L, Ganoza CA, Brohm C, Weiner J, Eisele B, Kaufmann SHE. Safety and immunogenicity of the recombinant BCG vaccine VPM1002 in a phase 1 open-label randomized clinical trial. Vaccine. 2013;31:1340–1348. doi: 10.1016/j.vaccine.2012.12.053. https://doi.org/10.1016/j.vaccine.2012.12.053. [DOI] [PubMed] [Google Scholar]
- Horton H, Thomas E, De Rosa SC. Optimization & Validation of an 8-colour ICS Assay To Quantify Antigen-Specific T Cells Induced by Vaccination. J Immunol Methods. 2007;323:39–54. doi: 10.1016/j.jim.2007.03.002. https://doi.org/10.1016/j.jim.2007.03.002.Optimization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James CA, Yu KKQ, Gilleron M, Prandi J, Yedulla VR, Moleda ZZ, Diamanti E, Khan M, Aggarwal VK, Reijneveld JF, Reinink P, Lenz S, Emerson RO, Scriba TJ, Souter MNT, Godfrey DI, Pellicci DG, Moody DB, Minnaard AJ, Seshadri C, Van Rhijn I. CD1b Tetramers Identify T Cells that Recognize Natural and Synthetic Diacylated Sulfoglycolipids from Mycobacterium tuberculosis. Cell Chem Biol. 2018 doi: 10.1016/j.chembiol.2018.01.006. https://doi.org/10.1016/j.chembiol.2018.01.006. [DOI] [PMC free article] [PubMed]
- Karadimitris A, Gadola S, Altamirano M, Brown D, Woolfson A, Klenerman P, Chen JL, Koezuka Y, Roberts IAG, Price DA, Dusheiko G, Milstein C, Fersht A, Luzzatto L, Cerundolo V. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci. 2001;98:3294–3298. doi: 10.1073/pnas.051604498. https://doi.org/10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmar AG, van Rhijn I, Cheng TY, Turner M, Seshadri C, Schiefner A, Kalathur RC, Annand JW, de Jong A, Shires J, Leon L, Brenner M, Wilson IA, Altman JD, Moody DB. CD1b tetramers bind αβ T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208:1741–1747. doi: 10.1084/jem.20110665. https://doi.org/10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasmar AG, Van Rhijn I, Magalhaes KG, Young DC, Cheng TY, Turner MT, Schiefner A, Kalathur RC, Wilson Ia, Bhati M, Gras S, Birkinshaw RW, Tan LL, Rossjohn J, Shires J, Jakobsen S, Altman JD, Moody DB. Cutting Edge: CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. J Immunol. 2013;191:4499–503. doi: 10.4049/jimmunol.1301660. https://doi.org/10.4049/jimmunol.1301660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, Mori L, Stenger S, De Libero G, Puzo G, Gilleron M. Mycolic Acids Constitute a Scaffold for Mycobacterial Lipid Antigens Stimulating CD1-Restricted T Cells. Chem Biol. 2009;16:82–92. doi: 10.1016/j.chembiol.2008.11.008. https://doi.org/10.1016/j.chembiol.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Ly D, Kasmar AG, Cheng TY, de Jong A, Huang S, Roy S, Bhatt A, van Summeren RP, Altman JD, Jacobs WR, Adams EJ, Minnaard AJ, Porcelli SA, Moody DB. CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med. 2013;210:729–741. doi: 10.1084/jem.20120624. https://doi.org/10.1084/jem.20120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahomed H, Hawkridge T, Verver S, Geiter L, Hatherill M, Abrahams D, Ehrlich R, Hanekom WA, Hussey GD. Predictive factors for latent tuberculosis infection among adolescents in a high-burden area in South Africa. Int J Tuberc Lung Dis. 2011;15:331–6. [PubMed] [Google Scholar]
- Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–54. doi: 10.1084/jem.192.5.741. https://doi.org/10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montamat-Sicotte DJ, Millington KA, Willcox CR, Hingley-Wilson S, Hackforth S, Innes J, Kon OM, Lammas DA, Minnikin DE, Besra GS, Willcox BE, Lalvani A. A mycolic acid-specific CD1-restricted T cell population contributes to acute and memory immune responses in human tuberculosis infection. J Clin Invest. 2011;121:2493–2503. doi: 10.1172/JCI46216. https://doi.org/10.1172/JCI46216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody DB, Guy MR, Grant E, Cheng TY, Brenner MB, Besra GS, Porcelli SA. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med. 2000;192:965–976. doi: 10.1084/jem.192.7.965. https://doi.org/10.1084/jem.192.7.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA. Structural requirements for glycolipid antigen recognition by CD1b- restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. https://doi.org/10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- Moody DB, Rosat J, Roura-mir C, Connor PBO, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB. T Cell Activation by Lipopetide antigens. Science. 2004;303:527–31. doi: 10.1126/science.1089353. https://doi.org/10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- Moody DB, Ulrichs T, Mühlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–888. doi: 10.1038/35009119. https://doi.org/10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods. 2009;6:497–499. doi: 10.1038/nmeth.1344. https://doi.org/10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TKA, Koets AP, Santema WJ, van Eden W, Rutten VPMG, Van Rhijn I. The mycobacterial glycolipid glucose monomycolate induces a memory T cell response comparable to a model protein antigen and no B cell response upon experimental vaccination of cattle. Vaccine. 2009;27:4818–4825. doi: 10.1016/j.vaccine.2009.05.078. https://doi.org/10.1016/j.vaccine.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. https://doi.org/10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. URL https://www.r-project.org/ [Google Scholar]
- Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–41. doi: 10.1126/science.1352912. https://doi.org/10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- Scriba TJ, Kaufmann SHE, Lambert PH, Sanicas M, Martin C, Neyrolles O. Vaccination against tuberculosis with whole-cell mycobacterial vaccines. J Infect Dis. 2016 doi: 10.1093/infdis/jiw228. https://doi.org/10.1093/infdis/jiw228. [DOI] [PubMed]
- Seshadri C, Lin L, Scriba TJ, Peterson G, Freidrich D, Frahm N, DeRosa SC, Moody DB, Prandi J, Gilleron M, Mahomed H, Jiang W, Finak G, Hanekom WA, Gottardo R, McElrath MJ, Hawn TR. T Cell Responses against Mycobacterial Lipids and Proteins Are Poorly Correlated in South African Adolescents. J Immunol. 2015;195:4595–4603. doi: 10.4049/jimmunol.1501285. https://doi.org/10.4049/jimmunol.1501285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva CL. Inflammation induced by mycolic acid-containing glycolipids of Mycobacterium bovis (BCG) Brazilian J Med Biol Res. 1985;18:327–335. [PubMed] [Google Scholar]
- Spertini F, Audran R, Chakour R, Karoui O, Steiner-Monard V, Thierry AC, Mayor CE, Rettby N, Jaton K, Vallotton L, Lazor-Blanchet C, Doce J, Puentes E, Marinova D, Aguilo N, Martin C. Safety of human immunisation with a live-attenuated Mycobacterium tuberculosis vaccine: A randomised, double-blind, controlled phase I trial. Lancet Respir Med. 2015;3:953–962. doi: 10.1016/S2213-2600(15)00435-X. https://doi.org/10.1016/S2213-2600(15)00435-X. [DOI] [PubMed] [Google Scholar]
- Stenger S, Mazzaccaro RJ, Uyemura K, Cho S, Barnes PF, Rosat JP, Sette A, Brenner MB, Porcelli SA, Bloom BR, Modlin RL. Differential effects of cytolytic T cell subsets on intracellular infection. Science. 1997;276:1684–7. doi: 10.1126/science.276.5319.1684. https://doi.org/10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]
- [accessed 2.1.18];Validation of Analytical Procedures: Text and Methodology Q2(R1) 2005 URL http://www.ich.org/fileadmin/Public_Web_Site/ICH_Products/Guidelines/Quality/Q2_R1/Step4/Q2_R1__Guideline.pdf.
- Van Rhijn I, Kasmar A, De Jong A, Gras S, Bhati M, Doorenspleet ME, De Vries N, Godfrey DI, Altman JD, De Jager W, Rossjohn J, Moody DB. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. 2013a;14:706–713. doi: 10.1038/ni.2630. https://doi.org/10.1038/ni.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn I, Ly D, Moody DB. CD1a, CD1b, and CD1c in immunity against mycobacteria. Adv Exp Med Biol. 2013b;783:181–197. doi: 10.1007/978-1-4614-6111-1_10. https://doi.org/10.1007/978-1-4614-6111-1_10. [DOI] [PubMed] [Google Scholar]
- Von Reyn CF, Mtei L, Arbeit RD, Waddell R, Cole B, MacKenzie T, Matee M, Bakari M, Tvaroha S, Adams LV, Horsburgh CR, Pallangyo K. Prevention of tuberculosis in Bacille Calmette-Guérin-primed, HIV-infected adults boosted with an inactivated whole-cell mycobacterial vaccine. AIDS. 2010;24:675–685. doi: 10.1097/QAD.0b013e3283350f1b. https://doi.org/10.1097/QAD.0b013e3283350f1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.