ABSTRACT
Fungi secrete a set of glycoside hydrolases and oxidoreductases, including lytic polysaccharide monooxygenases (LPMOs), for the degradation of plant polysaccharides. LPMOs catalyze the oxidative cleavage of glycosidic bonds after activation by an external electron donor. So far, only flavin-dependent oxidoreductases (from the auxiliary activity [AA] family AA3) have been shown to activate LPMOs. Here, we present LPMO activation by a pyrroloquinoline-quinone (PQQ)-dependent pyranose dehydrogenase (PDH) from Coprinopsis cinerea, CcPDH, the founding member of the recently discovered auxiliary activity family AA12. CcPDH contains a C-terminal family 1 carbohydrate binding module (CBM1), an N-terminal family AA8 cytochrome domain, and a central AA12 dehydrogenase domain. We have studied the ability of full-length CcPDH and its truncated variants to drive catalysis by two Neurospora crassa LPMOs. The results show that CcPDH indeed can activate the C-1-oxidizing N. crassa LPMO 9F (NcLPMO9F) and the C-4-oxidizing Neurospora crassa LPMO 9C (NcLPMO9C), that this activation depends on the cytochrome domain, and that the dehydrogenase and the LPMO reactions are strongly coupled. The two tested CcPDH-LPMO systems showed quite different efficiencies, and this difference disappeared upon the addition of free PQQ acting as a diphenol/quinone redox mediator, showing that LPMOs differ when it comes to their direct interactions with the cytochrome domain. Surprisingly, removal of the CBM domain from CcPDH had a considerable negative impact on the efficiency of the CcPDH-LPMO systems, suggesting that electron transfer in the vicinity of the substrate is beneficial. CcPDH does not oxidize cello-oligosaccharides, which makes this enzyme a useful tool for studying cellulose-oxidizing LPMOs.
IMPORTANCE Lytic polysaccharide monooxygenases (LPMOs) are currently receiving increasing attention because of their importance in degrading recalcitrant polysaccharides and their potential roles in biological processes, such as bacterial virulence. LPMO action requires an external electron donor, and fungi growing on biomass secrete various so-called glucose-methanol-choline (GMC) oxidoreductases, including cellobiose dehydrogenase, which can donate electrons to LPMOs. This paper describes how an enzyme not belonging to the GMC oxidoreductase family, CcPDH, can activate LPMOs, and it provides new insights into the activation process by (i) describing the roles of individual CcPDH domains (a dehydrogenase, a cytochrome, and a carbohydrate-binding domain), (ii) showing that the PDH and LPMO enzyme reactions are strongly coupled, (iii) demonstrating that LPMOs differ in terms of their efficiencies of activation by the same activator, and (iv) providing indications that electron transferring close to the substrate surface is beneficial for the overall efficiency of the CcPDH-LPMO system.
KEYWORDS: PQQ-dependent PDH, AA12, carbohydrate-binding module, lytic polysaccharide monooxygenase, LPMO, AA9, electron transfer
INTRODUCTION
Plant biomass is a promising raw material for industrial use because of its abundance in nature. However, industrial utilization of biomass via a biochemical conversion route is limited due to its resistance to enzymatic degradation (1). The recalcitrance of plant biomass resides in the plant cell wall, which is an insoluble and partly crystalline copolymeric structure composed of cellulose, hemicellulose, and lignin. The homopolymeric cellulose chains are organized into highly crystalline cellulose bundles, called cellulose microfibrils (2). Due to their compact structure, cellulose microfibrils are resistant to enzymatic depolymerization. Plant cell wall-degrading fungi, which are the main plant biomass decomposers in nature, express a set of glycoside hydrolases and oxidoreductases, including lytic polysaccharide monooxygenases (LPMOs), for the degradation of cellulose and other plant cell wall polysaccharides (3, 4).
LPMOs are copper-dependent enzymes that catalyze the oxidative cleavage of glycosidic bonds in various polysaccharides, such as cellulose (5–7), hemicelluloses (8–12), chitin (13), and starch (14). The known diversity in the substrate specificities of LPMOs and the abundances of LPMO-encoding genes in many microbes indicate that these enzymes are important in degradation of various biomasses (3, 4). LPMOs are currently classified into auxiliary activities (AA) families 9, 10, 11, 13, 14, and 15 in the Carbohydrate-Active enZymes (CAZy) database, which is based on the similarity of amino acid sequences (15). Fungal LPMOs belonging to the family AA9, also known as LPMO9s, contribute to the degradation of cellulose and hemicelluloses. Since these LPMOs accelerate the hydrolysis of cellulose by cellulases, they have recently attracted considerable attention as a key enzyme in efficient cellulose saccharification processes (16–19).
During its catalytic cycle, an LPMO requires an external electron donor for (re)activation (13, 20, 21). The electron donor can be a nonenzymatic reducing compound, such as ascorbic acid, lignin, and other plant biomass-derived phenols (13, 20, 22, 23). Alternatively, LPMOs can be activated by flavin-dependent oxidoreductases, directly or through plant-derived diphenols and quinones acting as redox mediators (17, 20, 24, 25). Flavoenzymes that have been shown to supply LPMOs with electrons are classified into the CAZy family AA3 and include cellobiose dehydrogenase (CDH; AA3_1), glucose dehydrogenase (AA3_2), and aryl-alcohol quinone oxidoreductase (AA3_2) (17, 20, 24–26). CDH, the first enzyme reported to be able to activate an LPMO (7, 17), is an extracellular flavocytochrome consisting of two domains: an N-terminal b-type heme-binding AA8 cytochrome domain and a C-terminal flavin adenine dinucleotide (FAD)-dependent AA3_1 dehydrogenase domain (27). Some CDHs have an additional family 1 carbohydrate-binding module (CBM1) at their C terminus (28). The AA3_1 dehydrogenase domain oxidizes cellobiose at the C-1 position to cellobiono-1,5-lactone, or longer cello-oligosaccharides to the corresponding lactones, via the reduction of the FAD cofactor (29). Then, in one scenario, two electrons from the FAD cofactor are sequentially transferred to an acceptor via the heme of the AA8 cytochrome b domain, which carries out two subsequent single-electron transfers (24, 30, 31). The external electron acceptor could be, for example, cytochrome c or an LPMO (24, 26, 31). In an alternative slower scenario, the FAD is reoxidized by direct electron transfer to molecular oxygen, leading to the production of hydrogen peroxide (20, 24, 32).
Recently, a new AA family, AA12, has been discovered, with the founding member being the pyrroloquinoline quinone (PQQ)-dependent pyranose dehydrogenase (PDH) from the plant saprophytic basidiomycete Coprinopsis cinerea (CcPDH), which is the first PQQ-dependent enzyme in eukaryotes (33). This enzyme has a three-domain structure similar to some CDHs. It is composed of an N-terminal heme b-binding AA8 cytochrome domain, a central PQQ-dependent AA12 dehydrogenase domain (instead of the AA3_1 cellobiose-oxidizing flavin domain in CDH), and a C-terminal CBM1. The PQQ cofactor binds with high affinity, and the binding constant, Kd, is 1.1 nM (33). The PQQ-dependent PDH domain shows oxidative activity toward d-glucosone (2-keto–d-glucose), l-fucose, and some rare pyranoses but is inactive toward glucose and cellobiose (33, 34). Of note, d-glucosone is the product of glucose oxidation by pyranose-2-oxidase, an enzyme found in several fungi. The N-terminal cytochrome domain of CcPDH exhibits 39% sequence identity with the cytochrome domain of PcCDH from the wood-rotting basidiomycete Phanerochaete chrysosporium, and spectral features, such as UV-Vis and resonance Raman spectra, are almost identical for CcPDH and PcCDH (34). The redox potentials of the cytochrome domains of the two enzymes are identical (130 mV, pH 7, versus standard hydrogen electrode [SHE]) (34, 35), indicating that, like CDH, CcPDH could also transfer electrons to LPMOs (Eo on the order of 250 mV versus SHE [20]).
Although there is no known role for CcPDH in plant cell wall degradation, such a role is suggested by the presence of the CBM1 and by the fact that the gene adjacent to the CcPDH-encoding gene encodes an LPMO (gene identification [ID], CC1G_09526). In the present study, we investigated the potential of CcPDH to drive catalysis by N. crassa LPMO 9C and N. crassa LPMO 9F (NcLPMO9C and NcLPMO9F, respectively), two well-characterized LPMOs from the ascomycete Neurospora crassa, and we compared CcPDH with ascorbic acid as an electron donor. Since, in contrast to CDH, CcPDH is inactive toward glucose and cello-oligosaccharides, reducing end-containing LPMO-generated products released from the cellulosic substrate are not oxidized by CcPDH. Thus, CcPDH is a good candidate to study the activity and product profiles of cellulose-active LPMOs. Furthermore, as shown here, product formation by CcPDH can be analyzed chromatographically, simultaneously with the analysis of LPMO-generated products, generating detailed insights into how the two enzymes work together.
RESULTS
Activation of LPMO9s by CcPDH.
In a previous study (36), we showed that the PQQ-dependent 2-keto–d-glucose dehydrogenase (2KGDH) from Pseudomonas aureofaciens, a bacterial homolog of CcPDH, oxidizes the aldehyde group of d-glucosone at the C-1 position, yielding 2-keto-d-gluconic acid, which is a precursor of erythorbic acid (also called isoascorbic acid). In the present study, we used external standards to identify the products generated by CcPDH from d-glucosone and l-fucose. The retention times of the reaction products generated from d-glucosone and l-fucose were 16.6 min and 8.9 min, i.e., identical to the retention times of 2-keto-d-gluconic acid and l-fuconic acid, respectively (see Fig. S1 in the supplemental material). Thus, as expected, CcPDH oxidizes the C-1 position of these substrates. In order to exclude the possibility that LPMOs may be activated by these reaction products of CcPDH, we incubated LPMOs with 2-keto-d-gluconic acid, l-fuconic acid, or products generated by CcPDH from after enzyme inactivation by boiling. None of these compounds were able to activate the LPMOs (Fig. S2 and S3). The control experiments depicted in Fig. S1 and S2 also showed that quantification of PDH action is possible in the case of d-glucosone as the substrate, since the oxidized product, 2-keto-d-gluconic acid, gives a clear sharp peak with a good response factor in the required concentration range (Fig. S1A). In the case of l-fucose, detection of l-fuconic acid was not sensitive enough in the required concentration range. In principle, the sharp l-fucose peak with a high response factor (Fig. S3A) would allow for the quantification of PDH action through measuring l-fucose consumption.
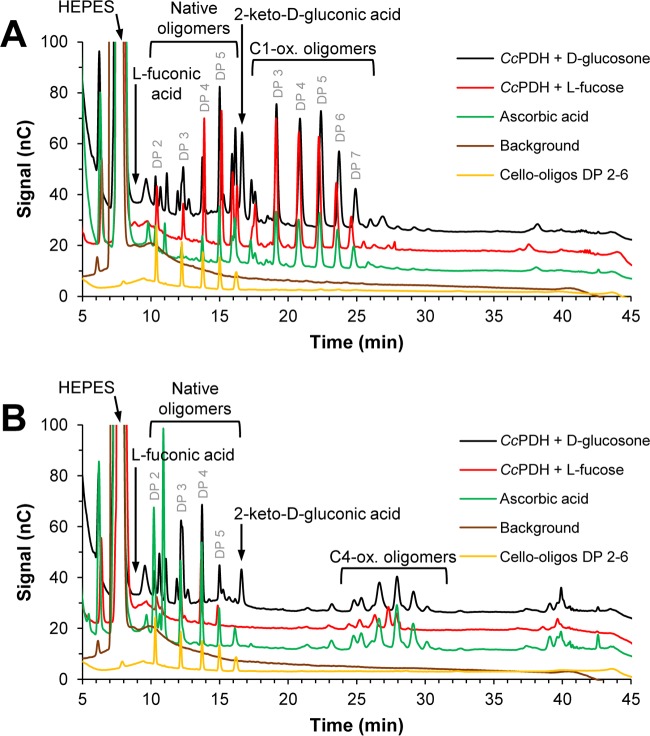
Figure 1 shows that when supplied with substrate, CcPDH can activate NcLPMO9C (NCU02916), a C-4-oxidizing LPMO with a C-terminal CBM1 domain, and NcLPMO9F (NCU03328), a C-1-oxidizing single-domain LPMO (8, 24, 37). LPMO9 activity was monitored by measuring the activity against phosphoric acid-swollen cellulose (PASC), and the released oligomeric products were detected by high-performance anion-exchange chromatography (HPAEC). In the presence of ascorbic acid, NcLPMO9F generated C-1-oxidized cello-oligosaccharides, while NcLPMO9C generated C-4-oxidized cello-oligosaccharides. Similar product profiles were observed when supplying the reaction mixtures with CcPDH and either d-glucosone or l-fucose (Fig. 1), showing that product specificity was not affected by varying the electron donor. Analysis of product formation over time (Fig. 2) revealed that NcLPMO9F performed slightly better when activated by CcPDH than when activated by ascorbic acid, whereas NcLPMO9C performed better when activated with ascorbic acid. Control reactions with free PQQ (without CcPDH) showed negligible (but larger than zero) amounts of LPMO products (Fig. 3), whereas the addition of PQQ-loaded CcPDH as such (i.e., without a PDH substrate) did lead to significant product formation (Fig. 3), explaining why the product formation curves shown in Fig. 2 start from product levels higher than 0 at time (t) = 0 (see Fig. 2 legend).
FIG 1.
Products generated by NcLPMO9F (A) and NcLPMO9C (B) from PASC in the presence of various electron donating systems. Reaction mixtures containing 0.2% (wt/vol) PASC in 20 mM HEPES buffer at pH 7.0 were incubated at 30°C for 10 min. The reaction mixtures contained either 1 mM ascorbic acid (green) or 1 μM CcPDH with 1 mM d-glucosone (black) or l-fucose (red) as the electron donating system. Control experiments with LPMO only, i.e., without any added component involved in electron donation, (brown) are also shown. Peak annotations are based on earlier work (see, e.g., Isaksen et al. [58]) and on control runs with standards as displayed in Fig. S1. The signal of amperometric detection is expressed in nanocoulombs (nC). Note that the response factor for l-fuconic acid is very low, explaining why there is no clearly visible peak. See Fig. S1 for more details. Numbers above peaks representing native or C-1-oxidized oligosaccharides indicate the number of sugars in the oligomer (DP, degree of polymerization); exact DP assignments for C-4-oxidized products were not possible. Cello-oligos, cello-oligosaccharides; ox., oxidizing.
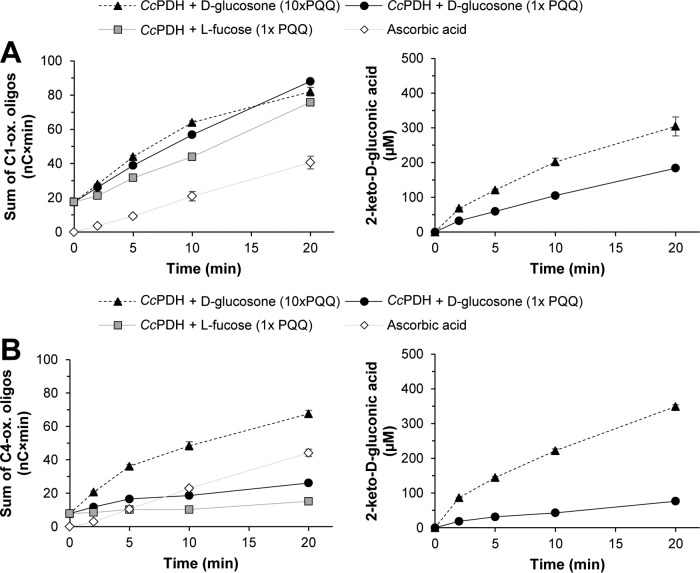
FIG 2.
Product accumulation by NcLPMO9F (A) and NcLPMO9C (B) in reactions with PASC, in the presence of various electron donating systems. Reaction mixtures containing 0.2% (wt/vol) PASC in 20 mM HEPES buffer at pH 7.0 were incubated at 30°C for 20 min. The reaction mixtures contained either 1 mM ascorbic acid (white diamond) or 1 μM CcPDH with 1 mM d-glucosone (black circle, solid line) or l-fucose (gray square) or 1 μM CcPDH with 9 μM additional PQQ and 1 mM d-glucosone (black triangle, dashed line) as electron donating system. At t = 0, reactions were started by adding ascorbic acid or the CcPDH substrate. In the case of the oxidized oligosaccharides (left graphs), the values provided represent the sum of the peak areas for all detectable individual oxidized species with different degrees of polymerization (3–7). CcPDH products generated from d-glucosone (right graphs) were quantified using 2-keto-d-gluconic acid standards. Control reaction mixtures containing no CcPDH did not show product increase over time. Control reaction mixtures containing PQQ-loaded CcPDH without its substrate showed some product formation (Fig. 3), which explains why LPMO products are observed at t = 0. The reactions were set up in such a way that maximal background levels of oxidized cello-oligosaccharides had been reached by t = 0 (for details, see Fig. 3). Reactions were carried out in triplicates; error bars show the standard deviation.
FIG 3.
Control experiments showing the background levels of oxidized cello-oligosaccharides due to LPMO activation by free PQQ or PQQ-saturated CcPDH variants in the absence of CcPDH substrates. The two panels show background levels of oxidized cello-oligosaccharides generated by NcLPMO9F (A) and NcLPMO9C (B) due to activation by free PQQ (white circles) and PQQ-saturated CcPDH variants (full-length, AA8-AA12-CBM1, black circles; two-domain, AA8-AA12, dark gray circles; single-domain, AA12, light gray circles). Reaction mixtures contained 0.2% (wt/vol) PASC, 1 μM LPMO, and 1 μM PQQ or PQQ-saturated CcPDH variant in 20 mM HEPES buffer at pH 7.0 and were incubated at 30°C. Note that the reaction mixtures did not contain a substrate for CcPDH. Reactions were initiated at t = 0 by the addition of PQQ or PQQ-saturated CcPDH variant.
We also tested the effect of additional PQQ on the LPMO-CcPDH system (Fig. 2). For both LPMOs, the addition of free PQQ, a potential redox mediator, enhanced LPMO activity, leading to an increase in the formation of oxidized products by the enzymes. Interestingly, free PQQ also promoted CcPDH activity (Fig. 2A and B, right), underpinning the strong coupling between the LPMO and the PDH reactions. The effect of adding free PQQ, in addition to PQQ-loaded CcPDH, on LPMO activity was clearly larger in the case of NcLPMO9C, which is the LPMO that seemed less well activated by CcPDH than by NcLPMO9F. It is worth noting that PDH activity is LPMO dependent (Fig. 2A and B, right) and that this dependency disappeared when additional PQQ was added. In the presence of additional PQQ, d-glucosone oxidation levels were similar in the two reactions. This suggests that the efficiency of electron transfer from CcPDH to an LPMO varies between LPMOs and that limitations in this transfer may be alleviated by redox mediators.
The functions of the AA8, AA12, and CBM1 domains in activating LPMOs.
In order to investigate whether CcPDH activates an LPMO only through its AA8 domain or also through its AA12 domain, we produced three variants of CcPDH, the full-length CcPDH (AA8-AA12-CBM1), the AA8-AA12 two-domain enzyme, and the AA12 single-domain enzyme, and studied their ability to oxidize d-glucosone and activate the LPMOs in reactions similar to those described above.
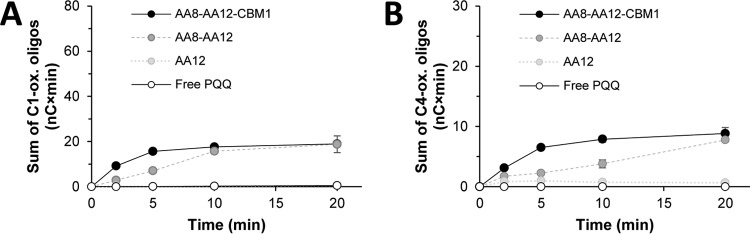
Figure 4 shows that the activity of the AA12 domain depended strongly on the presence of the AA8 domain. Removal of the AA8 domain almost completely abolished LPMO activity and AA12 activity. It has been shown previously that the presence of an electron acceptor, such as phenazine methosulfate (PMS), is crucial for the dehydrogenase activity of an isolated AA12 domain (38). The fact that the single AA12 domain oxidized d-glucosone to a much lower extent than the full-length enzyme shows that, in the absence of the AA8 domain, the presence of an electron-accepting LPMO is not sufficient to (efficiently) reoxidize the active site of the AA12 domain. Accordingly, Fig. 4 also shows that the single AA12 domain cannot activate the two LPMOs. Thus, with an LPMO being the only available electron acceptor, the AA8 domain is needed for mediating electron transfer from the PQQ cofactor in the AA12 domain to the LPMO, which activates the LPMO and is essential for reoxidizing CcPDH.
FIG 4.
The effect of individual CcPDH domains on CcPDH activity and on the activation of NcLPMO9F (A, C, and E, left graphs) and NcLPMO9C (B, D, and F, right graphs). (A to D) Curves for the LPMO (A and B) and the PDH (C and D) reactions, respectively. Progress curves for three CcPDH domain variants are shown: solid black line, full-length (AA8-AA12-CBM1); dashed dark gray line, two-domain (AA8-AA12); dotted light gray line, single-domain (AA12)variants. Reaction mixtures contained 0.2% (wt/vol) PASC, 1 μM LPMO, and 1 μM CcPDH variant in 20 mM HEPES buffer at pH 7.0 and were incubated at 30°C. Reactions were initiated at t = 0 by the addition of d-glucosone (1,000 μM), which initiates the CcPDH reaction and, thus, formation of 2-keto-d-gluconic acid. (E and F) Accumulation of oxidized cello-oligosaccharides (sum of peak areas; black bars) and 2-keto-d-gluconic acid (quantified using a standard; white bars) after 10 min. The values shown have been corrected for the product levels at t = 0 (see t = 20-min levels in Fig. 3 and legend to Fig. 2 for further explanation). Reactions were carried out in triplicate; error bars show the standard deviation.
Surprisingly, deletion of the CBM1, leaving an AA8-AA12 protein that would seem fully capable of electron transfer, also reduced the performances of both the PDH and the LPMO in reactions with PASC, indicating that electron transfer is impaired also in this case (Fig. 4E and F). This effect of deleting the CBM1 from CcPDH is similar for the two LPMOs and thus seems independent of the presence (NcLPMO9C) or absence (NcLPMO9F) of a CBM1 domain in the LPMO itself. Of note, the low-level background activation of LPMOs by CcPDH in the absence of substrate is also reduced upon removal of the CBM (Fig. 3). These observations suggest that adsorption of CcPDH to cellulose through the CBM1 domain, which would promote electron transfer to the LPMO to occur in the vicinity of the LPMO substrate, could enhance the efficiency of the electron transfer chain involving CcPDH, LPMO, and substrate.
Dose-response experiments.
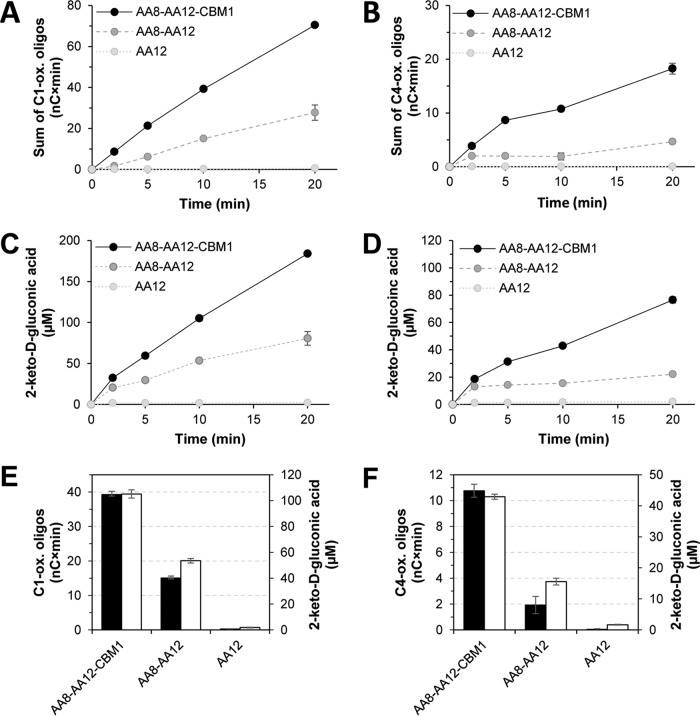
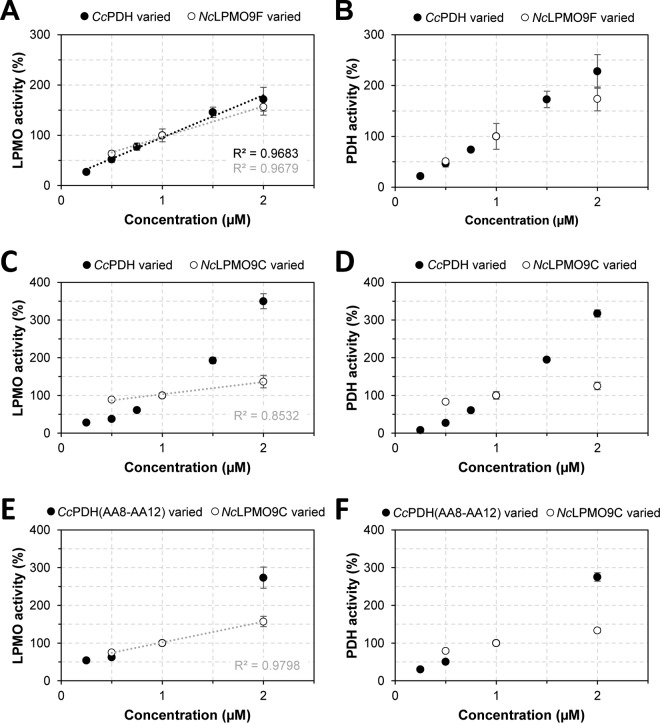
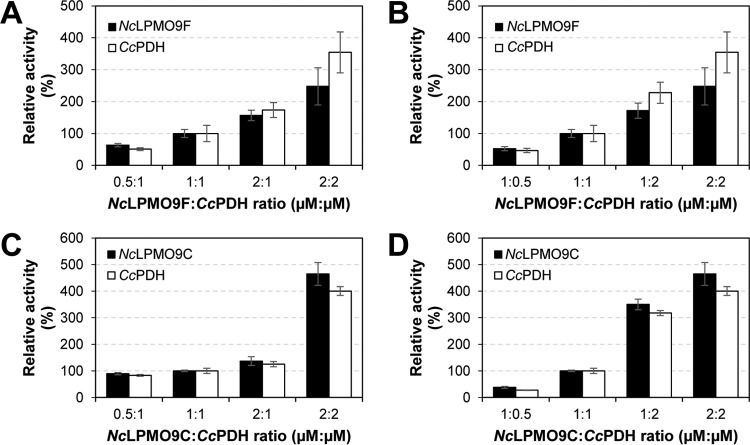
To gain further insight into the rate-limiting steps in the CcPDH-LPMO system, we coincubated NcLPMO9F or NcLPMO9C with CcPDH while varying the NcLPMO9-to-CcPDH ratios and keeping the other reaction parameters constant. In the reactions with NcLPMO9F and CcPDH, the amount of oxidized products released by the two enzymes depended equally on the two enzymes and correlated linearly and with their concentrations (Fig. 5A and B and 6A and B). The NcLPMO9C-CcPDH system showed a remarkably different pattern. While variation in the CcPDH concentration had a large and superstoichiometric effect on both the PDH and LPMO activities, the effects of varying the NcLPMO9C concentration were much smaller (Fig. 5C and D and 6C and D). Similar trends were observed when using the CBM1-free variant of CcPDH (Fig. 5E and F).
FIG 5.
The effect of varying enzyme ratios on LPMO (A, C, and E) and PDH (B, D, and F) activity. Reaction mixtures contained 0.2% (wt/vol) PASC and 1 mM d-glucosone in 20 mM HEPES buffer (pH 7.0) and were incubated at 30°C for 10 min. The coincubated enzymes were NcLPMO9F and CcPDH (A and B), NcLPMO9C and CcPDH (C and D), and NcLPMO9C and 2-domain CcPDH (AA8-AA12) (E and F). In the reaction, either the PDH concentration (black symbols) or the LPMO concentration (white symbols) was varied, while the concentration of the other enzyme was kept constant at 1 μM. (A, C, and E) Relative LPMO activities, which were quantified based on the accumulation of oxidized cello-oligosaccharides. (B, D, and F) Relative PDH activities, which were quantified based on the accumulation of 2-keto-d-gluconic acid.
FIG 6.
Effect of varying enzyme concentrations and ratios on LPMO and PDH activity. The graphs show a selection of the data shown in Fig. 5 and include one extra reaction (2 μM each enzyme; same reaction conditions). The coincubated enzymes were: (A and B) NcLPMO9F and CcPDH, primarily varying the concentration of NcLPMO9F (A) or CcPDH (B); and (C and D) NcLPMO9C and CcPDH, primarily varying the concentration of NcLPMO9C (C) or CcPDH (D). The black bars indicate LPMO activity, quantified by the accumulation of oxidized cello-oligosaccharide products. The white bars indicate PDH activity, quantified by the accumulation of 2-keto-d-gluconic acid.
DISCUSSION
Enzymatic electron donors for LPMO9s can act in different manners. Single-domain enzymes transfer electrons to LPMO9s through diphenol/quinone mediators and, albeit less efficiently, sometimes also directly (20, 25). Multidomain enzymes, such as MtCDH from Myriococcum thermophilum, can activate LPMO9s through diphenol/quinone mediators or through an appended AA8 cytochrome domain (7, 17, 20). Measurements of electron transfer by MtCDH have shown that the AA8 cytochrome domain is essential for electron transfer from MtCDH to an LPMO9, whereas electron transfer directly from the AA3_1 dehydrogenase domain is absent or very slow (24). Notably, the catalytic rates of LPMOs are orders of magnitude lower than the measured rates of CDH reoxidation by an LPMO (20), meaning that a lack of measurable electron transfer does not necessarily mean a complete inability to drive LPMO activity.
CcPDH has a multidomain structure similar to that of CDH, except that its central catalytic domain is a PQQ-dependent AA12 dehydrogenase instead of an FAD-dependent AA3_1 dehydrogenase domain. Figure 2 shows that when supplied with substrate, CcPDH indeed can drive the LPMO reaction, and it also shows that electron consumption by the LPMO reaction is required to maintain CcPDH activity. Importantly, a comparison of the results obtained with NcLPMO9C and NcLPMO9F showed that NcLPMO9F is more easily activated than NcLPMO9C. However, when supplying a potential redox mediator (free PQQ), the two CcPDH-LPMO systems work approximately equally quickly, as judged from the observed CcPDH activity (Fig. 2, right; note that the amount of added PQQ is much smaller than the amount of generated product, meaning that the PQQ acts as a mediator, not as a reductant). These observations indicate that in the absence of a mediator, direct reoxidation of CcPDH by NcLPMO9C is slower than reoxidation by NcLPMO9F. In this respect, it is worth noting that analyses of the rates of reoxidation of two CDHs from Neurospora crassa by LPMOs showed that, depending on the conditions and the CDH, those rates are up to one order of magnitude lower for NcLPMO9C than for NcLPMO9F (20) (NcLPMO9F gave the fastest kinetics of the four tested NcLPMOs). Thus, LPMOs seem to differ considerably when it comes to their ability to interact with the AA8 cytochrome domain attached to AA3 and AA12 dehydrogenases.
Figure 4 shows that the AA8 cytochrome domain is crucial in driving the LPMO reaction. It has previously been suggested that the heme b propionate of the AA8 domain in MtCDH (31.8% sequence identity with the AA8 domain of CcPDH) may have favorable interactions with the copper active site of NcLPMO9F (24), and nuclear magnetic resonance (NMR) experiments with NcLPMO9C and MtCDH have shown that there indeed are specific interactions between the cytochrome domain and the catalytic center of the LPMO (39). To date, the AA8 domain has been found in three types of fungal proteins, namely, AA3_1 CDH, AA12 PDH, and cellulose-binding cytochrome b562 (CBCyt b562; an N-terminal AA8 domain fused to a C-terminal CBM1), and these AA8 domains are highly similar to each other in terms of amino acid sequence and spectral, biochemical, and electrochemical features (34, 40, 41). PDHs, CDHs, and CBCyt b562 are extracellular proteins, can bind to cellulose, and often carry a cellulose-binding CBM1 module. This may be taken to suggest that these AA8 domains have evolved to enable interprotein electron transfer on cellulose surfaces in order to facilitate the extracellular redox pathways of fungal plant cell wall degradation.
Cellulose binding through a CBM1 enhances the efficiency of many plant cell wall-degrading enzymes, such as cellulases and hemicellulases, LPMOs, and CDHs (34, 42–45). On the other hand, the catalytic modules of these enzymes also have affinity for the substrate. Basidiomycetous CDHs, generally lacking a CBM1, possess a putative substrate-binding site located in the flavin-dependent AA3_1 domain (27, 46–48). The AA3_1 domains of ascomycetous CDHs lack this binding site (49), and these CDHs usually contain a C-terminal CBM1. A possible role of this CBM in promoting LPMO activation by CDH has not yet been assessed. Similarly to ascomycetous CDHs, CcPDH possesses a CBM1 at the C terminus, which enables its binding to both amorphous and crystalline celluloses (34). Figure 4 shows that the CBM1 domain of CcPDH is of considerable importance for the efficiency of the CcPDH-LPMO system, since the presence of CBM1 in CcPDH had a positive effect on both LPMO and CcPDH activities. This surprising and novel finding indicates that it is beneficial that electron transfer leading to the activation of the LPMO takes place in the vicinity of the crystalline cellulose surface. It is conceivable that LPMO action is enhanced by increasing the proximity between the substrate and the electron source for LPMOs. Likewise, CcPDH activity may be enhanced by the presence of an electron acceptor that will rapidly oxidize cellulose and be available for accepting new electrons. For the LPMO, activation close to the substrate also may have additional beneficial effects, since it has been shown that activated (reduced) LPMOs that are not bound to substrate suffer from oxidative self-inactivation (50). It is noteworthy that the effect of removing the CBM1 from CcPDH was largest for NcLPMO9C, which contains a CBM1, when acting on its preferred substrate, PASC. It is possible that LPMOs carrying a CBM1 benefit more from the presence of a CBM1 in CcPDH than single-domain LPMOs, as the CBM1 domain directs the PDH on parts of the cellulose surface where NcLPMO9C is associated, thus further increasing the interaction efficiency of the two enzymes.
Recently, it has been proposed that LPMOs may use H2O2 as a cosubstrate and even prefer H2O2 over O2 (51). In this scenario, stoichiometric and continuous transfer of electrons to the copper site in the LPMO is not necessary if H2O2 is supplied. Instead, the LPMO is primed by a one-electron reduction of the copper, after which the primed enzyme can carry out multiple reactions using H2O2. In the absence of externally supplied H2O2 but in the presence of reductant, H2O2 can be generated by the LPMO itself (52) or by reactions involving O2 and the reductant, or, as some would argue, the LPMOs can use another O2-based mechanism (21, 50). Although not very visible in Fig. 4, the AA12 domain can drive LPMO reactions at very low rates, analogous to what has been observed for certain flavin-dependent AA3_2 dehydrogenase domains (25). Slow reoxidation of the dehydrogenase by O2 and concomitant production of hydrogen peroxide, combined with LPMO reduction by low concentrations of free PQQ, or perhaps even directly by the AA12 domain itself, could explain why the AA12 domain alone can, although very slowly, drive LPMO reactions.
Our current view on LPMO catalysis is that in the absence of externally supplied H2O2 (as is the case for all reactions shown in this study) and in the presence of a reductant, the LPMO generates its own H2O2 from O2, which, notably, would require two electrons. Thus, no matter which LPMO mechanism one assumes, under the conditions used in this study, each LPMO catalysis consumes two electrons. This explains why the observed activities of CcPDH, which needs to get rid of two electrons in each cycle, and the LPMO, which needs to acquire two electrons in each cycle, are tightly coupled. A key remaining question for all scenarios without externally added H2O2 is how and when the second electron is transferred to the catalytic center, perhaps especially for scenarios where the LPMO is bound to substrate and has a less accessible copper site. It is worth noting that transfer of the second electron likely is rate limiting, since one-electron reduction of the copper in a free LPMO (i.e., transfer of the first electron) is orders of magnitude faster than LPMO catalysis driven by, e.g., ascorbic acid (20).
The experiments with varied enzyme ratios show large differences between the CcPDH-NcLPMO9F and the CcPDH-NcLPMO9C systems. In the CcPDH-NcLPMO9F system, the electron transfer and enzyme catalytic rates are apparently so high that, in the concentration range used in Fig. 5, the two enzymes limit each other. In other words, as shown in Fig. 5, increased amounts of reducing equivalents generated by increasing CcPDH concentrations can be absorbed by a stable amount of LPMO, while an increased demand for reducing equivalents by increasing amounts of LPMO can be supplied by a stable amount of CcPDH. For the CcPDH-NcLPMO9C system, the situation is very different. Increasing the concentration of NcLPMO9C only marginally increased LPMO and PDH activities, whereas increasing the concentration of CcPDH led to a superstoichiometric increase in LPMO and PDH activities. The observation regarding the concentration of CcPDH is not straightforward to explain but is likely due to a weak interaction between NcLPMO9C and CcPDH, as discussed above. In this case, electron transfer may be so slow that the first, presumably fast, reduction of the copper (the “priming” reduction) becomes rate limiting. Increasing the PDH concentration would then have a double effect, since more NcLPMO9C molecules will become catalytically competent whereas, at the same time, production of the cosubstrate H2O2 will increase.
Small molecular reductants, such as ascorbic acid, are regularly used as an electron donor for LPMOs, while such compounds are generally unstable, may lead to side reactions, and could lead to the inactivation of LPMOs at higher concentrations (26, 51). Studies on the activity of a chitin-active LPMO have shown that stable reaction kinetics are more easily obtained using CDH (with lactose as the substrate) to deliver electrons. Unfortunately, CDH interferes with the accurate determination of cello-oligosaccharides produced by LPMOs due to oxidation of the reducing ends of C-4-oxidized and native cello-oligosaccharides. CcPDH, on the other hand, is inactive against the most common sugars in biomass (such as d-glucose, as shown in Fig. S4, as well as d-xylose and d-mannose [33]) and will not oxidize the most common oligosaccharides produced by LPMO activity. Moreover, the reaction products of CcPDH (i.e., 2-keto-d-gluconic acid and, to a much lesser degree, l-fuconic acid) can be distinguished from oxidized cello-oligosaccharides in HPAEC-pulsed amperometric detection (HPAEC-PAD) analyses, which allow not only proper detection of oxidized cello-oligosaccharides but also simultaneous monitoring of dehydrogenase and LPMO activities. Thus, we believe that CcPDH will turn out to be a useful tool in future studies of LPMOs and associated redox enzyme systems involved in biomass degradation.
MATERIALS AND METHODS
Materials.
Pyrroloquinoline quinone (PQQ) disodium salt and l-fucose were purchased from Wako Pure Chemical Industries (Osaka, Japan). d-Glucosone (2-keto–d-glucose) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). 2-Keto-d-gluconic acid hemicalcium salt hydrate, l-fucono-1,4-lactone, cytochrome c from bovine heart, and Avicel PH 101 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Phosphoric acid-swollen cellulose (PASC) was prepared from Avicel as described by Wood (53).
Preparation of enzymes.
Full-length CcPDH (AA8-AA12-CBM1), truncated two-domain CcPDH (AA8-AA12), and the single-domain PQQ-dependent dehydrogenase domain (AA12) were heterologously expressed in the methylotrophic yeast Pichia pastoris and purified as described previously (33). AA8-AA12 and AA12 were subjected to additional purification using a Superdex 75 gel filtration column (GE Healthcare, Sweden) run in 20 mM HEPES (pH 7.0) and 150 mM NaCl. Recombinant NcLPMO9C and NcLPMO9F from Neurospora crassa were prepared according to Kittl et al. (52). Protein purity was confirmed by SDS-PAGE analysis. NcLPMO9C and NcLPMO9F were saturated with Cu(II) by incubating enzymes with an excess of CuSO4 (at a 3:1 molar ratio of copper/enzyme) for 30 min at room temperature, as described previously (55). After saturation, excess CuSO4 was removed using PD-10 desalting columns (GE Healthcare).
Holo-forms (i.e., PQQ- and Ca2+-saturated forms) of CcPDH and its truncated versions were prepared by incubating 20 μM enzyme with 200 μM PQQ and 2 mM CaCl2 for 30 min on ice, followed by removal of excess PQQ and CaCl2 using PD-10 desalting columns. PQQ saturation was confirmed by analyzing the enzyme activity of holo-CcPDH with and without the addition of extra (5 μM) PQQ and (50 μM) CaCl2. The enzyme activity of CcPDH was measured spectrophotometrically by monitoring the reduction of cytochrome c at 550 nm and 30°C (34). The reaction mixture contained 200 nM holo-enzyme, 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-NaOH (pH 7.0), 1 mM l-fucose, and 50 μM cytochrome c in a total volume of 500 μl. The addition of extra PQQ and CaCl2 to the putatively saturated enzyme had no effect on enzyme activity, indicating that the holo-enzyme was fully saturated (Fig. S5).
Protein concentrations were measured using the Bradford method (Bio-Rad's protein assay kit; Bio-Rad Laboratories, Hercules, CA, USA) with bovine serum albumin as a standard.
Activity assays.
For detecting LPMO activity, 1 μM LPMO was incubated with 0.2% (wt/vol) PASC in 20 mM HEPES buffer (pH 7.0) at 30°C for 10 min. All reaction mixtures contained 100 μM CaCl2. As an electron donor, either 1 mM ascorbic acid or 1 μM CcPDH holoenzyme supplemented with 1 mM d-glucosone or l-fucose was supplied, unless otherwise stated. Control reactions were set up in the same manner, leaving out one or more components, as indicated in the figure legends. In the dose-response experiments, the concentrations of NcLPMO9s and CcPDHs were varied between 0.25 and 2.0 μM. Reactions were stopped by boiling for 5 min. All reaction products were analyzed by high-performance anion-exchange chromatography (HPAEC) using a Dionex ICS 3000 system (Dionex, Sunnyvale, CA, USA) equipped with a CarboPac PA1 precolumn (2 by 50 mm), a CarboPac PA1 main column (2 by 250 mm), and a detector for pulsed amperometric detection (PAD), applying a 50-min gradient that has been described previously (56). Chromatograms were collected and analyzed with Chromeleon 7.0. In the dose-response experiments, the relative activity of CcPDH was calculated as the ratio of peak areas for 2-keto-d-gluconic acid in the given reaction sample and in the analogous sample from the reaction mixture containing 1 μM LPMO and 1 μM CcPDH. In the case of the LPMOs, the corresponding ratios were calculated for each oxidized oligosaccharide (with different degree of polymerization), and the mean of those ratios is presented as relative activity. In the case of NcLPMO9C, which produces large amounts of (partly on column-generated) native oligosaccharides (57), relative activities were calculated using peaks for C-4-oxidized products. The corresponding values were also calculated using peaks for the native products, which generally corresponded well with the values based on oxidized products, adding confidence to the results. All experiments were performed in triplicate.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bastien Bissaro for valuable discussions.
The work was carried out as part of the Biomim project funded by the Norwegian Research Council through grant 243663 and was partly cofunded by JSPS KAKENHI (grant 14J08518 to K.U. and 15H04526 to M.Y.).
We declare no conflicts of interest regarding the contents of this article.
A.V. and K.U. designed, performed, and analyzed the experiments. K.U. constructed vectors for expression of CcPDH domain variants. V.G.H.E. and M.Y. supervised the work and contributed to the design and analysis of the experiments. All authors contributed to writing the manuscript, reviewed the results, and approved the final version of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00156-18.
REFERENCES
- 1.Chundawat SP, Beckham GT, Himmel ME, Dale BE. 2011. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng 2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 2.Ramos LP. 2003. The chemistry involved in the steam treatment of lignocellulosic materials. Quím Nova 26:863–871. [Google Scholar]
- 3.Mba Medie F, Davies GJ, Drancourt M, Henrissat B. 2012. Genome analyses highlight the different biological roles of cellulases. Nat Rev Microbiol 10:227–234. doi: 10.1038/nrmicro2729. [DOI] [PubMed] [Google Scholar]
- 4.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VGH. 2012. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5:45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsberg Z, Vaaje-Kolstad G, Westereng B, Bunaes AC, Stenstrom Y, MacKenzie A, Sorlie M, Horn SJ, Eijsink VGH. 2011. Cleavage of cellulose by a CBM33 protein. Protein Sci 20:1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen JC, Johansen KS, Krogh KB, Jorgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A 108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips CM, Beeson WT, Cate JH, Marletta MA. 2011. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol 6:1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 8.Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WGT, Ludwig R, Horn SJ, Eijsink VGH, Westereng B. 2014. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci U S A 111:6287–6292. doi: 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou S, Fanuel M, Ropartz D, Rogniaux H, Gimbert I, Record E, Berrin JG. 2015. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol Biofuels 8:90. doi: 10.1186/s13068-015-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frommhagen M, Sforza S, Westphal AH, Visser J, Hinz SW, Koetsier MJ, van Berkel WJ, Gruppen H, Kabel MA. 2015. Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol Biofuels 8:101. doi: 10.1186/s13068-015-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nekiunaite L, Petrovic DM, Westereng B, Vaaje-Kolstad G, Hachem MA, Várnai A, Eijsink VGH. 2016. FgLPMO9A from Fusarium graminearum cleaves xyloglucan independently of the backbone substitution pattern. FEBS Lett 590:3346–3356. doi: 10.1002/1873-3468.12385. [DOI] [PubMed] [Google Scholar]
- 12.Kojima Y, Várnai A, Ishida T, Sunagawa N, Petrovic DM, Igarashi K, Jellison J, Goodell B, Alfredsen G, Westereng B, Eijsink VGH, Yoshida M. 2016. Characterization of an LPMO from the brown-rot fungus Gloeophyllum trabeum with broad xyloglucan specificity and its action on cellulose-xyloglucan complexes. Appl Environ Microbiol 82:6557–6572. doi: 10.1128/AEM.01768-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sorlie M, Eijsink VGH. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 14.Vu VV, Beeson WT, Span EA, Farquhar ER, Marletta MA. 2014. A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci U S A 111:13822–13827. doi: 10.1073/pnas.1408090111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen J-C, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, Lo Leggio L. 2010. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 17.Langston JA, Shaghasi T, Abbate E, Xu F, Vlasenko E, Sweeney MD. 2011. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl Environ Microbiol 77:7007–7015. doi: 10.1128/AEM.05815-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Arantes V, Pribowo A, Gourlay K, Saddler JN. 2014. Substrate factors that influence the synergistic interaction of AA9 and cellulases during the enzymatic hydrolysis of biomass. Energy Environ Sci 7:2308–2315. doi: 10.1039/C4EE00891J. [DOI] [Google Scholar]
- 19.Müller G, Várnai A, Johansen KS, Eijsink VGH, Horn SJ. 2015. Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol Biofuels 8:187. doi: 10.1186/s13068-015-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kracher D, Scheiblbrandner Felice SAK, Breslmayr E, Preims M, Ludwicka K, Haltrich D, Eijsink VGH, Ludwig R. 2016. Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 352:1098–1101. doi: 10.1126/science.aaf3165. [DOI] [PubMed] [Google Scholar]
- 21.Walton PH, Davies GJ. 2016. On the catalytic mechanisms of lytic polysaccharide monooxygenases. Curr Opin Chem Biol 31:195–207. doi: 10.1016/j.cbpa.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Westereng B, Cannella D, Agger JW, Jørgensen H, Larsen Andersen M, Eijsink VGH, Felby C. 2015. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep 5:18561. doi: 10.1038/srep18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frommhagen M, Koetsier MJ, Westphal AH, Visser J, Hinz SW, Vincken JP, van Berkel WJ, Kabel MA, Gruppen H. 2016. Lytic polysaccharide monooxygenases from Myceliophthora thermophila C1 differ in substrate preference and reducing agent specificity. Biotechnol Biofuels 9:186. doi: 10.1186/s13068-016-0594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan TC, Kracher D, Gandini R, Sygmund C, Kittl R, Haltrich D, Hallberg BM, Ludwig R, Divne C. 2015. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat Commun 6:7542. doi: 10.1038/ncomms8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garajova S, Mathieu Y, Beccia MR, Bennati-Granier C, Biaso F, Fanuel M, Ropartz D, Guigliarelli B, Record E, Rogniaux H, Henrissat B, Berrin JG. 2016. Single-domain flavoenzymes trigger lytic polysaccharide monooxygenases for oxidative degradation of cellulose. Sci Rep 6:28276. doi: 10.1038/srep28276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loose JS, Forsberg Z, Kracher D, Scheiblbrandner S, Ludwig R, Eijsink VGH, Vaaje-Kolstad G. 2016. Activation of bacterial lytic polysaccharide monooxygenases with cellobiose dehydrogenase. Protein Sci 25:2175–2186. doi: 10.1002/pro.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henriksson G, Pettersson G, Johansson G, Ruiz A, Uzcategui E. 1991. Cellobiose oxidase from Phanerochaete chrysosporium can be cleaved by papain into two domains. Eur J Biochem 196:101–106. doi: 10.1111/j.1432-1033.1991.tb15791.x. [DOI] [PubMed] [Google Scholar]
- 28.Henriksson G, Johansson G, Pettersson G. 2000. A critical review of cellobiose dehydrogenases. J Biotechnol 78:93–113. doi: 10.1016/S0168-1656(00)00206-6. [DOI] [PubMed] [Google Scholar]
- 29.Ayers AR, Ayers SB, Eriksson KE. 1978. Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur J Biochem 90:171–181. doi: 10.1111/j.1432-1033.1978.tb12588.x. [DOI] [PubMed] [Google Scholar]
- 30.Igarashi K, Momohara I, Nishino T, Samejima M. 2002. Kinetics of inter-domain electron transfer in flavocytochrome cellobiose dehydrogenase from the white-rot fungus Phanerochaete chrysosporium. Biochem J 365:521–526. doi: 10.1042/bj20011809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samejima M, Phillips RS, Eriksson KE. 1992. Cellobiose oxidase from Phanerochaete chrysosporium. Stopped-flow spectrophotometric analysis of pH-dependent reduction. FEBS Lett 306:165–168. [DOI] [PubMed] [Google Scholar]
- 32.Cameron MD, Aust SD. 2000. Kinetics and reactivity of the flavin and heme cofactors of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochemistry 39:13595–13601. doi: 10.1021/bi000862c. [DOI] [PubMed] [Google Scholar]
- 33.Matsumura H, Umezawa K, Takeda K, Sugimoto N, Ishida T, Samejima M, Ohno H, Yoshida M, Igarashi K, Nakamura N. 2014. Discovery of a eukaryotic pyrroloquinoline quinone-dependent oxidoreductase belonging to a new auxiliary activity family in the database of carbohydrate-active enzymes. PLoS One 9:e104851. doi: 10.1371/journal.pone.0104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda K, Matsumura H, Ishida T, Samejima M, Ohno H, Yoshida M, Igarashi K, Nakamura N. 2015. Characterization of a novel PQQ-dependent quinohemoprotein pyranose dehydrogenase from Coprinopsis cinerea classified into auxiliary activities family 12 in carbohydrate-active enzymes. PLoS One 10:e0115722. doi: 10.1371/journal.pone.0115722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Igarashi K, Verhagen MF, Samejima M, Schulein M, Eriksson KE, Nishino T. 1999. Cellobiose dehydrogenase from the fungi Phanerochaete chrysosporium and Humicola insolens. A flavohemoprotein from Humicola insolens contains 6-hydroxy-FAD as the dominant active cofactor. J Biol Chem 274:3338–3344. [DOI] [PubMed] [Google Scholar]
- 36.Umezawa K, Takeda K, Ishida T, Sunagawa N, Makabe A, Isobe K, Koba K, Ohno H, Samejima M, Nakamura N, Igarashi K, Yoshida M. 2015. A novel pyrroloquinoline quinone-dependent 2-keto-d-glucose dehydrogenase from Pseudomonas aureofaciens. J Bacteriol 197:1322–1329. doi: 10.1128/JB.02376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vu VV, Beeson WT, Phillips CM, Cate JHD, Marletta MA. 2014. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc 136:562–565. doi: 10.1021/ja409384b. [DOI] [PubMed] [Google Scholar]
- 38.Takeda K, Matsumura H, Ishida T, Yoshida M, Igarashi K, Samejima M, Ohno H, Nakamura N. 2016. pH-dependent electron transfer reaction and direct bioelectrocatalysis of the quinohemoprotein pyranose dehydrogenase. Biochem Biophys Res Commun 477:369–373. doi: 10.1016/j.bbrc.2016.06.096. [DOI] [PubMed] [Google Scholar]
- 39.Courtade G, Wimmer R, Rohr AK, Preims M, Felice AK, Dimarogona M, Vaaje-Kolstad G, Sorlie M, Sandgren M, Ludwig R, Eijsink VGH, Aachmann FL. 2016. Interactions of a fungal lytic polysaccharide monooxygenase with beta-glucan substrates and cellobiose dehydrogenase. Proc Natl Acad Sci U S A 113:5922–5927. doi: 10.1073/pnas.1602566113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida M, Ohira T, Igarashi K, Nagasawa H, Aida K, Hallberg BM, Divne C, Nishino T, Samejima M. 2001. Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris. Biosci Biotechnol Biochem 65:2050–2057. doi: 10.1271/bbb.65.2050. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida M, Igarashi K, Wada M, Kaneko S, Suzuki N, Matsumura H, Nakamura N, Ohno H, Samejima M. 2005. Characterization of carbohydrate-binding cytochrome b562 from the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 71:4548–4555. doi: 10.1128/AEM.71.8.4548-4555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomme P, Van Tilbeurgh H, Pettersson G, Van Damme J, Vandekerckhove J, Knowles J, Teeri T, Claeyssens M. 1988. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem 170:575–581. [DOI] [PubMed] [Google Scholar]
- 43.Ståhlberg J, Johansson G, Pettersson G. 1993. Trichoderma reesei has no true exo-cellulase: all intact and truncated cellulases produce new reducing end groups on cellulose. Biochim Biophys Acta 1157:107–113. doi: 10.1016/0304-4165(93)90085-M. [DOI] [PubMed] [Google Scholar]
- 44.Linder M, Teeri TT. 1997. The roles and function of cellulose-binding domains. J Biotechnol 57:15–28. doi: 10.1016/S0168-1656(97)00087-4. [DOI] [PubMed] [Google Scholar]
- 45.Borisova AS, Isaksen T, Dimarogona M, Kognole AA, Mathiesen G, Várnai A, Røhr AK, Payne CM, Sorlie M, Sandgren M, Eijsink VGH. 2015. Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J Biol Chem 290:22955–22969. doi: 10.1074/jbc.M115.660183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renganathan V, Usha SN, Lindenburg F. 1990. Cellobiose-oxidizing enzymes from the lignocellulose-degrading basidiomycete Phanerochaete chrysosporium: interaction with microcrystalline cellulose. Appl Microbiol Biotechnol 32:609–613. doi: 10.1007/BF00173735. [DOI] [Google Scholar]
- 47.Henriksson G, Salumets A, Divne C, Pettersson G. 1997. Studies of cellulose binding by cellobiose dehydrogenase and a comparison with cellobiohydrolase 1. Biochem J 324:833–838. doi: 10.1042/bj3240833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samejima M, Ohkubo T, Igarashi K, Isogai A, Kuga S, Sugiyama J, Eriksson K-EL. 1997. The behaviour of Phanerochaete chrysosporium cellobiose dehydrogenase on adsorption to crystalline and amorphous celluloses. Biotechnol Appl Biochem 25:135–141. [Google Scholar]
- 49.Harreither W, Sygmund C, Augustin M, Narciso M, Rabinovich ML, Gorton L, Haltrich D, Ludwig R. 2011. Catalytic properties and classification of cellobiose dehydrogenases from ascomycetes. Appl Environ Microbiol 77:1804–1815. doi: 10.1128/AEM.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bissaro B, Rohr AK, Muller G, Chylenski P, Skaugen M, Forsberg Z, Horn SJ, Vaaje-Kolstad G, Eijsink VGH. 2017. Oxidative cleavage of polysaccharides by monocopper enzymes depends on H2O2. Nat Chem Biol 13:1123–1128. doi: 10.1038/nchembio.2470. [DOI] [PubMed] [Google Scholar]
- 51.Bissaro B, Rohr AK, Skaugen M, Forsberg Z, Horn SJ, Vaaje-Kolstad G, Eijsink VGH. 2016. Fenton-type chemistry by a copper enzyme: molecular mechanism of polysaccharide oxidative cleavage. bioRxiv doi: 10.1101/097022. [DOI]
- 52.Kittl R, Kracher D, Burgstaller D, Haltrich D, Ludwig R. 2012. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol Biofuels 5:79. doi: 10.1186/1754-6834-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood TM. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol 160:19–25. doi: 10.1016/0076-6879(88)60103-0. [DOI] [Google Scholar]
- 54.Reference deleted.
- 55.Loose JS, Forsberg Z, Fraaije MW, Eijsink VGH, Vaaje-Kolstad G. 2014. A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett 588:3435–3440. doi: 10.1016/j.febslet.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 56.Westereng B, Agger JW, Horn SJ, Vaaje-Kolstad G, Aachmann FL, Stenstrom YH, Eijsink VGH. 2013. Efficient separation of oxidized cello-oligosaccharides generated by cellulose degrading lytic polysaccharide monooxygenases. J Chromatogr A 1271:144–152. doi: 10.1016/j.chroma.2012.11.048. [DOI] [PubMed] [Google Scholar]
- 57.Westereng B, Arntzen MO, Aachmann FL, Várnai A, Eijsink VGH, Agger JW. 2016. Simultaneous analysis of C1 and C4 oxidized oligosaccharides, the products of lytic polysaccharide monooxygenases acting on cellulose. J Chromatogr A 1445:46–54. doi: 10.1016/j.chroma.2016.03.064. [DOI] [PubMed] [Google Scholar]
- 58.Isaksen T, Westereng B, Aachmann FL, Agger JW, Kracher D, Kittl R, Ludwig R, Haltrich D, Eijsink VGH, Horn SJ. 2014. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem 289:2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.