Antibiotic nonsusceptibility was consistently associated with higher risks of recurrent bacteremia, but the estimated number of additional recurrent episodes in the Netherlands (40 per year) was rather limited.
Keywords: antibiotic resistance, recurrent bacteremia, population attributable effect, acquired antibiotic resistance
Abstract
Background
Direct health effects of antibiotic resistance are difficult to assess. We quantified the risk of recurrent bacteremia associated with resistance.
Methods
We extracted antimicrobial susceptibility testing data on blood isolates from the Dutch surveillance system for antimicrobial resistance between 2008 and 2017. First and first recurrent (4–30 days) bacteremia episodes were categorized as susceptible, single nonsusceptible, or co-nonsusceptible to third-generation cephalosporins without or with carbapenems (Enterobacteriaceae), ceftazidime without or with carbapenems (Pseudomonas species), aminopenicillins without or with vancomycin (Enterococcus species), or as methicillin-sensitive/-resistant S. aureus (MSSA/MRSA). We calculated risks of recurrent bacteremia after nonsusceptible vs susceptible first bacteremia, estimated the crude population attributable effect of resistance for the Netherlands, and calculated risks of nonsusceptible recurrent bacteremia after a susceptible first episode.
Results
Risk ratios for recurrent bacteremia after a single- and co-nonsusceptible first episode, respectively, vs susceptible first episode, were 1.7 (95% confidence interval [CI], 1.5–2.0) and 5.2 (95% CI, 2.1–12.4) for Enterobacteriaceae, 1.3 (95% CI, 0.5–3.1) and 5.0 (95% CI, 2.9–8.5) for Pseudomonas species, 1.4 (95% CI, 1.2–1.7) and 1.6 (95% CI, 0.6–4.2) for Enterococcus species, and 1.6 (95% CI, 1.1–2.4) for MRSA vs MSSA. The estimated population annual number of recurrent bacteremias associated with nonsusceptibility was 40. The risk of nonsusceptible recurrent bacteremia after a susceptible first episode was at most 0.4% (Pseudomonas species).
Conclusions
Although antibiotic nonsusceptibility was consistently associated with higher risks of recurrent bacteremia, the estimated annual number of additional recurrent episodes in the Netherlands (40) was rather limited.
Antibiotic resistance is considered a major threat for human health. Although infections caused by antibiotic-resistant bacteria have frequently been associated with increased morbidity, higher mortality, and enhanced healthcare costs, the direct and attributable effects of antibiotic resistance have been less accurately quantified. Detrimental health effects of antibiotic resistance may result from inappropriate empiric therapy, from reduced effectiveness of definite therapy with last-resort antibiotics, or a higher frequency of adverse events created by such antibiotics. Several studies have quantified the effects of antibiotic resistance, usually in patients with bacteremia, and mostly on survival (either 30 days after bacteremia or during hospitalization) and length of hospital stay [1–3]. Yet, associations between antibiotic resistance and patient outcome can be confounded and can be affected by competing events [4, 5]. Adjustment for patient-related and disease-associated factors typically reduces the observed crude associations between antibiotic resistance and patient outcome. As a result, the consequences of antibiotic resistance for patient health are not accurately quantified.
An important sequela of antibiotic resistance could be the occurrence of recurrent infection, either because of delays in providing appropriate antibiotic therapy or because of suboptimal definite antibiotic therapy. Furthermore, the risk of resistance development during antibiotic therapy and the subsequent risk of developing recurrent infections have never been quantified. In the current study, we quantified the association between antibiotic susceptibility and the occurrence of recurrent bacteremia, and to what extent antibiotic susceptibility had changed between first and recurrent infections.
METHODS
Setting
Since 1 January 2008, results of antimicrobial susceptibility testing, including underlying minimum inhibitory concentration (MIC) values and inhibition zone diameters, of isolates routinely tested in Dutch medical microbiology laboratories are collected in the Infectious Diseases Surveillance Information System–Antimicrobial Resistance (ISIS-AR) [6]. This surveillance system is a combined initiative of the Dutch Ministry of Health, Welfare and Sport and the Netherlands Society for Medical Microbiology, and is coordinated by the Centre for Infectious Disease Control at the National Institute for Public Health and the Environment. Data on blood cultures from 41 of 57 Dutch medical microbiology laboratories (72%) were available in ISIS-AR in May 2017 and were included in the current study. Four of these laboratories exclusively serve a university hospital, 1 exclusively serves general practices and long-term care facilities, and 36 serve both general hospitals and general practices. As both partners of ISIS-AR agreed that no identifiable personal data are collected, individual patient consent was not required for the current study.
Data Extraction and Preparation
On 17 May 2017, data on all blood isolates of Enterobacteriaceae, Pseudomonas species, Enterococcus species, and Staphylococcus aureus present in the database were extracted. After excluding polymicrobial episodes (defined as ≥2 isolates of different microorganisms in samples taken ≤1 day apart from the same patient, 12.6% of all isolates), first and recurrent episodes of bacteremia were identified. A first episode of bacteremia was defined as the first isolate from a blood culture per patient or an isolate not preceded by an isolate of the same species from the same patient in the previous 30 days. A recurrent episode was defined as an isolate of the same bacteria (at species level) from a blood culture taken between 4 and 30 days after the first episode. In patients with multiple recurrent episodes, only the first recurrent isolate was included. Isolates were categorized as either susceptible, single nonsusceptible, or co-nonsusceptible bacteremias based on their antibiotic susceptibility test results for the most frequently used antibiotics for the specific species. Categorization for susceptibility was based on reinterpretation of MIC values according to clinical breakpoints set by the European Committee on Antimicrobial Susceptibility Testing (version 7.0; 2017, EUCAST), using results from gradient tests above automated MIC determinations. Zone diameter values were excluded, as methods for disk diffusion are not harmonized across laboratories and exact inhibition zone diameters are often not reported to ISIS-AR. Enterobacteriaceae were categorized as single nonsusceptible if they were intermediate (I) or resistant (R) to third-generation cephalosporins (ceftriaxone/cefotaxime or ceftazidime), and as co-nonsusceptible when I/R to carbapenems (imipenem or meropenem) as well. Pseudomonas species were categorized as single nonsusceptible if I/R to ceftazidime, and as co-nonsusceptible when I/R to carbapenems (imipenem or meropenem) additionally. Enterococcus species were categorized as single nonsusceptible (ampicillin-resistant Entercocci [ARE]) when I/R to aminopenicillins (amoxicillin or ampicillin). When ARE were I/R to vancomycin as well, they were categorized as co-nonsusceptible. Staphylococcus aureus was categorized as methicillin-resistant S. aureus (MRSA) or methicillin-sensitive S. aureus (MSSA), based on (in hierarchical order) results from mecA or pbp2 confirmation tests, cefoxitin susceptibility (I/R), or flucloxacillin or oxacillin susceptibility (I/R to at least one), as reported by the laboratory.
Statistical Analysis
We calculated absolute and relative risks (including 95% confidence intervals [CIs]) of recurrent bacteremia in patients with single nonsusceptible and co-nonsusceptible vs susceptible first bacteremias for each of the 4 causative pathogen groups, using Poisson regression models in SAS software (version 9.4; SAS Institute, Cary, North Carolina). Furthermore, we estimated the crude attributable effect of antibiotic resistance per pathogen group, defined as the difference in absolute risk of recurrent episodes after a first episode caused by nonsusceptible strains (single and co-nonsusceptible episodes combined) minus the absolute risk after a first episode caused by susceptible strains, multiplied by the number of first episodes caused by nonsusceptible strains ([ARI/R − ARS] × number of first episodesI/R). We extrapolated these results to estimate the total number of recurrent episodes attributable to antibiotic resistance in the Netherlands. For that, we assumed inclusion of 60% of all blood culture results in ISIS-AR during the study period of 9.4 years (based on the average estimated hospital coverage in the Netherlands throughout the study period). Last, we calculated absolute risks of a recurrent bacteremia caused by a nonsusceptible pathogen after a first episode caused by a susceptible strain. To evaluate the impact of the arbitrarily chosen follow-up time of 30 days for recurrent bacteremia, we performed 2 sensitivity analyses using a maximum of 15 and 60 days of follow-up.
RESULTS
On 17 May 2017, the ISIS-AR database contained 170677 isolates of Enterobacteriaceae, Pseudomonas species, Enterococcus species, and S. aureus from monomicrobial blood cultures. A total number of 116720 (68.4%) fulfilled the criteria of first episode or first recurrent episode of bacteremia and were included in this study. Of a total of 75963 Enterobacteriaceae, results for ceftriaxone/cefotaxime/ceftazidime and imipenem/meropenem could not be interpreted according to EUCAST clinical breakpoints in 4450 (5.9%) and 4585 (6.0%) isolates, respectively, and original susceptibility as reported by the local laboratory was used instead. This also applied to 536 (11.7%) and 498 (10.3%) among 4844 Pseudomonas species isolates for ceftazidime and imipenem/meropenem, respectively, and to 3307 (28.0%) and 2416 (20.4%) among 11824 Enterococcus species isolates for amoxicillin/ampicillin and vancomycin, respectively.
Enterobacteriaceae
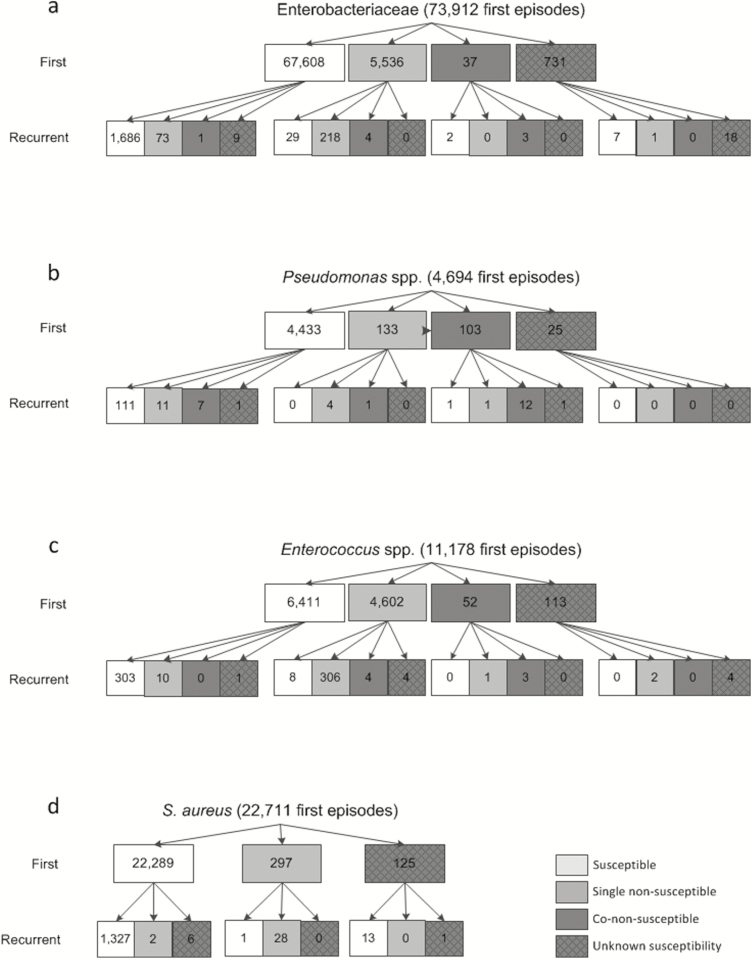
A number of 75963 Enterobacteriaceae bacteremia episodes were included, of which 73912 were classified as first episodes (97.3%) and 2051 (2.7%) as first recurrent episodes (Figure 1A). Overall, 5873 (7.7%) bacteremia episodes were caused by nonsusceptible Enterobacteriaceae. Co-nonsusceptibility to carbapenem antibiotics among nonsusceptible Enterobacteriaceae was observed in 37 of 5573 (0.7%) first episode isolates and in 8 of 300 (2.7%) isolates from recurrent episodes.
Figure 1.
Absolute number of first and recurrent bacteremia episodes between 1 January 2008 and 17 May 2017, by pathogen group and susceptibility category. A, Susceptible: susceptible to third-generation cephalosporins and carbapenems. Single nonsusceptible: (intermediate) resistant to third-generation cephalosporins, susceptible to carbapenems. Co-nonsusceptible: (intermediate) resistant to third-generation cephalosporins and carbapenems. B, Susceptible: susceptible to ceftazidime and carbapenems. Single nonsusceptible: (intermediate) resistant to ceftazidime, susceptible to carbapenems. Co-nonsusceptible: (intermediate) resistant to ceftazidime and carbapenems. C, Susceptible: susceptible to aminopenicillins and vancomycin. Single nonsusceptible: (intermediate) resistant to aminopenicillins, susceptible to vancomycin. Co-nonsusceptible: (intermediate) resistant to aminopenicillins and vancomycin. D, Susceptible: methicillin-sensitive Staphylococcus aureus. Single nonsusceptible: methicillin-resistant Staphylococcus aureus.
The absolute risk of recurrent bacteremia was 2.6% (1769/67608 episodes), 4.5% (251/5536 episodes), and 13.5% (5/37 episodes) for subjects with a first episode caused by either susceptible, single nonsusceptible, or co-nonsusceptible Enterobacteriaceae, respectively. Compared to susceptible first episodes, risk ratios for single and co-nonsusceptible first bacteremias were 1.7 (95% CI, 1.5–2.0) and 5.2 (95% CI, 2.1–12.4), respectively (Table 1). The absolute risk of a recurrent infection caused by nonsusceptible Enterobacteriaceae after a first episode caused by a susceptible isolate was 0.1% (74/67608 episodes; Figure 1A). The likelihood that recurrent bacteremia was caused by a nonsusceptible isolate was 87.9% (225/256 episodes) when the first episode was caused by a nonsusceptible isolate.
Table 1.
Susceptibility Distribution of First Bacteremias and Absolute and Relative Risk of Recurrent Bacteremia Following Single or Co-nonsusceptible First Bacteremia, Compared With Susceptible First Bacteremia
| First Episodes, No. (%) | Risk of Recurrent Episode by Susceptibility Status of First Episode (95% CI) |
||||||
|---|---|---|---|---|---|---|---|
| Pathogen (Group) | Susceptible | Single Nonsusceptible | Co-nonsusceptible | Risk | Susceptible | Single Nonsusceptible | Co-nonsusceptible |
| Enterobacteriaceae | 67608 (91.5) | 5536 (7.5) | 37 (0.1) | Absolute, % | 2.6 (2.5–2.7) | 4.5 (4.0–5.1) | 13.5 (5.6–32.5) |
| Relative | 1.7 (1.5–2.0) | 5.2 (2.1–12.4) | |||||
| Pseudomonas species | 4433 (94.4) | 133 (2.8) | 103 (2.2) | Absolute, % | 2.9 (2.5–3.5) | 3.8 (1.6–9.0) | 14.6 (8.8–24.2) |
| Relative | 1.3 (.5–3.1) | 5.0 (2.9–8.5) | |||||
| Enterococcus species | 6411 (57.4) | 4602 (41.2) | 52 (0.5) | Absolute, % | 4.9 (4.4–5.5) | 7.0 (6.3–7.8) | 7.7 (2.9–20.5) |
| Relative | 1.4 (1.2–1.7) | 1.6 (.6–4.2) | |||||
| Staphylococcus aureus | 22289 (98.1) | 297 (1.3) | Absolute, % | 6.0 (5.7–6.3) | 9.8 (6.8–14.1) | ||
| Relative | 1.6 (1.1–2.4) | ||||||
Abbreviation: CI, confidence interval.
Pseudomonas Species
A number of 4844 Pseudomonas species bacteremia episodes were identified; 4694 (96.9%) were classified as first episodes and 150 (3.1%) as first recurrent episodes (Figure 1B). Of all Pseudomonas species bacteremia episodes, 272 (5.7%) were caused by nonsusceptible Pseudomonas species. Among first episode isolates, 103 of 236 (43.6%) nonsusceptible Pseudomonas species were co-nonsusceptible to carbapenem antibiotics, as were 20 of 36 (55.6%) in recurrent episodes.
The absolute risk of recurrent bacteremia was 2.9% (130/4433 episodes), 3.8% (5/133 episodes), and 14.6% (15/103 episodes) for subjects with a first episode caused by susceptible, single nonsusceptible, or co-nonsusceptible Pseudomonas species, respectively. Risk ratios for single and co-nonsusceptible first bacteremias were 1.3 (95% CI, .5–3.1) and 5.0 (95% CI, 2.9–8.5), respectively, compared to susceptible first episodes (Table 1). The absolute risk of a recurrent infection caused by a nonsusceptible Pseudomonas species after a first episode caused by a susceptible strain was 0.4% (18/4433 episodes; Figure 1B). The likelihood that recurrent bacteremia was caused by a nonsusceptible strain was 90.0% (18/20 episodes) when the first episode was caused by a nonsusceptible strain.
Enterococcus Species
In total, 11824 Enterococcus species bacteremia episodes were included, of which 11178 (94.5%) were classified as first episodes and 646 (5.5%) as first recurrent episodes (Figure 1C). Of all Enterococcus species bacteremia episodes, 4980 (42.1%) were caused by nonsusceptible Enterococcus species (ARE). Among first episode isolates 52 of 4654 (1.1%) ARE were co-nonsusceptible to vancomycin, as were 7 of 326 (2.1%) in recurrent episodes.
The absolute risk of recurrent bacteremia was 4.9% (314/6411 episodes), 7.0% (322/4602 episodes), and 7.7% (4/52 episodes) for subjects with a first episode caused by susceptible, single nonsusceptible, or co-nonsusceptible Enterococcus species, respectively. Compared to susceptible first episodes, risk ratios for single and co-nonsusceptible first bacteremias were 1.4 (95% CI, 1.2–1.7) and 2.6 (95% CI, .6–4.2), respectively (Table 1). The absolute risk of a recurrent infection caused by a nonsusceptible strain after a first episode caused by a susceptible strain was 0.2% (10/6411 episodes; Figure 1C). The likelihood that recurrent bacteremia was caused by a nonsusceptible strain was 96.3% (314/326 episodes) when the first episode was caused by a nonsusceptible strain.
Staphylococcus aureus
A total of 24089 S. aureus bacteremia episodes were included, of which 22711 (94.3%) were classified as first episodes and 1378 (5.7%) as first recurrent episodes (Figure 1D). Overall, 327 (1.4%) bacteremia episodes were caused by MRSA.
The absolute risk of recurrent bacteremia was 6.0% (1335/22289 episodes) and 9.8% (29/297 episodes) for subjects with a first episode caused by MSSA or MRSA, respectively, yielding a risk ratio of 1.6 (95% CI, 1.1–2.4; Table 1). The absolute risk of a recurrent infection caused by MRSA after a first episode caused by MSSA was 0.01% (2/22289 episodes; Figure 1D). The likelihood that recurrent bacteremia was caused by MRSA was 96.6% (28/29 episodes) when the first episode was caused by MRSA.
Relative risks of recurrent bacteremia in each pathogen group were similar in sensitivity analyses using a maximum of 15 and 60 days of follow-up (Supplementary Table 1).
Population Attributable Effects
The absolute risk difference in the occurrence of recurrent bacteremia between first bacteremia episodes with susceptible and total nonsusceptible Enterobacteriaceae was 2.0% (Table 2). As the total number of nonsusceptible first episodes was 5536, this implies that the number of recurrent episodes attributable to nonsusceptibility was 110. Extrapolated to the total population, the estimated number of recurrent Enterobacteriaceae bacteremia episodes attributable to nonsusceptibility in the Netherlands was 183 between 1 January 2008 and 17 May 2017, or 19 per year. For Pseudomonas species, Enterococcus species, and S. aureus, the absolute risk difference was 5.5%, 2.1%, and 3.8%, respectively, yielding 22 (2 per year), 163 (17 per year), and 18 (2 per year) episodes attributable to antibiotic nonsusceptibility on the population level for the respective pathogens. For all species combined, the estimated number of recurrent bacteremia episodes attributable to nonsusceptibility in the period under study was 386 (40 per year).
Table 2.
Population Attributable Effect of Nonsusceptibility of First Bacteremia Episode on Recurrent Bacteremia
| Pathogen (Group) | Risk Difference for Recurrent Bacteremia Between Susceptible and Intermediate/ Resistant First Episodesa | Additional No. of Recurrent Episodes Associated With Nonsusceptibility in Study Populationb | Population Attributable Effect (per Year)c |
|---|---|---|---|
| Enterobacteriaceae | 2.0% | 110 | 183 (19) |
| Pseudomonas species | 5.5% | 13 | 22 (2) |
| Enterococcus species | 2.1% | 98 | 163 (17) |
| Staphylococcus aureus | 3.8% | 11 | 18 (2) |
Abbreviations: I/R, single or co-nonsusceptible; S, susceptible.
aAbsolute riskI/R – absolute riskS.
b(Absolute riskI/R – absolute riskS) × number of first episodesI/R.
c(Absolute riskI/R – absolute riskS) × number of first episodesI/R / national coverage proportion of 0.60.
DISCUSSION
In this study, the absolute risk of recurrent episodes of bacteremia was consistently higher among patients with bacteremia caused by nonsusceptible bacteria, compared with episodes caused by susceptible strains, with relative risks ranging from 1.4 for ARE to 5.2 for Enterobacteriaceae nonsusceptible to third-generation cephalosporins and carbapenems. The absolute risk of developing a recurrent episode of bacteremia ranged from 2.6% for susceptible Enterobacteriaceae, to 14.6% for Pseudomonas species nonsusceptible to ceftazidime and carbapenems. The estimated crude population attributable effect of nonsusceptibility among Enterobacteriaceae, Pseudomonas species, Enterococcus species, and S. aureus to first-line antibiotic treatment in the Netherlands was 40 recurrent episodes of bacteremia per year.
These findings add to those of a number of other recent studies quantifying the health effects of antibiotic resistance for individual patients. Several studies have demonstrated that bacteremia caused by third-generation cephalosporin-resistant Enterobacteriaceae is associated with a higher risk of hospital mortality or 30-day mortality and a longer hospital stay, compared with infections caused by susceptible bacteria [2, 3]. Likewise, bacteremia caused by MRSA has been associated with considerable higher risks of mortality compared with that caused by MSSA [7, 8]. Yet, more recent studies using methods to control for confounders and competing events reported lower attributable effects of methicillin resistance on patient outcome [2, 9–11]. The overarching conclusion of all these studies is that antibiotic resistance negatively affects patient outcome, but that application of more sophisticated techniques to adjust for confounding and competing events consistently leads to lower negative impacts than previously assumed. Increased mortality and prolonged hospital stay attributed to antibiotic nonsusceptibility may be partially explained by higher risks of recurrent bacteremia. This outcome has not been studied extensively. A population-based study in Denmark found a 25% higher risk of recurrent bacteremia within 2–365 days in patients who had received inappropriate antibiotic therapy, compared with appropriate therapy [12]. However, differences in definitions and methodology hamper a direct comparison of the results of that study and the current study.
In our study, the estimated crude population attributable effect of nonsusceptibility to first-line antibiotic treatment among all species combined is 40 recurrent episodes of bacteremia per year, of which almost half are caused by the widely considered low-grade enterococcal pathogens. This finding emphasizes that antibiotic resistance currently has—at most—a very minor impact on the population health in the Netherlands. This most likely results from effective infection control in Dutch hospitals and the continuing efforts made to reduce unnecessary and inappropriate antibiotic use [13, 14]. Naturally, numbers of recurrent bacteremia in general, and bacteremia attributable to antibiotic nonsusceptibility in particular, will be higher in countries with less effective infection control and more liberal antibiotic use. Yet, our analysis quantifies the risk of recurrent infection in individual patients, and we consider this risk to be mostly independent from the level of antibiotic resistance in other patients.
In our study, the absolute risk of developing recurrent infections with an antibiotic nonsusceptible variant, after a first bacteremia episode caused by a susceptible strain, was low. The highest risk (0.4%) was observed for bacteremia caused by Pseudomonas species, in which resistance to ceftazidime in Pseudomonas species may develop during treatment due to selection of mutants with changed porins and/or expressing efflux pumps. The transition from susceptibility to resistance to third-generation cephalosporins in Enterobacteriaceae requires the acquisition of either a plasmid with an extended-spectrum β-lactamase (ESBL) gene or overexpression of ampC mutations, which may largely depend on the presence of such genes in other bacteria of the microbiome. Yet, with current estimates of the prevalence of intestinal carriage with ESBL-producing bacteria of 5%–10% among Dutch inhabitants [15, 16], the observed absolute risk of 0.1% suggests that such within-host horizontal gene transfer events do not occur with high frequency. This also holds for S. aureus where a phenotype switch (from MSSA to MRSA) would require acquisition of the mecA gene. In the setting of Dutch hospitals, with low prevalence of MRSA, such a change in susceptibility would be more indicative of an infection caused by a different bacterial species, rather than a relapse of the initial infection. This is different for enterococci, where the prevalence of carriage with ARE in hospitalized patients is high (up to 35% in high-risk wards such as hematology and the intensive care unit [17]). Antibiotic treatment for susceptible enterococci may select for acquisition through cross-transmission of ARE carriage and subsequent infection, which occurred in 0.2% of all first bacteremia episodes in our study.
Strengths of our study include the large and complete data collection in ISIS-AR, allowing population-based estimates for rare but clinically relevant outcomes. Study limitations are those linked to population-based studies. First, as data were collected from different hospitals that all use their own patient identification system, recurrent bacteremias diagnosed in another hospital than the first episode may have been misclassified as first episodes. Second, misclassification may have occurred as a result of our choice for the definition of a recurrent episode (a positive culture within 4–30 days after the first positive culture). However, sensitivity analyses using 15 and 60 days as the maximum follow-up period did not change the results. Third, as we did not have information on the genome of the strains causing first and recurrent bacteremias, we could not differentiate whether recurrent episodes were due to relapse or reinfection. Therefore, our estimated proportions of recurrent bacteremias with a phenotype switch may also include episodes in which resistance was not acquired during treatment, but resulted from infection with a new genotype. Reinfection is also the most likely scenario in cases where the opposite occurred. For example, among first episodes caused by single-resistant Enterobacteriaceae, 29 of 251 (11.6%) of the recurrent episodes were caused by a susceptible strain. Last, absence of patient-related data precludes inferences on the causal nature of observations. Our findings confirm our hypothesis that antibiotic resistance is associated with a higher crude risk of recurrent bacteremia. Yet, in the absence of patient-related data (eg, comorbidities, source and severity of infection, treatment regimen) it is not possible to quantify the proportion of this increased risk that can be attributed to antibiotic resistance. On the one hand, patients with a primary episode caused by a nonsusceptible bacterium may have had more frequent and more prolonged hospital exposure and comorbidities, which could also increase risks of recurrent episodes. On the other hand, patients with nonsusceptible bacteremia might have had a higher risk of dying before developing a recurrent infection. More detailed studies are needed to quantify the attributable effects of antibiotic resistance on the occurrence of recurrent bacteremia.
The occurrence of recurrent bacteremia is a simple measure to quantify the consequences of antibiotic resistance. The next step should be to quantify the attribution of antibiotic resistance to the increased risk of recurrent bacteremia. Our study provides further evidence that Dutch patients are currently well protected against the detrimental consequences of antibiotic resistance, and exemplifies the power of large-scale data collection for quantifying the effects of antibiotic resistance.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
APPENDIX
Members of the ISIS-AR Study Group. J. W. T. Cohen Stuart, Department of Medical Microbiology, Noordwest Ziekenhuisgroep, Alkmaar. A. J. L. Weersink, Department of Medical Microbiology, Meander Medical Center, Amersfoort. C. M. J. E. Vandenbroucke-Grauls, Department of Medical Microbiology and Infection Control, VU University Medical Center, Amsterdam. C. E. Visser, Department of Medical Microbiology, Academic Medical Center, Amsterdam. M. L. van Ogtrop, Department of Medical Microbiology, Onze Lieve Vrouwe Gasthuis, Amsterdam. D. J. Kaersenhout, Department of Medical Microbiology, Slotervaart Hospital/Netherlands Cancer Institute, Amsterdam. M. Scholing, Public Health Laboratory, Public Health Service, Amsterdam. B. C. van Hees, Department of Medical Microbiology and Infection prevention, Gelre Hospitals, Apeldoorn. P. H. J. van Keulen, Department of Medical Microbiology, Bravis Hospital, Bergen op Zoom/Laboratory for Microbiology and Infection Control, Amphia Hospital, Breda. J. A. J. W. Kluytmans, Laboratory for Microbiology and Infection Control, Amphia Hospital, Breda. E. M. Kraan, Department of Medical Microbiology, IJsselland hospital, Capelle a/d IJssel. E. E. Mattsson, Department of Medical Microbiology, Reinier de Graaf Groep, Delft. F. W. Sebens, Department of Medical Microbiology, Deventer Hospital, Deventer. E. de Jong, Department of Medical Microbiology, Slingeland Hospital, Doetinchem. H. M. E. Frénay, Department of Medical Microbiology, Albert Schweitzer Hospital, Dordrecht. B. Maraha, Department of Medical Microbiology, Albert Schweitzer Hospital, Dordrecht. A. J. van Griethuysen, Department of Medical Microbiology, Gelderse Vallei Hospital, Ede. W. Silvis, Laboratory of Medical Microbiology and Public Health, Enschede. A. Demeulemeester, SHL-Groep, Etten-Leur. B. B. Wintermans, Department of Medical Microbiology, Admiraal De Ruyter Hospital, Goes. M. van Trijp, Department of Medical Microbiology and Infection Prevention, Groene Hart Hospital, Gouda. A. Ott, Department of Medical Microbiology, Certe, Groningen. J. P. Arends, Department of Medical Microbiology, University Medical Center, Groningen. G. A. Kampinga, Department of Medical Microbiology, University Medical Center, Groningen. D. Veenendaal, Regional Laboratory of Public Health, Haarlem. C. Hol, Department of Medical Microbiology, St Jansdal Hospital, Harderwijk. E. I. G. B. de Brauwer, Department of Medical Microbiology and Infection Control, Zuyderland Medical Centre, Heerlen. F. S. Stals, Department of Medical Microbiology and Infection Control, Zuyderland Medical Centre, Heerlen. L. J. Bakker, Department of Medical Microbiology, CBSL, Tergooi Hospital, Hilversum. J. W. Dorigo-Zetsma, Department of Medical Microbiology, CBSL, Tergooi Hospital, Hilversum. B. Ridwan, Department of Medical Microbiology, Westfriesgasthuis, Hoorn. J. H. van Zeijl, Izore Centre for Infectious Diseases Friesland, Leeuwarden. A. T. Bernards, Department of Medical Microbiology, Leiden University Medical Center, Leiden. S. Erkens-Hulshof, Department of Medical Microbiology, Maastricht University Medical Centre, Maastricht. B. M. de Jongh, Department of Medical Microbiology and Immunology, St Antonius Hospital, Nieuwegein. B. J. M. Vlaminckx, Department of Medical Microbiology and Immunology, St Antonius Hospital, Nieuwegein. M. H. Nabuurs-Franssen, Department of Medical Microbiology and Infectious Diseases, Canisius Wilhelmina Hospital, Nijmegen. S. Kuipers, Department of Medical Microbiology, Radboud University Medical Center, Nijmegen. B. M. W. Diederen, Department of Medical Microbiology, Bravis Hospital, Roosendaal/ZorgSaam Hospital Zeeuws-Vlaanderen, Terneuzen. D. C. Melles, Department of Medical Microbiology, Erasmus University Medical Center, Rotterdam/STAR Medical Diagnostic Center, Rotterdam. M. van Rijn, Department of Medical Microbiology, Ikazia Hospital, Rotterdam. S. Dinant, Department of Medical Microbiology, Maasstad Hospital, Rotterdam. O. Pontesilli, Department of Medical Microbiology, Maasstad Hospital, Rotterdam. P. de Man, Department of Medical Microbiology, Sint Franciscus Gasthuis, Rotterdam. N. Vaessen, Department of Medical Microbiology, Franciscus Vlietland Hospital, Schiedam. M. A. Leversteijn-van Hall, Department of Medical Microbiology and Infection Control, Alrijne Zorggroep/Bronovo Hospital, ‘s-Gravenhage. E. P. M. van Elzakker, Department of Medical Microbiology, Haga Hospital, ‘s-Gravenhage. A. E. Muller, Department of Medical Microbiology, MCH Westeinde Hospital, ‘s-Gravenhage. N. H. Renders, Department of Medical Microbiology and Infection Control, Jeroen Bosch Hospital, ‘s-Hertogenbosch. D. W. van Dam, Department of Medical Microbiology and Infection Control, Zuyderland Medical Centre, Sittard-Geleen. A. G. M. Buiting, Department of Medical Microbiology, St. Elisabeth Hospital, Tilburg. A. L. M. Vlek, Department of Medical Microbiology and Immunology, Diakonessenhuis, Utrecht. M. P. D. Deege, Department of Medical Microbiology, Saltro Diagnostic Centre, Utrecht. F. N. J. Frakking, Department of Medical Microbiology, University Medical Center Utrecht, Utrecht. I. T. M. A. Overdevest, Department of Medical Microbiology, PAMM, Veldhoven. R.W. Bosboom, Laboratory for Medical Microbiology and Immunology, Rijnstate Hospital, Velp. T. Trienekens, Department of Medical Microbiology, VieCuri Medical Center, Venlo. G. P. Voorn, Department of Medical Microbiology, Zuwe Hofpoort Hospital, Woerden. G. J. H. M. Ruijs, Laboratory of Medical Microbiology and Infectious Diseases, Isala Hospital, Zwolle. M. J. H. M. Wolfhagen, Laboratory of Medical Microbiology and Infectious Diseases, Isala Hospital, Zwolle. J. Alblas, W. Altorf–van der Kuil, L. Blijboom, S. Groenendijk, J. van Heereveld, R. Hertroys, J. C. Monen, D. W. Notermans, E. A. Reuland, M. I. van Triest, C. C. H. Wielders, Centre for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven.
Notes
Disclaimer. This study was conducted as part of author’s routine work and the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. ISIS-AR is supported by the Dutch Ministry of Health, Welfare and Sport.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Infectious Diseases Surveillance Information System–Antimicrobial Resistance (ISIS-AR) Study Group:
J W T Cohen Stuart, A J L Weersink, C M J E Vandenbroucke-Grauls, C E Visser, M L van Ogtrop, D J Kaersenhout, M Scholing, B C van Hees, P H J van Keulen, J A J W Kluytmans, E M Kraan, E E Mattsson, F W Sebens, E de Jong, H M E Frénay, B Maraha, A J van Griethuysen, W Silvis, A Demeulemeester, B B Wintermans, M van Trijp, A Ott, J P Arends, G A Kampinga, D Veenendaal, C Hol, E I G B de Brauwer, F S Stals, L J Bakker, J W Dorigo-Zetsma, B Ridwan, J H van Zeijl, A T Bernards, S Erkens-Hulshof, B M de Jongh, B J M Vlaminckx, M H Nabuurs-Franssen, S Kuipers, B M W Diederen, D C Melles, M van Rijn, S Dinant, O Pontesilli, P de Man, N Vaessen, M A Leversteijn-van Hall, E P M van Elzakker, A E Muller, N H Renders, D W van Dam, A G M Buiting, A L M Vlek, M P D Deege, F N J Frakking, I T M A Overdevest, R W Bosboom, T Trienekens, G P Voorn, G J H M Ruijs, M J H M Wolfhagen, J Alblas, W Altorf–van der Kuil, L Blijboom, S Groenendijk, J van Heereveld, R Hertroys, J C Monen, D W Notermans, E A Reuland, M I van Triest, and C C H Wielders
References
- 1. Dautzenberg MJ, Wekesa AN, Gniadkowski M, et al. Mastering Hospital Antimicrobial Resistance in Europe Work Package 3 Study Team The association between colonization with carbapenemase-producing Enterobacteriaceae and overall ICU mortality: an observational cohort study. Crit Care Med 2015; 43:1170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stewardson AJ, Allignol A, Beyersmann J, et al. The health and economic burden of bloodstream infections caused by antimicrobial-susceptible and non-susceptible Enterobacteriaceae and Staphylococcus aureus in European hospitals, 2010 and 2011: a multicentre retrospective cohort study. Euro Surveill 2016; 21. doi:10.2807/1560-7917.ES.2016.21.33.30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Kraker ME, Wolkewitz M, Davey PG, et al. Burden of antimicrobial resistance in European hospitals: excess mortality and length of hospital stay associated with bloodstream infections due to Escherichia coli resistant to third-generation cephalosporins. J Antimicrob Chemother 2011; 66:398–407. [DOI] [PubMed] [Google Scholar]
- 4. Wolkewitz M, Cooper BS, Bonten MJ, Barnett AG, Schumacher M. Interpreting and comparing risks in the presence of competing events. BMJ 2014; 349:g5060. [DOI] [PubMed] [Google Scholar]
- 5. Rottier WC, Ammerlaan HS, Bonten MJ. Effects of confounders and intermediates on the association of bacteraemia caused by extended-spectrum β-lactamase-producing Enterobacteriaceae and patient outcome: a meta-analysis. J Antimicrob Chemother 2012; 67:1311–20. [DOI] [PubMed] [Google Scholar]
- 6. Altorf-van der Kuil W, Schoffelen Annelot F, de Greeff Sabine C, Thijsen Steven FT, et al. National AMR Surveillance Study Group National laboratory-based surveillance system for antimicrobial resistance: a successful tool to support the control of antimicrobial resistance in the Netherlands. Euro Surveill 2017; 22 pii:17-00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blot SI, Vandewoude KH, Hoste EA, Colardyn FA. Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Arch Intern Med 2002; 162:2229–35. [DOI] [PubMed] [Google Scholar]
- 8. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 2003; 36:53–9. [DOI] [PubMed] [Google Scholar]
- 9. Ammerlaan H, Seifert H, Harbarth S, et al. European Practices of Infections with Staphylococcus aureus (SEPIA) Study Group Adequacy of antimicrobial treatment and outcome of Staphylococcus aureus bacteremia in 9 Western European countries. Clin Infect Dis 2009; 49:997–1005. [DOI] [PubMed] [Google Scholar]
- 10. Schweizer ML, Furuno JP, Harris AD, et al. Empiric antibiotic therapy for Staphylococcus aureus bacteremia may not reduce in-hospital mortality: a retrospective cohort study. PLoS One 2010; 5:e11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yaw LK, Robinson JO, Ho KM. A comparison of long-term outcomes after meticillin-resistant and meticillin-sensitive Staphylococcus aureus bacteraemia: an observational cohort study. Lancet Infect Dis 2014; 14:967–75. [DOI] [PubMed] [Google Scholar]
- 12. Gradel KO, Jensen US, Schønheyder HC, et al. Danish Collaborative Bacteraemia Network (DACOBAN) Impact of appropriate empirical antibiotic treatment on recurrence and mortality in patients with bacteraemia: a population-based cohort study. BMC Infect Dis 2017; 17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Troelstra A. Hospital infection control in the Netherlands. J Hosp Infect 2007; 65(Suppl 2):139–41. [DOI] [PubMed] [Google Scholar]
- 14. European Centre for Disease Prevention and Control. Summary of the latest data on antibiotic consumption in the European Union. Stockholm, Sweden: ECDC, 2015. [Google Scholar]
- 15. Huijbers PM, de Kraker M, Graat EA, et al. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in humans living in municipalities with high and low broiler density. Clin Microbiol Infect 2013; 19:E256–9. [DOI] [PubMed] [Google Scholar]
- 16. Reuland EA, Al Naiemi N, Kaiser AM, et al. Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in Amsterdam. J Antimicrob Chemother 2016; 71:1076–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Top J, Willems R, Blok H, et al. Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin Microbiol Infect 2007; 13:316–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.