Abstract
Purpose
To identify genetic risk factors contributing to central corneal thickness (CCT) in individuals from South India, a population with a high prevalence of ocular disorders.
Methods
One hundred ninety-five individuals from 15 large South Indian pedigrees were genotyped using the Omni2.5 bead array. Family-based association for CCT was conducted using the score test in MERLIN.
Results
Genome-wide association study (GWAS) identified strongest association for single nucleotide polymorphisms (SNPs) in the first intron of WNT7B and CCT (top SNP rs9330813; β = −0.57, 95% confidence interval [CI]: −0.78 to −0.36; P = 1.7 × 10−7). We further investigated rs9330813 in a Latino cohort and four independent European cohorts. A meta-analysis of these data sets demonstrated statistically significant association between rs9330813 and CCT (β = −3.94, 95% CI: −5.23 to −2.66; P = 1.7 × 10−9). WNT7B SNPs located in the same genomic region that includes rs9330813 have previously been associated with CCT in Latinos but with other ocular quantitative traits related to myopia (corneal curvature and axial length) in a Japanese population (rs10453441 and rs200329677). To evaluate the specificity of the observed WNT7B association with CCT in the South Indian families, we completed an ocular phenome-wide association study (PheWAS) for the top WNT7B SNPs using 45 ocular traits measured in these same families including corneal curvature and axial length. The ocular PheWAS results indicate that in the South Indian families WNT7B SNPs are primarily associated with CCT.
Conclusions
The results indicate robust evidence for association between WNT7B SNPs and CCT in South Indian pedigrees, and suggest that WNT7B SNPs can have population-specific effects on ocular quantitative traits.
Keywords: cornea central thickness, genetic association, quantitative trait, WNT7B, ocular PheWAS
Ocular quantitative traits such as central corneal thickness (CCT), axial length (AXL), and intraocular pressure are heritable intermediate phenotypes (endophenotypes) for common complex eye disorders such as keratoconus, myopia, and glaucoma.1 CCT is a highly heritable ocular quantitative trait with up to 95% of its phenotypic variance due to genetics.2 Thin CCT is related to several diseases of the cornea, especially keratoconus3 and brittle corneal syndrome.4 Very thin corneas are a hallmark of Ehlers Danlos,5 and thicker than normal corneas are found in patients with aniridia.6 Thinner than average CCT can influence development of primary open angle glaucoma7,8 with more severe disease evident in people with thinner corneas.9–11
CCT varies among ethnic populations with individuals of African descent having lower values than European Caucasians and East Asians.2,12,13 Genome-wide association studies (GWAS) in European Caucasians,14–16 Asians,14,17 and Hispanics18 have identified ZNF469, RXRA-COL5A1, COL8A2, and FOXO1 among others as important loci contributing to CCT. RXRA-COL5A1 and ZNF469 have been associated with CCT in most populations studied while the associations of other loci (COL8A2, FOXO1) may be restricted to specific populations.19 Recently, WNT7B single nucleotide polymorphisms (SNPs) have been associated with CCT in Latinos,20 and interestingly some of these same SNPs were associated with AXL and corneal curvature, traits influencing myopic refractive error, in a Japanese population.21
Few genetic studies of ocular quantitative traits have been completed in individuals from South India, a population with high prevalence of common ocular conditions, especially cataract and glaucoma.22–27 In Indian populations, CCT is thinner than the average values for Caucasians22 suggesting that CCT could be an important factor in the development of CCT-related common ocular disorders in this population. To identify genetic risk loci for CCT in South Indians, we completed a family-based association study using large pedigrees, many with consanguineous matings that are typical for this geographic region. For the top SNPs located in the WNT7B region, we also completed a phenome-wide association study (PheWAS) to examine the range of phenotypes associated with WNT7B SNPs in this South Indian population.
Materials and Methods
Pedigrees and Quantitative Traits
This study adhered to the tenets of the Declaration of Helsinki and has been reviewed and approved by the Institutional Review Boards of Massachusetts Eye and Ear Infirmary and Medical Research Foundation, Sankara Nethralaya, Chennai, India. After obtaining written informed consent, 197 individuals from 15 Indian pedigrees were recruited at Sankara Nethralaya, Chennai, India. CCT was measured by an ultrasonic pachymeter in triplicate and the average value was used. Methods to measure the other traits used in the PheWAS are described in the Supplementary Methods. Collections of samples for replication cohorts are described in the Supplementary Methods.
Genotyping and Quality Control (QC)
Genotyping for the South Indian families was performed at the Ocular Genomics Institute at the Massachusetts Eye and Ear Infirmary using the Illumina HumanOmni2.5-8 Beadchip kit (2,379,855 markers; Illumina, Inc., San Diego, CA, USA). Genotypes were called using GenomeStudio (v2011.1, Illumina, Inc.). The genetic sex of all individuals was consistent with the reported sex. Two samples were removed because genotyping call rates were <99%. The average call rate per sample was >99.8%. QC for 2,352,697 (98.9%) well-clustered SNPs was performed with PLINK (v1.07, provided in the public domain, http://pngu.mgh.harvard.edu/∼purcell/plink/).28 25,088 (1.1%) SNPs with call frequency <90% and 881,678 (37.5%) SNPs with minor allele frequency (MAF) <0.01 were removed from the analysis. 164,174 (7.0%) SNPs with Mendelian errors and 58,443 (2.5%) SNPs on chromosome X or Y, or on the mitochondrial chromosome were also excluded. After QC, 1,223,314 SNPs were included in the final analysis. Genotyping for replication cohorts is described in the Supplementary Methods.
Statistical Analysis
The kinship coefficients for pairwise relationships across pedigrees were estimated from the SNP data using the KING software (provided in the public domain, http://people.virginia.edu/∼wc9c/KING/index.html).29 The heritability for each trait was estimated with restricted maximum likelihood–based linear modeling in the GCTA software (provided in the public domain, http://gcta.freeforums.net/),30 taking into account all pedigree relationships simultaneously. Inverse-normal transformation of ranks was applied to CCT measurements before analysis. Age and sex were included as covariates in the association tests. The genome-wide association test was performed using the score test in MERLIN (v1.1.2, provided in the public domain, https://csg.sph.umich.edu/abecasis/Merlin/),31,32 which incorporated genetic relatedness based on the family structure. Because this program applies a restriction on pedigree size, 8 of the 15 pedigrees were split into nonoverlapping fragments of ≤18 bits using the PedSTR program (provided in the public domain, http://mga.bionet.nsc.ru/soft/PedStr/PedStr.tar.gz),33 which breaks inbreeding loops and identifies subpedigrees having the maximal total relationship between individuals of interest, resulting in a total of 26 effective subpedigrees used in the final analysis. To avoid an excess of false-positive results in regions of strong linkage, the likelihood-ratio test was performed to accurately evaluate the SNPs with suggestive association. The regional SNP association plot was generated using SNAP (provided in the public domain, http://archive.broadinstitute.org/mpg/snap/).34 The variance in CCT explained by all the SNPs in the Indian population was estimated using GCTA.30
Meta-analysis using the inverse-variance weighting method was done using both fixed-effects and random-effects models using Review Manager software (RevMan, version 5.3; Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014). The heterogeneity between data sets was evaluated by heterogeneity index (I2) and Cochran's Q statistic.35 Heterogeneity among data sets was further examined by evaluating differences in ethnicity (Indians, Latinos, or Europeans), study design (family-based design or population design), imputation quality score, age, and sex in meta-regression models using the R package “metafor.”36 Forest plots were generated using the R package “metafor.”36
PheWAS
Forty-five quantitative traits (including CCT) (Supplementary Table S1) were analyzed for association as described above. Methods for measuring each trait are described in the Supplementary Methods. The average value for each trait for both eyes was used for analysis. Age and sex were included as covariates in the association tests. The association tests were performed using the likelihood-ratio test in MERLIN (v1.1.2).31,32 The PheWAS plots were generated using the R package ggplot2 (provided in the public domain, https://www.r-project.org/).37 Phenotypes were grouped along the x-axis by categorization of ocular measures (Biometric traits, Corneal traits, Optic nerve traits, Refractive error traits). Each point in the plot represents the −log10(P) value of a trait measure in association analysis. The lower gray dashed line indicates P = 0.05. The upper black dashed line indicates a single-SNP Bonferroni correction P = 0.001 (0.05/45).
Power Analysis
Power analysis was performed using the Genetic Power Calculator (provided in the public domain, http://pngu.mgh.harvard.edu/∼purcell/gpc/).38 The total proportion of trait variance was derived from the estimated heritability of these ocular traits in the South Indian pedigrees. For CCT, AXL, and corneal curvature, heritability was 0.54, 0.84, and 0.82, respectively. The quantitative trait locus (QTL) increaser allele frequency was set to the same as the marker allele frequency. Linkage disequilibrium between the QTL and the marker was set at D' = 1.0. The sample size was set as 26 because a total of 26 effective subpedigrees were used in the final analysis. The sibling correlation was set as 0.5. The sibship size was set as 2. An additive effects only (1 df) test was used to calculate the power at the type I error rate of 5 × 10−8 for GWAS or 0.001 (0.05/45 traits) for PheWAS. Power results for all traits are listed in Supplementary Table S1.
Results
Study Sample
One hundred ninety-five individuals from 15 pedigrees (Supplementary Fig. S1) were recruited at Sankara Nethralaya Eye Hospital, Chennai, India for a family-based genetic association study. These pedigrees were unrelated to each other; the maximum kinship coefficient estimated from the SNP data across pedigrees was 0.0344. The pedigree size ranged from 2 to 26 members. Ten of the pedigrees included at least one consanguineous mating. Fifty-eight percent of the subjects were female and 42% male. The average age was 44.9 (±15.0) years and the age ranged from 16 to 85 years. These families were not ascertained on specific eye conditions. CCT was measured by an ultrasonic pachymeter in triplicate for each eye (Supplementary Methods) and the average value for both eyes was used (516.2 [±30.2] μm average; 433–608 μm range; Supplementary Table S1).
Genome-Wide Association Results for CCT
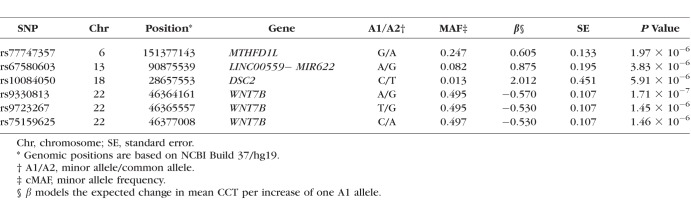
After QC, 1,223,314 SNPs were included in the genome-wide CCT analysis. The results for the family-based association test are shown in Supplementary Figure S2. The genomic inflation factor of 1.05 (QQ plot, Supplementary Fig. S3) suggested that population substructure or other confounding factors were not significant. Six SNPs located on chromosomes 6, 13, 18, and 22 showed suggestive evidence of association with CCT (P < 1.0 × 10−5; Table 1), with the top SNP (rs9330813, P = 1.7 × 10−7, β = −0.57, 95% confidence interval [CI]: −0.78 to −0.36 [A]) located in the first intron of WNT7B on chromosome 22 (Fig. 1). CCT association with rs9330813 was two orders of magnitude greater than any other SNP (Table 1) and accounted for 17% of the phenotypic variance in the South Indian families. WNT7B SNPs have previously only been associated with CCT in a Latino population (Mexican American Glaucoma Genetics Study [MAGGS]),20 and the top SNP in the Latino study (rs10453441) is 422 bp from rs9330813. rs10453441 is in moderate linkage disequilibrium with rs9330813 in the South Indian dataset (r2 = 0.55) and was nominally associated with CCT in the South Indian pedigrees (P = 5.85 × 10−4, Supplementary Table S2).
Table 1.
SNPs With P < 1.0 × 10−5 for Association With CCT in South Indian Pedigrees
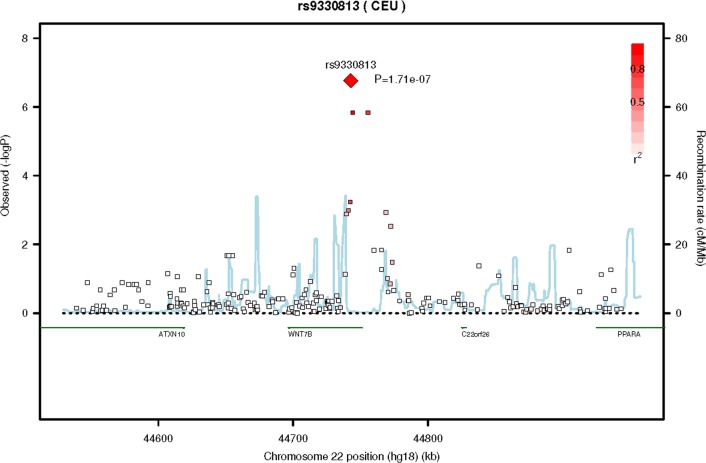
Figure 1.
Regional SNP association plot for the 22q13 region. A region of 408 kb around the top SNP (rs9330813) is displayed. The degree of linkage disequilibrium (LD) between the top SNP and any SNP tested is indicated by red shading. The recombination rate is displayed by a blue line with scale on the right-hand axis. Characterized genes in the region are represented with a green bar. The P value for rs9330813 (1.71 × 10−7) is shown as a red diamond.
To provide further support for the association of WNT7B with CCT in the South Indian pedigrees, we investigated the association of rs9330813 in the Latino study cohort as well as in four independent European data sets (Fig. 2). In addition, we investigated association of rs10453441 with CCT in an independent Singaporean Indian cohort, and five independent European data sets (Supplementary Fig. S4). The WNT7B SNPs were imputed from previous genotype data for the European cohorts. For both rs9330813 and rs10453441, association with CCT was evident with consistent direction of effects observed in all data sets with the exception of one European cohort for rs10453441 (Supplementary Fig. S4). For both SNPs, strongest association was observed for the South Indian and MAGGS (Latinos) data sets, with smaller effects in European cohorts (Fig. 2; Supplementary Fig. S4). Significant heterogeneity was detected among data sets, and ethnicity, study design, imputation quality score, and age and sex were evaluated for contribution. This analysis suggested that the heterogeneity was most likely caused by imputation quality and study design (meta-regression P = 0.0001 and P = 0.02, respectively). Limiting the meta-analysis to data sets with imputation scores >0.7 for each SNP reduced but did not completely eliminate heterogeneity (Fig. 2; Supplementary Fig. S4). Because of the residual heterogeneity reverse inverse weighted meta-analyses were completed using both fixed and random effects and investigated separately the data sets with imputation scores >0.7 for each SNP. Using the fixed effects model, significant association was observed for CCT and rs9330813 [A] (P = 1.7 × 10−9, β = −3.94, 95% CI: −5.23 to −2.66; Fig. 2), and rs10453441 [G] (P = 2.20 × 10−11, β = −3.11, 95% CI: −4.02 to −2.02; Supplementary Fig. S4). Evidence for association improved when only the data sets with imputation scores >0.7 were included in the meta-analyses: rs9330813[A] (P = 5.0 × 10−12, β = −5.59, 95% CI: −7.17 to −4.00; Fig. 2), and rs10453441 [G] (P = 5.3 × 10−12, β = −3.43, 95% CI: −4.40 to −2.45; Supplementary Fig. S4). Reduced but consistent association was observed using the random effects model for both SNPs: rs9330813 [A] (P = 7.0 × 10−3, β = −8.00, 95% CI: −13.85 to −2.15); rs10453441 [G] (P = 1.0 × 10−4, β = −3.44, 95% CI: −5.21 to −1.68).
Figure 2.
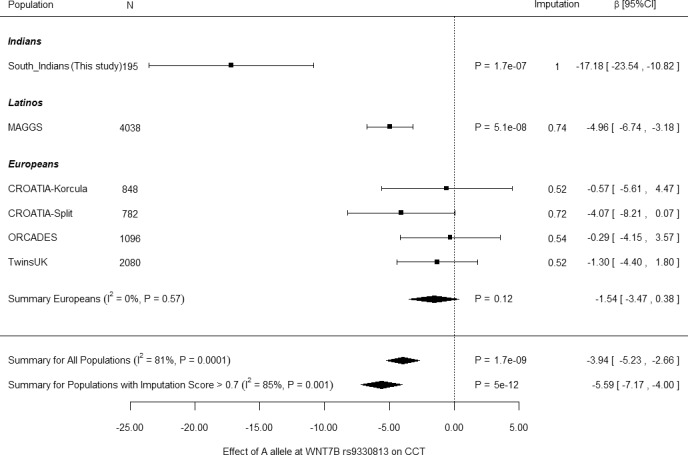
Meta-analysis for rs9330813 and CCT. Forest plot showing effect estimates for the South Indian pedigree, as well as for the replication effort. Pooled estimates for β and 95% CI were calculated by fixed-effects, inverse variance weighting meta-analysis. Reduced evidence of association but with similar effects was observed if the meta-analysis was calculated using random effects: P = 7.0 × 10−3, β = −8.00, 95% CI: −13.85 to −3.15. Individual data set results are indicated by black squares and summary values are indicated by black diamonds. ORCADES, Orkney Complex Disease Study; TwinsUK, UK Twin Study.
The top WNT7B SNP, rs9330813 is in strong equilibrium with rs9723267; r2 = 0.96 and 1.0 in the South Indian data set, 1000 Genomes (provided in the public domain, http://csg.sph.umich.edu/abecasis/MACH/download/1000G.2012-03-14.html), and Haploreg v.4.1 (provided in the public domain, http://www.broadinstitute.org/mammals/haploreg/haploreg.php), respectively, that disrupts a Rad21 binding motif and a CTCF (CCCTC-binding factor) binding site, as well as other transcription factor binding sites (RegulomeDB, provided in the public domain, http://regulome.stanford.edu/; Supplementary Fig. S5) suggesting a role in regulation of gene expression. The region of intron 1 that includes the WNT7B SNPs associated with CCT contains multiple DNaseI hypersensitivity sites and features of enhancers as annotated by ENCODE in multiple cell types (Supplementary Fig. S5).
In the South Indian family data set, we also replicated association (P < 0.005) with a number of loci previously associated with CCT including RXRA-COL5A1,16 ZNF469,15 GPR15,13 and GLT8D2,13 although none of these associations were as significant as those observed for the WNT7B SNPs in this population (Supplementary Table S3). It was estimated that 53.8% of the variance in CCT was explained by all the CCT-associated SNPs in this Indian population.
We also investigated the association of the WNT7B SNPs associated with CCT in this study with primary open angle glaucoma (POAG) in our NEIGHBORHOOD European Caucasian data set of 3853 cases and 33,480 controls.39 However, similar to other studies,14 we did not find evidence for association of these SNPs with POAG (P > 0.05).
PheWAS
Recently, SNPs also located in this same region of the first intron of WNT7B have been associated with two other ocular quantitative traits, corneal curvature and AXL, in a GWAS using a Japanese population.21 The lead SNP in the Japanese study, rs10453441, is the same SNP associated with CCT in the Latino study20 located 422 bp from rs9330813, the lead SNP in the South Indian pedigrees (Supplementary Fig. S5). To determine if the WNT7B association in our data set was specific for CCT, we performed an age- and sex-adjusted PheWAS (Phenotype-wide association study) using association data for 45 ocular quantitative traits measured in the same families used for the CCT analysis (see Supplementary Table S1 for complete list of traits), including AXL and corneal curvature, the two traits associated with the WNT7B SNP rs10453441 in the Japanese study.21 For the PheWAS, we investigated the top three WNT7B SNPs (rs9330813, rs9723267, and rs75159625) from our data (Supplementary Table S2) and also the top two SNPs in the Japanese study (rs10453441 and rs200329677). Four of these SNPs are preferentially associated with CCT in the South Indian sample (the remaining SNP, rs200329677, was not significantly associated with CCT or any other trait in this data set) (Fig. 3; Supplementary Fig. S6). In the South Indian data set, the PheWAS data did not support significant association of any WNT7B SNP with any trait other than CCT (P > 0.001) including AXL or corneal curvature as was observed in the Japanese study (Fig. 3; Supplementary Fig. S6) despite having sufficient power (>99.9%) for AXL and corneal curvature to detect the associations previously described (Supplementary Table S1).
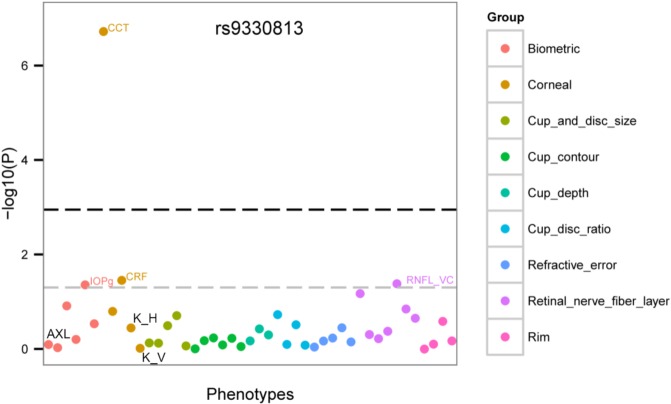
Figure 3.
PheWAS plot for the top SNP associated with CCT in the South Indian population (rs9330813). The association results for each measured trait (Supplementary Table S1) for this SNP were plotted with the phenotypes (ocular traits) grouped along the x-axis and the –log10(P) value for association analysis on the y-axis. The phenotype group is indicated by the color of the graph point as indicated by the side panel. The lower gray dashed line indicates P = 0.05. The upper black dashed line indicates a single-SNP Bonferroni correction for 45 traits, P = 0.001 (0.05/45). Other traits were not labeled in these figures due to limited space. Categories are grouped according to Supplementary Table S1. IOPg, intraocular pressure measured by Goldman applanation; CRF, corneal resistance factor; K_H, corneal curvature, horizontal; K_V, corneal curvature, vertical; RNFL_VC, retinal nerve fiber layer curvature as measured by the Heidelberg Retina Tomography and analyzed by using Glaucoma Probability Score (GPS).
Discussion
This is the first GWAS for CCT in individuals residing in Southern India, a population at increased risk for blinding ocular disorders.27,40 In this family-based study that included large consanguineous pedigrees, we identified association of CCT with WNT7B SNPs located in an apparent regulatory region likely to impact gene expression. Pedigrees with consanguineous matings are known to have added power for genetic studies of recessive traits. In this study, we have shown that consanguineous families can also provide genetic insights leading to discovery of loci for quantitative traits. The CCT boxplot for three genotypes of top SNP rs9330813 was consistent with an additive model in this South Indian data set (Supplementary Fig. S7). We estimated that we had at least 82% power to detect the associations between these WNT7B SNPs and CCT in this South Indian data set.
WNT7B codes for a member of the Wnt family of proteins that have critical roles in cell growth, patterning, and differentiation of multiple tissues and organs.41 The canonical WNT signaling pathway that includes WNT7b (the product of WNT7B) is known to contribute to stem cell proliferation in development.42 In the eye, WNT7B has been shown to have increased expression in the central cornea and may also be necessary for corneal limbal stem cell development.43 Interestingly, a rare exonic variant in another WNT family member, WNT10A, has also been associated with CCT in a quantitative trait study of European Caucasians.44
The WNT7B SNPs associated with CCT are located in the first intron of the gene in a region with multiple DNaseI hypersensitivity sites and enhancers as annotated by ENCODE. The top SNP is in strong linkage disequilibrium with rs9723267 that impacts Rad21 and CTCF (CCCTC-binding factor) binding sites. Rad21 is one of the subunits of the cohesin complex that together with CTCF associates with active enhancers and promoters forming long-range interactions important for gene regulation.45 Rad21 and CTCF activity is highest when a general transcription factor (TBP) binding site is also nearby46 as is the case in the WNT7B region associated with CCT (Supplementary Fig. S5), suggesting that genetic variants in this region could impact gene expression.
In addition to the association between WNT7B and CCT, we also confirmed association with several other loci previously associated with CCT in other populations, in particular ZNF469 and RXRA-COL5A1. Genomic association studies have now been completed for CCT in a variety of ethnic populations including European Caucasians,13–16 Asians,17 and Latinos.18,20 Evidence for association of CCT with ZNF469 and RXRA-COL5A1 has been found in most populations, while other CCT loci such as COL8A2, significantly associated in Asians,17 may be restricted to specific populations.19 Our study suggests that WNT7B is an important locus for CCT in the South Indian population.
WNT7B SNPs may also contribute to other ocular phenotypes. In a study conducted in Japanese, SNPs in the same genomic region associated with CCT in our study were associated with AXL and corneal curvature, ocular quantitative traits related to refractive error and myopia.21 We have previously measured 45 quantitative traits in the collection of Indian pedigrees used for this study including AXL, corneal curvature, and refractive error. This collection of quantitative trait data made it possible to complete an ocular PheWAS for the WNT7B SNPs associated with CCT in our study and the WNT7B SNPs associated with AXL and corneal curvature in the Japanese study. Understanding the range of phenotypic consequences of DNA sequence variants may provide insights into the mechanisms by which a variant or gene leads to disease. The PheWAS approach can test the association of a disease-associated variant with a broad range of phenotypes.47–49 We found that in the South Indian population, the WNT7B SNPs are specifically associated with CCT and did not show evidence of association with any other traits, including those related to myopia and refractive error. While the Japanese study did not specifically interrogate association with CCT, it appears that the WNT7B SNPs can be associated with additional or different traits in the Japanese population. The opportunity to complete a PheWAS to evaluate the association of the WNT7B SNPs with a broad range of ocular phenotypes was a strength of our study.
Conclusions
In summary, our family-based association analysis using South Indian pedigrees has identified WNT7B as a locus for CCT in this population and an ocular PheWAS conducted in the same data set showed that the WNT7B association is specific for this trait in these South Indian pedigrees. WNT7B is known to be associated with CCT in a Latino population20 but has not been previously shown to be a CCT locus in Asians or European Caucasians, suggesting that genomic studies in specific ethnic populations can uncover new loci for complex traits that provide additional insights into the underlying genetic architecture of these common conditions.
Supplementary Material
Acknowledgments
Supported by National Institutes of Health/National Eye Institute Grants R21EY018149 (JLW), R01EY027129 (JLW), P30EY014104 (JLW), R01EY022651 (XG), and P30EY001792 (XG).
Disclosure: B.J. Fan, None; X. Chen, None; N. Sondhi, None; P.F. Sharmila, None; N. Soumittra, None; S. Sripriya, None; S. Sacikala, None; R. Asokan, None; D.S. Friedman, None; L.R. Pasquale, None; X.R. Gao, None; L. Vijaya, None; J. Cooke Bailey, None; V. Vitart, None; S. MacGregor, None; C.J. Hammond, None; C.C. Khor, None; J.L. Haines, None; R. George, None; J.L. Wiggs, None
Appendix
Members of the Mexican American Glaucoma Genetic Study: R. Rand Allingham, X. Raymond Gao, Jim Gauderman, Michael Hauser, Jerome I. Rotter, Rohit Varma, and Janey Wiggs.
Members of the International Glaucoma Genetics Consortium: Tin Aung, Kathryn P. Burdon, Ching-Yu Cheng, Jamie E. Craig, Angela J. Cree, Puya Gharahkhani, Christopher J. Hammond, Alex W. Hewitt, René Höhn, Pirro Hysi, Adriana I. Iglesias Gonzalez, Jost Jonas, Anthony Khawaja, Chiea-Cheun Khor, Caroline C.W. Klaver, Francesca Pasutto, Stuart MacGregor, David Mackey, Paul Mitchell, Aniket Mishra, Calvin Pang, Louis R. Pasquale, Francesca Pasutto, Henriette Springelkamp, Gudmar Thorleifsson, Unnur Thorsteinsdottir, Cornelia M. van Duijn, Ananth Viswanathan, Veronique Vitart, Janey L. Wiggs, Robert Wojciechowski, Tien Wong, Terri L. Young, and Tanja Zeller.
Members of the NEIGHBORHOOD Consortium: Rand Allingham, Murray Brilliant, Don Budenz, Jessica Cooke Bailey, John Fingert, Douglas Gaasterland, Teresa Gaasterland, Jonathan L. Haines, Michael Hauser, Rob Igo, Jae Hee Kang, Peter Kraft, Richard Lee, Paul Lichter, Yutao Liu, Syoko Moroi, Louis R. Pasquale, Anthony Realini, Doug Rhee, Julia R. Richards, Robert Ritch, Joel Schuman, William K. Scott, Kuldev Singh, Arthur Sit, Douglas Vollrath, Robert N. Weinreb, Janey L. Wiggs, Gadi Wollstein, and Don Zack.
References
- 1. Charlesworth J, Kramer PL, Dyer T,et al. . The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. . 2010; 51: 3509– 3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dimasi DP, Burdon KP, Craig JE. . The genetics of central corneal thickness. . 2010; 94: 971– 976. [DOI] [PubMed] [Google Scholar]
- 3. Vincent AL, Jordan CA, Cadzow MJ, Merriman TR, McGhee CN. . Mutations in the zinc finger protein gene, ZNF469, contribute to the pathogenesis of keratoconus. . 2014; 55: 5629– 5635. [DOI] [PubMed] [Google Scholar]
- 4. Lu Y, Dimasi DP, Hysi PG,et al. . Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. . 2010; 6: e1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Villani E, Garoli E, Bassotti A,et al. . The cornea in classic type Ehlers-Danlos syndrome: macro- and microstructural changes. . 2013; 54: 8062– 8068. [DOI] [PubMed] [Google Scholar]
- 6. Brandt JD, Casuso LA, Budenz DL. . Markedly increased central corneal thickness: an unrecognized finding in congenital aniridia. . 2004; 137: 348– 350. [DOI] [PubMed] [Google Scholar]
- 7. Jiang X, Varma R, Wu S,et al.; for the Los Angeles Latino Eye Study Group. . Baseline risk factors that predict the development of open-angle glaucoma in a population: the Los Angeles Latino Eye Study. . 2012; 119: 2245– 2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gordon MO, Beiser JA, Brandt JD,et al. . The Ocular Hypertension Treatment Study: baseline factors that predict the onset of primary open-angle glaucoma. . 2002; 120: 714– 720. [DOI] [PubMed] [Google Scholar]
- 9. Shah H, Kniestedt C, Bostrom A, Stamper R, Lin S. . Role of central corneal thickness on baseline parameters and progression of visual fields in open angle glaucoma. . 2007; 17: 545– 549. [DOI] [PubMed] [Google Scholar]
- 10. Kniestedt C, Lin S, Choe J,et al. . Correlation between intraocular pressure, central corneal thickness, stage of glaucoma, and demographic patient data: prospective analysis of biophysical parameters in tertiary glaucoma practice populations. . 2006; 15: 91– 97. [DOI] [PubMed] [Google Scholar]
- 11. Jonas JB, Stroux A, Velten I, Juenemann A, Martus P, Budde WM. . Central corneal thickness correlated with glaucoma damage and rate of progression. . 2005; 46: 1269– 1274. [DOI] [PubMed] [Google Scholar]
- 12. Dimasi DP, Hewitt AW, Kagame K,et al. . Ethnic and mouse strain differences in central corneal thickness and association with pigmentation phenotype. . 2011; 6: e22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chua J, Tham YC, Liao J,et al. . Ethnic differences of intraocular pressure and central corneal thickness: the Singapore Epidemiology of Eye Diseases study. . 2014; 121: 2013– 2022. [DOI] [PubMed] [Google Scholar]
- 14. Lu Y, Vitart V, Burdon KP,et al. . Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. . 2013; 45: 155– 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu Y, Dimasi DP, Hysi PG,et al. . Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. . 2010; 6: e1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vitart V, Bencić G, Hayward C,et al. . New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. . 2010; 19: 4304– 4311. [DOI] [PubMed] [Google Scholar]
- 17. Vithana EN, Aung T, Khor CC,et al. . Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 2011; 20: 649– 658. [DOI] [PubMed] [Google Scholar]
- 18. Gao X, Gauderman WJ, Liu Y,et al. . A genome-wide association study of central corneal thickness in Latinos. . 2013; 54: 2435– 2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoehn R, Zeller T, Verhoeven VJ,et al. . Population-based meta-analysis in Caucasians confirms association with COL5A1 and ZNF469 but not COL8A2 with central corneal thickness. . 2012; 131: 1783– 1793. [DOI] [PubMed] [Google Scholar]
- 20. Gao X, Nannini DR, Corrao K,et al. for the International Glaucoma Genetics Consortium. . Genome-wide association study identifies WNT7B as a novel locus for central corneal thickness in Latinos. . 2016; 25: 5035– 5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyake M, Yamashiro K, Tabara Y,et al. . Identification of myopia-associated WNT7B polymorphisms provides insights into the mechanism underlying the development of myopia. . 2015; 6: 6689. [DOI] [PubMed] [Google Scholar]
- 22. Philomenadin FS, Asokan R, Visawanathan N, George R, Lingam V, Sarangapani S. . Genetic association of SNPs near ATOH7, CARD10, CDKN2B, CDC7 and SIX1/SIX6 with the endophenotypes of primary open angle glaucoma in Indian population. . 2015; 10: e0119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vijaya L, Rashima A, Panday M,et al. . Predictors for incidence of primary open-angle glaucoma in a South Indian population: the Chennai eye disease incidence study. . 2014; 121: 1370– 1376. [DOI] [PubMed] [Google Scholar]
- 24. Panday M, George R, Asokan R,et al. . Six-year incidence of ocular hypertension in a South Indian population: the Chennai eye disease incidence study. . 2015; 99: 604– 608. [DOI] [PubMed] [Google Scholar]
- 25. Bourne RR, Stevens GA, White RA,et al. . Causes of vision loss worldwide, 1990–2010: a systematic analysis. . 2013; 1: e339– e349. [DOI] [PubMed] [Google Scholar]
- 26. Vijaya L, Asokan R, Panday M,et al. . Baseline risk factors for incidence of blindness in a South Indian population: the Chennai Eye Disease Incidence Study. . 2014; 55: 5545– 5550. [DOI] [PubMed] [Google Scholar]
- 27. Vijaya L, George R, Asokan R, Velumuri L, Ramesh SV. . Prevalence and causes of low vision and blindness in an urban population: the Chennai Glaucoma Study. . 2014; 62: 477– 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Purcell S, Neale B, Todd-Brown K,et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. . 2007; 81: 559– 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. . Robust relationship inference in genome-wide association studies. . 2010; 26: 2867– 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang J, Benyamin B, McEvoy BP,et al. . Common SNPs explain a large proportion of the heritability for human height. . 2010; 42: 565– 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abecasis GR, Cherny SS, Cookson WO, Cardon LR. . Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002; 30: 97– 101. [DOI] [PubMed] [Google Scholar]
- 32. Chen WM, Abecasis GR. . Family-based association tests for genomewide association scans. . 2007; 81: 913– 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kirichenko AV, Belonogova NM, Aulchenko YS, Axenovich TI. . PedStr software for cutting large pedigrees for haplotyping, IBD computation and multipoint linkage analysis. . 2009; 73 pt 5: 527– 531. [DOI] [PubMed] [Google Scholar]
- 34. Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. . SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. . 2008; 24: 2938– 2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Higgins JP, Thompson SG, Deeks JJ, Altman DG. . Measuring inconsistency in meta-analyses. . 2003; 327: 557– 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Viechtbauer W. . Conducting meta-analyses in R with the metafor package. . 2010; 36: 1– 48. [Google Scholar]
- 37. Wickham H. . New York, NY: Springer-Verlag New York; 2009. [Google Scholar]
- 38. Purcell S, Cherny SS, Sham PC. . Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. . 2003; 19: 149– 150. [DOI] [PubMed] [Google Scholar]
- 39. Bailey JN, Loomis SJ, Kang JH,et al. . Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. . 2016; 48: 189– 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thulasiraj RD, Nirmalan PK, Ramakrishnan R,et al. . Blindness and vision impairment in a rural south Indian population: the Aravind Comprehensive Eye Survey. . 2003; 110: 1491– 1498. [DOI] [PubMed] [Google Scholar]
- 41. Bengoa-Vergniory N, Kypta RM. . Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. . 2015; 72: 4157– 4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Famili F, Brugman MH, Taskesen E, Naber BE, Fodde R, Staal FJ. . High levels of canonical Wnt signaling lead to loss of stemness and increased differentiation in hematopoietic stem cells. . 2016; 6: 652– 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakatsu MN, Ding Z, Ng MY, Truong TT, Yu F, Deng SX. . Wnt/β-catenin signaling regulates proliferation of human cornea epithelial stem/progenitor cells. . 2011; 52: 4734– 4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cuellar-Partida G, Springelkamp H, Lucas SE,et al. . WNT10A exonic variant increases the risk of keratoconus by decreasing corneal thickness. . 2015; 24: 5060– 5068. [DOI] [PubMed] [Google Scholar]
- 45. Seitan VC, Faure AJ, Zhan Y,et al. . Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. . 2013; 23: 2066– 2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roy S, Siahpirani AF, Chasman D,et al. . A predictive modeling approach for cell line-specific long-range regulatory interactions. . 2015; 43: 8694– 8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klarin D, Zhu QM, Emdin CA,et al. ; . for the CARDIoGRAMplusC4D Consortium. Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. . 2017; 49: 1392– 1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Denny JC, Bastarache L, Roden DM. . Phenome-wide association studies as a tool to advance precision medicine. . 2016; 17: 353– 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Denny JC, Bastarache L, Ritchie MD,et al. . Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. . 2013; 31: 1102– 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.