Abstract
CpG islands (CGIs) have long been implicated in the regulation of vertebrate gene expression. However, the involvement of CGIs in chromosomal architectures and associated gene expression regulations has not yet been thoroughly explored. By combining large-scale integrative data analyses and experimental validations, we show that CGIs clearly reconcile two competing models explaining nuclear gene localizations. We first identify CGI-containing (CGI+) and CGI-less (CGI−) genes are non-randomly clustered within the genome, which reflects CGI-dependent spatial gene segregation in the nucleus and corresponding gene regulatory modes. Regardless of their transcriptional activities, CGI+ genes are mainly located at the nuclear center and encounter frequent long-range chromosomal interactions. Meanwhile, nuclear peripheral CGI− genes forming heterochromatin are activated and internalized into the nuclear center by local enhancer–promoter interactions. Our findings demonstrate the crucial implications of CGIs on chromosomal architectures and gene positioning, linking the critical importance of CGIs in determining distinct mechanisms of global gene regulation in three-dimensional space in the nucleus.
INTRODUCTION
Three-dimensional (3D) chromosomal architecture plays critical roles in mammalian gene regulation. One model explaining nuclear gene localization is the chromosomal territory-extrusion model (CT-extrusion model) (1). In this model, each chromosome occupies its own territory, mainly located at the nuclear periphery and genes located in their own chromosome’s territory form silent heterochromatin. Activation of genes occurs through the internalization of genes into the nuclear center in concert with the conversion to euchromatin. Alternatively, other studies introduced a concept of nuclear subcompartments with specialized functions (nuclear subcompartment model) (2,3). According to this model, the nucleus contains hundreds of transcription factories where active transcriptional machineries are focally enriched, while Polycomb repressive complex (PRC) proteins aggregate to form Polycomb bodies. It was shown that genes remain repressed when they are located within Polycomb bodies (3,4) while localizing to the transcription factories when activated (2,5,6). However, it has long been overlooked that not only transcription factories (2,7) but also Polycomb bodies (3,8,9) are mainly detected within the nuclear center, rather than the periphery. Therefore, in terms of location of inactive genes, the two models explaining chromosomal architectures, CT-extrusion model and nuclear subcompartment model, are mutually incompatible, and neither of them can explain the general localization behaviors of the all genes within the genome. In parallel, it is unclear which genes are inactivated by heterochromatin formation and which genes are repressed by PRC.
To resolve these contradictions, we performed a large-scale integrative data analysis, particularly focusing on the implications of CpG islands (CGIs) in 3D chromosomal architectures. CGIs, originally defined based on the sequence characteristics of high-GC contents and CpG-dinucleotide frequencies (10–12), have been recently recognized as hotspots for global gene regulation (13–16). In mammalian genome, ∼60% of genes have CGIs near their promoters (CGI+ genes) while the other 40% do not (CGI− genes) (15,16). However, how CGI+ and CGI− genes are organized within nucleus has not been well understood.
MATERIALS AND METHODS
Definition of CGI± and CGI− genes
CGI-containing (CGI+) and CGI-less (CGI−) genes were defined as follows. We used experimentally validated CGI elements identified by CxxC-affinity purification followed by parallel sequencing (CAP-seq) data (17). In detail, we listed CxxC-affinity purified regions in sperm, blood and cerebellum (both in mouse and human) with a general ChIP-seq data analysis pipeline (see ChIP-seq, DamID-seq, MeDIP-seq and DNaseI-seq data analysis section in ‘Material and Methods’ section), and identified non-tissue specific consensus CxxC-domain binding regions (listed in Supplementary Table S3). For gene classification, genes surrounded by consensus CxxC binding regions (within ± 500 bp of the TSSs) were considered to be CGI+ genes, while genes without surrounding consensus CxxC binding regions were defined as CGI− genes (listed in Supplementary Table S4).
3D DNA-FISH (fluorescence in situ hybridization)
Fluorescence in situ hybridization (FISH) was performed as previously described using the Oligopaint technique (18) with modifications for the identification of exact 3D location of target loci. Fluorescence labeled FISH probe libraries were designed as either ssDNA 36mers (ATTO-550, Figure 2E and Supplementary Movie S1) or ssDNA 45mers (ATTO-550 and ATTO-488, Figure 2F) and synthesized by MYcroarray (Ann Arbor, MI, USA). Cells grown adherently were suspended by trypsinization and fixed using 4% PFA. To avoid loss of cells during the solution exchange and washing steps, suspended cells were stained with trypan blue whenever necessary. Single locus detection (Figure 2E) was done with 20 pmol probes in hybridization cocktail with 2× saline-sodium citrate buffer with 0.1% tween-20 (SSCT), 50% formamide, 10% (w/v) dextran sulfate and 10 μg RNase A. The target region for the CGI− gene cluster (Myosin heavy chain cluster in Figure 2E) was chr11:66,977,423–67,174,410 (n = 3,157), and the target region for the CGI+ cluster (Rbm24/Cap2 cluster in Figure 2E) was chr13:46,483,276–46,661,642 (n = 2,798). On the other hand, multiple loci detection (Figure 2F, ∼20 000 oligo library, target regions are listed in Supplementary Table S1) was done with 200 pmol probes in 2× SSCT, 50% formamide, 30% (w/v) dextran sulfate and 10 μg RNase A. After staining, cells were washed and resuspended in anti-fade mounting medium, and 10 μl of the cell resuspension was dropped onto a glass slide and gently covered by an 18 × 18mm coverslip. Nuclei were imaged with Zeiss LSM 710 Confocal Microscope with Z-stacks. To determine localization, loci detected within 10% of the longest diameter from nucleus periphery in reconstituted 3D-view images (for example, see Supplementary Movie S1) were considered as peripheral loci, while the rest were considered to be located at the nuclear center.
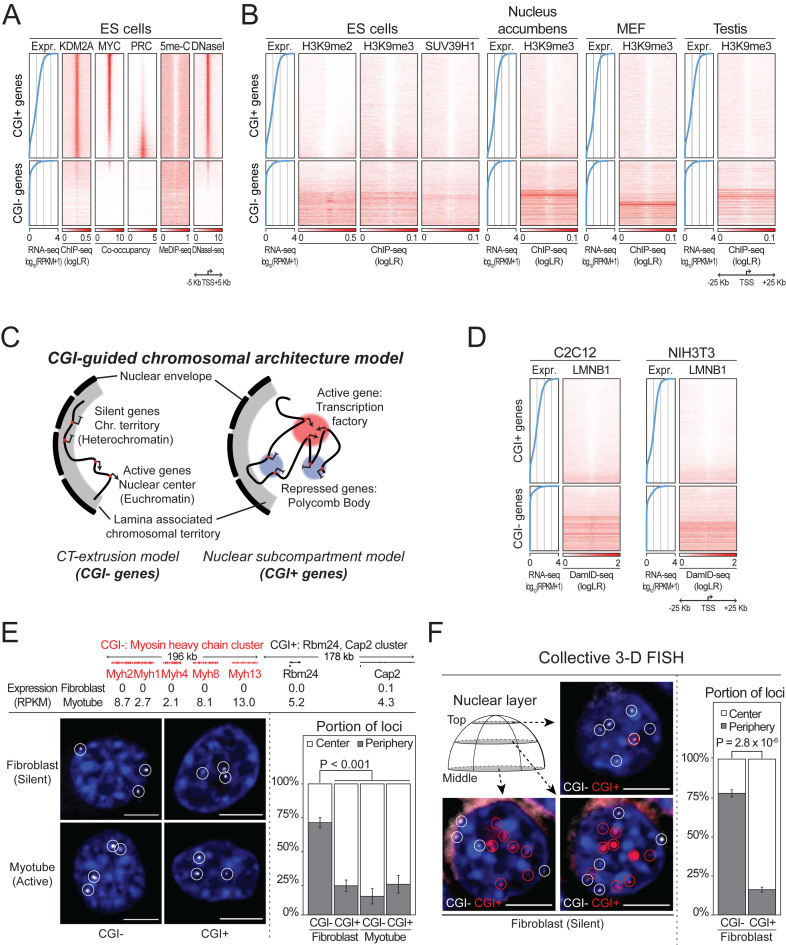
Figure 2.
Spatial segregation of CGI+ and CGI− genes. (A) CGI-centered regulation mechanisms in CGI+ genes, and (B) heterochromatin features in CGI− genes. All protein coding genes are sorted by their expression values (left blue line plot), and the genomic landscape of promoter surrounding regions are shown. (C) CGI-guided chromosomal architecture model. (D) Association of peripheral nuclear lamina (LMNB1) with silent CGI− genes, but not CGI+ genes. (E) Localization of single CGI+ (or CGI−) gene cluster in fibroblasts and myotubes, and (F) multiple silent CGI+/CGI− gene clusters in fibroblasts (eight clusters each) determined by 3D FISH. Percentages of gene loci located in the nuclear center compared to the periphery in each cell are shown in bottom right. P-values: Wilcoxon signed-rank tests. Scale bar: 5 μm. Error bars show S.E.M.
Selection of high-quality H3K9me2/3 ChIP-seq data
One of the most critical issues in ChIP-seq data analyses of heterochromatin regions is the contamination of active genomic regions, such as the promoters of housekeeping genes, during the ChIP process (19), which form high false positive signals. To circumvent this issue, we assessed H3K9me2/3 ChIP-seq data qualities in the published ChIP-seq data repository. Available mouse H3K9me2/3 ChIP-seq data deposited at the GEO website were listed and downloaded at 25 October 2015 and 29 September 2017 (total 287 experiments). Among them, the experiments done with in-house generated antibodies (12 experiments), experiments lacking antibody information (12 experiments), the experiments without author-provided control experiments (input or mock ChIP; 35 experiments) were not used for the further analyses. The quality of the remaining 228 experiments was assessed by two criteria: high signal to noise ratio (SNR) (16) and low false positive signal in active genomic regions. First, to assess SNR, H3K9me2/3 peaks of each experiment were called using histone modified region identification pipeline (‘–nomodel –nolambda’ option in MACS 1.4.2) using the High Performance Parallel Computing system (Texas Advanced Computing Center, the University of Texas at Austin). To monitor the background level, an SNR was calculated from duplicate read filtered bedGraph files generated by MACS 1.4.2 for each ChIP-seq data. After filtering out all high background data with a stringent filtering criterion of SNR 0.1, a total of 93 H3K9me2/3 ChIP-seq data were used for the further assessments. Second, to assess the false positive signal, the promoters of housekeeping genes were identified and used as the representative active genomic regions. For this, the 1524 pA+ RNA-seq profile (see RNA-seq data analysis in ‘Materials and Methods’ section; Supplementary Figure S6 and Table S5) was clustered with K-means clustering algorithm and 2442 genes that were active in all samples were identified, and the average H3K9me2/3 signal within the ± 500bp from the TSSs was calculated. For the reliable heterochromatin regions, Giemsa positive bands (gpos100 and gpos66) were used. As expected, a large portion of the experiments were contaminated with active genomic regions, and experiments whose average signal in Giemsa positive bands were at least threefold higher than the average signal in active regions were selected (39 experiments) and used for the further analyses shown in the examples of Figure 2B.
Gene frequency analyses in human G bands and isochores
For gene frequency analyses with regard to G bands (Supplementary Figure S2), human (hg19) cytoband lists were downloaded from the UCSC Genome Browser Database website (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/database/). Isochore lists were downloaded from the IsoFinder website (20) (http://bioinfo2.ugr.es:8080/isochores/human-isochores/).
Gene homology analysis (BLAST)
To measure the level of homologies between neighboring gene pairs in Figure 1C, we performed BLAST analysis using whole protein coding genes in the mouse genome. Since the BLAST E-values are sensitive to the target database size, we fixed the database to all proteins in the mouse genome and performed NxN BLASTP analysis using each single protein as query. Neighboring gene pairs with an E-value less than 1E-10 were considered homologous pairs.
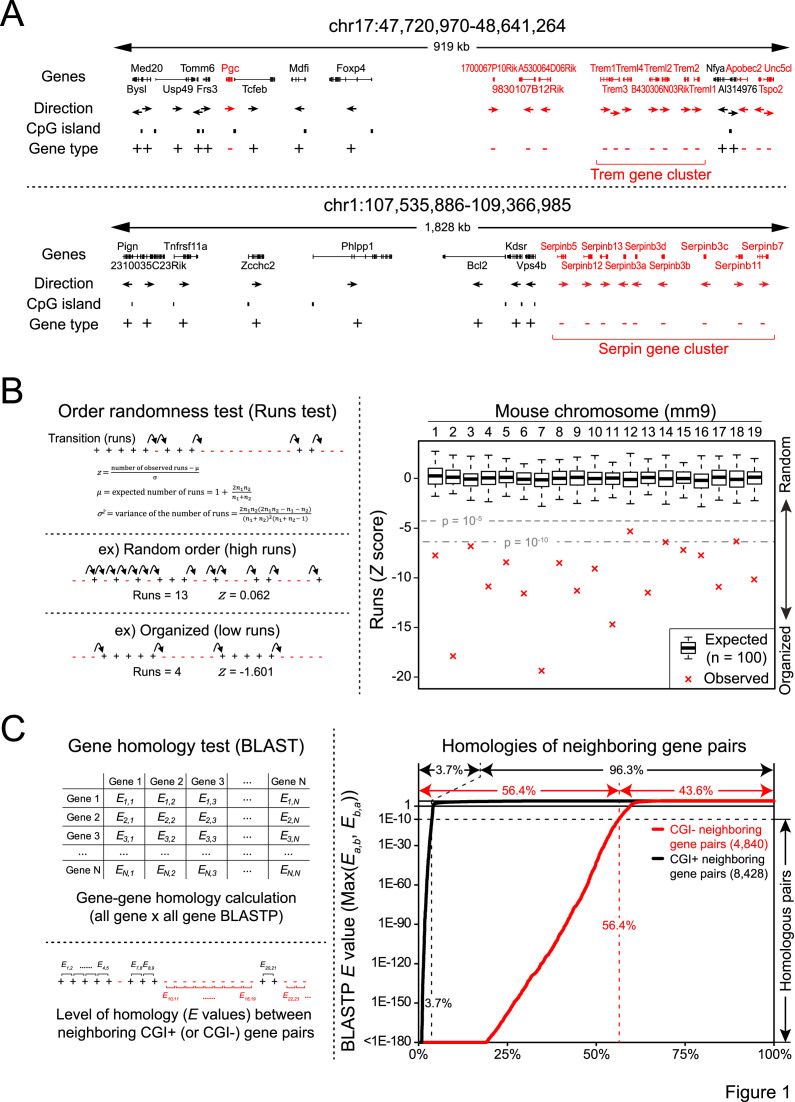
Figure 1.
Non-random linear separation of CGI+ and CGI− genes in the genome and juxtaposition of CGI− homologs. (A) Examples of CGI+ (black) and CGI− (red) gene arrangements in the mouse genome. Homologous genes that co-cluster together are indicated by red brackets. (B) Gene order randomness test (Runs test) of CGI+/CGI− genes in the mouse genome (right). Box plots show expected transition from random gene shuffling. Left panel shows schematic representation of the Runs test (top), examples of randomly arranged genes and well-organized genes (bottom). (C) Neighboring gene homology test between all neighboring CGI+ (black) and CGI− (red) gene pairs. Gene pairs are sorted by E-value in ascending manner.
Cell cultures
NIH-3T3 fibroblasts and C2C12 myoblasts were maintained in Dulbecco’s Modified Eagle Medium (DMEM; GIBCO) containing 10% fetal bovine serum (GIBCO). For myotube differentiation of C2C12 myoblasts, cells were grown to high confluence and then the media was switched to DMEM supplemented with 2% donor equine serum (HyClone) and 1 μM insulin (Sigma-Aldrich).
ChIP-seq, DamID-seq, MeDIP-seq and DNaseI-seq data analysis
ChIP-seq, DamID-seq, MeDIP-seq and DNaseI-seq data were downloaded from sequence read archive (SRA) from the National Center for Biotechnology Information (NCBI) database. FASTQ files were extracted with the SRA Toolkit version 2.5.5 and aligned using Bowtie 2.2.5 to the mouse genome (mm9, NCBI Build 37). To identify CTCF binding sites in mESC, CxxC binding regions for defining CGI+ and CGI− genes (see ‘Definition of CGI+ and CGI− genes’ in ‘Material and Methods’ section) and transcription factor (TF) binding sites (Figure 3E and F), model-based analysis for the ChIP-seq peak caller (MACS 1.4.2) (21) was used with a P-value cutoff of 1E-5. TF binding target genes were defined as genes with the TF binding within ±2 Kb of the TSS. Signal based analyses were done using duplicate filtered read pileup bedGraph files made from MACS 1.4.2. In order to summarize the ChIP signal enrichment over controls, the background subtracted bedGraph files with log likelihood ratios were made using MACS2 version 2.1.1 with ‘bdgcmp -m logLR’ command. For the data without control experiments (5me-C, DNaseI), the total area under the signal curve from bedGraph file was normalized to be one billion (1 × 109).
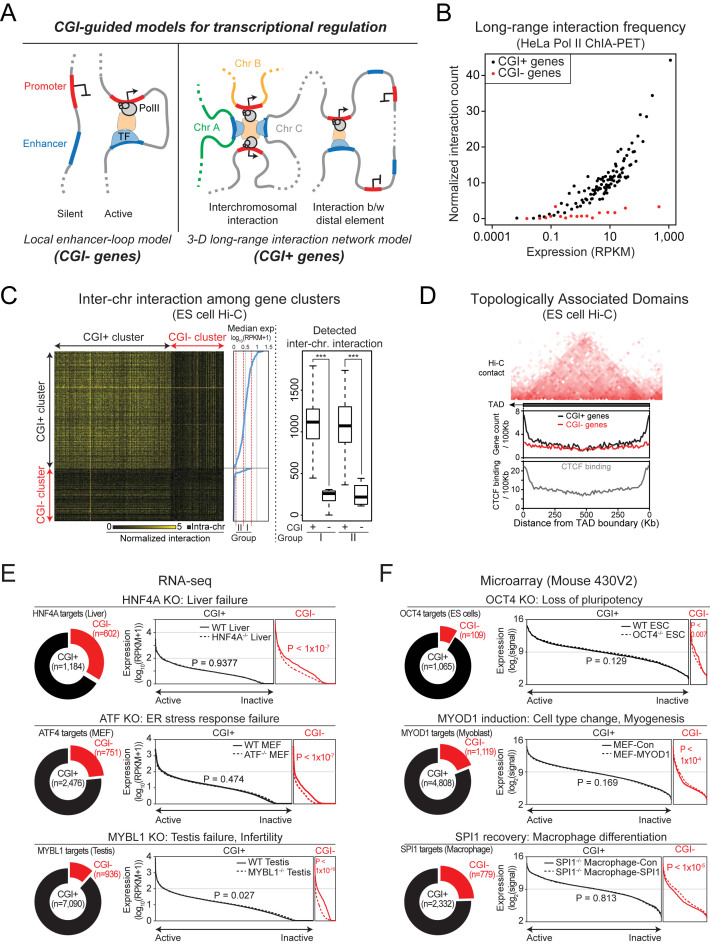
Figure 3.
Transcriptional regulation mechanisms of CGI+ and CGI− genes. (A) CGI-guided models for mammalian transcriptional regulation mechanisms. (B) Higher frequency of Pol II-mediated chromosomal long-range interactions in CGI+ genes compared to CGI− genes. Genes were sorted by expression level and binned into every 100 genes, and average expression and normalized long-range interaction count of bins are shown. (C) Frequency of inter-chromosomal interactions in CGI+ and CGI− gene clusters (Supplementary Table S2; sorted by their median expression) determined by mouse ES cell Hi-C data. Normalized total detected inter-chromosomal interaction counts in cluster groups are shown in right. ***P < 0.0001 from Wilcoxon signed-rank test. (D) CGI+ and CGI− gene frequencies as well as CTCF binding frequencies with regards to TAD structures. (E and F) TF binding target gene expression analysis upon TF perturbation. In each line plot, target genes are sorted by expression level. P-values are calculated from Kolmogorov–Smirnov tests.
RNA-seq analysis
RNA-seq data were downloaded from SRA. FASTQ files were aligned to the mouse genome (mm9, NCBI Build 37) using STAR version 2.4.2 (22). Gene expression was calculated as RPKM values using rpkmforgenes.py (23). Because the ranges of RPKM values span over three orders of magnitude and tend to give high random multiplicative error in high expression values, expression values were converted into log10 scale (log10(RPKM+1)) for graphical summarization. For a unified gene expression profile of diverse tissues and cell lines shown in Supplementary Figure S6, all available mouse poly-A positive RNA-seq data (3818 samples) were summarized and downloaded on 5 May 2015. The measurement of gene expression of these samples was done using high-performance parallel computing system (Texas Advanced Computing Center, the University of Texas at Austin). Among the 3,818 samples, excluding single cell RNA-seq or experiments whose expression verified gene counts are small (less than 5,000 genes with RPKM 0.5 or higher), 1,524 high quality RNA-seq data were used. The gene expression profile was summarized as log10 scale (log10(RPKM+1)) and neighboring gene expression similarities were monitored by calculating Pearson Correlation Coefficients. Manually curated sample information was summarized in Supplementary Table S5, and the expression profile was deposited in the Gene Expression Omnibus (accession number GSE80797).
Microarray analysis
As a unified gene expression profile of diverse tissues and cell lines shown in Supplementary Figure S6, microarray data from GNF (Genomics Institute of the Novartis Research Foundation) Mouse Gene Atlas V3 (GSE10246) (24) were used. To precisely monitor expression values, raw data files (.cel files) were background corrected and normalized with GC Robust Multi-array Average expression measure using sequence information (GCRMA) (25) methods to minimize the background signal originating from probe sequence or high GC content. For genes with multiple probe sets, only probes with maximal signal were used for further analyses. For the expression data shown in Figure 3F, downloaded .cel files were normalized with the Robust Multi-array Average (RMA) (26) method.
ChIA-PET, Hi-C, Repli-chip data analysis
For ChIA-PET data analysis (Figure 3B and Supplementary Figure S5), the long-range chromatin interaction lists detected in HeLa, HCT116, K562, and MCF7 cells (27) were downloaded from ENCODE website. CGI+ and CGI− genes were sorted by expression level and binned into every 100 genes. Average detected count of long-range interactions within ±3 Kb from the TSSs was measured.
For inter-chromosomal interaction frequency analysis using Hi-C in Figure 3C, aligned read pairs were downloaded from Gene Expression Omnibus (GEO; GSE35156). Among the reads, the pairs matching with the CGI+ or CGI− gene clusters at both ends were filtered, and the inter-chromosomal interaction counts between gene clusters were normalized with restriction enzyme site count (HindIII, NcoI) within gene clusters. For Hi-C PCA analysis in Supplementary Figure S4A, ‘runHiCpca.pl’ was used in the HOMER (Hypergeometric Optimization of Motif EnRichment) program suite (28). For gene count analysis with regards to topologically associated domain (TAD) in Figure 3D, TAD lists determined in mouse embryonic stem (ES) cells were downloaded and used (29).
For replication timing analysis in Supplementary Figure S4B, Repli-chip wavelet-smoothed signal data (30) were downloaded from ENCODE website. For each CGI+ and CGI− gene clusters, average replication timing values (log2[early S phase signal/late S phase signal]) were calculated.
RESULTS
Non-random arrangement of CGI+ and CGI− genes
We first focused on the arrangements of CGI+ and CGI− genes in chromosomes. Since modern genome sequences are the cumulative outcome of a series of chromosomal rearrangement events throughout generations (31), we speculated that the current gene arrangements may expose important information about the environments to which each gene has been exposed (32). One apparent pattern we observed from the visual inspection of the mouse genome was the non-random linear separation of CGI+ and CGI− genes: multiple CGI+ genes are co-clustered together, while CGI− genes are also gathered separately in other regions (Figure 1A). To further test whether this pattern is prevalent in the mammalian genome, we performed an order randomness test (Runs Test) (33) by simplifying gene arrangements in each chromosome into binarized gene orders (CGI+ or CGI−; Figure 1B). As a result, we found significant linear separations of CGI+ or CGI− genes in all mouse and most human chromosomes (Figure 1B and Supplementary Figure S1; see also Supplementary Figure S2).
We additionally observed that homologous CGI− genes, presumably formed by local gene duplication, are often clustered together (Trem/Serpin gene clusters in Figure 1A), while CGI+ genes are not. By assessing the homology between neighboring gene pairs (Figure 1C), we found that more than half (56.4%) of all CGI− neighboring gene pairs are highly homologous (BLAST E-value < 1E-10), while only 3.7% of CGI+ gene pairs are homologous. Notably, the local gene duplication patterns (Supplementary Figure S3) can be sequestered by chromosomal translocations (31,34), frequently occurring among spatially proximal regions (32,35). Therefore, our findings imply that CGI+ and CGI− gene classes are located within totally different nuclear environments: CGI+ genes might be spatially proximate to each other and encounter relatively frequent chromosomal contacts leading to subsequent rearrangements, while CGI− genes might be spatially segregated from CGI+ genes and experience fewer chromosomal interactions.
Effects of CGIs on chromatin status
To understand the localization behaviors of CGI+ and CGI− genes, we thoroughly investigated chromatin status using datasets generated from mouse ES cells (16,36–38). As shown in Figure 2A, promoters with CGI elements are occupied by KDM2A, a CxxC domain-containing protein, regardless of associated gene activities. As we previously reported, the occupancy patterns of MYC or PRC class DNA-binding proteins are also confined to CGI+ genes (16). Accordingly, CGI+ promoters constantly remain unmethylated (5me-C) and sustain chromatin accessibility to some degree (DNaseI), even when genes are repressed by PRC factors. Since TF occupancy, CpG hypomethylation, and chromatin accessibility are hallmarks of euchromatin (39), these results evidently illustrate that CGI+ genes are generally associated with euchromatin.
On the other hand, the promoters of CGI− genes are generally methylated (5me-C) and inaccessible (DNaseI). To test whether these genes form heterochromatin, we additionally monitored H3K9me2/3 signals, which are representative heterochromatin signatures, and the occupancy of SUV39H1, an H3K9 specific methyltransferase, using systematically selected high-quality ChIP-seq data attained from multiple mouse samples (‘Materials and Methods’ section) (40–45). The results revealed that the surrounding regions of silent CGI− promoters are largely associated with heterochromatin signatures compared to active CGI− genes (Figure 2B). This implies that CGI− genes are associated with heterochromatin, and can be converted to euchromatin upon activation. Importantly, CGI+ genes show largely depleted H3K9me2/3 and SUV39H1 signals regardless of their activities. These data clarify that PRC suppresses CGI+ gene expression while CGI− genes are inactivated by heterochromatin formation.
Spatial segregation of CGI+ and CGI− genes in the nucleus
Based on our findings along with prior observations that the heterochromatin is located at the nuclear periphery while euchromatin is located at the core of the nucleus (46), we hypothesized that CGI+ and CGI− genes are spatially segregated within the nucleus and propose a revised model for nuclear gene localizations (Figure 2C). In our model, CGI+ genes stay within the nuclear center regardless of their activities, co-localizing with either transcription factories or Polycomb bodies depending on their activities as explained in the nuclear subcompartment model. On the other hand, generally silent CGI− genes are positioned at the nuclear periphery, forming heterochromatin. Upon activation, they extrude into the nuclear center and form euchromatin, which fits into the CT-extrusion model.
To test our revised model, we examined chromosomal association with nuclear peripheral lamina (47) by assessing the association of lamin B1 (LMNB1) at the surrounding regions of CGI+ and CGI− promoters using published DamID-seq data (48). LMNB1 preferentially occupies the surrounding regions of silent CGI− promoters, suggesting that silent CGI− genes are located at the lamina-associated nuclear periphery (Figure 2D). The results also imply that active CGI− genes without LMNB1 association are located distantly from the nuclear periphery, and this fits into the CT-extrusion model. Notably, LMNB1 association is clearly depleted near almost all CGI+ gene promoters regardless of gene activities, implying their consistent localization at the nuclear center area as explained in the nuclear subcompartment model.
We further experimentally validated our model by performing FISH, mapping the nuclear position of CGI+ and CGI− gene clusters that are inactive in fibroblasts, but active in myotubes. As shown in Figure 2E (for 3D view, Supplementary Movie S1), the silent CGI− gene cluster was mainly detected at the nuclear periphery of fibroblasts, but at the nuclear center in myotubes. On the other hand, the CGI+ gene cluster was mostly found at the nuclear center in both fibroblasts and myotubes. We then monitored multiple gene clusters collectively using Oligopaint techniques (18). As the most important differences expected are localization behaviors between inactive CGI+ and CGI− genes (Figure 2C; center and periphery, respectively), we designed FISH probes targeting multiple CGI+ and CGI− gene clusters that are inactive in fibroblasts (eight clusters each; Supplementary Table S1). As shown in Figure 2F, we confirmed that silent CGI− gene clusters are generally located at the nuclear periphery, while inactive CGI+ gene clusters reside mainly at the nuclear center, clearly supporting our model of CGI-dependent spatial gene segregation (Figure 2C; see also Supplementary Figure S4).
CGI and transcriptional regulation mechanisms
We further questioned whether the different localization patterns of CGI+ and CGI− genes would resolve the discrepancy among the proposed models explaining transcriptional regulation mechanisms (Figure 3A). The most widely accepted transcriptional regulation mechanisms is the local enhancer-loop model proposed almost three decades ago (49), where enhancers occupied by TFs communicate with nearby promoters via chromosomal looping for gene activation. In this model, the expression levels of the target genes associated with a specific TF are expected to be directly affected by perturbation of the TF. However, TF target genes often do not show expected expression changes upon TF perturbations (50). Alternatively, recent high-throughput chromatin interaction analyses have revealed most enhancers are associated with promoters beyond the nearest ones, and multiple enhancers and promoters form complex 3D long-range interaction networks (3D long-range interaction network model) (51). This new model explains the inconsistencies between TF targets and expression changes upon perturbations of the TFs (50). Nevertheless, considering that the conventional local enhancer-loop model has been also supported by multiple examples (52–54), each model seems to explain the regulation of only limited sets of genes.
To test whether our finding of CGI-mediated gene segregation would explain the discrepancy between models, we measured the long-range interaction frequencies from surrounding regions of the CGI+ and CGI− gene promoters using Pol II ChIA-PET data (27). As shown in Figure 3B, CGI+ genes encounter more frequent long-range interactions than CGI− genes with similar activities (see also Supplementary Figure S5), indicating that nuclear central CGI+ genes are more strongly involved in Pol II-mediated chromosomal long-range interactions compared to CGI− genes, which are generally condensed as heterochromatin. We additionally monitored the relative frequencies of inter-chromosomal interactions using Hi-C data obtained from mouse ES cells (29). As shown in Figure 3C, CGI+ gene clusters show significantly higher frequencies of inter-chromosomal interactions than CGI− gene clusters. Surprisingly, the gene expression levels of the CGI+ gene clusters did not show any strong correlation with the inter-chromosomal interaction frequencies (Figure 3C, right box plot), suggesting that not only active, but also inactive CGI+ genes encounter frequent chromosomal long-range interactions, presumably mediated by Polycomb bodies (3,4).
We then questioned whether our observations of frequent long-range interactions among CGI+ genes agree with the structures formed by local genome entanglement: TADs defined from Hi-C data analysis (29). To delineate the relationship between local topological structures and CGIs, we investigated the location of CGI+ and CGI− genes with regards to the TADs. As shown in Figure 3D, CGI+ genes, but not CGI− genes, are largely enriched at each end of TADs (6.2-fold more than at the center of TADs). As a result, about 23.2% of total CGI+ genes are detected at the inter-TAD regions or the end of TADs (<20 Kb from the ends of TADs; 13.9% for CGI− genes). This is similar with the binding frequency of CTCF (3.5-fold more than at the center of TADs), which is also known to be involved in the chromosomal long-range interactions and responsible for the physical separation of chromosomal domains (29,55). These data show that the chromosomal long-range interactions of CGI+ genes strongly influence local genome structures, similarly to CTCF bindings.
Since CGI− genes are not highly involved in long-range chromosomal interactions (Figure 3B), we speculated that CGI− genes would be more dependent on the local enhancer-mediated regulations. We compared the ChIP-seq and the target gene expression data upon TF removal or induction. Although the majority of targets for each TF are CGI+ genes, not CGI− genes, the depletion or induction of each TF did not significantly affect the global expression levels of the CGI+ target genes (Figure 3E and F). On the other hand, CGI− target genes, which constitute only a minor portion of the total targets of the TF, showed significant changes in gene expression upon depletion or induction of the TF. Thus, CGI− genes respond more strongly to TF perturbation than CGI+ genes. Altogether, our findings strongly suggest that 3D long-range interaction network model explains the regulatory mode of CGI+ genes, while the regulation of CGI− genes fit squarely into local enhancer-loop model (Figure 3A).
DISCUSSION
In this study, we reveal CGI+ and CGI− genes are organized as separated clusters within the genome, and further show CGI-dependent chromatin architectures and CGI-associated global gene regulatory modes. Notably, our findings can explain the prior discrepancies among multiple models describing chromosomal architectures and transcriptional regulation as summarized in Table 1. These distinctions highlight the critical implications of CGIs on gene regulation which has never been clearly elucidated before.
Table 1. Summary of CGI-dependent chromosomal architectures and gene regulations.
| CGI− genes | CGI+ genes | |
|---|---|---|
| Gene regulation | Local enhancer-loop model | 3D long-range interaction network model |
| Chromatin status | Heterochromatin—Euchromatin | Euchromatin |
| Chromosomal architectures | Chromosomal territory (CT)-extrusion model | Nuclear subcompartment model |
| Default gene location in nucleus | Nuclear periphery (extruded into the center when activated) | Nuclear center (regardless of the gene activity) |
| Dimension of regulationa | Linear local regulation | 3D spatial regulation |
| Nuclear landscape modelb | Chromosomal territory-interchromatin compartment (CT-IC) model | Interchromatin network (ICN) model |
aSee also Supplementary Figure S6.
bDiscussed in Supplementary Figure S7.
Genome-wide linear organization of CGI+ and CGI− genes seems to be optimal for the cost-effective gene regulation. It is reasonable to speculate that the co-clustering patterns of CGI+ or CGI− genes (Figure 1) are beneficial for efficient spatial segregation (Figure 2), which in turn allows differential regulation without mutual interferences (Figure 3). The co-clustering patterns (Figure 1), as well as the similar directionality (Supplementary Figure S3) of the neighboring CGI− genes also seem advantageous for simultaneous tissue or stage-specific activation (Supplementary Figure S6) of the multiple CGI− genes with similar function (Figure 1C).
Interestingly, the key components of CGI-mediated dual mode gene regulation, PRC and heterochromatin-mediated repression mechanisms, are universally observed in a broad range of eukaryotes even in the species without CGIs (15,39,56,57). Therefore, it will be imperative to test whether there are distinct modes of gene regulation mediated by specific DNA elements corresponding to the mammalian CGIs in other eukaryotes.
DATA AVAILABILITY
The data reported in this paper are deposited at the Gene Expression Omnibus (accession number GSE80797).
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate Drs Edward Marcotte, Vishwanath Iyer (University of Texat at Austin), Jian Xu (University of Texas Southwestern) and Andrew J. Woo (University of Western Australia) for their critical comments on the manuscript, and Drs Ting Wu and Brian J. Beliveau (Harvard Medical School) for technical advice on Oligopaint technique. We thank the Texas Advanced Computing Center (TACC) for providing HPC resources.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [P20GM0103423 /P20GM104318 to S.B., R01GM112722 to J.K.]. Funding for open access charge: National Institutes of Health [P20GM104318].
Conflict of interest statement. None declared.
REFERENCES
- 1. Chubb J.R., Bickmore W.A.. Considering nuclear compartmentalization in the light of nuclear dynamics. Cell. 2003; 112:403–406. [DOI] [PubMed] [Google Scholar]
- 2. Sutherland H., Bickmore W.A.. Transcription factories: gene expression in unions?. Nat. Rev. Genet. 2009; 10:457–466. [DOI] [PubMed] [Google Scholar]
- 3. Pirrotta V., Li H.B.. A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 2012; 22:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bantignies F., Roure V., Comet I., Leblanc B., Schuettengruber B., Bonnet J., Tixier V., Mas A., Cavalli G.. Polycomb-dependent regulatory contacts between distant hox loci in drosophila. Cell. 2011; 144:214–226. [DOI] [PubMed] [Google Scholar]
- 5. Brown J.M., Green J., das Neves R.P., Wallace H.A.C., Smith A.J.H., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R. et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 2008; 182:1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buckley M.S., Lis J.T.. Imaging RNA Polymerase II transcription sites in living cells. Curr. Opin. Genet. Dev. 2014; 25:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iborra F.J., Pombo A., Jackson D.A., Cook P.R.. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J. Cell Sci. 1996; 109:1427–1436. [DOI] [PubMed] [Google Scholar]
- 8. Hernández-Muñoz I., Taghavi P., Kuijl C., Neefjes J., van Lohuizen M.. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol. Cell. Biol. 2005; 25:11047–11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van der Stoop P., Boutsma E.A., Hulsman D., Noback S., Heimerikx M., Kerkhoven R.M., Voncken J.W., Wessels L.F.A., van Lohuizen M.. Ubiquitin E3 ligase ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One. 2008; 3:e2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bird A., Taggart M., Frommer M., Miller O.J., Macleod D.. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985; 40:91–99. [DOI] [PubMed] [Google Scholar]
- 11. Gardiner-Garden M., Frommer M.. CpG Islands in vertebrate genomes. J. Mol. Biol. 1987; 196:261–282. [DOI] [PubMed] [Google Scholar]
- 12. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002; 16:6–21. [DOI] [PubMed] [Google Scholar]
- 13. Fouse S.D., Shen Y., Pellegrini M., Cole S., Meissner A., Van Neste L., Jaenisch R., Fan G.. Promoter CpG Methylation Contributes to ES Cell Gene Regulation in Parallel with Oct4/Nanog, PcG Complex, and Histone H3 K4/K27 Trimethylation. Cell Stem Cell. 2008; 2:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008; 454:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deaton A.M., Bird A.. CpG islands and the regulation of transcription. Genes Dev. 2011; 25:1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck S., Lee B.-K., Rhee C., Song J., Woo A.J., Kim J.. CpG island-mediated global gene regulatory modes in mouse embryonic stem cells. Nat. Commun. 2014; 5:5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Illingworth R.S., Gruenewald-Schneider U., Webb S., Kerr A.R.W., James K.D., Turner D.J., Smith C., Harrison D.J., Andrews R., Bird A.P.. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010; 6:e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beliveau B.J., Joyce E.F., Apostolopoulos N., Yilmaz F., Fonseka C.Y., McCole R.B., Chang Y., Li J.B., Senaratne T.N., Williams B.R. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:21301–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jain D., Baldi S., Zabel A., Straub T., Becker P.B.. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res. 2015; 43:6959–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oliver J.L., Carpena P., Hackenberg M., Bernaola-Galván P.. IsoFinder: Computational prediction of isochores in genome sequences. Nucleic Acids Res. 2004; 32:W287–W292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramsköld D., Wang E.T., Burge C.B., Sandberg R.. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009; 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lattin J.E., Schroder K., Su A.I., Walker J.R., Zhang J., Wiltshire T., Saijo K., Glass C.K., Hume D.A., Kellie S. et al. Expression analysis of G protein-coupled receptors in mouse macrophages. Immun. Res. 2008; 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu Z., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F.. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004; 99:909–917. [Google Scholar]
- 26. Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P.. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003; 4:249–264. [DOI] [PubMed] [Google Scholar]
- 27. Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012; 148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., Papp B., Fussner E., Bazett-Jones D.P., Plath K., Dalton S. et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010; 20:155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D.. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bickmore W.A., Teague P.. Influences of chromosome size, gene density and nuclear position on the frequency of constitutional translocations in the human population. Chromosome Res. 2002; 10:707–715. [DOI] [PubMed] [Google Scholar]
- 33. Wald A., Wolfowitz J., Wald2 A.. On a test whether two samples are from the same population. Ann. Math. Stat. 1940; 11:147–162. [Google Scholar]
- 34. Tillier E.R., Collins R.A.. Genome rearrangement by replication-directed translocation. Nat. Genet. 2000; 26:195–197. [DOI] [PubMed] [Google Scholar]
- 35. Branco M.R., Pombo A.. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006; 4:780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blackledge N.P., Zhou J.C., Tolstorukov M.Y., Farcas A.M., Park P.J., Klose R.J.. CpG islands recruit a histone H3 lysine 36 demethylase. Mol. Cell. 2010; 38:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen L., Wu H., Diep D., Yamaguchi S., D’Alessio A.C., Fung H.L., Zhang K., Zhang Y.. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013; 153:692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014; 515:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Henikoff S. Heterochromatin function in complex genomes. Biochim. Biophys. Acta. 2000; 1470:O1–O8. [DOI] [PubMed] [Google Scholar]
- 40. Maze I., Feng J., Wilkinson M.B., Sun H., Shen L., Nestler E.J.. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3035–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bulut-Karslioglu A., DeLaRosa-Velázquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galán C., Winter G.E., Engist B., Gerle B. et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014; 55:277–290. [DOI] [PubMed] [Google Scholar]
- 42. Pedersen M.T., Agger K., Laugesen A., Johansen J. V, Cloos P.A.C., Christensen J., Helin K.. The demethylase JMJD2C localizes to H3K4me3 positive transcription start sites and is dispensable for embryonic development. Mol. Cell. Biol. 2014; 34:1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pezic D., Manakov S.A., Sachidanandam R., Aravin A.A.. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 2014; 28:1410–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elsässer S.J., Noh K.M., Diaz N., Allis C.D., Banaszynski L.A.. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015; 522:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Meyenn F., Iurlaro M., Habibi E., Liu N.Q., Salehzadeh-Yazdi A., Santos F., Petrini E., Milagre I., Yu M., Xie Z. et al. Impairment of DNA methylation maintenance is the main cause of global demethylation in naive embryonic stem cells. Mol. Cell. 2016; 62:848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cremer T., Kurz A., Zirbel R., Dietzel S., Rinke B., Schrock E., Speicher M.R., Mathieu U., Jauch A., Emmerich P. et al. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb. Symp. Quant. Biol. 1993; 58:777–792. [DOI] [PubMed] [Google Scholar]
- 47. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., de Klein A., Wessels L., de Laat W. et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008; 453:948–951. [DOI] [PubMed] [Google Scholar]
- 48. Wu F., Yao J.. Spatial compartmentalization at the nuclear periphery characterized by genome-wide mapping. BMC Genomics. 2013; 14:591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang J.C., Giaever G.N.. Action at a distance along a DNA. Science. 1988; 240:300–304. [DOI] [PubMed] [Google Scholar]
- 50. Farnham P.J. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009; 10:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y., Wong C.-H.C.-H., Birnbaum R.Y., Li G., Favaro R., Ngan C.Y., Lim J., Tai E., Poh H.M., Wong E. et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013; 504:306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tolhuis B., Palstra R.J., Splinter E., Grosveld F., De Laat W.. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002; 10:1453–1465. [DOI] [PubMed] [Google Scholar]
- 53. Drissen R., Palstra R.J., Gillemans N., Splinter E., Grosveld F., Philipsen S., De Laat W.. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004; 18:2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rada-Iglesias A., Wallerman O., Koch C., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O.M., Ellis P.D. et al. Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Hum. Mol. Genet. 2005; 14:3435–3447. [DOI] [PubMed] [Google Scholar]
- 55. Lee B.K., Iyer V.R.. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 2012; 287:30906–30913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simon J.A., Kingston R.E.. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009; 10:697–708. [DOI] [PubMed] [Google Scholar]
- 57. Dumesic P.A., Homer C.M., Moresco J.J., Pack L.R., Shanle E.K., Coyle S.M., Strahl B.D., Fujimori D.G., Yates J.R., Madhani H.D.. Product binding enforces the genomic specificity of a yeast Polycomb repressive complex. Cell. 2015; 160:204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper are deposited at the Gene Expression Omnibus (accession number GSE80797).