Abstract
Non-coding RNAs (ncRNA) are involved in essential biological processes in all three domains of life. The regulatory potential of ncRNAs in Archaea is, however, not fully explored. In this study, RNA-seq analyses identified a set of 29 ncRNA transcripts in the hyperthermophilic archaeon Sulfolobus acidocaldarius that were differentially expressed in response to biofilm formation. The most abundant ncRNA of this set was found to be resistant to RNase R treatment (RNase R resistant RNA, RrrR(+)) due to duplex formation with a reverse complementary RNA (RrrR(−)). The deletion of the RrrR(+) gene resulted in significantly impaired biofilm formation, while its overproduction increased biofilm yield. RrrR(+) was found to act as an antisense RNA against the mRNA of a hypothetical membrane protein. The RrrR(+) transcript was shown to be stabilized by the presence of the RrrR(−) strand in S. acidocaldarius cell extracts. The accumulation of these RrrR duplexes correlates with an apparent absence of dsRNA degrading RNase III domains in archaeal proteins.
INTRODUCTION
Biofilm formation represents a prevalent strategy of microorganisms to persist in nature. Biofilms consist of surface-associated sessile communities encased in a self-produced extracellular matrix. Sessile cells within the biofilms differ substantially from their planktonic counterparts, particularly in regard to their increased resistance towards numerous environmental fluctuations such as temperature, pH, the presence of antibiotics and nutrient availability (1).
Although archaea ubiquitously colonize environments that are also inhabited by bacteria and eukaryotes, their molecular mechanisms involved in biofilm formation are just beginning to be unravelled (2–4). A first identification and characterization of six homologous transcriptional regulators related to the biofilm lifestyle of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius was recently reported (4). This study revealed that the gene abfR1 (for archaeal biofilm Regulator 1) encodes a wHTH transcriptional regulator that represses an extracellular polysaccharide substance (EPS) biosynthesis pathway during biofilm development and also promotes cell motility by controlling archaella (archaeal flagella) gene expression. As the archaeal biofilm lifestyle is tightly regulated at the transcriptional level, it is likely that non-coding RNA (ncRNA)-mediated post-transcriptional regulation occurs during biofilm formation of archaeal organisms.
Several ncRNAs associated with biofilm physiology have been identified and characterized in bacterial model organisms including Escherichia coli and Pseudomonas spp. (5,6). In these bacteria, ncRNAs link cell surface appendage (flagella and curli) production and biofilm formation, thereby adding a new layer of regulation to this process (5,7). The ncRNAs can interact with Hfq, a chaperone that facilitates bacterial RNA base-pairing with their targets (8). This mechanism influences the stability and processing of target transcripts resulting in modulated transcription, translation, mRNA stability and maintenance or in the silencing of gene expression. The chaperone Hfq belongs to a family of RNA-binding proteins, termed Sm-like (LSm), that have also been shown to bind small RNAs in archaea (9,10). Two different Sulfolobus solfataricus LSm proteins (Lsm1 and LSm2) have been characterized as heptameric ring assemblies that facilitate small RNA binding and interactions with components of the exosome involved in RNA degradation (11). The depletion of LSm1 activity in the halophilic archaeon Haloferax volcanii resulted in enhanced swarming capability, hinting at possible sRNA-mediated regulation of mobility in archaea (12).
In E. coli, for instance, the expression of the master biofilm transcriptional activator CsgD is partly controlled post-transcriptionally by two redundant small ncRNAs, OmrA and OmrB. Down-regulation of csgD occurs via direct antisense interactions between OmrA/B and the csgD 5′ untranslated region sequence (5′UTR). This subsequently leads to the down-regulation of curli genes, thereby impairing biofilm formation (13).
Over the last decade the use of RNA high-throughput sequencing (RNA-seq) has allowed for the identification of ncRNA transcripts with unknown functions in several archaeal organisms. For instance, in the crenarchaeon Sulfolobus solfataricus, 310 ncRNA genes were identified either in protein coding loci or intergenic regions (14). Similarly, in the euryarchaeon Methanosarcina mazei Gö1, 248 intergenic and antisense oriented ncRNA candidates were identified (15). RNA-seq analyses in Pyrococcus abyssi (16) and Haloferax volcanii have discovered hundreds of putative ncRNA genes in these euryarchaea (17,18).
The characterization of ncRNA molecules and their functional roles are just beginning to be explored for archaea. Thus, the ncRNAs’ regulatory mechanisms of action, as well as their mRNA or protein targets are most often not known. In S. solfataricus, ncRNA candidates are suggested to interact with the 3′ untranslated region (UTR) of the predicted target mRNAs; a feature which is reminiscent of miRNA-mediated mRNA regulation in eukaryotes. Nineteen antisense RNA transcripts against transposons, open reading frames and other ncRNAs were described in S. solfataricus (19,20). Genes involved in ion transport and metabolism were found to be involved in the generation of overlapping antisense transcripts, suggesting that antisense transcription might be a common regulatory mechanism for such genes in S. solfataricus (20). However, it still remains to be determined which of these ncRNAs are functionally important, and under what kind of conditions ncRNA-mediated gene regulation occurs. Intriguingly, the ncRNA Sso-155 was found to interact with the 5′ end of ORF SSO0118 (21), encoding the pili structural subunit UpsA of S. solfataricus, which plays an essential role during the initial attachment of S. solfataricus cells to abiotic surfaces when forming biofilms (22,23).
Antisense RNA interactions result in dsRNA formation. Archaeal dsRNA cleaving enzymes have not been identified. RNase III enzymes are found in Bacteria and Eukaryotes but appear to be absent in Archaea. It has been proposed that different dsRNA-specific nucleases must exist if dsRNA processing was a feature of the archaeal RNA metabolism (24).
Here, we compare the production of ncRNA transcripts during biofilm or planktonic growth of the archaeon S. acidocaldarius. An abundant double-stranded RNA was identified which was found to play a regulatory role during biofilm development. The stability of this RNA molecule was found to be modulated via duplex formation. The apparent absence of known dsRNase domains in archaeal proteins could promote regulatory RNA duplexes in S. acidocaldarius.
MATERIALS AND METHODS
S. acidocaldarius strains and growth conditions
Sulfolobus acidocaldarius MW001 (25) and the ΔrrrR markerless deletion mutant were aerobically grown at 76°C in Brock media (26), at pH 3 and supplemented with 0.1% (w/v) N-Z-Amine and 10 mg/ml uracil. Uracil was not added to the media for the cultivation of pyrEF disruption mutants. For transcript and protein overproduction experiments in S. acidocaldarius, 0.2% (w/v) dextrin was added to the media to induce expression. Growth progression was monitored by measurement of the optical density at 600 nm (OD600). All S. acidocaldarius deletion strains are described in Supplementary Table S2.
Biofilm culturing and confocal laser scanning microscopy (CLSM) analysis
Static biofilm cultures of S. acidocaldarius strains were grown in small Petri dishes (μ-dishes, 35 mm, Ibidi, Martinsried) in Brock media supplemented with 0.1% (w/v) N-Z amine, 0.2% (w/v) dextrin and 10 mg/ml of uracil when necessary. All strains were inoculated at OD600 = 0.01 and three biological replicates of each strain were grown for three days at 75°C. The medium was carefully exchanged every 24 h to ensure aerobic growth conditions and nutrient replenishment. Petri dishes were put in a specially designed metal box (25 cm L × 20 cm W × 20 cm D) filled with ∼500 ml of water in the bottom to minimize evaporation of the media, as described by Koerdt et al. (2).
Biofilm images were recorded on an inverted TCS-SP8 confocal microscope (Leica, Bensheim, Germany). As described by Koerdt et al. (27), DAPI (4,6-diamidino-2-phenylindole) was used to visualize the cells of the biofilm and fluorescently labelled lectins were employed to visualize the EPS (extracellular polymeric substances) of the biofilms. Fluorescein-conjugated concavalin A (ConA) (Invitrogen, Karlsruhe, Germany), was used to detect α-mannopyranosyl and a-glucopyranosyl residues, whereas IB4 was used (isolectin GS-IB4 from Griffonia simplicifolia, Invitrogen, Karlsruhe, Germany) for α-d-galactosyl residues. Images were then processed by using the IMARIS software package (Bitplane AG, Zürich, Switzerland).
Biofilm volumetric quantitation were performed using the biovolume determination tool of the IMARIS software package (Bitplane AG, Zürich, Switzerland). Volumes of the three channels were determined both separately and together. A total image's surface of 240 μm2 was taken to calculate the volume, which was expressed as μm3 × 106. Biofilms volumetric determination was performed in three biological replicates and nine images at different microscopy fields were recorded for each replicate.
Isolation of total RNA and small RNAs
Total RNA samples were isolated from 10 ml of exponentially growing shaking culture (OD600 = 0.4) and 40 ml of 3 days mature biofilm culture. To preserve RNA integrity, biofilm-containing petri dishes were cooled down on ice prior to isolation and shaking cultures were immediately harvested by centrifugation at 4°C. TRIzol reagent (Invitrogen) was used for total RNA isolation following manufacturer's instructions. For small RNAs isolation, S. acidocaldarius MW001 cells were harvested during logarithmic phase (OD600 0.4). A 15 ml cell pellet was lysed in a homogenizer and small RNAs (<200 nt) were isolated using the mirVana miRNA isolation kit (Ambion).
RNA-sequencing and differential expression analysis
For whole transcriptome RNA-seq analyses, three total RNA samples from independent cultures of each growth condition (biofilm or planktonic cells) were pooled in equal amounts to generate one mixed sample per condition. For whole small RNAs transcriptome generation, a biological duplicate was subjected to RNA-seq. Residual chromosomal DNA present in RNA samples was removed by RNase-free DNase I (Roche) treatment for 2 h at 37°C. Preparation of the RNA-Seq libraries and Illumina HiSeq2000 sequencing were performed at the Max-Planck Genome Centre, Cologne using the TruSeq RNA Library Prep Kit (Illumina) according to the manufacturer's protocol. For the small RNA data set, prepared with a TruSeq Small RNA Library Preparation Kit (Illumina), sequencing reads were trimmed by the removal of linker sequences and other sequences using a quality score limit of 0.05. The trimmed reads were mapped to the reference genome of S. acidocaldarius DSM639 (GenBank: NC_007181) using CLC Genomics Workbench 5.0 (CLC Bio, Aarhus, Denmark). The following mapping parameters were used: mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.5, similarity: 0.8.
For the whole transcriptome RNA-seq data from planktonic and biofilm cells, high quality reads (>Q30) were aligned against S. acidocaldarius DSM639 genome (GenBank: NC_007181) using Bowtie2 (28). Only perfect matches were considered. Reads mapping outside known coding genes regions (intergenic reads) were retrieved and assembled using BEDTools (29). Resulting assembled transcripts sequences were compared against all proteins available in NCBI database, using Blastx v2.2.28. Transcripts that did not retrieve significant hits with protein-encoding gene sequences were further analyzed using the Coding Potential Calculator (CPC) to determine whether they correspond to either non annotated protein-encoding genes or novel ncRNAs (30). The final set of transcripts without any coding potential was defined as novel intergenic non-coding transcripts.
The expression abundance of all intergenic non-coding transcripts was estimated in RPKM values using EDGE-pro (31). Transcripts with RPKM and counts below 10 were removed. Fold change between samples was manually calculated using RPKM values, after normalizing it by the total number of considered reads (32). Transcripts with a fold-change cutoff of 2.0 were considered as differentially expressed.
Quantitative RT-PCR
The preparation of the cDNA and the qRT-PCR were performed as described in Orell et al. (2013) (4). Cq values of each transcript of interest were standardized to the Cq value of the housekeeping gene saci0574 (secY) (33). qPCR reactions with DNA-free RNA as template were performed as control. Random hexamer oligonucleotides were used for cDNA preparation to quantify the expression of the potential RrrR target genes, whereas specific reverse oligonucleotides were employed for RrrR (+) and (−) strands cDNA synthesis. At least 3 biological replicates of each assessed condition and 2 technical replicates per qPCR reaction were analyzed. Oligonucleotides used for qPCR reactions are listed in Supplementary Table S1.
Promoter activity assay
Promoter activity assays for RrrR sense and antisense promoters as well as for their mutated versions were performed as described previously by Lassak et al. (34). All transcriptional promoter fusions were cloned into pSVA1431 (25) using SacII, NcoI restriction sites, thereby exchanging the mal promoter. The resulting plasmids (pACE113, pACE114, pACE115 and pACE116) and a promoter-less control plasmid (pSVA1614) were transformed into MW001 cells. Assays were performed in 96-well plates and the reactions were initiated by addition of 2 mM o-nitrophenyl-β-d-galactopyranosid (ONPG). All investigated strains were assayed in triplicates. The production of ONP was measured at 410 nm (35) over a period of 4 h at 42°C using an Infinite 200 plate reader (Tecan). Miller units were calculated as described previously (34).
In vitro transcription and RNAs substrate preparation
In vitro transcription reactions were basically performed as described by Richter at al. (2012) (36).Templates for in vitro transcription were obtained by cloning of each sense and antisense RrrR sequences with an upstream T7 RNA polymerase promoter sequence into pUC19 vector. After linearization of the plasmid with HindIII, in vitro transcription was performed in a final volume of 20 μl [40 mM HEPES–KOH (pH8.0); 22 mM MgCl2; 5 mM DTT; 1mM spermidine; 4mM UTP, CTP, GTP and 2 mM ATP; 20 U RNase Inhibitor; 1 mg T7 RNA polymerase; 1 mg linearized plasmid] at 37°C for 1 h. In vitro transcription of the sense RrrR transcript was performed in the presence of 2 μl of [α 32P]-ATP (800 Ci/mmol, 10 μCi/μl).
RNA in vitro transcripts were separated by denaturing PAGE (8 M urea; 1× TBE; 10% polyacrylamide), and full-length bands were cut out using sterile scalpels after brief autoradiographic exposure. The RNA was eluted from the gel piece using 500 ml RNA elution buffer [250 mM NaOAc, 20 mM Tris–HCl (pH 7.5), 1 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 0.25% SDS] and overnight incubation on ice. Precipitation of RNA was performed by adding two volumes EtOH (100%; ice cold) and 1/100 glycogen for 1 h at −20°C and subsequent washing with 70% EtOH of pelleted RNA.
An RrrR duplex (dsRrrR) was produced by hybridizing the [α 32P]-ATP labeled RrrR ssRNA sense transcript with the non-labeled RrrR ssRNA antisense transcript. Both RNA molecules were mixed in DEPC water in a 1:10 ratio (sense:antisense), incubated at 95°C during 5 min and subsequently cooled down to room temperature.
Low Molecular Weight RNA Marker, 10–100 nt (Affymetrix, cat# 76410) was 5′- radiolabeled in a 20 μl reaction volume as following: 10 μl of the RNA, 2 μl 10× T4 Polynucleotide Kinase (PNK) buffer (New England Biolabs (NEB)), 25 U T4 PNK (NEB) and 2.0 μl of [γ 32P]-ATP (800 Ci/mmol, 10 μCi/μl) at 37°C for 30 min.
Northern blot analyses
Semi-dry electrophoretic transfer was used to transfer RNA that was separated on denaturing polyacrylamide gels onto a positively charged nylon membrane (Roti®-Nylon plus, pore size 0.45 μm). Prior to the transfer, the membrane, the gels, as well as Whatman GB004, 3MM Paper were equilibrated in 1× TBE buffer for 5 min. The blot was assembled as a stack of 6× Whatman paper, nylon membrane, polyacrylamide gel, 6× Whatman paper and the transfer was performed for 2 h at 20 V. Subsequently, the RNA was UV-crosslinked to the membrane.
The membrane was pre-hybridized for 30 min at 42°C in ULTRAhyb-Oligo Hybridization Buffer (1 ml/10 cm2 membrane) to block non-specific binding sites. The 5′-terminal radiolabeled probes with complementarity to the RrrR(+) and RrrR(−) dúplex region (106 cpm/ml hybridization buffer) were applied to the hybridization buffer after incubation at 95°C for 5 min. Hybridization was performed over night at 42°C. The blot was washed twice, with 15 ml low stringency buffer (2× SSC, 0.1% SDS) and with 15 ml high stringency buffer (1× SSC, 0.1% SDS) for 30 min at 42°C each to remove unbound probe. Bands were visualized with a phosphorimager.
RNase R exonuclease assays
RNase R treatment was performed as described in Danan et al. (37). 60 U of recombinant Ribonuclease R (RNase R) from E. coli (Epicentre, cat# RNR07250) were added to 10 μg small RNA of S. acidocaldarius MW001 and the reaction was carried out in 10× reaction buffer at 37°C for 45 min. To remove the enzyme and salts from the reaction, ethanol precipitation was performed. For single strand RrrR and double stranded RrrR RNase R assays, 20 μl reactions were carried out as following: 2 μl 10× RNase R buffer (0.2 M Tris–HCl pH 8.0, 1 M KCl, 1 mM MgCl2), 20 U of RNase R and 20 000 cpm (41.3 ng) of [α 32P]-ATP labeled RrrR ssRNA sense transcript or [α32P]-ATP labeled RrrR dsRNA. When indicated, 5 μg of total protein cells extract of either S. acidocaldarius or E. coli DH5α were used instead of RNase R enzyme. Cell extracts were prepared by sonication in a buffer containing 50 mM Tris–HCl pH 8.0, 5 mM EDTA, 100 μg/μL PMSF. The reaction mix was incubated for 30 min at 37°C. Reactions containing total protein cells extract of S. acidocaldarius were incubated at 55°C. The digestions products were separated and visualized by denaturing PAGE electrophoresis (8 M urea; 1× TBE; 10% polyacrylamide).
Electrophoretic mobility shift assays (EMSA) with recombinant LSm proteins
The S. acidocaldarius genes coding for LSm1 (ORF saci1224, C-terminal His-tag) and LSm2 (ORF saci0799, C-terminal FLAG-tag) were cloned into the expression vector pCDF-Duet-1. The two proteins were produced in E. coli BL21AI. 1 mM IPTG was added at an OD600 of 0.6, followed by 3 h of incubation at 37°C. The pellet was disrupted by sonication and the cell lysate was heated to 70°C for 15 min. Denatured E. coli proteins were removed via centrifugation. The His-tagged Lsm1 protein was purified via Ni-NTA affinity chromatography (HisTrap HP; GE Healthcare) and eluted with a gradient of 10 to 500 mM imidazole using a fast protein liquid chromatography (FPLC) Äkta system (GE Healthcare). LSm2 was co-purified. EMSA experiments were performed with 9.3 nM radiolabeled RrrR(+) transcripts (10 000 cpm) that were mixed with 500 nM of recombinant Lsm1/Lsm2 in 10 μl EMSA binding buffer (100 mM Hepes pH 8.0, 50 mM NaCl, 5 mM MgCl2). Competition assays were performed by the addition of a 2-fold excess of unlabeled transcripts of the native saci0301 mRNA or a variant with a mutated 7 nt stretch of the interaction region (Supplementary Figure S4). The reactions were incubated for 10 min at 70°C and then separated by 8% native PAGE. The gel was exposed overnight to a phosphor screen and bands were visualized using a phosphorimager.
Construction of plasmids for in-frame gene deletion and in-trans-complementation
For the construction of the deletion mutant plasmid, the respective up- and down-stream flanking regions of the RrrR gene were PCR amplified from S. acidocaldarius genomic DNA using primer pairs as listed in Supplementary Table S1. The up- and down-stream flanking DNA regions were joined by means of overlap extension PCR using the outward bound primer of the respective primer pair. The overlap extension PCR products were restricted with PstI and BamHI and subsequently ligated into the plasmid pSVA406, containing the pyrEF cassette from S. solfataricus (25). This ligation yielded deletion plasmid pVT135 (see Supplementary Table S2 for details). For overexpression of RrrR(+) and (−) strand sequences in S. acidocaldarius MW001, each sequence was cloned into the S. acidocaldarius expression vector pSVA1431 (38) using NcoI and EagI restriction sites, which allows for maltose inducible expression of proteins. These constructs were termed pACE105 and pACE107, respectively (Supplementary Table S2). All constructs were sequenced to confirm their identity. The primer sequences are given in Supplementary Table S1. Recombinant plasmids were methylated using the E. coli strain ER1821 (Supplementary Table S2) before transformation into S. acidocaldarius cells to avoid cleavage by its restriction enzyme SuaI (39).
Construction of chromosomal deletion mutants
An in-frame markerless deletion mutant was generated for the non-coding RNA RrrR. To this end, methylated deletion mutant plasmid pVT135 was electroporated into MW001 as described by Wagner et al. (25). Integrants were selected on uracil selective gelrite plates after 5 days of incubation at 75°C and subsequently subjected to 5-FOA (100 μg/ml) gelrite plates to allow the excision of the DNA region containing the target gene. In frame markerless deletion mutants were confirmed by sequencing of PCR products that were obtained using primers binding at least 100 bp up and downstream of the respective primers used for the construction of the flanking regions for the deletion mutant plasmids.
Gene disruption by S. solfataricus pyrEF cassette exchange via homologous recombination was generated for saci0301. To this end, 50 bp of the up and downstream regions of the target gene were added to the 5′ and 3′ ends of the pyrEF cassette via PCR. S. acidocaldarius MW001 cells were electroporated with ∼300 ng of the PCR product. Transformed cells were selected on uracil selective gelrite plates after 5 days of incubation at 75°C. Obtained colonies were transferred to liquid Brock medium. Deletion mutants were confirmed by sequencing of PCR products that were obtained using primers that bound at least 100 bp up and downstream of the target gene and one reverse and one forward primer annealing in the pyrEF cassette sequence, respectively.
Computational target predictions
In silico target predictions for ncRNAs were performed using CopraRNA tool (40). For genome-wide target predictions of the RrrR(+) RNA sequence, the CopraRNA web-server 1.2.5 (wrapper 1.0.7.1) was used with using settings as following parameters: hybridization temperature: 75.0°C; window size: 150 nt; base pair distance: 100 nt and by extending the prediction area around the translational start site of the mRNAs from 75 nt to 100 nt. The genomes of S. acidocaldarius DSM639 (GenBank: NC_007181), S. solfataricus (GenBank: NC_002754) and S. tokodaii (GenBank: NC_003106) were used as reference.
RESULTS
Biofilm-associated noncoding RNA transcriptome profiling
RNA-seq data was obtained from S. acidocaldarius cells grown either as biofilms or as planktonic cultures to search for ncRNA transcripts. Sequencing reads that mapped within intergenic regions and did not represent previously predicted ncRNAs or transcripts of annotated genes were analyzed. To determine whether these reads belonged to ncRNAs or protein-coding genes, a transcript assembling approach was performed based on genomic coordinates clustering, using mergeBed of the Bedtools Package, as described by Quinlan and Hall (29). Transcripts that did not retrieve significant hits with protein-encoding gene sequences were further analyzed using the Coding Potential Calculator (CPC) to determine potential ncRNAs.
We then performed a comparative analysis of biofilm-associated and planktonically grown cells in order to determine the differential expression of the predicted ncRNAs. The expression profile was normalized by the total RPKM within each sample as described by Severino et al. (41). We determined that transcript levels of 29 ncRNAs were found to be significantly up- or down regulated in biofilm-associated S. acidocaldarius cells when compared to the planktonic transcriptome (Supplementary Table S3). This finding suggests that ncRNAs might be involved in S. acidocaldarius biofilm development regulation. A single ncRNA was found to be most abundant in both modes of growth (ncRNA239 in Supplementary Table S3). We determined that this ncRNA was upregulated 2.4-fold in the transcriptome of biofilm-associated cells. Therefore, we further investigated its potential regulatory role in S. acidocaldarius biofilm physiology.
Characterization of a double-stranded, RNase R resistant RNA (RrrR)
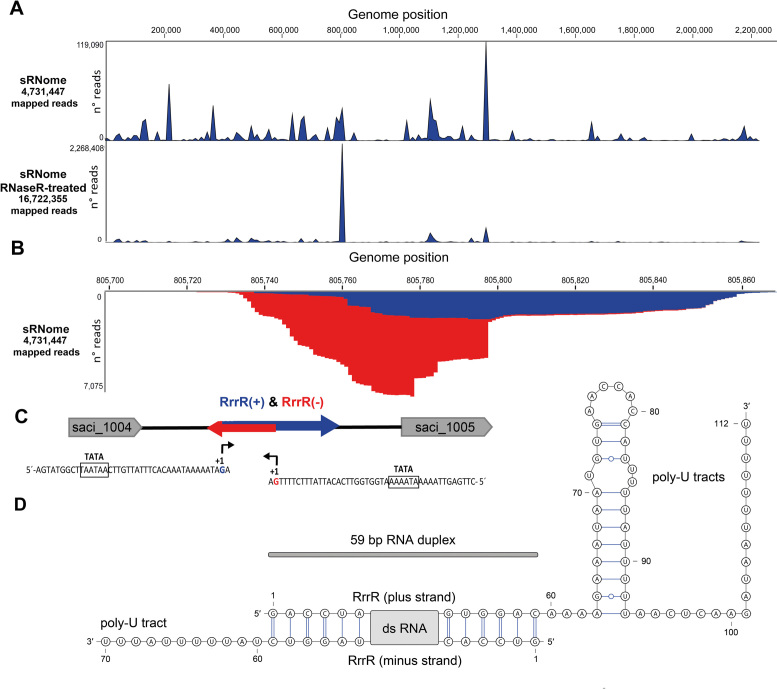
Close inspection of the RNA-seq data coverage of this abundant RNA identified significant amounts of reads for both directions (Figure 1). The most abundant 59 nt stretch was covered by reads in plus and minus direction. To identify double-stranded RNA molecules in S. acidocaldarius, small RNAs were isolated and treated with RNase R. RNase R is an E. coli 3′-to-5′ exoribonuclease which efficiently cleaves linear RNA species. However, circular or highly structured RNAs are not digested (42). RNA-seq analysis of the RNase R cleavage reaction products indicated a single major peak representing the described ncRNA239 (Figure 1). Consequently, this molecule was named RrrR (for RNase R resistant RNA). The gene for RrrR is located in the intergenic region between saci1004 (coding for a hypothetical protein) and saci1005 (coding for a PadR family transcription regulator). Both RrrR strands were also detected via Northern Blot analysis (Supplementary Figure S1). The RrrR sequence exhibits 100% conservation among ten sequenced S. acidocaldarius strains (DG1, Y14 13-1, Y14 16-22, Y14 20-20. Y14 18-5, NG05B CO5 07, GG12-C01-09, Ron12/I, N8, DSM639, SUSAZ), but BLAST analyses did not detect regions of significant similarity for other archaeal genomes. Genome alignments highlight a genome segment with three hypothetical protein-coding genes (saci1002-saci1004) and the RrrR gene located between a gene coding for tRNA-Leu (saci1000) and the saci1005 gene. This gene arrangement is characteristic for S. acidocaldarius species, but absent in other Sulfolobales genomes (e.g. S. solfataricus and S. tokodaii). The transcription start sites of both RrrR strands were mapped. Both transcripts start with a guanosine residue and potential TATA box promoter elements are located approximately 25 bp upstream of the transcription start site (Figure 1). Archaeal terminators are usually represented by poly-U stretches (43). In agreement, the RrrR(−) strand ends with a 11 nt overhang containing 9 uridine residues and the RrrR(+) strand has a 3′-terminal 52 nt extension beyond the duplex region. This 3′ end contains a potential hairpin structure and two poly-U stretches with 11 and 7 uridine residues, respectively (Figure 1). To verify the RrrR promoters, both potential promoter sequences were fused to the thermostable S. solfataricus lacS gene and β-galactosidase activity was measured. Both promoters provided strong β-galactosidase activity and mutation of the TATA box sequences abolished activity (Figure 2). The RrrR(−) strand promoter resulted in the highest β-galactosidase activity.
Figure 1.
Identification of RrrR transcripts. (A) Small RNAs from planktonic cells of S. acidocaldarius (sRNome) and an RNaseR-treated sRNome sample were subjected to Illumina RNA-seq. Genome-wide coverage of sequence reads is indicated. A single major peak corresponds to the RNase R resistant RNA RrrR. (B) The coverage plot histogram of the enriched RrrR region highlights mapped plus strand (+) sequencing reads (blue) as well as minus strand (−) sequencing reads (red). (C) A schematic representation of the RrrR genomic context is shown. TATA box sequences are located upstream of the transcriptional start sites (+1) for the plus transcript (blue arrow) and the minus transcript (red arrow). (D) Consensus secondary structure of the double-stranded RrrR transcripts.
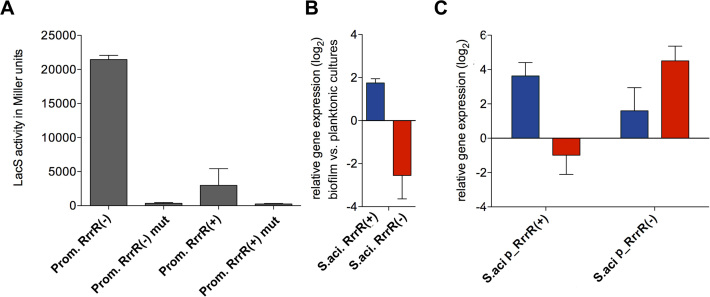
Figure 2.
Promoter activity assays and transcript level quantitation of RrrR. (A) Specific β-galactosidase activities of plasmids containing RrrR(+) and RrrR(−) promoter sequences were assayed for S. acidocaldarius MW001 cells during exponential growth (48h). A mutated TATA box version (Supplementary Table S1) was employed as negative control for each promoter. (B) Total RNA isolated from S. acidocaldarius MW001 grown either as biofilm or planktonic cultures were used for cDNA synthesis. qRT-PCR analysis was performed using specific primers for the RrrR(+) (blue bar) and RrrR(−) (red bar) transcripts. The values reflect the fold change in gene expression compared with cDNA prepared from exponential grown planktonic cells, which is designated as baseline. (C) Total RNA was isolated from cells grown as biofilms. Transcript levels of both RrrR(+) (blue bar) and RrrR(−) (red bar) were quantified by qRT-PCR from S. acidocaldarius MW001 recombinant strains overexpressing either RrrR(+) or RrrR(−). Relative transcript expression was normalized to the internal control gene secY. The means and standard deviation of three biological replicates are shown.
Quantification of qRT-PCR amplificates obtained with specific primers targeting the non-duplex RrrR overhangs verified that the RrrR(+) strand is upregulated in biofilm cells and revealed the RrrR(−) strand to be downregulated in biofilm-associated cells in comparison to planktonic cells (Figure 2). Next, we utilized qRT-PCR to assay changes in transcript abundance upon plasmid-borne overproduction of either RrrR strand. Overproduction of the RrrR(+) strand resulted in reduced numbers of RrrR(−) transcripts while overproduction of the RrrR(−) strand increased the abundance of RrrR(+) molecules (Figure 2). These results suggest that RrrR(−) molecules can stabilize the RrrR(+) strand.
Double-stranded RNA stability in S. acidocaldarius
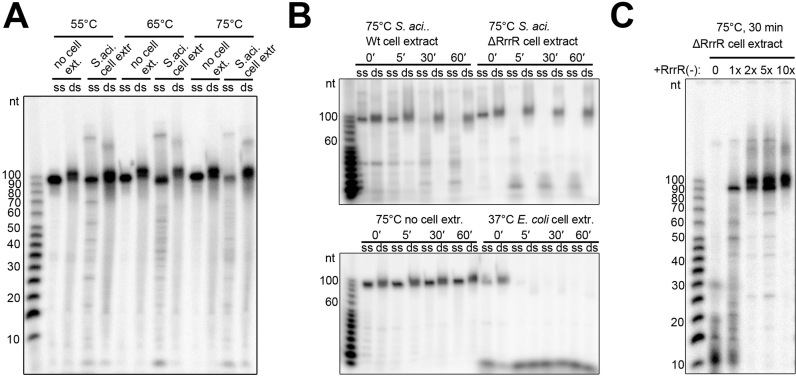
The enzymatic cleavage of dsRNAs by RNase III family enzymes plays a key function during the expression and regulation of cellular and viral genes in Bacteria and Eukaryotes. It is debated if RNase III-like activities are present in Archaea. Therefore, we aimed to investigate the stability of RrrR transcripts either as single-stranded or duplexed molecules in bacterial and archaeal cell extracts. First, we produced radioactively labelled RrrR transcripts and incubated them with RNase R. In agreement with our RNA-seq analyses, the single-stranded RrrR(+) molecules were degraded, while the 59 bp duplex portion of the hybridized RrrR strands remained protected (Supplementary Figure S2). Incubation of ssRrrR(+) or dsRrrR with E. coli cell lysate resulted in complete RNA degradation indicating the activity of effective RNases. Interestingly, incubation of the RNA molecules with S. acidocaldarius cell extract did not lead to effective dsRrrR degradation (Figure 3A and B). Single-stranded RrrR(+) transcripts were also not degraded after 5 min of incubation time and we hypothesized that the presence of RrrR(−) strands in the cell extract might stabilize the plus strand. Therefore, the experiment was repeated with a cell extract from a S. acidocaldarius ΔRrrR strain (see below). This cell extract did degrade the RrrR(+) strand in the absence of its duplex partner while the double-stranded RrrR remained stable (Figure 3B). Next, we added excess amounts of unlabeled RrrR(−) transcripts to these reactions and could show that these molecules stabilized RrrR(+) strands in the presence of S. acidocaldarius ΔRrrR cell extract (Figure 3C). Finally, we tested if the addition of excess amounts of unlabelled RrrR(+) molecules influences RrrR(+) degradation and observed stabilization of labelled RrrR(+) transcripts and the loss of upshifted bands (Supplementary Figure S3A). It is possible that these upshifted bands result from interactions with RNA-binding proteins (e.g. LSm proteins, see below) and unlabelled transcripts could compete with RNA-binding and degrading activities. We verified that cell extracts of both, S. acidocaldarius wild type and ΔRrrR strains, are equally capable of degrading a mRNA transcript (Supplementary Figure S3B).
Figure 3.
RrrR stability assays. (A) Radiolabeled single strand (ss) RrrR(+) and radiolabeled double-stranded (ds) RrrR were incubated with S. acidocaldarius MW001 cell extract for 30 min at different temperatures. (B) Transcripts of ssRrrR(+) and dsRrrR were incubated with different cell extracts at 75°C for the indicated time. (C) Transcripts of ssRrrR(+) are degraded by S. acidocaldarius ΔrrrR deletion strain cell extract (0). The addition of RrrR(−) transcripts (molar ratio 1:1, 1:2, 1:5, 1:10) prevents ssRrrR degradation. Five μg of total protein cell extract were added to all indicated assays. Digestion products were visualized on a 10% denaturating polyacrylamide gel. The sizes (in nucleotides) of the radiolabeled RNA ladder bands are provided.
Biofilm formation of S. acidocaldarius RrrR deletion mutant strains
To unravel the in vivo function of the RrrR molecule(s), a marker-less in-frame deletion mutant of the RrrR gene (ΔRrrR strain) was obtained in the uracil auxotrophic S. acidocaldarius reference strain MW0001 (25). Analysis of the growth curves in shaking cultures revealed no apparent growth phenotypes for the ΔRrrR strain (Supplementary Figure S4). In addition, no morphological defects could be observed when ΔRrrR strain cells were subjected to optical microscopic analysis.
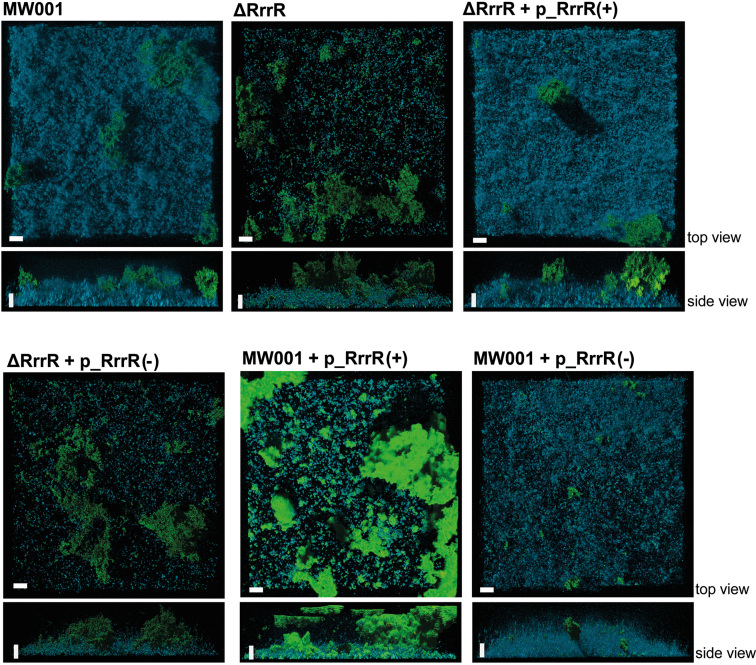
The ability of the ΔRrrR deletion strain to form static biofilms was then assessed and quantified by confocal laser scanning microscopy (CLSM) analysis. DAPI was used for visualization of cells within the biofilms while the presence of extracellular polysaccharide residues was detected using fluorescently labeled lectins that specifically bind mannose/glucose (ConA) and galactosyl sugar residues (IB4). After three days of static biofilm formation, the ΔRrrR deletion mutant strain exhibited significantly impaired biofilm formation (Figure 4). As described previously (4,22), the reference strain MW001 displayed a confluent dense biofilm morphology. The EPS pattern was characterized by a dominant ConA (mannose/glucose) signal, while the ΔRrrR strain showed remarkable morphological differences when forming biofilms. Cell colonization of the surface was strongly reduced resulting in a low DAPI signal (Figure 4). In addition, a biofilm volume of 2.51 μm3 × 106 was determined for the ΔRrrR strain, which represented a 40% decrease of the overall biofilm biomass in comparison to the reference strain MW001 (4.14 μm3 × 106) (Supplementary Table S4). The ratio of the three different signals was also altered in biofilms formed by the ΔRrrR strain. While the IB4 signal (indicating galactosyl sugar residues) did not show notable changes, the DAPI signal was found to be reduced by 13.7% and the ConA signal (indicating mannose/glucose) increased 12.4% in comparison to the MW001 strain signal ratio (Supplementary Table S4). These findings suggest that the deletion of the ncRNA RrrR has a clear impact on the formation of S. acidocaldarius biofilm communities.
Figure 4.
Confocal laser scanning microscopy analysis of biofilm formation by the S. acidocaldarius ΔRrrR deletion mutant and ΔRrrR complementation strains. Three days old S. acidocaldarius biofilm was subjeted to CLSM. The blue channel indicates DAPI-staining. The green channel represents the fluorescently labeld lectin ConA that binds to glucose and mannose residues. The lectin IB4, able to bind to a-galactosyl-residues, is assigned to the yellow channel. Overlay images of all three channels are shown. White bars indicate 20 μm length.
To further determine which RrrR transcript (plus or minus) is responsible for the biofilm formation phenotype, the ΔRrrR strain was complemented in trans with either RrrR plus or minus sequences. Each sequence was cloned into the expression plasmid pSVA1431 in order to control RrrR expression under the maltose inducible promoter. We determined that RrrR(+) and RrrR(−) transcripts were overproduced 17.85- and 18.74-fold, respectively in ΔRrrR complementation strains (Supplementary Figure S5). It was determined that the wild type biofilm phenotype was fully restored only when complementing the ΔRrrR strain with the RrrR(+) sequence (Figure 4). Quantification of the biofilm volume and the fluorescence signal profile revealed that the ΔRrrR strain with the RrrR(+) plasmid reached levels very close to the MW001 strain (4.05 μm3 × 106 versus 4.14 μm3 × 106) (Supplementary Table S4). In contrast, the ΔRrrR strain with the RrrR(−) plasmid did not restore the wild type biofilm phenotype and quantification of the fluorescence signals yielded levels that were similar to the ΔRrrR deletion strain (Figure 4, Supplementary Table S4). Hence, these complementation assays strongly suggest that the RrrR(+) transcript plays a regulatory role during biofilm development of S. acidocaldarius.
Additionally, we tested the impact of overproducing each RrrR transcript on the biofilm formation of wild type S. acidocaldarius to assess the interplay between the two RrrR transcripts. The overproduction of RrrR(+) resulted in a biofilm biomass volume that resembled the wild type strain's volume (Supplementary Table S4). However, a distinctive biofilm phenotype was observed which is characterized by increased EPS production, which was unevenly distributed as patchy areas at the top of the biofilm (Figure 4). This result suggests that the abundance of RrrR(+) transcripts might need to be tightly regulated in order to exert their regulatory role during biofilm formation. In agreement, the overproduction of the RrrR(−) transcript resulted in less robust biofilms (Figure 4). The overall biofilm biomass was reduced by 18% in the wild type strain with a RrrR(−) plasmid (Supplementary Table S4). This observation suggests that RrrR(−) is an ‘antisense’ RNA for RrrR(+).
Identification of potential target genes of RrrR
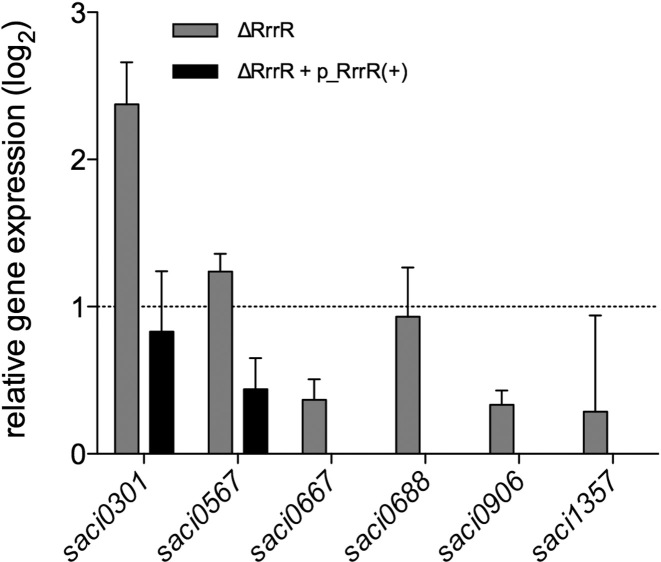
Based on our in vivo functional analyses we hypothesized that the RrrR(+)transcript might be regulating the abundance of mRNAs that are required for biofilm development of S. acidocaldarius. Hence, potential mRNA targets for RrrR(+) were predicted in silico using the CopraRNA tool (40). This analysis proposed 132 RrrR(+)-mRNA interaction possibilities (data not shown). Predicted interaction sites were mainly located between nucleotides 16 to 53 of RrrR(+), corresponding to the region of RrrR(+)/RrrR(−) base-paring (Figure 1). Six of the ten highest ranking candidates (Supplementary Table S5) were selected to experimentally analyze mRNA level changes in response to altered RrrR(+) levels via qRT-PCR. The expression levels of these six potential target genes were determined for 3-day-old biofilm communities of wild type cells and the ΔRrrR deletion strain. It was determined that the transcript abundances of the genes saci0301 (encoding a predicted membrane protein) and saci0567 (encoding a component of the crenarchaeal system for DNA exchange, Ced (44)) were significantly up-regulated in ΔRrrR biofilms (Figure 5). In addition, these transcript levels were nearly restituted to wild type levels when the ΔRrrR strain was complemented with RrrR(+) plasmid, which suggests that the mRNAs of these two genes are RrrR(+) interacting partners (Figure 5). We focused on the interactions between the saci0301 transcript and RrrR(+) (Supplementary Figure S6) and considered that LSm proteins could act as chaperones that mediate such RNA-RNA interactions. Therefore, we performed EMSA analyses and could show that a recombinant complex of LSm1 and LSm2 proteins binds RrrR(+) transcripts (Supplementary Figure S7). The LSm protein-induced shift was significantly reduced when saci0301 mRNA transcript was added and partly restored by a mutation of the proposed interaction region of RrrR(+) and saci0301 (Supplementary Figures S6 and S7).
Figure 5.
Relative transcript levels of potential RrrR target genes. qRT-PCR was used to assay gene expression of potential RrrR target genes (Supplementary Table S5) in the deletion strain ΔRrrR during biofilm growth. Relative transcript expression levels of each target gene were normalized to the internal control gene secY. The values reflect the fold changes of transcript levels in comparison to the reference strain MW001, which is designated as baseline. The means and standard deviation of three biological replicates are shown.
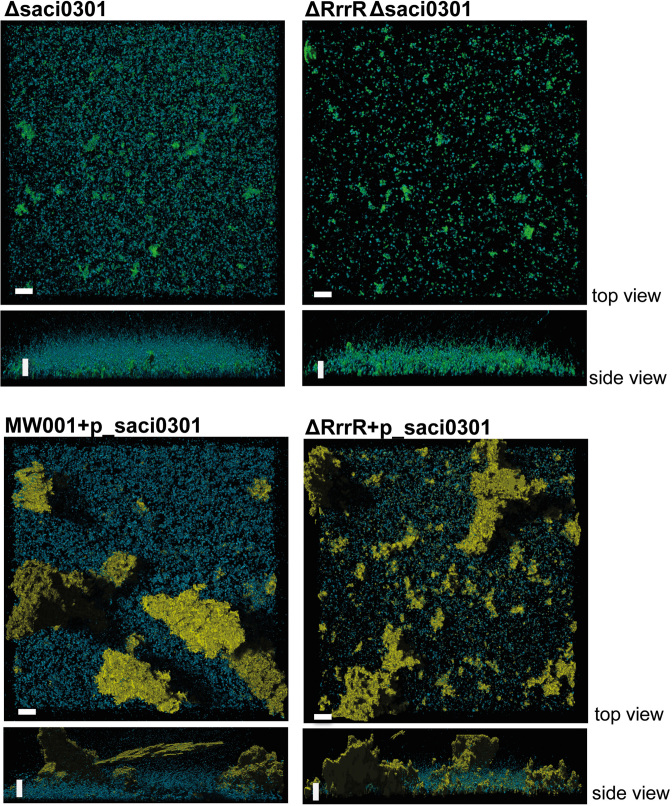
The saci0301 transcript levels were 21.3 fold up-regulated in biofilms-associated cells (Supplementary Table S5), corresponding to the highest gene expression difference between the two modes of growth. This could indicate that the predicted membrane protein encoded by the gene saci0301 plays an essential role during S. acidocaldarius biofilm formation. Thus, a double deletion mutant ΔRrrR-Δsaci0301 was generated. CLSM analysis determined that the biofilm phenotype of this strain resembled the one of the ΔRrrR mutant (Figure 6). This suggests that the absence of RrrR(+) might cause pleotropic effects during the biofilm development beyond the saci0301 mRNA–RrrR(+) interaction. The single deletion of saci0301 resulted in a diminished biofilm phenotype, indicating that the Saci0301 protein influences biofilm growth. This Δsaci0301 strain showed lower surface coverage and a reduced EPS pattern. The overproduction of Saci0301 either in ΔRrrR strain or wild type cells resulted in an augmented EPS pattern characterized by strongly increased production of galactosyl sugar residues (detected by IB4 lectin, yellow signal) (Figure 6). This observation suggests that Saci0301 may be involved in shaping the extracellular matrix of S. acidocaldarius biofilms.
Figure 6.
Confocal laser scanning microscopy analysis of biofilm formed by the S. acidocaldarius Δsaci0301 deletion mutant and ΔRrrR complementation strains. Three days old biofilm cultures were subjected to CLSM. Overlay images of three channels (see Figure 4) are shown.
DISCUSSION
Microbial biofilm development involves coordinated events that follow a regulated genetic program (45,46). Although archaea are frequently detected in biofilm communities, the molecular mechanisms that promote a sessile life-style are not yet fully understood (47). In this study, we describe the identification and the initial characterization of a double-stranded non-coding RNA (RrrR) from S. acidocaldarius that displays a potential regulatory role during biofilm development. Modulating the levels of the RrrR plus strand transcript, RrrR(+), resulted in distinct biofilm phenotypes. To our knowledge, this is the first report of a regulatory RNA involved in archaeal biofilm formation. The ΔRrrR strain biofilm morphology is reminiscent of the effects caused by the deletion of Saci1223, a transcriptional regulator that has been proposed to act as an biofilm activator of S. acidocaldarius (4). However, potential base-pairing interactions between the saci1223 mRNA and RrrR(+) are not evident. The increase of RrrR(+) transcript levels led to the secretion of larger amounts of EPS in wild type biofilms cultures. This finding may indicate that RrrR(+) plays a regulatory role in EPS biosynthesis pathways of S. acidocaldarius. Thus, exopolysaccharide production could be increased and/or glycosylation patterns of cell surface associated proteins could be changed. An exopolysaccharide biosynthesis pathway has not yet been characterized for archaeal organisms. A gene cluster (saci1904–1927) was identified in S. acidocaldarius which shows homologies to bacterial genes for exopolysaccharide biosynthesis and secretion (4). The mRNA transcript of these genes were not predicted as direct targets of RrrR(+). Our in silico target prediction approach in combination with in vivo mRNA level analyses propose that RrrR(+) interacts with the 3′-terminal portion of the saci0301 mRNA. LSm proteins could act as RNA chaperones to facilitate this interaction and to stabilize the RNA duplex. Saci0301 is a predicted membrane protein with no assigned function to date. Interestingly, saci0301 was found to be the most abundant up-regulated transcript in biofilm-associated cells in our RNA-seq transcriptome analyses, which may reflect its crucial role during S. acidocaldarius biofilm formation. In agreement, the Δsaci0301 deletion strain could not build robust biofilms, suggesting that this protein plays a significant role during the sessile life-style of S. acidocaldarius. It remains to be established how this hypothetical protein might be involved in shaping the extracellular matrix of archaeal cells and if additional targets of RrrR(+) exist. The increase of saci0301 transcript levels in the ΔRrrR strain suggests that RrrR(+) can act as a regulatory RNA for saci0301.
Regulatory RNAs have been discovered in several archaeal species and first target interactions have been characterized (48). The 3′ untranslated regions of mRNAs were identified as targets for sRNAs in Haloarchaea and Pyrobaculum species (48,49) and the 5′ region of mRNAs were found to be targeted in Methanosarcina mazei Gö1 (15). It should be noted that many Archaea contain mostly leaderless transcripts, which influences the range of interaction modes with target RNAs. The regulatory RrrR(+) transcript can form base-pairs with its own ‘antisense’ RrrR(−), resulting in double-strand formation between the two RrrR strands. Several interesting questions arise from this set-up. Our in vitro assays highlight that dsRNAs are stable in S. acidocaldarius cell extracts and that the ‘antisense’ RrrR(−) strand protects the single-stranded RrrR(+) from degradation. Thus, it is possible that dsRNA processing is not essential for archaeal organisms and RNA duplex formation can be utilized for RNA stabilization and titration. What is the role of the ‘antisense’ RrrR(−) transcript? We hypothesize that this RNA strand can be used to quickly and effectively titrate the amount of RrrR(+) transcripts, serving as an example of an ‘RNA sponge’ that sequesters regulatory RNAs of mRNAs. In Bacteria, cross talk of small RNAs with mRNA targets and sRNAs that act as target mimics has been described (50). Similar regulatory pathways are e.g. also found in Eukaryotes where long noncoding RNAs (lncRNAs) titrate away proteins and small regulatory RNAs and circular RNAs function as so-called sponges for regulatory RNAs (51–53). Recently, human cells have been found to contain natural double-stranded RNAs with potential regulatory functions (54). Here, sense-antisense transcription leads to the formation of natural double-stranded RNAs (ndsRNAs) that can localize in the nucleus and may participate in important biological processes in humans. Our data suggest that the presence of regulatory dsRNA could represent yet another shared feature between Eukaryotes and Archaea. The apparent absence of RNase III activity in Archaea highlights possibilities for using duplex formation as a general RNA stabilization mechanism in this domain of life.
DATA AVAILABILITY
Raw sequence read data for all the RNA-seq samples used are deposited at the NCBI′s Gene Expression Omnibus (55) and are accessible through GEO accession number: GSE99484.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Raul Arias for his kind help with the RNA-seq differential expression analysis and Michael Daume and Alexander Pastura for their help with the production of recombinant LSm1 and LSm2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [DFG RA 2169/3-1 to L.R.]; Comisión Nacional de Ciencia y Tecnología de Chile [FONDECYT 1130229 to A.O]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Costerton J.W., Lewandowski Z., Caldwell D.E., Korber D.R., Lappin-Scott H.M.. Microbial biofilms. Annu. Rev. Microbiol. 1995; 49:711–745. [DOI] [PubMed] [Google Scholar]
- 2. Koerdt A., Orell A., Pham T.K., Mukherjee J., Wlodkowski A., Karunakaran E., Biggs C.A., Wright P.C., Albers S.V.. Macromolecular fingerprinting of sulfolobus species in biofilm: a transcriptomic and proteomic approach combined with spectroscopic analysis. J. Proteome Res. 2011; 10:4105–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Losensky G., Jung K., Urlaub H., Pfeifer F., Frols S., Lenz C.. Shedding light on biofilm formation of Halobacterium salinarum R1 by SWATH-LC/MS/MS analysis of planktonic and sessile cells. Proteomics. 2017; 17:doi:10.1002/pmic.201600111 [DOI] [PubMed] [Google Scholar]
- 4. Orell A., Peeters E., Vassen V., Jachlewski S., Schalles S., Siebers B., Albers S.V.. Lrs14 transcriptional regulators influence biofilm formation and cell motility of Crenarchaea. ISME J. 2013; 7:1886–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mika F., Hengge R.. Small regulatory RNAs in the control of motility and biofilm formation in E. coli and Salmonella. Int. J. Mol. Sci. 2013; 14:4560–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petrova O.E., Sauer K.. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 2010; 192:5275–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghaz-Jahanian M.A., Khodaparastan F., Berenjian A., Jafarizadeh-Malmiri H.. Influence of small RNAs on biofilm formation process in bacteria. Mol. Biotechnol. 2013; 55:288–297. [DOI] [PubMed] [Google Scholar]
- 8. Vogel J., Luisi B.F.. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011; 9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fischer S., Benz J., Spath B., Maier L.K., Straub J., Granzow M., Raabe M., Urlaub H., Hoffmann J., Brutschy B. et al. The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010; 285:34429–34438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Toro I., Thore S., Mayer C., Basquin J., Seraphin B., Suck D.. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001; 20:2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martens B., Sharma K., Urlaub H., Blasi U.. The SmAP2 RNA binding motif in the 3′UTR affects mRNA stability in the crenarchaeum Sulfolobus solfataricus. Nucleic Acids Res. 2017; 45:8957–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maier L.K., Benz J., Fischer S., Alstetter M., Jaschinski K., Hilker R., Becker A., Allers T., Soppa J., Marchfelder A.. Deletion of the Sm1 encoding motif in the lsm gene results in distinct changes in the transcriptome and enhanced swarming activity of Haloferax cells. Biochimie. 2015; 117:129–137. [DOI] [PubMed] [Google Scholar]
- 13. Holmqvist E., Reimegard J., Sterk M., Grantcharova N., Romling U., Wagner E.G.. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010; 29:1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wurtzel O., Sapra R., Chen F., Zhu Y., Simmons B.A., Sorek R.. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010; 20:133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jager D., Sharma C.M., Thomsen J., Ehlers C., Vogel J., Schmitz R.A.. Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21878–21882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toffano-Nioche C., Ott A., Crozat E., Nguyen A.N., Zytnicki M., Leclerc F., Forterre P., Bouloc P., Gautheret D.. RNA at 92 degrees C: the non-coding transcriptome of the hyperthermophilic archaeon Pyrococcus abyssi. RNA Biol. 2013; 10:1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heyer R., Dorr M., Jellen-Ritter A., Spath B., Babski J., Jaschinski K., Soppa J., Marchfelder A.. High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol. 2012; 9:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Straub J., Brenneis M., Jellen-Ritter A., Heyer R., Soppa J., Marchfelder A.. Small RNAs in haloarchaea: identification, differential expression and biological function. RNA Biol. 2009; 6:281–292. [DOI] [PubMed] [Google Scholar]
- 19. Martens B., Manoharadas S., Hasenohrl D., Manica A., Blasi U.. Antisense regulation by transposon-derived RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO Rep. 2013; 14:527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu N., Li Y., Zhao Y.T., Guo L., Fang Y.Y., Zhao J.H., Wang X.J., Huang L., Guo H.S.. Identification and characterization of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. PLoS One. 2012; 7:e35306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang T.H., Polacek N., Zywicki M., Huber H., Brugger K., Garrett R., Bachellerie J.P., Huttenhofer A.. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005; 55:469–481. [DOI] [PubMed] [Google Scholar]
- 22. Henche A.L., Koerdt A., Ghosh A., Albers S.V.. Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ. Microbiol. 2012; 14:779–793. [DOI] [PubMed] [Google Scholar]
- 23. Zolghadr B., Klingl A., Koerdt A., Driessen A.J., Rachel R., Albers S.V.. Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 2010; 192:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nicholson A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscipl. Rev. RNA. 2014; 5:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wagner M., van Wolferen M., Wagner A., Lassak K., Meyer B.H., Reimann J., Albers S.V.. Versatile genetic tool box for the Crenarchaeote Sulfolobus acidocaldarius. Front. Microbiol. 2012; 3:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brock T.D., Brock K.M., Belly R.T., Weiss R.L.. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Archiv. Mikrobiol. 1972; 84:54–68. [DOI] [PubMed] [Google Scholar]
- 27. Koerdt A., Godeke J., Berger J., Thormann K.M., Albers S.V.. Crenarchaeal biofilm formation under extreme conditions. PLoS One. 2010; 5:e14104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L., Gao G.. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007; 35:W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Magoc T., Wood D., Salzberg S.L.. EDGE-pro: estimated degree of gene expression in prokaryotic genomes. Evol. Bioinformatics Online. 2013; 9:127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ribeiro-dos-Santos A., Khayat A.S., Silva A., Alencar D.O., Lobato J., Luz L., Pinheiro D.G., Varuzza L., Assumpcao M., Assumpcao P. et al. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PLoS One. 2010; 5:e13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van der Sluis E.O., Nouwen N., Koch J., de Keyzer J., van der Does C., Tampe R., Driessen A.J.. Identification of two interaction sites in SecY that are important for the functional interaction with SecA. J. Mol. Biol. 2006; 361:839–849. [DOI] [PubMed] [Google Scholar]
- 34. Lassak K., Neiner T., Ghosh A., Klingl A., Wirth R., Albers S.V.. Molecular analysis of the crenarchaeal flagellum. Mol. Microbiol. 2012; 83:110–124. [DOI] [PubMed] [Google Scholar]
- 35. Jonuscheit M., Martusewitsch E., Stedman K.M., Schleper C.. A reporter gene system for the hyperthermophilic archaeon Sulfolobus solfataricus based on a selectable and integrative shuttle vector. Mol. Microbiol. 2003; 48:1241–1252. [DOI] [PubMed] [Google Scholar]
- 36. Richter H., Zoephel J., Schermuly J., Maticzka D., Backofen R., Randau L.. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012; 40:9887–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Danan M., Schwartz S., Edelheit S., Sorek R.. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012; 40:3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berkner S., Wlodkowski A., Albers S.V., Lipps G.. Inducible and constitutive promoters for genetic systems in Sulfolobus acidocaldarius. Extremophiles. 2010; 14:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prangishvili D.A., Vashakidze R.P., Chelidze M.G., Gabriadze I.. A restriction endonuclease SuaI from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1985; 192:57–60. [DOI] [PubMed] [Google Scholar]
- 40. Wright P.R., Georg J., Mann M., Sorescu D.A., Richter A.S., Lott S., Kleinkauf R., Hess W.R., Backofen R.. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014; 42:W119–W123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Severino P., Oliveira L.S., Torres N., Andreghetto F.M., Klingbeil Mde F., Moyses R., Wunsch-Filho V., Nunes F.D., Mathor M.B., Paschoal A.R. et al. High-throughput sequencing of small RNA transcriptomes reveals critical biological features targeted by microRNAs in cell models used for squamous cell cancer research. BMC Genomics. 2013; 14:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hossain S.T., Malhotra A., Deutscher M.P.. How RNase R degrades structured RNA: role of the helicase activity and the S1 domain. J. Biol. Chem. 2016; 291:7877–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dar D., Prasse D., Schmitz R.A., Sorek R.. Widespread formation of alternative 3′ UTR isoforms via transcription termination in archaea. Nat. Microbiol. 2016; 1:16143. [DOI] [PubMed] [Google Scholar]
- 44. van Wolferen M., Wagner A., van der Does C., Albers S.V.. The archaeal Ced system imports DNA. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:2496–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Beloin C., Ghigo J.M.. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005; 13:16–19. [DOI] [PubMed] [Google Scholar]
- 46. Whiteley M., Bangera M.G., Bumgarner R.E., Parsek M.R., Teitzel G.M., Lory S., Greenberg E.P.. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001; 413:860–864. [DOI] [PubMed] [Google Scholar]
- 47. Orell A., Frols S., Albers S.V.. Archaeal biofilms: the great unexplored. Annu. Rev. Microbiol. 2013; 67:337–354. [DOI] [PubMed] [Google Scholar]
- 48. Prasse D., Ehlers C., Backofen R., Schmitz R.A.. Regulatory RNAs in archaea: first target identification in Methanoarchaea. Biochem. Soc. Trans. 2013; 41:344–349. [DOI] [PubMed] [Google Scholar]
- 49. Bernick D.L., Dennis P.P., Lui L.M., Lowe T.M.. Diversity of antisense and other non-coding RNAs in archaea revealed by comparative small RNA sequencing in four Pyrobaculum species. Front. Microbiol. 2012; 3:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Figueroa-Bossi N., Valentini M., Malleret L., Fiorini F., Bossi L.. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009; 23:2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 52. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 53. Wang K.C., Chang H.Y.. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011; 43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Portal M.M., Pavet V., Erb C., Gronemeyer H.. Human cells contain natural double-stranded RNAs with potential regulatory functions. Nat. Struct. Mol. Biol. 2015; 22:89–97. [DOI] [PubMed] [Google Scholar]
- 55. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence read data for all the RNA-seq samples used are deposited at the NCBI′s Gene Expression Omnibus (55) and are accessible through GEO accession number: GSE99484.