Abstract
While screening and treatment have sharply reduced breast cancer mortality in the past 50 years, more targeted diagnostic testing may improve the accuracy and efficiency of care. Our retrospective, age-matched, case-control study evaluated the differential value of mammography and genetic variants to predict breast cancer depending on patient age. We developed predictive models using logistic regression with group lasso comparing (1) diagnostic mammography findings, (2) selected genetic variants, and (3) a combination of both. For women older than 60, mammography features were most predictive of breast cancer risk (imaging AUC = 0.74, genetic variants AUC = 0.54, combined AUC = 0.71). For women younger than 60 there is additional benefit to obtaining genetic testing (imaging AUC = 0.69, genetic variants AUC = 0.70, combined AUC = 0.72). In summary, genetic testing supplements mammography in younger women while mammography appears sufficient in older women for breast cancer risk prediction.
Introduction
While breast cancer accounts for 30% of all new cancer diagnosis in women, the mortality rates have declined sharply in the past 50 years1. This has been largely attributed to early detection and treatment. The current standard of care is for women to be screened by mammography, with interest in moving towards screening based off estimates of women’s breast cancer risk. Improved prediction of breast cancer risk by further targeting screening with the tests demonstrating the highest sensitivity and specificity for a given population will further improve breast cancer care.
Early risk prediction models were based off demographic features, in particular patient age, hormonal risk factors, and mammographic breast density. Currently, there is increased optimism that we can more accurately characterize patient risk with advances in precision medicine that allow us to select the most predictive test for the patient. In particular, Genome Wide Association studies have allowed us to collect a set of single-nucleotide polymorphisms (SNPs) that are predictive of breast cancer risk2. These SNPs can be paired with mammographic features to improve the likelihood that a positive screen is a true positive3. The indications for genetic testing are not yet codified, therefore, it would be valuable to determine when these variables will improve breast cancer risk prediction.
In this paper, we aim to determine the next best test for breast cancer diagnosis in younger and older patients who were included in a personalized medicine data set. We hypothesize that while obtaining imaging and genetic variance improves the estimation of breast cancer risk compared to chance in all patient age groups. Furthermore, we predict that there are differences in the additive predictive value of genetic variants over and above mammography variables depending on patient age.
Methods
Subjects
This study includes subjects derived from the Marshfield Clinic Personalized Medicine Research Project (Marshfield PRMP), details of which have been previously published4. This registry included subjects residing in one of 19 zip codes surrounding Marshfield, Wisconsin who provided a blood sample for genetic testing, completed a brief questionnaire and gave permission to link this information with medical records. A case-control cohort of western European women was established by selecting patients from this registry who had received a diagnostic mammogram concerning for breast cancer, a breast biopsy within 12 months of the mammogram, and a blood plasma sample that could be assessed for genetic variants associated with an increased breast cancer risk. Cases in the study were women who were listed in the Marshfield cancer registry with a confirmed breast cancer diagnosis; controls were selected as women who had a benign breast biopsy result and did not have a breast cancer diagnosis in the Marshfield EMR. Cases and controls were age-matched such that each case had a control within 5 years of the age of each case. Exclusion criteria included cases with known BRCA1 or BRCA 2 mutations, as these are likely to dominate other predictive variables, nonwhite patients as the population did not have a sufficient number for appropriate distribution of nonwhites to race-match controls, and cases where BI-RADS features were missing. This resulted in 35 total subjects excluded, with 738 women included.
Features: mammography and genetic variants
One diagnostic mammogram for each case and each control was selected from within the 12 months prior to the biopsy. If multiple mammograms were available, the mammogram with the most suspicious features, closest in time to the biopsy, was selected5. Mammography features were drawn from the third edition of the Breast Imaging Reporting and Data System (BI-RADS) lexicon6. This lexicon standardizes mammography findings using descriptors that categorize breast density, abnormal features, and assessment categories. We utilized 49 hierarchical descriptors that are considered the most predictive of breast cancer7. BI-RADS descriptors included mass margins, microcalcification shape, micro-calcification morphology, architectural distortion and breast density, among others. These findings were extracted from the patients’ mammography reports using a parser and represented as a binary “present” or “not present”.
The Marshfield PRMP was one of five original biobanks in the eMERGE network funded by the National Human Genome Research Institute8. Plasma samples were sequenced on a Sequenom MassARRAY system. Genetic features included 77 common high-frequency/low-penetrance genetic variants that were identified by recent large-scale GWAS studies as having a higher prevalence in breast cancer cases than controls, and thus associated with increased breast cancer risk9. Risk alleles were those that had a higher prevalence in cases than in controls. The number of high-risk alleles was enumerated for each patient, where homozygotes could have up to two high-risk alleles, and heterozygotes up to one high-risk allele.
Model development and statistical analysis
We built breast cancer risk prediction models using a logistic regression with group lasso model [3] to assess the predictive power of imaging features and genetic variants. These models were developed utilizing solely mammography features, solely genetic variants and using both genetic variants and mammography.
The binomial logistic regression with group lasso is described in Fan et al (2016)3. A brief description of the model follows. For the binomial logistic regression model, we suppose that the response variable can take the value Y= {0, 1}. We can thus model
Given a sample {(xi, yi), i=1, 2…, N}, the objective function for the logistic regression with lasso is given by the negative binomial log - likelihood:
We note that within the mammography features there exists a natural group structure given by sub-characterizations of different major features3. Genetic variants also contain a group structure that can be characterized with hierarchical clustering3. To incorporate the group structure into the lasso logistic regression, we define the optimization problem for the group lasso logistic regression10,
where dg is the number of features (d) in group g, βg ∈ ℝdg is the corresponding coefficient vector in group g, λ1 > 0 is the tuning parameter and L(() is defined as the negative log - likelihood.
The models were applied to the mammography and genetic variant data set and fit with ten-fold cross validation. We generated receiver operating curves (ROC) that indicated the risk of a malignant breast lesion and used the area under the curves (AUC) to compare performance for two age groups: women age 29 to 59 years old and women aged 60 to 90 years old. This division allowed two sufficiently powered age groups that represented early breast cancer screening ages and later screening respectively. This method was repeated 100 times, and the mean AUC for each model was calculated, along with 95% confidence intervals (CI). A two-sided P value of <0.05 was the criterion for statistical significance. Statistical analysis and graphics were done in R 3.0.1 and R 3.3.111.
Institutional Review Board (IRB)
The Marshfield Clinic IRB approved the data collection and informed consent was obtained from participants. The Marshfield and University of Wisconsin IRBs approved this study. Additionally, Health Insurance Portability and Accountability Act compliance was maintained.
Results
We identified 362 cases and 376 controls, with an age range from 29 to 90 years old (Table 1). The subjects were predominantly Caucasian, with 4 subjects that were non-Caucasian or of unknown race in both the case and control groups. The subjects had a mean age of 62 years old, thus 323 subjects were in the 29 to 59 year old age group and 415 were in the 60 to 90 year old age group.
Table 1.
Number of cases and controls for different age groups.

We found that in older women (60 years and older), the mammography regression models and genetic variant regression models predicted breast cancer risk statistically significantly better than chance, with the mammography AUC = 0.744 (95% CI = 0.740 - 0.748) and the genetic variants AUC = 0.540 (95% CI = 0.532 - 0.549). The model using mammography features was statistically significantly superior to the model involving genetic features, with a performance that was also clinically significant (Figure 1). The combined model incorporating both imaging and genetic features performed statistically significantly better than the genetic variants only model (Figure 1). However, the mammography only model continued to perform statistically significantly better than the combined model (AUC = 0.713, 95% CI = 0.705 - 0.720).
Figure 1.
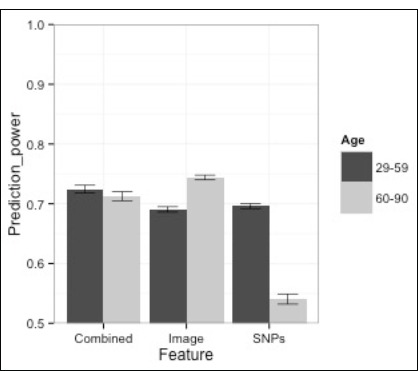
Prediction power (AUC) of imaging features, SNPs and combined model for different age groups.
We found that in younger women (less than 60 years old), evaluating breast cancer risk with either mammography variables or genetic variants was statistically significantly better than chance; mammography (AUC = 0.690, 95% CI = 0.686 - 0.695), genetic variants (AUC = 0.696, 95% CI = 0.692 - 0.700) (Table 2). However, their performances were similar (Figure 1). The combined model incorporating both imaging features and genetic variants performed statistically significantly better than the genetics variants only model and the mammography only model (combined AUC = 0.724, 95% CI = 0.718 - 0.731).
Table 2.
Area under the curve (AUC) and confidence intervels (CI) for models using imaging features genetic variants and combind, for youngers vs. older subjects.
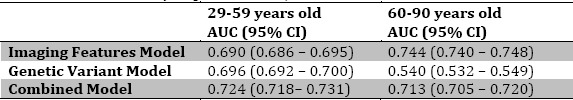
We found that similar features were selected in the combined model as compared to the imaging features alone and the genetic variants alone models. There was complete overlap in which mammography features were selected, with mass shape, mass margin, calcification distribution, architectural distortion, mass size, and breast density predictive of breast cancer risk. Some additional variants were selected by the genetic variants alone model, however these were not sufficiently predictive to be selected by the combined mammography and genetic model.
Discussion
Our study demonstrates the most valuable tests for evaluating the likelihood of breast cancer differs in younger (ages 29-59) as compared to older (ages 60 - 90) patients (Figure 2). For older patients, a logistic regression with group lasso model incorporating solely mammography features outperformed both a model with solely genetic features and a model combining mammography and genetic features. This indicates that for patients 60 and older, genetic variance will not improve risk prediction after mammography variables have been utilized. For younger patients, models based on either genetic variants or mammography features are comparable, while combining genetic variants and mammography improves performance. This indicates that for patients under age 60, acquiring genetic variants have the potential to improve breast cancer risk assessment.
Figure 2.
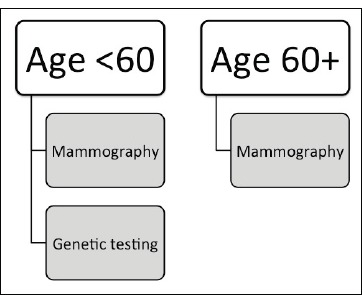
Clinical decision making model for maximal predictive power for breast cancer risk detection.
This study expands on previous work comparing the information utility of patient demographics and various tests in analyzing breast cancer risk. Burnside et al (2016)5 found that when comparing patient demographic features and, mammography features using a logistic regression model, mammography features (AUC = 0.689) was superior to both considering demographics (AUC = 0.598) and a model of 10 genetic variants (AUC = 0.601). A subsequent study using logistic regression with group lasso found that combining genetic testing and mammography features (AUC = 0.727) was superior to both mammography alone (AUC = 0.716) and a model of 77 genetic variants alone (AUC = 0.614)3. This study is a logical next step in outlining which patients may most benefit from supplementary genetic testing.
The group lasso model in this study takes advantage of the underlying structure information of both mammography features in the BI-RADS hierarchy, and extracted structure information in SNPs, as calculated by computing Euclidean distances12. Prior studies noted that encoding clinically relevant BI-RADS structure information as well as computationally extracted genetic structure information using a group lasso improves breast cancer prediction, in particular improving the performance of genetic features in combined model3. There are promising future directions of research with representations of biological dependencies using SNP linkage disequilibrium as encoded in haplotype maps (e.g. HapMap)13. The inclusion of structure representation in the model also aligns with the biologic basis for breast cancer development, as younger and older women are manifesting different risk factors.
Younger women are more likely to develop breast cancer due to an inherited predisposition to oncologic signaling pathways14. While high-penetrance variants such as ER/PR status, HER2 and BRCA genes have been commonly used to assess breast cancer risk, GWAS have identified low-penetrance SNPs not only associated with breast cancer risk, but with early onset and poorer prognosis15. It follows that information about genetic risk factors would be valuable in screening younger populations.
Our study builds on prior work using SNPS for breast cancer risk prediction and stratification by age. Mealiffe et al. [16] found that risk scores determined from genetic variants was independent from risk scores determined from the Gail model. However, their patients were all over fifty years old, and most over sixty years old. Darabi et al. [17] demonstrates how models incorporating genetic variants as a risk factor in addition to age increases the number of younger patients screened. Their model was also able to classify older patients as lower risk, aligning with our results demonstrating that while genetic variants are not the next best test for older patients, they are predictive of breast cancer risk.
There are several limitations to consider in this study. First, this study has a relatively small sample size, thus we needed to include clinical encounters over two decades (1989 - 2010) to generate a sufficient number of cases and controls. Second, due to the population used, this study was limited to only Caucasian women, and is thus not generalizable to other ethnic groups. Replication in a data set with broader ethnic variation would establish the generalizability of these results. Further, the development of BI-RADS lexicon and thus adherence to mammography descriptors has changed over this time period. Increased utilization of BI-RADS lexicon has been demonstrated to improve the predictive performance of these models5, and thus this study may underestimate the benefit of mammography alone.
The decision to pursue additional testing and treatment is challenging. Mammograms currently cost around one hundred dollars, and the cost of genetic testing varies from one hundred to thousands of dollars. Understanding the predictive power of imaging features and genetic variants in different age groups has the potential to aid clinicians in determining what tests can be used to improve information about the likelihood of malignancy.
Acknowledgements
The authors acknowledge the support from the National Institutes of Health (grants: U54AI117924, R01LM010921, K24CA194251, R01CA127379 and its supplement R01CA127379-03S1). We also acknowledge the Institute for Clinical and Translational Research (ICTR) supported by the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS) grant (UL1TR000427), the University of Wisconsin Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation, the University of Wisconsin Carbone Comprehensive Cancer Center Support grant (P30CA014520), and the University of Wisconsin School of Medicine and Public Health Departments of Radiology and Medical Physics. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Appendix
Area under the curve (AUC) and confidence intervals (CI) for models using imaging features, genetic variants and combined, for younger vs. older subjects with logistic regression with group lasso vs. with lasso.
2-sample t-tests at the 5% significance level were used to compare the mean AUC from the lasso method vs. the group lasso method. Compared to lasso, group lasso has statistically significant better performance for the Genetic Variant Models and statistically significant worse performance for the Combined Models. The performances of the Imaging Features Models, however, are not statistically significant different between the lasso vs. group lasso methods.
References
- 1.Committee on Practice Bulletins—Gynecology. Practice Bulletin Number 179: Breast Cancer Risk Assessment and Screening in Average-Risk Women. Obstet Gynecol. 2017 Jul;130(1):e1–e16. doi: 10.1097/AOG.0000000000002158. [DOI] [PubMed] [Google Scholar]
- 2.Wu Y, Abbey CK, Liu J, Ong I, Peissig P, Onitilo AA, Fan J, Yuan M, Burnside ES. Discriminatory power of common genetic variants in personalized breast cancer diagnosis. Proc SPIE Int Soc Opt Eng. 2016 Feb;27:9787. doi: 10.1117/12.2217030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan J, Wu Y, Yuan M, et al. Structure-Leveraged Methods in Breast Cancer Risk Prediction. Journal of machine learning research: JMLR. 2016;17:85. [PMC free article] [PubMed] [Google Scholar]
- 4.McCarty CA, Wilke RA, Giampietro PF, et al. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Med. 2005;2(1):49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 5.Burnside ES, Liu J, Wu Y, Onitilo AA, McCarty CA, Page CD, Peissig PL, Trentham-Dietz A, Kitchner T, Fan J, Yuan M. Comparing Mammography Abnormality Features to Genetic Variants in the Prediction of Breast Cancer in Women Recommended for Breast Biopsy. Acad Radiol. 2016 Jan;23(1):62–9. doi: 10.1016/j.acra.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.American College of Radiology. Breast Imaging Reporting And Data System (BI-RADS®) Reston VA: 2003
- 7.Wu Y, Alagoz O, Ayvaci MU, et al. A comprehensive methodology for determining the most informative mammographic features. J Digit Imaging. 2013;26:941–947. doi: 10.1007/s10278-013-9588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Page D, Peissig P, McCarty C, Onitilo AA, Trentham-Dietz A, Burnside E. New genetic variants improve personalized breast cancer diagnosis. AMIA Jt Summits Transl Sci Proc. 2014 Apr;7(2014):83–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Meier L, Van De Geer S, Bohlmann P. The Group Lasso for logistic regression. J. R. Statist. Soc. B. 2008;70:53–71. [Google Scholar]
- 11.R Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2013. R: A language and environment for statistical computing. 3.0.1. [Google Scholar]
- 12.Wang W. H. Kao, C. K. Hsiao. Using Hamming distance as information for SNP-sets clustering and testing in disease association studies. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0135918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Zhang C., McCarty C., Peissig P. L., Burnside E. S., Page D. Graphical-model based multiple testing under dependence, with applications to genome-wide association studies. In Proceedings of the 28th conference on uncertainty in artificial intelligence. 2012 [PMC free article] [PubMed] [Google Scholar]
- 14.Anders CK, Hsu DS, Broadwater G, Acharya CR, Foekens JA, Zhang Y, Wang Y, Marcom PK, Marks JR, Febbo PG, Nevins JR, Potti A, Blackwell KL. Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J Clin Oncol. 2008 Jul 10;26(20):3324–30. doi: 10.1200/JCO.2007.14.2471. [DOI] [PubMed] [Google Scholar]
- 15.Rafiq S, Tapper W, Collins A, Khan S, Politopoulos I, Gerty S, Blomqvist C, Couch FJ, Nevanlinna H, Liu J, Eccles D. Identification of inherited genetic variations influencing prognosis in early-onset breast cancer. Cancer Res. 2013 Mar 15;73(6):1883–91. doi: 10.1158/0008-5472.CAN-12-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mealiffe M, Stokowski RP, Rhees BK, Prentice RL, Pettinger M, Hinds DA. Assessment of clinical validity of a breast cancer risk model combining genetic and clinical information. J. Natl Cancer Inst. 2010 Nov 3;102(21):1618–1627. doi: 10.1093/jnci/djq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darabi H, Czene K, Zhao W, Liu J, Hall P, Humphreys K. Breast cancer risk prediction and individualized screening based on common genetic variation and breast density measurement. 2012;14(10):R25. doi: 10.1186/bcr3110. [DOI] [PMC free article] [PubMed] [Google Scholar]