Abstract
We characterized the enhancer landscape of 66 AML patients, identifying 6 novel subgroups and their associated regulatory loci. These subgroups are defined by their super-enhancer (SE) maps, orthogonal to somatic mutations, and are associated with distinct leukemic cell states. Examination of transcriptional drivers for these epigenomic subtypes uncovers a subset of patients with a particularly strong super-enhancer at the retinoic acid receptor alpha (RARA) gene locus. Presence of a RARA SE and concomitant high levels of RARA mRNA predisposes cell lines and ex vivo models to exquisite sensitivity to a selective agonist of RARα, SY-1425 (tamibarotene). Furthermore, only AML patient-derived xenograft (PDX) models with high RARA mRNA were found to respond to SY-1425. Mechanistically, we show that the response to SY-1425 in RARA-high AML cells is similar to that of APL treated with retinoids, characterized by the induction of known retinoic acid response genes, increased differentiation, and loss of proliferation.
Keywords: AML, Enhancer, RARα, SY-1425, MDS
INTRODUCTION
AML is the most common acute leukemia in adults (1) with an estimated 4 new cases diagnosed per 100,000 individuals in the United States in 2015 (SEER data, 2016(2)). Current treatment involves chemotherapy-based induction and consolidation with potential curative use of hematopoietic cell transplantation (HCT) in those eligible patients who respond to treatment. New therapeutic options are needed for AML patients given a 5-year overall survival of only 35–40%. Myelodysplastic syndrome (MDS) is a heterogeneous hematopoietic disease commonly associated with bone marrow failure. While early MDS is considered an indolent disease, the prognosis of patients with later stage MDS is poor and is associated with a low overall survival similar to AML. We sought to characterize the epigenomic circuitry and transcriptional state of leukemic cells to identify potential new treatments beyond those discovered by mutational analysis of protein coding genes.
To date, a large part of cancer research has focused on somatic mutations in protein coding regions to identify putative oncogenic drivers (3). Recently, large, highly active chromatin regions, named “super-enhancers” (SEs) have been described that define cell identity and cell state in normal and malignant cells, including cancer cells (4–6). While typical enhancers are short (~1 kb) stretches of DNA that regulate the expression of genes through the binding of transcription factors (TFs) and subsequent recruitment of RNA Pol II and associated proteins, SEs are larger in size (>20 kb on average) and are characterized by a much higher occupancy of transcriptional regulatory proteins (5,6). Previous work has shown that the histone three lysine-27 acetylation (H3K27ac) mark can be used as an efficient and robust means of super-enhancer demarcation (6–13). It has been shown that within a given cell, these few, highly active chromatin regions (<5% of all enhancers) tend to regulate key genes governing cell identity and phenotype. In tumor cells, this includes oncogene and non-oncogene drivers of the transformed state (7,8).
To define contributions of the non-coding genome in AML, we characterized the enhancer, super-enhancer, gene expression, and mutational landscapes of blasts from 66 AML patients and used this information to dissect transcriptional regulatory circuitry and predict therapeutic targets. De novo stratification of these enhancer profiles identified six distinct patient subgroups, independent of and orthogonal to mutational status. From this, we demonstrate that retinoic acid receptor alpha (RARα) occupies a critical node in the transcriptional network of a new subgroup of AML, putatively identifying a novel tumor cell liability that could be exploited therapeutically.
The selective and potent RARα agonist SY-1425 (tamibarotene) has been demonstrated to possess improved pharmacologic properties over first generation pan-retinoids such as all-trans retinoic acid (ATRA) (14–18). We found that SY-1425 potently and selectively inhibits the proliferation of AML cell lines that contain a strong SE at the RARA locus. This observation was confirmed in AML patient derived xenograft (PDX) models whereby RARA-high AML models are more sensitive to SY-1425 treatment than RARA-low AML models. We also observe RARA SEs and overexpression of RARA mRNA in MDS. These data provide the foundational support for investigating SY-1425 as a treatment option for AML and MDS patients with a strong RARA super-enhancer.
RESULTS
Identification and mapping of super-enhancers in AML
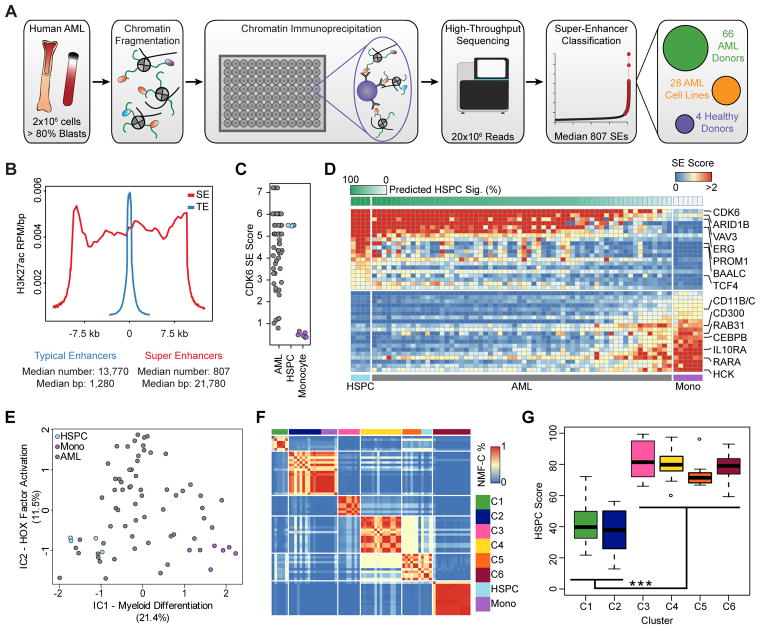
Using a high-throughput H3K27ac chromatin immunoprecipitation platform (Fig. 1A), we profiled the landscape of active enhancers in blasts from 66 AML donors, 28 AML cell lines, 4 FACS-purified HSPC samples, and 6 FACS-purified monocyte samples (Supplementary Fig. S1A). Four patient samples were excluded from the analysis due to low ChIP-seq quality. Using methods described previously (19), we identified individual enhancer loci and super-enhancers, revealing a median of 807 SEs per sample with a median length of 22 kb (Fig. 1B and Supplementary Fig. S1B–C) with enhancer ‘score’ reflecting both enhancer size and density of reads (see supplemental methods). As identified in previous studies, SEs are large, highly active regions of chromatin that regulate a small subset of genes critical to cell identity and state (4–6). This approach provided robust enhancer profiles with high technical replicate consistency (Supplementary Fig. S1D) and enabled the identification of a sample purity threshold (% AML blast cells) of 80% as a requirement for patient inclusion in this study (Supplementary Fig. S1E–G).
Figure 1.
Identification, refinement, and clustering of cancer-specific super-enhancers in human AML. A, Schematic of the high-throughput chromatin immunoprecipitation pipeline used to generate super-enhancer maps in human AML and normal blood cells. B, Metapeak representation of typical (TE - blue) and super (SE - red) enhancers in a single AML patient (SU223). H3K27ac ChIP-seq read density was calculated in each of 40 bins across each enhancer type (either typical or super) and plotted as the median enhancer width (21.7 kb), in addition to 40 bins in the 3kb upstream and 40 bins in the 3kb downstream of the enhancer. Reads in each bin were normalized by the total reads in the experiment (in millions) and to the size of the bin in base pairs. The average normalized read density in each bin across all SEs or TEs was plotted, and the x-axis was normalized by the relative median widths of SEs or TEs. C, Quantification of the SE linked to CDK6 for AML (top, gray), HSPC (middle, light blue), and monocyte (bottom, purple) samples. Ties are due to quantile normalization as CDK6 often has one of the top 4 strongest super-enhancers genome-wide. D, The twenty most HSPC-associated and the twenty most monocyte-associated SEs were used to estimate an HSPC signature through non-negative least squares. Primary AML samples are ordered by this estimated HSPC signature (top, green bar). The heatmap depicts SE scores for the HSPC-associated and monocyte-associated SEs used in the classification. E, Independent components analysis of SE data from AML, HSPCs, and monocytes. F, Heatmap shows the NMF consensus clustering distance (fraction of NMF iterations in which a pair of samples were grouped in the same cluster; blue - low, red - high) between each sample in this study. HSPCs (light blue) and monocytes (purple) are projected into the clustering without being allowed to contribute to the NMF. G, The estimated HSPC signature score (y axis) for each SE-based cluster is shown (*** p < 3×10−8).
To link SEs to genes, we utilized the correlation between SE score and H3K27ac abundance at promoters of genes within 500 kb of the enhancer, predicting a link when the p-value of the correlation was significant genome-wide, or when an SE lacked any significant correlation, linking it to the best correlated and nominally significant gene (Supplementary Fig. S2A–D; see supplementary methods). Most SEs are linked to multiple genes with this approach yielding on average 2.7 links per SE (Supplementary Fig. S2E). We then compared the SE to gene linking from this correlation method in primary AML patients to published chromatin conformation data from promoter capture Hi-C (PCHi-C) in monocytes (20) and found that 62% of SEs had validated linking.
Super-enhancer analysis reveals cell differentiation state and 6 distinct AML clusters
Previous work has identified the differentiation state of AML cells as a primary source of epigenetic variation (21–23). This is consistent with the observation that certain key HSPC enhancers, such as the SE linked to CDK6, show strong variability across our AML cohort (Fig. 1C). In an effort to assess how well SE maps can distinguish differentiation states in primary AML samples, we defined the extremes of myeloid differentiation by constructing SE maps from HSPCs and monocytes. We defined signatures of differentiation state based on the top 20 HSPC- and monocyte-specific SEs (Fig. 1D). Assessment of this enhancer-based HSPC signature within the 62 primary AML blasts shows a robust inverse correlation between HSPC-specific enhancers and monocyte-specific enhancers (Supplementary Fig. S3A). To quantify AML blast utilization of an HSPC-like enhancer signature, we derived a predicted HSPC score (Fig. 1D, green bar). This score defines the stem-like (high HSPC score) or differentiated (low HSPC score) state of a given AML sample. To determine if this differentiation state is linked to one of the major genome-wide enhancer programs, we employed independent components analysis (ICA) to find the two most pervasive enhancer signatures in the genome (Fig. 1E). The first IC, termed Myeloid Differentiation, accounted for, on average, 21.4% of the variance of SEs genome-wide, and correlated strongly with the predicted HSPC score (R2=0.85 Supplementary Fig. S3B). The second IC, termed HOX Factor Activation, accounted for an average of 11.5% of the variance, and correlated with enhancers associated with homeobox (HOX) genes, PBX3, and MEIS1, among others (Supplementary Fig. S3C). These TFs have been shown to co-occupy enhancers in murine models of leukemia, acting in concert to drive leukemogenesis and enforce immature differentiation states (24,25). Together, these two independent components accounted for 33% of the variance in the data, providing motivation for a higher-dimensional analysis.
To this end, we performed non-negative matrix factorization consensus clustering (NMF-C) (see supplemental methods) amongst the patient samples to group the patients by enhancer score. This approach identified 6 distinct subgroups of patients (referred to as clusters) in our AML cohort (Fig. 1F), each characterized by an unique combination of enhancer profiles (Supplementary Fig. S3D–E). While not included in the factorization, the HSPC and monocyte H3K27ac profiles were inserted into the clustering to illustrate their respective enhancer profiles (Fig. 1F, light blue and purple respectively). This demonstrates that cluster 2 was characterized by enhancers also found in monocytes, while cluster 5 utilized enhancers active in HSPCs. Additionally, the 6 clusters showed robust internal association by Pearson correlation of enhancer activity (Supplementary Fig. S3F). Despite clear separation in NMF-C, some clusters also showed strong inter-cluster correlation, with two groups of clusters: 1 and 2 and 3–6, showing high inter-cluster similarity (Supplementary Fig. S3F). This similarity is driven by the HSPC signature score of the samples within the clusters, with clusters 1 and 2 being far more monocyte-like than clusters 3–6 (Fig 1G, Supplementary Fig. S3G).
Characterization of distinct super-enhancer-based subgroups of AML
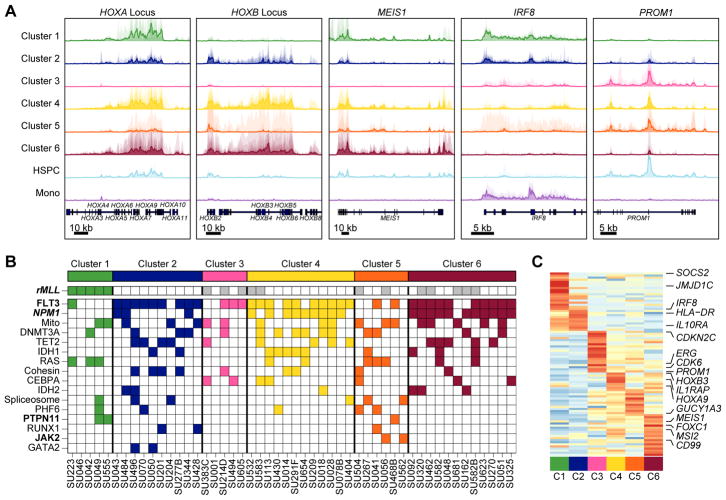
Using the clusters defined by overall SE distribution, we then focused on identifying the specific SE-linked genes forming the basis of these clusters. Prominent among these were SEs associated with known AML master TF genes (ex. HOXA/HOXB/MEIS1), an HSPC specific gene (PROM1), and a monocyte specific factor gene (IRF8) (Fig. 2A). These core transcription factors suggest that the clusters arise from shared underlying master regulatory circuitry.
Figure 2.
Super-enhancer-derived patient subgroups associate with genotype and survival. A, Metatrack plots for cluster-specific super-enhancer loci. Each individual sample in a cluster is shown as a transparent area plot and the median profile is represented over top by a thick line. The height of a given sample’s profile is determined by creating an H3K27Ac read density within the region and then scaling it to half the height of a similar region around MALAT1. B, AML samples were genotyped by either exome sequencing or targeted capture sequencing. Somatic non-synonymous mutations passing stringent filters (see methods) are displayed as colored blocks. Gray blocks indicate that the mutational status of that gene in the given patient is unknown. White blocks indicate that a mutation in that gene does not exist in the given patient. A bold mutation name indicates a nominally significant association (p < 0.05) between the mutation and the clustering (Fisher’s exact). Similarly, bold and italic font indicates significance after multiple hypothesis testing correction (p < 0.001). Only the most recurrent mutations (n > 2) are shown here. rMLL = MLL rearrangement; Mito = Mitochondrial genes. C, Cluster-specific SEs were determined by identifying the SEs with the largest dynamic range in the NMF basis matrix. The heatmap visualizes these 88 SEs as their median SE score per cluster (row normalized) and key linked genes are highlighted.
To assess the interplay between somatic mutations in protein coding genes and our SE-based clustering, we performed targeted genotyping on all samples to identify mutations in known leukemia-associated genes (Fig. 2B, Supplementary Table S1). The 130 genes sequenced included any gene found by whole exome sequencing to be mutated in more than 2% of AML cases (26) (Supplementary Table S1). These patient samples were also profiled for the presence of common prognostic cytogenetic abnormalities as part of routine clinical pathology tests. Comparing targeted genotyping and cytogenetics with our clustering, we observe that all patients with known MLL translocation are sorted into cluster 1 by their global enhancer profiles (Fig 2B, p < 1.6E–7). However, we note that 12 of our patients have not been genotyped for MLL translocation. Nevertheless, this suggests that MLL translocations may induce a unique epigenetic state that dominates the overall enhancer landscape of the given AML sample, not surprising given the function of MLL fusion proteins in mis-regulating DOT1L function leading to aberrant histone H3K79 methylation (27). While mutations in NPM1 and FLT3 are enriched in clusters 2, 3, 4, and 6, no cluster-specific associations were observed for mutations in DNMT3A, TET2, and IDH1 and IDH2.
To better understand the cluster-specific SEs driving our patient grouping, we visualized the median per-cluster SE score for the most influential SEs in the NMF decomposition (Fig. 2C). Clear cluster-specific SEs were dominated by genes with known roles in AML such as MEIS1, PROM1, SOCS2, and ERG among others (28,29). A pattern of cluster-specific SE usage emerged, with each cluster being regulated by a relatively compact number of individual super-enhancers.
Super-enhancer-defined patient clusters show differences in overall survival
To understand whether clustering of AML patient samples by enhancer landscape is biologically meaningful, we asked whether SE-defined clusters could be associated with other biological differences by bridging to a large patient data set (26). Our patient sample cohort showed a bias towards high blast count and FLT3-ITD positivity compared to other surveys of AML (30) (Supplementary Fig. S4A) and shows a median patient survival of 13.1 months from diagnosis (Supplementary Fig. S4B). However, the relatively small numbers of patients in the SE based clustering make robust correlations with clinical factors difficult. As an exploratory investigation, we attempted to bridge to well annotated expression based databases since ours was already the largest SE database in AML available. Thus, we developed a 1,710 gene transcriptional signature as a proxy for our SE profiling clusters and projected this scheme into the patients profiled by The Cancer Genome Atlas Consortium (Supplementary Fig. S4C–D, Supplementary Table S2, see methods). By expression, the TCGA clusters 1 and 2, which already showed some overlapping SEs, were combined in the classification as their transcriptional signatures were too similar to reliably distinguish their members on cross validation. While this analysis is not meant to replace established clinical AML prognostic categories or patient stratification, we note that enhancer clusters were associated with significantly different survival outcomes in univariate analysis (p<0.013, Supplementary Fig. S4E, Supplementary Table S3) which could not be accounted for by somatic mutation differences (Supplementary Fig. S4F). This suggests that enhancer profiles may provide orthogonal information to mutations in the coding region of the genome motivating future studies into the clinical relevance of enhancer landscapes.
An SE drives the RARA gene in a subset of AML
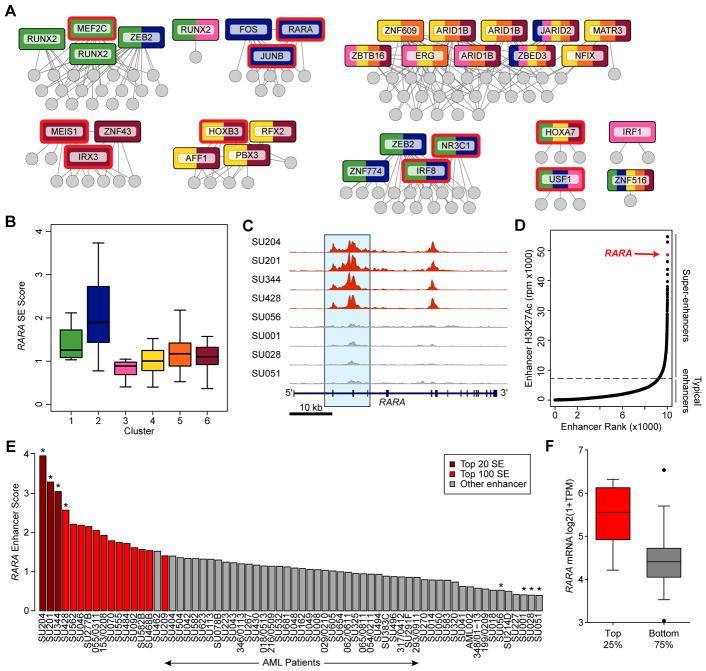
To identify targets for putative therapeutic intervention, we inferred a mutual information network of coordinated SEs across the clusters, and hypothesized that TFs in this network are key drivers of the transcriptional circuitry in these patients (Fig. 3A). Some TFs showed concomitant enrichment of their DNA binding motif in cluster-specific enhancers setting the foundation for the cell’s nuclear circuitry (Fig. 3A, red outline). One enhancer that was highly differential between patient samples is associated with the retinoic acid receptor α (RARA) gene (Fig. 3B–C, Supplementary Fig. S5A). In some cases, this enhancer was one of the strongest present genome-wide (Fig. 3D) and the most correlated enhancer with the RARA gene by expression. Overall, the RARA enhancer ranked among the top 100 enhancers in 25% of samples (Fig. 3E). For 59% of the samples, the RARA enhancer was classified as an SE, suggesting that the AML blasts from these individuals have committed a significant amount of transcriptional machinery to this locus. Indeed, RARA expression was significantly higher in the top 25% of samples in which the RARA enhancer ranked in the top 100 enhancers and was correlated with enhancer score overall (Fig 3F, Supplementary Fig. S5B). While translocations involving the RARA gene are common in acute promyelocytic leukemia (APL) (31), only two of the patients in our cohort harbored RARA fusions, and enhancer based clustering did not place these samples into cluster 2 where RARA SE samples were enriched (Fig. 3B, Supplementary Fig. S5A). The RARA SE itself is located over the gene body of RARA in AML, MDS, and normal monocytes (Fig. S5A&C) with acetylation spanning the active promoter and several introns and exons. Moreover, the size of the enhancer correlates with the expression of RARA in our patient samples further supporting a direct link of this overlapping enhancer and gene. Importantly, published PCHi-C data (20) validate the three-dimensional interaction of the RARA promoter with the RARA SE (Supplementary Fig. S5C).
Figure 3.
A subset of AML patients have a super enhancer at the RARA locus. A, Mutual information network of the SE score between cluster-specific SEs. SEs that are linked to TFs are named and displayed in boxes, with edges to the SEs with which they have high mutual information. Each box is colored by its contribution to each cluster, with multiple clusters indicated by color stripes. TFs with motifs that are enriched in SEs specific to the clusters indicated here over active background regions are outlined in red. Non-TF-linked SEs are indicated as gray circles. B, Boxplot showing the score of the RARA SE (y-axis) and corresponding cluster median (black bar), grouped by cluster membership (x-axis). C, H3K27ac ChIP-seq tracks at chr17:38,464,514–38,515,430 with large SEs shown in red and non-SE tracks shown in gray. D, Rank ordering of enhancers in a single patient sample (SU204). The SE linked to the RARA locus is indicated and is the third largest enhancer in this sample. E, Distribution of enhancer scores for the RARA enhancer across the 66 non-APL AML patient cohort. Crimson indicates samples where the RARA SE is amongst the top 20 largest enhancers in the sample; red indicates the same for the top 100 enhancers; gray is for remaining samples. The asterisks indicate the samples shown in the plot in (C). F, RARα mRNA levels (assessed by RNA-seq) in patient samples with differential RARA enhancer score. The top 25% of samples by enhancer score contain significantly higher RARα mRNA levels (p<0.0001, two tail T-test with Welch’s correction).
Given the similarities between AML and MDS, we also investigated the RARA locus in MDS and normal bone marrow CD34+ HSPCs. Analysis of a public data set (32) comparing gene expression in MDS CD34+ cells with normal CD34+ cells revealed that a subset of MDS cells have elevated RARA mRNA levels, similar to that seen in our AML dataset (Supplementary Fig. S5D). By analyzing bone marrow aspirates from two MDS patients, we detected RARA SEs with comparable score to those in AML (Supplementary Fig. S5E). In contrast, in normal bone marrow CD34+ cells, the RARA locus was not associated with an SE and H3K27ac levels were similar to AML blasts that do not contain a SE at this locus.
Overall, the fact that the RARA SE is one of the largest SEs detected in a subset of patient samples raises the hypothesis that these cells may be dependent on the function of overexpressed RARα.
An RARA SE predicts sensitivity in vitro to an RARα agonist in non-APL AML
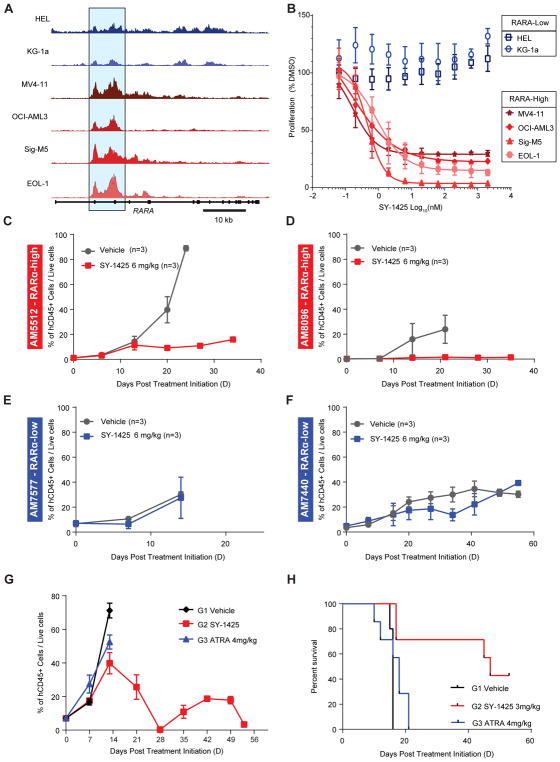
We asked whether the presence of a strong SE for RARA in non-APL AML cells could be predictive for sensitivity of such cells to RARα targeted compounds and identified a series of models to test this (Supplementary Fig. S5F). As in primary patient samples, the score of the RARA enhancer and the corresponding mRNA expression varied across a collection of non-APL AML cell lines (Fig. 4A, Supplementary Table S4). Four cell lines with strong enhancers (MV4–11, OCI-AML3, Sig-M5, and EOL1) and two with weak enhancers (HEL, and KG1a), hereafter referred to as RARA-high and -low, were used as initial models for differences in RARA SE score observed in our primary patient samples. RARα targeted agents were chosen for both selective agonism and antagonism. We first identified a potent and selective RARα agonist from the literature, tamibarotene (referred to as SY-1425 in the rest of the manuscript), which has previously been well validated as a selective and potent RARα agonist (14,16,17), and was further confirmed by us in biochemical and cellular assays (Supplementary Fig S6A–B).
Figure 4.
The presence of a RARA SE correlates with sensitivity to SY-1425 in non-APL AML cell lines and PDX models. A, H3K27ac tracks at the RARA locus for RARA-low cell lines in blue (HEL, KG1a) and RARA-high in red (MV411, AML3, EOL1, SigM5). Blue highlighted region indicates the RARA SE. B, Anti-proliferative response of non-APL AML cell lines to SY-1425 as assessed by ATPlite (PerkinElmer) for 2 RARA-low (blue) and 4 high (red). C, Effect of SY-1425 in AM5512 non-APL RARA-high PDX model. Tumor growth was monitored by measuring human CD45 positive cells in mouse circulation with treatment initiated 30 days after inoculation. D, Effect on percent human CD45 positive in peripheral blood of SY-1425 in AM8096 non-APL RARA-high PDX model. Treatment was initiated 43 days after inoculation. E, AM7577 non-APL RARA-low PDX model showing days post treatment initiation versus percent positive cells for human CD45. Treatment was initiated 36 days after inoculation. F, AM7440 non-APL RARA-low PDX model showing days post treatment initiation versus percent positive cells for human CD45. Treatment was initiated 105 days after inoculation. G, Comparison of ATRA (blue) with SY-1425 (red) and vehicle (black) in AM5512 non-APL RARA-high PDX model. Treatment was started 23 days after inoculation Mice were treated with 3mg/kg BID for SY-1425 or 4mg/kg BID for ATRA. H, Survival plot for study in (G) SY-1425 treated (red), ATRA (blue), or Vehicle (black). SY-1425 shows significant prolongation of survival (0.02 Mantel-Cox test).
RARA-high and low cell lines were treated with a range of concentrations of SY-1425 and assessed for proliferation (Fig. 4B). While SY-1425 was a potent inhibitor of proliferation in the RARA-high AML cell lines, with EC50s that ranged from 0.14 nM-0.9 nM, there was no effect on the RARA-low cell lines up to the highest concentration tested (2 μM). A broader panel of non-APL AML cell lines further confirmed the positive correlation of both RARA mRNA (11 cell lines, Supplementary Fig. S7A) and enhancer score (13 cell lines, Supplementary Fig. S7B) with SY-1425 sensitivity, with responders showing significantly higher RARA enhancer score (one-sided t-test, p=0.03), and a correlation of enhancer score and mRNA levels (Supplementary Fig S7C). To confirm that the effect of SY-1425 was indeed mediated by agonism of RARα, we tested the effect of a RARα selective antagonist, BMS195614 (18), alone or in combination with SY-1425 on AML cells. The antagonist did not show any anti-proliferative effect on RARA-high or RARA-low cell lines by itself (Supplementary Fig. S7D). Furthermore, co-administration of high concentrations of the antagonist with SY-1425 was found to completely rescue the effect of SY-1425 in the RARA-high cell line SigM5. Co-treatment with the RARα antagonist had no effect on growth inhibition by idarubicin, a DNA intercalator (Supplementary Fig. S7E). These results support the hypothesis that SY-1425 exerts its anti-proliferative effects on AML cells specifically through activation of RARα.
RARA mRNA levels correlate with SY-1425 sensitivity in vivo
To further strengthen our hypothesis that an SE at the RARA locus induces high levels of RARA transcripts, resulting in a cell state that is sensitive to a selective RARα agonist, we tested the effects of SY-1425 on four patient derived mouse xenograft models (PDXs) of AML. The four PDX models are disseminated AML leukemia models where treatment is initiated when a 2–7% AML blast burden is detected in the peripheral blood of inoculated mice. This usually occurs about 20–40 days after inoculation with human tumor cells depending on the model. Mice were treated daily with 6 mg/kg SY-1425 orally, a dose that was chosen to match the human exposure when APL patients are treated clinically with tamibarotene, and similarly studied for the non-selective retinoid ATRA (Supplementary Fig. S8A–B) (33,34). We used two PDX models (AM5512 and AM8096) where the tumor cells contained high levels of RARA mRNA and compared the effects of SY-1425 to two models where the tumor cells contained low levels of RARA mRNA (AM7577 and AM7440) (Supplementary Fig. S9A). We observed significant reductions of AML blasts (measured using anti-human CD45 antibody) in peripheral blood in the two RARA-high models (Fig. 4C–D) but no effect of SY-1425 in the two RARA-low models (Fig. 4E–F). The reduction in circulating tumor cells in the RARA-high models was also associated with blast reduction in bone marrow and spleen (Supplementary Fig. S9B–C) and led to significant prolongation of survival compared to vehicle in these aggressive models of human AML (Supplementary Fig. S9D–E). In contrast, in the models with low RARA mRNA expression, no blast reduction in the bone marrow and spleen or prolongation of survival was observed (Supplementary Fig. S9F–I). This suggests that elevated levels of RARA mRNA could be used as a correlated surrogate measurement for the RARA SE to detect cancer cells predisposed to respond to SY-1425 in vivo.
ATRA fails to show efficacy in RARA-high models
The less potent and less selective RAR agonist ATRA, used clinically for treating APL, has had limited success in non-APL AML (35,36). Indeed, ATRA was found to be less effective than SY-1425 in reducing proliferation of AML cell lines (Supplementary Fig. S10A). To explore whether the increased potency, selectivity, and significantly improved pharmacokinetics of SY-1425 vs. ATRA are relevant in vivo, we compared the effect of SY-1425 to ATRA in the RARA mRNA high PDX model AM5512. SY-1425 and ATRA were dosed twice daily with doses calculated to achieve exposure levels in mice similar to the human exposure observed in the clinic (33,34,37,38) (Supplementary Fig. S8A–B). SY-1425 greatly reduced tumor burden in the peripheral blood to <5% blasts, while ATRA treatment provided only a slight reduction in peripheral blast count compared to vehicle (Fig. 4G). Unlike SY-1425, the tumor burden in ATRA treated animals failed to decrease by two weeks, at which point the mice were sacrificed due to weight loss and progressive disease. SY-1425 also reduced spleen and bone marrow tumor burden compared to vehicle and ATRA (Supplementary Fig. S10B–C). Finally, SY-1425, but not ATRA, prolonged survival compared to vehicle (Fig. 4H). These data are consistent with the less potent anti-proliferative effect of ATRA on RARA-high cell lines compared to SY-1425 (7–21 fold less potent, Supplementary Fig. S10A) and improved pharmacology of SY-1425 including longer half-life, more stable repeat dosing exposure, and reduced sequestration.
SY-1425 induces differentiation in RARA-high models
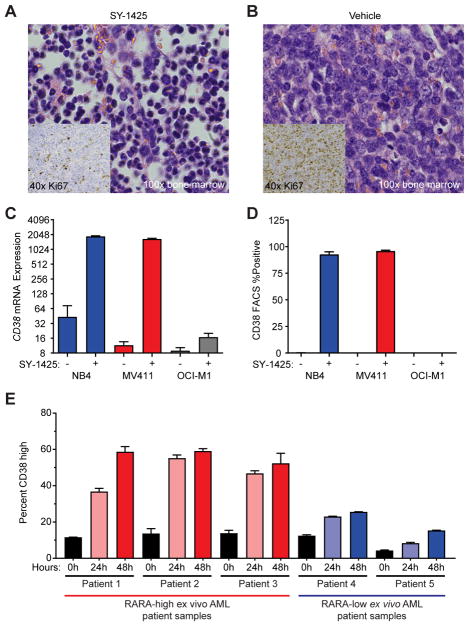
To further investigate the mechanism of action of SY-1425 in the RARA-high model (AM5512), a study was performed in which bone marrow and blood was sampled weekly, and analyzed by H&E staining and Ki67 IHC. Peripheral blood and bone marrow were found to closely mirror each other with respect to percent tumor burden (Supplementary Fig. S10D). At day 14, when the tumor burden in vehicle and treatment arms began to diverge, we observed marked morphologic differences between treated and untreated mice in both blood and bone marrow. In SY-1425 treated mice, we observed morphologic changes consistent with promotion of myeloid differentiation including banded nuclei and increased cytoplasm to nuclear area (Fig. 5A) compared to a high degree of poorly differentiated blasts in the vehicle treated mice (Fig. 5B). In addition, Ki67, a marker of cellular proliferation, was greatly reduced in the bone marrow of SY-1425 treated mice compared to those treated with vehicle (Fig. 5A–B inset).
Figure 5.
SY-1425 induces maturation in RARA-high AML. A–B, Histology from bone marrow for (A) SY-1425 treated and (B) Vehicle treated. H&E staining of bone marrow from mice at day 14 of treatment shown at 100x. Inset on each image is Ki67 staining from the same bone marrow and time at 40x. C, CD38 mRNA expression analyzed by microarray in an APL cell line (NB4 – blue), a RARA-high cell line (MV-411 – red), and a RARA-low cell line (OCI-M1 - gray). Error bars represent standard deviation of three biological replicates. D, CD38 protein expression analyzed by flow cytometry in an APL cell line (NB4 – blue), a RARA-high cell line (MV-411 – red), and a RARA-low cell line (OCI-M1 - gray). Error bars represent standard deviation of three biological replicates. E, Plot of CD38 high induction for three RARA-high AML patients (left, red) and two RARA-low (right, blue) at indicated time points. Samples were mononuclear cells from AML patients tested for ex vivo response to 50nM SY-1425.
To further investigate the hypothesis that SY-1425 induces differentiation in RARA-high, but not RARA-low AML cells, we analyzed the response to SY-1425 in a number of cell line models in more detail. We found that upon SY-1425 treatment, known granulocytic differentiation and retinoic acid response genes (39–42) were upregulated in the RARA-high cell line MV4–11, but not in the RARA-low cell line OCI-M1. In particular, CD38, which is a marker of early myeloid maturation, was found to be strongly induced in RARA-high, but not RARA-low cell lines at the mRNA (Fig. 5C) and protein level (Fig. 5D). Other myeloid differentiation markers such as ITGAX (CD11c) (Supplementary Fig. S11A), ITGAM (CD11b) (Supplementary Fig. S11B), and CD66 (Supplementary Fig. S11C) were more strongly induced in MV4–11 (RARA-high) and NB-4 (APL) than OCI-M1 (RARA-low).
We then sought to expand these findings beyond cell lines and analyzed 5 primary AML patient samples, three of which displayed high RARA mRNA levels (RARA-high) and two of which displayed low RARA mRNA levels (RARA-low). When treated ex vivo with 50 nM SY-1425, a similar trend of CD38 maturation marker induction in RARA-high, but not RARA-low cells was observed (Fig. 5E). While some AML is classified as CD38 positive on the basis of CD38 dim expression, these were gated on a high mean fluorescent intensity level which was achieved only after exposure to SY-1425. Comparing the induction in RARA-high vs. RARA-low finds a significantly greater percentage of CD38 high phenotype cells in the RARA-high patients (Supplementary Fig. S11D).
Transcriptional response in RARA-high cells treated with SY-1425 is similar to changes in APL cells treated with retinoids
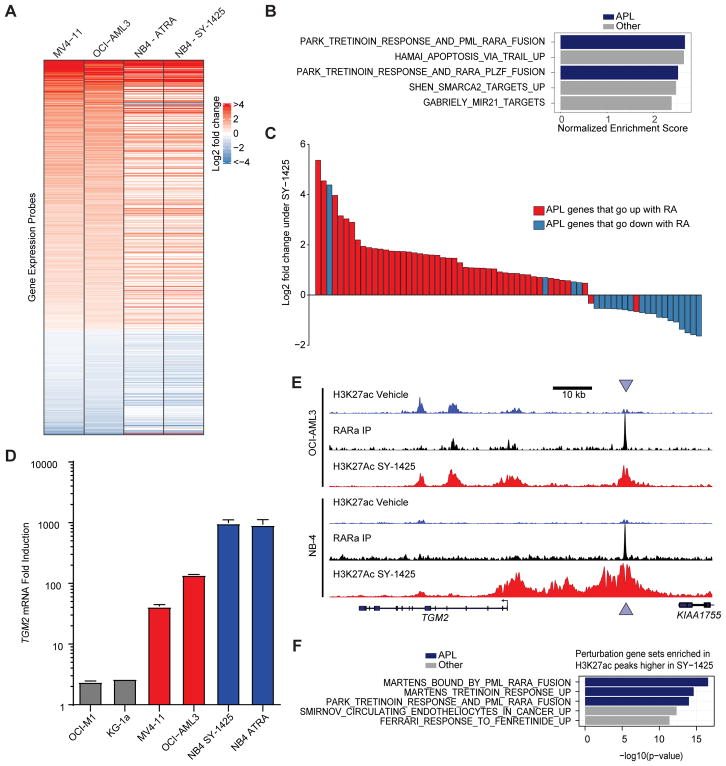
To investigate the similarity of SY-1425 response in RARA-high AML to retinoid response in APL, we assayed the gene expression of three RARA-high cell lines and the NB-4 APL cell line following treatment with either SY-1425 or vehicle. At a global level, the transcriptional changes observed after SY-1425 were broadly consistent between the RARA-high cell lines and NB-4, an APL cell line (Fig. 6A). We next performed gene set enrichment analysis (GSEA) to query whether the transcriptional response of RARA-high cell lines to SY-1425 resembles gene sets of chemical or genetic perturbations found in the literature. Two of the most highly associated gene sets (FDR<10−4) were relevant to acute promyelocytic leukemia (APL) pathogenesis or response to ATRA (tretinoin) (Fig. 6B, Supplementary Table S5). We next compared the transcriptional response of RARA-high cell lines to changes previously described in APL cells treated with ATRA ex vivo (43). We found that the genes differentially regulated in RARA-high AML cell lines have largely consistent directionality with those in ATRA treated APL patient samples (t-test p<10−16) (Fig. 6C). To further characterize the transcriptional response to SY-1425, we performed GSEA on pathways gene sets and found that immune cell pathways were induced and MYC target genes were down regulated (FDR<0.01, Supplementary Fig. S12A, Supplementary Table S6) consistent with the observed growth arrest and pro-differentiation effect. We conclude that the transcriptional response of APL cells to retinoids (ATRA or SY-1425) is similar to the response of RARA-high cell lines to SY-1425, suggesting an overlap in the mechanisms of action of these two agents in these two different forms of AML.
Figure 6.
SY-1425 shows similar response in RARA-high AML cell lines to APL. A, Gene expression response to SY-1425 (log2 fold-change) by Affymetrix probes in the APL cell line NB-4 and the RARA-high cell lines OCI-AML3 and MV4–11. Probes with FDR<0.01 in joint group of OCI-AML3 and MV4–11 (n=575) are shown. Probes are sorted by log2 fold-change in joint group of OCI-AML3 and MV4–11. B, Gene sets from the perturbation gene sets (MSigDB C2.CGP) that are enriched by GSEA in SY-1425 response in the RARA-high cell lines (top 5 gene sets by FDR, all of which are FDR<0.01). Gene sets referencing Tretinoin (ATRA) or APL are indicated in blue. X-axis indicates normalized enrichment score (NES). C, Log2 fold change of genes in RARA-high AML cell lines (OCI-AML3 and MV4–11) upon SY-1425. The 69 genes differentially expressed in both the RARA-high AML cell lines upon SY-1425 and the ex-vivo APL patient samples upon retinoic acid treatment (Meani et al. 2005) are shown. Colors indicate the direction of change in the APL patient samples. D, Gene expression fold induction of TGM2 by SY-1425 (AML cell lines) or SY-1425 and ATRA (NB-4). Gray bars indicate RARA-low lines, red bars indicate RARA-high lines, and blue bars indicate the APL cell line. E, H3K27ac and RARα ChIP-seq signal at the TGM2 locus (chr20:36,749,636–36,841,254) in OCI-AML3 (RARA-high AML) and NB-4 (APL). Arrows indicate RARα binding site. F, GREAT analysis of gene sets enriched in H3K27ac peaks that are up-regulated by SY-1425 in the RARA-high cell lines (OCI-AML3 and MV4–11). Top 5 (by FDR) perturbation gene sets are shown (full data in Supplementary Table S7).
To further study the changes in epigenetic cell states induced by SY-1425 we performed ChIP-seq for RARα and H3K27ac in cell lines representing APL, RARA-high AML, and RARA-low AML. Focusing on the TGM2 locus, a well described PML-RARα target gene (39) whose expression was induced in RARA-high AML and APL (Fig. 6D), we found an increase in H3K27ac in both NB-4 and the RARA- high line OCI-AML3 (Fig. 6E). There was a RARα binding site in both cell lines downstream of the promoter around which acetylation was induced (Fig. 6E). Globally, the majority of genes bound by RARα in NB-4 were also bound by RARα in the RARA-high cells and to a lesser degree in the RARA-low cells (Supplementary Fig. S12B). Since super-enhancer analysis has been hypothesized to reveal cell states (6,7), we also performed a clustering analysis of the differential SEs following SY-1425 treatment in all three cell types. This analysis revealed that the RARA-high cells clustered more closely with NB-4 than with the RARA-low cells (Supplementary Fig. S12C–D). To examine whether the H3K27ac peaks induced by SY-1425 regulate known sets of differentiation genes, we used the GREAT method (44), which detects enrichment of genes near genomic regions of interest. We found strong overlap between the genes near enhancers induced in the RARA-high cells and genes induced by ATRA and bound by the PML-RARα fusion protein in APL cell lines (39) (Fig. 6F, Supplementary Table S7). These results show that the response of enhancers in RARA-high AML cells to SY-1425 is similar to that associated with treatment of APL cells with ATRA.
SY-1425 exerts transcriptional and chromatin alterations in RARA-high cell lines
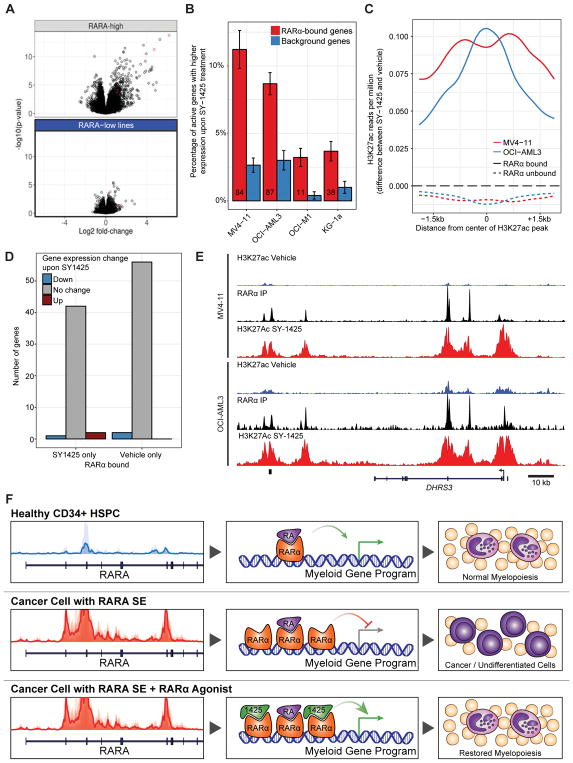
In addition to the similarities with APL, we sought to better understand the transcriptional and epigenomic consequences of SY-1425 treatment. Therefore, the global transcriptional changes and chromatin alterations were examined to study the relationship between RARα target genes, chromatin activation, and mRNA regulation. Of particular interest was whether RARα required redistribution to activate genes upon treatment or showed switch-like behavior at its original positions. We observed large changes after treatment with SY-1425 in both gene expression and enhancer score in the RARA-high lines, but little to no changes in the RARA-low cell lines, suggesting that the transcriptional and epigenomic changes are due to RARα cell state and correlate with cell line sensitivity to SY-1425 (Fig. 7A, Supplementary Fig. S13A–B). Further supporting the role of RARα in the SY-1425 response, we found that genes and enhancers bound by RARα in the vehicle condition were more likely to be induced by SY-1425 treatment (Fig. 7B–C, Supplementary Fig. S13C). To further test whether part of the mechanism of action of SY-1425 could be through redistribution of RARα among potential binding sites, we investigated the difference in RARα binding between SY-1425 and vehicle treatment conditions in the RARA-high AML cell lines OCI-AML3 and MV4–11. Large changes in RARα binding locations with SY-1425 treatment were not observed in either RARA-high AML cell line (only 1/9691 RARα peaks in OCI-AML3 and 480/14919 in MV4–11 changed). Examining the genes that were near the RARα binding sites that change in MV4–11, only three genes bound by RARα specifically upon treatment with SY-1425 were differentially expressed and only two genes bound by RARα specifically in the vehicle condition were differentially expressed (Fig. 7D). This supports the hypothesis that RARα undergoes a functional switch from repression to activation upon SY-1425 treatment rather than inducing a redistribution of RARα on the chromosome.
Figure 7.
SY-1425 induces transcriptional and epigenetic alterations. A, Volcano plots of gene expression response to SY-1425 by Affymetrix probes in RARA-high cell lines (OCI-AML3, MV4–11, and Sig-M5) and RARA-low cell lines (OCI-M1, KG-1a, Kasumi-1). Red points are probes that map to DHRS3. B, Percentage of genes in each set that are up-regulated by SY-1425 (FDR<0.05 and log2 fold change >1) in each cell line. RARα bound genes contain a RARα ChIP-seq peak (top 4000 peaks per cell line) within the gene regulatory region of the gene (promoter+10kb up to neighboring gene’s promoter). Background genes are drawn from genes with similar expression level to RARα bound genes. Error bars are from bootstrapping. Numbers in the RARα bound bars indicate the number of genes up-regulated and bound by RARα in that cell line. C, Meta-peak of H3K27ac peaks in the RARA-high cell lines by the difference in H3K27ac ChIP-seq reads per million in the SY-1425 and vehicle conditions (positive values indicate more signal in SY-1425 condition). D, Number of genes bound by RARα only in SY-1425 or vehicle conditions in MV4–11 cell line by whether their expression is differential between conditions. E, H3K27ac (blue: vehicle, red: SY-1425 50nM) and RARα (black) ChIP-seq signal at the DHRS3 locus (chr1:12,575,438–12,701,209) in RARA-high cell lines (OCI-AML3 and MV4–11). F, Proposed model of SY-1425 action in RARA-high AML. Normal CD34+ immature myeloid cell track at the RARA locus shows low acetylation leading to a balance of endogenous retinoic acid (RA) and RARα protein preceding normal development of granulocytes (top). Cancer cells with the SE at the RARA locus drive an excess RARα protein causing an imbalance of protein and ligand that favors the repressive form of RARα holding back maturation and enabling proliferation (middle). Treatment with SY-1425 strongly agonizes RARα to re-activate myeloid signaling and cause terminal differentiation of AML cells (bottom).
In order to find a pharmacodynamic marker for SY-1425 treatment, we investigated the gene expression changes in RARA-high cell lines upon treatment with SY-1425. DHRS3 showed the largest expression increase following SY-1425 treatment across RARA- high cell lines (Fig. 7A). This gene is implicated in endogenous retinoic acid metabolism and is responsive to retinoic acid levels (45–47). Furthermore, DHRS3 had multiple RARα binding sites both within and distal to the gene around which strong H3K27ac signals were observed following SY-1425 treatment (Fig. 7E). The positions of these RARα binding sites were largely unchanged upon SY-1425 treatment, supporting the hypothesis that new RARα binding sites are not required for the large induction of DHRS3 expression.
DISCUSSION
SE analysis identifies patient subsets and RARα as a therapeutic target in non-APL AML
Here we report the enhancer and transcriptional landscapes in 66 primary human AML patient samples. Using these patient-derived data sets, we identify super-enhancers that are active in AML and define the transcriptional regulatory circuits that govern this aggressive malignancy. As a comparison, we mapped the enhancer landscapes of healthy hematopoietic stem, progenitor cells, and monocytes to provide a framework for hematopoietic differentiation states. Super-enhancer analysis allows us to map the differentiation state of primary AML blast samples into the continuum of myeloid development. Applying non-negative matrix factorization to our SE patient dataset, we identified 6 distinct patient groups that were defined by the activity of enhancers driving expression of important transcription factor genes such as IRF8, PROM1, MEIS1, and HOXA/HOXB.
To assess a potential clinical relevance of our SE-defined patient clusters we made use of the well-curated transcriptomic and clinical data provided by The Cancer Genome Atlas. We projected our patient subgroup classification onto this large cohort to understand how enhancer landscapes might correlate with patient survival. This projection analysis demonstrated divergent overall survival between individual patient clusters. We note, however, that our AML cohort was biased toward high blast count AML which correlates with FLT3-ITD (30) mutation and this analysis should be validated in additional patient cohorts in the future. The FLT3-ITD, while more frequent overall, was not correlated with the RARA SE.
Since we also analyzed the somatic mutation spectrum in our patient samples, we asked how mutations are distributed throughout our clusters and whether a similar clustering could have been achieved through exonic mutation analysis. While some mutations were enriched or depleted in various clusters (for example no NPM1 mutations were found in cluster 5 or 3, and MLL translocations were enriched in cluster 1), other mutations such as those occurring in TET2, DNMT3A, and RAS, did not appear to be concentrated in a specific cluster. From this data, we conclude that clustering using SEs can provide orthogonal information that cannot be obtained by somatic mutational analysis alone.
SEs can also provide an avenue to identify novel druggable dependencies by finding those which are cancer specific or specific to certain tumor subtypes. By identifying SE associated, cluster-specific TFs we can begin to discern transcriptional regulatory circuitry. One SE-driven gene that appeared to be a key node in cluster 2 was the ligand-dependent transcription factor RARA. The RARA SE was exceptionally strong in many cluster 2 patients and RARα binding sites were enriched in cluster 2 SEs. The fact that this SE ranked as one of the largest enhancers in about 25% of the patient samples analyzed, while not present in normal, non-malignant CD34-positive blasts, raises the possibility that this RARα circuitry represents a novel tumor liability that could be pharmacologically exploited.
The RARA SE and associated high RARA mRNA predict response to SY-1425
Indeed, we found that a potent and selective RARα agonist (SY-1425) inhibited the proliferation of cell lines that contained the RARA SE, but not of cell lines that did not. This correlation was also true for RARA mRNA levels, presumably because SEs can drive high transcriptional activity for the genes they regulate (48). We verified this unique sensitivity of RARA-high AML cells in vivo in four AML patient derived xenograft models where two RARA-low models showed no response to SY-1425, whereas SY-1425 treatment led to a significant reduction of tumor burden and prolongation of survival in the two RARA-high models. This observation does not extend to the natural retinoid ATRA, presumably due to its lower potency, selectivity, and PK properties (37,49,50). Compared to SY-1425, ATRA has a significantly less favorable PK profile both in mice and humans due to the fact that, as a natural derivative of vitamin A, ATRA is degraded by CYP26A1 and has been reported to induce the expression of this metabolizing enzyme resulting in lower exposures upon repeat dosing (37,49). SY-1425, as a synthetic retinoid, is resistant to CYP26A1 degradation (50) and has reduced binding to other retinoic acid sequestration proteins.
SY-1425 exerts transcriptional and chromatin changes that lead to differentiation of RARA-high AML cells
Investigating the molecular mechanism of SY-1425 in non-APL AML cell lines, we found that SY-1425 induced profound transcriptional changes in RARA-high, but not RARA-low cell lines (Fig 7A). When we examined the distribution of RARα on chromatin and the alterations in H3K27ac before and after SY-1425 treatment, it became apparent that the chromatin location of RARα was not affected by treatment with SY-1425, rather, treatment tended to induce acetylation proximal to existing RARα ChIP-seq peaks. This is consistent with previously described activities of RARα, including acting in a transcriptional repressive complex while in the unliganded form and in a transcriptional activator complex when bound by an agonist (51). This supports a model in which chromatin bound RARα nucleates enhancer formation and gene expression in response to the ligand, SY-1425. Applying gene set enrichment analysis to the changed expression profile following SY-1425 treatment of RARA-high cells we found an induction of immune signaling pathways (integrin, protein secretion, complement, MHC pathways) and a reduction in MYC target genes. These findings are consistent with the decreased proliferation and pro-differentiation effects observed in cell lines and xenograft models following SY-1425 treatment. Specifically, we found myeloid differentiation RARα target genes, like ITGAX (encoding CD11c), ITGAM (encoding CD11b), and CD38 to be induced in RARA-high, but not RARA-low AML cell lines. Moreover, histology of the bone marrow from RARA-high PDX models treated with SY-1425 revealed morphological changes consistent with differentiation and a reduction in the proliferation marker Ki67 (Fig. 5 A and B).
The transcriptional response of RARA-high AML cells to SY-1425 is similar to the response of APL cells to retinoids
Since AML blasts that either contain a PML-RARA fusion (APL) or an SE at the RARA locus seem to both have a dependency on RARα, we compared the transcriptional response and epigenetic state in representative APL or RARA-high cell lines following RARα agonist treatment. We found the gene expression and enhancer responses of RARA-high AML cell lines to SY-1425 to be very similar to the responses of APL cells to ATRA or SY-1425. Gene sets of APL gene expression response to either treatment or genetic perturbation matched the response of RARA-high AML to SY-1425 at the gene expression and enhancer acetylation level. Across the genome, RARα binding was highly conserved between the APL cell line NB-4 and RARA-high AML cell lines and showed less similarity to the RARA-low AML cell lines. We conclude, that many aspects of the well-studied action of retinoids on APL cells (43,52–55) also apply to the treatment of RARA-high cells (defined here) with SY-1425. This includes the initial oncogenic cell state and the induction of differentiation and loss of proliferation after treatment with retinoids.
Proposed model of SY-1425 activity in non-APL AML
These findings support a model (Fig. 7F) where, just as in APL where the PML-RARA fusion protein acts as a transcriptional repressor driving the undifferentiated state of AML blasts, SE-induced overexpression of unliganded RARα acts as a transcriptional repressor of genes under the control of RARα. This assumes that the RARA SE and cell state are linked to super-physiological levels of RARα or local depletion of endogenous retinoic acid such that the balance is distorted. The SE may also favor a more repressive isoform of RARα by altering the transcriptional start site of the RARA gene as noted with RARα2 (56). When APL cells are treated with ATRA, the PML-RARA fusion protein is degraded and the remaining wildtype RARα is induced by ATRA to function as a transcriptional activator (35,57). Similarly, in the case of RARA-high AML cells, SY-1425 functions as an agonist turning RARα into a transcriptional activator. RARα binding to the genes and regulatory regions nucleates enhancer formation to drive upregulated expression of target genes in the presence of SY-1425. This potently reactivates differentiation pathways and drives the cells to terminal differentiation and growth arrest. Further validation of this model could be explored in future studies to demonstrate increased repressive complex association and activity with RARα in RARA-high cells. Alternatively, it could be conjectured that the RARA-high state merely potentiates the cells to gene activation without an active role in repression.
A biomarker directed clinical trial for SY-1425 in AML and MDS
In summary, through the analysis of super-enhancers in non-APL AML patient blasts, we have discovered a novel patient subgroup that is characterized by an SE at the RARA locus, predisposing cells to exquisite sensitivity to a potent and selective RARα agonist, SY-1425. SY-1425 acts through the induction of differentiation and arrest of proliferation in these cells employing a mechanism of action similar to that of retinoids in APL. While the discovery of this new cancer vulnerability was made possible by genome wide SE analysis, there are multiple transcriptional readouts associated with the RARA SE that now can be used clinically to identify this patient population.
Previous clinical studies of retinoids in non-APL AML employed a variety of patient selection methods, patient background types, and chemotherapy combinations (58–60). Individual response in these trials and case reports are consistent with the possibility of a therapeutic benefit of retinoids in a subset of AML and MDS patients distinct from APL. However, prior attempts may have suffered from the lack of an effective patient selection strategy. In the case of ATRA it is also possible that insufficient potency, non-selectivity over other RAR subtypes (in addition to other nuclear hormone receptors and sequestration proteins), short half-life, and diminished exposure after repeat dosing may have contributed to limited responses. Moreover, the increased reduction in blast count and survival seen with SY-1425 versus ATRA in our models is consistent with the results from clinical studies demonstrating increased CR rates of tamibarotene over ATRA in APL (33,38,61). The preclinical data presented here has led to the development of a clinical patient selection strategy utilizing an RARA SE based biomarker for patient selection in a Phase 2 study of SY-1425 in non-APL AML and MDS patients (NCT02807558).
METHODS
Sample collection and processing
Normal donor human blood cells were obtained fresh from AllCells (Alameda, CA) and FACS-enriched. Human AML samples were obtained from patients at the Stanford Medical Center with informed consent, according to Institutional Review Board (IRB)-approved protocols (Stanford IRB no 18329 and 6453). Mononuclear cells were isolated from each AML sample via Ficoll separation (GE Healthcare Life Sciences, Pittsburg, PA) and cryopreserved in liquid nitrogen in 90% FBS + 10% DMSO. All experiments presented here were conducted on freshly thawed cells. Criteria for inclusion of AML samples in this study were pre-established. Samples were included based on blast percentage at sample collection. For normal donors, no exclusion criteria were used. All AML samples were thawed in IMDM + 10% FBS + 200 U/ml DNase (Worthington Biochemical, Lakewood, NJ)
MDS samples
MDS patient samples were acquired from DVbiologics and processed as described in ChIP-seq methods. MDS sample 1 was from the bone marrow of a Caucasian female age 67. MDS sample 2 was from the bone marrow of a Caucasian female age 92.
Cell Culture
We obtained human AML cell lines OCI-AML3, Sig-M5, NOMO-1, and OCI-M1 from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ, Brunswick, Germany) We obtained other human AML cell lines (HL-60, MV-4–11, THP-1, KG-1a, HEL and Kasumi-1) from American Tissue Culture Collection (ATCC, Manassas, Virginia). All cell lines were obtained during 2014–2015. Cell lines were incubated at 37°C under 5% CO2, in RPMI 1640 plus 10% FBS and additional growth factors as necessary according to the tissue bank guidelines for each cell line. Mycoplasma testing was performed at least annually using the MycoAlert™ (Lonza) detection kit. Cell lines identity was confirmed at the time of study using the ATCC Human STR Profiling Cell Authentication Service. Cell passage number was maintained below 20 culture splits.
Cell Viability
Preparation Cell viability/proliferation studies were assays using ATPlite (Perkin Elmer) following the manufacturer’s instructions and analyzed using GraphPad Prism version 6.0. Extended description of study protocol is included in the Supplementary Methods.
ChIP-seq
Preparation and analysis of ChIP-seq are described in full detail in Supplementary Methods. Raw data has been submitted to the SRA under accession numbers SRP103200 (primary samples) and SRP103029 (cell lines).
RNA-seq
AML samples were homogenized in 1 mL Trizol and RNA isolated using RNAeasy (Ambion®, ThermoFisher, Waltham MA, USA) according to manufacturer instructions. RNA libraries were prepared and sequenced as described previously (5). Analysis of RNA-seq data is described in detail in Supplementary Methods. Raw data has been submitted to the SRA under accession number SRP103200.
Expression Studies in AML cell lines
Expression studies were performed using TRIzol (Thermo Fisher Scientific) for extraction, mirVana (Thermo Fisher Scientific) purification kits for RNA isolation, and were analyzed on Affymetrix PrimeView arrays. Full details of preparation and analysis are provided in the Supplementary Methods. Raw data has been submitted to GEO under the accession GSE97331.
In vivo studies
The PDX studies were performed at Crown Bioscience using their HuKemia™ systemic models. Raw RNA-seq data on the models was provided by Crown Bioscience and analyzed as per the RNA-seq methods above. FACS analysis was performed by Crown Biosciences. The study protocols and procedures involving the care and use of animals were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Crownbio prior to conduct. During the studies, the care and use of animals was conducted in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Slides for H&E and Ki67 were made from FFPE blocks from Crown Biosciences that were prepped and stained at the Specialized Histopathology Services – Longwood (DF/HCC, Brigham and Womens). Additional methods for in vivo studies provided in supplementary methods.
Supplementary Material
Identification and refinement of cancer-specific super-enhancers in human AML. A, H3K27Ac landscape at positive control locus MALAT1. Tracks show read density scale normalized ChIP-seq signal. B-C, Distribution of H3K27Ac signal across the 12,656 enhancers in a single AML patient sample (SU041 - left), and the 12,311 enhancers in a sample of healthy CD34+ HSPCs (right). A subset of the enhancers (red, 470 and 830 respectively) have exceptionally high H3K27Ac signal and are called super-enhancers by the ROSE algorithm. D, Heatmap illustrating Pearson correlation of replicate enhancer maps. All enhancers’ coordinates were merged across samples, and then all enhancers (including super-enhancers) were compared pairwise with Pearson correlation. The rows and columns are clustered with Euclidean distance and complete-linkage hierarchical clustering. The final two samples are CD34+ bone marrow cells from two separate healthy donors collected at separate locations. E, H3K27Ac ChIP-seq read density profiles across the admixture experiment samples. 11 Admix samples are shown (from 100% AML to 0% AML in steps of 10% from top to bottom). The three loci are “Ubiquitous”: the area over the ANKRD44 gene, “PBMC-specific”: the area around the IL10RA gene, and “AML-specific”: the area around the CDK6 gene. F, The percentage of PBMC (red) or AML (blue) -specific SEs recovered as super-enhancers are shown as a function of the percentage of blasts in each admixed sample. G, The percentage of PBMC (red) or AML (blue) -specific SEs recovered in the top 1000 enhancers (including SEs) are shown as a function of the percentage of blasts in each admixed sample.
Mapping of super-enhancers to genes. A, Schematic demonstrating the difficulty in assigning function to an enhancer in the absence of three-dimensional chromatin architecture data. B–D, Correlation of super-enhancer scores for an enhancer near the (B) NEDD9 (r2=0.75, p<2E−16), (C) ™EM170B (r2=0.45, p<5E−10), and (D) SMIM13 (r2=0.17, p<0.0014) genes with the promoter H3K27Ac signal of each gene across all AML patients. A p value cutoff of 1.8E−6 was used to determine whether a given enhancer acts on a given promoter. This is equivalent to 0.1 multiplied by the number of tested enhancer-promoter pairs genome-wide. E, Bar plot showing the number of SE-gene links per SE.
AML SE profiles show pronounced variation in enhancers related to myeloid differentiation and enable de novo stratification into six distinct epigenomic subgroups. A, Scatterplot showing the Euclidean distance of each sample from an HSPC centroid derived from the most HSPC- or monocyte-associated SEs from Figure 1D (x-axis), as well as the distance of each sample from the centroid of the monocyte samples (y-axis). B, Scatterplot showing the predicted HSPC signature from Figure 1D (x-axis) against the first independent component from the ICA in Figure 1E (y-axis) across all patient samples. C, Heatmap showing the super-enhancer score (with a ceiling at 2) of the top 40 ICA2-associated SEs by R2 value. The rows are clustered by Euclidean distance and complete-linkage hierarchical clustering, and the columns are ordered by the ICA2 loading for each sample (top row, blue to red). The bottom row shows the type of sample: HSPC, light blue; primary AML, gray; monocyte, purple. Example gene links for the SEs are displayed on the right. D, Coefficient matrix from the best (lowest reconstruction error) NMF. Cells show the non-negative weight put onto each factor for each sample. Samples are ordered by cluster label and rows (factors) are hierarchically clustered. E, Basis matrix from the best (lowest reconstruction error) NMF. Cells show the non-negative weight put into each factor for each SE. SEs (rows) and factors (columns) are ordered by hierarchical clustering with complete linkage. F, Pearson correlation heatmap showing positively (red) and negatively (blue) correlated samples by the top 200 most variable SEs with at least one strong patient score. Samples are ordered identically to the NMF-C distance matrix with the exception of the HSPC and monocyte samples. G, A scatter plot showing ICA1 vs ICA2 as in Figure 1E, colored by cluster (patient samples) or cell type (FACS-purified normal samples).
Recapitulation of SE-defined clusters in AML data from the TCGA. A, Bar graph showing the frequency of each mutation within our AML cohort. B, Overall survival within our cohort of 62 AML patients. Solid line: survival, dashed lines: confidence interval. C, Heatmap showing the 1,710 genes used in the nearest shrunken centroid classifier. Expression is row scaled by mean and standard deviation. Genes (rows) are ordered by which cluster they correspond to. Samples (columns) are ordered by cluster. D, Heatmap showing the contribution of each gene to each centroid. White indicates no contribution. Blue indicates a negative contribution and red indicates a positive contribution. E, Clustering was predicted for all non-M3 TCGA patient samples by scoring them against RNA-seq derived cluster centroids. Survival in the TCGA cohort significantly associates with projected cluster membership with a cox proportional hazards p value < 0.014. F, The mutation status of each of the TCGA AML samples is shown, ordered and colored by their predicted cluster. A bold mutation name indicates a nominally significant association (p < 0.05) between the mutation and the clustering (Fisher’s exact). Similarly, bold and italic font indicates significance after multiple hypothesis testing correction (p < 0.001). Only the most recurrent mutations (n > 2) are shown here. rMLL = MLL rearrangement; Mito = Mitochondrial genes.
An SE is present at the RARA locus in a subset of AML samples. A, Metatrack plots of the H3K27Ac landscape at the RARA locus in all AML clusters and healthy monocytes and HSPCs. B, Correlation of RARA enhancer score with RARα mRNA levels (by RNA-seq). Spearman rho ~ .47; p < 0.0008. C, Three-dimensional interactions in the local region surrounding the RARA gene locus from promoter capture Hi-C data. H3K27Ac ChIP-seq from one AML patient (SU070) and CTCF ChIP-seq from monocytes (Encode) are shown for reference. The RARA SE is highlighted by a blue box. Interactions of the RARA promoter with the RARA SE are shown in red. Only unique interactions with a CHiCAGO score > 5 are shown for clarity. Region represents chr17:38455078–38579467. D, Analysis of RARα mRNA from a public data set (1) reveals a significantly higher level of RARα mRNA in a subset of MDS patients blast cells compared to non-malignant CD34+ (two tail T-test with Welch’s correction). Both cohorts are CD34+ cells sorted from bone marrow aspirates. E, H3K27ac ChIP-seq tracks for two AML patients with RARA among their top 100 enhancers (crimson), two MDS patients with RARA among their top 100 enhancers (red), and two non-malignant blast cell samples from healthy donors (gray). F, Expression of mRNA by RNA-seq (log2 TPM) across a set of primary AML patients, AML cell lines, and AML PDXs.
SY-1425 is a selective and potent RARα agonist. A, Biochemical assay plot of SY-1425 (tamibarotene) concentration (log nM) versus degree of association between co-activator and RAR protein from FRET assay expressed as % maximal ATRA signal. Data are for tamibarotene effects on RARα (red circles), RARβ (blue squares), and RARγ (green triangles). The horizontal dashed line indicates the tamibarotene Emax on RARα and the vertical dashed line the EC50 value. B, Cellular reporter plot of degree of association between co-activator and RAR protein from FRET assay expressed as % maximal ATRA signal (Emax) induced by SY-1425 (tamibarotene) as a function of concentration (log nM). Data are for SY-1425 on RARα (red circles) and RARγ (green triangles).
Comparison of SY-1425 response to RARA expression and enhancer score and the effect of an RARα antagonist. Correlations of enhancer, mRNA, and sensitivity to SY-1425 are color coded by cell line as indicated. A, Plot of RARα mRNA for AML cell lines versus SY-1425 anti-proliferative effect. B, Plot of RARA enhancer score for AML cell lines versus SY-1425 anti-proliferative effect. C, Plot of normalized enhancer versus expression. D, Cellular proliferation by ATPlite in RARA-high MV4–11 (red, circles), RARA-high OCI-AML3 (crimson, triangles), RARA-low OCI-M1 (blue, diamonds) and RARA-low Kasumi-1 (black, squares). Open shapes for BMS195614, a selective RARα antagonist, and closed for SY-1425. E, Anti-proliferative effect of SY-1425 (red circles) or idarubicin (black squares) with (open symbol) or without (closed symbol) 1μM BMS195614, a RARα selective antagonist, on the RARA-high cell line SigM5.
SY-1425 in vivo pharmacokinetics. A, Chemical structure of SY-1425 and plot of time versus SY-1425 concentration in plasma from mice given 3mg/kg SY-1425 (black circle) vs human SY-1425 levels (white diamond) 6mg/m2. Tables of PK properties for SY-1425 in mice are shown (top right) with comparator data from Kanai et al. Hem. Int. 2014. B, Chemical structure of ATRA with PK plot. 3mg/kg SY-1425 (black circle), 4mg/kg ATRA (white diamond), or human ATRA levels 45mg/m2. Tables of PK properties for ATRA are shown (bottom right) with comparator data (2).
SY-1425 survival and tissue effects on AML PDX models. A, RARA mRNA levels for the four non-APL AML PDX models assessed by RNA-seq with RARA-high indicated in red and RARA-low indicated in blue. B–C, Tumor burden in blood, bone marrow, and spleen was assessed by human CD45 positive cell percentage by FACS for (B) AM5512, and (C) AM8096. SY-1425 treated (6 mg/kg) animals are shown in red to indicate RARA-high status while vehicle treated animals are shown in gray. D, Survival plot for AM5512 PDX model with significant prolongation of survival (0.03 Mantel-Cox test). E, Survival plot for AM8096 PDX model with significant prolongation of survival (0.02 Mantel-Cox test). F–G, Tumor burden in blood, bone marrow, and spleen was assessed by human CD45 positive cell percentage by FACS for (F) AM7577, and (G) AM7440. SY-1425 treated (6 mg/kg) animals are shown in blue to indicate RARA-low status while vehicle treated animals are shown in gray. H, Survival plot for AM7577 PDX model with no significant prolongation of survival (0.3 Mantel-Cox test). I, Survival plot for AM7440 PDX model.
SY-1425 shows greater potency than ATRA in vitro and in vivo. A, Plot of EC50 values for SY-1425 versus ATRA for AML cell lines with equipotency (solid), three fold stronger SY-1425 (dotted) and ten-fold stronger SY-1425 (dashed) lines indicated. B–C, Mice in AM5512 RARA-high PDX model were treated with 3mg/kg BID for SY-1425 or 4mg/kg BID for ATRA. Tumor burden in (B) spleen and (C) bone marrow are shown. Vehicle is shown in black; ATRA treatment in blue; and SY-1425 in dark red for mice sacrificed during the course of the study and light red for mice assessed at the end of two months of treatment. SY-1425 shows significant reduction versus ATRA (two tail T-test with Welch’s correction). D, Plot of percent human CD45 positive in vehicle (black) and SY-1425 3mg/kg BID (red) for peripheral blood (closed circle) or bone marrow (open squares).
Induction of markers of myeloid differentiation in RARA-high AML cell lines upon SY-1425 treatment. A–B, Induction of mRNA expression measured by microarray in cell lines treated with vehicle or SY-1425 50nM for 24 hours for (A) CD11c and (B) CD11b. C, FACS measurement of percent positive in vehicle or SY-1425 50nM 72 hours for CD66. The APL cell line NB4 is in blue, RARA-high cell line MV4–11 in red, and RARA-low cell line OCI-M1 in gray. D, Percentage of CD38 high cells in RARA-high versus RARA-low ex vivo samples after 48 hours. Significance by two tail T-test with Welch’s correction.
Comparison of SY-1425 expression and molecular response to APL. A, Canonical pathways (MSigDB C2.CP) and Hallmark (from MSigDB) gene sets enriched by GSEA in SY-1425 response in the RARA-high cell lines (Supplementary Table S6). Top 10 gene sets by FDR are shown. Positive normalized enrichment scores (NES) represent gene sets enriched in genes that are higher upon SY-1425 treatment, negative NES values represent gene sets enriched in genes that are lower upon SY-1425 treatment. B, Number of genes bound by RARα in NB-4 cell line in common with AML cell lines. Genes with RARα ChIP-seq peak (top 4000 peaks per cell line) in the gene regulatory region (promoter+1Mb up to neighboring gene’s promoter) are considered bound by RARα. NB4 bar indicates total number of genes bound in NB4. C, Heatmap of spearman correlation of super-enhancer (SE) response to SY-1425 in each cell line. Only SEs differential (FDR<0.05) in at least one cell line are included. Clustering is by Ward method. D, SE response to SY-1425 in NB-4 and RARA-high cell lines. Only SEs differential in NB-4 (FDR<0.05) (n=1967) are shown.
Supplementary Figure S13 SY-1425 expression and molecular response. A, Volcano plots of H3K27ac response to SY-1425 for the union of H3K27ac peaks in the vehicle and treatment conditions by cell line. B, Percentage of H3K27ac peaks in each cell line that are higher upon treatment with SY-1425. Red bars indicate RARA-high lines and blue bars indicate RARA-low lines. C, Percentage of H3K27ac peaks in each set that are up-regulated by SY-1425 (FDR<0.05 and log2 fold change >1) in each cell line. RARα bound peaks overlap a RARα ChIP-seq peak (top 4000 per cell line), and background peaks are drawn from peaks with similar H3K27ac level to RARα bound peaks. Error bars are from bootstrapping. Numbers in the RARα bound bars indicate the number of H3K27ac peaks up-regulated and bound by RARα in that cell line.
Supplementary Table S1: Sample mutations
Supplementary Table S2: RNA-seq signature and per-cluster weights
Supplementary Table S3: Median survival in TCGA data by predicted cluster
Supplementary Table S4: Enhancer, mRNA, and SY-1425 EC50 in AML cell lines
Supplementary Table S5: GSEA of transcriptional response to SY-1425 in RARA-high cell lines in perturbation gene sets
Supplementary Table S6: GSEA of transcriptional response to SY-1425 in RARA-high cell lines in hallmark and pathways gene sets
Supplementary Table S7: GREAT analysis of genes near H3K27ac peaks up-regulated by SY-1425 in RARA-high cell lines in perturbation gene sets
Significance.
We use the super-enhancer landscape of primary human AML to elucidate transcriptional circuitry and identify novel cancer vulnerabilities. A subset of patients were found to have an SE at RARA which is predictive for response to SY-1425, a potent and selective RARα agonist, in preclinical models, forming the rationale for its clinical investigation in biomarker-selected patients.
Acknowledgments
Thank you to Dr. Jason Marineau and Dr. Kevin Sprott for help on chemistry review and sourcing and Dr. Eric Olson for guidance. Thank you to Dr. Abner Louissaint for advice and interpretation of histopathology.
Grant Support
Supported by Stinehart-Reed Foundation (R.M.), Ludwig Center for Cancer Stem Cell Research (R.M.), NIH (R01CA18805 to R.M.). R.M. is a New York Stem Cell Foundation Robertson Investigator and Leukemia and Lymphoma Society Scholar.
Footnotes
Disclosure of Potential Conflicts of Interest
MRM, MLE, CF, EL, JTL, MGG, MWC, DS, DO, JTL, KA, and CCF are shareholders of Syros Pharmaceuticals. MRC, SMC, JLK and RM have no conflicts of interest to report.
References
- 1.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. The Lancet. 2013;381:484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance, Epidemiology, and End Results (SEER) Program Populations. National Cancer Institute, DCCPS, Surveillance Research Program; 1969–2015. ( www.seer.cancer.gov/popdata) released December 2016. [Google Scholar]
- 3.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer Genome Landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and Characterization of Super-Enhancer-Associated Dependencies in Diffuse Large B Cell Lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, et al. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol Cell. 2015;58:362–70. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SCJ, Erdos MR, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–62. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koues OI, Kowalewski RA, Chang L-W, Pyfrom SC, Schmidt JA, Luo H, et al. Enhancer Sequence Variants and Transcription-Factor Deregulation Synergize to Construct Pathogenic Regulatory Circuits in B-Cell Lymphoma. Immunity. 2015;42:186–98. doi: 10.1016/j.immuni.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–70. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, et al. Environment Drives Selection and Function of Enhancers Controlling Tissue-Specific Macrophage Identities. Cell. 2014;159:1327–40. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C-F, Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015;43:8183–203. doi: 10.1093/nar/gkv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delescluse C, Cavey MT, Martin B, Bernard BA, Reichert U, Maignan J, et al. Selective high affinity retinoic acid receptor alpha or beta-gamma ligands. Mol Pharmacol. 1991;40:556–562. [PubMed] [Google Scholar]
- 15.Miwako I, Kagechika H. Tamibarotene. Drugs Today. 2007;43:563. doi: 10.1358/dot.2007.43.8.1072615. [DOI] [PubMed] [Google Scholar]
- 16.Beard RL, Duong TT, Teng M, Klein ES, Standevan AM, Chandraratna RA. Synthesis and biological activity of retinoic acid receptor-α specific amides. Bioorg Med Chem Lett. 2002;12:3145–3148. doi: 10.1016/s0960-894x(02)00647-9. [DOI] [PubMed] [Google Scholar]
- 17.Umemiya H, Fukasawa H, Ebisawa M, Eyrolles L, Kawachi E, Eisenmann G, et al. Regulation of retinoidal actions by diazepinylbenzoic acids. 1 retinoid synergists which activate the RXR-RAR heterodimers. J Med Chem. 1997;40:4222–4234. doi: 10.1021/jm9704309. [DOI] [PubMed] [Google Scholar]
- 18.Gehin M, Vivat V, Wurtz J-M, Losson R, Chambon P, Moras D, et al. Structural basis for engineering of retinoic acid receptor isotype-selective agonists and antagonists. Chem Biol. 1999;6:519–29. doi: 10.1016/S1074-5521(99)80084-2. [DOI] [PubMed] [Google Scholar]
- 19.Whyte Wa, Orlando Da, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Cell. Vol. 153. Elsevier Inc; 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes; pp. 307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384. e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. 2016;127:42–52. doi: 10.1182/blood-2015-07-604512. [DOI] [PubMed] [Google Scholar]
- 23.Pastore F, Levine RL. Epigenetic regulators and their impact on therapy in acute myeloid leukemia. Haematologica. 2016;101:269–78. doi: 10.3324/haematol.2015.140822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Cuellar M-P, Steger J, Fuller E, Hetzner K, Slany RK. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox-induced murine leukemia. Haematologica. 2015;100:905–13. doi: 10.3324/haematol.2015.124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–98. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.TCGA Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama A. Transcriptional activation by MLL fusion proteins in leukemogenesis. Exp Hematol. 2017;46:21–30. doi: 10.1016/j.exphem.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining Roles for HOX and MEIS1 Genes in Induction of Acute Myeloid Leukemia. Mol Cell Biol. 2001;21:224–34. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrózek K, Whitman SP, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: A Cancer and Leukemia Group B study. J Clin Oncol. 2005;23:9234–42. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 30.Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17:1738–52. doi: 10.1038/sj.leu.2403099. [DOI] [PubMed] [Google Scholar]
- 31.Zhou G-B, Zhang J, Wang Z-Y, Chen S-J, Chen Z. Treatment of acute promyelocytic leukaemia with all-trans retinoic acid and arsenic trioxide: a paradigm of synergistic molecular targeting therapy. Philos Trans R Soc B Biol Sci. 2007;362:959–71. doi: 10.1098/rstb.2007.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerstung M, Pellagatti A, Malcovati L, Giagounidis A, Porta MGD, Jädersten M, et al. Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat Commun. 2015;6:5901. doi: 10.1038/ncomms6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobita T, Takeshita A, Kitamura K, Ohnishi K, Yanagi M, Hiraoka A, et al. Treatment with a new synthetic retinoid, Am80, of acute promyelocytic leukemia relapsed from complete remission induced by all-trans retinoic acid. Blood. 1997;90:967–973. [PubMed] [Google Scholar]
- 34.Kanai F, Obi S, Fujiyama S, Shiina S, Tamai H, Mochizuki H, et al. An open-label phase I/II study of tamibarotene in patients with advanced hepatocellular carcinoma. Hepatol Int. 2014;8:94–103. doi: 10.1007/s12072-013-9459-7. [DOI] [PubMed] [Google Scholar]
- 35.Johnson DE, Redner RL. An ATRActive future for differentiation therapy in AML. Blood Rev. 2015;29:263–8. doi: 10.1016/j.blre.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown G, Hughes P. Retinoid Differentiation Therapy for Common Types of Acute Myeloid Leukemia. Leuk Res Treat. 2012;2012:1–11. doi: 10.1155/2012/939021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muindi J, Frankel S, Miller WJ, Jakubowski A, Scheinberg DA, Young C, et al. Continuous treatment with all-trans retinoic acid causes a progressive reduction in plasma drug concentrations: implications for relapse and retinoid “resistance” in patients with acute promyelocytic leukemia [published erratum appears in Blood 1992 Aug 1; 80 (3): 855] Blood. 1992;79:299–303. [PubMed] [Google Scholar]
- 38.Shinagawa K, Yanada M, Sakura T, Ueda Y, Sawa M, Miyatake J, et al. Tamibarotene As Maintenance Therapy for Acute Promyelocytic Leukemia: Results From a Randomized Controlled Trial. J Clin Oncol. 2014;32:3729–35. doi: 10.1200/JCO.2013.53.3570. [DOI] [PubMed] [Google Scholar]
- 39.Martens JHA, Brinkman AB, Simmer F, Francoijs K-J, Nebbioso A, Ferrara F, et al. PML-RARα/RXR Alters the Epigenetic Landscape in Acute Promyelocytic Leukemia. Cancer Cell. 2010;17:173–85. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- 40.Mar JC, Quackenbush J. Decomposition of gene expression state space trajectories. PLoS Comput Biol. 2009;5:e1000626. doi: 10.1371/journal.pcbi.1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nasr R, Guillemin M-C, Ferhi O, Soilihi H, Peres L, Berthier C, et al. Eradication of acute promyelocytic leukemia-initiating cells through PML-RARA degradation. Nat Med. 2008;14:1333–42. doi: 10.1038/nm.1891. [DOI] [PubMed] [Google Scholar]
- 42.Jiao X, Hooper SD, Djureinovic T, Larsson C, Wärnberg F, Tellgren-Roth C, et al. Gene rearrangements in hormone receptor negative breast cancers revealed by mate pair sequencing. BMC Genomics. 2013;14:165. doi: 10.1186/1471-2164-14-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meani N, Minardi S, Licciulli S, Gelmetti V, Coco FL, Nervi C, et al. Molecular signature of retinoic acid treatment in acute promyelocytic leukemia. Oncogene. 2005;24:3358–68. doi: 10.1038/sj.onc.1208498. [DOI] [PubMed] [Google Scholar]
- 44.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams MK, Belyaeva OV, Wu L, Kedishvili NY. The Retinaldehyde Reductase Activity of DHRS3 Is Reciprocally Activated by Retinol Dehydrogenase 10 to Control Retinoid Homeostasis. J Biol Chem. 2014;289:14868–80. doi: 10.1074/jbc.M114.552257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Billings SE, Pierzchalski K, Butler Tjaden NE, Pang X-Y, Trainor PA, Kane MA, et al. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 2013;27:4877–89. doi: 10.1096/fj.13-227967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zolfaghari R, Chen Q, Ross AC. DHRS3, a retinal reductase, is differentially regulated by retinoic acid and lipopolysaccharide-induced inflammation in THP-1 cells and rat liver. AJP Gastrointest Liver Physiol. 2012;303:G578–88. doi: 10.1152/ajpgi.00234.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niederriter A, Varshney A, Parker S, Martin D. Super Enhancers in Cancers, Complex Disease, and Developmental Disorders. Genes. 2015;6:1183–200. doi: 10.3390/genes6041183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topletz AR, Tripathy S, Foti RS, Shimshoni JA, Nelson WL, Isoherranen N. Induction of CYP26A1 by Metabolites of Retinoic Acid: Evidence That CYP26A1 Is an Important Enzyme in the Elimination of Active Retinoids. Mol Pharmacol. 2015;87:430–41. doi: 10.1124/mol.114.096784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osanai M. Expression of the Retinoic Acid-Metabolizing Enzyme CYP26A1 Limits Programmed Cell Death. Mol Pharmacol. 2005;67:1808–17. doi: 10.1124/mol.104.005769. [DOI] [PubMed] [Google Scholar]
- 51.Niederreither K, Dollé P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–53. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 52.Liu M-J, Wang Z, Wu R-C, Sun S-Q, Wu Q-Y. Monitoring all-trans-retinoic acid-induced differentiation of human acute promyelocytic leukemia NB4 cells by Fourier-transform infrared spectroscopy. Leukemia. 2003;17:1670–4. doi: 10.1038/sj.leu.2403019. [DOI] [PubMed] [Google Scholar]
- 53.Yang L, Zhao H, Li S-W, Ahrens K, Collins C, Eckenrode S, et al. Gene expression profiling during all-trans retinoic acid-induced cell differentiation of acute promyelocytic leukemia cells. J Mol Diagn. 2003;5:212–221. doi: 10.1016/S1525-1578(10)60476-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamashev D. PML-RARA-RXR Oligomers Mediate Retinoid and Rexinoid/cAMP Cross-Talk in Acute Promyelocytic Leukemia Cell Differentiation. J Exp Med. 2004;199:1163–74. doi: 10.1084/jem.20032226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Idres N, Benoìt G, Flexor MA, Lanotte M, Chabot GG. Granulocytic differentiation of human NB4 promyelocytic leukemia cells induced by all-trans retinoic acid metabolites. Cancer Res. 2001;61:700–705. [PubMed] [Google Scholar]
- 56.Gianni M, Maddalena F, Marco B, Mami K, Adriana Z, Gabriela P, et al. RARα2 and PML-RAR similarities in the control of basal and retinoic acid induced myeloid maturation of acute myeloid leukemia cells. Oncotarget. 2016;8:37041–60. doi: 10.18632/oncotarget.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Licht JD. Reconstructing a disease: What essential features of the retinoic acid receptor fusion oncoproteins generate acute promyelocytic leukemia? Cancer Cell. 2006;9:73–74. doi: 10.1016/j.ccr.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 58.Coombs CC, Tavakkoli M, Tallman MS. Acute promyelocytic leukemia: where did we start, where are we now, and the future. Blood Cancer J. 2015;5:e304. doi: 10.1038/bcj.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schenk T, Stengel S, Zelent A. Unlocking the potential of retinoic acid in anticancer therapy. Br J Cancer. 2014;111:2039–45. doi: 10.1038/bjc.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garattini E, Bolis M, Garattini SK, Fratelli M, Centritto F, Paroni G, et al. Retinoids and breast cancer: From basic studies to the clinic and back again. Cancer Treat Rev. 2014;40:739–49. doi: 10.1016/j.ctrv.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Mi Y, Jiang B, Chen X, Ji C, Li Y, et al. Tamibarotene Compared to All-Trans Retinoic Acid (ATRA) As Add-on to Arsenic Trioxide (ATO) in Subjects with Relapsed Acute Promyelocytic Leukemia (APL) Blood. 2015;126:220. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification and refinement of cancer-specific super-enhancers in human AML. A, H3K27Ac landscape at positive control locus MALAT1. Tracks show read density scale normalized ChIP-seq signal. B-C, Distribution of H3K27Ac signal across the 12,656 enhancers in a single AML patient sample (SU041 - left), and the 12,311 enhancers in a sample of healthy CD34+ HSPCs (right). A subset of the enhancers (red, 470 and 830 respectively) have exceptionally high H3K27Ac signal and are called super-enhancers by the ROSE algorithm. D, Heatmap illustrating Pearson correlation of replicate enhancer maps. All enhancers’ coordinates were merged across samples, and then all enhancers (including super-enhancers) were compared pairwise with Pearson correlation. The rows and columns are clustered with Euclidean distance and complete-linkage hierarchical clustering. The final two samples are CD34+ bone marrow cells from two separate healthy donors collected at separate locations. E, H3K27Ac ChIP-seq read density profiles across the admixture experiment samples. 11 Admix samples are shown (from 100% AML to 0% AML in steps of 10% from top to bottom). The three loci are “Ubiquitous”: the area over the ANKRD44 gene, “PBMC-specific”: the area around the IL10RA gene, and “AML-specific”: the area around the CDK6 gene. F, The percentage of PBMC (red) or AML (blue) -specific SEs recovered as super-enhancers are shown as a function of the percentage of blasts in each admixed sample. G, The percentage of PBMC (red) or AML (blue) -specific SEs recovered in the top 1000 enhancers (including SEs) are shown as a function of the percentage of blasts in each admixed sample.
Mapping of super-enhancers to genes. A, Schematic demonstrating the difficulty in assigning function to an enhancer in the absence of three-dimensional chromatin architecture data. B–D, Correlation of super-enhancer scores for an enhancer near the (B) NEDD9 (r2=0.75, p<2E−16), (C) ™EM170B (r2=0.45, p<5E−10), and (D) SMIM13 (r2=0.17, p<0.0014) genes with the promoter H3K27Ac signal of each gene across all AML patients. A p value cutoff of 1.8E−6 was used to determine whether a given enhancer acts on a given promoter. This is equivalent to 0.1 multiplied by the number of tested enhancer-promoter pairs genome-wide. E, Bar plot showing the number of SE-gene links per SE.
AML SE profiles show pronounced variation in enhancers related to myeloid differentiation and enable de novo stratification into six distinct epigenomic subgroups. A, Scatterplot showing the Euclidean distance of each sample from an HSPC centroid derived from the most HSPC- or monocyte-associated SEs from Figure 1D (x-axis), as well as the distance of each sample from the centroid of the monocyte samples (y-axis). B, Scatterplot showing the predicted HSPC signature from Figure 1D (x-axis) against the first independent component from the ICA in Figure 1E (y-axis) across all patient samples. C, Heatmap showing the super-enhancer score (with a ceiling at 2) of the top 40 ICA2-associated SEs by R2 value. The rows are clustered by Euclidean distance and complete-linkage hierarchical clustering, and the columns are ordered by the ICA2 loading for each sample (top row, blue to red). The bottom row shows the type of sample: HSPC, light blue; primary AML, gray; monocyte, purple. Example gene links for the SEs are displayed on the right. D, Coefficient matrix from the best (lowest reconstruction error) NMF. Cells show the non-negative weight put onto each factor for each sample. Samples are ordered by cluster label and rows (factors) are hierarchically clustered. E, Basis matrix from the best (lowest reconstruction error) NMF. Cells show the non-negative weight put into each factor for each SE. SEs (rows) and factors (columns) are ordered by hierarchical clustering with complete linkage. F, Pearson correlation heatmap showing positively (red) and negatively (blue) correlated samples by the top 200 most variable SEs with at least one strong patient score. Samples are ordered identically to the NMF-C distance matrix with the exception of the HSPC and monocyte samples. G, A scatter plot showing ICA1 vs ICA2 as in Figure 1E, colored by cluster (patient samples) or cell type (FACS-purified normal samples).
Recapitulation of SE-defined clusters in AML data from the TCGA. A, Bar graph showing the frequency of each mutation within our AML cohort. B, Overall survival within our cohort of 62 AML patients. Solid line: survival, dashed lines: confidence interval. C, Heatmap showing the 1,710 genes used in the nearest shrunken centroid classifier. Expression is row scaled by mean and standard deviation. Genes (rows) are ordered by which cluster they correspond to. Samples (columns) are ordered by cluster. D, Heatmap showing the contribution of each gene to each centroid. White indicates no contribution. Blue indicates a negative contribution and red indicates a positive contribution. E, Clustering was predicted for all non-M3 TCGA patient samples by scoring them against RNA-seq derived cluster centroids. Survival in the TCGA cohort significantly associates with projected cluster membership with a cox proportional hazards p value < 0.014. F, The mutation status of each of the TCGA AML samples is shown, ordered and colored by their predicted cluster. A bold mutation name indicates a nominally significant association (p < 0.05) between the mutation and the clustering (Fisher’s exact). Similarly, bold and italic font indicates significance after multiple hypothesis testing correction (p < 0.001). Only the most recurrent mutations (n > 2) are shown here. rMLL = MLL rearrangement; Mito = Mitochondrial genes.
An SE is present at the RARA locus in a subset of AML samples. A, Metatrack plots of the H3K27Ac landscape at the RARA locus in all AML clusters and healthy monocytes and HSPCs. B, Correlation of RARA enhancer score with RARα mRNA levels (by RNA-seq). Spearman rho ~ .47; p < 0.0008. C, Three-dimensional interactions in the local region surrounding the RARA gene locus from promoter capture Hi-C data. H3K27Ac ChIP-seq from one AML patient (SU070) and CTCF ChIP-seq from monocytes (Encode) are shown for reference. The RARA SE is highlighted by a blue box. Interactions of the RARA promoter with the RARA SE are shown in red. Only unique interactions with a CHiCAGO score > 5 are shown for clarity. Region represents chr17:38455078–38579467. D, Analysis of RARα mRNA from a public data set (1) reveals a significantly higher level of RARα mRNA in a subset of MDS patients blast cells compared to non-malignant CD34+ (two tail T-test with Welch’s correction). Both cohorts are CD34+ cells sorted from bone marrow aspirates. E, H3K27ac ChIP-seq tracks for two AML patients with RARA among their top 100 enhancers (crimson), two MDS patients with RARA among their top 100 enhancers (red), and two non-malignant blast cell samples from healthy donors (gray). F, Expression of mRNA by RNA-seq (log2 TPM) across a set of primary AML patients, AML cell lines, and AML PDXs.
SY-1425 is a selective and potent RARα agonist. A, Biochemical assay plot of SY-1425 (tamibarotene) concentration (log nM) versus degree of association between co-activator and RAR protein from FRET assay expressed as % maximal ATRA signal. Data are for tamibarotene effects on RARα (red circles), RARβ (blue squares), and RARγ (green triangles). The horizontal dashed line indicates the tamibarotene Emax on RARα and the vertical dashed line the EC50 value. B, Cellular reporter plot of degree of association between co-activator and RAR protein from FRET assay expressed as % maximal ATRA signal (Emax) induced by SY-1425 (tamibarotene) as a function of concentration (log nM). Data are for SY-1425 on RARα (red circles) and RARγ (green triangles).
Comparison of SY-1425 response to RARA expression and enhancer score and the effect of an RARα antagonist. Correlations of enhancer, mRNA, and sensitivity to SY-1425 are color coded by cell line as indicated. A, Plot of RARα mRNA for AML cell lines versus SY-1425 anti-proliferative effect. B, Plot of RARA enhancer score for AML cell lines versus SY-1425 anti-proliferative effect. C, Plot of normalized enhancer versus expression. D, Cellular proliferation by ATPlite in RARA-high MV4–11 (red, circles), RARA-high OCI-AML3 (crimson, triangles), RARA-low OCI-M1 (blue, diamonds) and RARA-low Kasumi-1 (black, squares). Open shapes for BMS195614, a selective RARα antagonist, and closed for SY-1425. E, Anti-proliferative effect of SY-1425 (red circles) or idarubicin (black squares) with (open symbol) or without (closed symbol) 1μM BMS195614, a RARα selective antagonist, on the RARA-high cell line SigM5.
SY-1425 in vivo pharmacokinetics. A, Chemical structure of SY-1425 and plot of time versus SY-1425 concentration in plasma from mice given 3mg/kg SY-1425 (black circle) vs human SY-1425 levels (white diamond) 6mg/m2. Tables of PK properties for SY-1425 in mice are shown (top right) with comparator data from Kanai et al. Hem. Int. 2014. B, Chemical structure of ATRA with PK plot. 3mg/kg SY-1425 (black circle), 4mg/kg ATRA (white diamond), or human ATRA levels 45mg/m2. Tables of PK properties for ATRA are shown (bottom right) with comparator data (2).
SY-1425 survival and tissue effects on AML PDX models. A, RARA mRNA levels for the four non-APL AML PDX models assessed by RNA-seq with RARA-high indicated in red and RARA-low indicated in blue. B–C, Tumor burden in blood, bone marrow, and spleen was assessed by human CD45 positive cell percentage by FACS for (B) AM5512, and (C) AM8096. SY-1425 treated (6 mg/kg) animals are shown in red to indicate RARA-high status while vehicle treated animals are shown in gray. D, Survival plot for AM5512 PDX model with significant prolongation of survival (0.03 Mantel-Cox test). E, Survival plot for AM8096 PDX model with significant prolongation of survival (0.02 Mantel-Cox test). F–G, Tumor burden in blood, bone marrow, and spleen was assessed by human CD45 positive cell percentage by FACS for (F) AM7577, and (G) AM7440. SY-1425 treated (6 mg/kg) animals are shown in blue to indicate RARA-low status while vehicle treated animals are shown in gray. H, Survival plot for AM7577 PDX model with no significant prolongation of survival (0.3 Mantel-Cox test). I, Survival plot for AM7440 PDX model.
SY-1425 shows greater potency than ATRA in vitro and in vivo. A, Plot of EC50 values for SY-1425 versus ATRA for AML cell lines with equipotency (solid), three fold stronger SY-1425 (dotted) and ten-fold stronger SY-1425 (dashed) lines indicated. B–C, Mice in AM5512 RARA-high PDX model were treated with 3mg/kg BID for SY-1425 or 4mg/kg BID for ATRA. Tumor burden in (B) spleen and (C) bone marrow are shown. Vehicle is shown in black; ATRA treatment in blue; and SY-1425 in dark red for mice sacrificed during the course of the study and light red for mice assessed at the end of two months of treatment. SY-1425 shows significant reduction versus ATRA (two tail T-test with Welch’s correction). D, Plot of percent human CD45 positive in vehicle (black) and SY-1425 3mg/kg BID (red) for peripheral blood (closed circle) or bone marrow (open squares).
Induction of markers of myeloid differentiation in RARA-high AML cell lines upon SY-1425 treatment. A–B, Induction of mRNA expression measured by microarray in cell lines treated with vehicle or SY-1425 50nM for 24 hours for (A) CD11c and (B) CD11b. C, FACS measurement of percent positive in vehicle or SY-1425 50nM 72 hours for CD66. The APL cell line NB4 is in blue, RARA-high cell line MV4–11 in red, and RARA-low cell line OCI-M1 in gray. D, Percentage of CD38 high cells in RARA-high versus RARA-low ex vivo samples after 48 hours. Significance by two tail T-test with Welch’s correction.
Comparison of SY-1425 expression and molecular response to APL. A, Canonical pathways (MSigDB C2.CP) and Hallmark (from MSigDB) gene sets enriched by GSEA in SY-1425 response in the RARA-high cell lines (Supplementary Table S6). Top 10 gene sets by FDR are shown. Positive normalized enrichment scores (NES) represent gene sets enriched in genes that are higher upon SY-1425 treatment, negative NES values represent gene sets enriched in genes that are lower upon SY-1425 treatment. B, Number of genes bound by RARα in NB-4 cell line in common with AML cell lines. Genes with RARα ChIP-seq peak (top 4000 peaks per cell line) in the gene regulatory region (promoter+1Mb up to neighboring gene’s promoter) are considered bound by RARα. NB4 bar indicates total number of genes bound in NB4. C, Heatmap of spearman correlation of super-enhancer (SE) response to SY-1425 in each cell line. Only SEs differential (FDR<0.05) in at least one cell line are included. Clustering is by Ward method. D, SE response to SY-1425 in NB-4 and RARA-high cell lines. Only SEs differential in NB-4 (FDR<0.05) (n=1967) are shown.
Supplementary Figure S13 SY-1425 expression and molecular response. A, Volcano plots of H3K27ac response to SY-1425 for the union of H3K27ac peaks in the vehicle and treatment conditions by cell line. B, Percentage of H3K27ac peaks in each cell line that are higher upon treatment with SY-1425. Red bars indicate RARA-high lines and blue bars indicate RARA-low lines. C, Percentage of H3K27ac peaks in each set that are up-regulated by SY-1425 (FDR<0.05 and log2 fold change >1) in each cell line. RARα bound peaks overlap a RARα ChIP-seq peak (top 4000 per cell line), and background peaks are drawn from peaks with similar H3K27ac level to RARα bound peaks. Error bars are from bootstrapping. Numbers in the RARα bound bars indicate the number of H3K27ac peaks up-regulated and bound by RARα in that cell line.
Supplementary Table S1: Sample mutations
Supplementary Table S2: RNA-seq signature and per-cluster weights
Supplementary Table S3: Median survival in TCGA data by predicted cluster
Supplementary Table S4: Enhancer, mRNA, and SY-1425 EC50 in AML cell lines
Supplementary Table S5: GSEA of transcriptional response to SY-1425 in RARA-high cell lines in perturbation gene sets
Supplementary Table S6: GSEA of transcriptional response to SY-1425 in RARA-high cell lines in hallmark and pathways gene sets
Supplementary Table S7: GREAT analysis of genes near H3K27ac peaks up-regulated by SY-1425 in RARA-high cell lines in perturbation gene sets