Abstract
The Cas9 endonuclease can be programmed by guide RNA to introduce sequence-specific breaks in genomic DNA. Thus, Cas9-based approaches present a range of novel options for genome manipulation and precision editing. African trypanosomes are parasites that cause lethal human and animal diseases. They also serve as models for studies on eukaryotic biology, including ‘divergent’ biology. Genome modification, exploiting the native homologous recombination machinery, has been important for studies on trypanosomes but often requires multiple rounds of transfection using selectable markers that integrate at low efficiency. We report a system for delivering tetracycline inducible Cas9 and guide RNA to Trypanosoma brucei. In these cells, targeted DNA cleavage and gene disruption can be achieved at close to 100% efficiency without further selection. Disruption of aquaglyceroporin (AQP2) or amino acid transporter genes confers resistance to the clinical drugs pentamidine or eflornithine, respectively, providing simple and robust assays for editing efficiency. We also use the new system for homology-directed, precision base editing; a single-stranded oligodeoxyribonucleotide repair template was delivered to introduce a single AQP2 - T791G/L264R mutation in this case. The technology we describe now enables a range of novel programmed genome-editing approaches in T. brucei that would benefit from temporal control, high-efficiency and precision.
Introduction
The African trypanosomes of the Trypanosoma brucei group are transmitted by tsetse flies and cause lethal diseases in humans and livestock, known as sleeping sickness and nagana, respectively. Research on these protozoan parasites, and related parasitic trypanosomatids, often focuses on virulence mechanisms and druggable biology1. However, their divergent position within the eukaryotic phylogeny, and the experimental tractability of T. brucei in particular, means they are also of interest as model Excavates for studies on various aspects of eukaryotic biology, including novel biology not seen in other eukaryotic model systems. Unusual features include an almost complete lack of introns, widespread polycistronic transcription and trans-splicing of every mRNA2.
The ability to manipulate parasite genomes has had a huge impact on this field of research3. In T. brucei, DNA constructs, typically introduced by electroporation, can be used for gene knockout, gene knockdown exploiting the native RNA interference (RNAi) machinery, tagging native alleles or inducible expression of recombinant products; robust inducible expression4 has been of particular value. Sequences towards the ends of the linearised DNA constructs serve as templates for homologous recombination reactions. Thus, targeted integration depends upon the native homologous recombination machinery; microhomology-mediated end-joining (MMEJ)5 and single-strand annealing6 also operate in T. brucei, while non-homologous end-joining appears to be absent. Targeted integration occurs at low frequency, however (typically <1 × 10−5 electroporated cells), meaning that antibiotic selectable marker genes must be integrated at a modified site followed by selection for the desired recombinant cells. Since the T. brucei core genome is diploid7, gene disruption approaches typically require two rounds of electroporation and selection.
Chromosomal site-specific DNA double strand breaks (DSBs) can increase recombination efficiency in T. brucei, as demonstrated following inducible expression of the I-SceI meganuclease8. The same endonuclease can efficiently trigger inter-chromosomal homologous recombination5 or antigenic variation when breaks are targeted close to the expressed Variant Surface Glycoprotein gene9,10. For this approach, however, a specific 18-bp I-SceI cleavage site must first be engineered at the desired target site, which limits its versatility. Cas9 is an RNA-guided DNA endonuclease that can be programmed in a sequence-specific manner using a short (17–20 b) synthetic guide RNA sequence; specific targeting involves RNA:DNA base-pairing. The only additional requirement is that this ‘protospacer’ sequence must immediately precede a protospacer adjacent motif (PAM; at the DNA target site but not to be included in the gRNA). The PAM recognized by the most widely used Cas9 is 5′-NGG, for the type II system from Streptococcus pyogens11. It is this tremendous versatility that has made Cas9 repurposing so popular for genome manipulation and editing12. Cas9 systems function naturally as bacterial adaptive immune systems. In the bacterial cells, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), derived from invading phage DNA, produce CRISPR RNA (crRNA) which, together with a trans-activating crRNA (tracrRNA) program Cas9 (CRISPR associated protein 9) to cut both the RNA-complementary and non-complementary strands, producing DSBs in the target DNA, 3 bp upstream of the PAM. The crRNA and tracrRNA can be fused to generate a chimeric single guide RNA (sgRNA)11, such that a two-component system requires only Cas9 and an sgRNA for programmed editing in a heterologous system.
The application of Cas9-based genome editing to trypanosomatids has progressed rapidly in the last few years. For example, SpCas9 has been used to edit genes in T. brucei with a cloning-free approach, allowing for one-step gene knockout, but still requiring integration of antibiotic selectable markers at the target sites13. Another recent report describes the delivery of in vitro assembled ribonucleoproteins containing the smaller Staphylococcus aureus Cas9 for editing in trypanosomatid parasites; T. brucei, Trypanosoma cruzi and Leishmania major. In this case, a GFP-reporter was disrupted in >50% of targeted T. brucei cells14; the PAM recognized by SaCas9 is 5′-NNGRRT, where R represents A or G. SpCas9 has also been used to edit genes in the other trypanosomatids15, including those for which loss-of-function studies have previously been hampered, notably due to the absence of a native RNAi machinery16. Thus, these advances will have a major impact on tractability in terms of genetic manipulation in T. cruzi17–19, and Leishmania spp13,20–23.
Cas9-based approaches will also enable a range of new approaches in T. brucei. To date, however, an inducible Cas9 editing system has not yet been reported for any trypanosomatid. In T. brucei, double-allele editing without selectable markers has not yet been reported, double-allele editing efficiencies have not been determined and precision base editing is not readily achievable. We sought to address three specific issues to advance Cas9-based editing tools and approaches for T. brucei. Since current systems in protozoan parasites often use constitutive Cas9 expression, which can be toxic and continually risks off-target editing18,24, we first wanted to establish an inducible system. Second, we wanted to develop a system that allowed for DSB formation (and repair) in close to 100% of cells in the population in a week or less. Third, we wanted to use the system for template-directed precision base editing. By manipulating genes associated with resistance to clinical drugs, we report the successful attainment of all three goals.
Results
A system for inducible Cas9 expression and sgRNA expression in T. brucei
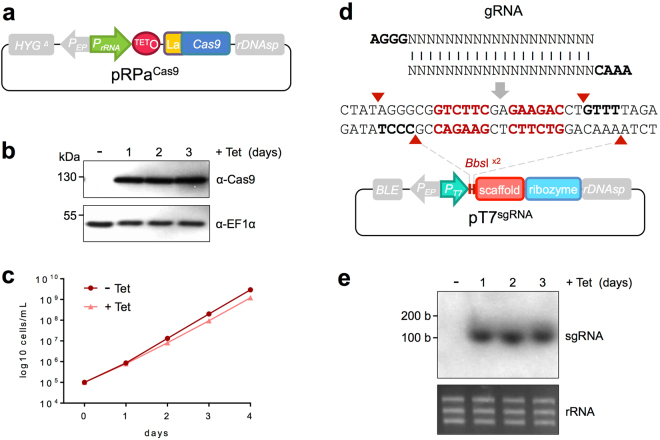
We assembled an expression construct with a tetracycline (Tet)-inducible ribosomal RNA (rRNA) promoter to drive SpCas9 expression in T. brucei. These pRPa constructs can be used in combination with T. brucei bloodstream-form 2T1 strains25, which express the Tet-repressor and have a partial hygromycin (HYG) drug-selectable marker gene at a non-transcribed rDNA spacer locus on chromosome 2; the expression construct contains a complementing portion of the HYG gene26. This allows for selection of hygromycin-resistant clones with expression cassettes reproducibly integrated at the same locus, a locus that supports robust and uniform Tet-inducible expression26. A synthetic SpCas9 gene with a La nuclear localisation signal27 at the N-terminus was codon-optimised for expression in T. brucei28. This gene was cloned in the pRPa expression construct and the resulting pRPaCas9 plasmid (Fig. 1a) was linearised and introduced into 2T1T7 cells; these cells also express the phage T7 polymerase to drive sgRNA expression (see below). Analysis of the resulting 2T1T7-Cas9 clones revealed tightly regulated and robust Cas9 expression (Fig. 1b, Supplementary Fig. 1). We observed a moderate reduction in growth rate associated with Cas9 induction (Fig. 1c), which could reflect off-target editing in the absence of sgRNA24.
Figure 1.
An inducible Cas9 system. (a) Schematic map of the pRPaCas9 construct. PrRNA, rRNA promoter; TETO, tetracycline operator; La, nuclear localisation signal. Other components are from the pRPa parent vector (see the text for more details). (b) The protein blots show tightly regulated and inducible Cas9 expression (predicted mass: 159.4 kDa). The EF1α panel provides a loading control. Multiple independent clones yielded similar results. (c) Cell growth with (+Tet) and without (−Tet) Cas9 induction. Error bars (SD) from triplicate counts are obscured by the data symbols. A second independent clone gave similar results. (d) Schematic map of the pT7sgRNA construct showing the gRNA cloning strategy using tandem BbsI sites. The sgRNA scaffold and HDV ribozyme are also shown. PT7, phage T7 promoter. Other components are from the pRPc6MYCn parent vector. (e) The RNA blot shows expression of the predicted 96 b sgRNAAQP2 that is dependent upon Cas9 expression. The rRNA panel provides a loading control. A second independent clone yielded similar results.
For expression of the sgRNA, a phage T7 promoter, a cloning site, the sgRNA scaffold and a hepatitis delta virus (HDV) ribozyme29 were synthesised in tandem (Fig. 1d). The cloning site comprises dual BbsI restriction enzyme cleavage sites, which allows for high-efficiency cloning of annealed oligonucleotide pairs (AGGG[N]20/AAAC[N]20, Fig. 1d) without introducing additional unnecessary flanking sequences. The HDV ribozyme is a self-cleaving RNA that liberates the upstream RNA. Thus, the ribozyme generates the 3′-end of the sgRNA. This synthetic cassette was used to generate pT7sgRNA that integrates at a second non-transcribed rDNA spacer locus (Fig. 1d); there are estimated to be 10–20 rDNA spacer loci in the T. brucei genome7.
We selected the aquaglyceroporin gene, AQP2, (Tb927.10.14170) as a suitable target for editing since this gene is involved in drug resistance and parasites are resistant to pentamidine when AQP2 is defective30. The full complement of >9,000 T. brucei protein-coding genes comprises 51% G + C content7 so suitable PAMs can be found approximately once every 8 bp in these sequences. We identified potential PAMs in AQP2, selected a suitable 20 b AQP2-specific gRNA sequence, annealed the cognate oligonucleotide pair and ligated the product to BbsI-digested pT7sgRNA. The resulting construct was linearised and introduced into 2T1T7-Cas9 cells. We analysed two independent Cas9/sgRNAAQP2 clones and observed robust and reproducible expression of sgRNA of the expected size (96 b), which was dependent upon Cas9 expression (Fig. 1e, Supplementary Fig. 1). This suggests that sgRNA is unstable in the absence of Cas9. Thus, sgRNA is efficiently processed by the HDV ribozyme and is specifically stabilised by Cas9, providing evidence for physical interaction of these two components and assembly of Cas9 ribonucleoproteins in T. brucei.
Tightly regulated and high-efficiency AQP2 editing
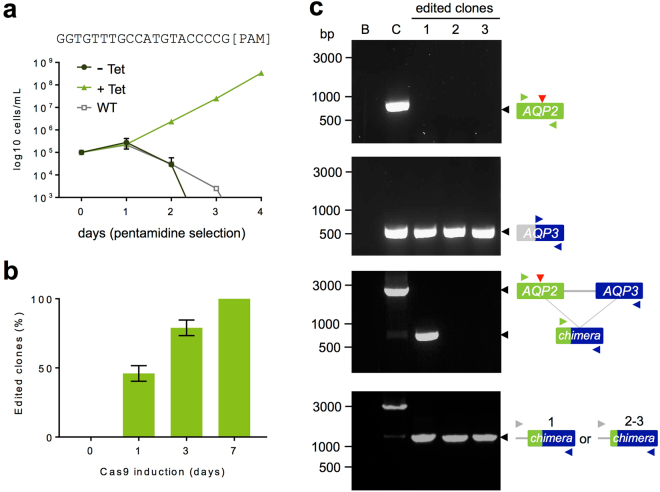
The T. brucei genome is diploid, meaning that Cas9-based editing for the vast majority of genes will involve targeting two identical sequences, ideally simultaneously in many cases. When AQP2 function is lost, the effective concentration of pentamidine to reduce growth by 50%, or the EC50, is approximately 15-fold greater than for wild-type cells30. Thus, pentamidine can be used to provide a robust phenotypic read-out and for quantitative analysis of bi-allelic editing. Using three independent Cas9/sgRNAAQP2 clones, we pre-induced Cas9 programmed to target AQP2 for three days, and then selected with pentamidine. The growth profiles indicated an absence of drug-resistance without induction and robust drug-resistance with induction (Fig. 2a).
Figure 2.
High-efficiency Cas9-based editing of the AQP2 gene. (a) Cumulative cell growth in the presence of 6 nM pentamidine (estimated 2 × EC50) following 3-days with (+Tet) or without (−Tet) Cas9 induced editing of AQP2. Three independent Cas9/sgRNAAQP2 clones and wild-type (WT) cells were analysed. Error bars, SD. The gRNA sequence is shown above the plot. (b) A cloning assay reveals efficient Cas9 editing. Cas9 was induced as indicated followed by sub-cloning in the absence of Tet. The sub-clones were then transferred to medium with or without 6 nM pentamidine and considered ‘edited’ if drug-resistant; twelve sub-clones were analysed from each of two independent Cas9/sgRNAAQP2 clones (n = 24 sub-clones) for each time-point. (c) The PCR assays reveal AQP2 editing in three independent drug-resistant clones. B, blank – no genomic DNA; C, control – wild-type genomic DNA. The AQP3 gene is adjacent to AQP2. The vertical (red) arrowheads indicate the Cas9 target site in AQP2. The horizontal (green, blue and grey) arrowheads indicate the primers used for the PCR-assays. Predicted wild-type AQP2 fragment, 724 bp; predicted AQP3 fragment, 508 bp; predicted wild-type AQP2-3 fragments in bottom two panels, 2,440 and 3,200 bp, respectively.
Encouraged by these results, using two independent Cas9/sgRNAAQP2 clones, we induced editing in the absence of pentamidine selection and sub-cloned the population either prior to induction, or one, three and seven days later. Under these conditions, un-edited, drug-sensitive cells remain viable and edited cells have no selective advantage. Twelve sub-clones from each clone and at each time-point were expanded under conditions where Cas9 was no longer induced and then tested for pentamidine resistance. This revealed remarkably tightly regulated editing (Fig. 2b). The absence of pentamidine-resistant sub-clones (0/24) from the uninduced population provided strong evidence for efficient temporal control, and this ‘baseline’ data allowed us to accurately assess editing during the induction time-course. This revealed efficiently induced editing, which increased from ~45% resistant clones after 1-day to ~80% after 3-days and reached the maximum 100% (24/24 sub-clones) after 7-days (Fig. 2b). This temporal response reflects the kinetics of Cas9 induction4, the Cas9 target search31 and DNA repair in T. brucei32, and may also be impacted by multiple rounds of single-allele break and error-free repair by homologous recombination. A series of PCR-based assays were used to assess the AQP2-3 locus in three independent resistant clones (Fig. 2c, Supplementary Fig. 1). AQP2 had indeed been disrupted (Fig. 2c, top panel) and replaced by AQP2-3 chimeras in all three clones (Fig. 2c, lower panels), similar to what has been seen previously in drug-resistant isolates from relapsed patients33,34. Cloning and sequencing of the products revealed recombination within a block of identity from 168–222 nt in AQP2 in clone 1, meaning that from 223 nt into the CDS is AQP3 sequence in the chimera, and recombination within a block of identity from 1–91 nt in AQP2 in clones 2–3, meaning that from 92 nt into the CDS is AQP3 sequence in these chimeras. The chimeras likely arise by single-strand annealing, whereby single-stranded regions extend to adjacent repeated sequences and complementary strands anneal, leading to repair and the loss of one repeat35; the PCR-assays indicate loss of ~1.8 kbp in each case (Fig. 2c, lower panels), which is the size of the semi-repetitive AQP2-3 region. Microhomology-mediated end-joining is more likely to be involved in repair at loci that lack extensive blocks of adjacent homology5. Thus, our inducible Cas9 editing system is tightly regulated and achieves temporally controlled and remarkably efficient bi-allelic editing.
Tightly regulated and high-efficiency AAT6 editing
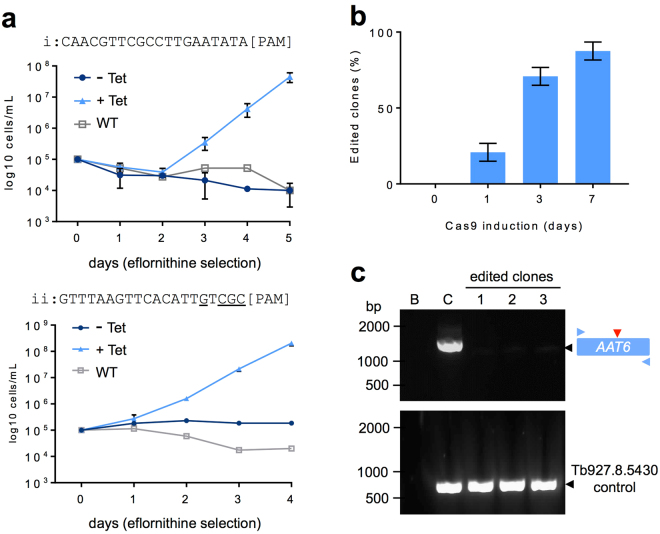
To determine whether the efficiency of our system could be extended to another native locus, and to further test sgRNA design for T. brucei, we selected a second gene, on a different chromosome, for analysis. Again exploiting a known drug-resistance mechanism, we selected the amino acid transporter, or AAT6 gene (Tb927.8.5450). Trypanosomes become resistant to the amino acid analogue, eflornithine when AAT6 is defective and the eflornithine EC50 is >40-fold greater than for wild-type cells in this case36. Thus, eflornithine can also be used to provide a robust phenotypic read-out for bi-allelic editing. Since a PAM-proximal GC-content of >50% (at least 4 of 6 bases) yields optimal efficiency gRNAs in other cell-types37, we cloned two 20 b AAT6-specific gRNA sequences in pT7sgRNA. One gRNA (i) has a PAM-adjacent GC content of 0/6 and the other (ii) has a PAM-adjacent GC content of 4/6; both had an overall GC-content of 40% (Fig. 3a). The resulting constructs were introduced into 2T1T7-Cas9 cells as above. Using two independent Cas9/sgRNAAAT6 clones for each sgRNA, we induced Cas9 programmed to target AAT6 for three days, followed by selection with eflornithine. In both cases, the growth profiles indicated an absence of resistance without induction and robust resistance with induction (Fig. 3a). We observed a striking difference between sgRNA-i and sgRNA-ii, however. As predicted by previous studies37, the sgRNA with the optimised PAM-proximal GC-content was associated with more efficient editing; with sgRNA-ii yielding drug-resistant cells more rapidly than sgRNA-i during selection (Fig. 3a).
Figure 3.
High efficiency Cas9-based editing of the AAT6 gene. (a) Cumulative cell growth in the presence of 270 µM eflornithine (estimated 10 × EC50) following 3-days with (+Tet) or without (−Tet) Cas9 induced editing of AAT6. Two independent Cas9/sgRNAAAT6 clones and wild-type (WT) cells were analysed for each gRNA (i and ii). Error bars, SD. The gRNA sequences are shown above the plot with PAM-adjacent G/C residues underlined (see text for details). As can be seen, eflornithine is a cytostatic rather than cytocidal drug. (b) A cloning assay reveals efficient Cas9 editing by gRNA-ii. Cas9 was induced as indicated followed by sub-cloning in the absence of Tet. The sub-clones were then transferred to medium with or without 270 µM eflornithine and considered ‘edited’ if drug-resistant. Twelve sub-clones were analysed from each of two independent clones (n = 24 sub-clones) for each time-point. (c) The PCR assays reveal AAT6 editing in three independent drug-resistant clones. B, blank – no genomic DNA; C, control – wild-type genomic DNA. The Tb927.8.5430 gene is close to AAT6 and serves as an unedited control. The vertical (red) arrowhead indicates the Cas9 target site. The horizontal (blue) arrowheads indicate the primers used for the AAT6 PCR-assay. Predicted wild-type AAT6 fragment, 1,467 bp; predicted wild-type Tb927.8.5430 fragment, 648 bp.
As above, and using the optimised sgRNA (AAT6-ii) we next induced editing in the absence of eflornithine and sub-cloned the populations. The proportions of eflornithine-resistant sub-clones revealed remarkably efficient editing, reaching ~90% after 7-days (Fig. 3b). PCR-based assays confirmed disruption of the AAT6 gene in three independent resistant clones (Fig. 3c, Supplementary Fig. 1). The process, from gRNA design to validation of edited clones can be performed in approximately three weeks. Thus, our tightly regulated inducible Cas9 editing system achieves rapid, high-efficiency, bi-allelic editing at native chromosomal loci.
Precise and ‘scar-less’ single base editing
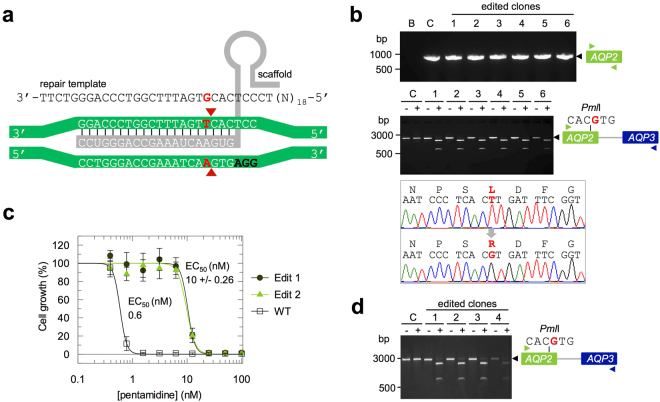
The analyses above demonstrate robust and efficient inducible editing. The edits described are site-specific and are sufficiently efficient that neither antibiotic selectable markers, nor exhaustive screening, are required to isolate edited clones. However, these edits typically result in deletions that may also affect neighbouring sequences or genes. We next sought to use this system for precision editing by co-opting the native homologous recombination machinery. This requires the introduction of a DSB adjacent to the edit site and the introduction of a DNA template containing the desired site-specific edit (Fig. 4a). We once again chose AQP2 as the target gene for this experiment. AQP2 is an atypical aquaglyceroporin that lacks many of the conserved pore-lining residues, including the ‘aromatic arginine’ motif30. A leucine residue is present at this position and a single L264R change generates a mutant AQP2 that fails to reverse pentamidine resistance when over-expressed in aqp2-null trypanosomes38. Cas9 editing technology now presents opportunities to test the behaviour of such mutant genes in their native context. Short homology flanks are sufficient for recombination in T. brucei13 and single-stranded oligonucleotides can be used to edit trypanosomatid14,22 and metazoan genomes39. We, therefore, used a new gRNA, cloned in the pT7sgRNA construct, that targets a sequence adjacent to the L264 codon (Fig. 4a). In this case, although the overall GC-content is 55%, the PAM-adjacent GC content of the new gRNA is 3/6 and may be suboptimal (Fig. 4a, see above).
Figure 4.
Precision base editing with Cas9. (a) The schematic illustrates the AQP2-editing strategy (one allele shown). The genomic DNA sequence is on a green background, the PAM is in bold black text, the gRNA is on a grey background and the arrowheads indicate the expected Cas9-induced break sites. The single-stranded repair template is centred on the editing site and shows the T-G edit in red letters. (b) Analysis of sub-clones from an edited and pentamidine-selected population. The PCR assays are as described in Fig. 2c. B, blank – no genomic DNA; C, control – wild-type genomic DNA. The AQP2-3 PCR products were also digested with PmlI (lower panel) and sequencing confirmed that all six clones had the templated T791G/L264R edit (one example shown, lower trace). (c) Dose-response curves for pentamidine. Both edited clones analysed display a drug-resistant phenotype. WT, wild-type cells. (d) Analysis of the four pentamidine-resistant sub-clones among 48 sub-clones from an edited population. The PCR assay and PmlI digest are as described in b.
We used this new ‘AQP2.2′ gRNA to assemble Cas9/sgRNAAQP2.2 clones. We next delivered the short (48 b) single-stranded oligonucleotide repair-template (Fig. 4a) immediately followed by induction of Cas9-based cleavage. The repair template carries the single edited base-change (Fig. 4a, red) and was delivered by electroporation while Cas9 was induced for only 24 h in this case, to maximise template-directed repair. Note that, despite the base-change, the edited alleles may remain substrates for re-cleavage, albeit at reduced efficiency.
We expected a number of possible outcomes within the resulting population of cells; no editing, non-templated deletions (as described above) or the desired edit. Un-edited cells remain pentamidine-sensitive and these were selected against by growth in pentamidine for three days, immediately following the 24 h induction period. The resulting pentamidine-resistant population was sub-cloned and six clones were subjected to an AQP2 PCR assay. In this case, all six clones analysed yielded an AQP2 PCR-product (Fig. 4b, upper panel, Supplementary Fig. 1), in contrast to the clones above that underwent non-templated repair (Fig. 2c). Using a second PCR assay, all six clones also yielded an AQP2-3 PCR-product that contained a new PmlI restriction site introduced by the single-base edit (Fig. 4b, middle panel, Supplementary Fig. 1). Sequencing of these products confirmed the desired T791G/L264R precision edit in every clone (Fig. 4b, lower panel; Supplementary Fig. 2). This suggests that the electroporated short single-stranded oligonucleotide is used remarkably efficiently as a template and that this particular homology-directed repair pathway is favoured over the error-prone single-strand annealing pathway. To assess the impact of expression of the AQP2 - L264R mutant in the native context, we determined the EC50 for pentamidine. This revealed robust resistance and an EC50 that was increased approximately 17-fold relative to the wild-type control (Fig. 4c).
The remarkable efficiency of precision base editing above suggested that we may be able to isolate precision edited clones without selection to enrich for the edited cells. We, therefore, repeated the experiment above but, instead of selecting against unedited cells by growth in pentamidine, we simply sub-cloned the population immediately following the 24 h induction period. Forty-eight clones were expanded for several days without further Cas9-induction and were then tested for pentamidine-resistance. We found four clones (~8%) to be resistant and, again using a PCR-assay and diagnostic digest (Fig. 4b), found that all four were precision-edited (Fig. 4d, Supplementary Fig. 1). These results suggest that, using the Cas9-editing system described here, it will be possible to isolate clones with precision-edits at different loci, following screening of a readily manageable number of clones.
Discussion
The versatility and accessibility of tools for genome editing have improved rapidly in recent years and Cas9-based approaches are central to this transition. Substantial progress has been made in developing these systems in trypanosomatids but an inducible Cas9 editing system has not yet been reported. Additionally, for T. brucei, double-allele editing has not been achieved without selectable markers nor have efficiencies been determined. Given these limitations, precision base editing is not readily achievable. To advance Cas9-based editing tools and approaches for T. brucei, we have established a highly efficient inducible system and have used the system for templated, homology-directed, precision base editing.
The system we describe incorporates several features that improve convenience and editing efficiency. In terms of convenience, the current strategy involves annealing a pair of oligonucleotides and ligating the product to the BbsI-digested pT7sgRNA construct prior to transfection. This cloning step is highly efficient, but sgRNA delivery can also be achieved in a cloning-free format. Such a cloning-free format, involving transient transfection of the sgRNA or an sgRNA-encoding PCR-product, may be suitable for strategies that do not require maximal-efficiency editing, such as when integrated antibiotic resistance markers are used to select for correctly edited cells13. In terms of the editing efficiency of our system, several features likely contribute; first, Cas9 was codon-optimised to increase expression in T. brucei28. Second, Cas9 and the sgRNA are transcribed from strong rDNA and T7 promoters, respectively. Third, the T7 polymerase, BbsI-cloning approach and ribozyme all help to produce sgRNAs with minimal superfluous sequence, likely enhancing assembly with Cas9. In addition, tightly regulated temporal control of Cas9 expression increases the versatility of the system, also minimising off-target edits and allowing for phenotypic analysis of edited cells without potential interference from continuously expressed Cas9. Finally, and as in metazoan cells37, our findings support the view that gRNA sequences achieve optimal efficiency cleavage in T. brucei when the PAM-adjacent GC-content is >50%.
A major source of off-target edits may involve the non-specific association of Cas9 with native RNA. Indeed, expression of Cas9 in the absence of sgRNA has been reported to be toxic in T. cruzi18 and Toxoplasma gondii, and a “decoy” sgRNA has been used in Toxoplasma to reduce these effects24. The current system should minimise such problems in T. brucei since we constitutively transcribe the sgRNA and induce a tightly regulated Cas9 gene, which stabilises the sgRNA. Thus, cells are not exposed to Cas9 in the absence of sgRNA. Titration of Cas9-dosage can dramatically increase cleavage specificity and reduce off-target effects in metazoan cells40 and the inducible system now presents an opportunity to titrate Cas9 expression in T. brucei simply by reducing the tetracycline concentration in the growth medium4 or by inducing Cas9 for only a short period of time. Temporally constrained exposure to Cas9 is also important for precision base editing since precision-edited sequences may still be potential substrates for re-cleavage37,40–42. It will be worthwhile to test other versions of Cas9 in T. brucei. For example, HypaCas9 (SpCas9: N692A/M694A/Q695A/H698A) displays reduced off-target editing43 while xCas9 variants display broader PAM compatibility44.
Further optimisation of specific editing and minimising off-target edits can also be achieved by modifying the gRNA. In metazoan cells, gRNA sequences can be truncated by 2–3 nt to reduce off-target cleavage without substantially reducing on-target efficiency45 and these gRNAs can be further optimised to improve specific activity37,40,41. On the other hand, trypanosomatid genomes are relatively small at ~30 Mbp, likely presenting relatively few potential off-target sites. In addition, homologous recombination is the dominant repair pathway in T. brucei5 such that low-frequency off-target edits in one allele will typically be reversed, since allelic templated, homology-directed repair will be favoured over error-prone repair by end-joining. In circumstances where off-target edits must be avoided, genome sequencing and profiling of potential off-target sites will remain an option.
Established tools for the genetic dissection of T. brucei already present a range of options. Cas9-based approaches present alternative strategies and, perhaps more importantly, present completely new opportunities, for increased throughput and for marker-free and precision editing, in particular. In terms of alternative strategies, RNAi is currently the favoured approach for conditional loss-of-function studies in T. brucei, particularly for many likely essential genes. Similarly, native, single-allele tagging for protein localisation studies is readily achievable using selectable marker based constructs in T. brucei46 and can now be considered alongside (double-allele) Cas9-based editing options. Clearly, editing efficiency need not be high if selectable markers are to be used to enrich for cells with a desired, non-lethal edit. When an edit is lethal on the other hand, the desired editing will fail to yield long-term viable recombinant cells. In these circumstances, editing that is both inducible and close to 100% efficiency, will be a major advantage since it will facilitate the study of phenotypes associated with lethal edits that precede cell death. High efficiency will also be important for other marker-free or high-throughput approaches. In these cases, high validation rates for a desired edit will substantially reduce the number of clones that must be screened and will improve coverage across large numbers of editing sites, respectively.
Key advantages of precision editing without selectable markers are that the (double-allele) edit is generated in one step in a native context, retaining native flanking sequences that may control expression level or timing during the cell cycle or the life cycle. A recombinant AQP2 - L264R mutant was previously ectopically overexpressed in an aqp2 null background, for example38. The pentamidine resistance phenotype associated with AQP2 - L264R expression is confirmed here but the prior ectopic expression approach required three genetic manipulation steps and was also associated with risks of potential artefacts associated with non-physiological expression. Thus, the current editing approach adds an important new ‘one-step, precision’ option to the genetic manipulation ‘toolbox’ for T. brucei. The efficiency reported here indicates that it is now feasible to obtain cells with edited loci without selection, by screening manageable numbers of clones.
The Cas9-based editing system we describe for T. brucei incorporates key features, including robust, temporal control of Cas9 and sgRNA expression. We show that null-clones (both alleles disrupted) can be generated at exceptionally high frequency without selection. Future applications will likely extend beyond precision edits, deletions and insertions to high-throughput approaches and the repurposing of nuclease-dead Cas9 for targeting other functionalities to specific genomic loci12.
Materials and Methods
T. brucei strains and growth conditions
Bloodstream form T. brucei, Lister 427 (MITat 1.2), clone 221a cells, 2T1 and 2T1T7 cells25 and their derivatives were grown in HMI-11 medium. Cumulative growth rates were monitored by splitting to 1 × 104 cells/ml and by counting daily. Tetracycline (Sigma) was applied at 1 µg/mL. Pentamidine isethionate was from Sigma and eflornithine was from Sanofi Aventis.
T. brucei genetic manipulation and plasmid construction
T. brucei were genetically manipulated using electroporation as described25. To generate the pRPaCas9 construct, the SpCas9 sequence was codon optimised and synthesised (Genscript) with a La nuclear localisation signal27 at the N-terminus. The synthetic Cas9 cassette, digested with HindIII/BglII, was ligated to pRPac6MYCn 26 [Addgene ID:69242] digested with HindIII/BamHI. The resulting pRPaCas9 construct was linearised with AscI prior to electroporation; 2T1T7-Cas9 parasites were selected with hygromycin at 2.5 µg/mL and checked for sensitivity to puromycin to indicate correct construct integration. To generate the pT7sgRNA construct, the BLA resistance cassette in pRPc6MYCn 26 [Addgene ID:69243] was replaced by a BLE cassette following NgoMIV/BstZ17I digestion. A synthetic sequence comprising a T7 promoter, tandem BbsI sites, the sgRNA scaffold and an HDV ribozyme was then used to replace the expression cassette from pRPc6MYCn, following NheI/ApaI digestion. Potential gRNA sequences were identified either manually or using the LeishGEdit.net online tool13, and then selected based on PAM-adjacent GC-content37. The gRNA oligonucleotide pairs were FwgAQP2 (AGGGGGTGTTTGCCATGTACCCCG) and RvgAQP2 (AAACCGGGGTACATGGCAAACACC); FwgAAT6i (AGGGGTTTAAGTTCACATTGTCGC) and RvgAAT6i (AAACGCGACAATGTGAACTTAAAC); FwgAAT6ii (AGGGCAACGTTCGCCTTGAATATA) and RvgAAT6ii (AAACTATATTCAAGGCGAACGTTG) and, for the ‘AQP2.2′ precision edit, FwgAQP2c (AGGGCCTGGGACCGAAATCAAGTG) and RvgAQP2c (AAACCACTTGATTTCGGTCCCAGG); the overhanging ends that facilitate cloning are in bold. Oligonucleotide pairs were annealed by heating to 70 °C for 3 min followed by slow cooling. Annealed pairs were ligated to BbsI-digested pT7sgRNA and constructs were confirmed by sequencing. pT7sgRNA constructs containing specific gRNA sequences were linearised with NotI prior to electroporation and parasites were selected with phleomycin at 2 µg/mL. 40 µg of single-stranded oligonucleotide (GTCTCCCCTTGCGATGAATCCCTCACGTGATTTCGGTCCCAGGGTCTT) was used as a repair template, which was resuspended in 10 μL of H2O and delivered by electroporation as described above; the edited base is in bold.
DNA analysis
DNA was extracted from T. brucei using a DNeasy blood and tissue Kit (Qiagen). The primer pairs used for the PCR assays were FwAQP2only (GAGCGGTGGGATGCAGATGTAC) and RvAQP2only (CCCCGAGAAGGATCGCACCG), FwAQP3only (GTGTGGTCGCCACGGTTATC) and RvAQP3only (GCGTAACCCGTTGAGTAACCG), FwAAT6only (ATGAGAGAGCCGATACAAACTTCAAC) and RvAAT6only (TCAGAGTTCAGCAATGACGCTG), Fw5430ATG (ATGGGCAACAACGGAAGTAGC) and Rv5430STOP (CTATGCCGCCAGTCGAACAC). The FwAQP2only or Fw14180 (CCAAAATCAGCGGGTTCACTGA) primers were combined with RvAQP3only for the AQP2-3 PCR assays (lower panels in Fig. 2c).
RNA analysis
For northern blotting, RNA was extracted from T. brucei using an RNeasy Mini Kit (Qiagen). 1.5 µg of RNA was resolved on formaldehyde agarose gels in MOPS buffer and transferred to nylon membranes by capillary blotting. A digoxigenin (DIG)-labelled riboprobe was hybridized to the membrane and the transcript was detected using an anti-DIG antibody (Roche) with a chemiluminescent CDP-star substrate (Roche). rRNA stained with ethidium-bromide was used as loading control.
Protein analysis
Total T. brucei extracts equivalent to 1 × 106 cells were separated on SDS-polyacrylamide gels and subjected to western blotting analysis according to the manufacturers’ instructions (BioRad). Blots were blocked in 5% milk in PBS-T and washes were performed in PBS-T (0.05% Tween). Blots were then cut in two, and the upper part (above 70KDa according to the pre-stained protein ladder) was probed with 1/1000 α-Cas9 primary antibody (Abcam) and 1/5000 α-mouse secondary antibody (Bio-Rad). As a loading control, the lower part of each blot was incubated with 1/20000 α-EF1α (Merck Millipore) and 1/5000 α-mouse secondary antibody (Bio-Rad).
Drug sensitivity analysis
EC50 analysis was performed using the AlamarBlue method as described47; drug exposure was for 72 h and incubation with resazurin (Sigma-Aldrich) was for 5–6 h. Plates were read on an Infinite 200 Pro plate-reader (Tecan).
Data Availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank M. Pawlowic for comments on the draft manuscript. This work was funded by a Wellcome Trust Senior Investigator Award to D.H. (100320/Z/12/Z) and a Wellcome Trust Centre Award (203134/Z/16/Z).
Author Contributions
E.R. performed the experiments and prepared Figures 1–4. E.R. and D.H. designed the experiments and analysed the data. L.J. performed preliminary experiments. J.K. contributed to plasmid construction. D.H. supervised the study and wrote the original draft. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26303-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Field MC, et al. Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need. Nat Rev Microbiol. 2017;15:217–231. doi: 10.1038/nrmicro.2016.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clayton CE. Gene expression in Kinetoplastids. Curr Opin Microbiol. 2016;32:46–51. doi: 10.1016/j.mib.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Meissner M, Agop-Nersesian C, Sullivan WJ., Jr. Molecular tools for analysis of gene function in parasitic microorganisms. Appl Microbiol Biotechnol. 2007;75:963–975. doi: 10.1007/s00253-007-0946-4. [DOI] [PubMed] [Google Scholar]
- 4.Wirtz E, Clayton C. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science. 1995;268:1179–1183. doi: 10.1126/science.7761835. [DOI] [PubMed] [Google Scholar]
- 5.Glover L, McCulloch R, Horn D. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res. 2008;36:2608–2618. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glover L, Horn D. Locus-specific control of DNA resection and suppression of subtelomeric VSG recombination by HAT3 in the African trypanosome. Nucleic Acids Res. 2014;42:12600–12613. doi: 10.1093/nar/gku900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 8.Glover L, Horn D. Site-specific DNA double-strand breaks greatly increase stable transformation efficiency in Trypanosoma brucei. Mol Biochem Parasitol. 2009;166:194–197. doi: 10.1016/j.molbiopara.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boothroyd CE, et al. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glover L, Alsford S, Horn D. DNA break site at fragile subtelomeres determines probability and mechanism of antigenic variation in African trypanosomes. PLoS Pathog. 2013;9:e1003260. doi: 10.1371/journal.ppat.1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 13.Beneke T, et al. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R Soc Open Sci. 2017;4:170095. doi: 10.1098/rsos.170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soares Medeiros LC, et al. Rapid, selection-free, high-efficiency genome editing in protozoan parasites using CRISPR-Cas9 ribonucleoproteins. MBio. 2017;8:e01788–17. doi: 10.1128/mBio.01788-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander N, Chiurillo MA, Docampo R. Genome editing by CRISPR/Cas9: A game change in the genetic manipulation of protists. J Eukaryot Microbiol. 2016;63:679–690. doi: 10.1111/jeu.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lye LF, et al. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6:e1001161. doi: 10.1371/journal.ppat.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander N, Li ZH, Niyogi S, Docampo R. CRISPR/Cas9-induced disruption of paraflagellar rod protein 1 and 2 genes in Trypanosoma cruzi reveals their role in flagellar attachment. MBio. 2015;6:e01012. doi: 10.1128/mBio.01012-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng D, Kurup SP, Yao PY, Minning TA, Tarleton RL. CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. MBio. 2014;6:e02097–02014. doi: 10.1128/mBio.02097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romagnoli BAA, et al. Improvements in the CRISPR/Cas9 system for high efficiency gene disruption in Trypanosoma cruzi. Acta Trop. 2018;178:190–195. doi: 10.1016/j.actatropica.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Duncan SM, Jones NG, Mottram JC. Recent advances in Leishmania reverse genetics: Manipulating a manipulative parasite. Mol Biochem Parasitol. 2017;216:30–38. doi: 10.1016/j.molbiopara.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Sollelis L, et al. First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell Microbiol. 2015;17:1405–1412. doi: 10.1111/cmi.12456. [DOI] [PubMed] [Google Scholar]
- 22.Zhang WW, Lypaczewski P, Matlashewski G. Optimized CRISPR-Cas9 genome editing for Leishmania and its use to target a multigene family, induce chromosomal translocation, and study DNA break repair mechanisms. mSphere. 2017;2:e00340–16. doi: 10.1128/mSphere.00340-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang WW, Matlashewski G. CRISPR-Cas9-mediated genome editing in Leishmania donovani. MBio. 2015;6:e00861. doi: 10.1128/mBio.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sidik SM, et al. A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell. 2016;166:1423–1435 e1412. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alsford S, Kawahara T, Glover L, Horn D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol Biochem Parasitol. 2005;144:142–148. doi: 10.1016/j.molbiopara.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alsford S, Horn D. Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol Biochem Parasitol. 2008;161:76–79. doi: 10.1016/j.molbiopara.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchetti MA, Tschudi C, Kwon H, Wolin SL, Ullu E. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J Cell Sci. 2000;113:899–906. doi: 10.1242/jcs.113.5.899. [DOI] [PubMed] [Google Scholar]
- 28.Horn D. Codon usage suggests that translational selection has a major impact on protein expression in trypanosomatids. BMC Genomics. 2008;9:2. doi: 10.1186/1471-2164-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 30.Baker N, et al. Aquaglyceroporin 2 controls susceptibility to melarsoprol and pentamidine in African trypanosomes. Proc Natl Acad Sci USA. 2012;109:10996–11001. doi: 10.1073/pnas.1202885109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DL, et al. Kinetics of dCas9 target search in Escherichia coli. Science. 2017;357:1420–1424. doi: 10.1126/science.aah7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marin PA, da Silva MS, Pavani RS, Machado CR, Elias MC. Recruitment kinetics of the homologous recombination pathway in procyclic forms of Trypanosoma brucei after ionizing radiation treatment. Sci Rep. 2018;8:5405. doi: 10.1038/s41598-018-23731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graf FE, et al. Chimerization at the AQP2-AQP3 locus is the genetic basis of melarsoprol-pentamidine cross-resistance in clinical Trypanosoma brucei gambiense isolates. Int J Parasitol Drugs Drug Resist. 2015;5:65–68. doi: 10.1016/j.ijpddr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graf FE, et al. Aquaporin 2 mutations in Trypanosoma brucei gambiense field isolates correlate with decreased susceptibility to pentamidine and melarsoprol. PLoS Negl Trop Dis. 2013;7:e2475. doi: 10.1371/journal.pntd.0002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent IM, et al. A molecular mechanism for eflornithine resistance in African trypanosomes. PLoS Pathog. 2010;6:e1001204. doi: 10.1371/journal.ppat.1001204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren X, et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J, et al. Pentamidine is not a permeant but a nanomolar inhibitor of the Trypanosoma brucei aquaglyceroporin-2. PLoS Pathog. 2016;12:e1005436. doi: 10.1371/journal.ppat.1005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paix A, et al. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Natl Acad Sci USA. 2017;114:E10745–E10754. doi: 10.1073/pnas.1711979114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu JH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dean S, Sunter JD, Wheeler RJ. TrypTag.org: A trypanosome genome-wide protein localisation resource. Trends Parasitol. 2017;33:80–82. doi: 10.1016/j.pt.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raz B, Iten M, Grether-Buhler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 1997;68:139–147. doi: 10.1016/S0001-706X(97)00079-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.