Abstract
Cellular differentiation comprises a progressive, multistep program that drives cells to fabricate a tissue with specific and site distinctive structural and functional properties. Cartilage constitutes one of the potential differentiation lineages that mesenchymal stem cells (MSCs) can follow under the guidance of specific bioactive agents. Single agents such as transforming growth factor beta (TGF-β) and bone morphogenetic protein 2 in unchanging culture conditions have been historically used to induce in vitro chondrogenic differentiation of MSCs. Despite the expression of traditional chondrogenic biomarkers such as type II collagen and aggrecan, the resulting tissue represents a transient cartilage rather than an in vivo articular cartilage (AC), differing significantly in structure, chemical composition, cellular phenotypes, and mechanical properties. Moreover, there have been no comprehensive, multicomponent parameters to define high-quality and functional engineered hyaline AC. To address these issues, we have taken an innovative approach based on the molecular interrogation of human neonatal articular cartilage (hNAC), dissected from the knees of 1-month-old cadaveric specimens. Subsequently, we compared hNAC-specific transcriptional regulatory elements and differentially expressed genes with adult human bone marrow (hBM) MSC-derived three-dimensional cartilage structures formed in vitro. Using microarray analysis, the transcriptome of hNAC was found to be globally distinct from the transient, cartilage-like tissue formed by hBM-MSCs in vitro. Specifically, over 500 genes that are highly expressed in hNAC were not expressed at any time point during in vitro human MSC chondrogenesis. The analysis also showed that the differences were less variant during the initial stages (first 7 days) of the in vitro chondrogenic differentiation program. These observations suggest that the endochondral fate of hBM-MSC-derived cartilage may be rerouted at earlier stages of the TGF-β-stimulated chondrogenic differentiation program. Based on these analyses, several key molecular differences (transcription factors and coded cartilage-related proteins) were identified in hNAC that will be useful as molecular inductors and identifiers of the in vivo AC phenotype. Our findings provide a new gold standard of a molecularly defined AC phenotype that will serve as a platform to generate novel approaches for AC tissue engineering.
Keywords: : mesenchymal stem cells, neonatal articular cartilage, transcriptional profile, cartilage tissue engineering
Introduction
The current state of tissue-engineered cartilage repair still falls short of preclinical and clinical expectations. Treatment protocols for articular cartilage (AC) defects are available using animals as preclinical models.1,2 However, such approaches have not yielded a technology that is successful in clinical translation. While cell-based therapies (microfacture, autologous chondrocyte implantation) are used to promote AC repair,3 none has been proven successful long term by restoring the original AC structure. Mesenchymal stem cells (MSCs) are at the forefront as therapeutic tools for regenerative medicine applications. During the last decade, the description of their medicinal properties involving trophic and immunomodulatory activities4 has been added to the original data documenting the multipotential capacity of adult MSCs5,6; this multipotentiality can be exploited for tissue engineering purposes.7
Tissue-engineered cartilage that originates from adult human bone marrow (hBM)-MSCs was reported by us many years ago.8,9 However, the usefulness of hBM-MSCs as components for tissue-engineered replacement units has been de-emphasized despite their well-established chondrogenic potential. Now, it is clear that the logic used in the past for human MSC (hMSC) chondrogenesis was inadequate. This original logic that used a single bioactive agent such as transforming growth factor beta (TGF-β), without the dynamic exposure to other bioactive agents, failed to initiate the recapitulation of the complex microenvironmental signals and dynamic morphogenetic events that mesenchymal progenitors are exposed to during native AC development.10,11 Therefore, the resulting in vitro hBM-MSC-derived cartilage differs significantly from in situ AC in terms of structure, chemical composition, cell phenotype, and function.
A transient cartilage typical of endochondral processes such as embryonic bone formation and adult fracture healing, rather than permanent hyaline AC, appears to be the differentiation pathway that hBM-MSCs follow under current induction protocols.12–14 This differentiation capacity, which serves as the conceptual basis for several clinical treatments for AC defects, ultimately results in cartilage-like structures quite different from the native AC in a number of parameters.15 In addition, the endochondral program dictates that the ultimate cellular phenotype is of a hypertrophic nature, which is recognized as a sign of degenerative cartilage states (i.e., osteoarthritic cartilage).16
It is important to emphasize that there is not clear evidence in vitro or in vivo about the innate capability (or incapability) of hBM-MSCs to make AC, which may depend on the induction protocols that are currently used.13,17 In this regard, we have made progress in modulating this unwanted hypertrophic phenotype by exposing differentiating hBM-MSCs to a sequential regimen of growth factors, reminiscent of embryonic processes in which one stimulus primes the cells for the activity of a subsequent one.18 Although many of the molecular players involved in chondrogenic differentiation of MSCs have been identified, a comprehensive understanding of control elements involved in the chondrogenic program, and the particular gene signature in each lineage stage, may help to guide the cells to escape their endochondral fate and form a functional hyaline AC phenotype.
Realistically, we are still far from developing efficient therapeutic clinical applications for the regeneration of hyaline AC with hMSCs. If MSCs have the potential to form a tissue that resembles native AC, the microenvironmental conditions required for MSCs to differentiate into a suitable chondrocytic phenotype, both in vitro and in vivo, will have to be optimized. Current evidence shows that the inductive and formative conditions must be developmentally oriented,17,19–22 incorporating principles of “Developmental Engineering” to mimic the embryonic development of a suitable chondroprogenitor.23,24 This approach implies that both the dynamics of the differentiation program and the initial receptivity of cells must be properly taken into consideration. To accomplish this, our understanding of the control elements of the complex multistep lineage process involved in hyaline AC development, including the distinctive cellular and molecular signatures specific for both the initial and the sequentially formed tissue, must be refined.
We address these issues by interrogating, at the molecular level, human neonatal articular cartilage (hNAC), which had been dissected from the knees of deidentified 1-month-old cadaveric specimens. In addition, in vitro formed, hBM-MSC-derived three-dimensional (3D) cartilage structures are comparatively interrogated with the aim of identifying specific transcriptional regulatory elements and proteins that are differentially expressed. Gene expression clustering analysis included several other neonatal knee tissues (i.e., meniscus, synovial membrane, tendon, among others). This allowed us to perform a comprehensive identification of differentially regulated genes across these tissues and compare them with in vitro hMSC-derived cartilage structures.
Importantly, we set up the early neonatal AC as our gold standard, given that this tissue will greatly expand physically while mechanically supporting and adapting from low-stress to high-stress loading. We propose that these are perfect parameters for tissue-engineered and implantable cartilage. Consistent with this approach, it has been recognized that neonatal articular chondrocytes have superior capabilities to in vitro differentiate into cartilage-like tissue compared with adult chondrocytes and MSCs.25–28
Methods
Tissue dissection
hNAC from both femoral condyle and tibial plateau, along with other intra-articular tissues, was carefully dissected from both knees of deidentified 1-month-old cadaveric specimens (n = 2) procured from consented guardians of the human tissue donors. The samples were dissected sequentially, minimizing “contamination” from surrounding tissues, by leaving wide tissue margins between them. The tissue samples were stored at −80°C suspended in RNAlater reagent (Qiagen, Valencia, CA) to preserve RNA during the final processing before microarray analysis. Adult hip cartilage (n = 1) was dissected from a femoral head discarded during surgery from a consented, deidentified adult diagnosed with advanced osteoarthritis (OA), and provided by the Human Tissue Procurement Facility of Case Western Reserve University. The cartilage tissue was similarly stored at −80°C in RNAlater reagent (Qiagen).
Cell cultures
Cultures of hBM-MSC from two healthy deidentified adult volunteer donors were established as previously described.29 The BM was collected using a procedure reviewed and approved by the University Hospitals of Cleveland Institutional Review Board; informed consent was obtained from all deidentified donors. Cells were expanded in Dulbecco's modified Eagle's medium-low glucose with fetal bovine serum 10% (selected batch)30 supplemented with 10 ng/mL fibroblast growth factor (FGF)2.18
In vitro hBM-MSC chondrogenic differentiation
hBM-MSCs were cultured in cell aggregates (3D pellets) in complete chondrogenic medium (DMEM-high glucose supplemented with 1% ITS+, 10−7 M dexamethasone, 1 mM sodium pyruvate, 120 mM ascorbic acid-2 phosphate, 100 mM nonessential amino acids, and 10 ng/mL TGF-β1).8,9 Chondrogenic pellets were harvested at different time points (3, 7, 10, 14, 21, and 28 days) for RNA extraction and microarray analysis. A day 0 sample was also included that corresponds to hMSCs in monolayer culture before chondrogenic induction.
RNA preparation from tissues
Total RNA was prepared from tissues in RNAlater (Qiagen) after the addition of TRIzol (Life Technologies) and homogenized with a Polytron (PT-MR2100; Polytron Corp.). Following the transfer of processed sample to Qiagen RNeasy mini columns, the extraction of total RNA was performed according to the manufacturer's instructions. For hBM-MSC-derived cartilage, aggregates were homogenized with RNase-free disposable Pellet-Pestles® (Kimble-Chase, TN), digested on-column with DNase-I, and purified with the RNeasy mini kit. RNA concentrations were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific).
Microarray analysis
Whole-genome expression analysis was carried out using Illumina (CA) Human Ref-8v3 or Human HT-12 v4 BeadArrays™. For the Illumina BeadArrays, total RNA was linearly amplified and biotin labeled using Illumina TotalPrep™ kits (Life Technologies, CA); cRNA was quality controlled using an Agilent 2100 bioanalyzer. The cRNA was hybridized to Illumina BeadChips™, processed and read using a BeadStation™ array reader according to the manufacturer's instructions (Illumina). Values of less than 130 relative fluorescence units were considered to be nonspecific background signals.
Data analysis
To investigate how gene expression varied across the samples, unsupervised sample clustering in Cluster 3.0 (bonsai.ims.u-tokyo.ac.jp/∼mdehoon/software/cluster) was performed. In this analysis, samples were grouped according to their expression profile using only differentially expressed genes (>2.0-fold) from all samples and among all experimental conditions. Unsupervised hierarchical clustering was performed and heat maps and dendrograms were generated in Java Treeview (jtreeview.sourceforge.net) and MultiExperiment Viewer (MeV).
Principal component analysis (PCA) algorithm applied to all samples was performed in Chipster 3.6.3 and MeV 4.8. The statistical analysis of the microarray data was based on the normalized mean expression values per probe at six time points with two replicates at each time point (two different hMSC donors, two hNAC donors, 12 observations per probe).
Significance analysis of microarray (SAM) method was used to identify differentially expressed genes.31 SAM computes a statistic di for each gene i, measuring the strength of the relationship between gene expression and the response variable. It uses repeated permutations of the data to determine if the expression of any genes is significantly related to the response.31
Pathway analysis
WebGestalt (Web Gene Set Analysis Toolkit) Pathway Commons enrichment analysis was performed on 373 differentially expressed genes (significant genes based on SAM) identified by microarray analysis to identify significantly enriched pathways. Significantly enriched pathways were identified using the human genome as a reference set. Default hypergeometric and multiple testing statistical analysis methods were used with a significance level of 0.001. Output was restricted to pathways with a multiple testing p-value <0.05 and an overlap of at least two genes between the pathway term gene list and differentially expressed gene list.32
Immunohistochemistry
Tissues and chondrogenic pellets were fixed, paraffin embedded, and sectioned. Adjacent sections were stained with toluidine blue O to evaluate proteoglycan content. For type II collagen (Col2), matrilin-1, and unique cartilage matrix-associated protein (UCMA) immunohistochemistry, sections were deparaffinized and rehydrated followed by antigen retrieval (20 min in 10 mg/mL proteinase-K [Roche] in Tris-ethylenediaminetetraaceticacid buffer for at 37°C) and endogenous peroxidase activity blocked with a 30-min exposure to 3% hydrogen peroxide at room temperature. Nonspecific binding sites were blocked with 2.5% normal horse serum, and sections were incubated overnight with mouse monoclonal antibodies. Sections were incubated with anti-mouse Ig (ImmPRESS™ polymerized reporter enzyme staining system; Vector Laboratories, CA) for 30 min followed by a short incubation with ImmPACT™ red peroxidase substrate (Vector Laboratories, CA).
Results
Significant transcriptional differences between in vitro hMSC-derived 3D cartilage and hNAC
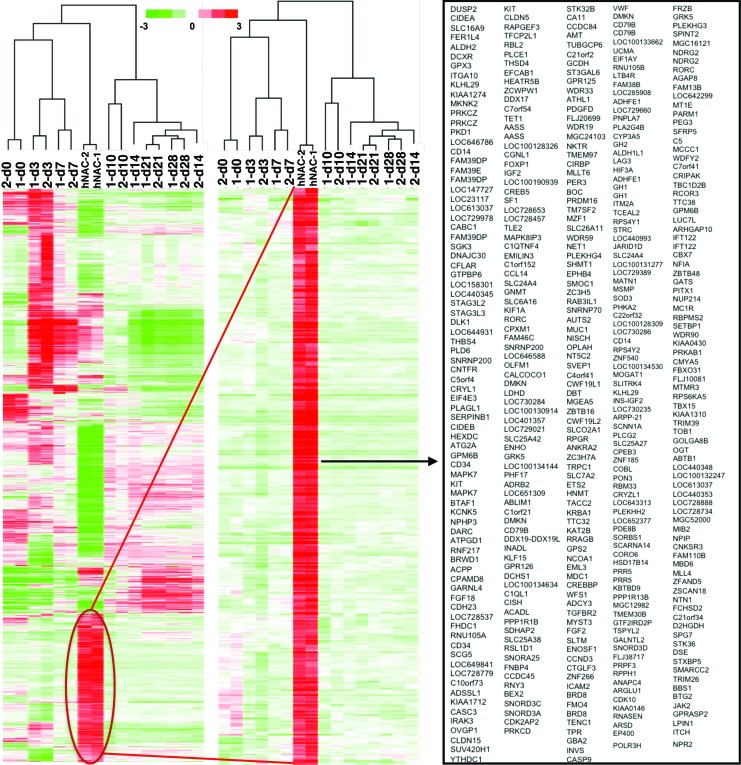
The transcriptomes of hNAC and 3D cartilage-like tissue formed by hBM-MSCs in vitro were found to be globally distinct based on microarray gene expression analysis (twofold change and p < 0.05). Importantly, over 500 genes that are highly expressed in hNAC in vivo were not expressed at any time point during in vitro hMSC chondrogenesis (Fig. 1). Further biostatistical analysis showed that those overall differences are less pronounced at the initial stages (first 7 days) of hMSC in vitro 3D chondrogenic differentiation (Fig. 2). This stage-dependent effect suggests that the osteochondral fate of hMSC-derived cartilage may be rerouted during earlier phases of in vitro chondrogenesis.
FIG. 1.
Hierarchical clustering analysis of hMSC-derived 3D cartilage transcriptome compared to hNAC. hMSC in vitro chondrogenic differentiation program (d3–d28); MSCs before chondrogenesis (d0). 3D, three-dimensional; hMSC, human mesenchymal stem cell; hNAC, human neonatal articular cartilage. Color images available online at www.liebertpub.com/tea
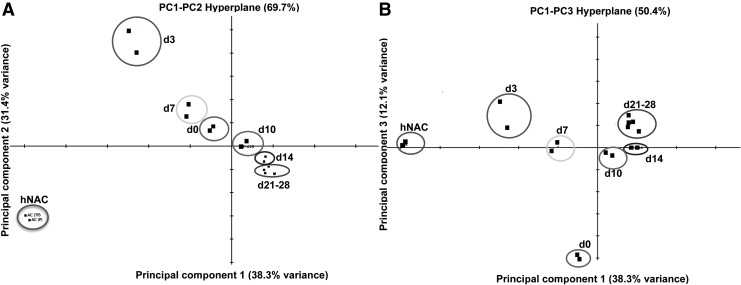
FIG. 2.
PCA of in vitro hBM-MSC-derived 3D cartilage compared to hNAC. The two major principal components explain 69.7% of the expression variance (A). The PC1–PC3 hyperplane shows a departure of the transcriptional profile of hMSC chondrogenesis from the hNAC profile (B). Data from two different donors cluster together within the analysis. hMSC in vitro chondrogenic differentiation program (d3–d28); hMSCs before chondrogenesis (d0). PCA, principal component analysis; hBM, human bone marrow.
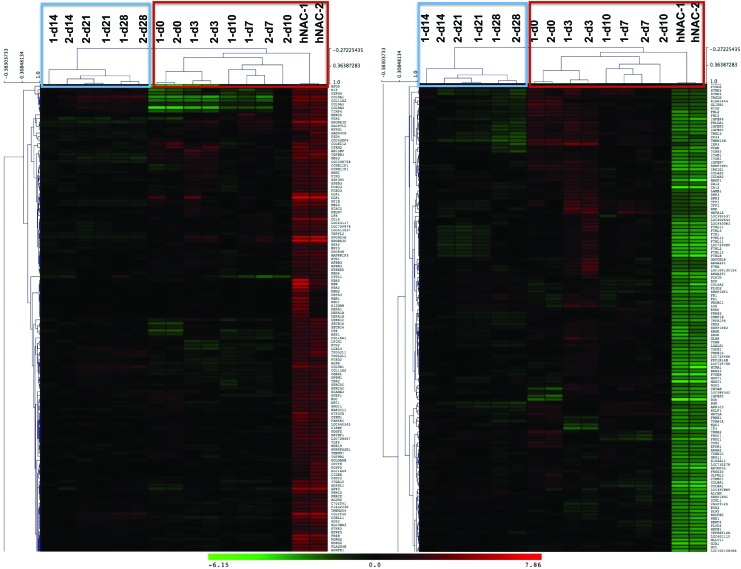
A PCA was performed based on our microarray data to establish the relationship between hMSC-derived cartilage and hNAC using all genes in the microarray data set. PCA is a mathematical algorithm that allows a dimensionality reduction of large data sets, typically composed by several different variables, through the identification of specific directions called principal components. This reduction, however, retains most of the variance within the data set. This dimensionality reduction makes it possible to visually assess similarities and differences between samples and to determine whether samples cluster together (similar) or cluster far from each other (different).33 In our data set, the two major principal components explain 69.7% of the expression variance (Fig. 2A). There is a clear departure between hMSCs (d0) and hMSC-derived 3D cartilage at different time points during the differentiation program (d3–d28) with hNAC in the first and second components (Fig. 2A). However, a close relationship between hNAC and the earlier time points during hMSC chondrogenesis (d3 and d7) is observed in the first and third components hyperplane (Fig. 2B). Average linkage hierarchical clustering of in vitro hMSC-derived 3D cartilage compared to hNAC revealed a clear separation between early (d0–d10) and late (d14–d28) time points of the chondrogenic program based exclusively on significant genes in the microarray data set (according to SAM, Tables 1 and 2). Furthermore, hNAC clustered together with the earlier in vitro 3D cartilage time points (Fig. 3). This analysis reveals that hMSC-derived cartilage transcriptome progressively differs from hNAC with some similarities exclusively at early stages of chondrogenesis (d3 and d7).
Table 1.
Significant Upregulated Genes Differentially Expressed in Human Neonatal Articular Cartilage Compared to Human Mesenchymal Stem Cell-Derived Three-Dimensional Cartilage
| Gene | dExp | d | Fold change | Gene | dExp | d | Fold change |
|---|---|---|---|---|---|---|---|
| MSMP | −0.38 | 2.98 | 170.4 | SCNN1A | −0.14 | 1.69 | 17.8 |
| CD79B | −0.31 | 2.70 | 98.9 | RNU105B | −0.13 | 1.69 | 17.2 |
| SNORD3C | −0.32 | 2.10 | 58.0 | TCEAL2 | −0.13 | 1.73 | 16.9 |
| RPS4Y1 | −0.21 | 2.45 | 56.1 | ALDH1L1 | −0.12 | 1.70 | 16.3 |
| UCMA | −0.19 | 2.37 | 53.6 | MATN1 | −0.12 | 1.66 | 15.7 |
| ITM2A | −0.20 | 2.40 | 50.9 | SOD3 | −0.12 | 1.67 | 15.5 |
| HBB | −0.21 | 1.69 | 45.8 | SPINT2 | −0.12 | 1.60 | 15.1 |
| SNORD3A | −0.45 | 1.89 | 42.8 | GRK5 | −0.16 | 1.58 | 14.7 |
| DLK1 | −0.26 | 1.90 | 41.3 | H19 | −0.34 | 1.32 | 14.4 |
| CD79B | −0.19 | 2.20 | 40.8 | SLITRK4 | −0.11 | 1.59 | 14.3 |
| HBA2 | −0.20 | 1.58 | 38.0 | COBLL1 | −0.18 | 1.42 | 14.3 |
| GPX3 | −0.63 | 1.93 | 36.7 | NFIB | −0.16 | 1.43 | 14.0 |
| PPP1R1B | −0.17 | 2.00 | 29.1 | COL11A2 | −0.24 | 1.31 | 13.8 |
| FRZB | −0.20 | 1.90 | 26.8 | STK32B | −0.13 | 1.44 | 13.5 |
| C2orf40 | −0.18 | 1.73 | 26.4 | TGFBR3 | −0.19 | 1.27 | 12.2 |
| CD14 | −0.17 | 1.76 | 22.2 | PHF17 | −0.13 | 1.47 | 12.0 |
| NDRG2 | −0.16 | 1.68 | 20.7 | ITGA10 | −0.28 | 1.39 | 11.6 |
| PNPLA7 | −0.16 | 1.78 | 19.5 | COL9A1 | −0.13 | 1.21 | 10.0 |
| SNORD3D | −0.33 | 1.49 | 19.2 | COL9A2 | −0.34 | 1.15 | 9.6 |
| PEG3 | −0.16 | 1.71 | 18.9 | FGF18 | −0.08 | 1.28 | 9.4 |
Analysis performed using a two-class unpaired SAM (Mev). Genes with a >9 fold change are shown. Gene names in bold letters have or may have a role during chondrogenesis.
dExp, expected score; d, observed score; SAM, significance analysis of microarray.
Table 2.
Significant Downregulated Genes in Human Neonatal Articular Cartilage Compared to Human Mesenchymal Stem Cell-Derived Three-Dimensional Cartilage
| Gene | dExp | d | Fold change | Gene | dExp | d | Fold change | Gene | dExp | d | Fold change |
|---|---|---|---|---|---|---|---|---|---|---|---|
| COL10A1 | −2.37 | −20.98 | 0.0075 | INSIG1 | −0.19 | −11.47 | 0.0682 | FTHL3 | −0.44 | −8.27 | 0.1173 |
| PENK | −2.16 | −13.24 | 0.0087 | FTHL8 | −1.88 | −7.74 | 0.0686 | PMEPA1 | −0.02 | −5.69 | 0.1177 |
| SPP1 | −0.79 | −13.47 | 0.0123 | COL8A1 | −0.26 | −7.14 | 0.0743 | FTH1 | 0.17 | −7.40 | 0.1180 |
| TACSTD2 | −0.64 | −9.91 | 0.0144 | FABP5 | 0.29 | −6.07 | 0.0762 | FAM180A | 3.02 | −7.25 | 0.1191 |
| CLEC3A | −1.69 | −16.20 | 0.0148 | FNDC1 | 0.13 | −7.87 | 0.0790 | CRYAB | −1.29 | −6.48 | 0.1199 |
| IBSP | −0.44 | −11.62 | 0.0188 | ID3 | −0.28 | −9.44 | 0.0793 | HSPB1 | −0.54 | −5.04 | 0.1199 |
| DPT | −0.82 | −6.77 | 0.0203 | GJA1 | −0.38 | −7.64 | 0.0795 | GLRX | −0.51 | −6.51 | 0.1248 |
| DCN | −1.65 | −8.11 | 0.0273 | CXCL14 | 0.84 | −9.00 | 0.0803 | TGFBI | −1.98 | −5.70 | 0.1291 |
| PSAT1 | −0.03 | −12.04 | 0.0286 | THY1 | 0.54 | −9.10 | 0.0839 | SERF2 | −0.74 | −6.38 | 0.1293 |
| LUM | −0.26 | −11.16 | 0.0289 | KDELR3 | 0.35 | −7.15 | 0.0842 | FTHL11 | −0.59 | −6.78 | 0.1315 |
| SLC7A5 | −0.05 | −7.76 | 0.0313 | FTHL12 | −1.85 | −7.39 | 0.0845 | SERF2 | −0.74 | −6.38 | 0.1293 |
| CXCL13 | 0.11 | −8.05 | 0.0316 | ASNS | −0.02 | −6.86 | 0.0862 | FTHL11 | −0.59 | −6.78 | 0.1315 |
| ALPL | −0.24 | −7.01 | 0.0367 | PTGES | 0.06 | −6.18 | 0.0878 | NDUFB3 | 0.31 | −5.65 | 0.1329 |
| MGST1 | 0.05 | −5.99 | 0.0385 | KDELR3 | 0.03 | −8.21 | 0.0934 | MT1G | −0.02 | −6.46 | 0.1334 |
| P4HA2 | −0.30 | −10.34 | 0.0474 | FTHL12 | −1.71 | −7.41 | 0.0978 | PTH1R | −1.05 | −7.92 | 0.1344 |
| LOX | −1.64 | −6.66 | 0.0502 | FTHL2 | −1.46 | −8.35 | 0.0985 | VKORC1 | 0.26 | −7.19 | 0.1369 |
| PANX3 | −0.02 | −8.35 | 0.0540 | CA12 | −0.27 | −7.54 | 0.0990 | FNDC1 | 0.78 | −7.05 | 0.1370 |
| MATN3 | −0.72 | −8.84 | 0.0553 | C1QTNF3 | 1.67 | −7.13 | 0.1017 | DRD4 | 0.30 | −6.33 | 0.1388 |
| PPIC | 0.04 | −10.47 | 0.0556 | CRYGS | −0.25 | −7.65 | 0.1019 | TMSB10 | −1.97 | −5.99 | 0.1390 |
| CSRP2 | 0.32 | −10.00 | 0.0566 | ANGPTL2 | 0.10 | −5.17 | 0.1030 | THBS2 | −0.25 | −5.02 | 0.1394 |
| RAB31 | −0.51 | −10.21 | 0.0582 | ANGPTL7 | −0.26 | −5.59 | 0.1048 | COL6A3 | −1.36 | −5.51 | 0.1476 |
| RASL11B | 0.29 | −9.10 | 0.0585 | NME1 | −0.06 | −5.20 | 0.1059 | PPP1R14B | 0.74 | −6.94 | 0.1507 |
| FABP5 L2 | −0.36 | −6.05 | 0.0610 | ANXA2P1 | −1.39 | −6.95 | 0.1064 | GNG11 | 0.37 | −5.54 | 0.1516 |
| FN1 | 1.61 | −5.78 | 0.0614 | EPDR1 | 0.49 | −7.73 | 0.1074 | CALU | 0.31 | −6.65 | 0.1664 |
| ALCAM | 4.33 | −6.92 | 0.0619 | PLOD2 | 0.03 | −5.17 | 0.1077 | CALD1 | −0.03 | −5.63 | 0.1682 |
| PPIC | 0.05 | −9.65 | 0.0639 | APCDD1 L | 0.18 | −5.02 | 0.1108 | DCN | 0.07 | −4.91 | 0.1685 |
| SERPINF1 | 0.41 | −6.51 | 0.0659 | SCD | −0.53 | −6.99 | 0.1117 | TMEM4 | 0.57 | −5.89 | 0.1770 |
| COL1A1 | −1.86 | −6.12 | 0.0659 | SERPINE1 | −0.27 | −5.31 | 0.1122 | FGFR3 | 0.16 | −5.45 | 0.1818 |
| POSTN | 0.24 | −6.96 | 0.0664 | FABP5 L2 | 1.82 | −5.68 | 0.1159 | OSTC | 0.02 | −5.47 | 0.1849 |
Analysis performed using a two-class unpaired SAM (Mev). Gene names in bold letters have or may have a role during chondrogenesis.
FIG. 3.
Average linkage hierarchical clustering of in vitro hMSC-derived 3D cartilage compared to hNAC. The analysis was based only on significant genes in the microarray data set (according to significance analysis of microarray). Color images and readable text are available online at www.liebertpub.com/tea
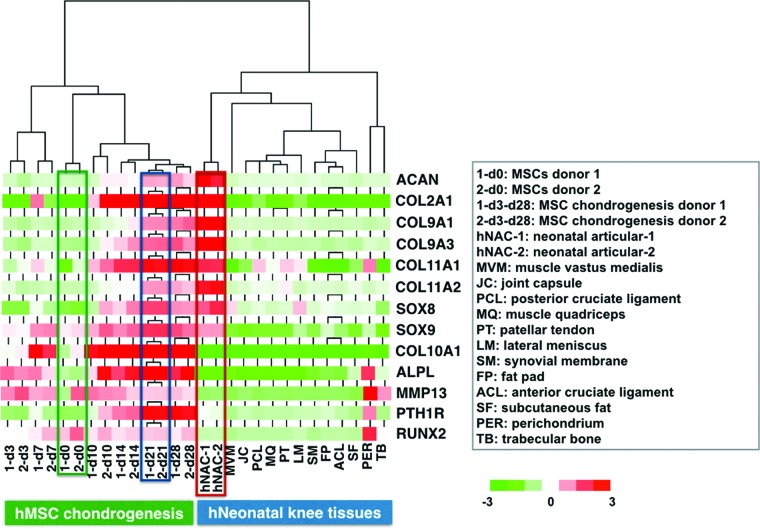
Exclusive transcriptional profile of hNAC compared to in vitro hMSC-derived 3D cartilage and other neonatal knee tissues
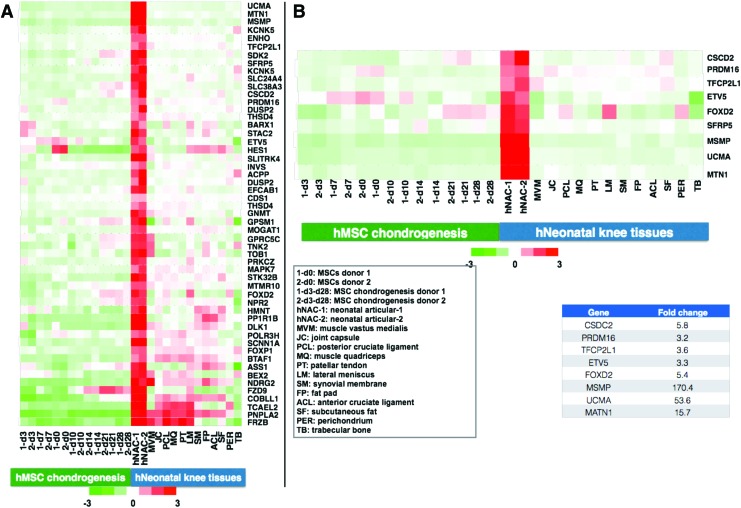
In an effort to narrow the number of genes that are exclusively expressed in hNAC, a similar hierarchical cluster analysis was performed to compare not only with hMSC-derived but also with other neonatal tissues, including muscle, ligament, tendon, meniscus, synovium, fat pad, trabecular bone, and perichondrium (Fig. 4). The goal of this comprehensive search is to define a molecular target or gold standard transcriptional profile for the characterization of native human AC for tissue engineering approaches. Based on this analysis, we narrowed the number of differently expressed genes to ∼10% of the total genes found to be upregulated in hNAC compared to hMSC 3D chondrogenesis (Fig. 4). From this set of genes, we focused on genes that had DNA binding domains to identify transcription factors that were exclusively expressed in hNAC. In addition, we also identified genes that coded for proteins with potential roles in cartilage biology that will serve as a molecular profile (read-out) to categorize an engineered 3D cartilage tissue with a potential AC phenotype (Fig 4B).
FIG. 4.
Identification of genes that are almost exclusively expressed in hNAC. Other neonatal knee tissues were included in the analysis with the objective of narrowing the set of genes that will be used to describe the criteria or gold standard for developing a tissue-engineered AC from hMSCs. (A) A total of 53 genes were selected that are highly expressed in hNAC compared to in vitro hMSC-derived 3D cartilage and other knee tissues. (B) From this list of genes, specific control elements (transcription factors) were selected that may be involved in the development of native AC and coded proteins that will be used as markers that define a molecular criterion or new gold standard. AC, articular cartilage. Color images available online at www.liebertpub.com/tea
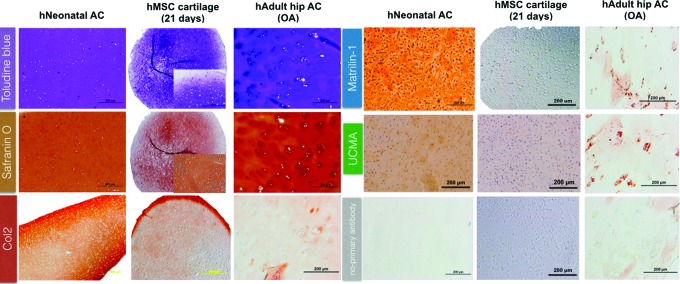
Histomorphological characterization of in vitro hMSC-derived 3D cartilage, healthy hNAC, and diseased AC
In addition to the transcriptome analysis, a comparative histomorphological assessment was performed between in vitro hMSC-derived 3D cartilage and native human AC at two extremes of the spectrum: healthyhNAC and cartilage from a patient with OA undergoing hip arthroplasty. hNAC presented a homogenous metachromatic stain with toluidine blue with nonhypertrophic chondrocytes evenly dispersed through the tissue. This is in contrast to the observed clustered and hypertrophic appearance of chondrocytes in adult OA hip cartilage34 and the in vitro MSC-derived 3D cartilage (hMSC pellet at 21 days) with all of the cells observed to be situated in large lacunae (Fig. 5). These observations suggest that hMSC-derived 3D cartilage differs from hNAC both transcriptionally and also structurally by the hypertrophic appearance of the chondrocytes that is reminiscent of old or degenerative cartilage.
FIG. 5.
Histological differences between hNAC, hMSC-derived 3D cartilage, and human adult diseased AC. Paraffin-embedded sections were stained with toluidine blue and safranin O to assess relative glycosaminoglycan/proteoglycan content in samples. A more homogeneous distribution of cartilage extracellular matrix is observed in hNAC compared to hMSC-derived 3D cartilage and adult cartilage. For immunohistochemistry, sections were incubated with anti-mouse Ig (ImmPRESS™ polymerized reporter enzyme staining). High expression of collagen type 2 is observed in hNAC compare to the other cartilage tissues. Expression of matrilin-1 and unique cartilage matrix-associated proteins was detected in hNAC and not in hMSC-derived 3D cartilage, which correlated with the microarray data. Some small degree of expression was found in adult diseased cartilage. Color images available online at www.liebertpub.com/tea
To validate the results from the microarray, we stained the three types of cartilages with antibodies for matrilin-1 and UCMA. The gene expression analysis for these molecules showed that they are exclusively expressed in hNAC and not in hMSC-derived cartilage. Both matrilin-1 and UCMA have been studied in the context of cartilage biology,35,36 however, this is the first time that they have been shown by transcriptome analysis to be exclusively expressed in AC, therefore have included them as part of the molecular targeted and read-out of AC. As expected, both proteins are only expressed in hNAC and not in hMSC-derived 3D cartilage. An intracellular staining using the anti-UCMA antibody was observed within hMSC cartilage aggregates; however, UCMA was not detected within the hMSC-derived 3D cartilage extracellular matrix (ECM). In addition, weak staining was observed in adult osteoarthritic cartilage specimen.
Expression of “classical” cartilage markers does not constitute a specific read-out of native AC
An in silico analysis of the microarray data shows that common cartilage markers are expressed in both hMSC-derived 3D cartilage and hNAC. As expected, the expression patterns between both types of cartilages are dissimilar, most notably with markers associated with hypertrophic cartilage such as Col10, Runx2, ALPL, PTHR1, and matrix metalloproteinase (MMP)-13 (Fig. 6). On the contrary and also expected, the “classical” cartilage markers such as Sox9, Sox8, Col9, Col2, and aggrecan (ACAN) were differentially expressed but only at early stages of in vitro hMSC-derived 3D cartilage when cells are entering into the chondrogenic lineage. From d10 thereafter, and especially at d21–d28, the expression was very similar between tissue-cultured specimens.
FIG. 6.
Differential expression of common cartilage genes. Unsupervised hierarchical clustering of differentially expressed genes between undifferentiated hMSC (d0) and during 3D chondrogenesis (at 3, 7, 10, 14, 21, and 28 days), hNAC, and other neonatal knee tissues. Expression profiles were clustered by average linkage hierarchical clustering; common chondrogenesis genes are shown. The color scale of standardized signal intensities in the microarray extends from bright green (downregulation) to bright red (upregulation). Color images available online at www.liebertpub.com/tea
In most studies in which hMSC-derived 3D cartilage is generated for AC tissue engineering, the outcome analysis is often limited to examining the selected profile of such common markers. Often, the expression of only these markers is misleadingly interpreted as being indicative of successful generation of AC. According to our results, these “classical” markers for “hyaline type” of cartilage cannot be used to distinguish AC from growth plate cartilage,37 or from in vitro hMSC-derived 3D cartilage. Therefore, using a comparative microarray analysis of dissected hNAC and in vitro hBM-MSC-derived cartilage, we conclude that the classical markers of cartilage are not sufficient to classify the resulting cartilage tissue as comparable to AC (Fig. 6).
As shown by us and others,18 in vitro hMSC-derived cartilage expresses high levels of Col10, which indicates the endochondral cartilage differentiation potential of these induced hMSCs. As a control, we analyzed the expression of these markers in other knee tissues (Fig. 6) and showed the specific expression could be observed only in cartilage tissue. These observations also document that, despite the expression profile of common cartilage markers, hMSC-derived transient cartilage is distinct from native permanent AC. In fact, it has been suggested that MSCs do not form AC at all.15 Based on these observations, we propose that a new molecular target (gold standard) of cartilage markers can be based on the transcriptional profile of hNAC.
Transcriptional changes that occur during hMSC chondrogenic differentiation indicate an early departure from the native AC lineage pathway
The sequential changes that occur during the hMSC chondrogenic differentiation program were studied using bioinformatic analysis of the microarray data based on the normalized mean expression values per probe at six time points (two different hMSC donors, 12 observations per probe). To identify subgroups of probes with similar expression profiles over time, PCA of the covariance matrix was carried out on the mean expression value for each probe at each time point. The first three components explain 87.2% of the data variance (Fig. 7). Component 1 describes a general level of gene expression; component 2 shows changes on gene expression during hMSC in vitro chondrogenic differentiation.37 The highest level of expression was observed at day 0 (nondifferentiated hMSCs), as expected. Component 3 shows an elevation in gene expression around the first week of differentiation, showing that the higher transcriptional changes occur during the first week of the differentiation program (Fig. 7A). Component 2 is more likely to contain genes associated with the loss of hMSC characteristics. Component 3 is more likely to contain genes related to the gain of a chondrocyte phenotype with the peak occurring at the end of the first week of differentiation.
FIG. 7.
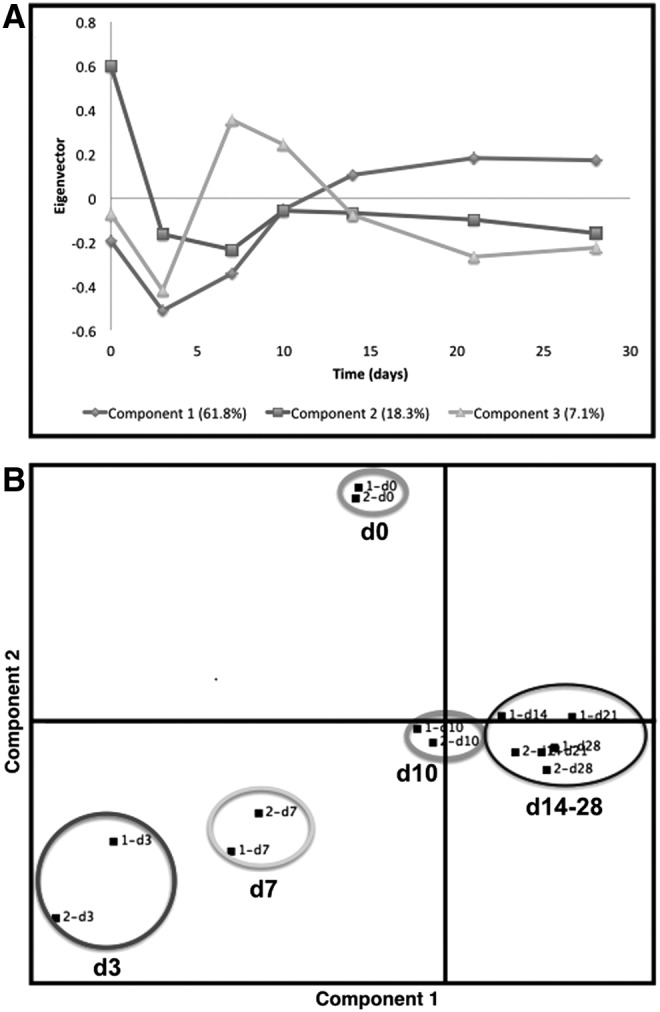
PCA of microarray data at different time points during hMSC (day 0 → day 28) pellet chondrogenesis. PCA of the covariance matrix was carried out on the mean expression value for each probe at each time point. (A) The first three components explain 87.2% of the data variance. Component 1 represents the general level of expression, Component 2 the degree of change from the undifferentiated state, and Component 3 most likely represents gene expression related to the gain of a chondrogenic phenotype. (B) Components 1 and 2 hyperplane (80.1% of the variance in the data set) shows distinct separation between undifferentiated hMSCs (day 0) and in vitro 3D chondrogenesis (d3 → day 28).
The cluster structure within the hyperplane formed by components 1 and 2, which capture most of the variance in the data set, shows distinct separation between undifferentiated hMSCs (day 0) and time points during chondrogenesis (d3 → day 28) (Fig. 7B). Within these time points, days 3 and 7 group closer together with days 10, 14, 21, and 28 forming another distinct cluster group. This analysis provides evidence that the gene expression profile during the first week differs greatly with the profile of the following 3 weeks of the chondrogenic differentiation program. Directional distances on the PCA dimensions also indicate a continuum in the direction of changes that is consistent with differentiation progression. In addition, both donors clustered together, indicating that there is a strong homogeneity in their gene expression pattern at each different time point (Fig. 7B).
Based on the initial analysis of the microarray data, it is possible to observe that the transcriptional profile at day 3 (d3) of the chondrogenic program seems to differ less with the profile of hNAC in comparison with later time points (Fig. 2B). To study the detailed differential gene expression within the chondrogenic program, a thorough breakdown of the data was performed using statistical analysis of microarray data (SAM). This analysis allows the identification of genes that are upregulated or downregulated during each measured stage of the chondrogenic program (Table 3). Clearly, the higher upregulation of several genes was found when hMSCs (d0) enter the initial stages of the differentiation program (d0 → d3). No cartilage-/chondrogenic-related genes were found to be significantly upregulated during this initial stage.
Table 3.
Transcriptional Changes During Human Mesenchymal Stem Cell Three-Dimensional Chondrogenesis
| d0 → d3 | d3 → d7 | d7 → d10 | d10 → d14 | d14 → d21 | d21 → d28 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Fold change | Gene | Fold change | Gene | Fold change | Gene | Fold change | Gene | Fold change | Gene | Fold change |
| Significant upregulated genes comparing each time point during chondrogenesis | |||||||||||
| RHBDL2 | 56.0 | APOD | 11.6 | None | EPYC | 5.10 | CXCL14 | 4.07 | ORM1 | 6.17 | |
| SERPINA3 | 44.0 | GPAM | 9.3 | LECT1 | 4.75 | MMP10 | 3.80 | SLC38A4 | 2.36 | ||
| IL8 | 43.1 | RASL11B | 7.1 | KLK4 | 4.26 | ANGPTL7 | 3.40 | MT1H | 2.19 | ||
| PTGR2 | 42.5 | ANGPTL2 | 6.9 | CLEC3A | 4.05 | COL9A1 | 2.96 | APOD | 2.15 | ||
| RN7SK | 41.3 | MAT2A | 6.7 | MATN3 | 3.86 | FOXA2 | 2.79 | CXCL13 | 2.07 | ||
| CYCSL1 | 39.1 | COL10A1 | 6.2 | ANGPTL7 | 3.43 | IBSP | 2.64 | FTHL3 | 2.04 | ||
| LOC389765 | 38.2 | SCRG1 | 6.2 | OGN | 2.78 | COL11A2 | 2.59 | ORM1 | 6.17 | ||
| LOC729120 | 36.8 | CXCL13 | 6.1 | EPYC | 5.10 | F13A1 | 2.55 | SLC38A4 | 2.36 | ||
| FLJ40722 | 35.9 | PSAT1 | 5.5 | FMOD | 2.48 | MT1H | 2.19 | ||||
| LOC730993 | 35.5 | HAPLN1 | 5.3 | CSPG4 | 2.47 | ||||||
| GSTTP2 | 34.5 | FOXO1 | 5.3 | C2orf82 | 2.45 | ||||||
| C11orf63 | 33.8 | ID3 | 5.2 | PTH1R | 2.44 | ||||||
| TPR | 31.5 | AKR1C2 | 5.2 | COL9A2 | 2.44 | ||||||
| FAM73A | 31.4 | ID1 | 4.9 | ENPP1 | 2.38 | ||||||
| MBTD1 | 30.8 | SPP1 | 4.8 | IFITM5 | 2.36 | ||||||
| ZNF557 | 29.6 | PCNA | 4.7 | AEBP1 | 2.35 | ||||||
| MIR1974 | 29.1 | SPP1 | 4.7 | EFHD1 | 2.31 | ||||||
| RN7SK | 28.1 | RNU4-1 | 4.1 | CAPN6 | 2.31 | ||||||
| EEF1A2 | 26.8 | NOP56 | 4.1 | PRELP | 2.25 | ||||||
| ANKRD44 | 24.8 | HNRNPAB | 4.0 | MIA | 2.23 | ||||||
| GCLM | 24.0 | HNRPM | 3.9 | NGF | 2.21 | ||||||
| PRO1853 | 23.1 | MRFAP1 L1 | 3.8 | PMP2 | 2.21 | ||||||
| PRG4 | 22.7 | HSPA6 | 3.8 | CHST3 | 2.20 | ||||||
| Significant downregulated genes comparing each time point during chondrogenesis | |||||||||||
| FOSB | 0.06 | IL8 | 0.08 | PTGR2 | 0.09 | RN7SK | 0.09 | AKR1C3 | 0.40 | IL11 | 0.28 |
| CCL2 | 0.08 | PRG4 | 0.09 | FAM73A | 0.10 | RN5S9 | 0.18 | HSPE1 | 0.40 | IER3 | 0.28 |
| ARHGEF3 | 0.10 | TPR | 0.09 | RHBDL2 | 0.10 | HSPA6 | 0.21 | ANGPT1 | 0.43 | TUBB3 | 0.29 |
| ODZ4 | 0.10 | LBP | 0.11 | MBTD1 | 0.10 | RNU4-2 | 0.23 | C7orf10 | 0.44 | IFITM5 | 0.32 |
| CD248 | 0.11 | CHI3 L1 | 0.12 | GSTTP2 | 0.10 | RNU4-1 | 0.25 | GPAM | 0.45 | DBNDD1 | 0.33 |
| THBS1 | 0.12 | SFRP4 | 0.12 | FLJ40722 | 0.12 | RHBDL2 | 0.27 | TMSL3 | 0.46 | CRYGS | 0.33 |
| PDE7B | 0.12 | IER3 | 0.12 | KIAA1751 | 0.12 | KIAA1666 | 0.28 | CA12 | 0.34 | ||
| ANGPTL2 | 0.13 | C20orf186 | 0.13 | ZNF557 | 0.12 | FLJ46309 | 0.32 | CDC20 | 0.36 | ||
| ID1 | 0.13 | MIR1974 | 0.13 | IL17RD | 0.13 | FOXQ1 | 0.32 | TOP2A | 0.41 | ||
| MAT2A | 0.14 | MT3 | 0.13 | MIR886 | 0.13 | ARL16 | 0.33 | MATN4 | 0.41 | ||
| THBS2 | 0.15 | PTGS2 | 0.14 | FCAR | 0.13 | CAMK2 N1 | 0.35 | PTGS2 | 0.41 | ||
| NRP1 | 0.16 | STC1 | 0.15 | SLC5A8 | 0.14 | ZNF682 | 0.35 | ANGPTL4 | 0.42 | ||
| ID3 | 0.17 | C7orf68 | 0.15 | CYCSL1 | 0.14 | RNU6-1 | 0.36 | TAGLN | 0.44 | ||
| FASN | 0.17 | ANXA2P1 | 0.16 | ANKRD44 | 0.14 | RNU6-15 | 0.36 | MATN4 | 0.44 | ||
| KIAA1644 | 0.18 | SLC2A1 | 0.16 | XRCC2 | 0.14 | CYCSL1 | 0.37 | TYMS | 0.45 | ||
| PDGFRA | 0.18 | EFEMP1 | 0.16 | PRO1853 | 0.14 | FLJ40722 | 0.38 | KCNS1 | 0.46 | ||
| SFRS5 | 0.20 | LOX | 0.16 | SSTR2 | 0.15 | PNPT1 | 0.39 | PANX3 | 0.46 | ||
| STEAP1 | 0.20 | LOC387763 | 0.16 | CREB1 | 0.15 | RNU1G2 | 0.39 | BAPX1 | 0.46 | ||
| C14orf4 | 0.21 | HLA-DRA | 0.16 | DTWD2 | 0.15 | RNU1-5 | 0.39 | BEND5 | 0.46 | ||
| H3F3B | 0.22 | LOC730167 | 0.16 | DENR | 0.16 | RNU1-3 | 0.40 | SLC13A5 | 0.47 | ||
| SLC1A3 | 0.22 | PTGS2 | 0.17 | DEM1 | 0.16 | TMEM158 | 0.40 | PHLDA1 | 0.47 | ||
| EPDR1 | 0.23 | PPIB | 0.17 | LEP | 0.16 | CCBE1 | 0.41 | PRC1 | 0.47 | ||
Analysis of the filtered data showing changes in gene expression accumulated between each time point during the chondrogenic program. Significant up- and downregulated genes were analyzed for each time frame. Gene names in bold letters have or may have a role during chondrogenesis.
Upregulated genes such as interleukin 8 (IL-8), prostaglandin reductase 2, and proteoglycan 4 (Prg4) are highlighted (Table 3), which may have an important role during chondrogenesis and AC development.38–40 Downregulated genes may also be critical for the molecular definition of the new criteria or gold standard for AC tissue engineering. Rho guanine nucleotide exchange factor 3, which encodes a guanine nucleotide exchange factor that specifically activates members of the Rho GTPase family, has an important role in bone biology41; thrombospondin 1 that encodes for an adhesive glycoprotein that mediates cell-to-cell and cell-to-matrix interactions and is differently expressed during cartilage development is a potential new target for OA.42,43
Through the first week of chondrogenesis (d3 → d7), Col10a1 is upregulated and Prg4 is downregulated. These early changes appear to be the hallmark of the departure from the native AC lineage pathway. According to the analysis (Table 3), the time frame from d7 through d14 (d7 → d14) can be interpreted to mean that cells enter a stationary transcriptional phase characterized by a small number of upregulated genes (Table 3). Interestingly, the most chondrogenic-active phase (based on specific expression of cartilage markers) is found during the d14 → d21 time frame with significant upregulation of genes such as Mmp10, Col9a1, Col11a2, among others. Adding Fgf9 or Fgf18 at day 14 of the differentiation program, and not at the beginning (d0), produces a strong anabolic effect and a decrease in the hypertrophic molecular phenotype18; this argues that this phase of the chondrogenic program is the most responsive to exogenous stimuli and may be used as a target for designing novel and dynamic strategies to generate good-quality AC derived from hMSCs.
Analysis of the time-course data set shows that the most significant changes in gene expression occur from day 0 to 7 and, separately, from day 14 to 21 of the chondrogenic program. The changes in gene expression observed during the above time frames are consistent with the observation that cells undergo a transition from a state of high developmental plasticity to a state of determination, specification, and ultimately terminal differentiation.44 Moreover, the changes observed from day 14 to 21 are consistent with chondrogenic differentiation as many common chondrogenesis markers are overexpressed during this time frame (Table 3).
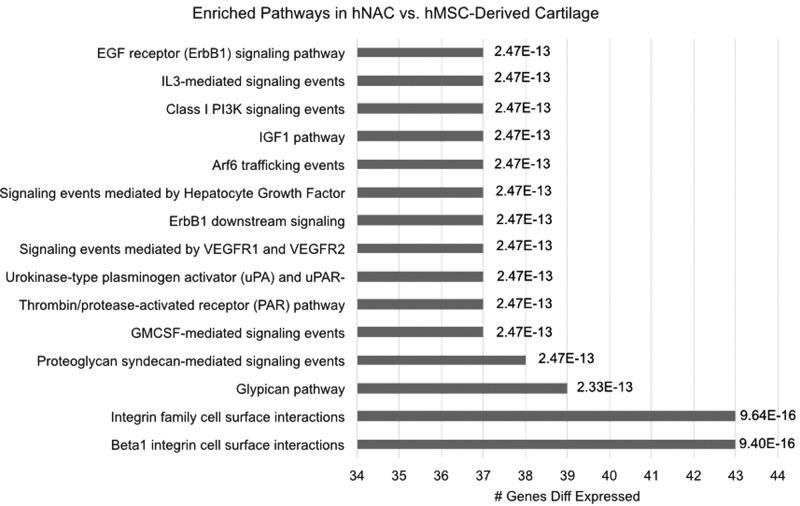
Key pathways are enriched in hNAC compared to hMSC-derived cartilage
To further dissect the significance of gene expression differences between hNAC and hMSC-derived 3D cartilage and to identify key biological pathways for AC formation in vivo, we performed pathway analysis using differentially expressed genes in hNAC compared to hMSC 3D chondrogenesis. Accordingly, 182 upregulated (>5-fold) and 191 downregulated (<0.024-fold) genes were included in this stringent analysis to identify key pathways. We found several pathways to be differentially enriched in hNAC compared to hMSC chondrogenesis (Fig. 8). Among the identified pathways, our analysis showed a strong enrichment of the integrin pathways in hNAC compared to hMSC-derived cartilage.
FIG. 8.
Pathway analysis of differentially expressed genes in hNAC versus hMSC-derived 3D cartilage. Pathway names are listed on the Y-axis, the percentage of genes affected in each pathway is indicated on the X-axis, and the multiple testing p-value is given at the end of each bar.
Discussion
A major milestone was achieved by Yoo et al. when hMSCs were successfully guided to enter the chondrogenic lineage with TGF-β.8,9 This differentiation capacity, which serves as the conceptual basis for several clinical treatments for AC defects, ultimately results in cartilage-like structures quite different from native AC in a number of parameters. Evidently, TGF-β is a required chondroinductive bioactive agent and is the only known effective inducer of the chondrogenic program in MSCs45; however, it is not sufficient and may not be adequate to induce an AC phenotype from hMSCs.15,46–48 For instance, it has been reported that TGF-β suppresses precartilage condensation.46
Chondrogenesis during embryonic development is a multistep, multicomponent, and dynamic process. In sharp contrast, the standard in vitro culture conditions for the differentiation of hMSCs into 3D cartilage as first described by Yoo et al.8 are unchanging with regard to exogenously added stimulatory factors. Only recently, it was shown that the sequential exposure of hMSCs to a particular set of fibroblast growth factors produces distinct effects depending on the stage at which they are added.18 For instance, by adding FGF9 during earlier time points of the differentiation program (day 0–7), a negative effect on differentiation and cartilage ECM deposition could be observed. On the contrary, adding FGF9 later at day 14 generated an anabolic effect on ECM production while delaying the appearance of a hypertrophic phenotype.18 These time-regulated effects were correlated with the expression profile of FGF-receptors 1 and 3 and their downstream intracellular signaling. In this regard, the details of the molecular transition within the AC chondrogenic program are of primary interest.
We propose that the ultimate goal for a successful tissue engineering approach to regenerate AC is to fabricate a cartilaginous tissue with similar characteristics as hNAC. In addition, the results of this study will be useful for further optimization of current chondrogenic differentiation protocols.
In this study, Illumina chips (representing >50,000 genes) were used to identify genes differentially expressed during hBM-MSC chondrogenesis. The analysis was performed with FGF2-expanded hMSCs (d0) and different time points during the chondrogenic differentiation program (d3–d28). The goal was to determine the prominent up- and downregulated genes between the various time points. The key comparator for these measurements was the data obtained with native hNAC (Tables 1 and 2). Gene expression clustering analysis, including other neonatal knee tissues (i.e., meniscus, synovial membrane, tendon, among others) from the same donor, allowed a comprehensive evaluation of differentially regulated genes across these tissues and compared them with in vitro hMSC-derived 3D cartilage structures. The assumption is that these differentially expressed genes could serve as the molecular signature or gold standard to guide the bioactive factor-assisted transcriptional profile of hMSC-derived cartilage.
As a first approximation to establish a molecular guide for these developmental transitions, we have chosen 1-month-old hNAC because it has the following characteristics: (i) cells are committed to an AC lineage; (ii) these cells must have a large expansion capacity and will expand to facilitate the formation of this tissue to reach the adult size; and finally, (iii) the instructions to bring them into a more mature phenotype must already exist, in part, as encoded in their transcriptional profile with its control elements (transcription factors) determining what the tissue and its functional identity will become (i.e., the gold standard). In this study, comprehensive gene expression profiling of hNAC and in vitro hBM-MSC-derived 3D cartilage tissue has been used to systematically investigate the presence of factors that are differentially expressed in both types of cartilage tissues.
Currently, only a handful of studies are available on the potential of juvenile chondrocytes and their comparison with adult chondrocytes. As far as we know, this is the first study using a hNAC tissue. Furthermore, the election of using hNAC as a reference standard builds on previous reports showing greater intrinsic capability of juvenile chondrocytes to expand, produce ECM, upregulate cartilage markers, and secrete paracrine factors that stimulate stem cell function.27,49–52 Taken together, the properties of neonatal chondrocytes may provide a more efficient way to generate functional cartilage compared to adult cells for the generation of tissue-engineered cartilage by recapitulating the natural development and growth processes. Although multiple studies have documented the potential of juvenile chondrocytes, the molecular basis of such regeneration has not been explored. Part of the challenge for the lack of such studies is the difficulty in obtaining healthy human cartilage especially from young donors.
Previous studies have shown that hMSC transcriptome is significantly different from that of native chondrocytes.53 In this regard, the question can be asked: are hMSCs intrinsically restricted from differentiating into hyaline AC-producing chondrocytes? One assessment strategy is to use a molecular snapshot of hNAC as a principal means of comparison. In this study, this approach was validated as a criteria or gold standard molecular read-out for high-quality AC. Importantly, we confirmed that the presence of standard markers (Sox9/8, Col2/9/10, and ACAN) is not sufficient to define implantation release criteria, as they also appear in failed engineered cartilage constructs. Therefore, these markers are necessary but not sufficient to predict ultimate success or failure of tissue-engineered cartilage. It has been shown that expression of Sox9, both at the mRNA and protein levels, is not a predictor of successful chondrogenesis.54 Thus, the expression of other control elements is needed to assess the quality of tissue-engineered cartilage.
The analysis performed here provided a screened list of novel markers and control elements that are presumed to have a critical role during the generation of native AC. Molecular differences between hNAC and hBM-MSC-derived 3D cartilage at various time points (d0–d28) were identified, focusing on genes not expressed at any time in vitro hBM-MSC-derived cartilage. Based on this analysis, specific gene “signatures” that hMSC must acquire during differentiation to form AC that has, at least, a molecular signature of native AC were recognized. From a list of ∼500 genes differentially expressed, 53 genes were identified in hNAC that were not expressed at any time point during the hMSC chondrogenic program (d3–d28) or in undifferentiated FGF2-expanded hMSCs (d0) and in other neonatal knee tissues.
A set of genes not previously described as chondrogenic markers was also identified, suggesting that additional novel markers that have not been implicated in chondrogenic differentiation exist and deserve additional investigation. Moreover, nine genes (five transcription factors and four structural proteins) were identified as prospective molecular signature of AC phenotype based on fold changes between AC and hMSC-derived pellets and their associated function in chondrogenesis. Kubo et al. found that the molecular signature of BM-derived hMSC is determined by transcription factors that are expressed selectively in these cells in contrast to other cells such as fibroblasts.55 In agreement with this study, transcription factors such as PR domain zinc finger protein 16 and Ets variant 5) are expressed by FGF2-expanded hMSCs and, according to our data, are highly expressed in hNAC. However hMSCs rapidly lose the expression of these factors during in vitro chondrogenesis.
The presented data also indicate that hMSC transcriptome is more similar to the hNAC transcriptome in the first days of in vitro chondrogenic differentiation (d3–d7). This suggests that the osteochondral fate of hMSC-derived cartilage may be rerouted during earlier phases of the chondrogenic program. It has been reported that progenitor cells in the superficial layer of human AC exist and that these cells express Prg4.11,12 Interestingly, we found that Prg4 is highly expressed at day 3 of the in vitro MSC chondrogenic program. Given this observation and based on our biostatistical analysis (Fig. 2), it can be suggested that the transcriptome of hMSC-derived cartilage at day 3 is similar to that of AC progenitors in hNAC. However, this is something that needs to be further studied.
Pathway analysis showed a strong enrichment of the integrin pathways in hNAC, compared to hMSC-derived 3D cartilage. Although the molecular mechanisms of integrins in articular chondrocytes is not yet completely understood, studies have demonstrated that cell signaling mediated by integrins regulates several key chondrocyte functions, including differentiation, matrix production and remodeling, mechanical stimulation, and cell survival.56 In particular, β1 integrin-mediated adhesion plays crucial roles in cartilage morphology and function regulating chondrocyte physiology and ECM assembly.57,58
According to our data, the regulation of integrin pathways may be a key factor in the generation of AC from hMSC chondrogenesis. It is important to consider that this regulation must be strictly controlled since integrin pathway deregulation is likewise involved in OA.56 It has been shown that the collagen binding α1β1 integrin is expressed at a low level on chondrocytes of the normal AC, but its expression is upregulated during OA.56 The epidermal growth factor receptor (EGFR) signaling pathway activation has been shown to produce an anabolic effect in AC that is followed by an accelerated catabolic effect if sustained activation is present.59 This suggests that this pathway may be initially active during AC development and then downregulated in developed AC. Chondrocyte-specific EGFR signaling is an important regulator of hypertrophy and growth plate development.60 Since this pathway is upregulated in hNAC according to our data, its involvement during MSC chondrogenesis may shed light into hypertrophy and cartilage degradation mechanisms.
Recently, it was reported that IL-3-mediated signaling, which is also found upregulated in our analysis, upregulates the expression of Sox9 and collagen type II and downregulates the expression of matrix-degrading enzymes such as MMP-1, MMP-3, and MMP-13 in chondrocytes.61 The insulin growth factor I pathway has been shown to play multiple roles in cartilage development such as differentiation and maintenance of articular chondrocytes and regulation of cartilage ECM synthesis.62,63 Signaling by hepatocyte growth factor has been shown to be important in the regulation of chondrogenesis and endochondral ossification events in mouse64,65; however, its specific role in cartilage development and chondrogenic differentiation is understudied.
The vascular endothelial growth factor (VEGF) pathway has been extensively studied in relation to its role in angiogenesis; however, its role in chondrogenesis and AC development has not been fully elucidated. VEGF and its receptors have been found to play a role in OA.66 Interestingly, one study found that VEGF and its receptors are expressed in osteoarthritic and growing AC, but not in healthy adult AC.67 Syndecan and glypican pathways have been shown to be involved in cartilage remodeling and pathological processes.68 However, their specific roles in AC development are still not well understood.
The study of the pathways identified in this study is the subject of further investigational efforts in our laboratories to test the various conclusions of criteria uncovered by this transcriptome analysis.
Conclusions
In this report, a specific molecular phenotype of high-quality AC is described as a gold standard for tissue-engineered AC that could fulfill the high standards that are required for a successful clinical application. These results will enable a clear molecular roadmap to generate proper articular hyaline cartilage from a variety of stem cell and reprogramming technologies. In addition, they will be extremely valuable in setting the read-out to generate robust hyaline AC phenotypes after successful multifactor-directed lineage progression following temporal and molecular induction. This will allow a consistent method to identify and control the development of AC phenotypes during lineage progression in vitro.
We expect that these results will provide sufficient output data to help develop the new technology required to fulfill the expectations that adult human BM-derived hMSCs can be used in culture to engineer a chondrogenic tissue with properties optimized to regenerate hyaline AC in situ. In addition, these data will be useful to identify a comprehensive read-out of a successful AC generation in vitro using hMSCs.
Acknowledgments
We thank AlloSource (Centennial, CO) for their support of our research through tissue donation and sincerely thank the families who have so selflessly made these donations possible. We thank Amad Awadallah for the histological processing of the samples and optimizing the immunostaining procedures, Donald Lennon for critical reading of the manuscript, Margie Harris for hMSC preparations, and L. David and E. Virginia Baldwin Fund and NIH/NIBIB grants 1R01EB020367 and 1P41EB021911 for support.
Disclosure Statement
No competing financial interests exist with the CWRU authors. All the authors from BioTime group receive salary and other benefits from BioTime.
References
- 1.Bornes T.D., Adesida A.B., and Jomha N.M. Mesenchymal stem cells in the treatment of traumatic articular cartilage defects: a comprehensive review. Arthritis Res Ther 16, 432, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu C.R., Szczodry M., and Bruno S. Animal models for cartilage regeneration and repair. Tissue Eng Part B Rev 16, 105, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makris E.A., Gomoll A.H., Malizos K.N., Hu J.C., and Athanasiou K.A. Repair and tissue engineering techniques for articular cartilage. Nat Rev Rheumatol 11, 21, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei X., Yang X., Han Z.P., Qu F.F., Shao L., and Shi Y.F. Mesenchymal stem cells: a new trend for cell therapy. Acta Pharmacol Sin 34, 747, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caplan A.I. Mesenchymal stem cells. J Orthop Res 9, 641, 1991 [DOI] [PubMed] [Google Scholar]
- 6.Caplan A.I. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J Cell Physiol 213, 341, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Marion N.W., and Mao J.J. Mesenchymal stem cells and tissue engineering. Methods Enzymol 420, 339, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo J.U., Barthel T.S., Nishimura K., Solchaga L., Caplan A.I., Goldberg V.M., and Johnstone B. The chondrogenic potential of human bone-marrow-derived mesenchymal progenitor cells. J Bone Joint Surg Am 80, 1745, 1998 [DOI] [PubMed] [Google Scholar]
- 9.Johnstone B., Hering T.M., Caplan A.I., Goldberg V.M., and Yoo J.U. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res 238, 265, 1998 [DOI] [PubMed] [Google Scholar]
- 10.Khan I.M., Redman S.N., Williams R., Dowthwaite G.P., Oldfield S.F., and Archer C.W. The development of synovial joints. Curr Top Dev Biol 79, 1, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Caldwell K.L., and Wang J. Cell-based articular cartilage repair: the link between development and regeneration. Osteoarthritis Cartilage 23, 351, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller M.B., and Tuan R.S. Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum 58, 1377, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelttari K., Winter A., Steck E., Goetzke K., Hennig T., Ochs B.G., Aigner T., and Richter W. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum 54, 3254, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Steck E., Fischer J., Lorenz H., Gotterbarm T., Jung M., and Richter W. Mesenchymal stem cell differentiation in an experimental cartilage defect: restriction of hypertrophy to bone-close neocartilage. Stem Cells Dev 18, 969, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Somoza R.A., Welter J.F., Correa D., and Caplan A.I. Chondrogenic differentiation of mesenchymal stem cells: challenges and unfulfilled expectations. Tissue Eng Part B Rev 20, 596, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Kraan P.M., and van den Berg W.B. Chondrocyte hypertrophy and osteoarthritis: role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage 20, 223, 2012 [DOI] [PubMed] [Google Scholar]
- 17.Hellingman C.A., Koevoet W., and van Osch G.J.V.M. Can one generate stable hyaline cartilage from adult mesenchymal stem cells? A developmental approach. J Tissue Eng Regen Med 6, e1, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Correa D., Somoza R.A., Lin P., Greenberg S., Rom E., Duesler L., Welter J.F., Yayon A., and Caplan A.I. Sequential exposure to fibroblast growth factors (FGF) 2, 9 and 18 enhances hMSC chondrogenic differentiation. Osteoarthritis Cartilage 23, 443, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L., Bluguermann C., Kyupelyan L., Latour B., Gonzalez S., Shah S., Galic Z., Ge S.D., Zhu Y.H., Petrigliano F.A., Nsair A., Miriuka S.G., Li X.M., Lyons K.M., Crooks G.M., McAllister D.R., Van Handel B., Adams J.S., and Evseenko D. Human developmental chondrogenesis as a basis for engineering chondrocytes from pluripotent stem cells. Stem Cell Rep 1, 575, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitsillides A.A., and Beier F. Cartilage biology in osteoarthritis-lessons from developmental biology. Nat Rev Rheumatol 7, 654, 2011. (Erratum: Nat Rev Rheumatol 9, 64, 2013) [DOI] [PubMed] [Google Scholar]
- 21.Gadjanski I., Spiller K., and Vunjak-Novakovic G. Time-dependent processes in stem cell-based tissue engineering of articular cartilage. Stem Cell Rev Rep 8, 863, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centola M., Tonnarelli B., Scharen S., Glaser N., Barbero A., and Martin I. Priming 3D cultures of human mesenchymal stromal cells toward cartilage formation via developmental pathways. Stem Cells Dev 22, 2849, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenas P., Moos M., and Luyten F.P. Developmental engineering: a new paradigm for the design and manufacturing of cell-based products. Part II: from genes to networks: tissue engineering from the viewpoint of systems biology and network science. Tissue Eng Part B Rev 15, 395, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Ingber D.E., Mow V.C., Butler D., Niklason L., Huard J., Mao J., Yannas I., Kaplan D., and Vunjak-Novakovic G. Tissue engineering and developmental biology: going biomimetic. Tissue Eng 12, 3265, 2006 [DOI] [PubMed] [Google Scholar]
- 25.Adkisson H.D., Gillis M.P., Davis E.C., Maloney W., and Hruska K.A. In vitro generation of scaffold independent neocartilage. Clin Orthop Relat Res S280, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Adkisson H.D., Milliman C., Zhang X., Mauch K., Maziarz R.T., and Streeter P.R. Immune evasion by neocartilage-derived chondrocytes: implications for biologic repair of joint articular cartilage. Stem Cell Res 4, 57, 2010 [DOI] [PubMed] [Google Scholar]
- 27.Adkisson H.D.t., Martin J.A., Amendola R.L., Milliman C., Mauch K.A., Katwal A.B., Seyedin M., Amendola A., Streeter P.R., and Buckwalter J.A. The potential of human allogeneic juvenile chondrocytes for restoration of articular cartilage. Am J Sports Med 38, 1324, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai J.H., Kajiyama G., Smith R.L., Maloney W., and Yang F. Stem cells catalyze cartilage formation by neonatal articular chondrocytes in 3D biomimetic hydrogels. Sci Rep 3, 3553, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennon D.P., and Caplan A.I. Isolation of human marrow-derived mesenchymal stem cells. Exp Hematol 34, 1604, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Lennon D.P., Haynesworth S.E., Bruder S.P., Jaiswal N., and Caplan A.I. Human and animal mesenchymal progenitor cells from bone marrow: identification of serum for optimal selection and proliferation. In Vitro Cell Dev Anim 32, 602, 1996 [Google Scholar]
- 31.Tusher V.G., Tibshirani R., and Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98, 5116, 2001. (Erratum: Proc Natl Acad Sci U S A 98, 10515, 2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B., Kirov S., and Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33, W741, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringner M. What is principal component analysis? Nat Biotechnol 26, 303, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Lotz M.K., Otsuki S., Grogan S.P., Sah R., Terkeltaub R., and D'Lima D. Cartilage cell clusters. Arthritis Rheum 62, 2206, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X.M., Birk D.E., and Goetinck P.F. Mice lacking matrilin-1 (cartilage matrix protein) have alterations in type II collagen fibrillogenesis and fibril organization. Dev Dyn 216, 434, 1999 [DOI] [PubMed] [Google Scholar]
- 36.Surmann-Schmitt C., Dietz U., Kireva T., Adam N., Park J., Tagariello A., Oennerfjord P., Heinegard D., Schlotzer-Schrehardt U., Deutzmann R., von der Mark K., and Stock M. Ucma, a novel secreted cartilage-specific protein with implications in osteogenesis. J Biol Chem 283, 7082, 2008 [DOI] [PubMed] [Google Scholar]
- 37.van Gool S.A., Emons J.A., Leijten J.C., Decker E., Sticht C., van Houwelingen J.C., Goeman J.J., Kleijburg C., Scherjon S.A., Gretz N., Wit J.M., Rappold G., Post J.N., and Karperien M. Fetal mesenchymal stromal cells differentiating towards chondrocytes acquire a gene expression profile resembling human growth plate cartilage. PLoS One 7, e44561, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caron M.M., Emans P.J., Sanen K., Surtel D.A., Cremers A., Ophelders D., van Rhijn L.W., and Welting T.J. The role of prostaglandins and COX-enzymes in chondrogenic differentiation of ATDC5 progenitor cells. PLoS One 11, e0153162, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozhemyakina E., Zhang M., Ionescu A., Ayturk U.M., Ono N., Kobayashi A., Kronenberg H., Warman M.L., and Lassar A.B. Identification of a Prg4-expressing articular cartilage progenitor cell population in mice. Arthritis Rheumatol 67, 1261, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon D.S., Lee K.M., Kim S.H., Kim S.H., Jung Y., Kim S.H., Park K.H., Choi Y., Ryu H.A., Choi W.J., and Lee J.W. Synergistic action of IL-8 and bone marrow concentrate on cartilage regeneration through upregulation of chondrogenic transcription factors. Tissue Eng Part A 22, 363, 2016 [DOI] [PubMed] [Google Scholar]
- 41.Mullin B.H., Mamotte C., Prince R.L., and Wilson S.G. Influence of ARHGEF3 and RHOA knockdown on ACTA2 and other genes in osteoblasts and osteoclasts. PLoS One 9, e98116, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson R., Norris E.L., Brachvogel B., Angelucci C., Zivkovic S., Gordon L., Bernardo B.C., Stermann J., Sekiguchi K., Gorman J.J., and Bateman J.F. Changes in the chondrocyte and extracellular matrix proteome during post-natal mouse cartilage development. Mol Cell Proteomics 11, M111.014159, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calamia V., Mateos J., Fernandez-Puente P., Lourido L., Rocha B., Fernandez-Costa C., Montell E., Verges J., Ruiz-Romero C., and Blanco F.J. A pharmacoproteomic study confirms the synergistic effect of chondroitin sulfate and glucosamine. Sci Rep 4, 5069, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James C.G., Appleton C.T., Ulici V., Underhill T.M., and Beier F. Microarray analyses of gene expression during chondrocyte differentiation identifies novel regulators of hypertrophy. Mol Biol Cell 16, 5316, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diederichs S., Zachert K., Raiss P., and Richter W. Regulating chondrogenesis of human mesenchymal stromal cells with a retinoic acid receptor-beta inhibitor: differential sensitivity of chondral versus osteochondral development. Cell Physiol Biochem 33, 1607, 2014 [DOI] [PubMed] [Google Scholar]
- 46.Jin E.J., Park K.S., Kim D., Lee Y.S., Sonn J.K., Jung J.C., Bang O.S., and Kang S.S. TGF-beta3 inhibits chondrogenesis by suppressing precartilage condensation through stimulation of N-cadherin shedding and reduction of cRREB-1 expression. Mol Cells 29, 425, 2010 [DOI] [PubMed] [Google Scholar]
- 47.Fang J., Xu L., Li Y., and Zhao Z. Roles of TGF-beta 1 signaling in the development of osteoarthritis. Histol Histopathol 31, 1161, 2016 [DOI] [PubMed] [Google Scholar]
- 48.Shen J., Li S., and Chen D. TGF-beta signaling and the development of osteoarthritis. Bone Res 2, pii: , 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acosta F.L., Jr., Metz L., Adkisson H.D., Liu J., Carruthers-Liebenberg E., Milliman C., Maloney M., and Lotz J.C. Porcine intervertebral disc repair using allogeneic juvenile articular chondrocytes or mesenchymal stem cells. Tissue Eng Part A 17, 3045, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonasia D.E., Martin J.A., Marmotti A., Amendola R.L., Buckwalter J.A., Rossi R., Blonna D., Adkisson H.D.T., and Amendola A. Cocultures of adult and juvenile chondrocytes compared with adult and juvenile chondral fragments: in vitro matrix production. Am J Sports Med 39, 2355, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smeriglio P., Lai J.H., Dhulipala L., Behn A.W., Goodman S.B., Smith R.L., Maloney W.J., Yang F., and Bhutani N. Comparative potential of juvenile and adult human articular chondrocytes for cartilage tissue formation in three-dimensional biomimetic hydrogels. Tissue Eng Part A 21, 147, 2015 [DOI] [PubMed] [Google Scholar]
- 52.Taylor S.E., Lee J., Smeriglio P., Razzaque A., Smith R.L., Dragoo J.L., Maloney W.J., and Bhutani N. Identification of human juvenile chondrocyte-specific factors that stimulate stem cell growth. Tissue Eng Part A 22, 645, 2016 [DOI] [PubMed] [Google Scholar]
- 53.Zhai L.J., Zhao K.Q., Wang Z.Q., Feng Y., and Xing S.C. Mesenchymal stem cells display different gene expression profiles compared to hyaline and elastic chondrocytes. Int J Clin Exp Med 4, 81, 2011 [PMC free article] [PubMed] [Google Scholar]
- 54.Diederichs S., Gabler J., Autenrieth J., Kynast K.L., Merle C., Walles H., Utikal J., and Richter W. Differential regulation of SOX9 protein during chondrogenesis of induced pluripotent stem cells versus mesenchymal stromal cells: a shortcoming for cartilage formation. Stem Cells Dev 25, 598, 2016 [DOI] [PubMed] [Google Scholar]
- 55.Kubo H., Shimizu M., Taya Y., Kawamoto T., Michida M., Kaneko E., Igarashi A., Nishimura M., Segoshi K., Shimazu Y., Tsuji K., Aoba T., and Kato Y. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry. Genes Cells 14, 407, 2009 [DOI] [PubMed] [Google Scholar]
- 56.Loeser R.F. Integrins and cell signaling in chondrocytes. Biorheology 39, 119, 2002 [PubMed] [Google Scholar]
- 57.Gao Y., Liu S.Y., Huang J.X., Guo W.M., Chen J.F., Zhang L., Zhao B., Peng J., Wang A.Y., Wang Y., Xu W.J., Lu S.B., Yuan M., and Guo Q.Y. The ECM-cell interaction of cartilage extracellular matrix on chondrocytes. Biomed Res Int 648459, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L.Q., Zhao G.Z., Xu X.Y., Fang J., Chen J.M., Li J.W., Gao X.J., Hao L.J., and Chen Y.Z. Integrin-beta 1 regulates chondrocyte proliferation and apoptosis through the upregulation of GIT1 expression. Int J Mol Med 35, 1074, 2015 [DOI] [PubMed] [Google Scholar]
- 59.Shepard J.B., Jeong J.W., Maihle N.J., O'Brien S., and Dealy C.N. Transient anabolic effects accompany epidermal growth factor receptor signal activation in articular cartilage in vivo. Arthritis Res Ther 15, R60, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X., Zhu J., Li Y., Lin T., Siclari V.A., Chandra A., Candela E.M., Koyama E., Enomoto-Iwamoto M., and Qin L. Epidermal growth factor receptor (EGFR) signaling regulates epiphyseal cartilage development through beta-catenin-dependent and -independent pathways. J Biol Chem 288, 32229, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kour S., Garimella M.G., Shiroor D.A., Mhaske S.T., Joshi S.R., Singh K., Pal S., Mittal M., Krishnan H.B., Chattopadhyay N., Ulemale A.H., and Wani M.R. IL-3 decreases cartilage degeneration by downregulating matrix metalloproteinases and reduces joint destruction in osteoarthritic mice. J Immunol 196, 5024, 2016 [DOI] [PubMed] [Google Scholar]
- 62.Martin J.A., Scherb M.B., Lembke L.A., and Buckwalter J.A. Damage control mechanisms in articular cartilage: the role of the insulin-like growth factor I axis. Iowa Orthop J 20, 1, 2000 [PMC free article] [PubMed] [Google Scholar]
- 63.Oh C.D., and Chun J.S. Signaling mechanisms leading to the regulation of differentiation and apoptosis of articular chondrocytes by insulin-like growth factor-1. J Biol Chem 278, 36563, 2003 [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi A., Amano O., Tani Y., Nakamura T., Iseki S., and Tomita K. Hepatocyte growth factor regulates the proliferation and differentiation of cartilage in developing forelimb of mouse embryos in vitro. Biomed Res Tokyo 25, 219, 2004 [Google Scholar]
- 65.Amano O., Koshimizu U., Nakamura T., and Iseki S. Enhancement by hepatocyte growth factor of bone and cartilage formation during embryonic mouse mandibular development in vitro. Arch Oral Biol 44, 935, 1999 [DOI] [PubMed] [Google Scholar]
- 66.Hamilton J.L., Nagao M., Levine B.R., Chen D., Olsen B.R., and Im H.J. Targeting VEGF and its receptors for the treatment of osteoarthritis and associated pain. J Bone Miner Res 31, 911, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lingaraj K., Poh C.K., and Wang W. Vascular endothelial growth factor (VEGF) is expressed during articular cartilage growth and re-expressed in osteoarthritis. Ann Acad Med Singapore 39, 399, 2010 [PubMed] [Google Scholar]
- 68.Pap T., and Bertrand J. Syndecans in cartilage breakdown and synovial inflammation. Nat Rev Rheumatol 9, 43, 2013 [DOI] [PubMed] [Google Scholar]