Abstract
For the first time, molecular phylogenetic data on the peculiar diaporthalean genus Caudospora are available. Macro- and microscopic morphology and phylogenetic multilocus analyses of partial nuc SSU-ITS-LSU rDNA, cal, ms204, rpb1, rpb2, tef1 and tub2 sequences revealed two distinct species of Caudospora, which are described and illustrated by light and scanning electron microscopy. Caudospora iranica is described as a new species from corticated dead twigs of Quercus sp. collected in Iran. It differs from the generic type, C. taleola, mainly by coarsely verrucose ascospores. The asexual morph of C. taleola on natural substrate is described and illustrated. Caudospora taleola is neotypified, and it is recorded from Iran for the first time. Phylogenetic analyses of a multigene matrix containing a representative selection of Diaporthales from four loci (ITS, LSU rDNA, rpb2 and tef1) revealed a placement of Caudospora within Sydowiellaceae.
Keywords: Ascomycota, Diaporthales, new species, phylogenetic analysis, Sydowiellaceae, taxonomy
Starbäck (1889) introduced the genus Caudospora for a single peculiar species, C. taleola, which is characterized by perithecia immersed in a brown corticolous pseudostroma delimited by a distinct blackish zone, a whitish ectostromatic disc bearing few converging dark ostioles, cylindrical asci and hyaline, two-celled ascospores with an appendage at each end and 2–3 median appendages. The single known species, C. taleola, is confined to various species of Quercus spp., where it is widely distributed in Europe and North America (Rogers 1984). Ecologically, C. taleola is a saprotroph or weak pathogen of oaks, which can cause canker diseases (Phillips & Burdekin 1992).
Whereas morphological features clearly place C. taleola within Diaporthales, its generic as well as familial affiliation within the order remained controversial. Munk (1957) provided a detailed morphological description, noted that the genus is very distinct and found a high similarity of its centrum to that of Sydowiella fenestrans (Sydowiellaceae). Wehmeyer (1933) included Caudospora in the genus Diaporthe, whereas Müller & Arx (1962) transferred it to Hercospora. This generic classification was accepted by Kobayashi (1970) and Barr (1978). Rogers (1984) re-examined C. taleola from natural substrate and characterized the anamorph in pure culture. He concluded that Caudospora should not be considered synonymous with Hercospora because of its verrucose ascospores and appendages. Moreover, the anamorph of H. tiliae, the type species of Hercospora, is rabenhorstia-like whereas that of C. taleola is phomopsis-like. Rogers (1984) concluded that C. taleola is related to certain species of Diaporthe (D. leiphaemia var. raveneliana) based on the verrucose ascospores and anamorph. However, this connection has never been proven in molecular studies, and it is also doubtful whether D. leiphaemia var. raveneliana really belongs to Diaporthe. Until now, in the lack of sequence data the phylogenetic position of Caudospora within Diaporthales is uncertain.
During a survey of diatrypaceous fungi in Iran, a collection resembling Caudospora was revealed from dead corticated twigs of Quercus sp., which markedly differed from C. taleola in coarsely verrucose ascospores. Several fresh collections of C. taleola from Austria, France and Iran provided the opportunity for pure culture isolation and sequencing to clarify the phylogenetic affinities of Caudospora and the species status of the Iranian collection with verrucose ascospores, the results of which are here presented.
Materials and methods
Isolates and specimens
All isolates used in this study originated from ascospores of fresh specimens. Numbers of strains including NCBI GenBank accession numbers of gene sequences generated in the present study are listed in Tables 1 and 2. Strain acronyms other than those of official culture collections are used here primarily as strain identifiers throughout the work. Details of the specimens used for morphological investigations are listed in the Taxonomy section under the respective descriptions. In addition, the following strain of Hapalocystis berkeleyi was sequenced as outgroup taxon: UK, England, London, Royal Botanic Gardens Kew (RGB), on corticated branches of Platanus ×hispanica, 11 November 2008, H. Voglmayr D84 (WU 39959, living culture CBS 124568). Isolates were deposited in the Iranian Fungal Culture Collection of the Iranian Research Institute of Plant Protection (IRAN…C) and at the Westerdijk Fungal Biodiversity Centre, Utrecht, The Netherlands (CBS culture collection), and dry specimens in the Herbarium of Iranian Research Institute of Plant Protection (IRAN…F) and the Fungarium of the Department of Botany and Biodiversity Research, University of Vienna (WU).
Tab. 1.
Strains and NCBI GenBank accessions used in the phylogenetic analyses of the combined SSU-ITS-LSU-cal-ms204-rpb1-rpb2-tef1-tub1 matrix. All sequences were generated during the present study.
| Taxon | Strain | Voucher | Country | Host/substrate | GenBank accession numbers |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSU-ITS-LSU | cal | ms204 | rpb1 | rpb2 | tef1 | tub2 | |||||
| Caudospora iranica | D189 = IRAN 2552 C = CBS 143507 | IRAN 16716 F, WU 39950 | Iran | Quercus sp. | MG495960 | MG495951 | MG495970 | MG495979 | MG495988 | MG495997 | MG496004 |
| C. taleola | D185 = CBS 143508 | WU 39951 | Austria | Quercus robur | MG495961 | MG495952 | MG495971 | MG495980 | MG495989 | MG495998 | MG496005 |
| C. taleola | D186 | WU 39952 | Austria | Quercus robur | MG495962 | MG495953 | MG495972 | MG495981 | MG495990 | MG495999 | MG496006 |
| C. taleola | D187 | WU 39953 | Austria | Quercus robur | MG495963 | MG495954 | MG495973 | MG495982 | MG495991 | MG496000 | MG496007 |
| C. taleola | D188 = IRAN 2551 C | IRAN 16715 F, WU 39954 | Iran | Quercus sp. | MG495964 | MG495955 | MG495974 | MG495983 | MG495992 | MG496001 | MG496008 |
| C. taleola | D197 | WU 39955 | France | Quercus robur | MG495965 | MG495956 | MG495975 | MG495984 | MG495993 | MG496002 | MG496009 |
| C. taleola | D198 | WU 39956 | France | Quercus robur | MG495966 | MG495957 | MG495976 | MG495985 | MG495994 | – | MG496010 |
| C. taleola | D216 | WU 39957 | Austria | Quercus petraea | MG495967 | MG495958 | MG495977 | MG495986 | MG495995 | – | MG496011 |
| Hapalocystis berkeleyi | D84 = CBS 124568 | WU 39959 | UK | Platanus ×hispanica | MG495968 | MG495959 | MG495978 | MG495987 | MG495996 | MG496003 | MG496012 |
Tab. 2.
Strains and NCBI GenBank accessions used in the phylogenetic analyses of the combined ITS-LSU-rpb2-tef1 matrix. Sequences in bold were generated during the present study.
| Taxon | Strain | Voucher | Host/substrate | GenBank accession numbers |
|||
|---|---|---|---|---|---|---|---|
| ITS | LSU | rpb2 | tef1 | ||||
| Alnecium auctum | CBS 124263 | WU 30206 | Alnus glutinosa | KF570154 | KF570154 | KF570170 | KF570200 |
| Ambarignomonia petiolorum | CBS 121227 | BPI 844274 | Liquidambar styraciflua | EU254748 | EU255070 | EU219307 | EU221898 |
| Amphiporthe hranicensis | CBS 119289 | BPI 843515 | Tilia platyphyllos | EU199178 | EU199122 | EU199137 | – |
| Apiognomonia errabunda | CBS 109747 | AR 2813 | Fagus sylvatica | DQ313525 | NG_027592 | DQ862014 | DQ313565 |
| Apoharknessia insueta | CBS 111377 | CPC 1451 | Eucalyptus pellita | JQ706083 | AY720814 | – | – |
| Asterosporium asterospermum | CBS 112404 | – | Fagus sylvatica | – | AB553745 | – | – |
| Auratiopycnidiella tristaniopsidis | CBS 132180 | CBS H-20932 | Tristaniopsis laurina | JQ685516 | JQ685522 | – | – |
| Cainiella johansonii | Kruys 731 | UPS F-567263 | Dryas octopetala | JF701922 | JF701920 | – | – |
| Caudospora iranica | D189 = IRAN 2552 C | IRAN 16716 F | Quercus sp. | MG495960 | MG495960 | MG495988 | MG495997 |
| Caudospora taleola | D185 | WU 39951 | Quercus robur | MG495961 | MG495961 | MG495989 | MG495998 |
| Caudospora taleola | D188 = IRAN 2551 C | IRAN 16715 F | Quercus sp. | MG495964 | MG495964 | MG495992 | MG496001 |
| Calosporella innesii | AR 3639 | BPI 840945 | Acer pseudoplatanus | JF681965 | EU683071 | – | – |
| Celoporthe dispersa | CBS 118781 | PREM 58897 | Syzygium cordatum | AY214316 | HQ730854 | – | HQ730841 |
| Chapeckia nigrospora | CBS 125532 | BPI 863766 | Betula sp. | JF681957 | EU683068 | – | – |
| Chrysocrypta corymbiae | CBS 132528 | CPC 19279 | Corymbia sp. | JX069867 | JX069851 | – | – |
| Chrysoporthe cubensis | CBS 118654 | PREM 58788 | Eucalyptus sp. | JN942342 | JN940856 | – | GQ290137 |
| Coniella fragariae | CBS 172.49 | CBS H-10697 | Forest soil | AY339317 | AY339282 | KX833472 | KX833663 |
| Coniella obovata | CBS 111025 | CBS H-22703 | Leaf litter | AY339313 | KX833409 | KX833497 | KX833692 |
| Coniella straminea | CBS 149.22 | STE U 3932 | Fragaria sp. | AY339348 | AY339296 | KX833506 | KX833704 |
| Coryneum depressum | AR 3897 | BPI 843585 | Quercus cerris | – | EU683074 | – | – |
| Coryneum modonium | AR 3558 | BPI 749131 | Castanea sativa | – | EU683073 | – | – |
| Coryneum umbonatum | AR 3541 | BPI 872021 | Quercus cerris | – | EU683072 | – | – |
| Crinitospora pulchra | CBS 138014 | CBS H-21729 | Mangifera indica | KJ710466 | KJ710443 | – | – |
| Cryphonectria parasitica | ATCC 38755 | – | Castanea dentata | AY141856 | EU199123 | – | EU222014 |
| Cryptodiaporthe aesculi | CBS 109765 | BPI 748430 | Aesculus hippocastanum | EU199179 | AF408342 | EU199138 | – |
| Cryptosporella hypodermia | CBS 116866 | BPI 748432 | Ulmus minor | EU199181 | AF408346 | EU199140 | – |
| Cytospora chrysosperma | CFCC 89630 | BJFC-S978 | Salix psammophila | KF765674 | KF765690 | KF765706 | KP321972 |
| Cytospora hippophaes | CFCC 89640 | – | Hippophae rhamnoides | KF765682 | KF765698 | KF765714 | KP310865 |
| Cytospora nivea | CFCC 89643 | – | Salix psammophila | KF765685 | KF765701 | KF765717 | KP310863 |
| Diaporthe eres | CBS 138594 | BPI 892912 | Ulmus sp. | KJ210529 | – | – | KJ210550 |
| Discula destructiva | CBS 109771 | BPI 1107757 | Cornus nuttallii | EU199186 | AF408359 | EU199144 | – |
| Disculoides eucalypti | CBS 132183 | CBS H-20935 | Eucalyptus sp. | JQ685517 | JQ685523 | – | – |
| Ditopella ditopa | CBS 109748 | BPI 748439 | Alnus glutinosa | DQ323526 | EU199126 | EU199145 | – |
| Endothia gyrosa | CBS 112915 | – | Quercus palustris | AF046905 | AY194114 | – | – |
| Erythrogloeum hymenaeae | CBS 132185 | CBS H-20937 | Hymenaea courbaril | JQ685519 | JQ685525 | – | – |
| Gaeumannomyces graminis | CBS 235.32 | – | Oryza sativa | JX134669 | JX134681 | – | JX134695 |
| Gnomonia gnomon | CBS 199.53 | – | Corylus avellana | DQ491518 | AF408361 | EU219295 | EU221885 |
| Gnomoniopsis racemula | CBS 121469 | BPI 871003 | Epilobium angustifolium | EU254841 | EU255122 | EU219241 | EU221889 |
| Greeneria uvicola | FI1 2007 | – | Vitis sp. | HQ586009 | GQ870619 | – | – |
| Hapalocystis berkeleyi | CBS 124568 | WU 39959 | Platanus ×hispanica | MG495968 | MG495968 | MG495996 | MG496003 |
| Harknessia eucalypti | CBS 342.97 | – | Eucalyptus regnans | AY720745 | AF408363 | – | – |
| Harknessia molokaiensis | CBS 114877 | – | Eucalyptus robusta | AY720749 | AY720842 | – | – |
| Harknessia weresubiae | CBS 113075 | CBS H-9903 | Eucalyptus sp. | AY720741 | AY720835 | – | – |
| Hercospora tiliae | CBS 109746 | BPI 748440 | Tilia tomentosa | – | AF408365 | – | – |
| Immersiporthe knoxdaviesiana | CBS 132862 | PREM 60740 | Rapanea melanophloeos | JQ862770 | JQ862760 | – | – |
| Juglanconis appendiculata | CBS 142389 | WU 35960 | Juglans nigra | KY427145 | KY427145 | KY427195 | KY427214 |
| Juglanconis juglandina | ME23 | WU 35965 | Juglans nigra | KY427150 | KY427150 | KY427203 | KY427222 |
| Juglanconis pterocaryae | MAFF 410079 | TFM FPH 3373 | Pterocarya rhoifolia | KY427155 | KY427155 | KY427205 | KY427224 |
| Kohlmeyeriopsis medullaris | CBS 117849 | – | Juncus roemerianus | KM484852 | FJ176854 | – | – |
| Lamproconium desmazieri | AR 3525 | BPI 748445 | Tilia sp. | – | AF408372 | – | – |
| Luteocirrhus shearii | CBS 130776 | PERTH 08439362 | Banksia baxteri | KC197021 | KC197019 | – | – |
| Macrohilum eucalypti | CBS 118551 | – | Eucalyptus sp. | DQ195781 | DQ195793 | – | – |
| Melanconiella chrysodiscosporina | CBS 125597 | WU 31859 | Carpinus betulus | JQ926238 | JQ926238 | JQ926310 | JQ926376 |
| Melanconiella hyperopta | CBS 132231 | WU 31836 | Carpinus betulus | JQ926285 | JQ926285 | JQ926351 | JQ926418 |
| Melanconiella spodiaea | SPOD | WU 31854 | Carpinus betulus | JQ926300 | JQ926300 | JQ926366 | JQ926433 |
| Melanconis alni | CBS 122310 | BPI 872035 | Alnus viridis | EU199195 | EU199130 | EU199153 | – |
| Melanconis marginalis | CBS 109744 | BPI 748446 | Alnus rubra | EU199197 | AF408373 | EU219301 | EU221991 |
| Melanconis stilbostoma | D143 | WU 35970 | Betula pendula | KY427156 | KY427156 | KY427206 | KY427225 |
| Occultocarpon ailaoshanense | CBS 129147 | BPI 879254 | Alnus nepalensis | JF779848 | JF779852 | JF779857 | JF779862 |
| Ophiodiaporthe cyatheae | BCRC 34961 | HAST 1364 | Cyathea lepifera | JX570889 | JX570891 | JX570893 | KC465406 |
| Ophiognomonia melanostyla | CBS 128482 | BPI 879257 | Tilia cordata | JF779850 | JF779854 | JF779858 | – |
| Phaeocytostroma ambiguum | CBS 128562 | – | Zea mays | FR748042 | FR748101 | – | FR748074 |
| Phaeodiaporthe appendiculata | CBS 123821 | WU 32449 | Acer campestre | KF570156 | KF570156 | – | – |
| Pleuroceras tenellum | CBS 121082 | BPI 871059 | Acer rubrum | EU199199 | EU255202 | EU199155 | EU221907 |
| Prosopidicola mexicana | CBS 113529 | CBS-H 7948 | Prosopis glandulosa | AY720709 | – | – | – |
| Pseudoplagiostoma eucalypti | CBS 124807 | CBS H-20303 | Eucalyptus urophylla | GU973512 | GU973606 | – | GU973542 |
| Pseudoplagiostoma oldii | CBS 124808 | CBS H-20300 | Eucalyptus camaldulensis | GU973534 | GU973609 | – | GU973564 |
| Pseudoplagiostoma variabile | CBS 113067 | CBS H-20304 | Eucalyptus globulus | GU973536 | GU973611 | – | GU973566 |
| Pustulomyces bambusicola | MFLUCC 11-0436 | MFLU 13–0369 | Bambusa sp. | KF806752 | KF806753 | – | KF806755 |
| Rossmania ukurunduensis | AR 3484 | BPI 747566 | Acer ukurunduense | – | EU683075 | – | – |
| Rostraureum tropicale | CBS 115757 | PREM 57519 | Terminalia ivorensis | AY167436 | AY194092 | – | – |
| Sillia ferruginea | CBS 126567 | BPI 843619 | Corylus avellana | JF681959 | EU683076 | – | – |
| Sirococcus tsugae | CBS 119626 | BPI 871167 | Tsuga mertensiana | EU199203 | EU199136 | EU199159 | EF512534 |
| Stegonsporium acerophilum | CBS 117025 | WU 28050 | Acer saccharum | EU039982 | EU039993 | KF570173 | EU040027 |
| Stenocarpella macrospora | CBS 117560 | MRC 8615 | Zea mays | FR748048 | EU754219 | – | – |
| Stilbospora macrosperma | CBS 115073 | WU 24708 | Carpinus betulus | EU039965 | MG495969 | KF570195 | EU039999 |
| Sydowiella fenestrans | CBS 125530 | BPI 843503 | Chamerion angustifolium | JF681956 | EU683078 | – | – |
Light microscopy
Dead corticated branches were examined with the aid of an Olympus SZH or a Nikon SMZ 1500 stereomicroscope. Microscopic observations were made in tap water except where noted. Sections were cut manually with a sterilized razorblade and microscope mounts were examined with a Nikon 80i equipped with a Canon digital camera or a Zeiss Axio Imager.A1 equipped with a Zeiss Axiocam 506 colour digital camera. Measurements were taken with the help of DinoCapture 2.0 or Zeiss ZEN Blue Edition softwares. Measurements are reported as maxima and minima in parentheses and the range representing the mean plus and minus the standard deviation of a number of measurements given in parentheses; in addition, the means (x̄) are given.
Scanning electron microscopy (SEM)
For SEM of ascospores, perithecial contents were re-hydrated in a drop of distilled water on a cover slide and thoroughly cleaved with a fine forceps and preparation needles to release the ascospores from the asci. After evaporation of the water at room temperature, the cover slides were mounted on Cambridge stubs, sputter coated with gold, and examined in a Jeol JSM-6390 scanning electron microscope at 10 kV.
Pure culture isolation
Single ascospore isolates were made by spreading ascospores on 1.5 % potato dextrose agar (PDA, Difco), or on 2 % corn meal agar (CMA, Sigma-Aldrich) plus 2 % w/v dextrose (CMD) supplemented with 200 mg/l penicillin G and streptomycin sulphate (Sigma-Aldrich). Germinated ascospores were then transferred to PDA or CMD plates, sealed with laboratory film and incubated at room temperature. Colony morphology, colour (Rayner, 1970) and growth rate were determined on PDA at 24 °C.
DNA extraction, PCR and sequencing
Growth of liquid cultures and extraction of genomic DNA was done according to Voglmayr & Jaklitsch (2011), using the DNeasy Plant Mini Kit (QIAgen GmbH, Hilden, Germany). The following loci were sequenced for phylogenetic analyses: The complete ITS region and D1 and D2 domains of 28S nuc rDNA region (ITS-LSU) were amplified with primers V9G (Hoog & Gerrits van den Ende 1998) and LR5 (Vilgalys & Hester 1990); a ca 0.6–0.7 kb fragment of the calmodulin (cal) gene with primers CAL-228F (Carbone & Kohn 1999) and CAL2Rd (Groenewald et al. 2013); a ca 1 kb fragment of the guanine nucleotide-binding protein subunit beta (ms204) gene with primers MS-E1F1n1 (5’ AAGGGNACYCTSGAGGGCCAC 3’) and MS-E5R2n (5’ CCASAGCATGGTGGTRCCRTC 3’); a ca 1.1 kb fragment of the RNA polymerase II subunit 1 (rpb1) gene with primers RPB1-6R1asc (Hofstetter et al. 2007) and RPB1-Af (Stiller & Hall 1997); a ca 1.1 kb fragment of the RNA polymerase II subunit 2 (rpb2) gene with primers dRPB2-5f and dRPB2-7r (Voglmayr et al. 2016a); a ca 1.3–1.5 kb fragment of the translation elongation factor 1-α (tef1) gene with primers EF1728F (Carbone & Kohn 1999) and TEF1LLErev (Jaklitsch et al. 2005) or EF1-2218R (Rehner & Buckley 2005); and a ca 1.5 kb fragment of the beta tubulin (tub2) gene with primers T1 and T22 (O’Donnell & Cigelnik 1997). PCR products were purified using an enzymatic PCR cleanup (Werle et al. 1994) as described in Voglmayr & Jaklitsch (2008). DNA was cycle-sequenced using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit v. 3.1 (Applied Biosystems, Warrington) and the PCR primers; in addition, the following primers were used: ITS-LSU region: ITS4 (White et al. 1990), LR3 (Vilgalys & Hester 1990); tub2: BtHV2r (Voglmayr et al. 2016b, 2017) and BtHVf (5’ AACTGGGCMAAGGGYCAYTACAC 3’). Sequencing was performed on an automated DNA sequencer (ABI 3730xl Genetic Analyzer, Applied Biosystems). GenBank accession numbers of the newly generated sequences are given in Tab. 1.
Phylogenetic analyses
All alignments were produced with the server version of MAFFT (http://mafft.cbrc.jp/alignment/server/), the E-INS-i iterative refinement method and a gap opening penalty of 1.4 for the ITS-LSU and tef1, and with default settings for the other loci. All alignments were checked and refined using BioEdit version 7.0.9.0 (Hall 1999).
To reveal the phylogenetic position of Caudospora within Diaporthales, ITS-LSU, rpb2 and tef1 sequences of two accessions of C. taleola and of the new Iranian species sequenced in the present study were aligned to representative sequences of Diaporthales selected and downloaded from GenBank according to the phylogeny of Senanayake et al. (2017a). The finial matrix contained 75 representatives of Diaporthales, with Kohlmeyeriopsis medullaris and Gaeumannomyces graminis (Magnaporthales) selected as outgroups (Voglmayr et al. 2017); the strain details and GenBank accession numbers of sequences are given in Tab 2. The resulting combined four-loci sequence matrix contained 5188 nucleotide positions (2148 from ITS-LSU, 1210 from rpb2, 1830 from tef1).
For phylogenetic relationships within Caudospora, a combined matrix of nine loci was produced, containing 8683 nucleotide positions (1665 from SSU-ITS-LSU, 710 from cal, 1017 from ms204, 1151 from rpb1, 1160 from rpb2, 1433 from tef1, 1547 from tub2). According to the results of phylogenetic analyses of the four-loci matrix, Hapalocystis berkeleyi was selected as outgroup.
Maximum likelihood (ML) analyses were performed with RAxML (Stamatakis 2006) as implemented in raxmlGUI 1.3 (Silvestro & Michalak 2012), using the ML + rapid bootstrap setting and the GTRGAMMA substitution model with 1000 bootstrap replicates. The matrix was partitioned for the individual gene regions, and substitution model parameters were calculated separately for them.
Maximum parsimony (MP) analyses were performed with PAUP v. 4.0a157 (Swofford 2002). All molecular characters were unordered and given equal weight; analyses were performed with gaps treated as missing data; the COLLAPSE command was set to MINBRLEN. For the combined four-loci matrix, a parsimony ratchet approach was applied. A nexus file was prepared using PRAP v. 2.0b3 (Müller 2004), implementing 1000 ratchet replicates with 25 % of randomly chosen positions upweighted to 2, which was then run with PAUP. The resulting best trees were subsequently loaded in PAUP and subjected to heuristic search with TBR branch swapping (MULTREES option in effect, steepest descent option not in effect). Bootstrap analysis with 1000 replicates was performed using 5 rounds of replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping during each bootstrap replicate, with each replicate limited to 1 million rearrangements. MP analysis of the combined nine-loci matrix was done using 1 000 replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping. Bootstrap analyses with 1 000 replicates were performed in the same way, but using 10 rounds of random sequence addition and subsequent TBR branch swapping during each bootstrap replicate.
Results
Molecular phylogeny
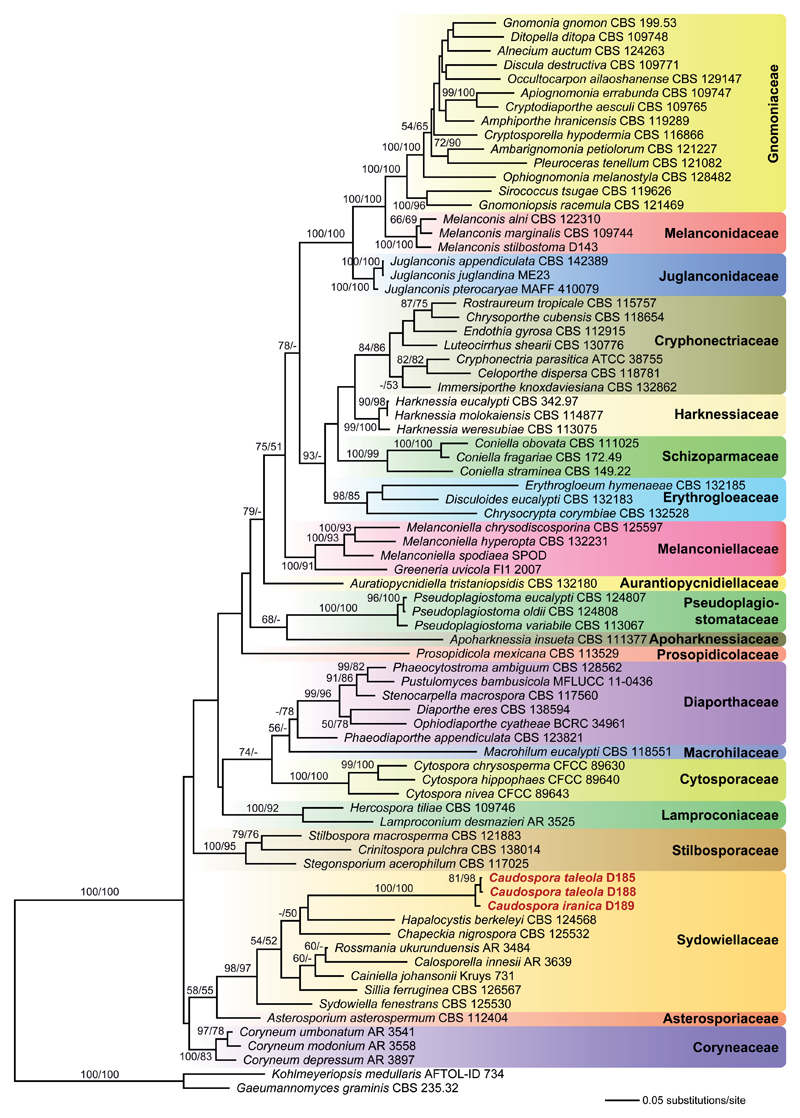
Of the 5188 nucleotide positions included in the four-loci sequence matrix, 1838 were parsimony informative (590 from ITS-LSU, 566 from rpb2, 682 from tef1). The best ML tree (lnL = –48924.555) revealed by the RAxML analysis is shown as phylogram in Fig. 1. Maximum parsimony analyses revealed 12 MP trees 9903 steps long (not shown). In both ML and MP analyses, Caudospora is placed within the Sydowiellaceae with high support, but its closest relatives remain uncertain due to lack of bootstrap support of most nodes within Sydowiellaceae.
Fig. 1.
Phylogram of the best maximum likelihood tree (lnL = -48924.555) revealed by RAxML from an analysis of the combined ITS-LSU-rpb2-tef1 matrix of 75 selected members of Diaporthales, with two members of Magnaporthaceae (Gaeumannomyces graminis, Kohlmeyeriopsis medullaris) selected as outgroup. Familial classification follows Senanayake et al. (2017a). Caudospora (bold) is revealed as a member of Sydowiellaceae with high support. ML and MP bootstrap support above 50 % are given above or below the branches.
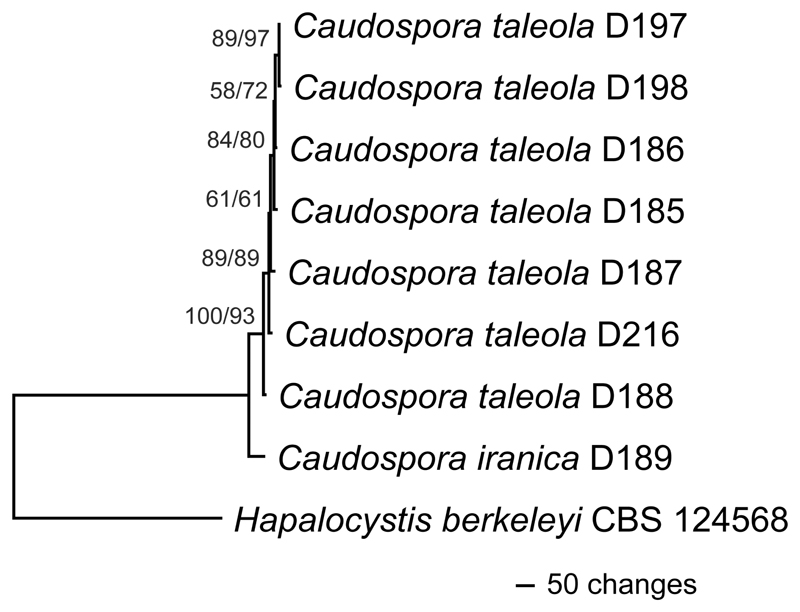
Of the 8683 nucleotide positions included in the nine-loci sequence matrix, 72 were parsimony informative (6 from SSU-ITS-LSU, 5 from cal, 15 from ms204, 9 from rpb1, 14 from rpb2, 15 from tef1, 8 from tub2). The MP analysis revealed a single tree 1559 steps long (Fig. 2); it is identical in topology to the best ML tree (lnL = –18010.352) revealed by RAxML. Sister group relationship of Caudospora iranica to C. taleola received maximum and high (93 %) bootstrap support in MP and ML analyses (Fig. 2), respectively, confirming their status as distinct species.
Fig. 2.
Phylogram of the single MP tree 1559 steps long, obtained by PAUP from an analysis of the combined nine-loci matrix (SSU-ITS-LSU, cal, ms204, rpb1, rpb2, tef1, tub2) of Caudospora, with Hapalocystis berkeleyi selected as outgroup according to Fig. 1. MP and ML bootstrap support above 50 % are given at the first and second position, respectively, above or below the branches.
Taxonomy
Caudospora iranica Mehrabi & Voglmayr, sp. nov. – Figs. 3, 5a, b, e.
MycoBank no.: MB 823458
Type. – IRAN. East Azerbaijan Province, Arasbaran, on dead branches of Quercus sp., 11 July 2015, leg. M. Mehrabi (IRAN 16716 F holotype; WU 39950 isotype; ex-holotype culture IRAN 2552 C, ex-isotype culture CBS 143507).
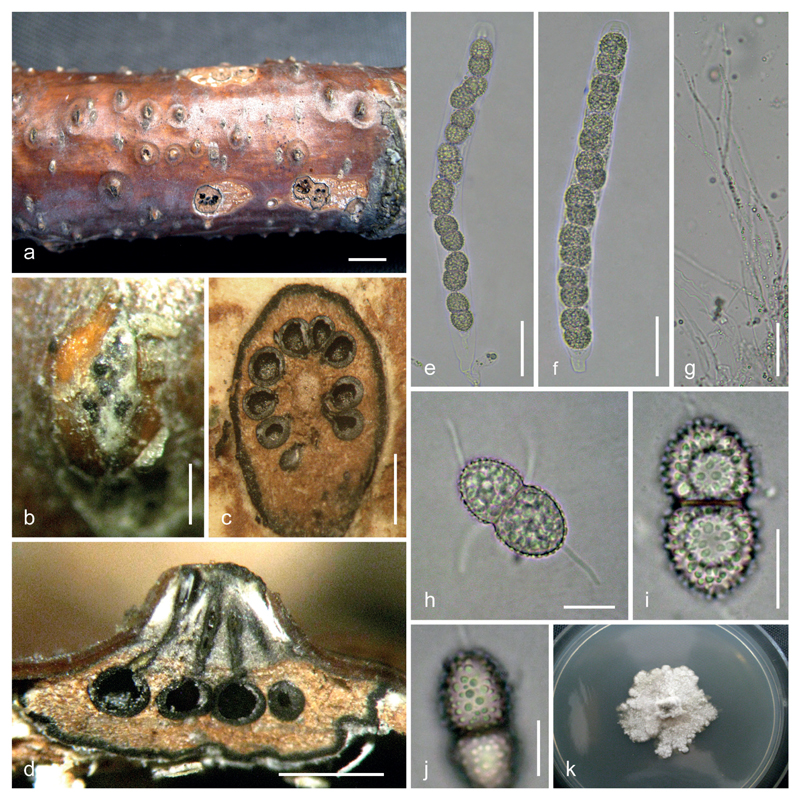
Description. – Pseudostromata 1–3.5 mm diam., immersed in the bark of dead branches (ca. 1.5 cm thick), erumpent, circular to irregular, separate, scattered, sometimes confluent, delimited from surrounding bark by a black line, the latter visible on the bark surface. Ectostromatic discs white, convex, circular to oval, tissue between ostiolar necks distinctly gray to dark grey, powdery. Central column whitish to grey. – Ostioles dark, at the same level as the disc surface, rarely projecting, opening separately. – Perithecia 2–7 per stroma, 300–700 μm diam., subglobose to ovoid, black, monostichous, arranged circinately in brown entostromata, with long ostiolar necks converging towards the ectostromatic disc. – Paraphyses elongate, hyaline, filiform, septate, evanescent. – Asci (170)181–211(230) × 15–18 μm (x̄ = 196 × 16.7 μm; n=20), cylindrical, with short or obsolete stalks and a conspicuous apical ring, containing eight uniseriate ascospores. – Ascospores (18)21–27(32) × (8)10.5–13(15) μm (x̄ = 23.7 × 11.7 μm; n=42), l/w = (1.6)1.8–2.3(2.5), broadly ellipsoid, two-celled, constricted at the septum, hyaline or pale yellowish, coarsely verrucose, in SEM with isolated warts 0.4–1 μm diam., with a tubular gelatinous appendage 10–17 × 1–1.3 μm long at each end and 2–3 (rarely 4) similar median tubular gelatinous appendages arising from near the septum.
Cultural characteristics. – Colonies felty, with irregular margins, first white, later primrose (23,,b) at the surface, often deeply immersed into the agar, reverse primrose, becoming olivaceous black (27,,,,m) in the dark at 24 °C, reaching the margin of a 30 mm Petri-dish after 30 days at 24 °C. No asexual morph seen.
Etymology. – Referring to its origin from Iran.
Habitat. – On dead corticated twigs of Quercus sp.
Distribution. – Only known from the type collection in Iran.
Notes. – Caudospora iranica resembles C. taleola in stroma and ascoma morphology and ascospore appendages. However, there are pronounced differences between these species in the ascospore ornamentation. In C. iranica, the surface of the ascospore wall is covered with prominent isolated warts up to 1 μm diam. that are easily visible by light microscopy, whereas in the latter a finely verruculose ornamentation is scarcely visible in light microscopy without staining; the much smaller (up to 0.4 μm diam.) and densely disposed warts are clearly revealed only in SEM. In addition, asci (196 × 16.7 vs. 162 × 12.1 μm on average) and ascospores (23.7 × 11.7 vs. 23.9 × 9 μm on average) of C. iranica are wider than those of C. taleola.
Caudospora taleola (Fr.) Bih. K. Svenska Vetensk Akad. Handl., Afd. 15(2): 11 (1889). – Figs. 4, 5c, d, f–m.
Basionym. – Sphaeria taleola Fr., Syst. mycol. (Lundae) 2(2): 391 (1823).
Synonyms. – Aglaospora taleola (Fr.) Tul. & C. Tul., Select. fung. carpol. (Paris) 2: 168 (1863).
Chorostate taleola (Fr.) Traverso, Fl. ital. crypt. 1(2): 212 (1906).
Diaporthe taleola (Fr.) Sacc., Atti Soc. Veneto-Trent. Sci. Nat., Padova, Sér. 4 4: 112 (1875).
Hercospora kornhuberi Bäumler, Annln K. K. naturh. Hofmus. Wien 13: 440 (1898).
Hercospora taleola (Fr.) E. Müll., in Müller & von Arx, Beitr. Kryptfl. Schweiz 11(no. 2): 728 (1962).
Melanconis taleola (Fr.) Speg., Atti Soc. Crittogam. Ital., Série 2 3(1): 54 [no. 77] (1881).
Valsa taleola (Fr.) Fr., Summa veg. Scand., Sectio Post. (Stockholm): 391 (1849).
Type. – AUSTRIA. Oberösterreich, Raab, Wetzlbach, on dead branches of Quercus robur, 24 December 2015, leg. H.Voglmayr (WU 39951, neotype here designated, MBT381444; ex-neotype culture D185 = CBS 143508)
Description. – Pseudostromata 1–4 mm diam., immersed in the bark of dead branches (1–3 cm diam.), flat or erumpent, circular to irregular, separate, scattered, sometimes confluent, delimited from surrounding bark by a black line. Ectostromatic discs white, flat, circular to ovoid, tissue between ostiolar necks distinctly gray to dark grey, powdery. Central column whitish to grey. – Ostioles dark, at the same level as the disc surface, rarely projecting, opening separately, sometimes necks of perithecia united below the disc and discharging through a single ostiole. – Perithecia 2–16 per stroma, 300–700 μm diam., subglobose to ovoid, black, monostichous, arranged circinately in brown entostromata, with long ostiolar necks converging towards the ectostromatic disc. – Paraphyses elongate, hyaline, filiform, septate, evanescent. – Asci (140)145–180(207) × 10–13 μm (x̄ = 163 × 12.1 μm; n = 20), cylindrical, with short or obsolete stalks and a conspicuous apical ring, containing eight uniseriate ascospores. – Ascospores (17)20–28(31) × (6.5)8–10(11) μm (x̄ = 23.9 × 9 μm; n = 30), l/w = (2.1)2.3–3.0(3.4), ellipsoid, two-celled, constricted at the septum, hyaline, appearing smooth in light microscopy but distinctly verrucose in SEM with densely disposed warts 0.1–0.4 μm diam., with a hyaline tubular gelatinous appendage 6–17 × 1–1.5 μm long at each end and 2–3 (rarely 4) similar median tubular gelatinous appendages arising from near the septum.
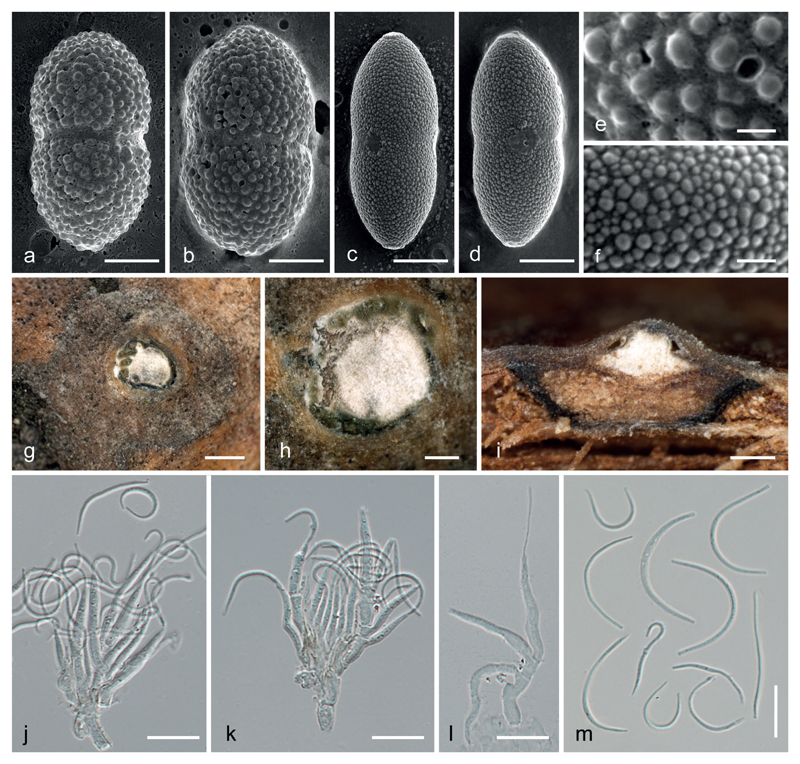
Asexual morph on natural substrate acervular, phomopsis-like. – Conidiomata ca. 1 mm diam., visible as darker spots in surface view, showing the same organisation as the sexual morph, i.e. a central whitish stromatic column with a white ectostromatic disc and a brown pseudostroma delimited from surrounding bark by a black line, preceding the sexual morph; conidiogenous region in section pale olivaceous green. – Conidiophores branched, hyaline, septate, formed on the upper margin of the central column. – Conidiogenous cells phialidic, (6.5)11.5–18.5(26) × (1.4)1.7–2.5(3.5) μm (x̄ = 14.8 × 2.1 μm; n = 70), hyaline, smooth. – Conidia (15)17–25 (32) × 0.7–1.1(1.6) μm (x̄ = 21 × 0.9 μm; n=50), l/w = (14.8)18.2–29.7(37.2), filiform with variable shape from more or less straight, sigmoid, falcate, semicircular to circular, unicellular, hyaline, smooth.
Cultural characteristics. – Colonies felty, irregular, often deeply immersed into the agar, white at the surface and primrose (23,,b) to olivaceous black (27,,,,m) at the reverse in the dark at 24 °C, reaching the margin of 30 mm Petri-dish after 30 days at 24 °C.
Habitat. – On dead corticated twigs of Quercus sp.
Distribution. – Widely distributed in Europe, Asia, North America.
Other specimens examined. – AUSTRIA. Oberösterreich, St. Willibald, Gr. Sallet, on dead branches of Quercus robur, 26 December 2015, leg. H. Voglmayr (WU 39952, culture D186); St. Willibald, Geitzedt, on dead branches of Quercus robur, 31 December 2015, leg. H. Voglmayr (WU 39953, culture D187); Niederösterreich, Mannersdorf im Leithagebirge, Schweingraben, on dead branches of Quercus petraea, 12. March 2016, leg. H. Voglmayr (WU 39957, culture D216). FRANCE. Ardèche (07), Saint-Jean-Roure, Mandon, on dead branches of Quercus robur, 27 December 2015, leg. A. Gardiennet A.G. 15171 (WU 39955, culture D197); Saint-Jean-Roure, Grange de Sagne, on dead branches of Quercus robur, 28 December 2015, leg. A. Gardiennet A.G. 15173 (WU 39956, culture D198). IRAN. East Azerbaijan Province, Arasbaran, on dead branches of Quercus sp., 11 July 2015, leg. M. Mehrabi (IRAN 16715 F, WU 39954, culture IRAN 2551 C).
Notes. – No type specimen of Sphaeria taleola is preserved in the Fries collection at UPS (A. Kruys, pers. comm.), which necessitates neotypification. We here select a well-developed, abundant Austrian collection from the most likely original host, Quercus robur, as neotype, for which a culture and sequence data are available.
To our knowledge, C. taleola has not been previously been recorded from Iran; it is not listed in Ershad (2009). For comparison with C. iranica, see Notes there.
Discussion
Although morphologically undoubtedly a member of Diaporthales (Kirk et al. 2008), the phylogenetic affiliation and familial classification of Caudospora remained unclear up to now in the lack of DNA sequence data. Based on similarities in stromata and perithecia, Müller & Arx (1962) classified Caudospora in Hercospora (Lamproconiaceae), which was challenged by Rogers (1984), who recognized it to be substantially different from Hercospora in verrucose ascospores and a phialidic phomopsis-like asexual morph, vs. the holoblastic conidiation of the rabenhorstia-like asexual morph in Hercospora. However, although Rogers (1984) noted some similarities to the quercicolous Diaporthe leiphaemia var. raveneliana in verrucose ascospore ornamentation, he considered taxonomic rearrangements premature due to insufficient data. In our phylogenies, Caudospora is clearly revealed as a member of Sydowiellaceae (Fig. 1). Remarkably, already Munk (1957) noted a high similarity of centrum morphology of Caudospora to that of Sydowiella fenestrans, to which it is closely related.
After transfer of several lineages originally classified in other families of Diaporthales (e.g. Gnomoniaceae, Melanconidaceae), the Sydowiellaceae have in recent years become a rather heterogeneous assemblage which is difficult to define morphologically (Rossman et al. 2007). It currently comprises about 15 genera occurring as saprotrophs, endophytes and parasites of various herbaceous and woody hosts (Senanayake et al. 2017b). Most genera and species are characterized by reduced, poorly developed stromata containing few to a single perithecium, but there are few exceptions like Sillia and Chapeckia with well-developed multiperitheciate stromata. Also Caudospora has well-developed pseudostromata, which are delimited from the surrounding host tissue by a conspicuous black line, which is so far unique in Sydowiellaceae.
The molecular data reveal that Caudospora iranica and C. taleola are closely related, and they share a similar ecology and the same host genus, Quercus. However, they can be easily separated by ascospore morphology. Caudospora iranica has coarsely verrucose ascospores with isolated warts (Figs. 3h–j, 5a, b, e), whereas the ascospores of C. taleola appear smooth in light microscopy (Fig. 4g, h) but are shown to be finely verrucose in SEM (Fig. 5c, d, f). Ascospore ornamentation of C. taleola revealed by SEM in our investigations matches that illustrated in Rogers (1984: fig. 5) from a North American collection.
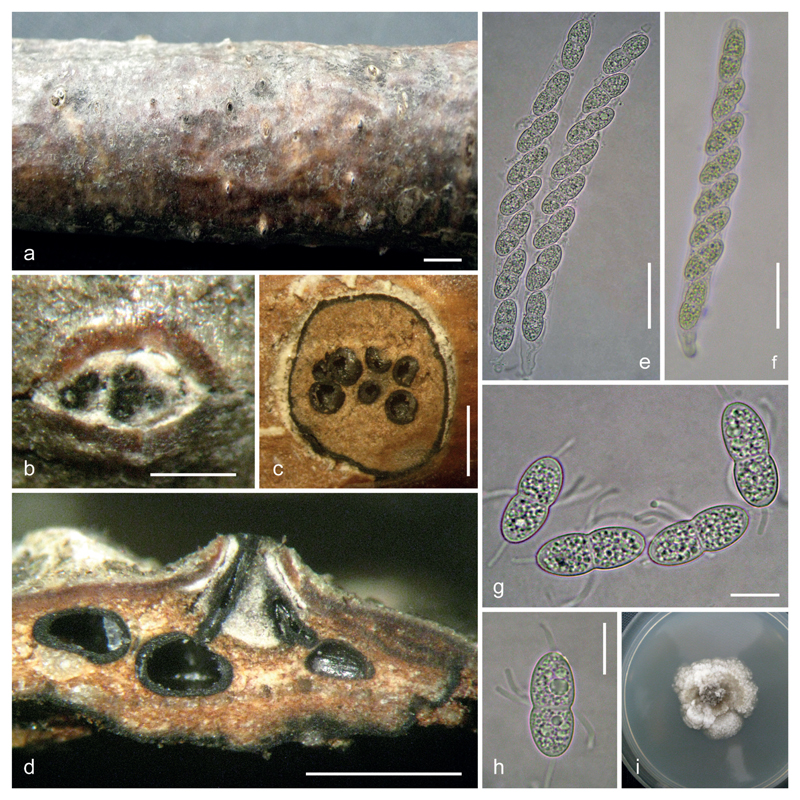
Fig. 3.
Caudospora iranica (IRAN 16716 F, holotype). a. stromata on dead branch of Quercus sp.; b. ectostromatic disc; c. transverse sections of a pseudostroma; d. vertical section of a pseudostroma; e, f. asci; g. paraphyses; h–j. ascospores, showing coarsely verrucose ornamentation; k. culture on PDA. Bars: a 3 mm; b 300 μm; c, d 1 mm; e–g 30 μm; h–j 10 μm.
Fig. 5.
a-f. SEM pictures of ascospores of Caudospora iranica (a, b, e) and C. taleola (c, d, f), showing the coarsely and finely verrucose ascospore ornamentation in C. iranica and C. taleola, respectively; the median smooth circular areas in c, d. represent the scars of median gelatinous appendages. g–m. Asexual morph of C. taleola. g, h. conidiomata in transverse section, showing the central stromatic column and olive-green conidial masses; i. conidiomata in vertical section, showing pseudostromata delimited by a black line, the central stromatic column and the olive-green conidial masses produced on the stromatic column; j–l. branched conidiophores with phialides producing filiform conidia (in j. with released elongate and circular conidia); m. conidia. a, b, e. IRAN 16716 F (holotype of C. iranica). c, d, f–m. WU 39951 (neotype of C. taleola). Bars: a–d 5 μm; e, f 1 μm; g 500 μm; h, i 200 μm; j–m 10 μm.
Fig. 4.
Caudospora taleola (IRAN 16715 F). a. stromata on dead branch of Quercus sp.; b. ectostromatic disc; c. transverse sections of a pseudostroma; d. vertical section of a pseudostroma (showing two united necks under ectostromatic disc); e, f. asci; g, h. ascospores; i. culture on PDA. Bars: a 3 mm; b 300 μm; c, d 1 mm; e, f 30 μm; g, h 10 μm.
To our knowledge, the asexual morph of C. taleola is here described and illustrated for the first time in detail from natural substrate. We did not observe an asexual morph in our cultures, but Rogers (1984) obtained a phomopsis-like asexual morph on oatmeal agar, with unicellular, sigmoid to falcate hyaline conidia (5)10–17 × 1–1.5 μm in size produced on phialides. He compared it with the descriptions of Myxosporium taleola and Libertella taleola, which were formally described by Saccardo (1884) as putative asexual morphs of C. taleola, based on data reported by Tulasne & Tulasne (1863) and Fuckel (1871) from associations of sexual and asexual morphs on natural substrate. Rogers (1984) found some differences of his strain in conidial size; especially conidial width was substantially narrower in his isolate than that reported for Myxosporium taleola (16 × 8 μm) and Libertella taleola (20–30 × 4 μm). However, he was unable to resolve whether these differences related to strain differences, differences caused by conidiation in pure culture vs. on natural substrate, or erroneous connections of asexual morphs of species other than C. taleola. The asexual morph from natural substrate investigated by us agrees well with that reported by Rogers (1984) in its phialidic conidiogenous cells produced on short, branched conidiophores, conidial shape and conidial width, but our conidia are somewhat longer (15–32 vs. (5)10–17 μm in Rogers 1984), which may be caused by the different substrate. It therefore seems likely that the erroneous conidial width reported by Tulasne & Tulasne (1863) and Fuckel (1871) is due to misidentification.
Acknowledgements
The financial support by the Austrian Science Fund (FWF; project P27645-B16) to HV is gratefully acknowledged. Cordial thanks are due to Susi Sontag for help with SEM, Alain Gardiennet for providing fresh specimens and to Åsa Kruys (UPS) for information about the presence of a type specimen of Sphaeria taleola in the Herb. Fries.
References
- Barr ME. Mycologia Memoir. Vol. 7 J. Cramer; Lehre, Germany: 1978. The Diaporthales in North America. [Google Scholar]
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- Ershad D. Fungi of Iran. 3rd edn. Iranian Research Institute of Plant Protection; Tehran, Iran: 2009. [Google Scholar]
- Fuckel KWGL. Symbolae Mycologicae. Erster Nachtrag. Jahrbücher des Nassauischen Vereins für Naturkunde. 1871;25–26:289–346. [Google Scholar]
- Groenewald JZ, Nakashima C, Nishikawa J, Shin HD, Park JH, et al. Species concepts in Cercospora: spotting the weeds among the roses. Studies in Mycology. 2013;75:115–170. doi: 10.3114/sim0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis, program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hofstetter V, Miadlikowska J, Kauff F, Lutzoni F. Phylogenetic comparison of protein-coding versus ribosomal RNA-coding sequence data: A case study of the Lecanoro-mycetes (Ascomycota) Molecular Phylogenetics and Evolution. 2007;44:412–426. doi: 10.1016/j.ympev.2006.10.016. [DOI] [PubMed] [Google Scholar]
- de Hoog GS, Gerrits van den Ende AHG. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses. 1998;41:183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia. 2005;97:1365–1378. doi: 10.3852/mycologia.97.6.1365. [DOI] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. Dictionary of the fungi. 10th edn. CAB International; Wallingford: 2008. [Google Scholar]
- Kobayashi T. Taxonomic studies of Japanese Diaporthaceae with special reference to their life-histories. Bulletin of the Government Forest Experimental Station Meguro. 1970;226:1–242. [Google Scholar]
- Müller E, von Arx JA. Die Gattungen der didymosporen Pyrenomyceten. Beiträge zur Kryptogamenflora der Schweiz. 1962;11(2):1–922. [Google Scholar]
- Müller K. PRAP - calculation of Bremer support for large data sets. Molecular Phylogenetics and Evolution. 2004;31:780–782. doi: 10.1016/j.ympev.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Munk A. Danish Pyrenomycetes. A preliminary flora. Dansk Botanisk Arkiv. 1957;17(1):1–491. [Google Scholar]
- O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- Phillips DH, Burdekin DA. Diseases of oak (Quercus spp.) In: Phillips DH, Burdekin DA, editors. Diseases of forest and ornamental trees. Palgrave Macmillan; London: 1992. pp. 241–258. [Google Scholar]
- Rayner RW. A Mycological colour chart. CMI and British Mycological Society; Kew, Surrey, UK: 1970. [Google Scholar]
- Rehner SA, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- Rogers JD. Caudospora taleola: Anamorph and systematic position. Mycotaxon. 1984;21:475–484. [Google Scholar]
- Rossman AY, Farr DF, Castlebury LA. A review of the phylogeny and biology of the Diaporthales. Mycoscience. 2007;48:135–144. [Google Scholar]
- Saccardo PA. Sylloge Fungorum. Vol. 3 Padova: 1884. [Google Scholar]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchi-kumbura SSN, Jeewon R, Phillips AJL, Bhat JD, Perera RH, Li QR, Li WJ, Tangthirasunun N et al. Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology. 2017a;86:217–296. doi: 10.1016/j.simyco.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Maharachchikumbura SSN, Jeewon R, Promputtha I, Al-Sadi AM, Camporesi E, Hyde KD. Morphophylogenetic study of Sydowiellaceae reveals several new genera. Mycosphere. 2017b;8:172–217. [Google Scholar]
- Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity and Evolution. 2012;12:335–337. [Google Scholar]
- Stamatakis E. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Starbäck K. Ascomyceter från Öland och Östergötland. Bihang till Kungliga svenska Vetenskaps-Akademiens Handlingar. 1889;15(2):1–28. [Google Scholar]
- Stiller JW, Hall BD. The origin of red algae: implications for plastid evolution. Proceedings of the National Academy of Sciences, USA. 1997;94:4520–4525. doi: 10.1073/pnas.94.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* 4.0b10: phylogenetic analysis using parsimony (*and other methods) Sinauer Associates; Sunderland, Massachusetts: 2002. [Google Scholar]
- Tulasne LR, Tulasne C. Selecta Fungorum Carpologia. Vol. 2 Paris: 1863. [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Akulov OY, Jaklitsch WM. Reassessment of Allantonectria, phylogenetic position of Thyronectroidea, and Thyronectria caraganae sp. nov. Mycological Progress. 2016a;15:921–937. doi: 10.1007/s11557-016-1218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Castlebury LA, Jaklitsch W. Juglanconis gen. nov. on Juglandaceae, and the new family Juglanconidaceae (Diaporthales) Persoonia. 2017;38:136–155. doi: 10.3767/003158517X694768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Gardiennet A, Jaklitsch WM. Asterodiscus and Stigmatodiscus, two new apothecial dothideomycete genera and the new order Stigmatodiscales. Fungal Diversity. 2016b;80:271–284. doi: 10.1007/s13225-016-0356-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM. Prosthecium species with Stegonsporium anamorphs on Acer. Mycological Research. 2008;112:885–905. doi: 10.1016/j.mycres.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Voglmayr H, Jaklitsch WM. Molecular data reveal high host specificity in the phylogenetically isolated genus Massaria (Ascomycota, Massariaceae) Fungal Diversity. 2011;46:133–170. [Google Scholar]
- Wehmeyer LE. University of Michigan Scientific Series. Vol. 10 University of Michigan Press; Ann Arbor: 1933. The genus Diaporthe Nitschke and its segregates. [Google Scholar]
- Werle E, Schneider C, Renner M, Völker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Research. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. Academic Press; San Diego: 1990. pp. 315–322. [Google Scholar]