Abstract
Nanopore-based sensors for nucleic acid sequencing and single-molecule detection typically employ pore-forming membrane proteins with hydrophobic external surfaces, suitable for insertion into a lipid bilayer. In contrast, hydrophilic pore-containing molecules, such as DNA origami, have been shown to require chemical modification to favor insertion into a lipid environment. In this work, we describe a strategy for inserting polar proteins with an inner pore into lipid membranes, focusing here on a circular 12-subunit assembly of the thermophage G20c portal protein. X-ray crystallography, electron microscopy, molecular dynamics, and thermal/chaotrope denaturation experiments all find the G20c portal protein to have a highly stable structure, favorable for nanopore sensing applications. Porphyrin conjugation to a cysteine mutant in the protein facilitates the protein’s insertion into lipid bilayers, allowing us to probe ion transport through the pore. Finally, we probed the portal interior size and shape using a series of cyclodextrins of varying sizes, revealing asymmetric transport that possibly originates from the portal’s DNA-ratchet function.
Keywords: portal protein, single molecule, protein nanopore, electrical detection, lipid bilayer, porphyrin, electroosmosis
Graphical Abstract

In recent years, pore-forming biological macromolecules have found various uses as tools for direct DNA sequencing,1–5 molecular sensing,6–13 molecular sizing, 14–16 monitoring enzymatic reactions,17–21 and protein characterization,22–33 including detection of post-translational modifications.34,35 In a typical experiment, a voltage bias is applied between two electrolyte compartments insulated by a membrane that contains a single nanopore. The bias generates a steady-state ionic current that reports on ion flow through the pore. Macromolecules that partition into the pore constriction transiently block the ion current; the current blockage indirectly reports on the physical and chemical properties of the macromolecules.36 To date, most nanopore sensing studies employed pore forming toxins and outer membrane proteins that contained a β-barrel. These proteins insert into planar lipid bilayer membranes, which makes them perfect candidates for sensing applications. However, while other, non β-barrel types of protein assemblies may offer superior analyte recognition properties, using them for nanopore sensing experiments is tempered by the need of stable insertion into lipid bilayers.
One class of noninserting ringlike proteins are viral portal proteins, circular structures through which the viral genome is pumped into a preformed protein shell, or capsid, during the packaging process.37 These natural DNA pores seated in the portal vertex of the icosahedral capsid38–42 (Figure 1) are usually assembled from 12 identical subunits, arrayed in a toroid-like structure with a central pore. Translocation of viral DNA through the pore is driven by a powerful motor that continues to function despite an enormous capsid pressure that builds up during the packaging process.43 Motor-driven DNA packaging is thought to be assisted by an asymmetric internal pore shape that acts as a dynamic unidirectional valve to retain packaged DNA in the phage head.38,44–46 The encapsulated DNA is maintained under pressure within the mature particle until host cell infection, during which conformational changes in the particle lead to its ejection through the portal vertex into the target cell.37 To date, the main ring-shaped portal assembly studied using single-channel ion current measurements is the heavily mutated portal protein (also called “connector”) from bacteriophage phi29,47,48 in which the loops that make up the pore constriction are either flexible and/or exhibit conformational variability.40 Recent work49,50 has also explored the use of the portal assembly from bacteriophage SPP1 which can insert spontaneously into lipid bilayers to form a pore with a well-defined internal structure.38 However, this protein exhibits oligomeric state variations where 12-, 13-, or 14-subunit oligomers create a mixture of nanopores with different internal dimensions.
Figure 1.
Route to studying transport through the G20c portal protein. The bacteriophage DNA translocation motor is formed when the large terminase (magenta) is assembled onto the portal protein (dark blue) vertex of the viral capsid (example shown for T4 phage EMD 1572). The motor translocates viral genomic DNA (graphical representation not to scale, light blue) into the capsid. The thermostable portal assembly from bacteriophage G20c (PDB code 4zjn; dark blue) is shown schematically with inserted DNA (gray, not to scale). Protein engineering through a point mutation (mutated protein shown in light blue with introduced cysteine residues in orange) allows attachment of a maleimide–porphyrin lipid anchor (orange) to facilitate insertion into a lipid membrane and single-channel measurements. Internal point mutations (not shown) afford pore size/shape control.
In this work, we characterize the transport through the portal protein from a thermophilic bacteriophage G20c that infects the bacteria Thermus thermophilus.51 The X-ray structure of this protein (PDB code 4zjn) revealed a symmetrical arrangement of 12 subunits in a circular assembly. All residues lining the internal channel of this protein have clear electron density, defining the central pore that is only 1.8 nm in diameter at its narrowest portion (Figure 2). Thus, compared to previously characterized portal proteins, this protein is better suited for structure-based reprogramming of its inner tunnel properties through introduction of point mutations. To investigate the transport properties of the portal pore, we developed a facile molecular anchor route toward inserting the portal into a lipid bilayer, allowing ionic current measurements. Complementary molecular dynamics (MD) simulations characterized the structure of the interface between the protein and a lipid bilayer, elucidating the amount of leakage current that flows through the interface. To demonstrate control over the portal’s properties, we altered the diameter of its narrowest section through point mutations in the tunnel loop and characterized the mutant proteins by X-ray crystallography, MD simulations, and ionic current recordings. Finally, we probed the inner diameter of the altered portal proteins using several cyclodextrin variants that differed by their physical dimensions, revealing the preferred direction of driven molecular transport through the portal corroborated by the all-atom MD method.
Figure 2.
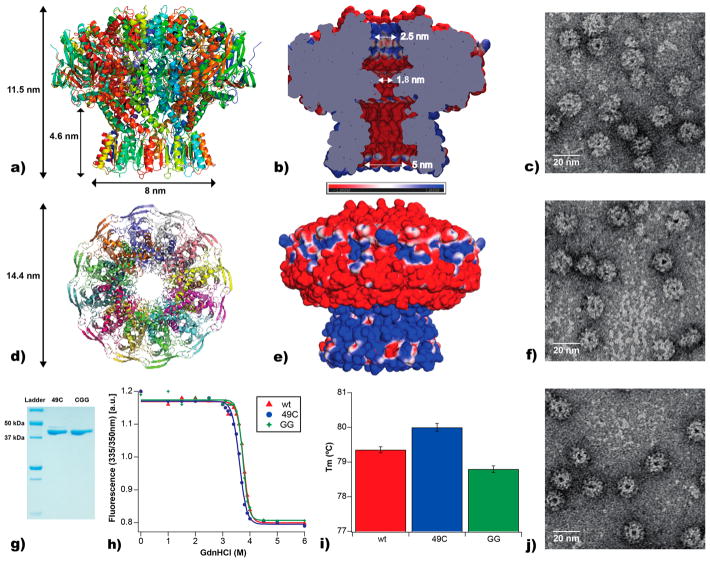
Structure of G20c portal proteins and stability. (a) Side view of a cartoon depiction of the G20c portal protein (PDB 4zjn). (b) Slice through the middle of the G20c diagram showing the electrostatic potentials inside the tunnel from −1 (red) to +1 kT/e (blue) represented by the scale bar and, (e) on the outside of the pore. (d) Top view of a cartoon depiction of the G20c portal protein. (c) Transmission electron microscopy of negatively stained wt, (f) single mutant 49C, (j) and triple mutant CGG portal proteins. (g) 12% SDS-PAGE of purified recombinant 49C and CGG portal proteins. (h) wt (red), 49C (blue), and GG (double mutant, green) unfolding equilibrium transition assessed by measuring the change in tryptophan fluorescence emission ratio of 335/350 nm (excitation wavelength: 280 nm) as a function of GdnHCl concentration in 1 M NaCl, 20 mM Tris pH 7.5. (i) Melting temperatures of wt (red), 49C (blue), and CGG (green) portal proteins deduced by Thermofluor assay in 1 M NaCl, 20 mM Tris pH 7.5.
RESULTS AND DISCUSSION
G20c Portal Protein Assembles into a Stable Cylindrical Dodecamer
The previously determined X-ray structure of the G20c portal protein52 (PDB code 4zjn, Figure 2a,d) revealed a symmetrical circular assembly of 12 monomers that is shaped like a champagne cork with a central hole running through it (Figure 2b). Akin to portal proteins from other viruses, the narrowest portion of the internal tunnel is defined by the tunnel loop. Twelve such loops, one from each subunit, pack neatly next to each other forming a tight and well-defined 1.8 nm aperture of the tunnel (Figure 2b,d). Tight packing of monomers, stabilized by 36 hydrogen bonds and eight salt bridges per monomer–monomer interface, leaves no side voids that would connect the central channel with the outer solvent (Figure 2b). Analysis of the electrostatic potentials of the solvent accessible surfaces of the portal assembly, including the tunnel, reveals a charged protein nanopore (Figure 2b, e). The external surface is essentially bipolar, with a negatively charged top, or “cap”, consisting of the crown and wing subdomains (Supporting Information Figure S1), and a positively charged stem (stem and clip subdomains). The surface of the internal pore is negatively charged along its length, except for the top ~3 nm region that is somewhat positively charged (Figure 2b).
To study the properties of the portal, we have made several specific mutations in the protein sequence. First, to increase the pore diameter we replaced two bulky amino acids in the narrowest constriction of the internal tunnel loops with glycines (V325G/I328G double mutant: GG). Initial low-resolution X-ray data on the GG mutant confirmed that this protein also forms 12-mer assemblies (Supporting Information Figure S1), and a complementary high-resolution X-ray structure of the related single V325G mutant protein showed that this substitution did not affect the oligomeric state and conformation of the assembly, with a backbone α-carbon (Cα) root-mean-square deviation (RMSD) of 0.38 Å between the wild type and mutant proteins. Although the tunnel loops (residues 312–328) are stabilized by extensive van der Waals (vdW) interactions, the 1.90 Å resolution structure of the V325G portal variant showed that the substitution did not affect their overall position (Cα RMSD = 0.1 Å) and resulted in the predicted increase in tunnel diameter at residue 325, from 1.8 to 2.3 nm (vdW 1.5 to 1.9 nm; Supporting Information Figure S1). These data suggest that the double glycine substitution in the tunnel loops would result in a similar additional increase in pore diameter at the I238 position.
Given the polarity of the external surface, engineering a protein that is amenable to lipid bilayer insertion is a challenging task. Here, we explored the feasibility of a chemical biology approach based on cysteine–maleimide conjugation, since the wild-type portal protein is cysteine-free. We therefore introduced a cysteine mutation at the underside of the wing subdomain near the cap-stem junction (L49C; Figure 1 and Supporting Information Figure S1d). The resulting dodecamer contains 12 such cysteine residues displayed in a ring around the top of the stem (Figure 1 and Supporting Information Figure S1e).
Electron microscopy images showed that the engineered portal proteins (single mutant, L49C and triple mutant L49C/ V325G/I328G, hereafter referred to as 49C and CGG, respectively) maintained the dodecameric assembly (Figure 2c,f,j). Thermal and chaotrope tolerance of the unmodified protein and the engineered derivatives were explored under guanidinium hydrochloride and thermal denaturing conditions. We found that the mutations did not reduce the chemical stability, with all variants remaining folded, as determined by tryptophan fluorescence spectroscopy, until the addition of >3 M denaturant (Figure 2h). Likewise, all variants exhibited similar thermal stability with no significant change in melting temperature (~79–80 °C; Figure 2i).
Modification of Portal Protein for Insertion into Lipid Bilayers Using a Maleimide–Porphyrin Conjugate
Porphyrin-functionalized DNA oligomers have been previously used to facilitate insertion of DNA-based nanopores into lipid bilayer membranes.53–59 Porphyrin conjugation to peptides and polypeptides is also commonly used in medicine for targeted photodynamic therapy.60–62 In this work, we conjugated a maleimide–porphyrin derivative through sulfhydryl chemistry to the engineered cysteine residues of the 49C and CGG mutant proteins (Supporting Information Figure S1e).
We performed analytical gel filtration to compare the porphyrin-coupled protein with the unlabeled protein, the two distinguished by absorbance peaks at 280 and 410 nm for tryptophan and porphyrin, respectively. The data revealed a significant population of dodecamers with an average of one porphyrin moiety covalently attached (Figure 3a and b). However, this population is likely to be a heterogeneous mixture of differentially labeled portal assemblies, including unlabeled dodecamers and those conjugated with one or more porphyrin moieties. Both variants of porphyrin-portal (49C and CGG) exhibited similar hydrodynamic characteristics to their unlabeled counterparts, although a small increase in the proportion of double-dodecamers were observed in the porphyrin-labeled samples. Notably, large aggregates of porphyrin that contains a low level of protein were observed to elute in the void volume of the column. Analysis of the UV–vis spectra of the fractions comprising the main dodecameric peak of the porphyrin-modified protein revealed a small peak at ~410 nm, characteristic of porphyrin in water,63 and a larger peak at 280 nm, indicative of aromatic amino acid residues (Figure 3c). Negative-stained TEM imaging of both labeled proteins revealed characteristic ring-shaped proteins (Figure 3d and e) are formed, indicating that assembly was not mitigated by either the addition of the porphyrin tag or the internal CGG mutations.
Figure 3.
Modification of portal protein with maleimide–porphyrin conjugate for incorporation into lipid bilayers. Analytical size-exclusion chromatography of 49C (a) and CGG (b) mutant portal proteins before (1,2 dashed lines) and after (3,4 solid lines) conjugation with maleimide–porphyrin followed at 280 (1,3 black) and 410 nm (2,4 red). (c) UV–vis absorbance spectra of the main peak fractions for 49C (green) and CGG (blue) mutants after maleimide–porphyrin conjugation and analytical gel filtration chromatography. (d) Negative-stain transmission electron micrographs of the 49C (d) and CGG (e) mutant proteins after maleimide–pophyrin conjugation.
Electrical Properties of the Portal Protein Inserted into a Lipid Bilayer
We used single-channel electrical recording to characterize the ion-transport properties of the portal protein embedded into a lipid bilayer (Figure 4a). In these experiments, portal protein that was preconjugated to maleimide–porphyrin was added to the cis chamber, and the current across the membrane was monitored as a small voltage was applied across it (~100 mV). Insertion of a portal protein into the lipid bilayer resulted in discrete stepwise increases of the ionic current (Figure 4b). The unitary current in 1 M NaCl, 20 mM Tris pH 7.5, was 96.4 ± 3.7 and 107.4 ± 4.5 pA, respectively, for 49C and CGG portal proteins at an applied voltage of +100 mV. The corresponding conductance for the 49C and CGG portals was 0.96 ± 0.03 nS and 1.07 ± 0.04 nS, respectively. Control experiments with wild-type portal protein did not result in self-insertion into the lipid bilayer, even after several hours (Supporting Information Figure S2), indicating that insertion of the labeled proteins must be driven by maleimide–porphyrin tags. Figure 4c shows a ~1 min current trace for a single 49C assembly during which the entire portal dodecamer exits from the lipid bilayer. Notwithstanding, the labeled portals remained stably inserted into the lipid bilayer for long periods of time, from a few minutes to hours (Supporting Information Figure S3). We propose that the positioning of the cysteine mutation under the wing subdomain of the portal, in proximity to the top of its stem, leads to directional insertion of the porphyrin-labeled portal protein, without a need for other insertion methods that include nickel–NTA (nitrilotriacetic acid) beads or lipid vesicle formation.47–50 In order to accurately measure the conductance of a single portal protein, we recorded current–voltage (IV) curves for both 49C and CGG portals (Figure 4d). The IV curves indicate slight rectification of the current at positive biases, which could be expected given the asymmetric distribution of the electrical charges along the portal surface and the differences in the pore geometry (2.5/5 nm diameter of the cis/trans entrance). The average conductance of a single 49C and CGG portal is 0.95 ± 0.02 nS and 1.14 ± 0.11 nS, respectively, representing a 17% increase for the CGG mutant, as compared to 49C portal variant. This increase in conductance suggests that mutation of the amino acid side chain does indeed increase the aperture of the internal constriction.
Figure 4.
Electrical properties of the bacteriophage G20c portal inserted into a lipid bilayer. (a) Schematic of the ion–current measurement setup. One G20c portal protein is inserted into a suspended lipid bilayer via maleimide–porphyrin tags (red). An electrical potential is applied via two Ag/AgCl electrodes, which induces a current of Na+ and Cl− ions through the nanopore (1 M NaCl, 20 mM Tris pH 7.5). (b) Typical current trace and the current histogram showing insertion of individual CGG portal channels into a lipid membrane. Data were collected at +100 mV. The average current value is 107.4 ± 4.5 pA for a single pore insertion, 222.8 ± 6.5 pA for two pores, and 325.7 ± 8.1 pA for three pores. (c) A typical current trace recorded through a single 49C portal at ±100 mV, showing pore expunction from the lipid membrane at ~57 s. The average current value is −98.0 ± 3.2 pA at −100 mV and 96.4 ± 3.7 pA at +100 mV. (d) Current–voltage (IV) curves of 49C (red) and CGG (blue) portals fitted to average data from eight independent recordings. The error bars represent a standard deviation from the mean curve.
One drawback that plagues DNA origami-based nanopores is leakage of ion current through the DNA structure.55,64 Since our protein is not designed to be a lipid-embedded channel, we probed the microscopic structure of the interface between the lipid bilayer and the protein and independently assessed the protein’s ionic conductance using MD simulations. All-atom models of the G20c portal protein embedded in a DOPC lipid bilayer (Figure 5a, top) were constructed using the crystal structure of the protein (see the Methods). When simulated in 1 M NaCl bulk electrolyte and, subsequently, in the lipid-bilayer environment, the protein structure remained stable, with RMSD values saturating at 1.7 A, Figure 5b. Figure 5c illustrates the evolution of the lipid–protein interface observed during the 150 ns equilibration. Starting from a typical bilayer configuration, the lipid head groups rearranged to form a toroidal pore near the stem, or trans, exit of the portal’s channel. However, no major rearrangement of lipid head groups was observed near the cap-stem junction. Visual inspection revealed the absence of water molecules at the protein–lipid interface at the cap-stem junction (Figure 5a, bottom). In the case of DNA pores embedded in lipid membranes by means of porphyrin or cholesterol anchors,55,59 water-filled toroidal pores were found to carry a significant fraction of the transmembrane ionic current. In contrast, the structure of the lipid bilayer around the G20c portal appears not to feature a continuous water path from one side of the membrane to the other along the outer surface of the protein.
Figure 5.
Molecular dynamics simulation of G20c portal ionic conductance. (a) Simulation system consisting of the protein channel, shown as a cut-away molecular surface, embedded in a lipid bilayer membrane (cyan) via porphyrin moieties (orange). A white semitransparent surface shows the extent of the solvent (1 M NaCl); green and purple spheres represent the chloride and sodium ions, respectively. The system contains 792391 atoms. A bottom panel shows a zoomed-in view of the equilibrated lipid–protein interface, where water molecules are shown explicitly as red (oxygen) and white (hydrogen) spheres. (b) The root-mean-square deviation (RMSD) of the protein Cα atoms from their crystallographic coordinates during the equilibration simulations. The black and red lines correspond to simulations carried out in bulk electrolyte and lipid bilayer environments. The data were sampled every 4.8 ps. (c) A set of cross sections illustrating development of the lipid–protein interface during the equilibration simulation. Blue and green color maps specify local density of the protein’s α-carbon and lipids’ phosphorus atoms, respectively. Each cross-section represents a time average of 4.8 ps sampled coordinate frames. (d) Total charge transported through the channel by various ionic species versus simulation time. The slope of each line gives the average ionic current. The simulations were performed under a transmembrane bias of ±100 mV. Solid and dashed lines illustrate the simulated currents for 49C and CGG portal channels. The plots were obtained by integration of the ionic current versus simulation time; the ionic current data were sampled every 4.8 ps and averaged in 2.4 ns blocks prior to integration. (e) Simulated conductance of 49C and CGG channels. The conductance values were scaled by the ratio of the experimentally measured (7.43 S/m) and simulated (11.56 S/m) bulk conductivity of 1 M NaCl. Error bars represent standard errors. (f) Ionic selectivity of 49C and CGG variants of the channel defined by the ratio of chloride to sodium currents. (g) Steady-state local densities of lipids (all non-hydrogen atoms, green color scale), protein (all non-hydrogen atoms, blue color scale), and ionic current (streamlines, purple-red-yellow color scale). The arrows indicate the direction of the local ionic current flux, and the color indicates the flux’s magnitude. The maps were computed from a 30 ns long MD trajectory at a + 100 mV bias sampled with a frequency of 48 ps, radially averaged about the z-axis to improve the resolution.
To independently assess the channel’s conductance and determine the fraction of the current that passes through the central pore, the equilibrated structure was simulated under a transmembrane bias of ±100 mV for 30 ns (see the Methods for details). The plots of the resulting integrated currents (Figure 5d) indicate a steady flow of ions at both biases; the slope of each dependence gives the average current. Dividing the current by the applied bias, we find the raw simulated conductance of 2.3 ± 0.8 and 3.6 ± 0.9 nS for 49C and CGG pores (Supporting Information Figure S4). Scaled by the ratio of the simulated and experimentally measured bulk conductivity of 1 M NaCl, the simulated conductance values are within error of the experimental values (Figure 5e), although the simulations appear to somewhat overestimate the ionic conductance. We attribute the latter to lower than bulk concentration of ions within the nanopore volume (Supporting Information Figure S5) and the ion-concentration dependence of the simulated-to-measured bulk conductivity ratio.65
The current through both 49C and CGG portal channels is carried predominantly by chloride ions (Figure 5f), with the ion selectivity being milder for the CGG pore. The plot of local ion currents (Figure 5g) indeed shows very small currents at the interface of the protein and the lipid bilayer. By integrating the local currents within and outside the central pore of the protein, we find the current through the lipid–protein interface to contribute at most 3% of the total transmembrane current.
Probing CGG Portal Protein Size and Interactions with α-, β-, and γ-cyclodextrins
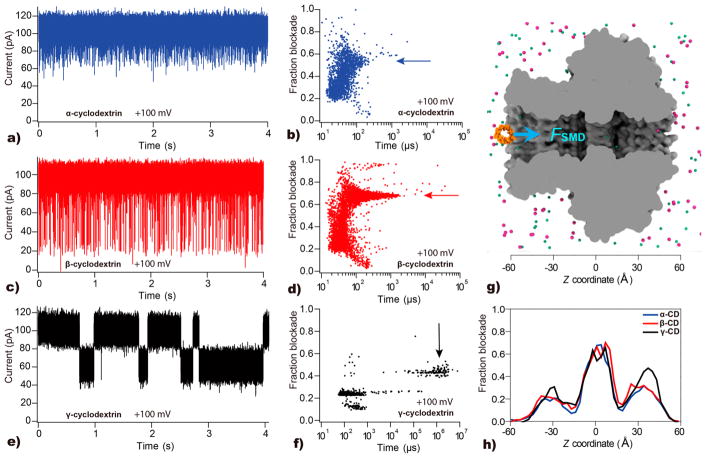
To experimentally probe the size of the narrowest constriction of the CGG portal protein, we performed electrical recordings of the ion current through the portal in the presence of cyclodextrins (CD), neutral cyclic glucose oligosaccharides. The three variants of CD, α-CD, β-CD, and γ-CD, have outer diameters of 1.37, 1.53, and 1.69 nm, respectively,66 which are comparable with the tunnel constriction dimensions (~2.3 nm, based on crystallography). Addition of 0.16 mM α, β and γ-CD to both chambers produced reversible partial blockades of the ionic current at +100 mV (Figure 6a,c,e; note that further experiments where CD was added to one chamber are described below). Analysis of the traces yielded the scatter plots of the dwell times versus the current blockades, which are shown on the right of each respective trace (Figure 6b,d,f). While the distribution encompassed both short and long events, longer dwell times are characteristic of translocation associated interaction and subsequent analysis was focused on these events (see Supporting Information Figure S6 for details on event frequency).
Figure 6.
Interaction of α-, β-, and γ-CDs with the CGG portal protein. (a) Current vs time trace recorded through a single CGG portal pore at +100 mV in the presence of 0.16 mM α-CD in both chambers. (b) Scatter plot of fraction blockade versus time for α-CD at +100 mV. (c, d) Same as in panel a and b, respectively, but in the presence of β-CD. The calculated standard deviation for the noise is 8.3 pA (see Methods). (e, f) Same as in panel a and b, respectively, but in the presence of γ-CD. Arrows represent the population of longer-lived events in each respective experiment. (g) Steered MD simulation of CD transport through the CGG portal. The protein channel (gray) is shown as a cutaway molecular surface, the α-CD is in orange, chloride and potassium ions are in green and purple, respectively, water molecules not shown for clarity. CD molecules were pulled along the axis of the channel using the constant velocity SMD protocol67 (see the Methods). (h) Blockade current through the portal channel for different placements of the CD variants. The currents were computed using a theoretical model based on the position dependence of the electrolyte conductivity68 (see the Methods for details).
The nature of interactions of the CDs with the portal can be described using simple on–off binding rate equations, as previously described by Bayley and co-workers.69 The dissociation (koff) and association (kon) rates of a CD–portal complex can provide the dissociation constant (Kd), using the equations koff = 1/τt and kon = 1/(cτi), where c represents the concentration of CD, τt is the average dwell-time, and τi is the mean interevent time, as previously described.70 Prior to applying these equations, we verified that (1) increasing the concentration α-CD resulted in a linear increase in event frequency (Supporting Information Figure S7a) and (2) event dwell times are independent of the α-CD concentration (Supporting Information Figure S7c). This analysis yields dissociation constants at +100 mV of Kd = 15.85 ± 2.87 M and 56.36 ± 9.80 M for α-CD and β-CD, respectively. Therefore, we can conclude that the interactions between the CGG pore between α-CD and β-CD are weak reversible bimolecular interactions.
Next, to provide a microscopic interpretation of the CD translocation experiments, we used a harmonic spring potential to move α-, β-, and γ-CD through the central pore of the CGG portal with a constant velocity, Figure 6g; see the Methods for simulation details. As expected, the forced translocation of larger molecules required more work done by the spring force (Supporting Information Figure S8). Using the ensemble of conformation observed during the force translocation simulations and a theoretical model of blockade current,68 we evaluated the fractional current blockade produced by different CD variants as a function of their position within the nanopore, Figure 6h. Interestingly, the fractional current blockade produced by different CD variants does not show a considerable dependence on the CD type, indicating that variation in the fractional blockade observed in experiment is produced by differential placement of CDs within the G20c pore. Using the fractional blockade map of the CGG portal, Figure 6h, we can assign long-duration blockade events seen in Figure 6b and d to α-CD and β-CD residence, respectively, at the constriction of the portal (fractional blockade ~0.6). The two shallower fractional blockades produced by γ-CD (at 0.25 and 0.4, Figure 6f) can be assigned to γ-CD interaction with the narrowest sections of the stem (z = −30 Å) and cap (z = 40 Å) parts of the portal, respectively. Finally, using the experimental values of the fractional blockade currents for α-CD and β-CD, we obtain estimated pore diameters of 1.98 ± 0.01 nm and 1.86 ± 0.02 nm, respectively (see Supporting Information S9), values that are significantly smaller than the value predicted from the crystal structure (~2.3 nm), as previously seen for α-hemolysin.71 We note, however, that these observations are consistent with the internal pore diameter (~1.9 nm) determined considering the van der Waals radii of the portal Cα atoms at 325G.
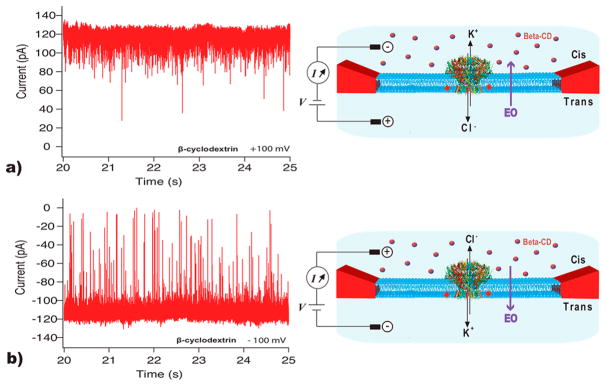
To gain insight into the capture mechanism of the neutral CD molecules into our portal channel, we have performed measurements that probe electro-osmotic flow (EOF),69,72–76 by analyzing the long-lived blockade events for β-CD and the CGG pore. First, we investigated the direction of the EOF in the CGG portal protein by adding β-CDs only to the cis compartment and measuring current traces at both positive and negative bias (Figure 7). While at +100 mV we only observed a few characteristic current blockades (Figure 7a), a drastic increase in the event rate at −100 mV was observed (Figure 7b). This observation can be attributed to EOF through the pore governed by K+ flow, in good agreement with the negative internal charges within the pore and extensive negative isocontours of the cap (Figure 2b and Supporting Information Figure S10).
Figure 7.
Electroosmotically driven β-CD transport through the CGG portal. (a) Current vs time trace of a single CGG pore at +100 mV in the presence of 0.16 mM β-CD in cis compartment only, and scheme showing the direction of the electro-osmotic flux from trans to cis compartment. (b) Current vs time trace of a single CGG pore at −100 mV, in the presence of 0.16 mM β-CD in cis compartment only and scheme showing the direction of the electro-omotic flux from cis to trans compartment. Experiments were conducted in 1 M KCl, 20 mM Tris pH 7.5. The calculated standard deviation of the noise in these experiments is 8.3 pA (see the Methods).
We further characterized the dynamics of β-CD transport for both sides of the pore (cis side entrance for negative bias and trans side entrance for positive bias). Interestingly, we find that the mean fractional current blockade values are voltage dependent for negative bias, whereas they remain constant for all positive bias values (Figure 8a–g). We determined the interaction dwell times (Figure 8h) by probing the characteristic time scale of the longer events in the distribution, as a function of the applied voltage. We observe that events are faster in the positive voltage direction (trans to cis) and slower in the negative direction (cis to trans), and further, transport is slower as the bias magnitude is increased in the cis to trans direction, with reduced fractional blockade values. To rationalize this asymmetric transport mechanism, we analyzed the structure of the internal loops that define the narrowest constriction of the pore, which serves an apparent dynamic function as a unidirectional valve (molecular ratchet) that retains packaged DNA in the phage head.38,44–46 Structural data on SPP1 portal suggested that the tunnel loops could be displaced along the tunnel axis along the trajectory of DNA translocation.38 The loop movements toward the capsid would slightly open the tunnel (increase its diameter) facilitating DNA translocation, while their movement in the opposite direction would narrow the channel. Thus, the tunnel loops may act as a molecular diaphragm that closes around DNA to prevent its leakage.
Figure 8.
Asymmetric interactions of β-CD with the CGG portal protein (a–f) Scatter plots of fraction blockade versus dwell time (bias values indicated in legends). (g) Mean fraction blockade of the long-lived events (>200 μs) as a function of the applied voltage. The error bars represent error over one recording. (h) Characteristic dwell times of the long-lived events as a function of the applied voltage in the presence of 0.16 mM β-CD in both chambers, cis side entrance for negative bias and trans side entrance for positive bias. The error bars represent error over one recording. (i) Force applied to β-CD during constant velocity SMD simulation of β-CD transport through the CGG portal in the cap-to-stem and stem-to-cap directions. The force plots were obtained by differentiating the work plots shown in the Figure S8. The z-coordinate is defined graphically in Figure 6g.
Interestingly, the plot of the SMD force applied during the forced permeation of β-CD through the CGG portal, Figure 8i, gives further support to the asymmetric transport hypothesis. Thus, the peak force required to move β-CD through the portal in the stem-to-cap direction is about 30% less than in the reverse direction, in qualitative agreement with the interpretation of the measurement. Note that the high absolute values of the SMD force are caused by the high speed of the β-CD transport realized in the SMD simulations.67 Using a ratchet analogy to describe the portal channel, transit of β-CD from trans to cis (positive voltage) is facilitated by the molecular diaphragm, whereas transit in the opposite direction (negative voltage) is not. One interpretation of the longer dwell times with increasing negative voltage values is that a β-CD interacts with the portal more favorably as voltage is increased or that the tunnel loops have restricted motion in this direction (toward the stem), consistent with structural observations.38
CONCLUSION
We have demonstrated here that chemically labeling a cysteine residue of an engineered G20c portal protein with a maleimide–porphyrin molecule facilitates its directional insertion into a planar lipid bilayer. Using electrical measurements, corroborated with MD simulations, we find that, unlike DNA origami pores embedded in lipid bilayers,55,59 the portal–lipid interface is not leaky, and >95% of ions are transported through the central portal channel. Engineering the inner channel of the portal by replacing residues with glycines results in pores with a larger constriction, as indicated by current–voltage measurements, MD simulations, and X-ray crystallography. The electroosmotic transport measurements using β-CD molecules point to a negatively charged inner pore consistent with the portal’s structure (Supporting Information Figure S10). However, electroosmotically assisted β-CD transport through the portal revealed slower transport in the cis to trans direction than the trans to cis direction, suggesting asymmetry in the inner portal channel architecture and/or biased motion of one or more structural features that form the pore surface. This asymmetry points to a possible mechanism for allowing DNA to be packaged into the capsid while preventing its uncontrolled escape, as previously suggested in other work.38,44–46 In summary, our simple approach for single-channel probing of the portal structure required minimal protein engineering, and provided valuable insight into the dynamic function of the internal pore, which complements X-ray crystallographic observations. This simple method could be used to engineer other nonmembrane inserting ring-shaped proteins, such as the trp attenuation protein (TRAP),77 for potential use as nanosensors. Future work will focus on further reprogramming the structural and electrostatic properties of the G20c portal protein for molecular sensing of charged species such as DNA and other biopolymers.
METHODS
Cloning, Expression, And Purification of G20c Portal Proteins
The DNA encoding for G20c portal protein (residues 25–438) was amplified by PCR using Phusion high fidelity DNA polymerase (New England Biolabs), and cloned into the YSBL-Lic+ expression plasmid78 encoding an N-terminal 3C protease cleavable hexahistidine tag using the HiFi DNA assembly master mix (New England Biolabs). All mutant variations of the wild-type (WT) protein were produced using a variation of the linear exponential PCR and ligase-dependent production of closed circular plasmid DNA using Phusion DNA polymerase (New England Biolabs). Briefly, primer sets were designed to introduce the mutation(s) and amplify the entire plasmid (see Supporting Information S11) by PCR, after which the DNA product was purified using a PCR cleanup kit (Thermofisher). Phosphorylation and ligation of the amplified DNA ends and Dpn I digest of the template plasmid was achieved in a 3 h reaction at 37 °C, containing 1X Cutsmart buffer (New England Biolabs), 1 mM ATP, 10 mM DTT, and 1 unit each of Dpn I, T4 polynucleotide kinase, T4 ligase (New England Biolabs). Ligated closed circular plasmid DNA was transformed into competent DH5α cells. Mutants were screened by colony PCR and confirmed by DNA sequencing in both directions.
Wild type and mutant proteins G20c WT (WT) and G20c V325G_I328G (GG)) were expressed and purified from E. coli BL21 (DE3) pLys S cells. Proteins containing cysteine mutants, G20c-L49C (49C) and G20c-L49C_V325G_I328G (CGG) were expressed and purified from the SHuffle (New England Biolabs) expression strain.
Protein expression and purification was conducted as described52 in LB (Melford) containing 35 μg/mL kanamycin and 50 μg/mL chloramphenicol. Briefly, 10 mL of an overnight culture was inoculated into 1L of LB (containing antibiotics) and incubated at 37 °C until the OD600 reached 0.8, followed by induction overnight at 16 °C with 0.5 mM IPTG when the cells were harvested by centrifugation at 4000 rpm for 30 min and the pellets snap frozen in liquid nitrogen and stored at −80 °C until use. Proteins (49C and CGG) expressed in Shuffle cells were incubated at 30 °C before and after induction. Cell pellets were thawed and resuspended in 5 mL/g of lysis buffer (50 mM Tris pH 8, 1 M NaCl, 10 mM imidazole, 100 mM AEBSF, 10 mg/mL lysozyme) and lysed by sonication on ice. The lysate was clarified by centrifugation at 15000 rpm for 30 min, filtered through a 0.22 μm membrane before loading on a HisTrap FF 5 mL (GE Healthcare Life Sciences). The His-tagged G20c protein was eluted using a gradient to 100% Buffer B (1 M NaCl, 50 mM Tris pH 7.5, 500 mM imidazole) over 10 column volumes. Fractions of the purified protein were pooled, buffer exchanged into 50 mM Tris pH 8, 500 mM NaCl 50 mM potassium glutamate, and the histidine tag removed by 3C protease digestion at rt overnight. The cleaved protein was then further purified over a HisTrap FF 5 mL. Fractions containing cleaved G20c protein were pooled, concentrated, and finally purified on a 16/ 600 Superose 6 (GE healthcare Life Sciences) gel filtration column in 20 mM Tris pH8, 1 M NaCl, 50 mM potassium glutamate, concentrated by ultrafiltration using a 100 kDa cutoff filter (Vivaspin) to ~4 mg/mL for biophysical experiments or ~10 mg/mL for crystallographic studies, snap frozen on liquid nitrogen and stored at −80 °C. Proteins containing the L49C mutation were purified in buffers containing 2 mM DTT.
Crystallization and X-ray Data Collection
The purified V325G mutant protein (in 1 M NaCl, 20 mM Tris pH 7.5) was concentrated to ~11 mg/mL for crystallization by sitting drop vapor diffusion. The best crystals, which diffracted to 1.90 Å, were obtained after 5 days in 0.1 M imidazole · HCl pH 8.0 with 30% (w/v) MPD and 10% (w/v) PEG 4000, and belonged to the R3 space group with a = 223, c = 116 Å (hexagonal setting). Using a similar approach, the V325G I328G variant (in 0.5 M NaCl, 20 mM Tris pH 7.5, and concentrated to ~12 mg/mL) yielded the best crystals after 7 days with the reservoir solution containing 0.2 M NH4Cl and 40% (v/v) MPD. These belonged to the P212121 space group with a = 158, b = 192, c = 251. X-ray diffraction data were collected using synchrotron radiation at 0.97883 Å wavelength and 100 K, at the I24 beamline of Diamond Light Source. The data were processed using DIALS.79 The V325G and V325G/I328G portal protein crystals diffracted to 1.90 and 3.50 Å, respectively, and the self-rotation function of the V325G/I328G variant was calculated using MOLREP.80
X-ray Structure Determination
The crystal structure of the V325G mutant was determined using the CCP4 software suite.81 Phases were calculated using data previously collected for the selenomethionine derivative of the wild-type G20C portal protein (PDB code 4zjn). The selenomethionine crystal belongs to the P4212 space group with a = 156, c = 116 Å, and diffracted to ~2.4 Å. Heavy atom sites were found using SHELXC/SHELXD, phases were calculated using SHELXE82 and a selenomethionine model was autobuilt using Buccaneer.83 Data reduction for the V325G mutant protein using AIMLESS84 confirmed the R3 space group. The selenomethionine protein structure chain A coordinates were used as a search model for molecular replacement using Phaser,85 by searching for 4 subunits per asymmetric part. Given the presence of the 3-fold rotational symmetry in the R3 space group, this corresponds to the 12-fold symmetry of the portal protein. The final model was obtained from an iterative process of refinement using REFMAC586 and manual model building using Coot87 (refinement statistics listed in Supporting Information S12). The quality of this model was analyzed using MolProbity.88 Display images were prepared using Chimera89 and PyMol (Schrödinger LLC).
Analysis of Subunit–Subunit Interfaces and Electrostatic Surface Potential
Direct intersubunit hydrogen bonds and salt bridges were identified using the FindHbond function of Chimera.89 A 4.0 Å cutoff was applied for salt bridges,90 whereas hydrogen bonds were identified by considering a maximum length of 3.3 Å between donors and acceptors, and by relaxing angles 8° away from the geometry criteria described by Mills and Dean.91 Surface electrostatic potentials (Supporting Information Figure S10) were calculated (at 150 mM NaCl, pH 7.0) under the SWANSON force field ±1 kT/e using APBS.92–94
Electron Microscopy
Grids for transmission electron microscopy were prepared by negative staining, using 2% uranyl acetate solution 0.1 mg/mL protein in 1 M NaCl 20 mM Tris pH 7.5. Images were recorded at 120kx magnification, on a TEM JEOL JEM-1010 80 kV instrument.
Denaturant-Induced Unfolding Transition
A Fluoromax-4 (Horiba Scientific) fluorimeter was used for all tryptophan fluorescence measurements. G20c contains 6 tryptophan residues. Samples were incubated 24 h at 20 °C using 0.5 μM of G20c in 1 M NaCl, 20 mM Tris pH 7.5 plus concentrations of Gdn-HCl varying from 0 to 6M. The data were collected at an excitation wavelength of 280 nm. The integration time was 1 s.
Thermofluor Assays
Thermofluor assays were carried out in 96-well plates using a Mx3005P QPCR system by adding 10 μL of 0.25 mg/mL protein, 10 μL of buffer (1 M NaCl, 20 mM Tris pH 7.5), and 5 μL of 5× SYPRO Orange dye95–97 to each well. The melting temperature was determined by fitting the data with a five-parameter sigmoid using the Levenberg–Marquadt algorithm.
Maleimide–Porphyrin Synthesis
The synthesis was based on previous work published by Liu et al.98 Protoporphyrin IX (100 mg, 0.17 mmol), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC, 32.6 mg, 0.17 mmol), and N-hydroxysuccinimide (NHS, 20 mg, 0.17 mmol) were dissolved in 10 mL of dimethylformamide (DMF) and stirred for 2 h at room temperature. N-(2-Aminoethyl) maleimide trifluoroacetate in 2 mL of DMF was added. The reaction mixture was allowed to stir overnight. The product was purified by silica chromatography to give a red solid.
Maleimide–Porphyrin Conjugation with Portal Protein
The method used in this step is adapted from previous worked published by Milgrom et al.99 In summary, DTT was removed before maleimide–porphyrin conjugation reaction using Zeba Spin Desalting Columns (Thermo Scientific). Porphyrin–maleimide dissolved in DMSO was added to the purified portal protein in 20-fold molar excess. The vial was completely flushed with N2 gas and kept at rt overnight. Nonconjugated porphyrin–maleimide molecules were separated from conjugated portal protein using a Superdex 75 16/ 600 (GE healthcare Life Sciences) gel filtration column following absorbance at 280 and 410 nm.
Analytical Size-Exclusion Chromatography and UV–vis Absorbance Spectroscopy
Labeled proteins and their unlabeled counterparts were compared by analytical size-exclusion chromatography on a Superose 6 10/300 column (GE healthcare Life Sciences), equilibrated with 20 mM Tris pH 8, 1 M NaCl, and 1 mM TCEP, using an AKTA PURE fast protein purification system (GE healthcare Life Sciences). Each sample (100 μL, 13–130 μM) was loaded onto the column using 500 μL loops and eluted at a linear flow rate of 0.5 mL/min. Eluted products were monitored for optical density at the wavelengths for the absorbance maxima for both protein (280 nm) and porphyrin (410 nm). The elution volumes for the observed peaks were compared to those of known high molecular weight standards used to calibrate the column. Absorbance profiles were normalized using the molar extinction coefficients of 620560 and 80000 M−1 cm−1 for portal dodecamers and porphyrin in aqueous buffers,63 respectively, and plotted using proFit (Quantum Soft). Fractions comprising the labeled dodecameric assemblies were pooled and the absorbance spectral profile measured over 200–800 nm in a 1 cm quartz cuvette and Cary 100 UV–vis spectrophotometer (Agilent).
Electrical Detection and Data Acquisition
Membrane lipid bilayers were made according to previously described methods.100 In brief, a film of a 1% solution of diphytanoyl-phosphatidylcholine-lecithin (Avanti) in decane was spread across a 150 μm diameter hole drilled in a polysulfone wall separating the two compartments of a chamber. Each compartment contained 1 mL of 1 M NaCl, 20 mM tris, pH 7.5 or 1 M KCl, 20 mM Tris pH 7.5. After thinning of the decane film and formation of a planar lipid bilayer, dodecameric recombinant G20c from a stock solution was added into the cis compartment.
The ionic current through a single G20c portal protein was measured using an Axopatch 200B amplifier (Molecular Devices). Data were filtered at 10 kHz and acquired at 250 kHz using the DigiData 1200 digitizer with a custom National Instruments LabVIEW program. Data was processed and events were detected using MOSAIC101 as follows. The values for the open pore current (Io) and the standard deviation of the noise (σ) was extracted (108 and 8.3 pA, respectively, for β-CD, Figure S13). The threshold (Th) applied in Mosaic to separate events from the noise is given by Th = Io – 3σ (Th = 83 pA for β-CD, Figure S13). An example of this data processing is provided for β-CD experiments in Supporting Information Figure S13.
Molecular Dynamics Simulations
All simulations were performed using the molecular dynamics program NAMD2,102 periodic boundary conditions, and a 2 fs time step. The CHARMM36 force field103 was used to describe proteins, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) lipids, CDs, TIP3P water, and ions. Cgenff server103,104 was used to generate CHARMM-compatible parameters for porphyrin moieties. The CUFIX corrections were applied to improve description of charge–charge interactions.105,106 RATTLE107 and SETTLE108 algorithms were applied to describe covalent bonds that involved hydrogen atoms. Particle-Mesh-Ewald (PME)109 algorithm was used to evaluate the long-range electrostatic interaction on a 1 Å-spaced grid; the full electrostatics calculation was performed every three timesteps. van der Waals interactions were evaluated using a smooth 10–12 Å cutoff.
An all-atom model of the G20c protein was constructed starting from its crystallographic structure, Protein Data Bank entry 4ZJN. Residues missing in the crystallographic structure were added using the psfgen module of VMD.110 The structure was then aligned to be coaxial with the z-axis of our coordinate system. One M NaCl solution was added using the solvate and autoionize plugins of VMD, and a small number of additional ions were added to make the final system of 838565 atoms electrically neutral. Following assembly, the system was minimized in 9600 steps using the conjugate gradient method and then equilibrated for 50 ns at a constant number of atoms, pressure and temperature (NPT) ensemble. During the initial stage of equilibration, all non-hydrogen atoms of the protein were restrained to their initial coordinates using harmonic potentials; the spring constant of the potentials decreased from 1.0 to 0.1 to 0.01 kcal/(mol Å2) in ~4 ns steps. The system was simulated in the absence of any restraints for 40 ns. The Nose–Hoover Langevin piston pressure control111,112 was used to maintain the pressure of the system at 1 atm by adjusting the system’s dimension. Langevin thermostat113 was applied to all the heavy atoms of the system with a damping coefficient of 0.1 ps−1 to maintain the system temperature at 295 K.
A model of the G20c protein embedded in a lipid membrane was constructed by combining the equilibrated structure of the protein with a 21 nm × 21 nm patch of a pre-equilibrated DOPC bilayer. Prior to protein insertion, 12 porphyrin moieties were added to the equilibrated structure via a harmonic bond (2.9 Å bond length and 1 kcal/(mol Å2) spring constant) between the sulfur atom of each Cys49 residue and a carbon atom of each porphyrin, mimicking the chemical bond realized in experiment. The porphyrins were initially positioned below the G20c cap, in the region where the DOPC patch would be placed; the plane of the porphyrins’ aromatic rings was normal to the plane of the bilayer. The lipid bilayer membrane was aligned with the x–y plane and shifted along the z-axis to have the Cys49 residues of the protein were located within the same plane as the head groups of the nearest lipid leaflet. Lipid and water molecules that overlapped with the protein and the porphyrins were removed. One M NaCl solution was added above and below the membrane, producing an electrically neutral system containing 792391 atoms. Following a 9600-step minimization, the system was equilibrated for ~150 ns in the NPT ensemble; fluctuation of the system’s dimensions within the plane of the bilayer were coupled by a constant factor. During the initial state of equilibration, all non-hydrogen atoms of the protein and the porphyrins were harmonically restrained to their initial coordinates; the strength of each harmonic restrain was 0.1 kcal/ (molÅ 2) for the first 2.4 ns of equilibration and 0.01 kcal/(molÅ 2) for the subsequent 8.4 ns. The system was simulated without any restrained for another 140 ns. The final equilibrated conformation was used make the V325G/I328G mutant structure. The mutant structure was equilibrated for 2 ns in the constant ratio NPT ensemble. The simulations under a transmembrane bias were performed in a constant number of particle, volume and temperature (NVT) ensemble. For the NVT simulations, the system’s dimensions were set to the average dimensions observed within the last 24 ns of the NPT equilibration. To produce a transmembrane bias of ±100 mV, an external electric field was applied along the z-axis (normal to the membrane); the strength of the electric field was computed as E = −VLz, where Lz is the dimension of the simulated system in the direction of the applied electric field.65
Prior to ionic current calculations, frames from the MD trajectory were aligned using protein coordinates to correct for the drift in the x-y plane and lipid bilayer coordinates to correct for the drift along the z-axis. The ionic current was calculated as
where δzj(t) is the displacement of ion j along the z direction during the time interval δt = 4.8 ps and qj is the charge of ion j. To minimize the effect of thermal noise, the current was calculated within an Lz = 30 Å thickness slab centered at the midplane of the lipid bilayer membrane (the slab spanned the entire simulation system in the x–y plane). The calculations of local densities and ion fluxed were performed on a 50 × 50 × 76 cubic grid using a previously described method.55,59,114
Steered molecular dynamics (SMD)67 was used to probe translocation of α, β-, and γ-CDs through the G20c portal. The initial coordinates for the CD variants were taken from PDB structures 5E6Y, 5E6Z, and 2ZYK. Each CD molecule was combined with the equilibrated structure of the G20c protein; the structure did not contain porphyrin anchors nor did it contain the lipid bilayer. Each CD molecule was initially placed ~1.0 nm away from an entrance of the portal channel. KCl solution (1 M) was added, producing an electrically neutral system of approximately 396800 atoms. The systems were minimized and equilibrated for 9.6 ns following the same protocols as the bulk equilibration of G20c; all non-hydrogen atoms of the protein were restrained to their initial coordinates using harmonic potentials with the 0.01 kcal/(molÅ2) spring constant. The SMD simulations were carried out in the NVT ensemble. The center of mass (CoM) of each CD molecule was tethered to a harmonic spring of 1.0 kcal/(mol Å2) spring constant. The other end of the spring was moved along the z-axis (the symmetry axis of the G20c pore) with the speed of 1 nm/ns; the CD’s CoM was also radially restrained (spring constant of 1.0 kcal/(mol Å2)) to remain at the axis of the pore. After 13 ns, the CDs passed through the channel and the SMD simulations were repeated in the reverse direction. In addition to SMD pulling of CDs through the G20c pore, they were also pulled through bulk 1 M KCl solution. The average force required to move α-, β-, and γ-CDs through bulk solution at 1 nm/ns was approximately 5pN.
Ionic current blockade produced by the presence of the CD variants in the G20c pore was computed using a theoretical model described elsewhere.68 Briefly, for each microscopic configuration realized during the SMD simulations, we computed a 3D distance map at 1 Å resolution that specified, for each location within the G20c pore volume, the nearest distance to the protein or CD surface. The distance map was used to compute local ionic conductivity within the G20c pore; the pore’s conductance was determined by applying the Ohm law. When applied to the open-pore 49C and CGG systems, the model yielded the raw (unscaled) conductance values of 2.7 and 3.5 nS, respectively, which is in excellent agreement with the results of brute-force all-atom simulations.
Supplementary Material
Acknowledgments
This work was supported by grants from the following agencies: Bilateral United Kingdom Biotechnology and Biological Sciences Research Council - United States National Science Foundation Lead Agency Pilot Program (BB/N018729/1 and NSF-1645671 to A.A.A. and M.W.), Wellcome Trust (098230, 101528 to A.A.A.), the National Institutes of Health (R01-HG007406 and P41-GM104601 to A.A.), and The Royal Society (International Exchange Award IE150888 to B.C. and S.J.G.). We acknowledge Johan Turkenburg and Sam Hart for X-ray data collection at Diamond Light Source, UK, and Diamond Light Source for access to the I24 beamline (Proposal No. mx-13587) that contributed to the results presented here. Supercomputer time was provided through the XSEDE Allocation Grant No. MCA05S028 and the Blue Waters petascale supercomputer system at the University of Illinois at Urbana–Champaign. W.S. acknowledges financial support from the China Scholarship Council (CSC201506090040) and the National Natural Science Foundation of China (Grant No. 51435003).
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b06980.
Additional figures, list of primers, and X-ray data (PDF)
References
- 1.Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, Gundlach JH. Nanopore DNA Sequencing with MspA. Proc Natl Acad Sci U S A. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain M, Fiddes IT, Miga KH, Olsen HE, Paten B, Akeson M. Improved Data Analysis for the MinION Nanopore Sequencer. Nat Methods. 2015;12:351–356. doi: 10.1038/nmeth.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loman NJ, Quick J, Simpson JT. A Complete Bacterial Genome Assembled De Novo Using Only Nanopore Sequencing Data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 4.Madoui MA, Engelen S, Cruaud C, Belser C, Bertrand L, Alberti A, Lemainque A, Wincker P, Aury JM. Genome Assembly Using Nanopore-Guided Long and Error-Free DNA Reads. BMC Genomics. 2015;16:327. doi: 10.1186/s12864-015-1519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O’Grady J. MinION Nanopore Sequencing Identifies the Position and Structure of a Bacterial Antibiotic Resistance Island. Nat Biotechnol. 2015;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 6.Howorka S, Siwy Z. Nanopore Analytics: Sensing of Single Molecules. Chem Soc Rev. 2009;38:2360–2384. doi: 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- 7.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc Natl Acad Sci U S A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Stochastic Sensing of Organic Analytes by a Pore-Forming Protein Containing a Molecular Adapter. Nature. 1999;398:686–690. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 9.Guan X, Gu LQ, Cheley S, Braha O, Bayley H. Stochastic Sensing of TNT with a Genetically Engineered Pore. ChemBioChem. 2005;6:1875–1881. doi: 10.1002/cbic.200500064. [DOI] [PubMed] [Google Scholar]
- 10.Mohammad MM, Iyer R, Howard KR, McPike MP, Borer PN, Movileanu L. Engineering a Rigid Protein Tunnel for Biomolecular Detection. J Am Chem Soc. 2012;134:9521–9531. doi: 10.1021/ja3043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wescoe ZL, Schreiber J, Akeson M. Nanopores Discriminate Among Five C5-Cytosine Variants in DNA. J Am Chem Soc. 2014;136:16582–16587. doi: 10.1021/ja508527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasianowicz JJ, Balijepalli AK, Ettedgui J, Forstater JH, Wang H, Zhang H, Robertson JWF. Analytical Applications for Pore-Forming Proteins. Biochim Biophys Acta, Biomembr. 2016;1858:593–606. doi: 10.1016/j.bbamem.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Ettedgui J, Kasianowicz JJ, Balijepalli A. Single Molecule Discrimination of Heteropolytungstates and Their Isomers in Solution with a Nanometer-Scale Pore. J Am Chem Soc. 2016;138:7228–7231. doi: 10.1021/jacs.6b02917. [DOI] [PubMed] [Google Scholar]
- 14.Baaken G, Halimeh I, Bacri L, Pelta J, Oukhaled A, Behrends JC. High-Resolution Size-Discrimination of Single Nonionic Synthetic Polymers with a Highly Charged Biological Nanopore. ACS Nano. 2015;9:6443–6449. doi: 10.1021/acsnano.5b02096. [DOI] [PubMed] [Google Scholar]
- 15.Robertson JWF, Rodrigues CG, Stanford VM, Rubinson KA, Krasilnikov OV, Kasianowicz JJ. Single-Molecule Mass Spectrometry in Solution Using a Solitary Nanopore. Proc Natl Acad Sci U S A. 2007;104:8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piguet F, Ouldali H, Discala F, Breton MF, Behrends JC, Pelta J, Oukhaled A. High Temperature Extends the Range of Size Discrimination of Nonionic Polymers by a Biological Nanopore. Sci Rep. 2016;6:38675. doi: 10.1038/srep38675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fennouri A, Daniel R, Pastoriza-Gallego M, Auvray L, Pelta J, Bacri L. Kinetics of Enzymatic Degradation of High Molecular Weight Polysaccharides Through a Nanopore: Experiments and Data-Modeling. Anal Chem. 2013;85:8488–8492. doi: 10.1021/ac4020929. [DOI] [PubMed] [Google Scholar]
- 18.Fennouri AA, Przybylski CC, Pastoriza-Gallego MM, Bacri LL, Auvray LL, Daniel RR. Single Molecule Detection of Glycosaminoglycan Hyaluronic Acid Oligosaccharides and Depolyme-rization Enzyme Activity Using a Protein Nanopore. ACS Nano. 2012;6:9672–9678. doi: 10.1021/nn3031047. [DOI] [PubMed] [Google Scholar]
- 19.Luchian T, Shin SH, Bayley H. Kinetics of a Three-Step Reaction Observed at the Single-Molecule Level. Angew Chem, Int Ed. 2003;42:1926–1929. doi: 10.1002/anie.200250666. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, Boersma AJ, Boudreau MA, Cheley S, Daltrop O, Li J, Tamagaki H, Bayley H. Semisynthetic Nanoreactor for Reversible Single-Molecule Covalent Chemistry. ACS Nano. 2016;10:8843–8850. doi: 10.1021/acsnano.6b04663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willems K, Van Meervelt V, Wloka C, Maglia G. Single-Molecule Nanopore Enzymology. Philos Trans R Soc, B. 2017;372:20160230. doi: 10.1098/rstb.2016.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammad MM, Movileanu L. Protein Sensing with Engineered Protein Nanopores. Methods Mol Biol. 2012;870:21–37. doi: 10.1007/978-1-61779-773-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nivala J, Mulroney L, Li G, Schreiber J, Akeson M. Discrimination Among Protein Variants Using an Unfoldase-Coupled Nanopore. ACS Nano. 2014;8:12365–12375. doi: 10.1021/nn5049987. [DOI] [PubMed] [Google Scholar]
- 24.Nivala J, Marks DB, Akeson M. Unfoldase-Mediated Protein Translocation Through an A-Hemolysin Nanopore. Nat Biotechnol. 2013;31:247–250. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Larrea D, Bayley H. Multistep Protein Unfolding During Nanopore Translocation. Nat Nanotechnol. 2013;8:288–295. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Larrea D, Bayley H. Protein Co-Translocational Unfolding Depends on the Direction of Pulling. Nat Commun. 2014;5:4841–4841. doi: 10.1038/ncomms5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oukhaled G, Mathe J, Biance AL, Bacri L, Betton JM, Lairez D, Pelta J, Auvray L. Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys Rev Lett. 2007;98:158101–158101. doi: 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- 28.Merstorf C, Cressiot B, Pastoriza-Gallego M, Oukhaled A, Betton JM, Auvray L, Pelta J. Wild Type, Mutant Protein Unfolding and Phase Transition Detected by Single-Nanopore Recording. ACS Chem Biol. 2012;7:652–658. doi: 10.1021/cb2004737. [DOI] [PubMed] [Google Scholar]
- 29.Payet L, Martinho M, Pastoriza-Gallego M, Betton JM, Auvray L, Pelta J, Mathé J. Thermal Unfolding of Proteins Probed at the Single Molecule Level Using Nanopores. Anal Chem. 2012;84:4071–4076. doi: 10.1021/ac300129e. [DOI] [PubMed] [Google Scholar]
- 30.Pastoriza-Gallego M, Rabah L, Gibrat G, Thiebot B, van der Goot FG, Auvray L, Betton JM, Pelta J. Dynamics of Unfolded Protein Transport Through an Aerolysin Pore. J Am Chem Soc. 2011;133:2923–2931. doi: 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- 31.Pastoriza-Gallego M, Breton MF, Discala F, Auvray L, Betton JM, Pelta J. Evidence of Unfolded Protein Translocation Through a Protein Nanopore. ACS Nano. 2014;8:11350–11360. doi: 10.1021/nn5042398. [DOI] [PubMed] [Google Scholar]
- 32.Cressiot B, Braselmann E, Oukhaled A, Elcock AH, Pelta J, Clark PL. Dynamics and Energy Contributions for Transport of Unfolded Pertactin Through a Protein Nanopore. ACS Nano. 2015;9:9050–9061. doi: 10.1021/acsnano.5b03053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biesemans A, Soskine M, Maglia G. A Protein Rotaxane Controls the Translocation of Proteins Across a ClyA Nanopore. Nano Lett. 2015;15:6076–6081. doi: 10.1021/acs.nanolett.5b02309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen CB, Rodriguez-Larrea D, Bayley H. Single-Molecule Site-Specific Detection of Protein Phosphorylation with a Nanopore. Nat Biotechnol. 2014;32:179–181. doi: 10.1038/nbt.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wloka C, Van Meervelt V, van Gelder D, Danda N, Jager N, Williams CP, Maglia G. Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore. ACS Nano. 2017;11:4387–4394. doi: 10.1021/acsnano.6b07760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmerberg J, Parsegian VA. Polymer Inaccessible Volume Changes During Opening and Closing of a Voltage-Dependent Ionic Channel. Nature. 1986;323:36–39. doi: 10.1038/323036a0. [DOI] [PubMed] [Google Scholar]
- 37.Casjens SR, Gilcrease EB. Determining DNA Packaging Strategy by Analysis of the Termini of the Chromosomes in Tailed-Bacteriophage Virions. Methods Mol Biol. 2009;502:91–111. doi: 10.1007/978-1-60327-565-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural Framework for DNA Translocation via the Viral Portal Protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olia AS, Prevelige PE, Johnson JE, Cingolani G. Three-Dimensional Structure of a Viral Genome-Delivery Portal Vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossmann MG, Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, et al. Structure of the Bacteriophage |[Phis]|29 DNA Packaging Motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlova EV, Gowen B, Dröge A, Stiege A, Weise F, Lurz R, van Heel M, Tavares P. Structure of a Viral DNA Gatekeeper at 10 a Resolution by Cryo-Electron Microscopy. EMBO J. 2003;22:1255–1262. doi: 10.1093/emboj/cdg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The Structure of an Infectious P22 Virion Shows the Signal for Headful DNA Packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 43.Casjens SR. The DNA-Packaging Nanomotor of Tailed Bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 44.Grimes S, Ma S, Gao J, Atz R, Jardine PJ. Role of Φ29 Connector Channel Loops in Late-Stage DNA Packaging. J Mol Biol. 2011;410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geng J, Fang H, Haque F, Zhang L, Guo P. Three Reversible and Controllable Discrete Steps of Channel Gating of a Viral DNA Packaging Motor. Biomaterials. 2011;32:8234–8242. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar R, Grubmüller H. Phi29 Connector-DNA Interactions Govern DNA Crunching and Rotation, Supporting the Check-Valve Model. Biophys J. 2016;110:455–469. doi: 10.1016/j.bpj.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji Z, Wang S, Zhao Z, Zhou Z, Haque F, Guo P. Fingerprinting of Peptides with a Large Channel of Bacteriophage Phi29 DNA Packaging Motor. Small. 2016;12:4572–4578. doi: 10.1002/smll.201601157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P. Translocation of Double-Stranded DNA Through Membrane-Adapted Phi29 Motor Protein Nanopores. Nat Nanotechnol. 2009;4:765–772. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, Zhou Z, Zhao Z, Zhang H, Haque F, Guo P. Channel of Viral DNA Packaging Motor for Real Time Kinetic Analysis of Peptide Oxidation States. Biomaterials. 2017;126:10–17. doi: 10.1016/j.biomaterials.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Z, Ji Z, Wang S, Haque F, Guo P. Oriented Single Directional Insertion of Nanochannel of Bacteriophage SPP1 DNA Packaging Motor Into Lipid Bilayer via Polar Hydrophobicity. Biomaterials. 2016;105:222–227. doi: 10.1016/j.biomaterials.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu R-G, Jenkins HT, Chechik M, Blagova EV, Lopatina A, Klimuk E, Minakhin L, Severinov K, Greive SJ, Antson AA. Viral Genome Packaging Terminase Cleaves DNA Using the Canonical RuvC-Like Two-Metal Catalysis Mechanism. Nucleic Acids Res. 2017;45(6):3580–3590. doi: 10.1093/nar/gkw1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams LS, Levdikov VM, Minakhin L, Severinov K, Antson AA. 12-Fold Symmetry of the Putative Portal Protein From the Thermus Thermophilus Bacteriophage G20C Determined by X-Ray Analysis. Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2013;69:1239–1241. doi: 10.1107/S174430911302486X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howorka S. Building Membrane Nanopores. Nat Nanotechnol. 2017;12:619–630. doi: 10.1038/nnano.2017.99. [DOI] [PubMed] [Google Scholar]
- 54.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Göpfrich K, Li CY, Ricci M, Bhamidimarri SP, Yoo J, Gyenes B, Ohmann A, Winterhalter M, Aksimentiev A, Keyser UF. Large-Conductance Transmembrane Porin Made From DNA Origami. ACS Nano. 2016;10:8207–8214. doi: 10.1021/acsnano.6b03759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burns JR, Stulz E, Howorka S. Self-Assembled DNA Nanopores That Span Lipid Bilayers. Nano Lett. 2013;13:2351–2356. doi: 10.1021/nl304147f. [DOI] [PubMed] [Google Scholar]
- 57.Burns JR, Göpfrich K, Wood JW, Thacker VV, Stulz E, Keyser UF, Howorka S. Lipid-Bilayer-Spanning DNA Nanopores with a Bifunctional Porphyrin Anchor. Angew Chem, Int Ed. 2013;52:12069–12072. doi: 10.1002/anie.201305765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seifert A, Göpfrich K, Burns JR, Fertig N, Keyser UF, Howorka S. Bilayer-Spanning DNA Nanopores with Voltage-Switching Between Open and Closed State. ACS Nano. 2015;9:1117–1126. doi: 10.1021/nn5039433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Göpfrich K, Li CY, Mames I, Bhamidimarri SP, Ricci M, Yoo J, Mames A, Ohmann A, Winterhalter M, Stulz E, et al. Ion Channels Made From a Single Membrane-Spanning DNA Duplex. Nano Lett. 2016;16:4665–4669. doi: 10.1021/acs.nanolett.6b02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hudson R, Carcenac M, Smith K, Madden L, Clarke OJ, Pèlegrin A, Greenman J, Boyle RW. The Development and Characterisation of Porphyrin Isothiocyanate-Monoclonal Antibody Conjugates for Photoimmunotherapy. Br J Cancer. 2005;92:1442–1449. doi: 10.1038/sj.bjc.6602517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donald PJ, Cardiff RD, He DE, Kendall K. Monoclonal Antibody-Porphyrin Conjugate for Head and Neck Cancer: the Possible Magic Bullet. Otolaryngol –Head Neck Surg. 1991;105:781–787. doi: 10.1177/019459989110500603. [DOI] [PubMed] [Google Scholar]
- 62.Alonso CMA, Palumbo A, Bullous AJ, Pretto F, Neri D, Boyle RW. Site-Specific and Stoichiometric Conjugation of Cationic Porphyrins to Antiangiogenic Monoclonal Antibodies. Bioconjugate Chem. 2010;21:302–313. doi: 10.1021/bc9003537. [DOI] [PubMed] [Google Scholar]
- 63.Hasan T, Ortel B, Solban N, Pogue B. Photodynamic Therapy of Cancer. Cancer Med. 2003;7:537–48. [Google Scholar]
- 64.Yoo J, Aksimentiev A. Molecular Dynamics of Membrane-Spanning DNA Channels: Conductance Mechanism, Electro-Osmotic Transport, and Mechanical Gating. J Phys Chem Lett. 2015;6:4680–4687. doi: 10.1021/acs.jpclett.5b01964. [DOI] [PubMed] [Google Scholar]
- 65.Aksimentiev AA, Schulten KK. Imaging Alpha-Hemolysin with Molecular Dynamics: Ionic Conductance, Osmotic Permeability, and the Electrostatic Potential Map. Biophys J. 2005;88:3745–3761. doi: 10.1529/biophysj.104.058727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gidwani B, Vyas A. A Comprehensive Review on Cyclodextrin-Based Carriers for Delivery of Chemotherapeutic Cytotoxic Anticancer Drugs. BioMed Res Int. 2015;2015:198268. doi: 10.1155/2015/198268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isralewitz B, Izrailev S, Schulten K. Binding Pathway of Retinal to Bacterio-Opsin: a Prediction by Molecular Dynamics Simulations. Biophys J. 1997;73:2972–2979. doi: 10.1016/S0006-3495(97)78326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Si W, Aksimentiev A. Nanopore Sensing of Protein Folding. ACS Nano. 2017;11:7091–7100. doi: 10.1021/acsnano.7b02718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu LQ, Bayley H. Interaction of the Noncovalent Molecular Adapter, Beta-Cyclodextrin, with the Staphylococcal Alpha-Hemolysin Pore. Biophys J. 2000;79:1967–1975. doi: 10.1016/S0006-3495(00)76445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oukhaled A, Bacri L, Pastoriza-Gallego M, Betton JM, Pelta J. Sensing Proteins Through Nanopores: Fundamental to Applications. ACS Chem Biol. 2012;7:1935–1949. doi: 10.1021/cb300449t. [DOI] [PubMed] [Google Scholar]
- 71.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 72.Piguet F, Discala F, Breton MF, Pelta J, Bacri L, Oukhaled A. Electroosmosis Through Alpha-Hemolysin That Depends on Alkali Cation Type. J Phys Chem Lett. 2014;5:4362–4367. doi: 10.1021/jz502360c. [DOI] [PubMed] [Google Scholar]
- 73.Gu LQ, Cheley S, Bayley H. Electroosmotic Enhancement of the Binding of a Neutral Molecule to a Transmembrane Pore. Proc Natl Acad Sci U S A. 2003;100:15498–15503. doi: 10.1073/pnas.2531778100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boukhet M, Piguet F, Ouldali H, Pastoriza-Gallego M, Pelta J, Oukhaled A. Probing Driving Forces in Aerolysin and A-Hemolysin Biological Nanopores: Electrophoresis Versus Electro-osmosis. Nanoscale. 2016;8:18352–18359. doi: 10.1039/c6nr06936c. [DOI] [PubMed] [Google Scholar]
- 75.Wong CTA, Muthukumar M. Polymer Translocation Through A-Hemolysin Pore with Tunable Polymer-Pore Electrostatic Interaction. J Chem Phys. 2010;133:045101. doi: 10.1063/1.3464333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhamidimarri SP, Prajapati JD, Van den Berg B, Winterhalter M, Kleinekathöfer U. Role of Electroosmosis in the Permeation of Neutral Molecules: CymA and Cyclodextrin as an Example. Biophys J. 2016;110:600–611. doi: 10.1016/j.bpj.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. Structure of the Trp RNA-Binding Attenuation Protein, TRAP, Bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 78.Bonsor D, Butz SF, Solomons J, Grant S, Fairlamb IJS, Fogg MJ, Grogan G. Ligation Independent Cloning (LIC) as a Rapid Route to Families of Recombinant Biocatalysts From Sequenced Prokaryotic Genomes. Org Biomol Chem. 2006;4:1252–1260. doi: 10.1039/b517338h. [DOI] [PubMed] [Google Scholar]
- 79.Waterman DG, Winter G, Gildea RJ, Parkhurst JM, Brewster AS, Sauter NK, Evans G. Diffraction-Geometry Refinement in the DIALS Framework. Acta Crystallogr, D, Struct Biol. 2016;72:558–575. doi: 10.1107/S2059798316002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 81.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr, Sect D: Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sheldrick GM. A Short History of SHELX. Acta Crystallogr, Sect A: Found Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 83.Cowtan K. The Buccaneer Software for Automated Model Building. 1. Tracing Protein Chains. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 84.Evans PR, Murshudov GN. How Good Are My Data and What Is the Resolution? Acta Crystallogr, Sect D: Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser Crystallographic Software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr, Sect D: Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 88.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 90.Barlow DJ, Thornton JM. Ion-Pairs in Proteins. J Mol Biol. 1983;168:867–885. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- 91.Mills JE, Dean PM. Three-Dimensional Hydrogen-Bond Geometry and Probability Information From a Crystal Survey. J Comput-Aided Mol Des. 1996;10:607–622. doi: 10.1007/BF00134183. [DOI] [PubMed] [Google Scholar]
- 92.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swanson JMJ, Wagoner JA, Baker NA, McCammon JA. Optimizing the Poisson Dielectric Boundary with Explicit Solvent Forces and Energies: Lessons Learned with Atom-Centered Dielectric Functions. J Chem Theory Comput. 2007;3:170–183. doi: 10.1021/ct600216k. [DOI] [PubMed] [Google Scholar]
- 94.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an Automated Pipeline for the Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boivin S, Kozak S, Meijers R. Optimization of Protein Purification and Characterization Using Thermofluor Screens. Protein Expression Purif. 2013;91:192–206. doi: 10.1016/j.pep.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 96.Niesen FH, Berglund H, Vedadi M. The Use of Differential Scanning Fluorimetry to Detect Ligand Interactions That Promote Protein Stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 97.Steinberg TH, Haugland RP, Singer VL. Applications of SYPRO Orange and SYPRO Red Protein Gel Stains. Anal Biochem. 1996;239:238–245. doi: 10.1006/abio.1996.0320. [DOI] [PubMed] [Google Scholar]
- 98.Liu F, Ni ASY, Lim Y, Mohanram H, Bhattacharjya S, Xing B. Lipopolysaccharide Neutralizing Peptide-Porphyrin Conjugates for Effective Photoinactivation and Intracellular Imaging of Gram-Negative Bacteria Strains. Bioconjugate Chem. 2012;23:1639–1647. doi: 10.1021/bc300203d. [DOI] [PubMed] [Google Scholar]
- 99.Milgrom LR, O’Neill F. Towards Synthetic-Porphyrin/ Monoclonal Antibody Conjugates - ScienceDirect. Tetrahedron. 1995;51:2137–214. [Google Scholar]
- 100.Montal M, Mueller P. Formation of Bimolecular Membranes From Lipid Monolayers and a Study of Their Electrical Properties. Proc Natl Acad Sci U S A. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Forstater JH, Briggs K, Robertson JWF, Ettedgui J, Marie-Rose O, Vaz C, Kasianowicz JJ, Tabard-Cossa V, Balijepalli A. MOSAIC: a Modular Single-Molecule Analysis Interface for Decoding Multistate Nanopore Data. Anal Chem. 2016;88:11900–11907. doi: 10.1021/acs.analchem.6b03725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable Molecular Dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. CHARMM General Force Field: a Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu W, He X, Vanommeslaeghe K, MacKerell AD. Extension of the CHARMM General Force Field to Sulfonyl-Containing Compounds and Its Utility in Biomolecular Simulations. J Comput Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoo J, Aksimentiev A. Improved Parametrization of Li+, Na +, K+, and Mg2+ Ions for All-Atom Molecular Dynamics Simulations of Nucleic Acid Systems. J Phys Chem Lett. 2012;3:45–50. [Google Scholar]
- 106.Yoo J, Aksimentiev A. Improved Parameterization of Amine-Carboxylate and Amine-Phosphate Interactions for Molecular Dynamics Simulations Using the CHARMM and AMBER Force Fields. J Chem Theory Comput. 2016;12:430–443. doi: 10.1021/acs.jctc.5b00967. [DOI] [PubMed] [Google Scholar]
- 107.Andersen OS. Ion Movement Through Gramicidin a Channels. Studies on the Diffusion-Controlled Association Step. Biophys J. 1983;41:147–165. doi: 10.1016/S0006-3495(83)84416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miyamoto S, Kollman PA. Settle - an Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 109.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 110.Humphrey W, Dalke A, Schulten K. VMD: Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 111.Martyna GJ, Tobias DJ, Klein ML. Constant-Pressure Molecular-Dynamics Algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 112.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 113.Brünger AT. X-PLOR: Version 3.1: a System for X-Ray Crystallography and NMR. Yale University Press; New Haven: 1992. [Google Scholar]
- 114.Li CY, Hemmig EA, Kong J, Yoo J, Hernández-Ainsa S, Keyser UF, Aksimentiev A. Ionic Conductivity, Structural Deformation, and Programmable Anisotropy of DNA Origami in Electric Field. ACS Nano. 2015;9:1420–1433. doi: 10.1021/nn505825z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.