Abstract
The majority of mutations studied in animal models are designated as recessive based on the absence of visible phenotypes in germline heterozygotes. Accordingly, genetic studies primarily rely on homozygous loss-of-function to determine gene requirements, and a conceptually-related ‘two-hit model’ remains the central paradigm in cancer genetics. Here we investigate pathogenesis due to somatic mutation in epithelial tissues, a process that predominantly generates heterozygous cell clones. To study somatic mutation in Drosophila, we generated inducible alleles that mimic human Juvenile polyposis-associated BMPR1A mutations. Unexpectedly, four of these mutations had no phenotype in heterozygous carriers but exhibited clear tissue-level effects when present in somatic clones of heterozygous cells. We conclude that these alleles are indeed recessive when present in the germline, but nevertheless deleterious when present in heterozygous clones. This unforeseen effect, deleterious heteromosaicism, suggests a ‘one-hit’ mechanism for disease initiation that may explain some instances of pathogenesis associated with spontaneous mutation.
Research organism: D. melanogaster
Introduction
Genomes are inherently unstable, an attribute that drives evolution at the population level but simultaneously underlies the incidence of spontaneous disease (Veltman and Brunner, 2012; Frank, 2014; Campbell et al., 2015; Acuna-Hidalgo et al., 2016; Forsberg et al., 2017; Stenson et al., 2017). In humans, DNA replication error is estimated to induce mutations at a frequency of 10−9 per replicative cycle such that newborns already exhibit substantial somatic mosaicism at birth (Lynch, 2010; Frank, 2014; Campbell et al., 2015; Fernández et al., 2016; Forsberg et al., 2017; Machiela and Chanock, 2017). Additional somatic mutations accumulate over time, resulting in the age-dependent incidence of various pathologies, including cancer, X-linked disorders, and neurodevelopmental disease (Veltman and Brunner, 2012; Deng et al., 2014; Frank, 2014; Fernández et al., 2016; Machiela and Chanock, 2017). Indeed, it is estimated that spontaneous DNA replication error accounts for two-thirds of cancer mutations (Tomasetti et al., 2017). Nevertheless, little is known about the underlying cellular mechanisms by which somatic mutations trigger disease, primarily because of the technical difficulty of inducing specific mutations in endogenous loci during development of model animals.
Results and discussion
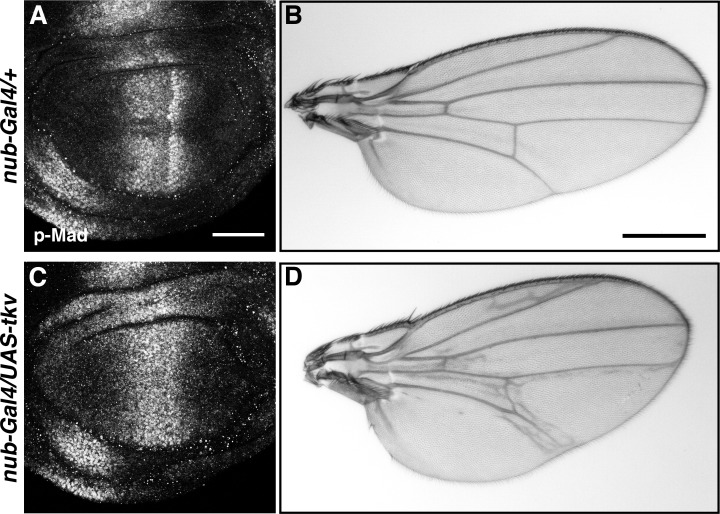
To investigate the phenotypic consequences of disease-associated alleles in vivo, previous studies have frequently employed methods that drive aberrant protein expression under the control of non-endogenous promoter elements. The Drosophila Gal4/UAS system, for example, provides a highly flexible platform to drive gene expression in vivo (Brand and Perrimon, 1993). This methodology can effectively model tissue mosaicism for putative disease alleles, but results can be difficult to interpret due to the reliance on gene overexpression through exogenous regulatory elements. Indeed, even wild-type alleles expressed at non-physiological levels can produce aberrant phenotypes (Figure 1—figure supplement 1) (Rørth, 1996; Grieder et al., 2007; Akiyama et al., 2008).
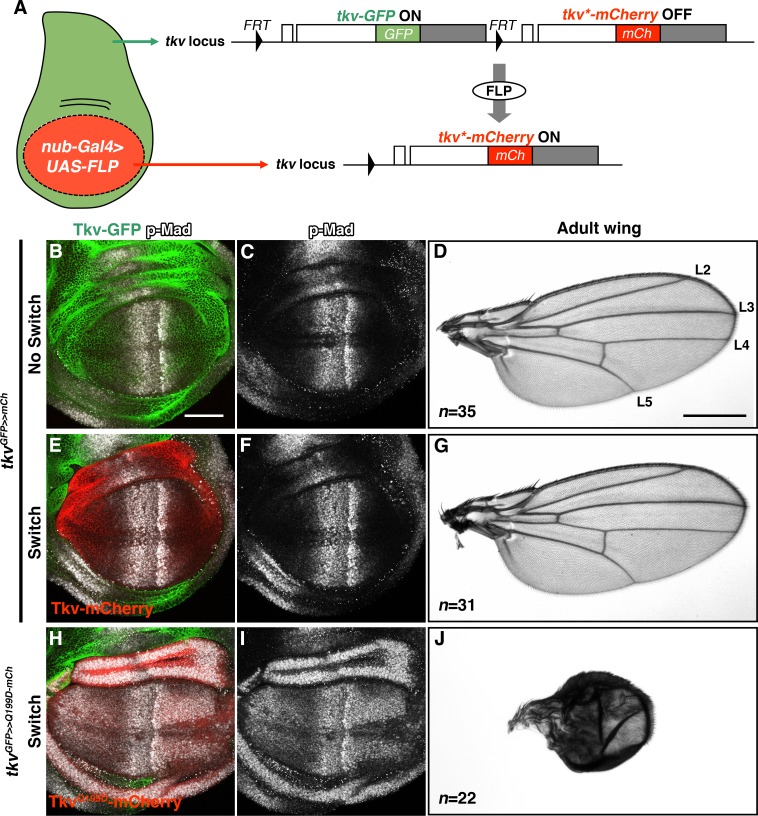
To more rigorously investigate the effects of disease-associated mutations in vivo, we used CRISPR/Cas9-dependent genome editing to establish a Flippase (FLP) recombination-dependent allele switch system. Using human disease-associated alleles of the Drosophila Bone Morphogenetic Protein Receptor 1A (BMPR1A) homologue thickveins (tkv) as a model, our approach permits the inducible conversion of the wild-type locus to a specific mutant allele (Figure 1A; Figure 1—figure supplement 2A). In Drosophila, signaling through BMPR1A/Tkv is required to control growth and direct cell fates in the developing wing imaginal disc. The primary ligand, Decapentaplegic (DPP), is expressed in a narrow band of cells at the anterior-posterior compartment boundary, leading to a downstream phospho-Mothers against dpp (p-Mad) activity gradient in the center of the disc that functions to establish presumptive wing vein regions (Figure 1B–D) (Lecuit et al., 1996; Nellen et al., 1996; Affolter and Basler, 2007; Restrepo et al., 2014; Akiyama and Gibson, 2015b; Akiyama and Gibson, 2015a).
Figure 1. Inducible tkv allele switch system.
(A) Schematic illustration of a FLP/FRT-mediated tkv allele switch. mCherry-tagged Tkv expression was specifically induced in the wing pouch via FLP/FRT-mediated recombination that eliminates tkv-GFP from the locus. White and grey boxes represent tkv coding and 3’UTR sequences, respectively. (B–G) tkvGFP>>mCherry heterozygous animal before and after allele switch. (B, C, E, F) GFP (green) and mCherry (red) represent endogenous Tkv expression in the developing wing disc. BMP/DPP activity was detected by anti-p-Mad staining (grey). (D, G) Both Tkv-GFP and Tkv-mCherry expressing animals developed normal adult wings. Longitudinal veins L2-L5 are indicated. (H–J) Wing pouch-specific allele switch of the constitutively active form of Tkv, TkvQ199D. TkvQ199D-mCherry (red) expression led to ectopic p-Mad activation (grey, H, I), resulting in wing malformation (J). Scale bars: 50 μm for (B), and 0.5 mm for (D). Anterior is oriented to the left in all wing disc figures and to the top in adult wing images.
Figure 1—figure supplement 1. Wing patterning defects caused by tkv overexpression.
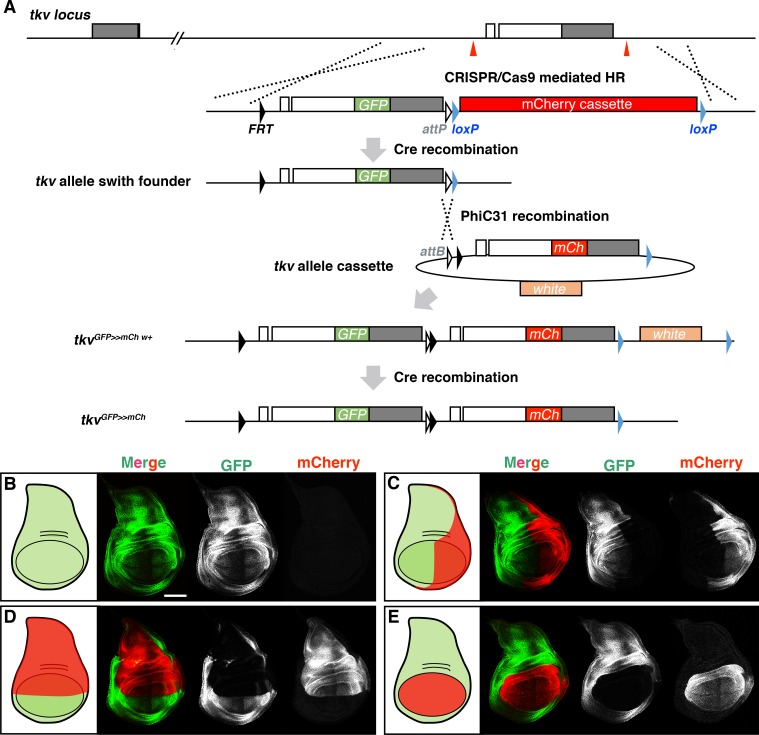
Figure 1—figure supplement 2. Spatial regulation of tkv allele switch.
We first engineered a switchable control allele, tkvGFP>>mCherry, which carries tandem duplications of the wild-type tkv coding sequence. Each tkv copy is tagged with a distinctive fluorophore such that Green Fluorescent Protein (GFP) labels the 5’ allele and mCherry labels the 3’ copy. FRT sites flank the tkv-GFP coding exons to allow recombinogenic removal of tkv-GFP and thus a switch to expression of tkv-mCherry within the endogenous regulatory architecture (Figure 1A; Figure 1—figure supplement 2A). Under normal conditions, transcription terminates before the tkv-mCherry coding sequence and only Tkv-GFP is expressed (Figure 1B–D; Figure 1—figure supplement 2B). Marked allele switch events can thus be induced in precise spatio-temporal patterns using controlled FLP expression to excise tkv-GFP (Figure 1E–G; Figure 1—figure supplement 2C–E). In control experiments, we detected no adverse effects of tkvGFP>>mCherry (green) conversion to tkv>mCherry (red). In fact, while UAS-tkv overexpression in the wing pouch using nub-Gal4 caused abnormal phenotypes (Figure 1—figure supplement 1), induction of Tkv-mCherry in the nub-Gal4 domain had no detectable effect on development (Figure 1B–G).
To further validate the allele switch methodology, we generated a transgenic line that permits inducible in-locus expression of TkvQ199D, a constitutively active form of the receptor (Hoodless et al., 1996). As expected, w; tkvGFP>>Q199D-mCherry/nub-Gal4; UAS-FLP/+ wing discs exhibited ectopic BMP/DPP activity throughout the Nub expression domain, resulting in both wing disc and adult wing abnormalities (Figure 1H–J). Taken together, these results demonstrate the utility of an in-locus allele switch system for elucidating the in vivo consequences of spontaneous disease-linked mutations.
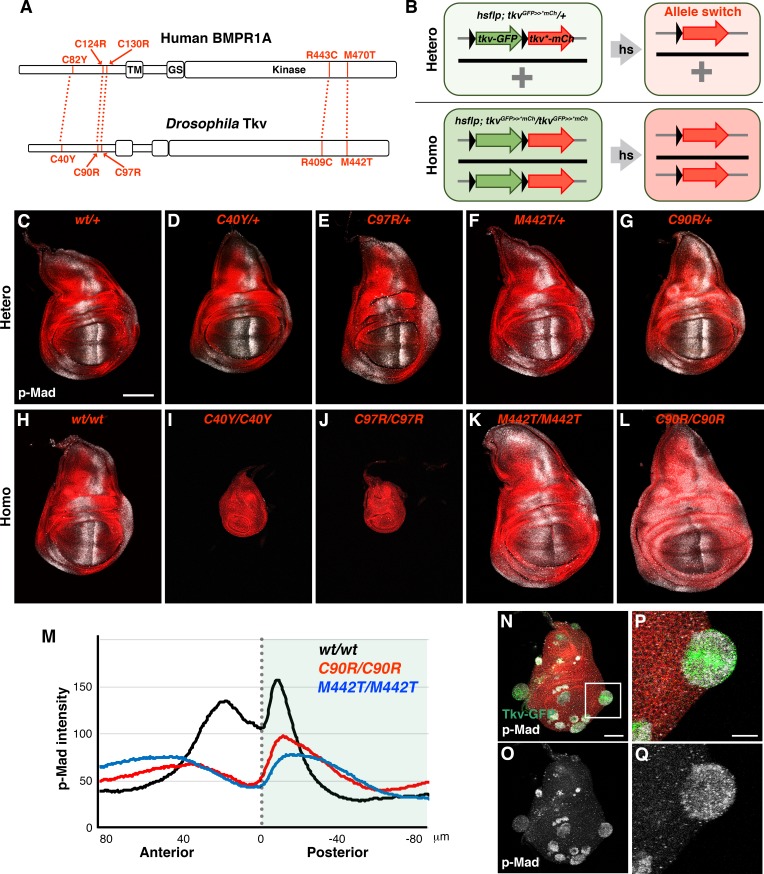
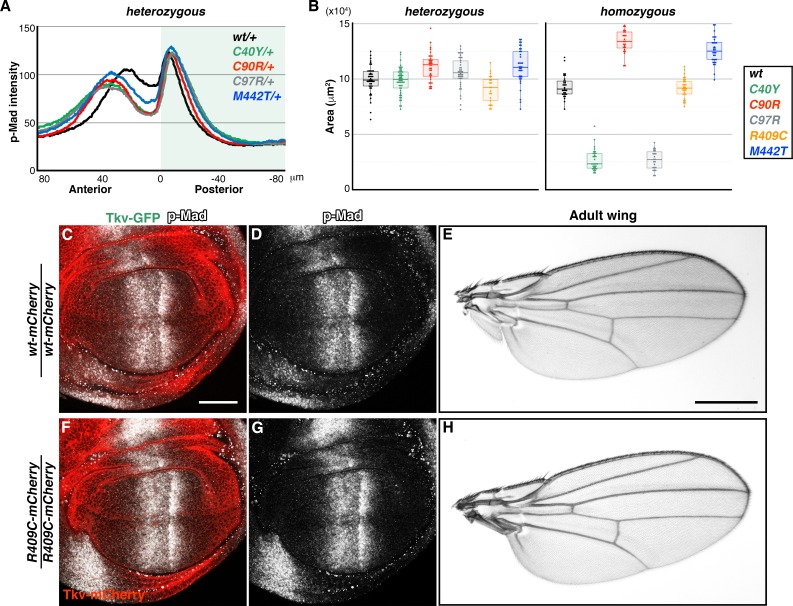
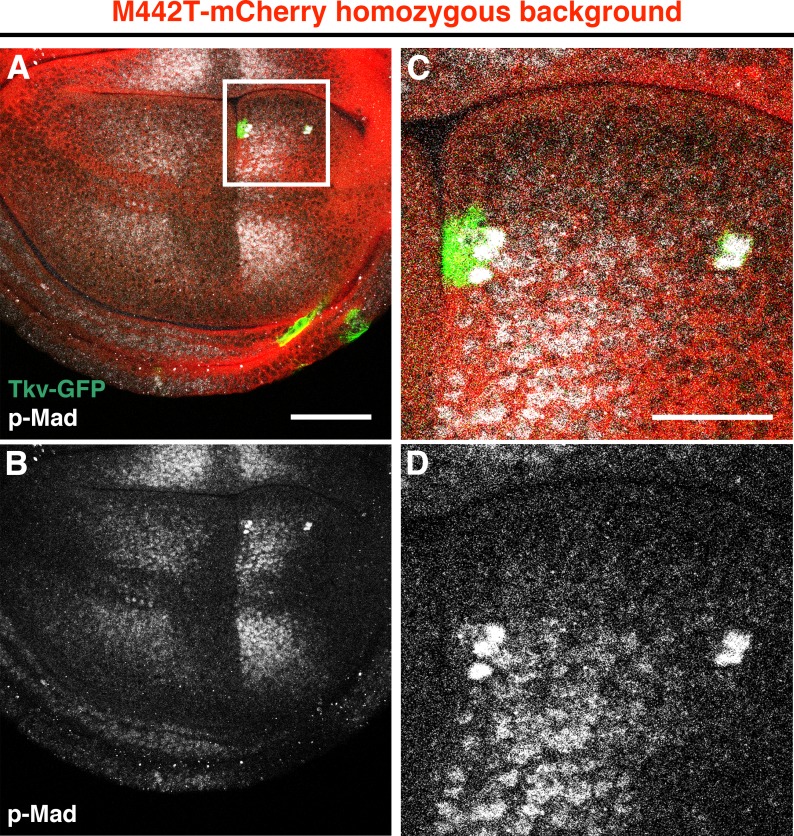
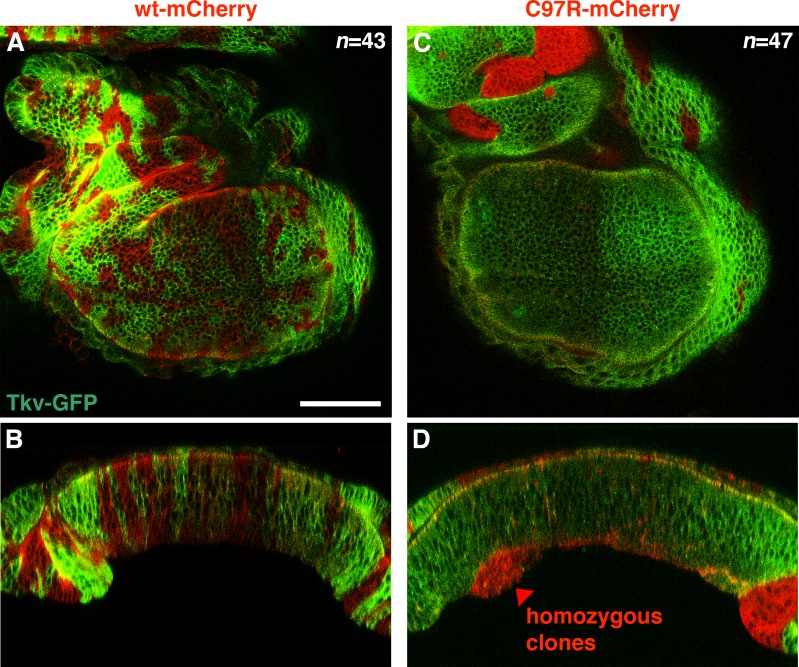
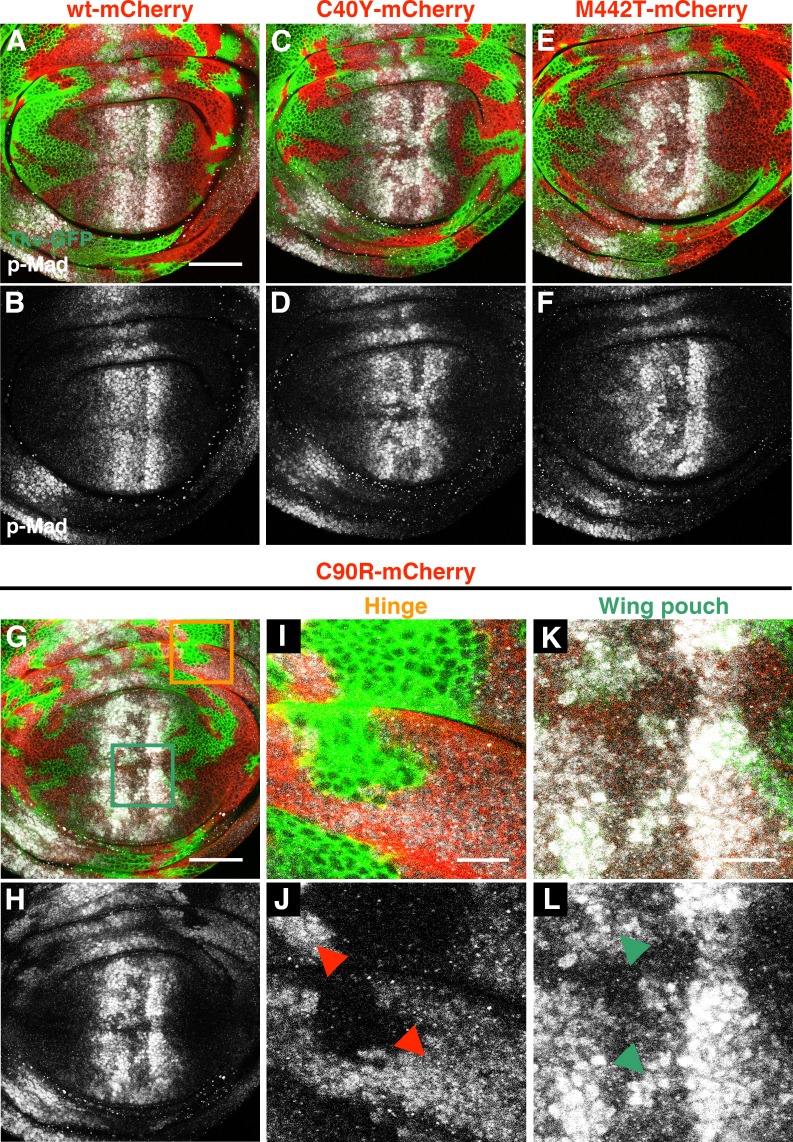
In humans, BMPR1A is known as a causative gene for Juvenile polyposis syndrome (JPS), a condition which predisposes patients to the development of gastrointestinal cancer (Figure 2A) (Howe et al., 2001; Sayed et al., 2002; Greenman et al., 2007; Hardwick et al., 2008). Although several BMPR1A point mutations have been identified in JPS tissue samples, precisely how each lesion influences receptor activity remains unclear. To investigate this, we generated allele switch transgenic strains for five JPS-linked point mutations at sites that are conserved in Drosophila Tkv (Figure 2A). In order to uniformly induce either hetero- or homozygosity throughout all cells of developing wing discs, larvae of tkv allele switch transgenics carrying hs-flp were subjected to a prolonged 1 hr heat shock at 72 hr after egg laying (AEL) (Figure 2B–L; Figure 2—figure supplement 1A and B). We found that these mutations fell into three functional classes: no effect, loss-of-function, and context-dependence. First, although the BMPR1AR443C mutation was detected in JPS patients (Sayed et al., 2002; Greenman et al., 2007), the corresponding tkv mutation produced no obvious phenotypes in either the hetero- or homozygous condition (Figure 2—figure supplement 1B–H). In a second allele class, we observed the expected loss-of-function effects. While heterozygosity for C40Y, C97R and M442T mutations produced normal wings and similarly mild effects on the p-Mad activity gradient (Figure 2D–F; Figure 2—figure supplement 1A and B), wing discs homozygous for these mutations displayed distinct phenotypes (Figure 2I–K; Figure 2—figure supplement 1B). tkvC40Y-mCherry and tkvC97R-mCherry homozygosity resulted in small wing discs with no detectable p-Mad activity, consistent with strong loss-of-function alleles (Figure 2I and J; Figure 2—figure supplement 1B). In contrast, tkvM442T-mCherry homozygous discs were larger and featured a broadened p-Mad activity gradient (Figure 2K and M; Figure 2—figure supplement 1B), perhaps due to reduced receptor binding and increased BMP/DPP ligand availability. Supporting this model, occasional residual clones expressing wild-type Tkv-GFP exhibited ectopic signaling activity in tkvM442T-mCherry homozygous wing discs (Figure 2—figure supplement 2). Similarly, residual wild-type clones within tkvC97R-mCherry mutant wing discs showed high levels of BMP activity and frequently produced distinctly rounded outgrowths (n = 6/8 discs; Figure 2N–Q).
Figure 2. Distinct wing disc phenotypes caused by JPS-associated tkv mutations.
(A) Human BMPR1A and Drosophila Tkv protein structures. Conserved JPS-associated mutations are indicated. (B) Scheme for the induction of tkv hetero- and homozygous mutant cells by heat shock. (C–L) wild-type and mutant forms of Tkv-mCherry (red) together with p-Mad (grey) expression in either hetero- (C–G) or homozygous (H–L) wing discs. Scale bars: 100 μm. (M) Averaged p-Mad intensity plot profiles for wild-type tkvmCherry (n = 16), tkvC90R-mCherry (n = 15) and tkvM442T-mCherry (n = 16) homozygous wing discs. 0 indicates the compartment boundary position (posterior is shaded). (N–Q) High levels of BMP activity observed in residual wild-type clones within predominantly tkvC97R-mCherry homozygous wing discs. Ectopic p-Mad expression (grey) is detected in rounded clusters of wild-type cells (green in N, P). (P and Q) show magnified images of the boxed region in (N). Scale bars: 50 μm for (N), 20 μm for (P). Anterior is oriented to the left side of all images.
Figure 2—figure supplement 1. Phenotypes of tkv mutations.
Figure 2—figure supplement 2. Strong BMP activation in cells expressing wild-type Tkv within the tkvM442T background.
In a third and final allele class, we found that the tkvC90R-mCherry variant possessed distinct signaling capabilities depending on the wing disc location. While tkvC90R-mCherry homozygosity resulted in weaker p-Mad expression in the wing pouch, the same mutation activated BMP signaling in the hinge region in both hetero- and homozygous discs (Figure 2G,L and M; n = 35 and 32 discs, respectively). Altogether, these results indicate that JPS-associated tkv mutations present a variety of wing disc growth phenotypes linked to distinct effects on signaling activity. In addition, we identified an intriguing new allele, tkvC90R, which either promotes or represses signaling activity in a position-dependent manner.
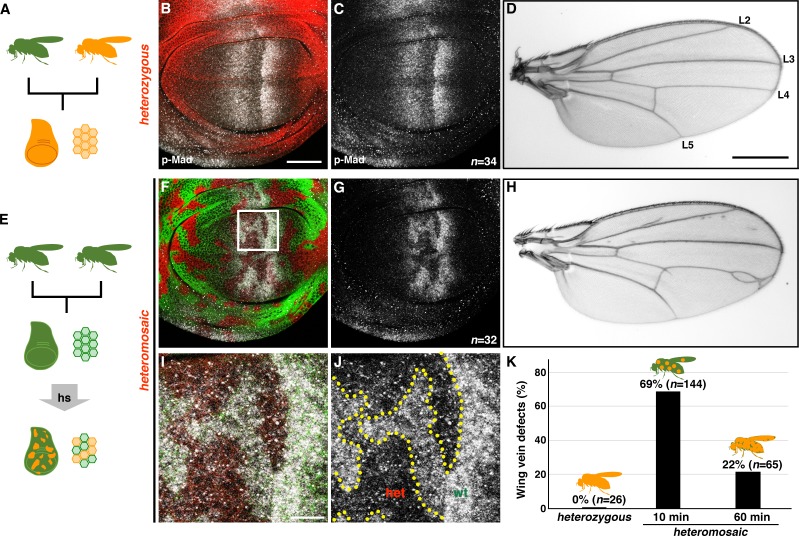
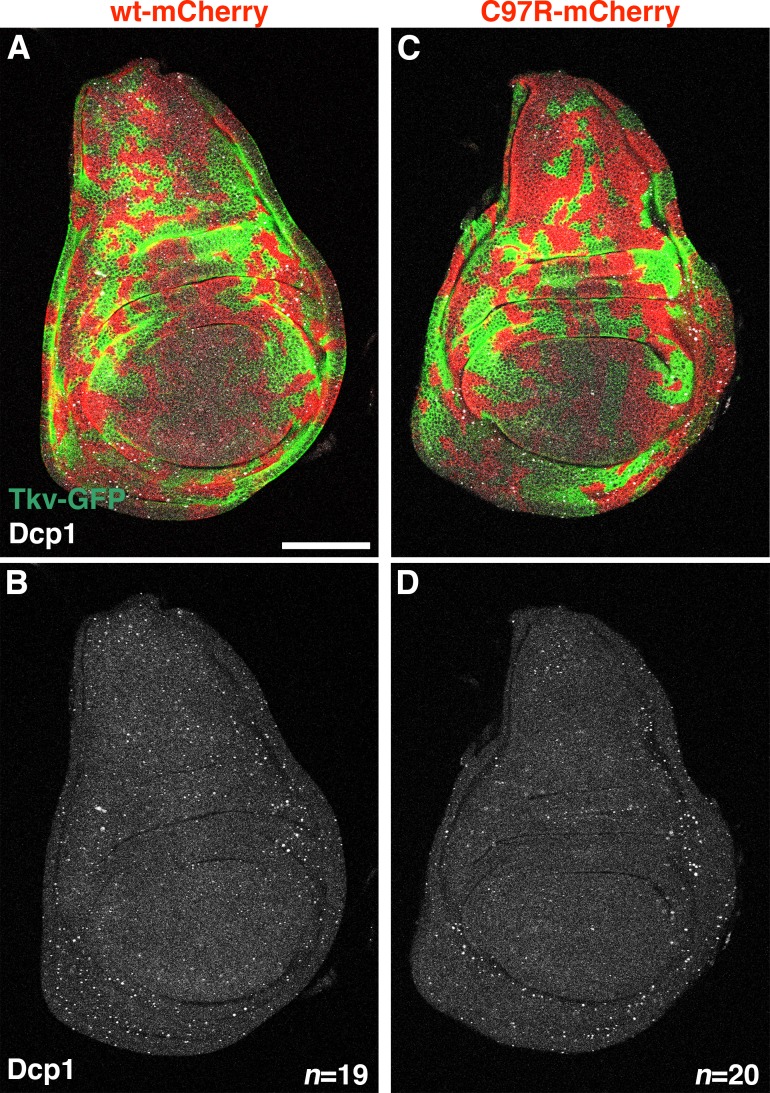
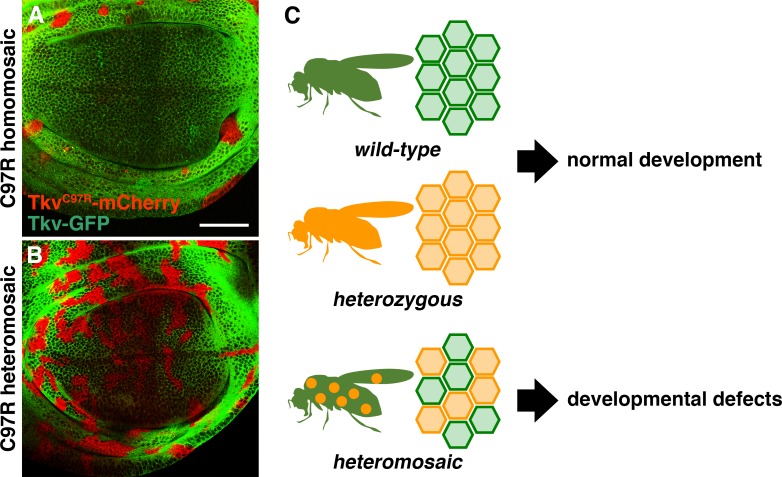
Human disease alleles, such as those identified in JPS patients, can be categorized according to either spontaneous or inherited origins. Leveraging the allele switch system, we next sought to model the cellular etiology associated with somatic mutations in JPS-associated alleles (Figure 3). Consistent with the results of uniformly-induced tkvC97R heterozygosity during larval development (Figure 2E; Figure 2—figure supplement 1A and B), germline tkvC97R heterozygotes exhibited little effect on p-Mad expression and developed normal adult wings (Figure 3A–D and K). Strikingly, however, when we induced sporadic tkvC97R heterozygous clones at 72 hr AEL, experimental animals showed aberrant p-Mad expression in wing discs and patterning defects in adult wings (Figure 3E–K). At the cellular level, clones of tkvC97R heterozygous cells exhibited strongly reduced BMP/DPP signaling activity and increased p-Mad levels in adjacent wild-type cells (Figure 3I and J). These results demonstrate the unexpectedly detrimental effect of mosaic heterozygosity for a classically recessive allele, an effect we define as deleterious heteromosaicism.
Figure 3. tkv heterozygous mosaicism disrupts wing pattern formation.
(A–D) Germline inheritance of the recessive tkvC97R-mCherry mutation (A). TkvC97R-mCherry (red) and p-Mad (grey) expression in a heterozygous wing disc (B, C). tkvC97R heterozygotes developed normal wings (D). Positions of longitudinal vein L2-L5 are shown. (E–J) Generating somatic clones of tkvC97R heterozygous cells by heat shock (E). wild-type and tkvC97 heterozygous cells are shown by green and red, respectively (F, G, I, J). tkvC97R-mCherry heteromosaicism disrupted the p-Mad activity gradient (grey) and caused wing vein patterning defects (H). (I, J) Magnified images of the boxed area in (F). A similar effect on p-Mad activity was observed in male wing discs, although adult males exhibited milder wing phenotypes. Scale bars: 50 μm for (B), 0.5 mm for (D), and 10 μm for (I). Anterior is oriented to the left side of wing disc images and to the top side of adult wing pictures. (K) Quantification of adult wing phenotypes in tkvC97R-mCherry heterozygous and heteromosaic animals. Longer heat shock generated more uniformly heterozygous cell populations and reduced mosaicism, rescuing wing vein phenotypes.
Figure 3—figure supplement 1. Basal extrusion of tkvC97R homozygous mutant clones from the wing epithelia.
Figure 3—figure supplement 2. tkv heteromosaicism does not induce cell death.
Figure 3—figure supplement 3. Additional JPS-associated tkv mutations exhibit deleterious heteromosaicism.
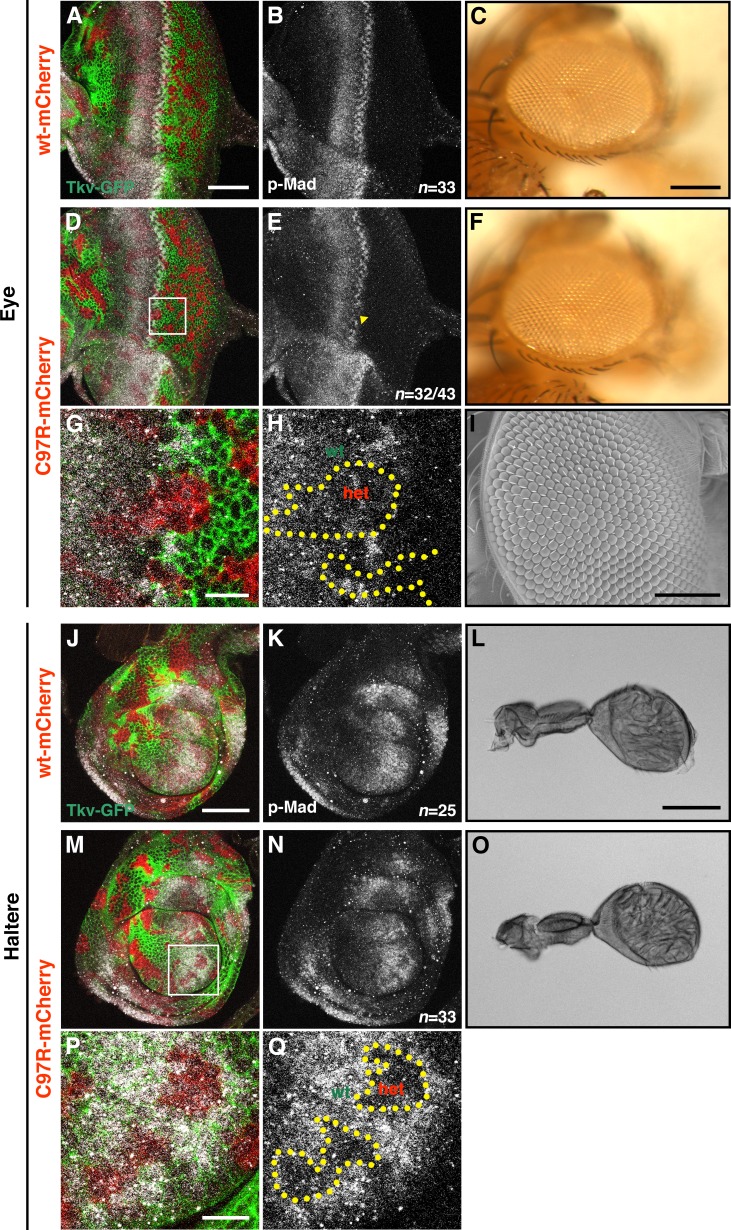
Figure 3—figure supplement 4. Effects of tkvC97R heterozygous mutant clones in the developing eye and haltere discs.
To confirm that mosaicism itself was the cause of abnormal wing vein patterning, we more uniformly induced tkvC97R-mCherry heterozygous cells throughout the wing disc by increasing the duration of heat shock from 10 to 60 min. Indeed, longer duration heat shock increased the number of heterozygous cells present in developing discs, and rescued the wing vein phenotypes associated with heterozygous mosaicism (Figure 3K). This indicates that deleterious heteromosaic effects derive from the clonal confrontation between heterozygous and wild-type cells.
Cell autonomous abrogation of BMP signaling typically leads to clone extrusion and apoptosis (Adachi-Yamada and O'Connor, 2002; Gibson and Perrimon, 2005; Shen and Dahmann, 2005). Consistent with these observations, the majority of homozygous tkvC97R clones were eliminated from the wing epithelia (Figure 4A; Figure 3—figure supplement 1). However, neither form of cell elimination was observed in wing discs carrying tkvC97R heterozygous clones (Figure 3F,G,I and J; Figure 4B; Figure 3—figure supplement 2). We infer that BMP signal reduction in heterozygous cells was adequate to cause deleterious effects but insufficient to trigger cell removal. Further, consistent with a general phenomenon not restricted to tkvC97R-mCherry, heterozygous cell clones for other recessive tkv alleles elicited similar phenotypes (Figure 3—figure supplement 3). In agreement with previous results (Figure 2G and L), we also found that clones heterozygous for tkvC90R-mCherry disrupted p-Mad activity in the developing wing blade region but ectopically activated BMP/DPP signaling in the presumptive hinge territory (n = 26/27 discs; Figure 3—figure supplement 3G–L). Lastly, we also examined for the effects of deleterious heteromosaicism in other tissues. Although tkvC97R heterozygous clones influenced BMP/DPP activity in both eye and haltere discs at the level of Mad phosphorylation, the adult structures developed normally with no obvious defect (Figure 3—figure supplement 4). These findings suggest a tissue-specific susceptibility to the same recessive mutations in heterozygous cell clones.
Figure 4. Deleterious heteromosaicism.
(A, B) tkvC97R-mCherry homozygous clones were eliminated from wing disc epithelia (A), while heterozygous cells were retained (B). Scale bars: 50 μm. Anterior is to the left. (C) While wild-type animals and heterozygous carriers both develop normally, animals carrying heterozygous somatic clones exhibit developmental defects.
Recent advances in genome-wide association studies report a substantial accumulation of somatic mutations within individuals, resulting in genetic heterogeneity (Frank, 2014; Campbell et al., 2015; Fernández et al., 2016; Forsberg et al., 2017; Machiela and Chanock, 2017). This mutational load is associated with the initiation and progression of a number of diseases, including cancer and X-linked disorders (Veltman and Brunner, 2012; Deng et al., 2014; Fernández et al., 2016; Tomasetti et al., 2017). Here, we have established an inducible allele switch system that allows us to study specific disease-associated point mutations within endogenous loci (Figures 1 and 2; Figure 1—figure supplement 2; Figure 2—figure supplements 1 and 2). For the first time, genome engineering methods make it possible to model the occurrence of spontaneous disease mutations and investigate their biological consequences in living tissue (Figures 3 and 4; Figure 3—figure supplements 2–4). Using this approach, we have uncovered a unique facet of tissue genetics whereby classically recessive alleles can trigger aberrant development when present in heterozygous cell clones, a phenomenon we call deleterious heteromosaicism (Figures 3 and 4; Figure 3—figure supplements 2 and 3). Since animals uniformly heterozygous for the same alleles are phenotypically normal, heteromosaic phenotypes likely emerge from local disparities between wild-type and heterozygous cells. Cellular heterogeneity associated with homozygous mutant clones is known to perturb tissue integrity by causing cyst formation, cell elimination and apoptosis (Adachi-Yamada and O'Connor, 2002; Gibson and Perrimon, 2005; Shen and Dahmann, 2005; Hogan et al., 2009; Bielmeier et al., 2016). In contrast, tkv deleterious heteromosaic clones showed no evidence of abnormal tissue architecture or cell death (Figures 3 and 4; Figure 3—figure supplements 2 and 3). Thus, although tkv heterozygous cells disrupted normal wing pattern formation, they did not strongly influence growth and appear to escape from homeostatic surveillance mechanisms such as cell competition (Di Gregorio et al., 2016).
A prevailing model in cancer genetics, the two-hit hypothesis requires that two independent mutations arise in a single tumor suppressor gene (Nordling, 1953; Knudson, 1971). Assuming mutagenesis by random DNA replication error, the probability of two independent mutations hitting the same gene within the same cell lineage is relatively low. Here, we report that a single somatic mutation in a putative tumor suppressor causes abnormal development by disrupting the homogeneity of BMPR1A-mediated cellular communication at the tissue level (Figures 3 and 4; Figure 3—figure supplements 2 and 3). Given that a number of disease-linked mutations are known in major signaling pathway components, we speculate that deleterious heteromosaicism is not limited to the BMP signaling pathway. Further, we propose that investigation into the unique facets of tissue genetics has the potential to shed new light on the mechanisms of how disease-linked recessive mutations impact human health.
Materials and methods
Generation of tkv allele switch transgenic lines
Two tkv sgRNA DNA constructs were generated using the primers listed in Supplementary file 1. tkv sgRNA DNA construct 1 (primers 1 and 2), and tkv sgRNA DNA construct 2 (primers 3 and 4), were annealed and cloned into the BbsI site of pBFv-U6.2 (Kondo and Ueda, 2013). A ubi-mCherry selection marker was generated as previously described (Akiyama and Gibson, 2015a). To obtain a donor DNA construct for generating a tkv allele switch founder transgenic line, six PCR fragments were prepared using primers 5 to 16 in Supplementary file 1. The PCR products were combined via Gibson assembly (NEB).
A DNA mixture containing two tkv sgRNA DNAs and the donor plasmid (250 ng/μl for each) was injected into the posterior side of embryos expressing Cas9 controlled by the nos promoter (Kondo and Ueda, 2013). Transgenic flies were selected by mCherry expression and further confirmed by DNA sequencing. Finally, the selection cassette was removed via Cre/loxP-mediated recombination, resulting in the tkv allele switch founder (see Figure 1—figure supplement 2A).
To generate tkv allele cassettes, a tkv-mCherry DNA fragment with 5’ EcoRI and 3’ AscI sites was constructed by Gibson assembly using primers 17 to 22 in Supplementary file 1. After Gibson assembly, a second round of PCR was conducted using primers 17 and 22 (Supplementary file 1). The resulting PCR product was cloned into pCRII blunt TOPO (Thermo Fisher Scientific).
Each tkv point mutation was induced with the Q5 Site-Directed Mutagenesis Kit (NEB), using primers 23 to 34 described in Supplementary file 1. After DNA sequencing, pCRII blunt TOPO tkv*-mCherry DNAs were digested by EcoRI and AscI, and cloned into w + attB (addgene #30326), resulting in w + attB tkv*-mCherry.
To establish tkv* allele switch transgenic lines, w + attB tkv*-mCherry DNA constructs were injected into the posterior regions of embryos obtained from a cross of nos-phiC31 int. NLS (on X, Bloomington #34770) and the w; tkv allele switch founder. Transformants were screened by the presence of red eye color (see Figure 1—figure supplement 2A).
Heat shock
To mimic induction of somatic mutation in vivo, FLP expression was induced at 37°C for 10 (Figure 3F–J; Figure 3—figure supplements 1–4), 15 (Figure 2N–Q), 30 (Figure 2—figure supplement 2), or 60 (Figure 2C–L; Figure 2—figure supplement 1A and B) minutes at 72 hr AEL.
tkv* allele switch heterozygous cross (Figure 2C–G: Figure 3F–K; Figure 2—figure supplement 1A and B; Figure 3—figure supplements 2–4): w, hs-flp x w; tkv* allele switch/CyO tkv allele switch homozygous cross (Figure 2H–Q; Figure 2—figure supplement 1B; Figure 2—figure supplement 2): w, hs-flp; tkv* allele switch/CyO,sChFP x w, hs-flp/Y; tkv* allele switch/CyO,sChFP
tkv allele switch in heterozygous backgrounds (Figure 4A and B; Figure 3—figure supplement 1): w; tkv*-mCherry/CyO,twi-Gal4, UAS-GFP x w, hs-flp/Y; tkv* allele switch/CyO,sChFP
Gal4/UAS
For region-specific tkv allele switching, FLP expression was induced using nub-GAL4 (Calleja et al., 1996), hh-Gal4 (Mullor and Guerrero, 2000), and ap-GAL4 (Calleja et al., 1996) for FLP/FRT-mediated recombination.
nub-GAL4
tkv allele switch cross: w; nub-Gal4/CyO; UAS-FLP/TM6C x w; tkvGFP>>mCherry/CyO (Figure 1E–G; Figure 1—figure supplement 2E) tkvQ199D allele switch cross: w; nub-Gal4/CyO; UAS-FLP/TM6C x w; tkvGFP>>Q199D-mCherry/CyO (Figure 1H–J)
hh-GAL4
w; hh-Gal4,UAS-FLP/TM6B x w; tkvGFP>>mCherry/CyO (Figure 1—figure supplement 2C)
ap-GAL4
w; ap-Gal4/CyO; UAS-FLP/TM6C x w; tkvGFP>>mCherry/CyO (Figure 1—figure supplement 2D)
Immunohistochemistry and imaging
Third-instar wing discs were dissected from female larvae and stained with rabbit anti-pSmad3 (EP823Y, 1:1,000, abcam) (Akiyama et al., 2012; Akiyama and Gibson, 2015a) and rabbit anti-cleaved Drosophila Dcp-1 (Asp216, 1:500, Cell Signaling Technology) (Harris et al., 2016; Verghese and Su, 2016). Alexa conjugated secondary antibodies (1:500, Invitrogen) were used to detect primary antibodies. Can Get Signal Immunostain Solution B (TOYOBO, New York, NY) was used to dilute primary antibodies. Images of wing discs and adult female wings were obtained using a Leica TCS SP5 confocal microscope and a Leica CTR 5000, respectively.
Image analysis
To generate p-Mad intensity plot profiles, all images were collected at the same confocal setting and analyzed using the RGB profiler of FIJI. The ‘Measure’ function of FIJI was used to analyze sizes of wing discs.
Acknowledgements
We thank A Ikmi for extensive discussion and suggestions, and H Nakato and the Bloomington Stock Center for fly stocks. We thank W Fropf, L Ellington and E Bauerly for a critical reading of the manuscript, H Yin for administrative support, and members of the Gibson lab for discussions and advice. This work was supported by funding from the Stowers Institute for Medical Research and NIH R01 GM111733 to MCG.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Matthew C Gibson, Email: mg2@stowers.org.
Jody Rosenblatt, University of Utah, United States.
Funding Information
This paper was supported by the following grants:
Stowers Institute for Medical Research to Matthew C Gibson.
National Institutes of Health GM111733 to Matthew C Gibson.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Investigation, Writing—original draft, Writing—review and editing.
Investigation.
Conceptualization, Funding acquisition, Investigation, Writing—original draft, Writing—review and editing.
Additional files
EcoRI and AscI sites are underlined in 17 and 22, respectively. Lowercase characters indicate point mutations.
Data availability
All primary data is available at the Stowers Institute Original Data Repository (https://www.stowers.org/research/publications/libpb-1261)
The following dataset was generated:
Akiyama T, author; User S, author; Gibson M, author. LIBPB-1261. 2018 https://www.stowers.org/research/publications/libpb-1261 Available at the Stowers Institute Original Data Repository.
References
- Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biology. 2016;17:241. doi: 10.1186/s13059-016-1110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi-Yamada T, O'Connor MB. Morphogenetic apoptosis: a mechanism for correcting discontinuities in morphogen gradients. Developmental Biology. 2002;251:74–90. doi: 10.1006/dbio.2002.0821. [DOI] [PubMed] [Google Scholar]
- Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nature Reviews Genetics. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Gibson MC. Decapentaplegic and growth control in the developing Drosophila wing. Nature. 2015a;527:375–378. doi: 10.1038/nature15730. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Gibson MC. Morphogen transport: theoretical and experimental controversies. Wiley Interdisciplinary Reviews: Developmental Biology. 2015b;4:99–112. doi: 10.1002/wdev.167. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Developmental Biology. 2008;313:408–419. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, Marqués G, Wharton KA. A large bioactive BMP ligand with distinct signaling properties is produced by alternative proconvertase processing. Science Signaling. 2012;5:ra28. doi: 10.1126/scisignal.2002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielmeier C, Alt S, Weichselberger V, La Fortezza M, Harz H, Jülicher F, Salbreux G, Classen AK. Interface contractility between differently fated cells drives cell elimination and cyst formation. Current Biology. 2016;26:563–574. doi: 10.1016/j.cub.2015.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Campbell IM, Shaw CA, Stankiewicz P, Lupski JR. Somatic mosaicism: implications for disease and transmission genetics. Trends in Genetics. 2015;31:382–392. doi: 10.1016/j.tig.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: diverse patterns in development, tissues and disease. Nature Reviews Genetics. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio A, Bowling S, Rodriguez TA. Cell competition and its role in the regulation of cell fitness from development to cancer. Developmental Cell. 2016;38:621–634. doi: 10.1016/j.devcel.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Fernández LC, Torres M, Real FX. Somatic mosaicism: on the road to cancer. Nature Reviews Cancer. 2016;16:43–55. doi: 10.1038/nrc.2015.1. [DOI] [PubMed] [Google Scholar]
- Forsberg LA, Gisselsson D, Dumanski JP. Mosaicism in health and disease - clones picking up speed. Nature Reviews Genetics. 2017;18:128–142. doi: 10.1038/nrg.2016.145. [DOI] [PubMed] [Google Scholar]
- Frank SA. Somatic mosaicism and disease. Current Biology. 2014;24:R577–R581. doi: 10.1016/j.cub.2014.05.021. [DOI] [PubMed] [Google Scholar]
- Gibson MC, Perrimon N. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science. 2005;307:1785–1789. doi: 10.1126/science.1104751. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder NC, Charlafti I, Kloter U, Jäckle H, Schäfer U, Gehring WJ. Misexpression screen in Drosophila melanogaster aiming to reveal novel factors involved in formation of body parts. Genetics. 2007;175:1707–1718. doi: 10.1534/genetics.106.064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JC, Kodach LL, Offerhaus GJ, van den Brink GR. Bone morphogenetic protein signalling in colorectal cancer. Nature Reviews Cancer. 2008;8:806–812. doi: 10.1038/nrc2467. [DOI] [PubMed] [Google Scholar]
- Harris RE, Setiawan L, Saul J, Hariharan IK. Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. eLife. 2016;5:e11588. doi: 10.7554/eLife.11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C, Dupré-Crochet S, Norman M, Kajita M, Zimmermann C, Pelling AE, Piddini E, Baena-López LA, Vincent JP, Itoh Y, Hosoya H, Pichaud F, Fujita Y. Characterization of the interface between normal and transformed epithelial cells. Nature Cell Biology. 2009;11:460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Haerry T, Abdollah S, Stapleton M, O'Connor MB, Attisano L, Wrana JL. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/S0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nature Genetics. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- Knudson AG. Mutation and cancer: statistical study of retinoblastoma. PNAS. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Lynch M. Rate, molecular spectrum, and consequences of human mutation. PNAS. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ. The ageing genome, clonal mosaicism and chronic disease. Current Opinion in Genetics & Development. 2017;42:8–13. doi: 10.1016/j.gde.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullor JL, Guerrero I. A gain-of-function mutant of patched dissects different responses to the hedgehog gradient. Developmental Biology. 2000;228:211–224. doi: 10.1006/dbio.2000.9862. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/S0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Nordling CO. A new theory on cancer-inducing mechanism. British Journal of Cancer. 1953;7:68–72. doi: 10.1038/bjc.1953.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. PNAS. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo S, Zartman JJ, Basler K. Coordination of patterning and growth by the morphogen DPP. Current Biology. 2014;24:R245–R255. doi: 10.1016/j.cub.2014.01.055. [DOI] [PubMed] [Google Scholar]
- Sayed MG, Ahmed AF, Ringold JR, Anderson ME, Bair JL, Mitros FA, Lynch HT, Tinley ST, Petersen GM, Giardiello FM, Vogelstein B, Howe JR. Germline SMAD4 or BMPR1A mutations and phenotype of juvenile polyposis. Annals of Surgical Oncology. 2002;9:901–906. doi: 10.1007/BF02557528. [DOI] [PubMed] [Google Scholar]
- Shen J, Dahmann C. Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science. 2005;307:1789–1790. doi: 10.1126/science.1104784. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. The human gene mutation database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Human Genetics. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355:1330–1334. doi: 10.1126/science.aaf9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nature Reviews Genetics. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Verghese S, Su TT. Drosophila Wnt and STAT define apoptosis-resistant epithelial cells for tissue regeneration after irradiation. PLoS Biology. 2016;14:e1002536. doi: 10.1371/journal.pbio.1002536. [DOI] [PMC free article] [PubMed] [Google Scholar]