Abstract
Purpose of review
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB), remains a major public health threat globally. Several lines of evidence support a role for host genetic factors in resistance/susceptibility to TB disease and MTB infection. However, results across candidate gene and genome-wide association studies (GWAS) are largely inconsistent, so a cohesive genetic model underlying TB risk has not emerged.
Recent Findings
Despite the difficulties in identifying consistent genetic associations, genetic studies of TB and MTB infection have revealed a few well-documented loci. These well validated genes are presented in this review, but there remains a large gap in how these genes translate into better understanding of TB. To address this, we present a pathway based extension of standard association analyses, seeding the results with the best validated genes from candidate gene and GWAS studies.
Summary
Several pathways were significantly enriched using pathway analyses that may help to explain population patterns of TB risk. In conclusion, we advocate for novel approaches to the study of host genetic analysis of TB that extend traditional association approaches.
Keywords: Tuberculosis genetics, Tuberculosis resistance, Tuberculosis pathways and disease
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB), is a global public health problem with an estimated 1 in 3 to 1 in 4 people being infected by MTB [1, 2]. However, less than 10% of those infected by MTB develop active TB disease. In 2014, 9.6 million new cases were diagnosed, many (12%) due to coinfection with the human immunodeficiency virus (HIV), and there were 1.4 million deaths attributed to TB [3].
Initially, TB was considered to be a familial disease, as it was realized early on that close relatives of TB cases were at higher risk of TB than similarly close unrelated contacts. Twin studies estimated the concordance of TB among monozygotic twins to be between 32-62%, and between dizygotic twins to be between 14-18% [4, 5]. These observations of co-occurrence in families and concordance decreasing with degree of relatedness led to genetic studies to identify genes associated with TB. These included candidate gene studies (reviewed in [6–8]) and more recently genome-wide studies [9] (summarized below). As yet, no consistent genetic model underlying TB risk has emerged. This is likely to be due to multiple genes affecting TB risk, and/or each gene or set of genes promoting risk in different people. However, it is possible that several genes residing in the same or related pathways modulate TB risk. Therefore, in this review, we take a pathway approach to dissecting the host genetics of TB in order to shed light on potentially validated findings and pathways that could be exploited for functional studies. Using genomic studies to reveal putative pathways that are dysregulated in TB may be a particularly efficient means to develop more efficacious vaccines and treatments.
Epidemiology
The development of TB disease requires exposure, host genetic, and environmental factors that promote susceptibility. Exposure to an active TB case is required for transmission of MTB, and several epidemiologic studies have identified specific characteristics of the susceptible host and infectious TB case that facilitate the acquisition of MTB infection and development of TB disease [10–12]. In immunocompetent hosts, MTB organisms may be eliminated early by the innate immune system or, as in the majority of individuals, contained as a low-organism load in asymptomatic latent infection; latent infection develops into active disease in about 10% of people over their lifetimes. Because only a minority of MTB-infected individuals go on to develop active TB disease, host biologic factors are likely involved in progression vs long-term control or disease.
HIV is a prominent factor involved in global TB epidemiology, but the Population Attributable Fraction (PAF) for HIV is only 11% of TB cases in the world globally [13]. Even in TB/HIV high burden countries, the majority of TB patients are HIV-negative, e.g. in Guinea Bissau, one of the WHO TB/HIV high burden countries, HIV-1 co-infection is prevalent in one in four patients [14]. Hence, immunocompromising factors may play a role, but only a partial role in TB; host genetic factors may play a modifying role, although the PAF for host genetics on TB risk is unknown. The complexity of the host-pathogen interaction is considerable. involving modified antigen-expression by the dormant pathogen [15]. Proper control of the dormant MTB is, therefore, likely to be influenced by the genetic makeup of the host.
TB burden is clearly the worst in sub-Saharan Africa, East Asia, and to a lesser degree in the former Soviet Republics. The global distribution of TB is also manifested through the genetic diversity of MTB itself, as phylogenetic analysis of sequence variation in MTB shows geographic clustering of MTB lineages [16–18]. Clearly, both human genetic and MTB genetic variation have roles in the global distribution of TB.
Previous genetic studies of TB
Robust genetic associations – Candidate Gene Studies
In recent years, numerous candidate gene studies have been conducted to assess host genetic factors in TB risk. As with studies of other diseases, the early studies focused on one gene at a time, with only a few polymorphisms within each gene. Even after the advent of high-throughput genotyping technology, many studies continued in this vein, thus contributing limited amounts to our knowledge of the genetic underpinnings of TB. Given clear differences in incidence by geography, early studies that were only conducted in a handful of global settings could not necessarily be generalized across continents. However, with time, the geographic scope of research expanded, allowing increased potential to assess replicability across diverse ethnic backgrounds and to determine if universal susceptibility factors exist. We review here both robust candidate gene results and discuss why significant associations with candidate gene associations across multiple studies may not replicate well in other studies. Genome-wide association studies (GWAS) are also reported in the section below.
To identify robust risk loci for this review, we searched for genetic associations using the HuGE Navigator (https://phgkb.cdc.gov/HuGENavigator/home.do), a web-based application that queries population-based epidemiologic studies utilizing a machine learning algorithm to systematically select relevant studies. Indexing based on GeneID and MeSH terms, along with manual review, ensured appropriateness for inclusion in the database. Here, all abstracts identified by Phenopedia, an extension of HuGE Navigator that reports gene-disease associations organized by disease, were compiled for ‘Pulmonary Tuberculosis’ (PTB). In addition, studies co-authored by members of our groups were also included [19–29]. Using manual review of each article, we identified the specific variant(s) and gene(s) associated with PTB risk, as well as the direction of effect. In addition, manual review excluded articles reporting solely genetic associations with PTB severity or extrapulmonary TB. Other exclusion criteria included the presence of extrapulmonary cases in the study population as well. We included genes that showed a significant association with PTB in at least 5 publications, whether significant single locus main effects, effects detected by interactions among loci (epistasis), diplotype associations, haplotype associations, or genetic risk score associations were reported (Table 1).
Table 1. Summary of Candidate Gene Associations by Type of Association.
Candidate genes with at least five studies reporting positive association of variants with PTB according to the HuGE Navigator literature search or publication by our groups are reported below. For each gene, the number of studies reporting a specific type of association, including main effects, interactive effects, haplotype associations, and other associations, are reported, along with the total number of studies reporting any type of association for a given gene. Within each of these four types of associations, studies could report than one type of association.
| Number of Studies | |||||
|---|---|---|---|---|---|
| Gene | Main Effects | Interactive Effects | Haplotype Association | Othera | Total |
| CD209/DC-SIGN | 3 | 2 | 0 | 0 | 5 |
| HLA-A | 4 | 0 | 3 | 0 | 6 |
| HLA-DQB1 | 6 | 0 | 2 | 0 | 7 |
| HLA-DRB1 | 12 | 1 | 4 | 0 | 15 |
| IFNG | 8 | 0 | 1 | 2 | 9 |
| IFNGR1 | 4 | 1 | 1 | 0 | 5 |
| IL10 | 6 | 0 | 0 | 1 | 6 |
| MBL2 | 3 | 0 | 2 | 4 | 7 |
| MCP1 | 7 | 1 | 2 | 1 | 9 |
| NRAMP1/SLC11A1 | 12 | 0 | 5 | 2 | 13 |
| TLR2 | 7 | 1 | 1 | 1 | 8 |
| TLR4 | 4 | 3 | 1 | 1 | 7 |
| TLR9 | 3 | 2 | 0 | 2 | 6 |
| TNFA | 7 | 0 | 1 | 1 | 8 |
| VDR | 12 | 2 | 5 | 2 | 15 |
other includes both genetic risk scores and diplotype associations.
A total of 15 genes met these criteria, with main effects representing the majority of the associations. However, all of the included genes also demonstrated other types of associations in addition to main effects. Additionally, genetic variants with main effects were reported according to their gene identity (Table 2). HLA-DRB1 and VDR had the most reported studies, with 15 unique studies recorded for each of these genes (Table 1), but aside from a few reports from South Africa, there were no reported associations between HLA and TB in Sub-Saharan African populations. A SNP in NRAMP1, rs17235416, was identified as associated with PTB risk in eight studies in a variety of ethnic populations. However, many of specific genetic variants found to associate with PTB risk were reported by only one study. Notably, when multiple studies reported an association for a given polymorphism, the direction of effect was not always consistent between studies. For example, although rs4804803 in CD209 associated with PTB risk in both an Iranian and a West African population, the minor allele associated with protection and risk, respectively (Table 2). Similar to the main effects, most of the haplotype associations were reported by only one study. In addition, HLA-DRB1 had the most PTB associated haplotypes reported (Table 3). However, relatively few associations for the genes in Table 2 were reported in African populations, where the burden of TB is greatest. This suggests one of several possibilities. The genetic effects could differ, environmental factors, including sanitation and malnutrition, involved in Africa are more pronounced thereby possibly overwhelming genetic effects, or as is the case these populations have been severely understudied.
Table 2. Studies Reporting Significant Main Effects for Candidate Gene Studies.
Genetic variants associating with PTB risk as main effects are reported by gene if five or more studies reported any type of significant association, including main effects, genetic risk score, haplotype, and SNP-SNP interaction associations. For each genetic variant, study population and direction of effect relative to the minor allele are reported unless noted otherwise.
| Gene | SNPs/Genetic Locus | Population | Reference and Effecta |
|---|---|---|---|
| CD209/DC-SIGN | |||
|
| |||
| rs4804803 | Iranian | [89] + | |
| West African | [27] − | ||
|
|
|||
| rs735240 | Indonesian | [90] − | |
|
| |||
| HLA-A | |||
|
| |||
| HLA-A*02 | Chinese | [91] + | |
|
|
|||
| HLA-A*24 | Chinese | [91] + | |
|
|
|||
| HLA-A*26 | Iranian | [92] + | |
|
|
|||
| HLA-A*32 | Indian (Sahariya) | [93] + | |
|
|
|||
| HLA-A-R114 | Greek | [94] + | |
|
| |||
| HLA-DQB1 | |||
|
| |||
| HLA-DQB1*02 | Polish | [95] − | |
|
|
|||
| HLA-DQB1*0201 | Polish | [96] − | |
|
|
|||
| HLA-DQB1*0301 | Thai | [97] − | |
|
|
|||
| HLA-DQB1*0303 | Indian | [98] − | |
|
|
|||
| HLA-DQB1*05 | Pole | [95] + | |
|
|
|||
| HLA-DQB1*0502 | Thai | [97] + | |
|
|
|||
| HLA-DQB1*050301 | Indian | [98] + | |
|
|
|||
| HLA-DQB1*0601 | Indian | [98] + | |
| Korean | [99] + | ||
|
|
|||
| HLA-DQB1*1502 | Indian | [100] − | |
|
|
|||
| HLA-DQB1*1601 | Polish | [96] + | |
|
| |||
| HLA-DRB1 | |||
|
| |||
| HLA-DRB1*04 | Brazilian | [101] + | |
|
|
|||
| HLA-DRB1*04:07:01 | Amazonian | [102] − | |
|
|
|||
| HLA-DRB1*04:11:01 | Amazonian | [102] + | |
|
|
|||
| HLA-DRB1*04:92 | Amazonian | [102] + | |
|
|
|||
| HLA-DRB1*0601 | Korean | [99] + | |
|
|
|||
| HLA-DRB1*07 | Iranian | [92] + | |
|
|
|||
| HLA-DRB1*0803 | Korean | [99] + | |
|
|
|||
| HLA-DRB1*1302 | South African | [103] + | |
|
|
|||
| HLA-DRB1*14 | Portuguese | [104] + | |
|
|
|||
| HLA-DRB1*15 | Brazilian | [101] − | |
| Chinese | [105] + | ||
| Indian (Sahariya) | [93] + | ||
|
|
|||
| HLA-DRB1*1502 | Indian | [101] + | |
|
|
|||
| HLA-DRB1*16 | Chinese | [91] + | |
| Indian (Sahariya) | [93] − | ||
|
|
|||
| HLA-DRB1*1601 | Polish | [96] + | |
|
|
|||
| HLA-DRB1-N37 | Greek | [94] + | |
|
| |||
| IFNG | |||
|
| |||
| CA repeats | Chinese | [106] -e | |
|
|
|||
| rs1861493 | Indian | [107] − | |
|
|
|||
| rs1861494 | Chinese | [108] − | |
| Indian | [107] − | ||
|
| |||
| rs2069705 | West African | [28] − | |
|
|
|||
| rs2069718 | West African | [28] − | |
|
|
|||
| rs2430561 | Iranian | [109] − | |
| Sicilian | [110] − | ||
| Spaniard | [111] − | ||
| Tunisian | [112] − | ||
|
| |||
| IFNGR1 | |||
|
| |||
| -56 T/C | West African | [28] + | |
|
|
|||
| rs1327475 | Chinese | [113] + | |
|
|
|||
| rs2234711 | Chinese | [113] + | |
|
|
|||
| rs7749390 | Chinese | [113] − | |
|
|
|||
| rs489243 | Ugandan | [24] − | |
|
|
|||
| CA repeats | Indonesian | [114] +e | |
| Ugandans | [24] + | ||
|
| |||
| IL10 | |||
|
| |||
| rs1800871 | Chinese | [115] − | |
|
|
|||
| rs1800872 | Chinese | [106] − | |
| Korean | [116] − | ||
|
|
|||
| rs1800896 | Cambodian | [117] + | |
| Korean | [118] − | ||
| Sicilian | [119] b | ||
|
| |||
| MBL2 | |||
|
| |||
| Exon 1 ”AO” | Brazilian | [120] + | |
| Brazilian | [121] + | ||
|
|
|||
| rs11003125 | Chinese | [122] − | |
|
|
|||
| rs1800451 | Brazilian | [120] + | |
|
|
|||
| rs7096206 | Brazilian | [120] + | |
|
| |||
| MCP1 | |||
|
| |||
| -11822 G/A | Ghanaian | [123] − | |
|
|
|||
| rs1024611 | Chinese | [124] + | |
| Chinese | [125] + | ||
| Ghanaian | [126] − | ||
| Ghanaian | [123] − | ||
| Korean | [127] + | ||
| Mexican | [127] + | ||
| Moroccan | [128] − | ||
| Tunisian | [129] + | ||
|
|
|||
| -1549 A/T | Ghanaian | [123]b | |
|
|
|||
| rs2857656 | Ghanaian | [126] − | |
| Ghanaian | [123] − | ||
|
|
|||
| +900 C/T | Ghanaian | [123]b | |
|
|
|||
| rs3917887 | Ghanaian | [126] − | |
|
|
|||
| +5356 C/T | Ghanaian | [123] − | |
|
| |||
| NRAMP1/SLC11A1 | |||
|
| |||
| (GT)n Allele 2 | Gambian | [130] +f | |
|
|
|||
| (GT)n Allele 3 | Gambian | [130] -g | |
| Polish | [131] +h | ||
|
|
|||
| 5′(GT)n Allele 9 | South African | [132]i | |
|
|
|||
| rs17221959 | Indian | [133] − | |
|
|
|||
| rs17235409 | Cambodian | [117]b | |
| Chinese | [134] +c | ||
| Chinese | [135] + | ||
| Peruvian | [136] b | ||
| Tunisian | [137] + | ||
|
|
|||
| rs17235416 | Cambodian | [117] b | |
| Chinese | [135] + | ||
| Chinese | [122] + | ||
| Indian | [133] + | ||
| Indonesian | [138] + | ||
| South African | [132] + | ||
| Tunisian | [137] + | ||
| Chinese | [134] +c | ||
|
|
|||
| rs3731865 | Chinese | [139] + | |
| Indian | [133] + | ||
| Peruvian | [136] + | ||
|
| |||
| TLR2 | |||
|
| |||
| -196 to -174 del | Pakistan | [140] + | |
| Guinea-Bissauan | [21] − | ||
|
|
|||
| GTn repeats in intron 2 | Chinese | [141] +j | |
| Chinese | [141] -k | ||
|
|
|||
| rs121917864 | Tunisian | [142] + | |
|
|
|||
| rs3804099 | Chinese | [143] +d | |
| Iranian | [144] + | ||
| Moroccan | [145]b | ||
|
| |||
| TLR4 | |||
|
| |||
| rs11536889 | Moroccan | [145] + | |
|
|
|||
| rs1927906 | Sudanese | [146] + | |
|
|
|||
| rs1927911 | Sudanese | [146] + | |
|
|
|||
| rs4986790 | Indian | [147] + | |
| Tanzanian | [148] + | ||
|
|
|||
| rs4986791 | Indian | [147] + | |
|
|
|||
| rs5030725 | Sudanese | [146] + | |
|
|
|||
| rs7869402 | Sudanese | [146] + | |
|
| |||
| TLR9 | |||
|
| |||
| rs352139 | Chinese | [108] + | |
|
|
|||
| rs352140 | Aché | [149] + | |
| Chinese | [108] + | ||
|
|
|||
| rs352143 | Vietnamese | [150] + | |
|
| |||
| TNF | |||
|
| |||
| rs1799724 | Chinese | [151] − | |
|
|
|||
| rs1800629 | Chinese | [152] + | |
| Colombian | [153] − | ||
| Iranian | [154] + | ||
| Mozambican | [155] + | ||
| Sicilian | [119] + | ||
|
|
|||
| rs1800630 | Chinese | [151] + | |
|
|
|||
| rs361525 | Colombian | [153] + | |
| Iranian | [156] − | ||
|
| |||
| VDR | |||
|
| |||
| rs1544410 | Indian | [157] + | |
| Indonesian (Batak) | [158] − | ||
| Iranian | [154] − | ||
| Iranian | [159] + | ||
| Iranian | [160]− | ||
|
|
|||
| rs2228570 | Chinese | [122] + | |
| Chinese | [135] + | ||
| Indians | [161] + | ||
| Indians | [162] + | ||
| Iranian | [160] − | ||
|
|
|||
| rs731236 | Chinese | [122] − | |
| Indian | [163] − | ||
| Indian | [164] + | ||
| Indian | [162] + | ||
| Iranian | [159] + | ||
|
|
|||
| rs7975232 | Indian | [163] + | |
| Indian | [162] − | ||
| West African | [26] − | ||
When the minor allele associated with increased risk, + was reported for effect, while - was reported when the minor allele associated with decreased risk. Direction of effect was reported based on allelic and/or genotypic associations.
Denotes associations where only heterozygotes associated with PTB risk.
Indicates the association with PTB risk was only found in women.
Indicates the association with PTB risk was only found in men.
Direction of effect is relative to non-12 CA repeats
Direction of effect is relative to non-allele 2
Allele 3 associated with decreased risk
Allele 3 associated with increased risk
Direction of effect is relative to non 5′ (GT)9 alleles
S/M genotype associated with increased risk
S/L genotype associated with decreased risk
Table 3. Studies Reporting Significant Haplotype Associations by Gene.
For each association the genetic variants composing the haplotype, the study population, and the reference are reported. Inclusion criteria for a particular gene to be located in this table included at least 5 studies reporting genetic variation within that gene In the situation genetic variants composing a haplotype were located within multiple genes, such as the HLA region, the haplotype association was only reported for one gene.
| Gene | Haplotype | Population | Reference |
|---|---|---|---|
| HLA-A | |||
|
| |||
| HLA-A2-DRB1*1502 | Indian | [100] | |
|
|
|||
| HLA-A*24-B*40-DRB1*15 | Indian (Sahariya) | [93] | |
|
|
|||
| HLA-A*02-B*40-DRB1*16 | Indian (Sahariya) | [93] | |
|
|
|||
| HLA-A*02-B*40-DRB1*03 | Indian (Sahariya) | [93] | |
|
| |||
| HLA-DQB1 | |||
|
| |||
| HLA-DQB1*301-304 | South African | [103] | |
|
|
|||
| HLA-DQB1*0601-DPB1*0201 | Indian | [98] | |
|
| |||
| HLA-DRB1 | |||
|
| |||
| HLA-DRB1*1101-1121-DQB1*05 | South African | [103] | |
|
|
|||
| HLA-DRB1*16-DQB1*05 | Polish | [96] | |
|
|
|||
| HLA-DRB1*04-DQB1*03 | Polish | [96] | |
|
|
|||
| HLA-DRB1*1601-DQB1*0502 | Polish | [96] | |
|
|
|||
| HLA-DRB1*11-DQB1*03 | Polish | [96] | |
|
|
|||
| HLA-DRB1*1502-DQB1*0601-DPB1*201 | Indian | [98] | |
|
|
|||
| HLA-B40-DRB1*1501 | Indian | [100] | |
|
|
|||
| HLA-B40-DRB1*04 | Indian | [100] | |
|
|
|||
| HLA-DRB1*1502-DPB1*0201 | Indian | [98] | |
|
|
|||
| HLA-DRB1*1502-DQB1*0601-DPB1*0201 | Indian | [98] | |
|
| |||
| IFNG | |||
|
| |||
| rs1861493-rs1861494 | Indian | [107] | |
|
| |||
| IFNGR1 | |||
|
| |||
| rs2234711-rs3799488 | Chinese | [113] | |
|
| |||
| MBL2 | |||
|
| |||
| rs11003125-rs7095891-rs7096206-rs1800450 | Chinese | [122] | |
|
|
|||
| rs7096206-rs1800450 | Chinese | [165] | |
|
| |||
| MCP1 | |||
|
| |||
| rs1024611-rs2857656-rs3917887 | Ghanaian | [126] | |
|
|
|||
| rs1024611-rs2857656 | Ghanaian | [123] | |
|
| |||
| NRAMP1/SLC11A1 | |||
|
| |||
| rs17235416-rs3731865-rs17221959 | Indian | [133] | |
| Peruvian | [136] | ||
| Iranian | [154] | ||
|
|
|||
| rs17235416-rs17221959 | Tunisian | [137] | |
|
| |||
| TLR2 | |||
|
| |||
| rs4696480-rs3804099-rs5743699 | Moroccan | [145] | |
|
|
|||
| rs4696480-rs1898830-(-196 to -174 ins/del)-rs3804100 | Taiwanese | [166] | |
|
| |||
| TLR4 | |||
|
| |||
| rs10116253-rs10759932-rs1927911-rs1927907-rs5030717-rs5030725 | Sudanese | [146] | |
|
|
|||
| rs7869402-rs1927906-rs7044464 | Sudanese | [146] | |
|
| |||
| TNFA | |||
|
| |||
| HLA-B17- rs361525 | Indian | [167] | |
|
|
|||
| HLA-B17- rs1800629 | Indian | [167] | |
|
|
|||
| rs1800629-rs361525 | Colombian | [153] | |
|
| |||
| VDR | |||
|
| |||
| rs2228570-rs1544410-rs7975232-rs731236 | Indian | [162] | |
| Moroccan | [145] | ||
| South African | [103] | ||
|
|
|||
| rs2228570- rs1544410-rs7975232 | Indian | [162] | |
|
|
|||
| rs1544410-rs7975232-rs731236 | Indian | [162] | |
| Indian | [164] | ||
|
|
|||
| rs1544410-rs7975232 | Indian | [162] | |
|
|
|||
| rs2228570-rs1544410 | Indian | [162] | |
As noted above [30], there has been very limited replication of genetic associations across studies. Possible reasons for this include:
Population genetic differences across populations: It is well established that linkage disequilibrium (LD) patterns differ greatly globally. This is especially true in Africa, where LD generally is the lowest [31–33] and TB burden is the greatest. Therefore, if a SNP under examination is not the causal SNP, LD differences among populations will decrease the probability of detecting an association even with a common functional variant. This is compounded by studies that only examine a few polymorphisms within a gene and do not cover the genetic variation sufficiently. Therefore, the SNPs may not be highly correlated with the same functional variant in diverse populations. This is compounded when there is allelic heterogeneity across populations.
Differences in study design: Both case and control definitions vary widely by research setting. Some TB cases are defined strictly using microbiological confirmation, while others use a smattering of clinical criteria to define TB (reviewed in [30]). Controls are often defined even more inconsistently. Some controls are known to be exposed to an active TB case, and thus have the opportunity to develop TB; others are population-based and have not been clinically characterized at all. These potential misclassifications can result in a bias towards the null hypothesis. Some studies have been family-based, while others used a traditional case-control design. The advantage of family-based studies of TB is that exposure is more certain even if variable [34, 35].
Gene-gene and gene-environment interactions: A number of studies have suggested that TB susceptibility genes interact with each other (Table 1) [7, 20, 22, 25, 36, 37]. When interaction exists between two genes, it is possible that significant main effects may not be observed due to different allele frequencies at the second gene or environmental exposure [38]. This is a generic danger for studies that examine one gene at a time. Further complicating gene discovery, a few studies have indicated an interaction between human genes and MTB lineage [39, 40], further complicating identification of human genes. If significant genetic effects only exist in the context of a specific MTB lineage, non-replication across populations may be due to strong gene-environment factors.
Role of GWAS vs candidate gene studies
To date, nine GWAS studies have been published in PTB (summarized in Table 1 of Uren et al [9]). A 2010 study discovered an association in a gene desert on chromosome 18q11.2 in a combined Ghanaian, Gambian and Malawian cohort [41]. Availability of 1000 Genomes Project data allowed the authors to impute SNPs into the Ghanaian cohort and identify a genome-wide significant association for a locus 46 kb downstream of WT1 [42]. This association was replicated in Gambian, Indonesian and Russian populations [42] and also by an independently conducted GWAS in an admixed population in South Africa [43]. The South African GWAS also detected loci on chromosomes 14q24.2 and 11q21-q22 that were just below genome-wide significance [43]. A case-control GWAS in a Russian population replicated the chromosome 11 locus, but not the chromosome 18 locus; it also detected significant association with the ASAP1 gene [44]. While a GWAS conducted in a Moroccan population did not detect any genome-wide significant associations, it replicated results from chromosomes 11 and 18 at a nominal significance (p<0.05) [45]. A GWAS in an Indonesian population did not detect any loci that were significant after multiple testing correction, although it did identify suggestively associated loci involved in immune signaling; the previous GWAS associations were not explicitly tested in this study [46]. A GWAS conducted in Icelandic, Russian, and Croatian populations identified significant association with the HLA region [47], but did not replicate the loci on chromosomes 11 and 18. This is possibly because these previously associated variants are rarer in European populations. An analysis that stratified young versus older subjects with TB in Indonesia and Japan detected a significant locus on chromosome 20q12 associated in the younger onset group [48]. Finally, our GWAS of TB in HIV-infected subjects identified a significant association at 5q33.3, and haplotype analyses suggested that this association is due to the IL12B gene [49].
Reasons for failure to replicate GWAS results include the reasons as noted above as for targeted candidate gene studies. Overall, such limitations have impeded progress in identifying key genetic associations for TB, but if we treat association studies as hypothesis generating exercises, they still provide an important way to gain insight about TB pathogenesis; with careful study design, genome-wide studies can provide entry points for learning about TB biology.
Pathway enrichment from gene associations
With few exceptions [21, 23, 50], most previous genetic association studies have not considered genes as part of pathways Using the 15 genes and 36 genes identified by candidate literature review and GWAS literature review (the latter as described in Table 1 and do overlap with some of the candidate genes [9]), respectively, we used Ingenuity Pathway Analysis (IPA) (Qiagen) to determine if any pathways were enriched using all genes from our list. IPA mapped all genes from our provided list onto expert-curated canonical pathways from the Ingenuity Knowledge Base. A right-tailed Fisher’s exact test using a Benjamini-Hochberg multiple testing corrected p-value of 0.05 determined whether the association between our literature generated list and a particular canonical pathway was significant compared to random chance.
IPA identified several enriched pathways, the majority of which were involved in immune response or processes (Figure 1; Supplemental Table 1). In total 71 pathways were found to be significantly enriched. The most significant pathway (p < 10−10) was altered T cell and B cell signaling in Rheumatoid Arthritis, of which our literature generated gene list represented 12.2% of the genes contained in the pathway (Figure 1 and Supplemental Table 1). While this top pathway is labeled as being associated with Rheumatoid Arthritis (RA), it is well known that the same inflammatory pathways involved in RA are also involved infectious disease response. The next few top pathways also reflected major components of the immune response to TB, such as T cells and other cells involved in pathogen recognition and the innate and adaptive immune response, and Th1 and Th2 cytokine response. Of course, as with all analyses of this type, pathway enrichment may be driven by common genes across pathways, and this is the case here as well.
Figure 1. Ten Most Significantly Associated Canonical Pathways from IPA.
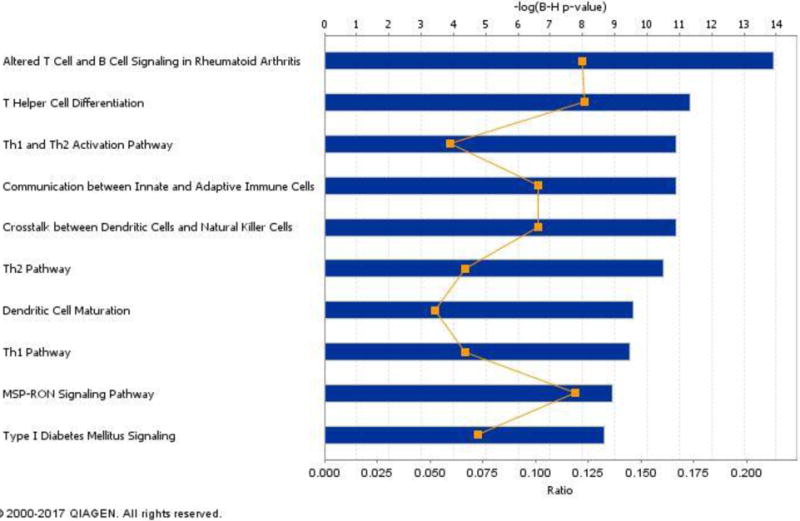
IPA determined the association between canonical pathways and the provided literature-based gene list. Here the top 10 associated pathways are shown. For each canonical pathway, the significance of association for a canonical pathway is depicted as the blue bar that is the -log(p-value) for the Benjamini-Hochberg corrected p-value. The ratio of genes represented within the provided list vs all genes contained within a pathway are shown as a gold point for each pathway.
While this recognition of genetic influences on the innate and adaptive immune response is not novel in itself, this analysis provides three novel insights into the genetics of response to TB. First, there may be other genes within these pathways, perhaps with smaller effect sizes, that are also important in TB genetics. Such smaller non-significant effect sizes would reflect the newly proposed “omnigenic” theory of complex trait genetics [51]. Second, we may find that the disruption of entire pathways is what contributes to TB susceptibility, not single genes. This line of thinking matches the approach of transcriptomic studies (summarized below). Third, it is noteworthy that a significantly associated pathway is diabetes signaling, and there is a well-established comorbidity between diabetes and TB. Therefore, approaches incorporating broader biological perspectives such as pathways may help to enhance our understanding of TB risk through how it may relate to other diseases.
Role of transcriptomic studies
Another approach to examine the role of host genomics in TB risk has been through gene expression studies, most of which have focused on identifying biomarkers that uniquely characterize TB cases [52, 53] [54] [55–59]. As with genetic studies, these studies have differed widely in their choice of comparison group (household contacts, latent MTB infection (LTBI), and/or uncharacterized healthy subjects). They have also differed widely in the number of gene transcripts interrogated. Some studies reported a transcriptional signature involving the type I/II interferon pathways [52, 58] [54] [59], while the others either did not compare their signatures to those previously published or did not describe the genes that composed these signatures. Some studies demonstrated that the signature observed in newly diagnosed TB cases normalized during or after treatment [52, 54] [59], validating that those genes were expressed in newly diagnosed TB but did not establish a causal relationship. One prospective study identified a signature that occurred prior to the development of TB that resolved post-treatment, and went on to show that SNPs within CCL1, one of the differentially expressed genes, were associated with TB in a case-control study [54]. The aforementioned studies used peripheral blood, which is attractive for a biomarker. Thuong et al. [60] took a different approach, by obtaining monocytes from circulating blood, stimulating them with MTB in vitro, and comparing the transcriptional responses to MTB between samples from cases and controls. This may more accurately reflect the in vivo response, but is not as translatable as a whole blood biomarker.
While the specific results of all these studies are quite different, two broad conclusions can be drawn. First, the usual focus on one gene at a time may be inadequate to accurately assess risk. The challenge of transcriptional studies is that this approach is not easily translatable to the field, even if it represents excellent biomarkers. Second, by examining the entire transcriptome instead of single genes, there are more likely to be consistent findings in pathways of interest than by simply assessing individual genes or transcripts. A more comprehensive review of transcriptional effects can be found in a review by Orlova and Schurr in this same issue.
Lessons learned from Mendelian immuodeficiencies and on family studies
Mendelian genetics has provided a proof of concept for susceptibility to mycobacterial infection and disease by demonstrating a role for gene variants. A monogenic etiology due to rare mutations with strong phenotypic effects, has been observed in some children with severe or disseminated mycobacterial infections and can inform studies of the complexity of genetic risk for TB. To date, the immuno-deficiencies (PIDs) associated with increased risk for mycobacterial infection and disease (including TB) in children have been primarily associated with defects in the IFN-γ signaling pathway. [61]. This Mendelian Susceptibility to Mycobacterial Diseases, OMIM209950 (MSMD), is a rare (10−5−10−6) and highly heterogeneous condition identified in families with parental consanguinity [62]. Molecular studies of MSMD revealed 18 genetic forms that associated with IFNGR1, IFNGR2, STAT1, IRF8, CYBB, IL12B, IL12RB1, NEMO and ISG15 genes, that all are part of the IL12/IFN-γ signaling pathways [61]. Mutations in these nine genes vary and exhibit incomplete penetrance that can translate into partial or complete loss of function. A general consistency among these phenotypes is impairment of IFN-γ function that affects activity of macrophages and dendritic cells for anti-mycobacterial defenses and antigen processing. Of relevance to more complex forms of resistance to TB as well as to therapeutic strategies, IL12B (IL-12 p40) or IL12RB1 (its receptor) mutations do not produce enough IFN-γ, and benefit from human recombinant cytokine treatment. Not surprisingly, IFN-γ treatment has no effect in persons with mutations in IFNGR1 and IFNGR2 [63]. Clearly, molecular dissection of MSMD has been crucial in defining the central role of the IL-12/IFNγ axis and associated genetic network in controlling/determining mycobacterial infection and disease, and has also provided therapeutic opportunities.
Resistance vs. susceptibility as a measured phenotype
An alternative approach to studying the genomics of TB susceptibility that we have successfully taken is to study the genetics of extreme resistance instead of the development of disease. Specifically, HIV-infected individuals living in TB-endemic settings are at high risk for developing TB. The importance of this is borne out by the fact that TB is the number one killer of HIV-infected individuals. Therefore, individuals who are HIV-infected but either resistant to disease or resistant to MTB display protection from MTB infection and its resulting disease. Additionally, genes that confer this resistance may help identify novel therapeutics more easily than genetic factors that increase susceptibility; adding something is easier than subtracting it in vivo. We have used this strategy to differentiate between TB cases and controls despite immunosuppression to shed light on innate immune factors that influence resistance [49]. We identified a locus near IL12B, a previously described candidate gene that has been shown to affect response to TB in mice as well as define highly susceptible families that have an IL12B knockout as well as among persons with MSMD. We have also studied persistently tuberculin skin test negativity despite close and prolonged exposure to active TB cases [64, 65] – while the focus here is on infection and not disease, this resistance phenotype may help identify novel targets for vaccine development and host directed therapies [66].
Necessary future directions
Need for biological validation
The majority of TB genetic association studies fail to identify the functional consequences of the associated polymorphisms. A few exceptions have yielded insight into TB biology. For example, a candidate gene study of CD1a illustrated how SNPs in this gene associate with markers of T cell response [67]. Similarly, a case-control study of TOLLIP showed association with TB, levels of mRNA expression of that gene, and IL6 production [68]. Gene expression studies indicated that the sodium butyrate pathway was associated with persistent TST negativity. Follow-up studies demonstrated that sodium butyrate and histone deacetylase inhibitors were associated with immunological response to MTB in vitro [66]. While these studies had smaller sample sizes, they often included independent case-control replication sets with functional validation relevant to TB biology. Another approach to functional validation are eQTL studies, where the association between genetic variants and RNA expression levels are demonstrated. This has proven quite insightful into TB biology [69, 70].
Need for novel pathway approaches
As mentioned above, examination of pathways instead of single gene effects has revealed novel therapeutic insight for TB [66]. Network approaches may be better for discovery and characterization of gene-gene interactions and pathways associated with disease than standard analytical approaches, because variants may have unremarkable individual effects but instead affect the phenotype through gene-gene interactions [71, 72]. In fact, the integration of data generated across multiple platforms to disentangle multifactorial diseases was suggested years ago [73], but rarely executed [74, 75].
Need for thorough epidemiology/clinical characterization
As we have reviewed previously [30], variability in diagnostic criteria and documentation of MTB exposure in controls can contribute to the inconsistency in the TB genetics literature. This also likely contributes to the variability across GWAS studies summarized above - differences in diagnostic criteria, local prevalence of TB, and lack of documented exposure in controls all may explain the inability to replicate across studies. However, our GWAS of HIV-infected populations in Uganda and Tanzania showed great consistency across those two populations [49], likely due to the thorough clinical characterization of the study subjects, and longitudinal follow-up of controls over several years to exclude the development of TB. Future studies should aim to use strict criteria such as the CDC/ATS criteria for diagnosis of TB [76] and include efforts to quantify exposure in controls. In our study of resistance to MTB infection, we carefully quantified exposure to an index TB case using extensive epidemiologic and clinical data [35]. An epidemiologic risk score can be used to determine whether control subjects were highly exposed to an infectious TB case, but resisted MTB infection or disease. There are other ways to demonstrate a high level of MTB exposure; for example, South African miners who work in poorly ventilated environments and are heavily exposed to MTB, can resist MTB infection and disease for years, if not decades [77].
The importance of HIV
Although globally 1 in 8 new TB cases are in people with HIV and 1 in 4 HIV-infected individuals die due to TB [2, 75, 78], examining the genetic risk for TB in HIV-infected individuals is uncommon. In fact, all of the aforementioned GWAS for TB excluded HIV-infected individuals, except ours [49]. Thus, the majority of genetic studies of TB disregarded the potential modification of the relationship between host genetic variation and risk of TB by HIV infection. We have previously shown a significant interaction between TNFR1 alleles and HIV status [24], reinforcing the importance of assessing genetic risk of TB in the context of HIV. As previously shown, novel insights into TB genetic resistance and susceptibility can be gained by focusing on TB-HIV co-infected individuals; additional studies using this approach are warranted.
Need to study host-pathogen interaction in diverse populations
One additional set of genetic factors that may affect risk of developing TB are external to the human genome. Host-pathogen interaction or co-evolution defined as “reciprocal, adaptive genetic changes in interacting host and pathogen species” [79] may affect MTB pathogenesis. Co-evolution of host and pathogen has been hypothesized to account for some disease pathogenesis variation and the discrepancy between exposure and disease in several infectious diseases, including TB [80]. Specifically, some MTB strains are highly infectious - but only in certain hosts, where the pathogenicity is modulated by host genetic variation for which ethnicity has been used as a surrogate measure of host genetics [81–85]. This is supported by the observation that there may be an association of MTB strains with host ethnicity [83]. The historical co-occurrence of humans and MTB and their co-migration out of Africa supports a long-standing relationship that provides the ideal condition for co-evolution, leading to reduced pathogenicity [16]. Host-pathogen co-evolution in TB is additionally supported by animal models [86, 87]. The existence of co-evolved genes can significantly affect our ability to identify loci in both species, which interact to affect disease risk or severity. A gene in one study may associate with TB because of the MTB strain, while it may not in places where the MTB strain differs. In a similar vein, we have recently shown that the disruption of co-evolution between Helicobacter pylori ancestry and human ancestry increases gastric disease severity in a diverse cohort of Colombians, providing proof of principle that co-evolution can be detected genomewide [88].
Conclusions
While numerous studies have been done on the human genetics of susceptibility to MTB, the inconsistencies across studies warrant new approaches to studying the genetics of TB. Family-based studies provide an opportunity for documented exposure and powerful genetic epidemiological approaches for rare variant mapping. Rigorous clinical characterization is essential. Where single gene approaches have fallen short, new approaches using pathway and/or polygenic/omnigenic approaches have the potential to reveal new insight into the complex host-pathogen interaction between MTB and humans. Finally, functional characterization and validation of findings in diverse populations are essential for genetic findings to turn into useful tools for risk stratification and development of novel vaccine and therapeutic targets.
Supplementary Material
References
- 1.Houben RM, Dodd PJ. The Global Burden of Latent Tuberculosis Infection: A Re-estimation Using Mathematical Modelling. PLoS medicine. 2016;13(10):e1002152. doi: 10.1371/journal.pmed.1002152. Epub 2016/10/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. Jama. 1999;282(7):677–86. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 3.Organization. WH. Global tuberculosis control: epidemiology, strategy, financing: WHO report 2009. Geneva: World Health Organization; 2009. 2009. [Google Scholar]
- 4.Comstock G. Tuberculosis in twins: a re-analysis of the Prophit Study. Am Rev Resp Dis. 1978;117:621–4. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 5.Kallmann F, Reisner D. Twin studies on the significance of genetic factors in tuberculosis. Am Rev Tuberculosis. 1943;47:549–74. [Google Scholar]
- 6.Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90(2):71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun. 2012;80(10):3343–59. doi: 10.1128/IAI.00443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abel L, El-Baghdadi J, Bousfiha AA, Casanova JL, Schurr E. Human genetics of tuberculosis: a long and winding road. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2014;369(1645):20130428. doi: 10.1098/rstb.2013.0428. Epub 2014/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uren C, Henn BM, Franke A, Wittig M, van Helden PD, Hoal EG, et al. A post-GWAS analysis of predicted regulatory variants and tuberculosis susceptibility. PloS one. 2017;12(4):e0174738. doi: 10.1371/journal.pone.0174738. Epub 2017/04/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guwattude D, Nakakeeto M, Jones-Lopez E, Maganda A, Chiunda A, Mugerwa R, et al. Tuberculosis in household contacts of infectious cases in Kampala, Uganda. Am J Epidemiol. 2003;158:887–98. doi: 10.1093/aje/kwg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lienhardt C, Fielding K, Sillah J, Tunkara A, Donkor S, Manneh K, et al. Risk factors for tuberculosis infection in sub-Saharan Africa: A contact study in The Gambia. American journal of respiratory and critical care medicine. 2003;168:448–55. doi: 10.1164/rccm.200212-1483OC. [DOI] [PubMed] [Google Scholar]
- 12.Lienhardt C, Sillah J, Fielding K, Donkor S, Manneh K, Warndorff D, et al. Risk factors for tuberculosis infection in children in contact with infectious tuberculosis cases in The Gambia, West Africa. Pediatrics. 2003;111:e608–e14. doi: 10.1542/peds.111.5.e608. [DOI] [PubMed] [Google Scholar]
- 13.Lonnroth K, Castro KG, Chakaya JM, Chauhan LS, Floyd K, Glaziou P, et al. Tuberculosis control and elimination 2010-50: cure, care, and social development. Lancet. 2010;375(9728):1814–29. doi: 10.1016/S0140-6736(10)60483-7. [DOI] [PubMed] [Google Scholar]
- 14.Lemvik G, Rudolf F, Vieira F, Sodemann M, Ostergaard L, Rodrigues A, et al. Decline in overall, smear-negative and HIV-positive TB incidence while smear-positive incidence stays stable in Guinea-Bissau 2004–2011. Trop Med Int Health. 2014;19(11):1367–76. doi: 10.1111/tmi.12378. [DOI] [PubMed] [Google Scholar]
- 15.Gengenbacher M, Kaufmann SH. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev. 2012;36(3):514–32. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nature genetics. 2013;45(10):1176–82. doi: 10.1038/ng.2744. Epub 2013/09/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7(5):328–37. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 18.Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nature genetics. 2016;48(12):1535–43. doi: 10.1038/ng.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris GA, Edwards DR, Hill PC, Wejse C, Bisseye C, Olesen R, et al. Interleukin 12B (IL12B) genetic variation and pulmonary tuberculosis: a study of cohorts from The Gambia, Guinea-Bissau, United States and Argentina. PloS one. 2011;6(2):e16656. doi: 10.1371/journal.pone.0016656. Epub 2011/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velez Edwards DR, Tacconelli A, Wejse C, Hill PC, Morris GA, Edwards TL, et al. MCP1 SNPs and pulmonary tuberculosis in cohorts from West Africa, the USA and Argentina: lack of association or epistasis with IL12B polymorphisms. PloS one. 2012;7(2):e32275. doi: 10.1371/journal.pone.0032275. Epub 2012/03/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velez DR, Wejse C, Stryjewski ME, Abbate E, Hulme WF, Myers JL, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet. 2010;127(1):65–73. doi: 10.1007/s00439-009-0741-7. Epub 2009/09/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu T, Chen Y, Kiralis JW, Collins RL, Wejse C, Sirugo G, et al. An information-gain approach to detecting three-way epistatic interactions in genetic association studies. J Am Med Inform Assoc. 2013;20(4):630–6. doi: 10.1136/amiajnl-2012-001525. Epub 2013/02/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velez DR, Hulme WF, Myers JL, Weinberg JB, Levesque MC, Stryjewski ME, et al. NOS2A, TLR4, and IFNGR1 interactions influence pulmonary tuberculosis susceptibility in African-Americans. Hum Genet. 2009;126(5):643–53. doi: 10.1007/s00439-009-0713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein CM, Zalwango S, Chiunda AB, Millard C, Leontiev DV, Horvath AL, et al. Linkage and association analysis of candidate genes for TB and TNFalpha cytokine expression: evidence for association with IFNGR1, IL-10, and TNF receptor 1 genes. Hum Genet. 2007;121(6):663–73. doi: 10.1007/s00439-007-0357-8. Epub 2007/04/14. [DOI] [PubMed] [Google Scholar]
- 25.Collins RL, Hu T, Wejse C, Sirugo G, Williams SM, Moore JH. Multifactor dimensionality reduction reveals a three-locus epistatic interaction associated with susceptibility to pulmonary tuberculosis. BioData Min. 2013;6(1):4. doi: 10.1186/1756-0381-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bornman L, Campbell SJ, Fielding K, Bah B, Sillah J, Gustafson P, et al. Vitamin D receptor polymorphisms and susceptibility to tuberculosis in West Africa: a case-control and family study. J Infect Dis. 2004;190(9):1631–41. doi: 10.1086/424462. [DOI] [PubMed] [Google Scholar]
- 27.Vannberg FO, Chapman SJ, Khor CC, Tosh K, Floyd S, Jackson-Sillah D, et al. CD209 genetic polymorphism and tuberculosis disease. PloS one. 2008;3(1):e1388. doi: 10.1371/journal.pone.0001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooke GS, Campbell SJ, Sillah J, Gustafson P, Bah B, Sirugo G, et al. Polymorphism within the interferon-gamma/receptor complex is associated with pulmonary tuberculosis. American journal of respiratory and critical care medicine. 2006;174(3):339–43. doi: 10.1164/rccm.200601-088OC. [DOI] [PubMed] [Google Scholar]
- 29.Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, Aaby P, et al. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immun. 2007;8(6):456–67. doi: 10.1038/sj.gene.6364410. Epub 2007/07/06. [DOI] [PubMed] [Google Scholar]
- 30.Stein CM. Genetic epidemiology of tuberculosis susceptibility: impact of study design. PLoS Pathog. 2011;7(1) doi: 10.1371/journal.ppat.1001189. %20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science (New York, NY) 2002;298(5602):2381–5. doi: 10.1126/science.1078311. Epub 2002/12/21. [DOI] [PubMed] [Google Scholar]
- 32.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, et al. The genetic structure and history of Africans and African Americans. Science (New York, NY) 2009;324(5930):1035–44. doi: 10.1126/science.1172257. Epub 2009/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3(8):611–21. doi: 10.1038/nrg865. Epub 2002/08/03. [DOI] [PubMed] [Google Scholar]
- 34.Stein CM, Hall NB, Malone LL, Mupere E. The household contact study design for genetic epidemiological studies of infectious diseases. Front Genet. 2013;4:61. doi: 10.3389/fgene.2013.00061. Print;%2013.:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma N, Zalwango S, Malone LL, Nsereko M, Wampande EM, Thiel BA, et al. Clinical and epidemiological characteristics of individuals resistant to M. tuberculosis infection in a longitudinal TB household contact study in Kampala, Uganda. BMC Infect Dis. 2014;14:352. doi: 10.1186/1471-2334-14-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White MJ, Tacconelli A, Chen JS, Wejse C, Hill PC, Gomes VF, et al. Epiregulin (EREG) and human V-ATPase (TCIRG1): genetic variation, ethnicity and pulmonary tuberculosis susceptibility in Guinea-Bissau and The Gambia. Genes Immun. 2014;15(6):370–7. doi: 10.1038/gene.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velez DR, Hulme WF, Myers JL, Stryjewski ME, Abbate E, Estevan R, et al. Association of SLC11A1 with tuberculosis and interactions with NOS2A and TLR2 in African-Americans and Caucasians. Int J Tuberc Lung Dis. 2009;13(9):1068–76. [PMC free article] [PubMed] [Google Scholar]
- 38.Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PloS one. 2009;4(6):e5639. doi: 10.1371/journal.pone.0005639. Epub 2009/06/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caws M, Thwaites G, Dunstan S, Hawn TR, Lan NT, Thuong NT, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4(3):e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salie M, van der Merwe L, Moller M, Daya M, van der Spuy GD, van Helden PD, et al. Associations between human leukocyte antigen class I variants and the Mycobacterium tuberculosis subtypes causing disease. J Infect Dis. 2014;209(2):216–23. doi: 10.1093/infdis/jit443. Epub 2013/08/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Thye T, Vannberg FO, Wong SH, Owusu-Dabo E, Osei I, Gyapong J, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42(9):739–41. doi: 10.1038/ng.639. This paper was the first GWAS study of tuberculosis susceptibility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thye T, Owusu-Dabo E, Vannberg FO, van CR, Curtis J, Sahiratmadja E, et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nature genetics. 2012;44(3):257–9. doi: 10.1038/ng.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chimusa ER, Zaitlen N, Daya M, Moller M, Van Helden PD, Mulder NJ, et al. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum Mol Genet. 2014;23(3):796–809. doi: 10.1093/hmg/ddt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtis J, Luo Y, Zenner HL, Cuchet-Lourenco D, Wu C, Lo K, et al. Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nature genetics. 2015;47(5):523–7. doi: 10.1038/ng.3248. Epub 2015/03/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grant AV, Sabri A, Abid A, Abderrahmani RI, Benkirane M, Souhi H, et al. A genome-wide association study of pulmonary tuberculosis in Morocco. Hum Genet. 2016;135(3):299–307. doi: 10.1007/s00439-016-1633-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Png E, Alisjahbana B, Sahiratmadja E, Marzuki S, Nelwan R, Balabanova Y, et al. A genome wide association study of pulmonary tuberculosis susceptibility in Indonesians. BMC medical genetics. 2012;13(5):5. doi: 10.1186/1471-2350-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sveinbjornsson G, Gudbjartsson DF, Halldorsson BV, Kristinsson KG, Gottfredsson M, Barrett JC, et al. HLA class II sequence variants influence tuberculosis risk in populations of European ancestry. Nat Genet. 2016;48(3):318–22. doi: 10.1038/ng.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahasirimongkol S, Yanai H, Mushiroda T, Promphittayarat W, Wattanapokayakit S, Phromjai J, et al. Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J Hum Genet. 2012;10 doi: 10.1038/jhg.2012.35. [DOI] [PubMed] [Google Scholar]
- 49*.Sobota RS, Stein CM, Kodaman N, Scheinfeldt LB, Maro I, Wieland-Alter W, et al. A Locus at 5q33.3 Confers Resistance to Tuberculosis in Highly Susceptible Individuals. Am J Hum Genet. 2016;98(3):514–24. doi: 10.1016/j.ajhg.2016.01.015. This paper examined the resistance to tuberculosis in highly susceptible HIV positive subjects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall NB, Igo RP, Jr, Malone LL, Truitt B, Schnell A, Tao L, et al. Polymorphisms in TICAM2 and IL1B are associated with TB. Genes Immun. 2015;16(2):127–33. doi: 10.1038/gene.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyle EA, Li YI, Pritchard JK. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169(7):1177–86. doi: 10.1016/j.cell.2017.05.038. Epub 2017/06/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–7. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bloom CI, Graham CM, Berry MP, Wilkinson KA, Oni T, Rozakeas F, et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PloS one. 2012;7(10):e46191. doi: 10.1371/journal.pone.0046191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387(10035):2312–22. doi: 10.1016/S0140-6736(15)01316-1. This paper identified a gene expression signature in a prospective cohort associated with risk of developing active tuberculosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. The New England journal of medicine. 2014;370(18):1712–23. doi: 10.1056/NEJMoa1303657. Epub 2014/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaforou M, Wright VJ, Oni T, French N, Anderson ST, Bangani N, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS medicine. 2013;10(10):e1001538. doi: 10.1371/journal.pmed.1001538. Epub 2013/10/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12(1):15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 58.Maertzdorf J, Weiner J, 3rd, Mollenkopf HJ, Bauer T, Prasse A, Muller-Quernheim J, et al. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(20):7853–8. doi: 10.1073/pnas.1121072109. Epub 2012/05/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ottenhoff TH, Dass RH, Yang N, Zhang MM, Wong HE, Sahiratmadja E, et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PloS one. 2012;7(9):e45839. doi: 10.1371/journal.pone.0045839. Epub 2012/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thuong NT, Dunstan SJ, Chau TT, Thorsson V, Simmons CP, Quyen NT, et al. Identification of tuberculosis susceptibility genes with human macrophage gene expression profiles. PLoS Pathog. 2008;4(12):e1000229. doi: 10.1371/journal.ppat.1000229. Epub 2008/12/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boisson-Dupuis S, Bustamante J, El-Baghdadi J, Camcioglu Y, Parvaneh N, El Azbaoui S, et al. Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol Rev. 2015;264(1):103–20. doi: 10.1111/imr.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin Immunol. 2014;26(6):454–70. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alangari AA, Al-Zamil F, Al-Mazrou A, Al-Muhsen S, Boisson-Dupuis S, Awadallah S, et al. Treatment of disseminated mycobacterial infection with high-dose IFN-gamma in a patient with IL-12Rbeta1 deficiency. Clin Dev Immunol. 2011;2011:691956. doi: 10.1155/2011/691956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64*.Stein CM, Zalwango S, Malone LL, Won S, Mayanja-Kizza H, Mugerwa RD, et al. Genome scan of M. tuberculosis infection and disease in Ugandans. PLoS ONE. 2008;3(12):e4094. doi: 10.1371/journal.pone.0004094. This study was the first to identify putative loci for persistent skin test negative individuals in the face of documented and continuous MTB exposure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sobota RS, Stein CM, Kodaman N, Maro I, Wieland-Alter W, Igo RP, Jr, et al. A chromosome 5q31.1 locus associates with tuberculin skin test reactivity in HIV-positive individuals from tuberculosis hyper-endemic regions in east Africa. PLoS genetics. 2017;13(6):e1006710. doi: 10.1371/journal.pgen.1006710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Seshadri C, Sedaghat N, Campo M, Peterson G, Wells RD, Olson GS, et al. Transcriptional networks are associated with resistance to Mycobacterium tuberculosis infection. PloS one. 2017;12(4):e0175844. doi: 10.1371/journal.pone.0175844. This paper studied gene expression networtks that associated with resistance to tubrculin skin test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seshadri C, Shenoy M, Wells RD, Hensley-McBain T, Andersen-Nissen E, McElrath MJ, et al. Human CD1a deficiency is common and genetically regulated. Journal of immunology. 2013;191(4):1586–93. doi: 10.4049/jimmunol.1300575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shah JA, Vary JC, Chau TT, Bang ND, Yen NT, Farrar JJ, et al. Human TOLLIP regulates TLR2 and TLR4 signaling and its polymorphisms are associated with susceptibility to tuberculosis. J Immunol. 2012;189(4):1737–46. doi: 10.4049/jimmunol.1103541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci U S A. 2012;109(4):1204–9. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacis A, Nedelec Y, Barreiro LB. When genetics meets epigenetics: deciphering the mechanisms controlling inter-individual variation in immune responses to infection. Current opinion in immunology. 2014;29:119–26. doi: 10.1016/j.coi.2014.06.002. Epub 2014/07/02. [DOI] [PubMed] [Google Scholar]
- 71.McKinney BA, Pajewski NM. Six Degrees of Epistasis: Statistical Network Models for GWAS. Front Genet. 2011;2:109. doi: 10.3389/fgene.2011.00109.:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jia P, Liu Y, Zhao Z. Integrative pathway analysis of genome-wide association studies and gene expression data in prostate cancer. BMC Syst Biol. 2012;6(Suppl 3):S13. doi: 10.1186/1752-0509-6-S3-S13. Epub;%2012 Dec 17.:S13-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iyengar SK, Elston RC. The genetic basis of complex traits: rare variants or “common gene, common disease”? Methods Mol Biol. 2007;376(71–84):71–84. doi: 10.1007/978-1-59745-389-9_6. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y, Patel S, Nibbe R, Maxwell S, Chowdhury SA, Koyuturk M, et al. Systems biology analyses of gene expression and genome wide association study data in obstructive sleep apnea. Pac Symp Biocomput. 2011:14–25. doi: 10.1142/9789814335058_0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flexner C. HIV drug development: the next 25 years. Nat Rev Drug Discov. 2007;6(12):959–66. doi: 10.1038/nrd2336. [DOI] [PubMed] [Google Scholar]
- 76.Diagnostic Standards and Classification of Tuberculosis in Adults and Children. This official statement of the American Thoracic Society and the Centers for Disease Control and Prevention was adopted by the ATS Board of Directors, July 1999. This statement was endorsed by the Council of the Infectious Disease Society of America, September 1999. American journal of respiratory and critical care medicine. 2000;161(4 Pt 1):1376–95. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- 77.Hanifa Y, Grant AD, Lewis J, Corbett EL, Fielding K, Churchyard G. Prevalence of latent tuberculosis infection among gold miners in South Africa. Int J Tuberc Lung Dis. 2009;13(1):39–46. Epub 2008/12/25. [PubMed] [Google Scholar]
- 78.Kaufmann SH, McMichael AJ. Annulling a dangerous liaison: vaccination strategies against AIDS and tuberculosis. Nat Med. 2005;11(4 Suppl):S33–S44. doi: 10.1038/nm1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nature genetics. 2002;32(4):569–77. doi: 10.1038/ng1202-569. Epub 2002/11/29. [DOI] [PubMed] [Google Scholar]
- 80.Kodaman N, Sobota RS, Mera R, Schneider BG, Williams SM. Disrupted human-pathogen co-evolution: a model for disease. Front Genet. 2014;5:290. doi: 10.3389/fgene.2014.00290. Epub 2014/09/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pareek M, Evans J, Innes J, Smith G, Hingley-Wilson S, Lougheed KE, et al. Ethnicity and mycobacterial lineage as determinants of tuberculosis disease phenotype. Thorax. 2013;68(3):221–9. doi: 10.1136/thoraxjnl-2012-201824. Epub 2012/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6(12):e311. doi: 10.1371/journal.pbio.0060311. Epub 2008/12/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Di Pietrantonio T, Schurr E. Host-pathogen specificity in tuberculosis. Adv Exp Med Biol. 2013;783:33–44. doi: 10.1007/978-1-4614-6111-1_2. Epub 2013/03/08. [DOI] [PubMed] [Google Scholar]
- 84*.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2869–73. doi: 10.1073/pnas.0511240103. Epub 2006/02/16. This study demonstrated distinct MTB lineages, defined by geography, and showed that in cosmopolitan populations infection was more likely in sympatric host populations. These results provided evidence for historical co-adaptation between human and MTB by geographic commensalism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gagneux S. Host-pathogen coevolution in human tuberculosis. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012;367(1590):850–9. doi: 10.1098/rstb.2011.0316. Epub 2012/02/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kato-Maeda M, Shanley CA, Ackart D, Jarlsberg LG, Shang S, Obregon-Henao A, et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin Vaccine Immunol. 2012;19(8):1227–37. doi: 10.1128/CVI.00250-12. Epub 2012/06/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87*.Di Pietrantonio T, Hernandez C, Girard M, Verville A, Orlova M, Belley A, et al. Strain-specific differences in the genetic control of two closely related mycobacteria. PLoS Pathog. 2010;6(10):e1001169. doi: 10.1371/journal.ppat.1001169. Epub 2010/11/10. Based on findings suggestive of human-pathogen co-evolution, this study examined the strain effects in mouse lines. It showed a clear relationship between host genetics and MTB strain-specifc pathogenisis, indicative of co-evolution between host and pathogen for MTB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kodaman N, Pazos A, Schneider BG, Piazuelo MB, Mera R, Sobota RS, et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(4):1455–60. doi: 10.1073/pnas.1318093111. Epub 2014/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Naderi M, Hashemi M, Taheri M, Pesarakli H, Eskandari-Nasab E, Bahari G. CD209 promoter-336 A/G (rs4804803) polymorphism is associated with susceptibility to pulmonary tuberculosis in Zahedan, southeast Iran. J Microbiol Immunol. 2014;47(3):171–5. doi: 10.1016/j.jmii.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 90.Kobayashi K, Yuliwulandari R, Yanai H, Lien LT, Nguyen TLH, Hijikata M, et al. Association of CD209 polymorphisms with tuberculosis in an Indonesian population. Hum Immunol. 2011;72(9):741–5. doi: 10.1016/j.humimm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 91.Cai SJ, Su G, Li H, Xie B, Luo JM. Profiling of human leukocyte antigens in Eales disease and tuberculosis. International ophthalmology. 2013;33(5):475–9. doi: 10.1007/s10792-013-9718-6. Epub 2013/02/07. [DOI] [PubMed] [Google Scholar]
- 92.Amirzargar AA, Yalda A, Hajabolbaghi M, Khosravi F, Jabbari H, Rezaei N, et al. The association of HLA-DRB, DQA1, DQB1 alleles and haplotype frequency in Iranian patients with pulmonary tuberculosis. Int J Tuberc Lung Dis. 2004;8(8):1017–21. [PubMed] [Google Scholar]
- 93.Mishra G, Kumar N, Kaur G, Jain S, Tiwari PK, Mehra NK. Distribution of HLA-A, B and DRB1 alleles in Sahariya tribe of North Central India: An association with pulmonary tuberculosis. Infect Genet Evol. 2014;22:175–82. doi: 10.1016/j.meegid.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 94.Magira EE, Papasteriades C, Kanterakis S, Toubis M, Roussos C, Monos DS. HLA-A and HLA-DRB1 amino acid polymorphisms are associated with susceptibility and protection to pulmonary tuberculosis in a Greek population. Hum Immunol. 2012;73(6):641–6. doi: 10.1016/j.humimm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 95.Dubaniewicz A, Moszkowska G, Szczerkowska Z, Hoppe A. Analysis of DQB1 allele frequencies in pulmonary tuberculosis: preliminary report. Thorax. 2003;58(10):890–1. doi: 10.1136/thorax.58.10.890. Epub 2003/09/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dubaniewicz A, Moszkowska G, Szczerkowska Z. Frequency of DRB1-DQB1 two-locus haplotypes in tuberculosis: preliminary report. Tuberculosis. 2005;85(4):259–67. doi: 10.1016/j.tube.2004.12.003. Epub 2005/06/17. [DOI] [PubMed] [Google Scholar]
- 97.Vejbaesya S, Chierakul N, Luangtrakool K, Srinak D, Stephens HA. Associations of HLA class II alleles with pulmonary tuberculosis in Thais. European journal of immunogenetics: official journal of the British Society for Histocompatibility and Immunogenetics. 2002;29(5):431–4. doi: 10.1046/j.1365-2370.2002.00352.x. Epub 2002/10/03. [DOI] [PubMed] [Google Scholar]
- 98.Selvaraj P, Raghavan S, Swaminathan S, Alagarasu K, Narendran G, Narayanan PR. HLA-DQB1 and-DPB1 allele profile in HIV infected patients with and without pulmonary tuberculosis of south India. Infect Genet Evol. 2008;8(5):664–71. doi: 10.1016/j.meegid.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 99.Kim HS, Park MH, Song EY, Park H, Kwon SY, Han SK, et al. Association of HLA-DR and HLA-DQ genes with susceptibility to pulmonary tuberculosis in Koreans: preliminary evidence of associations with drug resistance, disease severity, and disease recurrence. Hum Immunol. 2005;66(10):1074–81. doi: 10.1016/j.humimm.2005.08.242. Epub 2006/01/03. [DOI] [PubMed] [Google Scholar]
- 100.Raghavan S, Selvaraj P, Swaminathan S, Alagarasu K, Narendran G, Narayanan PR. Haplotype analysis of HLA-A, -B antigens and-DRB1 alleles in south Indian HIV-1-infected patients with and without pulmonary tuberculosis. Int J Immunogenet. 2009;36(3):129–33. doi: 10.1111/j.1744-313X.2009.00835.x. [DOI] [PubMed] [Google Scholar]
- 101.Lucena-Silva N, Baliza MD, Martins AES, Deghaide NHS, Teixeira KM, Rodrigues LC, et al. Relatedness and HLA-DRB1 typing may discriminate the magnitude of the genetic susceptibility to tuberculosis using a household contact model. J Epidemiol Commun H. 2010;64(6):513–7. doi: 10.1136/jech.2008.086801. [DOI] [PubMed] [Google Scholar]
- 102.Souza de Lima D, Morishi Ogusku M, Porto Dos Santos M, de Melo Silva CM, Alves de Almeida V, Assumpcao Antunes I, et al. Alleles of HLA-DRB1*04 Associated with Pulmonary Tuberculosis in Amazon Brazilian Population. Plos One. 2016;11(2):e0147543. doi: 10.1371/journal.pone.0147543. Epub 2016/02/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lombard Z, Dalton DL, Venter PA, Williams RC, Bornman L. Association of HLA-DR, -DQ, and vitamin D receptor alleles and haplotypes with tuberculosis in the Venda of South Africa. Hum Immunol. 2006;67(8):643–54. doi: 10.1016/j.humimm.2006.04.008. Epub 2006/08/19. [DOI] [PubMed] [Google Scholar]
- 104.Duarte R, Carvalho C, Pereira C, Bettencourt A, Carvalho A, Villar M, et al. HLA class II alleles as markers of tuberculosis susceptibility and resistance. Rev Port Pneumol. 2011;17(1):15–9. doi: 10.1016/s0873-2159(11)70005-8. [DOI] [PubMed] [Google Scholar]
- 105.Shi GL, Hu XL, Yang L, Rong CL, Guo YL, Song CX. Association of HLA-DRB alleles and pulmonary tuberculosis in North Chinese patients. Genetics and molecular research: GMR. 2011;10(3):1331–6. doi: 10.4238/vol10-3gmr1132. Epub 2011/07/14. [DOI] [PubMed] [Google Scholar]
- 106.Hu Y, Wu LL, Li DG, Zhao Q, Jiang WL, Xu B. Association between cytokine gene polymorphisms and tuberculosis in a Chinese population in Shanghai: a case-control study. Bmc Immunol. 2015;16 doi: 10.1186/S12865-015-0071-6. doi: Artn 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abhimanyu Bose M, Jha P, Consortium IGV Footprints of genetic susceptibility to pulmonary tuberculosis: Cytokine gene variants in north Indians. Indian J Med Res. 2012;135(5):763–70. [PMC free article] [PubMed] [Google Scholar]
- 108.Yang Y, Li X, Cui W, Guan L, Shen F, Xu J, et al. Potential association of pulmonary tuberculosis with genetic polymorphisms of toll-like receptor 9 and interferon-gamma in a Chinese population. BMC infectious diseases. 2013;13:511. doi: 10.1186/1471-2334-13-511. Epub 2013/11/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hashemi M, Sharifi-Mood B, Nezamdoost M, Moazeni-Roodi A, Naderi M, Kouhpayeh H, et al. Functional polymorphism of interferon-gamma (IFN-gamma) gene +874T/A polymorphism is associated with pulmonary tuberculosis in Zahedan, Southeast Iran. Prague medical report. 2011;112(1):38–43. Epub 2011/04/08. [PubMed] [Google Scholar]
- 110.Lio D, Marino V, Serauto A, Gioia V, Scola L, Crivello A, et al. Genotype frequencies of the +874T-->A single nucleotide polymorphism in the first intron of the interferon-gamma gene in a sample of Sicilian patients affected by tuberculosis. European journal of immunogenetics: official journal of the British Society for Histocompatibility and Immunogenetics. 2002;29(5):371–4. doi: 10.1046/j.1365-2370.2002.00327.x. Epub 2002/10/03. [DOI] [PubMed] [Google Scholar]
- 111.Lopez-Maderuelo D, Arnalich F, Serantes R, Gonzalez A, Codoceo R, Madero R, et al. Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. American journal of respiratory and critical care medicine. 2003;167(7):970–5. doi: 10.1164/rccm.200205-438BC. [DOI] [PubMed] [Google Scholar]
- 112.Ben Selma W, Harizi H, Bougmiza I, Hannachi N, Ben Kahla I, Zaieni R, et al. Interferon gamma +874T/A polymorphism is associated with susceptibility to active pulmonary tuberculosis development in Tunisian patients. DNA and cell biology. 2011;30(6):379–87. doi: 10.1089/dna.2010.1157. Epub 2011/02/22. [DOI] [PubMed] [Google Scholar]
- 113.Lu JQ, Pan HQ, Chen YZ, Tang SW, Feng Y, Qiu SS, et al. Genetic polymorphisms of IFNG and IFNGR1 in association with the risk of pulmonary tuberculosis. Gene. 2014;543(1):140–4. doi: 10.1016/j.gene.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 114.Sahiratmadja E, Baak-Pablo R, Visser AW, Alisjahbana B, Adnan I, van Crevel R, et al. Association of polymorphisms in IL-12/IFN-gamma pathway genes with susceptibility to pulmonary tuberculosis in Indonesia. Tuberculosis. 2007;87(4):303–11. doi: 10.1016/j.tube.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 115.Feng FM, Liu XX, Sun YH, Zhang P, Sun SF, Zhang B, et al. Independent and joint effects of the IL-6 and IL-10 gene polymorphisms in pulmonary tuberculosis among the Chinese Han population. Genetics and Molecular Research. 2014;13(3):7766–72. doi: 10.4238/2014.September.26.14. [DOI] [PubMed] [Google Scholar]
- 116.Shin HD, Park BL, Kim LH, Cheong HS, Lee IH, Park SK. Common interleukin 10 polymorphism associated with decreased risk of tuberculosis. Exp Mol Med. 2005;37(2):128–32. doi: 10.1038/emm.2005.17. [DOI] [PubMed] [Google Scholar]
- 117.Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. Journal of Infectious Diseases. 2002;186(10):1463–8. doi: 10.1086/344891. [DOI] [PubMed] [Google Scholar]
- 118.Oh JH, Yang CS, Noh YK, Kweon YM, Jung SS, Son JW, et al. Polymorphisms of interleukin-10 and tumour necrosis factor-alpha genes are associated with newly diagnosed and recurrent pulmonary tuberculosis. Respirology. 2007;12(4):594–8. doi: 10.1111/j.1440-1843.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 119.Scola L, Crivello A, Marino V, Gioia V, Serauto A, Candore G, et al. IL-10 and TNF-alpha polymorphisms in a sample of Sicilian patients affected by tuberculosis: implication for ageing and life span expectancy. Mech Ageing Dev. 2003;124(4):569–72. doi: 10.1016/S0047-6374(03)00038-1. [DOI] [PubMed] [Google Scholar]
- 120.da Cruz HLA, da Silva RC, Segat L, de Carvalho MSZDG, Brandao LAC, Guimaraes RL, et al. MBL2 gene polymorphisms and susceptibility to tuberculosis in a northeastern Brazilian population. Infect Genet Evol. 2013;19:323–9. doi: 10.1016/j.meegid.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 121.Capparelli R, Iannaccone M, Palumbo D, Medaglia C, Moscariello E, Russo A, et al. Role Played by Human Mannose-Binding Lectin Polymorphisms in Pulmonary Tuberculosis. Journal of Infectious Diseases. 2009;199(5):666–72. doi: 10.1086/596658. [DOI] [PubMed] [Google Scholar]
- 122.Wu LL, Deng HJ, Zheng YH, Mansjo M, Zheng XB, Hu Y, et al. An association study of NRAMP1, VDR, MBL and their interaction with the susceptibility to tuberculosis in a Chinese population. Int J Infect Dis. 2015;38:129–35. doi: 10.1016/j.ijid.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 123.Thye T, Nejentsev S, Intemann CD, Browne EN, Chinbuah MA, Gyapong J, et al. MCP-1 promoter variant-362C associated with protection from pulmonary tuberculosis in Ghana, West Africa. Hum Mol Genet. 2009;18(2):381–8. doi: 10.1093/hmg/ddn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen LM, Tong X, Li XB, Wu ZJ, Zhang YG. The association between the-2518A/G polymorphism in the MCP-1 gene and the risk of pulmonary tuberculosis in Sichuan Chinese population. Eur Rev Med Pharmaco. 2015;19(4):563–6. [PubMed] [Google Scholar]
- 125.Shi GL, Yang L, Sun Y, Yin YJ, Song CX. MCP-1 gene polymorphisms in North Chinese patients with pulmonary tuberculosis. Genetics and Molecular Research. 2015;14(2):4035–40. doi: 10.4238/2015.April.27.18. [DOI] [PubMed] [Google Scholar]
- 126.Intemann CD, Thye T, Forster B, Owusu-Dabo E, Gyapong J, Horstmann RD, et al. MCP1 haplotypes associated with protection from pulmonary tuberculosis. Bmc Genet. 2011;12 doi: 10.1186/1471-2156-12-34. doi: Artn 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Flores-Villanueva PO, Ruiz-Morales JA, Song CH, Flores LM, Jo EK, Montano M, et al. A functional promoter polymorphism in monocyte chemoattractant protein-1 is associated with increased susceptibility to pulmonary tuberculosis. J Exp Med. 2005;202(12):1649–58. doi: 10.1084/jem20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arji N, Busson M, Iraqi G, Bourkadi JE, Benjouad A, Boukouaci W, et al. The MCP-1 (CCL2)-2518 GG genotype is associated with protection against pulmonary tuberculosis in Moroccan patients. J Infect Dev Countr. 2012;6(1):73–8. doi: 10.3855/jidc.1925. [DOI] [PubMed] [Google Scholar]
- 129.Ben-Selma W, Harizi H, Boukadida J. MCP-1 -2518 A/G functional polymorphism is associated with increased susceptibility to active pulmonary tuberculosis in Tunisian patients. Mol Biol Rep. 2011;38(8):5413–9. doi: 10.1007/s11033-011-0695-4. [DOI] [PubMed] [Google Scholar]
- 130.Awomoyi AA, Marchant A, Howson JMM, McAdam KPWJ, Blackwell JM, Newport MJ Interleukin-10, polymorphism in SLC11A1 (formerly NRAMP1), and susceptibility to tuberculosis. Journal of Infectious Diseases. 2002;186(12):1808–14. doi: 10.1086/345920. [DOI] [PubMed] [Google Scholar]
- 131.Dubaniewicz A, Jamieson SE, Dubaniewicz-Wybieralska MD, Fakiola M, Miller EN, Blackwell JM. Association between SLC11A1 (formerly NRAMP1) and the risk of sarcoidosis in Poland. Eur J Hum Genet. 2005;13(7):829–34. doi: 10.1038/sj.ejhg.5201370. [DOI] [PubMed] [Google Scholar]
- 132.Hoal EG, Lewis LA, Jamieson SE, Tanzer F, Rossouw M, Victor T, et al. SLC11A1 (NRAMP1) but not SLC11A2 (NRAMP2) polymorphisms are associated with susceptibility to tuberculosis in a high-incidence community in South Africa. Int J Tuberc Lung Dis. 2004;8(12):1464–71. Epub 2005/01/08. [PubMed] [Google Scholar]
- 133.Singh A, Gaughan JP, Kashyap VK. SLC11A1 and VDR gene variants and susceptibility to tuberculosis and disease progression in East India. Int J Tuberc Lung D. 2011;15(11):1468–74. doi: 10.5588/ijtld.11.0089. [DOI] [PubMed] [Google Scholar]
- 134.Leung KH, Yip SP, Wong WS, Yiu LS, Chan KK, Lai WM, et al. Sex- and age-dependent association of SLC11A1 polymorphisms with tuberculosis in Chinese: a case control study. BMC infectious diseases. 2007;7:19. doi: 10.1186/1471-2334-7-19. Epub 2007/03/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu W, Cao WC, Zhang CY, Tian L, Wu XM, Habbema JDF, et al. VDR and NRAMP1 gene polymorphisms in susceptibility to pulmonary tuberculosis among the Chinese Han population: a case-control study. Int J Tuberc Lung D. 2004;8(4):428–34. [PubMed] [Google Scholar]
- 136.Taype CA, Castro JC, Accinelli RA, Herrera-Velit P, Shaw MA, Espinoza JR. Association between SLC11A1 polymorphisms and susceptibility to different clinical forms of tuberculosis in the Peruvian population. Infect Genet Evol. 2006;6(5):361–7. doi: 10.1016/j.meegid.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 137.Ben-Selma W, Harizi H, Letaief M, Boukadida J. Age- and gender-specific effects on NRAMP1 gene polymorphisms and risk of the development of active tuberculosis in Tunisian populations. Int J Infect Dis. 2012;16(7):E543–E50. doi: 10.1016/j.ijid.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 138.Nugraha J, Anggraini R. Nramp1 Polymorphism and Susceptibility to Lung Tuberculosis in Surabaya, Indonesia. Se Asian J Trop Med. 2011;42(2):338–41. [PubMed] [Google Scholar]
- 139.Qu YB, Tang YX, Cao DZ, Wu F, Liu J, Lu GL, et al. Genetic polymorphisms in alveolar macrophage response-related genes, and risk of silicosis and pulmonary tuberculosis in Chinese iron miners. Int J Hyg Envir Heal. 2007;210(6):679–89. doi: 10.1016/j.ijheh.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 140.Khan AU, Aslam MA, Hussain I, Naz AG, Rana IA, Ahmad MM, et al. Role of Toll-like receptor 2 (-196 to -174) polymorphism in susceptibility to pulmonary tuberculosis in Pakistani population. Int J Immunogenet. 2014;41(2):105–11. doi: 10.1111/iji.12086. Epub 2013/09/04. [DOI] [PubMed] [Google Scholar]
- 141.Xue Y, Jin L, Li AZ, Wang HJ, Li M, Zhang YX, et al. Microsatellite polymorphisms in intron 2 of the toll-like receptor 2 gene and their association with susceptibility to pulmonary tuberculosis in Han Chinese. Clin Chem Lab Med. 2010;48(6):785–9. doi: 10.1515/Cclm.2010.154. [DOI] [PubMed] [Google Scholar]
- 142.Ben-Ali M, Barbouche MR, Bousnina S, Chabbou A, Dellagi K. Toll-like receptor 2 Arg677Trp polymorphism is associated with susceptibility to tuberculosis in Tunisian patients. Clin Diagn Lab Immun. 2004;11(3):625–6. doi: 10.1128/Cdli.11.3.625-626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhao Y, Bu H, Hong K, Yin H, Zou YL, Geng SJ, et al. Genetic polymorphisms of CCL1 rs2072069 G/A and TLR2 rs3804099 T/C in pulmonary or meningeal tuberculosis patients. International journal of clinical and experimental pathology. 2015;8(10):12608–20. Epub 2016/01/02. [PMC free article] [PubMed] [Google Scholar]
- 144.Naderi M, Hashemi M, Hazire-Yazdi L, Taheri M, Moazeni-Roodi A, Eskandari-Nasab E, et al. Association between toll-like receptor2 Arg677Trp and 597T/C gene polymorphisms and pulmonary tuberculosis in Zahedan, Southeast Iran. Braz J Infect Dis. 2013;17(5):516–20. doi: 10.1016/j.bjid.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Arji N, Busson M, Iraqi G, Bourkadi JE, Benjouad A, Bouayad A, et al. Genetic diversity of TLR2, TLR4, and VDR loci and pulmonary tuberculosis in Moroccan patients. J Infect Dev Countr. 2014;8(4):430–40. doi: 10.3855/jidc.3820. [DOI] [PubMed] [Google Scholar]
- 146.Zaki HY, Leung KH, Yiu WC, Gasmelseed N, Elwali NEM, Yip SP. Common polymorphisms in TLR4 gene associated with susceptibility to pulmonary tuberculosis in the Sudanese. Int J Tuberc Lung D. 2012;16(7):934–40. doi: 10.5588/ijtld.11.0517. [DOI] [PubMed] [Google Scholar]
- 147.Najmi N, Kaur G, Sharma SK, Mehra NK. Human Toll-like receptor 4 polymorphisms TLR4 Asp299Gly and Thr399Ile influence susceptibility and severity of pulmonary tuberculosis in the Asian Indian population. Tissue Antigens. 2010;76(2):102–9. doi: 10.1111/j.1399-0039.2010.01481.x. [DOI] [PubMed] [Google Scholar]
- 148.Ferwerda B, Kibiki GS, Netea MG, Dolmans WMV, van der Ven AJ. The toll-like receptor 4 Asp299Gly variant and tuberculosis susceptibility in HIV-infected patients in Tanzania. Aids. 2007;21(10):1375–7. doi: 10.1097/Qad.0b013e32814e6b2d. [DOI] [PubMed] [Google Scholar]
- 149.Lindenau JD, Salzano FM, Hurtado AM, Hill KR, Hutz MH. The role of variants from the innate immune system genes in tuberculosis and skin test response in a Native American population. Hum Immunol. 2016;77(10):981–4. doi: 10.1016/j.humimm.2016.06.017. Epub 2016/06/28. [DOI] [PubMed] [Google Scholar]
- 150.Graustein AD, Horne DJ, Arentz M, Bang ND, Chau TT, Thwaites GE, et al. TLR9 gene region polymorphisms and susceptibility to tuberculosis in Vietnam. Tuberculosis. 2015;95(2):190–6. doi: 10.1016/j.tube.2014.12.009. Epub 2015/01/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ma MJ, Xie LP, Wu SC, Tang F, Li H, Zhang ZS, et al. Toll-like receptors, tumor necrosis factor-alpha, and interleukin-10 gene polymorphisms in risk of pulmonary tuberculosis and disease severity. Hum Immunol. 2010;71(10):1005–10. doi: 10.1016/j.humimm.2010.07.009. Epub 2010/07/24. [DOI] [PubMed] [Google Scholar]
- 152.Fan HM, Wang Z, Feng FM, Zhang KL, Yuan JX, Sui H, et al. Association of TNF-alpha-238G/A and 308 G/A Gene Polymorphisms with Pulmonary Tuberculosis among Patients with Coal Worker’s Pneumoconiosis. Biomed Environ Sci. 2010;23(2):137–45. doi: 10.1016/S0895-3988(10)60043-8. [DOI] [PubMed] [Google Scholar]
- 153.Correa PA, Gomez LM, Cadena J, Anaya JM. Autoimmunity and tuberculosis. Opposite association with TNF polymorphism. J Rheumatol. 2005;32(2):219–24. [PubMed] [Google Scholar]
- 154.Merza M, Farnia P, Anoosheh S, Varahram M, Kazampour M, Pajand O, et al. The NRAMPI, VDR and TNF-alpha Gene Polymorphisms in Iranian Tuberculosis Patients: The Study on Host Susceptibility. Braz J Infect Dis. 2009;13(4):252–6. doi: 10.1590/s1413-86702009000400002. [DOI] [PubMed] [Google Scholar]
- 155.Mabunda N, Alvarado-Arnez LE, Vubil A, Mariamo A, Pacheco AG, Jani IV, et al. Gene polymorphisms in patients with pulmonary tuberculosis from Mozambique. Mol Biol Rep. 2015;42(1):71–6. doi: 10.1007/s11033-014-3741-1. [DOI] [PubMed] [Google Scholar]
- 156.Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, Hajabdolbaghi M, et al. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw. 2006;17(2):84–9. [PubMed] [Google Scholar]
- 157.Joshi L, Ponnana M, Penmetsa SR, Nallari P, Valluri V, Gaddam S. Serum Vitamin D Levels and VDR Polymorphisms (BsmI and FokI) in Patients and their Household Contacts Susceptible to Tuberculosis. Scand J Immunol. 2014;79(2):113–9. doi: 10.1111/sji.12127. [DOI] [PubMed] [Google Scholar]
- 158.Sinaga BY, Amin M, Siregar Y, Sarumpaet SM. Correlation between Vitamin D receptor gene FOKI and BSMI polymorphisms and the susceptibility to pulmonary tuberculosis in an Indonesian Batak-ethnic population. Acta medica Indonesiana. 2014;46(4):275–82. Epub 2015/01/31. [PubMed] [Google Scholar]
- 159.Banoei MM, Mirsaeidi MS, Houshmand M, Tabarsi P, Ebrahimi G, Zargari L, et al. Vitamin D receptor homozygote mutant tt and bb are associated with susceptibility to pulmonary tuberculosis in the Iranian population. Int J Infect Dis. 2010;14(1):E84–E5. doi: 10.1016/j.ijid.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 160.Marashian SM, Farnia P, Seyf S, Anoosheh S, Velayati AA. Evaluating the role of vitamin D receptor polymorphisms on susceptibility to tuberculosis among iranian patients: a case-control study. Tuberkuloz ve toraks. 2010;58(2):147–53. Epub 2010/09/25. [PubMed] [Google Scholar]
- 161.Rathored J, Sharma SK, Singh B, Banavaliker JN, Sreenivas V, Srivastava AK, et al. Risk and outcome of multidrug-resistant tuberculosis: vitamin D receptor polymorphisms and serum 25(OH)D. Int J Tuberc Lung Dis. 2012;16(11):1522–8. doi: 10.5588/ijtld.12.0122. Epub 2012/09/20. [DOI] [PubMed] [Google Scholar]
- 162.Sharma PR, Singh S, Jena M, Mishra G, Prakash R, Das PK, et al. Coding and non-coding polymorphisms in VDR gene and susceptibility to pulmonary tuberculosis in tribes, castes and Muslims of Central India. Infect Genet Evol. 2011;11(6):1456–61. doi: 10.1016/j.meegid.2011.05.019. Epub 2011/06/08. [DOI] [PubMed] [Google Scholar]
- 163.Panwar A, Garg RK, Malhotra HS, Jain A, Singh AK, Prakash S, et al. 25-Hydroxy Vitamin D, Vitamin D Receptor and Toll-like Receptor 2 Polymorphisms in Spinal Tuberculosis: A Case-Control Study. Medicine. 2016;95(17):e3418. doi: 10.1097/MD.0000000000003418. Epub 2016/04/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alagarasu K, Selvaraj P, Swaminathan S, Narendran G, Narayanan PR. 5 ′ Regulatory and 3 ′ Untranslated Region Polymorphisms of Vitamin D Receptor Gene in South Indian HIV and HIV-TB Patients. J Clin Immunol. 2009;29(2):196–204. doi: 10.1007/s10875-008-9234-z. [DOI] [PubMed] [Google Scholar]
- 165.Liu W, Zhang F, Xin ZT, Zhao QM, Wu XM, Zhang PH, et al. Sequence variations in the MBL gene and their relationship to pulmonary tuberculosis in the Chinese Han population. Int J Tuberc Lung D. 2006;10(10):1098–103. [PubMed] [Google Scholar]
- 166.Chen YC, Hsiao CC, Chen CJ, Chin CH, Liu SF, Wu CC, et al. Toll-like receptor 2 gene polymorphisms, pulmonary tuberculosis, and natural killer cell counts. BMC medical genetics. 2010;11:17. doi: 10.1186/1471-2350-11-17. Epub 2010/02/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Selvaraj P, Sriram U, Mathan Kurian S, Reetha AM, Narayanan PR. Tumour necrosis factor alpha (-238 and -308) and beta gene polymorphisms in pulmonary tuberculosis: haplotype analysis with HLA-A, B and DR genes. Tuberculosis. 2001;81(5–6):335–41. doi: 10.1054/tube.2001.0307. Epub 2002/01/22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


