Abstract
For over 40 years, EBV infection has been implicated in the etiology of a variety of lymphoid malignancies with the exceptional ability to drive resting B cells to continuously proliferate by successfully overriding cellular apoptotic stimuli. EBV utilizes the normal physiology of B-cell differentiation to persist within the memory B-cell pool of the immunocompetent host and subsequently establishes a lifelong latent infection. During latency, out of a subset of viral genes expressed, EBNA-3C is one of the essential antigens required for in vitro primary B-cell transformation. EBNA-3C acts as a transcriptional coregulator by interacting with various cellular and viral factors. For the last 10 years, we have been actively engaged in discerning the biological significance of these interactions and revealed that EBNA-3C primarily targets two important cellular pathways – cell cycle and apoptosis. This review aims to summarize our current knowledge on EBNA-3C-mediated functions and describe how EBNA-3C seizes these cellular pathways that eventually promote B-cell lymphomagenesis. A scrupulous understanding of the critical relationship between EBNA-3C and these cellular machineries will not only aid in elucidating EBV pathogenesis, but also largely facilitate the development of novel diagnostic, as well as therapeutic, strategies against a vast range of EBV-associated B-cell lymphomas.
Keywords: apoptosis, cell cycle, cyclins, E2F1, EBNA-3C, EBV, p53, ubiquitin–proteasome pathway
EBV: the first human tumor virus
EBV, or human herpes virus 4, which is perhaps the most intensively studied member of the γ-Herpesviridae subfamily, was first detected by Michael Anthony Epstein and Yvonne Barr in cells derived from Burkitt’s lymphoma (BL), a B-cell-derived childhood malignancy that is endemic in Equatorial Africa. Subsequent studies have demonstrated that it is the causative agent in most cases of infectious mononucleosis. Currently, it is estimated that greater than 95% of the world’s population is asymptomatically infected with this virus. However, EBV has been demonstrated to be associated with numerous human neoplasms including hematopoietic, epithelial and mesenchymal tumors (Box 1) [1–4]. EBV is typically transmitted through salivary contact. While initial replication occurs in oro-pharyngeal epithelial cells, EBV preferentially infects B lymphocytes and in vitro infection can result in immortalization of resting B cells, known as lymphoblastoid cell lines (LCLs) [1,4,5]. In order to penetrate the B lymphocytes, EBV-encoded envelope glycoprotein, GP350, directly interacts with the type 2 complement receptor, CD21 [6]. After primary infection, EBV can persist in its latent form within the memory B cells for the host’s lifetime and is an approximately 182-kb long dsDNA virus. The genome is linear in the virus particle, but circularizes in the infected nuclei [1,6].
Box 1. EBV-associated cancers.
B-cell malignancies in the immunocompromised host
AIDS-associated B-cell lymphomas
Post-transplantation lymphoproliferative disorder
Lymphomatoid granulomatosis
Severe combined immunodeficiency-associated B-cell lymphomas
Wiskott–Aldrich syndrome-associated B-cell lymphomas
X-linked lymphoproliferative disorder-associated B-cell lymphomas
Kaposi’s sarcoma-associated herpesvirus-positive primary effusion lymphoma and its solid variant
B-cell malignancies in the immunocompetent host
Burkitt’s lymphoma
Classic Hodgkin’s lymphoma
T-cell malignancies
Extranodal NK and T-cell lymphoma
Hemophagocytic syndrome T-cell lymphomas
Epithelial cell malignancies
Nasopharyngeal carcinoma
Hepatocellular carcinoma
Gastric carcinoma
EBV-associated lymphomas are linked to latent infection
While B cells are largely nonpermissive for virus replication, they readily express a set of viral genes that are collectively known as the latency genes that differentiate from the much more numerous lytic genes expressed during productive infection [1]. In lytic infection, EBV-encoded genes selectively replicate virion components including viral DNA genomes and proteins. In latent infection, EBV-encoded genes are six nuclear antigens (EBNA-1, -2, -3A, -3B, -3C and -LP), three membrane-associated proteins (LMP-1, -2A and -2B), two small noncoding RNAs (EBER1 and EBER2) and BARTs [1,5]. Based on the expression pattern of these latent genes the latency program can be divided into many subgroups, which is believed to have evolved in order to maintain episomal persistence and enable the virus to evade adaptive immune response and immune surveillance [3]. Latently infected B-cells express one of four EBV latency programs that appear to reflect the adaptation of the virus to different stages of B-cell activation and differentiation [7,8]. Healthy EBV carrier populations contain approximately one to 50 virus-infected B lymphocytes per million cells in the peripheral blood, which are phenotypically indistinguishable from the long-lived memory cells [9]. These cells either express a ‘latency 0’ program, characterized by a complete silencing of the viral genome, or ‘latency I’ program, in which LMP-2A, or together with EBNA-1 expression is detected. In the absence of a complete immune response, as observed in the case of in vitro or in vivo immunocompromised patient’s samples, the EBV-infected B cells express all latency proteins, known as ‘latency III’ or ‘growth program’ [10]. An intermediate form of latency program is also characterized with the expression of EBNA-1 along with the three LMPs [10]. This is known as either ‘latency II’ or ‘rescue program’ [10]. The relevance of these discrete latency programs has been strongly supported by a series of studies on numerous EBV-associated lymphoproliferative disorders [11]. For example, latency program III is expressed in the immunoblast-like cells of EBV, associated with lymphoproliferative disorders arising in organ and bone marrow transplant recipients, HIV patients and in vitro transformed LCLs, whereas latency program I is found in EBV-carrying BLs that are phenotypically similar to memory B lymphocytes. Latency II program is associated with Hodgkin’s lymphomas [6,11].
The underlying mechanisms of EBV-induced B-lymphocyte growth transformation expressing a type III latency program have been under intensive investigation. These mechanisms are particularly relevant in order to uncover the roles of the essential latent gene products in controlling the early stages of primary infection and in developing lymphoproliferative disorders in immunocompromised individuals, such as patients with HIV infection and organ transplantation [9–11]. It is well established that EBV-latent gene products can drive oncogenesis [6,8,11]. However, recent studies have also implicated the EBV lytic cycle in the development of B-cell lymphomas, in the context of active host immune response [12,13]. In vitro genetic engineering studies using recombinant viruses from a number of different groups, showed that four nuclear antigens, including EBNA-1, -2, -3A and -3C, along with LMP-1, out of 11 latent transcripts are required for efficient B-cell transformation in vitro [1]. On the contrary, recent work by Hertle et al. showed that EBNA-3A may not be essential for B-cell transformation, as infection with an EBNA-3A mutant virus can still initiate cell cycle entry and proliferation of primary human B cells; however, the cells exhibited reduced cell proliferation rates with increased apoptosis [14]. A comprehensive summary of these essential latent gene products and their proposed functions in the B-cell transformation process is tabulated in Table 1.
Table 1.
Four EBV-encoded absolute essential latent antigens and some of their major functions in naive B-cell transformation.
| Latent antigens | Associated functions | Linked with other human cancers | Ref. |
|---|---|---|---|
| EBNA-1 | Assists in EBV episome maintenance and replication, blocks interaction between HAUSP and p53 to facilitate p53 degradation | BL, HD, AAL and PTLD | [151] |
| EBNA-2 | Transcriptional coactivator that upregulates expression of viral (LMP1) and cellular (c-myc) genes, interacts with RBP-Jκ and Notch signaling pathway | AAL and PTLD | [45,152] |
| EBNA-3C | Overrides both G1/S and G2/M cell cycle blockage, binds to RBP-Jκ and regulates viral gene transcription from Cp promoter, promotes LMP-1 expression, blocks p53-, E2F1- and Bim-mediated apoptotic cell deaths, enhances kinase activity of both cyclin D1/CDK6 and cyclin A/CDK2 complexes, induces degradation of p27KIP1 and pRb tumor-suppressor proteins, stabilizes c-Myc, cyclin D1 and MDM2, manipulates host chromatin remodeling machinery, interacts with and negatively modulates Nm23-H1-mediated metastatic suppression, and together with Nm23-H1, regulates Necdin-mediated transcriptional repression and antiangiogenic activities. Binds to MRS18-2 and directs it to the nucleus, where it functions as a pRb inhibitory factor | AAL and PTLD | [15,16,61,64,90,97,108] |
| LMP-1 | Mimics CD40 ligand binding signal, stimulates bcl-2 and a20 expression to block apoptosis, acts as a constitutively active receptor for stimulating many cellular genes, regulates NF-κB, JAK/STAT, ERK MAPK, IRF and Wnt signaling pathways | HD, AAL and PTLD | [153] |
AAL: AIDS-associated lymphoma; BL: Burkitt’s lymphoma; HD: Hodgkin’s disease; PTLD: Post-transplant lymphoproliferative disease.
EBNA-3C: an essential EBV-encoded multifunctional protein
Historical perspective
The EBNA-3 proteins were first identified in latently infected B-cell cultures through immunoblotting with the patient’s sera after prior EBV exposure or rheumatoid arthritis [15,16]. In addition to the previously identified latent proteins EBNA-1, EBNA-2 and LMP-1, latently infected B lymphocytes were also demonstrated to express an additional nuclear protein, which was initially named EBNA-3, approximately 140 kDa in size [15,16]. The EBNA-3 nuclear proteins, which include EBNA-3A, -3B and -3C, are encoded by three tandemly arranged genes in the viral episome, each containing a short 5′ and long 3′ exon [16]. Originally, EBNA-3 proteins were determined to be a single polypeptide in all EBV-infected B-cell lymphoma lines and encoded from the BamHI-E fragment rightward open reading frame 1 (BERF1) region of the viral episome using bacterial expression system. In addition, transfection of the BERF1 region cloned in a eukaryotic expression vector into rodent cells also produced an approximately 130 kDa protein that specifically immunoblotted with affinity-purified EBNA-3 human antisera and was found to be localized in the nucleus using an immunofluorescence study [16]. Interestingly, several of the human antisera samples tested reacted with EBV-latent proteins somewhat larger than EBNA-3 protein, suggesting that there may be a family of potential EBNA-3 proteins in B cells [15,16]. Subsequent studies demonstrated that the most rightward short and long BamHI-E open reading frames, BERF3 and BERF4, encoded a 155 kDa protein, later named EBNA-3C. The results showed that subcellular localization of this protein is also restricted to the nucleus and is present in all latently infected B cells with an intact EBV epi-some [15,16]. However, by contrast, one of the EBV-positive B-cell lines, Raji, did not express EBNA-3C as there is a deletion of the BERF4 region [15]. EBNA-3B was identified in a similar fashion, with the expression of a 165-kDa nuclear protein [15,16]. In parallel studies, other laboratories have also found multiple BamHI-E DNA fragments transfected into Cos-1 cells yielded EBNA-3B (or EBNA-4) and EBNA-3C (or EBNA-6) proteins through western blot using EBV-exposed human antisera [15,16].
Structural & functional similarities among EBNA-3 proteins
In general, all EBNA-3 proteins are large, hydrophilic, proline-rich and positively charged, sharing a similar genetic structure and a limited, but significant, amino acid sequence homology that is approximately 40%, suggesting that they perhaps evolved by a series of gene duplication events [17,18]. In addition, these proteins share more than 70% sequence homology between EBV type-1 and -2 strains [19]. EBNA-3 proteins possess a number of similar structural motifs, which include a binding site for a Notch signaling mediator and a highly potent transcriptional regulator RBP-Jκ, a leucine zipper motif, an acidic domain, proline- and glutamine-rich repeats and several arginine and lysine residues known to be important for nuclear localization [18]. Studies evaluating the functional domains of the EBNA-3A protein found that the amino terminus (residues 147–157) contains a nuclear localization sequence (NLS; RDRRRNPASR). Further computational studies, as well as wet laboratory experiments with green fluorescent protein-tagged EBNA-3A constructs, have shown an additional five NLS located throughout the protein. Similar experiments demonstrated that EBNA-3B also contains four potential NLS domains, but only two (residues 160–166 and 867–873) appear to be functional, while EBNA-3C contains five predicted NLS domains, three of which appear to be functional, positioned at residues 72–80, 412–418 and 939–945 [20–22].
Together with other EBNAs, all EBNA-3 transcripts are alternatively spliced from large mRNAs initiated at the Cp latency promoter [4]. In general, LCLs express very few copies of these transcripts per cell, suggesting that their expression is tightly regulated, whereas the EBNA-3 proteins are relatively stable [23,24]. Despite their partial sequence homology [18], EBNA-3 proteins were shown to have significant functional similarities. For example, they all form a complex with RBP-Jκ, a major Notch signaling modulator and a sequence-specific DNA-binding transcription factor that specifically upregulates EBNA-2-mediated transcriptional activation of several viral (LMP-1/-2) or cellular (CD23) genes [15,16,23,24]. Molecular genetics studies inserting stop codons into viral genes have been employed to evaluate the individual function of EBNA-3 proteins. Interestingly, insertion of stop codon mutations in the EBNA-3A and EBNA-3C genes, but not in the EBNA-3B gene, negatively affected the ability of the recombinant virus to transform quiescent B lymphocytes. This indicates that both EBNA-3A and -3C are essential for in vitro B-cell transformation, whereas EBNA-3B is dispensable for this process [15,16,25–29]. However, recently a similar group has extended their work using EBNA-3A deletion mutant virus and further demonstrated that EBNA-3A may not be completely essential for B-cell transformation [14]. Interestingly, the EBNA-3 family of proteins, collectively with EBNA-3B, appear to be the primary antigenic targets for cytotoxic T-cell responses against immortalized B cells [4], suggesting a critical role for each of these proteins during primary EBV infection and provides a possible mechanism for treatment of EBV-associated B-cell lymphomas.
EBNA-3C: a major transcriptional regulator
EBNA-3C (or EBNA-6) was the fifth EBV-encoded latent antigen identified in virus-transformed cell lines [30]. It is a 992 amino acid protein with a molecular weight of approximately 160 kDa on SDS-PAGE analysis [31]. As aforementioned, subcellular localization of EBNA-3C was shown to be exclusively in the nucleus in a distinct punctate pattern throughout different stages of the cell cycle [20]. Initial immunofluo-rescence studies revealed that EBNA-3C can colocalize with survival of motor neuron protein (SMN), which is part of a large complex that plays an important role in small nuclear ribo-nucleoprotein assembly, pre-mRNA splicing, as well as gene transcription [32,33]. The functional significance of these interactions has not been properly described, but they suggested that EBNA-3C may be involved in RNA processing or transcriptional regulation. It is still unclear whether EBNA-3C has any direct role in regulating RNA turnover, whereas a huge amount of data generated by the tremendous efforts of different groups, including our group for the last 15 years, showed that EBNA-3C is a potent transcriptional coregulator that interacts with numerous cellular proteins (Figure 1), essentially involved in regulating multiple important cellular pathways, including cell cycle and apoptosis (Figures 2 & 3).
Figure 1. Known interacting domains of EBNA-3C with various cellular proteins.
EBNA-3C is a large nuclear protein consisting of 992 amino acid residues. The protein contains three NLS located at residues 72–80, 412–418 and 939–945. Besides NLSs, the protein contains many canonical domains, namely an acidic domain, leucine zipper motif, proline-rich domain and transcriptional activation as well as repression domains. EBNA-3C interacts with an array of transcription factors, cell cycle regulatory proteins, chromatin remodeling enzymes and ubiquitin–proteasome machinery. The interaction has been observed using both in vitro and in vivo methods. Refer to Table 2 for EBNA-3C-mediated deregulation of functional activities of these interacting proteins. Dotted circles represent possible complex formation. HDAC: Histone deacetylase; NLS: Nuclear localization signal.
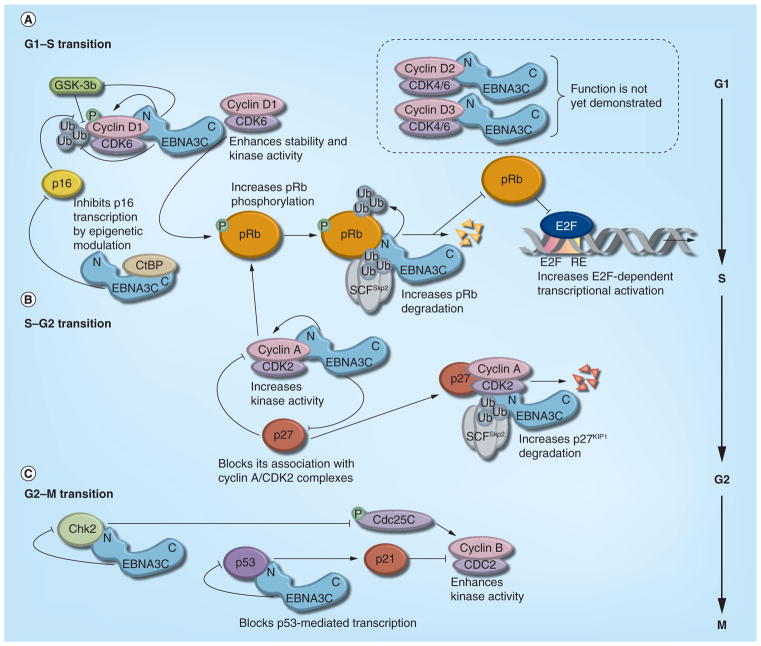
Figure 2. EBNA-3C accelerates cell cycle activities by overriding multiple checkpoints.
(A) EBNA-3C forms a complex with cyclin D1/CDK6 and enhances its stability through inhibiting both polyubiquitination and GSK3β-mediated phosphorylation. EBNA-3C further enhances the kinase activity of the the cyclin D1/CDK6 complex and recruits its activity to facilitate the ubiquitination and subsequent degradation of the hyperphosphorylated form of pRb, which, in turn, releases E2F transcription factor from an inhibitory constraint and enables the expression of genes required for G1–S-phase transition. In addition, EBNA-3C coupled with CtBP negatively regulates transcriptional expression of p16INK4A, which is a specific CDK inhibitor of the cyclin D1/CDK6 complex. EBNA-3C was also shown to form complexes with cyclin D2 and D3. However, whether EBNA-3C has any role in regulating their activity during the cell cycle is largely unknown. (B) EBNA-3C also forms a direct complex with cyclin A/CDK2, blocks p27KIP1-mediated suppression and subsequently increases its kinase activity to enhance pRb phosphorylation status. EBNA-3C recruits SCFskp2 E3 ligase activity to both pRb and p27KIP1 for facilitating their ubiquitin–proteasome-mediated degradation. Altogether, these activities contribute to both the G1–S and S–G2 transition of the cell cycle. (C) By regulating both Chk2 and p53-mediated activities, EBNA-3C indirectly enhances kinase activity of the cyclin B1/CDC2 complex. EBNA-3C blocks p53-dependent transcriptional activation of p21CIP1, which negatively regulates cyclin B1 and activity. EBNA-3C directly interacts with Chk2, which results in the inactivation of Cdc25c through phosphorylation at S216 sequestration in the cytoplasm. The resulting effect leads to the kinase activation of cyclin B/CDC2 and subsequent cell cycle progression through the G2/M stage.
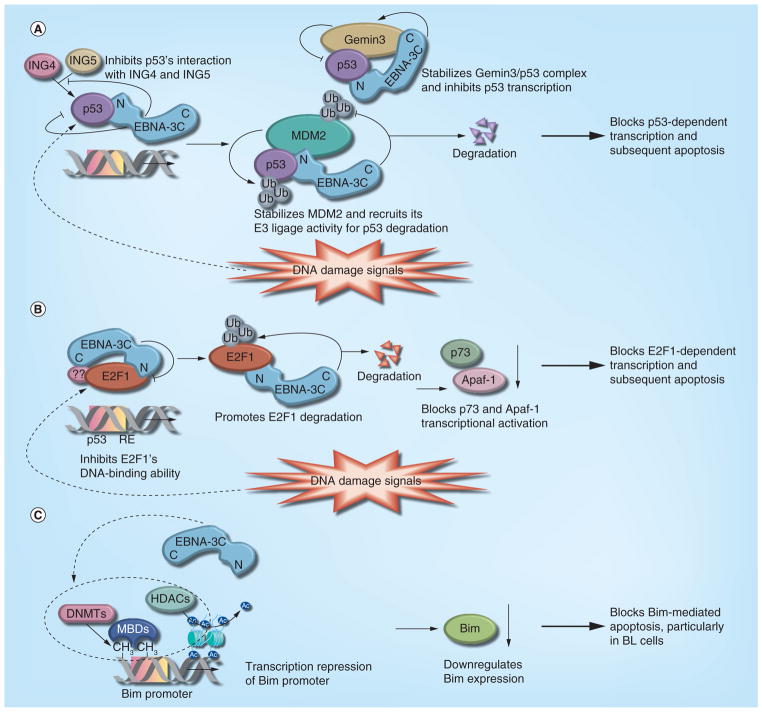
Figure 3. EBNA-3C blocks both p53- and E2F1-mediated apoptotic cell death.
Schematic representations of EBNA-3C-mediated p53- and E2F1-mediated transcriptional as well as apoptotic regulation. (A) In response to genotoxic stress, p53 achieves its antiproliferative properties through its action as a DNA-binding transcriptional activator to induce numerous downstream target genes involved in cell cycle arrest and apoptosis. EBNA-3C potentially inhibits p53-mediated transcriptional activity via forming a stable complex with p53, which subsequently blocks its DNA-binding ability. In response to genotoxic stress, both ING4 and ING5, members of the inhibitor of growth family of proteins, trigger a p53-mediated antiproliferative effect. EBNA-3C potentially inhibits both ING4- and ING5-mediated p53 transcriptional activity via displacing the interaction between p53 and ING4 or ING5. EBNA-3C forms a ternary complex with MDM2 and p53, and recruits MDM2 E3 ligase activity towards p53 to enhance its proteolysis. In addition, EBNA-3C stabilizes Gemin3 and enhances a stable complex formation between Gemin3 and p53, which further inhibits p53-mediated transcriptional activity and cell apoptosis. (B) In response to DNA damage signals, E2F1 is stabilized and transcriptionally activates proapoptotic genes p73 and Apaf-1, which eventually induces cellular apoptosis. By forming a stable complex with E2F1, EBNA-3C inhibits its DNA-binding activity and transcriptional activation of both p73 and Apaf-1. In addition, EBNA-3C specifically targets E2F1 for a ubiquitin–proteasome-mediated degradation, which altogether delays apoptotic response in EBV-transformed B cells. (C) Particularly in BL development, EBNA-3C plays a critical role by regulating expression of Bim tumor-suppressor protein through epigenetic modification. However, the precise mechanism is not clear yet.
BL: Burkitt’s lymphoma; DNMT: DNA methyltransferase; HDAC: Histone deacetylase; MBD: Methyl-binding protein; RE: Response element.
EBNA-3C as a transcriptional activator
Although genetic analysis clearly showed a critical role for EBNA-3C in EBV-induced growth transformation of primary B lymphocytes [26,27], there is very little previous evidence to explain the biochemical function of this protein, particularly in regulating gene transcription. The functional property of EBNA-3C was first described as a result of experiments designed to study the effects of stable latent gene expression on the EBV-negative B lymphocytes surface protein expression pattern [15,16,34]. EBNA-3C was shown to upregulate CD21 expression, which functions as an EBV receptor, on the surface of BJAB cells, an EBV-negative BL cell line [34,35]. The effects of EBNA-3C on CD21 expression levels suggested that EBNA-3C may function as a transcriptional activator and a putative activation domain within EBNA-3C (residues 724–826) was subsequently identified in Gal4-fusion-protein assays [35,36]. Primary amino acid sequence analysis of EBNA-3C further revealed that the protein contains a number of domains commonly found in transcription factors. These include a proline-rich and glutamine–proline-rich domain and a leucine zipper motif [15,16]. Interestingly, even today the precise mechanism by which EBNA-3C upregulates CD21 expression levels remains elusive, as EBNA-3C was not able to transactivate the CD21 promoter directly in reporter gene assays [36]. However, a number of studies have since documented increased gene expression or enhanced transcription of certain genes, including both viral and cellular, in response to EBNA-3C expression. For example, stable ectopic expression of EBNA-3C in the EBNA-3C-deleted EBV-positive Raji cell line, significantly increases expression levels of the viral LMP-1 protein, cellular cytoskeletal protein vimentin and the B-cell activation antigen CD23 gene expressions [37,38]. EBNA-3C expression alone also transactivates the LMP-1 promoter, where it significantly accelerates EBNA-2-mediated LMP-1 promoter transcriptional activation, signifying EBNA-3C as a potent transcriptional activator [37]. Later, the ability of EBNA-3C-dependent transcriptional activation of LMP-1 promoter has been shown to be mediated through the binding site for the Spi-1/Spi-B (PU.1) cellular DNA-binding protein and interactions with EBNA-3C [39]. Supporting the role of EBNA-3C as a transcriptional activator, the histone acetyltrans-ferase (HAT) coactivator p300 has also been shown to associate with EBNA-3C, along with the proliferation-associated protein prothymo-sin-α [40–42], suggesting that EBNA-3C may mediate its transcriptional activation effects through chromatin modification. EBNA-3C interacts with prothymosin-α through amino acid residues 366–400, and with p300 through its amino- and carboxy-termini [40]. A trans-activation domain rich in glutamine and pro-line residues that has similarities to the cellular transcription factor Sp1, has been mapped to EBNA-3C residues 724–826 in Gal4 activation assays [15,16,43]. It is important to note that apart from a weak transcriptional activation of LMP-1 promoter [37,39], no other direct effects of EBNA-3C on both viral and cellular promoters have been clearly documented. The precise mechanism(s) through which EBNA-3C potentially regulates gene transcription is not yet fully understood concerning whether it has a direct or indirect impact on a particular gene. In fact, after much effort over the last two decades, researchers have clearly documented that much of this transcriptional regulation by EBNA-3C is indirect rather than direct. It is, therefore, extremely important to identify cellular factors that are specifically modulated by EBNA-3C. Through utilization of a vast range of proteomics and transcriptomics analyses, as well as genetically engineered recombinant viruses lacking specific domains of EBNA-3C, it was shown that direct or indirect protein–protein interactions include an array of cotran-scriptional factors and most importantly post-translational as well as post-transcriptional modifications, largely involved in EBNA-3C-mediated transcriptional activities, which eventually regulates multiple cellular pathways. EBNA-3C interacting proteins and their subsequent effect on biological functions, particularly in B-cell transformation, are summarized in Figure 1 and Table 2.
Table 2.
EBNA-3C-interacting cellular partners and their biological consequences.
| Interacting cellular proteins | Experimental method(s) | Functional deregulation | Ref. |
|---|---|---|---|
| RBP-Jκ (CBF-1) | Coimmunoprecipitation using both EBNA-3C-stable as well as EBV-transformed cells | Competes with EBNA-2 for the interaction with RBP-Jκ and subsequently downregulates EBNA-2-mediated transactivation of the EBV Cp promoter for EBNA expression. The interaction has been shown to be responsible for in vitro B-cell transformation and EBNA-3C-mediated growth maintenance of LCLs | [47,154] |
| p53 | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Blocks p53-dependent transcriptional activation and subsequent apoptotic induction | [98] |
| E2F1 | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Inhibits its DNA-binding ability and, as a result, blocks E2F1 transcriptional activity, as well as apoptotic induction, in response to DNA damage. In addition, EBNA-3C enhances E2F1 degradation by recruiting ubiquitin–proteasome machinery | [97] |
| Cyclin A | Yeast two-hybrid, in vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Enhances kinase activity of cyclin A/CDK2 complex and rescues p27-mediated inhibition of cyclin A/CDK2 kinase activity by decreasing the molecular association between cyclin A and p27 in EBV-transformed cells | [95,96] |
| Cyclin E | In vitro using GST-fused protein incubated with radiolabeled EBNA-3C | No direct correlation has been depicted. It is speculated that EBNA-3C also regulates cyclin E/CDK2 activity during S phase | [95] |
| Cyclin D1 | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Stabilizes via blocking polyubiquitination levels, increases kinase activity of cyclin D1/CDK6 complex, thereby increasing the phosphorylation status of pRb tumor-suppressor protein, which, in turn, facilitates its ubiquitin–proteasome-dependent degradation. EBNA-3C interaction with cyclin D1 also results in nuclear accumulation of cyclin D1 | [90,95] |
| Cyclin D2 | Coimmunoprecipitation using ectopically expressed cells | No functional correlation has been established | [90] |
| Cyclin D3 | Coimmunoprecipitation using ectopically expressed cells | No functional correlation has been established | [90] |
| MDM2 | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Enhances MDM2 stabilization through blocking its ubiquitin–proteasome-mediated proteolysis or recruiting deubiquitination activity. EBNA-3C also recruits MDM2 E3 ligase activity towards p53 for facilitating its degradation | [31] |
| pRb | In vitro using GST-fused proteins and coimmunoprecipitation using ectopically expressed cells | Is destabilized in an ubiquitin–proteasome-dependent pathway by recruiting SCFSkp2 E3 ligase activity | [87] |
| Chk2 | In vitro using GST-fused proteins and coimmunoprecipitation using ectopically expressed cells | Releases G2/M cell cycle blockage induced by nocodazole. The interaction results in a predominant phosphorylation of residue and subsequently sequesters in the Cdc25c at S216 cytoplasm through interaction with 14-3-3 | [113] |
| GADD34 | Yeast two-hybrid and coimmunoprecipitation using ectopically expressed cells | Promotes eIF2-α and, at the same phosphorylation at S51 time, blocks XBP1 activation and ATF6 cleavage of unfolded protein response components | [155] |
| Spi-1/Spi-B | In vitro GST pulldown using radiolabeled EBNA-3C protein | Recruits Spi-1/Spi-B transcription factors to enhance EBNA-2-mediated LMP-1 promoter activation in a RBP-Jκ-independent manner | [39] |
| SCFSkp2 molecules | In vitro using GST-fused protein incubated with radiolabeled EBNA-3C proteins and coimmunoprecipitation using ectopically expressed cells | Recruits SCFSkp2 E3 ligase activity towards many tumor-suppressor proteins, including p27KIP1 and pRb to enhance their ubiquitin–proteasome-dependent proteolysis | [93] |
| c-Myc | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Increases its stability as well as c-Myc-mediated transcriptional activation | [92] |
| Prothymosin-α | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Through interaction with prothymosin-α, EBNA-3C recruits p300 HAT activity, as well as other basal transcription factors, to modulate gene transcription | [40,156] |
| CtBP | Coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | With the interaction with CtBP, EBNA-3C aids in chromatin remodeling and epigenetic suppression of p16INK4A gene expression | [17,54] |
| NM23-H1 | Yeast two-hybrid, in vitro GST pulldown and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Enhances nuclear localization, modulates its transcriptional activity as well as cell migration ability. EBNA-3C induces metastasis in a nude mouse model in cooperation with Nm23-H1 | [61,62,157] |
| Gemin3/DP103 | Yeast two-hybrid, in vitro using GST-fused proteins and coimmunoprecipitation using ectopically expressed as well as EBV-transformed cells | Stabilizes Gemin3 and promotes a complex formation between Gemin3 and p53, which, in turn, blocks p53-mediated transcriptional activation as well as apoptosis | [134,137] |
| p300 | In vitro using GST-fused proteins incubated with radiolabeled EBNA-3C proteins | Regulates p300- and prothymosin-α-mediated acetylation of histone molecules in the nucleosomes and results in transcriptional activation. However, a precise functional consequence has not been demonstrated | [40,156] |
| HDAC1 | In vitro using GST-fused proteins and coimmunoprecipitation using ectopically expressed cells | HDAC1 was shown to play a crucial role in EBNA-3C-mediated transcription repression. However, a direct biological phenomenon has not yet been described | [41,42] |
| HDAC2 | GST pulldown and coimmunoprecipiation experiments using stable cells as well as LCLs | It has been suggested that prothymosin-α plays an important role in regulating the association between EBNA-3C and HDAC molecules, as well as corepressor complexes including mSinA and NCoR | [42] |
| mSin3A | GST pulldown and coimmunoprecipiation experiments using stable cells as well as LCLs | Forms a transcriptional repressor complex. However, a direct functional correlation has not been described yet | [42] |
| NCoR | GST pulldown and coimmunoprecipiation experiments using stable cells as well as LCLs | Like mSin3A, NCoR presents in the same transcriptional repressor complex and a direct functional correlation has not been found | [42] |
| ING4 | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | EBNA-3C blocks its interaction with p53 and thereby inhibits p53-mediated transcriptional as well as apoptotic activities | [99] |
| ING5 | In vitro using GST-fused proteins and coimmunoprecipitation using both ectopically expressed as well as EBV-transformed cells | Similar to ING4, EBNA-3C inhibits its interaction with p53, and as a result, blocks p53-mediated transcriptional as well as apoptotic activation | [99] |
| GSK-3β | Coimmunoprecipitation using ectopically expressed cells | Inhibits phosphorylation of cyclin D1 at T286 and subsequently accelerates nuclear accumulation of cyclin D1, which contributes to its increased stability | [90] |
| SUMO-1/3 | Yeast two-hybrid, GST pulldown and coimmunoprecipitation using ectopically expressed cells | Recruits SUMO-1 and -3 activities for LMP-1 promoter activation coupled with EBNA-2 | [37,158] |
| MRS18–2 | Yeast two-hybrid, GST pulldown and coimmunoprecipitation using LCLs | Interacts with and promotes nuclear localization, resulting in the disruption of pRb–E2F1 complexes | [88] |
GST: Glutathione S-transferase; HAT: Histone acetyltransferase; HDAC: Histone deacetyltransferase; LCL: Lymphoblastoid cell line.
EBNA-3C as a potent transcriptional repressor
The most important transcriptional regulation by EBNA-3C was first documented after identification of RBP-Jκ (or CBF-1) as an intermediate cellular DNA-binding protein involved in EBNA-2-mediated transactivation to its targeted promoters [17,23]. EBNA-2 stimulates transcription from Cp latency promoter to synthesize all EBNA-encoding messages, and activates the LMP promoters and the promoters of the cellular genes CD23, CD21, c-fgr and c-myc [44,45]. As similar to EBNA-3C, EBNA-2 was also shown to be unable to interact with DNA directly and so regulates its responsive promoters through associations with multiple cellular transcription factors, including RBP-Jκ and PU.1 (Spi-1/Spi-B) [39,46,47]. Soon after the discovery of RBP-Jκ as an EBNA-2-associated cotranscriptional regulator, it was also shown to interact with EBNA-3C, as well as with other EBNA-3 proteins both in vivo and in vitro [22,48]. Interestingly, the interaction between EBNA-3C and RBP-Jκ seems to block interaction of RBP-Jκ to either its responsive DNA element or EBNA-2, suggesting that EBNA-3C acts as a functional antagonist of EBNA-2 and forms a negative regulatory feedback loop to finely tune latent gene expression in EBV-transformed cells [27,47]. In addition to its function as a passive repressor of EBNA-2-mediated transcriptional activation, EBNA-3C was also shown to act as an active repressor [15,16]. The repression domains of EBNA-3C was subsequently mapped to a region rich in acidic and proline amino acids (residues 280–525), which is referred to as ‘strong repression domain’, and another relatively weaker repression domain was found to be located at the C-terminal region, rich in proline and glutamine amino acids (residues 580–992) [15,16]. Collectively, these studies have clearly established a dual role in EBNA-3C-mediated activities in order to modulate gene transcription by acting as both a transactivator as well as a transrepressor. The exact molecular mechanisms that direct the switch between EBNA-3C-regulated gene activation and repression are not fully known yet, but may include several post-translational and post-transcriptional modifications or recruitment of different transcriptional cofactors, such as RBP-Jκ, a downstream effector in the Notch signaling pathway. Fascinatingly, it has been shown that the interaction between EBNA-2 and RBP-Jκ appears to be functionally analogous as compared with the binding of the intracellular domains of activated Notch receptors (Notch-IC) with RBP-Jκ [45]. Binding of Notch receptors to RBP-Jκ mitigates RBP-Jκ-mediated repression of its targeted downstream promoters to activate gene transcription involved in myoblast differentiation [49]. In EBV-transformed cells, EBNA-2 substitutes for the Notch1 intracellular domain and blocks its mediated differentiation process, which is negatively controlled further by interaction between RBP-Jκ and EBNA-3 family proteins [45]. The EBNA-3 proteins interact with RBP-Jκ via residues in their amino termini, as demonstrated by a series of truncation experiments from several different groups [50]. Although the precise amino acids sequence required for the strongest interactions remains controversial, most studies agree that the binding region lies within the amino terminus homology domain of these proteins [50].
Role of EBNA-3C in chromatin remodeling as a mechanism of transcriptional repression
Consistent with its function as a potent transcriptional repressor, work from our laboratory clearly demonstrated that EBNA-3C forms stable complexes with several transcriptional core-pressors and chromatin modification enzymes both in cells and in vitro [40,42]. The human genome is packaged as chromatin, which contains dsDNA wrapped with core histones [51,52]. Covalent post-translational modifications of these histones play a vital role in regulating gene transcription through varying chromatin structure [51,52]. Acetylation of core histones by HATs leads to transcriptional activation by loosening the chromatin and nucleosomal structure [53]. By contrast, deacetylation of core histone molecules by histone deacetylases (HDACs), leads to chromatin condensation and thereby transcriptional silencing [53]. In addition, modified histones also can recruit multiple transcription factors and chromatin remodeling complexes to control transcriptional activity [52]. Initial studies demonstrated that EBNA-3C may recruit HDAC activities by a transcriptional repression mechanism, as a pan-HDAC inhibitor tricho-statin A inhibits EBNA-3C-mediated transcriptional repression from Cp latent promoter in transient transfection assays [41]. Subsequently, we have shown that EBNA-3C can form stable complexes with multiple HDACs and HATs, in association with other transcriptional cofactors, including prothymosin-α, mSin3A and NCoR in EBNA-3C-expressing B cells [40,42]. Furthermore, the association of EBNA-3C with HATs, HDACs and corepressors in EBV-infected B cells strengthens the argument for a role of EBNA-3C in both transcriptional activation as well as repression, which is possibly dependent on certain stimuli. EBNA-3C forms a separate complex with acetylases and deacetylases in which prothymosin-α appears to play an important role in stabilization, as well as activation, of these complexes [40,42]. The signaling events that dictate EBNA-3C-mediated transcriptional activation or repression mechanism are still not clear. However, it is suggested that the associated functions are perhaps controlled by cell cycle events, as well as contributions from other cellular and viral antigens involved in regulating cell growth and proliferation. Nevertheless, additional studies focusing on these particular cellular events are essential to shed light on the distinctive roles of EBNA-3C and prothymosin-α complexes and their connections to regulate gene transcription.
Another protein, CtBP or C-terminal binding protein, was also identified as a cellular factor of EBNA-3C-mediated transcriptional repression mechanism [54]. CtBP was initially demonstrated as an interacting cellular partner with the C-terminal domain of adenovirus-encoded E1A oncoprotein that potentially represses E1A-mediated transformation and metastatic progression [55]. Although the precise molecular mechanism by which CtBP blocks gene transcription is unknown, it has been demonstrated that CtBP alters chromatin structure through recruiting HDACs-1, -4, -5 and -7, as well as the transcriptional repressor Sin3A [54]. CtBP interacts with these various cellular transcription factors, including E1A, through a conserved Pro–X–Asp–Leu–Ser (PLDLS) interaction domain [55]. Similarly, EBNA-3C has also been shown to interact with CtBP via this PLDLS motif that was found to be located at amino acids 728–732, a region previously shown to possess transcriptional repression activity in GAL4 fusion protein assays (within residues 580–992) [54]. Additionally, deletion of the PLDLS motif within this region was also shown to ablate EBNA-3C-mediated transcriptional repression [54]. These results further confirm the importance of the C-terminal domain of EBNA-3C in transcriptional repression. However, disruption of the CtBP interaction motif showed little or no effect on the ability of full-length EBNA-3C to repress transcription, suggesting that the N-terminal repression domain is much stronger as compared with the C-terminal repression domain [54]. Importantly, disruption of the PLDLS motif impaired the activity of EBNA-3C in transformation assays with Ha-Ras, indicating that the interaction with CtBP may have wider functional consequences [54]. Subsequently, two nonconsensus bipartite CtBP binding sites (ALDLS, residues 857–861 and VLDLS, residues 886–890) have been identified in the C-terminal region of EBNA-3A [17,56]. Similar to EBNA-3C, this region was shown to be required for EBNA-3A-mediated immortalization of rat embryonic fibroblasts in cooperation with Ha-Ras, as well as contributing to the transcriptional repression activity of EBNA-3A [56]. Overall, these studies signify the importance of CtBP during EBV-mediated B-cell transformation and, therefore, could offer a potential therapeutic lead for the development of future drug discovery against EBV-associated multiple B-cell lymphomas.
EBNA-3C regulates the metastatic potential of Nm23-H1 through alteration of its transcriptional activity
Nm23-H1 was discovered as the first anti-metastatic cellular factor more than 20 years ago [57–59]. Since then, a great deal of work has successfully contributed to the understanding of its association with the development of a number of human cancers and its role in various cellular signaling pathways, including cell proliferation and apoptosis in B cells [59,60]. The nm23 gene family encodes a closely related group of nucleoside diphosphate kinases, of which eight members (Nm23-H1–H8) have been identified so far in humans [59]. The nm23-h1 gene product Nm23-H1 is the best characterized member of this family of proteins [59]. A growing body of evidence suggests that altered Nm23-H1 expression, both at protein as well as transcript levels, are directly linked with cancer progression [58,61]. These results, however, are contradictory to each other. While an inverse association between Nm23-H1 expression and the metastatic potential was observed for breast, hepatocellular, colon, esophageal and ovarian cancer, a positive correlation has also been demonstrated for several other cancer types, including cervical and hematologic malignancies [61]. Moreover, a dual functional role for Nm23-H1 expression has been suggested during cancer development [58]. The primary tumors are coupled with elevated Nm23-H1 expression, whereas a drastic downregulation was observed during later stages of cancer development with aggressive metastatic potential [58]. The pathogenesis associated with Nm23-H1-mediated deregulation could, therefore, be more tumor-stage specific rather than simple metastatic suppression. A thorough investigation of the regulatory mechanisms that governs differential Nm23-H1 expression and subsequent outcomes would certainly enhance our current understanding of Nm23-H1-associated type-specific cancer development and future therapeutic strategies.
Along these lines, a number of recent studies have clearly suggested a critical role for Nm23-H1 in the suppression of many tumor virus-induced cell migration and subsequent cancer propagation apparently mediated by direct protein–protein interactions between Nm23-H1 and tumor virus-encoded essential antigens [58]. EBNA-3C is one of the best studied Nm23-H1 binding proteins [62]. We initially identified Nm23-H1 as a binding partner for EBNA-3C in a yeast two-hybrid screening using C-terminal domain (residues 366–992) of EBNA-3C as bait [62]. Subsequently, the binding site was found to be located within the glutamine- and proline-rich domains (residues 657–675) of EBNA-3C using in vitro studies [58,59]. The binding studies were further confirmed using transiently transfected cells, EBNA-3C expressing stable cell lines and in vitro EBV-transformed cells [16,58]. Interestingly, EBNA-3C was found to reverse Nm23-H1-mediated inhibition of cell migration when coexpressed with Nm23-H1 in a breast carcinoma cell line and an EBV-negative BL cell line, indicating that EBNA-3C may act to promote metastasis in EBV-positive tumors by modulating Nm23-H1 activities [16,58]. This interaction between Nm23-H1 and EBNA-3C was shown to result in an increase in transcriptional activity on a responsive promoter [16,58]. For example, Nm23-H1 tethered to DNA by a Gal4 DNA-binding domain can activate transcription from a basal promoter at relatively low levels, whereas in the presence of EBNA-3C expression, a substantial increase in transcriptional activity was observed [16,58]. This clearly suggests that Nm23-H1 possesses transcriptional regulatory activity. Later, follow-up studies from our group demonstrated that EBNA-3C coupled with Nm23-H1 regulates gene transcription from multiple promoters, including Cox-2, αv integrin and MMP-9 [16,58,59,61]. Interestingly, the presence of EBNA-3C mediates the subcellular localization of Nm23-H1 from a mostly cytoplasmic to a predominantly nuclear signal. These studies suggest that EBNA-3C reverses the antimigratory effects of Nm23-H1 in vitro, but increases its ability to activate multiple gene transcriptions, which ultimately helps in aberrant B-cell proliferation and metastasis.
Recent studies have provided further insight into the functional consequences of the interaction between Nm23-H1 and EBNA-3C. The significance of this interaction was determined in the nude mice model using cancer cells expressing EBNA-3C and Nm23-H1 [61]. These in vivo studies showed that EBNA-3C promoted the growth of transformed cells in the absence of immune surveillance [61]. However, their expressions have been shown to be less important at the later stage of tumor progression [61]. This is in agreement with our initial study in which we tested the expression levels of Nm23-H1 at both protein and transcript levels using EBV-negative and -positive cells and found no significant difference [62]. Typically, EBNA-3C is expressed in lymphomas associated with AIDS patients or immunocompromised post-transplant patients [1]. However, EBNA-3C has so far not been detected in other EBV-associated carcinomas, including NPC, gastric carcinomas and Hodgkin’s lymphomas [1]. Although these post-transplant tumors are reported to be highly invasive, it is difficult to determine whether this is a consequence of EBNA-3C expression or occurs as a result of the expression of other viral oncoproteins that have been also shown to promote tumor invasion and metastatic potential [58] through deregulation of many metastasis suppressor genes. The reason that EBNA-3C is required for B-cell transformation [1,5], is that effects due to EBNA-3C expression are likely manifested during early steps after infection in EBV-associated malignancies, including B-cell lymphomas prior to its downregulation at a later stage of latent infection. Moreover, identification of a subset of BL tumors that express EBNA-3 family proteins, but not EBNA-2 or LMPs, has clearly justified a critical role for these proteins in order to block apoptosis and subsequently promote tumorigenesis [16,63]. The effects of EBNA-3 proteins on the metastatic potential of these tumors are yet to be examined. In nude mice experiments, the expression of Nm23-H1 alone critically affected the growth of cancer cells and suppressed their metastatic potential [58,61]. This effect was rescued by the expression of EBNA-3C along with another EBV-encoded essential latent antigen, EBNA-1 [61]. However, the prometastatic potential of EBNA-3C was found to be higher when compared with EBNA-1, which triggered a dramatic immune response, as indicated by increased spleen size and development of ascites in nude mice [61]. This study was the first in vivo report that directly linked tumor virus-encoded antigens with metastasis and, at least in part, widens the range of potential drug targets. The underlying molecular mechanisms by which these viral oncoproteins function as prometastatic factors are still been investigated to improve targeted therapies against EBV-associated B-cell lymphomas.
Later, in our laboratory, an amino acid sequence Blast analysis of the EBNA-3C-interacting domain demonstrated a significant sequence homology to Necdin, a member of the MAGE family of proteins known to regulate multiple cellular processes, including cell cycle regulation and apoptosis [64]. The most significant structural feature of the MAGE proteins is in a large central MAGE homology domain flanked by variable amino- and carboxy-terminal domains [65]. The most critical biological consequences of Necdin are its negative impact on cell proliferation and its anti-angiogenic activity [65]. Necdin functions as a potent transcriptional repressor either through direct binding with DNA at guanosine clusters within the promoter region of target genes or through its interaction with other major transcription factors, including p53, E2F1 and Hif-1α [58,64,66]. It has also been reported that EBV-transformed B lymphocytes show a higher methylation status within the CpG islands of the Necdin promoter compared with primary lymphocytes [64]. This suggests that the EBV latent antigens may be involved in regulation of Necdin-mediated functions, possibly related to cell cycle regulation and apoptosis. Subsequently, our results showed that EBNA-3C, together with Nm23-H1, modulates the biological functions of Necdin in EBV-infected cells [64]. Our results clearly demonstrated that the Necdin expression level is significantly lower in EBV-positive cells than that in EBV-negative cells [64]. This effect may be the result of increased methylation and epigenetic silencing of the Necdin promoter. Whether or not EBNA-3C can recruit methyltransferase activities in downregulating Necdin expression is currently under investigation in our laboratory. Previous studies have shown that Necdin is predominantly localized to the cytoplasm and is translocated to the nucleus under certain physiological conditions to exert its transcriptional activity [64]. EBNA-3C, together with Nm23-H1, was further shown to affect the subcellular localization of Necdin [64]. Interestingly, both EBNA-3C and Nm23-H1 were able to rescue not only Necdin-mediated transcriptional repression, but also its growth suppression and anti-angiogenic effects on cancer cells [64]. This study suggests a novel role for Necdin in regulation of downstream cellular targets in development of virus-associated human cancers. Nevertheless, future studies need to be pursued to provide a better understanding of the effects of Necdin on the cell cycle regulatory machinery, as well as their modulation by EBV antigens during B-cell transformation.
EBNA-3C critically engages cell cycle regulators
A general overview of the mammalian cell cycle
The cell cycle is delicately controlled by complex mechanisms integrating many proteins to ensure correct cell division with duplication of the cellular genome into daughter cells. It is characterized by four major steps, which include DNA replication (S or synthesis phase), segregation of replicated chromosomes into two separate daughter cells (M or mitosis phase), G1 and G2, which both represent gaps in the cell cycle that occur between S and M phases. G1, S, G2 and M phases are the traditional subdivisions of a typical mammalian cell cycle [67,68]. However, cells in G1, prior to entry for DNA replication, exist as a resting state referred to as the G0 phase [67,68]. The transition from one cell cycle phase to another is critically regulated by four major families of proteins, such as cyclins, CDKs, CDK inhibitors (CDKIs) and pocket proteins [69]. So far, nine CDKs and 16 cyclins have been identified and a specific combination of a particular CDK with its regulatory cyclin molecule forming an active complex is required for cell cycle progression at each stage [67,68]. While CDKs are expressed throughout the cell cycle, cyclin levels fluctuate during the cell cycle as a result of coordinated synthesis and ubiquitin–proteosome-mediated degradation to ensure the correct temporal activation of each CDK [70]. The first cyclins to be expressed following mitogenic or growth factor stimulation are the D-type cyclins (D1, D2 and D3), which form active holoenzymes with CDK4 and CDK6 and allow cells to leave from G0 phase to enter into the G1 phase. Subsequently, CDK2 plays a crucial role in order to complete the G1 phase and initiate S phase. CDK2 is sequentially activated by the E-type cyclins (E1, E2 and E3) and A-type cyclins (A1, A2 and A3) during the G1–S-phase transition, as well as in the S-phase progression. In late G2 and early M, cyclin A forms complexes with CDK1/CDC2 to facilitate entry into M phase. Cyclin B is subsequently expressed in late S phase and G2, and through interaction with CDK1/CDC2 further regulates M phase [67,68,70,71].
Regulation of cell cycle activities
Cancer development critically depends on the subtle balance between cell proliferation and apoptosis-mediated cell death [68,71–73]. The integrity of CDKI-cyclins/CDK-Rb pocket proteins-E2F family cascade is thought to be a major determinant in regulating cell fate. Besides cyclin binding, CDK activity is additionally regulated by phosphorylation and dephosphorylation on conserved threonine and tyrosine residues [70,71]. The kinase activity of cyclin/CDK complexes can be further negatively controlled by CDKIs [70,74]. These are small proteins that fall into two distinct classes, the INK4 family and Cip/Kip family [74]. The INK4 family CDKIs, which include p15INK4b, p16INK4a, p18INK4c and p19INK4d, specifically inhibit CDK4/6-dependent kinase activity, particularly at G1 phase, whereas the Cip/Kip family CDKIs, such as p21WAF1/CIP1, p27KIP1 and p57KIP2 block CDK2/CDK4-mediated activities [74]. While p21WAF1/CIP1 and p27KIP1 act as both promoters and inhibitors of cyclin/CDK kinase activity, p21WAF1/CIP1 is the only CDKI that is capable of binding to all the cyclin/CDK complexes involved in cell cycle progression [74]. CDKIs are also regulated by internal and external signals. For example, the master regulator of apoptosis, p53, transcriptionally controls the gene expression of p21WAF1/CIP1 in response to DNA damage signals or withdrawal of growth factors, and TGF-β, a major tumor suppressor, plays a crucial role in regulating the expression and activation of both p15INK4b and p27KIP1, which ultimately results in arresting cell cycle at G1 phase [74].
In addition to CDKIs, pRb and the related ‘pocket’ proteins p107 and p130 are some of the major negative regulators in controlling mammalian cell cycle progression [75]. These pocket proteins, in their active hypophosphorylated state as found in quiescent cells (G0 phase), block cell cycle progression through interactions with the E2F family of transcription factors [76,77]. In response to mitogenic stimuli, active cyclin/CDK complexes phosphorylate pRb, resulting in dissociation from E2F that promotes E2F-mediated gene transcription involved in both DNA replication (i.e., DHFR, PCNA and Orc) and S-phase entry (i.e., cyclin E and cyclin A) [76,77]. It has been suggested that hypophosphorylated pRb blocks E2F transcriptional activity through recruitment of the repression complexes containing HDACs and the chromatin remodeling protein SWI/SNF to the E2F responsive promoters required for S-phase entry [76,77]. Cyclin/CDK complexes have been shown to inhibit the binding between pRb and these chromatin remodeling enzymes. Several lines of evidence suggest that, in addition to pRb, other pocket proteins, p130 and p107 are also actively engaged in regulation of G1 phase [75].
In order to ensure correct DNA replication, as well as to block cellular transformation processes, cells have intrinsic properties to control cell cycle progression at various ‘checkpoints’ or ‘restriction points’, which is defined as a point of no return before entering into either the S or M phases [78,79]. These checkpoints are largely controlled by the CDKIs and their upstream regulators [74,78,79]. Checkpoints are activated owing to either improper entry into S or M phases or if damage to either the replicated genome or mitotic spindle is detected. It is, therefore, conceivable that abrogation of these checkpoints can directly contribute to aberrant cell proliferation and, thus, cancer development. In response to DNA damage, p53 is stabilized and subsequently transcriptionally activates p21WAF1/CIP1, which, in turn, initiates cell cycle arrest at the G1–S-phase transition [80]. Although the molecular mechanism of DNA damage-induced checkpoint during S phase is not clearly elucidated, a number of recent studies have shown that ATM-mediated phosphorylation of NBS1 is essential to induce cell cycle arrest at S phase. In response to DNA damage during G2, cell cycle arrest occurs owing to temporal activation of ATM/ATR and downstream kinases, Chk1 and Chk2, in a p53-dependent or -independent manner [81,82]. The activated Chk2 phosphorylates and inactivates Cdc25C, which subsequently binds to 14-3-3 protein that sequesters Cdc25C in the cytoplasm, blocking the activation of cyclin B1/CDK1, and resulting in cell cycle arrest at G2/M checkpoint [83]. In response to DNA damage signals, p53 transcriptionally activates 14–3–3σ, which, in turn, accelerates the interaction between cyclin B and 14-3-3σ to facilitate nuclear exclusion [82]. p53 also enhances the dissociation of the CDK1–cyclin B1 complex through transcriptional induction of Gadd45 [82]. Spindle checkpoint is an evolutionarily conserved mitotic regulatory mechanism that delays anaphase until all chromosomes become aligned at the spindle equator in metaphase [84]. This checkpoint is essential for preventing inappropriate chromosome segregation and aneuploidy. Major proteins that are known to regulate spindle checkpoint include Mad1, Mad2, Bub1, BubR1, Bub3 and Mps1 [84,85].
Role of EBNA-3C in manipulating G1–S phase
Tumor viruses have developed numerous sophisticated strategies to ensure the continuous cell proliferation of latently infected cells [5]. Functional alterations of several important components in the regulatory circuit of the mammalian cell cycle, as described above, are most noticeable features in human tumor virus-mediated lymphomagenesis [5].
The initial clue demonstrating a possible link between EBNA-3C and deregulated cell cycle activity came from the study in which an EBNA-3C-deficient EBV-infected BL cell line shows a gradual decrease in LMP-1 expression level when cell cycle is arrested at G1 phase [34]. However, re-expression of EBNA-3C in these arrested cells promotes LMP-1 expression, as well as induces the hyperphosphorylated form of pRb, suggesting a potential role for EBNA-3C in regulating G1–S-phase transition [34]. In connection to these findings, subsequent studies demonstrate that EBNA-3C can disrupt normal cell cycle regulatory mechanisms. In general, tumor virus-encoded transforming antigens, such as SV40 large T antigen, adenovirus E1A or HPV E7, can readily deregulate cell cycle activities and assist the cellular transformation process through targeting major tumor-suppressor proteins [5]. Similarly, EBNA-3C has also been shown to function as a dominant EBV-encoded oncoprotein that can cooperate with activated Ha-Ras to transform rodent embryonic fibroblasts through blocking cyclin D-dependent kinase inhibitor p16INK4A-mediated growth suppressive activities [26,86]. Similarly to two other potent viral oncoproteins, HPV E7 and adenovirus E1A, EBNA-3C was also shown to interact with pRb in vitro and promote E2F-dependent transcriptional activities [86]. Collectively, these results suggest that EBNA-3C may be critically involved in disrupting the cyclin/CDK-pRb-E2F signaling pathway at the G1 restriction point to enhance cell cycle activities [86]. Later, our group among others have shown that EBNA-3C forms a stable complex with pRb [87,88]. However, unlike HPV E7 and adenovirus E1A, EBNA-3C does not have any activity on the other pocket proteins, p107 and p130 [87]. In this study, we also showed that EBNA-3C recruits SCFSkp2 E3-ubiquitin ligase activity to enhance pRb degradation [87]. On the contrary, another group showed that EBNA-3C accelerated the hyperphosphorylated inactive state of pRb [89]. In addition, we recently showed that EBNA-3C-mediated phosphorylation of pRb is an event prior to initiation of pRb proteolysis [90]. These observations altogether provide a direct molecular link of EBNA-3C-associated deregulation of cell cycle activities at the G1–S transition. Two integral components of SCFSkp2 ubiquitin ligase, Skp1 and Skp2, have been initially identified as S-phase kinase-associated proteins to be associated with the cyclin A/CDK2 complex in tumor cells [91]. Several lines of evidence have shown that this complex plays a central role in regulating the stability of many important cell cycle proteins including p27KIP1, E2F1 and c-Myc [91–94]. Interestingly, in subsequent studies, we have shown that EBNA-3C can regulate the stability of these proteins by modulating Skp2 function [93]. For example, EBNA-3C facilitates p27KIP1 degradation in an ubiquitin–proteasome-dependent manner [93]. This regulation appears to take place at the level of p27KIP1 binding to cyclin A/CDK2 complex, providing a potential mechanism by which EBNA-3C disrupts p27KIP1 inhibitory activity and eventually stimulates cyclin A-dependent kinase activity in order to support S-phase entry as well as progression through G2 phase [93]. As described earlier, cyclins/CDK complexes, including cyclin D1/CDK4/6, cyclin A/CDK2 and cyclin E/CDK2, promote cell cycle progression at G1–S/G2 phases via phosphorylation of pRb, thereby disrupting the pRb-E2F repressor complex [71]. Surprisingly, in our studies, EBNA-3C was indeed shown to interact with all of these cyclin/CDK complexes independently [90,95,96], providing an overall complex mechanistic pathway by which EBNA-3C targets the cell cycle repository to facilitate B-cell transformation. In an initial study from our laboratory, a yeast two-hybrid screen with the C-terminal domain (residues 890–992) of EBNA-3C as bait, identified cyclin A as a potential EBNA-3C-interacting partner, which was subsequently confirmed by both in vitro and in vivo studies using LCLs [96]. Additionally, we have shown that EBNA-3C expression led to a decrease in the association between cyclin A and its specific CDKI p27KIP1 [96]. In an attempt to better understand the functional relationship between EBNA-3C and cyclin A in the context of cell cycle deregulation, a predominant cyclin A binding motif was found to be located at the N-terminal region of EBNA-3C (residues 130–159), a stretch of amino acid sequence that is also conserved in other -3C homologous proteins in both Baboon and Rhesus lymphocryptoviruses [95]. Interestingly, this particular small region of EBNA-3C lies within the EBNA-3 homology domain located at residues 90–320, whereas experimentally, only EBNA-3B, but not EBNA-3A, showed moderate binding activity towards cyclin A [95]. The interaction between EBNA-3C residues 130–159 and cyclin A was shown to be at least partially dependent on the α1 helix of the conserved cyclin box, an important determinant for binding to p27KIP1 [95]. Moreover, this α–1 helix contains a hydrophobic amino acid patch that specifically serves as a platform to recruit substrates, including p107, E2F1, p27KIP1 and p21CIP1, for CDK2-mediated phosphorylation [95]. In a recent study using a recombinant EBV-expressing, conditionally active EBNA-3C, Maruo et al. further demonstrated the importance of this particular N-terminal region in maintaining LCL outgrowth [27]. In addition, several independent studies from our group also showed that this region of EBNA-3C specifically interacts with many important cellular proteins, such as the master regulator of apoptosis, p53, and its interacting proteins MDM2, ING4 and ING5, as well as E2F1 and c-Myc [31,92,97–99]. Therefore, this short stretch of EBNA-3C domain is of particular significance in deregulating cell proliferation of EBV-infected cells. Interestingly, earlier second-site recombination experiments have demonstrated that introduction of a stop codon at residue 365 of EBNA-3C abolished EBV mediated naive B-cell transformation in vitro, suggesting that expression of the N-terminal alone is not enough to mimic all functions of the wild-type EBNA-3C protein. Furthermore, in coinfected LCLs containing wild-type as well as recombinant N-terminal domain of EBNA-3C (residues 1–365), the recombinant EBNA-3C expression levels declined in continuously growing cultures, indicating that the N-terminal domain may act as a negative regulatory feedback to modulate EBNA-3C function, when necessary. In agreement with these findings our results also demonstrated that this N-terminal EBNA-3C domain negatively regulates cyclin A-dependent kinase activity [95], potentially providing a selective pressure to lose this region in recombinant LCLs by sequestering cyclin A into nonfunctional complexes.
Initial reports have suggested that immortalization of naive B lymphocytes by EBV is associated with the transcriptional activation of the cyclin D2 gene but not cyclin D1 [100,101]. However, a number of follow-up studies by different groups showed significant changes in cyclin D1 protein expression in multiple LCLs, as well as in EBV-positive SCID mice lymphomas [90,102,103]. In spite of initial studies regarding the cyclin D1 expression in EBV-positive B-lymphoma cells, it is clear from the above discussed facts that in order to deregulate the mammalian cell cycle at G1/S checkpoint, EBNA-3C critically manipulates the putative cyclin/CDK-pRb-E2F signaling pathway [27,87,95,96]. Additionally, in cyclin A-related studies we clearly demonstrated an in vitro complex formation between EBNA-3C residues 130–159 and cyclin D1 [95]. Nevertheless, this finding led us to investigate the molecular association between EBNA-3C and cyclin D1 complexes in cells to obtain a more in-depth understanding of EBNA-3C-mediated deregulation of cyclin D1 activity, which will lead to further understanding of the basic mechanism by which EBV regulates the mammalian cell cycle, particularly at the G1–S phase. Whether the association between EBNA-3C with different cyclin molecules is cell cycle dependent and how one gets replaced by another substrate, which may depend on a specific stage of the cell cycle, ultimately triggering aberrant cell proliferation in EBV-transformed cells, is currently under investigation. An initial study indicated that different cyclin D proteins may possess distinct biological activities at a specific step of B-cell differentiation, and their expression may be differentially influenced upon EBV infection [101]. In connection to this, our results also demonstrated that in vitro, EBV infection in naive B cells, as well as in EBV-positive BL cell lines, resulted in significant upregulation of all the D-type cyclins at the protein level [90]. On the contrary, initial attempts from different laboratories were not entirely intended to describe a possible oncogenic role for cyclin D1 during EBV-mediated B-lymphocyte transformation. In our study, we clearly demonstrated that EBNA-3C expression can lead to a significant upregulation of cyclin D1 protein level without affecting its transcription [90]. Cyclin D1 protein level was shown to be elevated in multiple cancer types without manipulating its genetic structure [104], indicating that increased stability of cyclin D1 protein could be a potential mechanism for its deregulated activities at G1–S-phase transition of the cell cycle. Cyclin D1 expression is strictly cell cycle dependent and its expression is affected by the ubiquitin-mediated proteolysis machinery as well as subcellular localization [105]. In normal cells, during G1–S-phase transition and in cancer cells for elevated cell cycle activities, cyclin D1 nuclear localization was accelerated via either decreased proteolysis in the cytoplasm or inhibition of GSK-3β-mediated phosphorylation function at T286 [106]. Our results portrayed a model where EBNA-3C plays a dual role in increasing nuclear localization of cyclin D1 by blocking its polyubiquitination level, as well as inhibiting GSK-3β-mediated kinase activity [90]. However, we cannot eliminate other possibilities by which EBNA-3C may also facilitate cyclin D1 nuclear import. Moreover, cyclin D1 coupled with its kinase partners CDK4 (or CDK6) plays a central role in the coordination of cell cycle progression at the G1–S transition by integrating the control of pRb phosphorylation with the transcriptional activity of E2F transcription factors [90,104]. As previously discussed, EBNA-3C increases the kinase activity of cyclin A/CDK2 complex and recruits an E3 ligase activity to enhance pRb proteolysis [87,96]. Similarly, in this report we have shown that EBNA-3C can also increase the kinase activity of cyclin D1/CDK6 complex, which acts as a prerequisite of pRb polyubiquitination and subsequent degradation [90]. Overall, our results pointed towards a general model in which EBNA-3C promotes pRb hyperphosphorylation and degradation by regulating the kinase activity of multiple kinase complexes, particularly by increasing the stability of cyclin D1 protein. Since elevation of cyclin D1 protein level is directly linked to malignancy and diagnostic index of multiple cancer types, the degradation mechanisms, particularly the ubiquitin–proteasome pathway, could be modulated as a potential therapeutic strategy. Along these lines, our data also suggested that cyclin D1 could be used as a legitimate drug target in numerous EBV-associated B-cell lymphomas.
EBNA-3C efficiently disrupts the block to the G2/M phase
A number of independent studies have clearly suggested that EBNA-3C not only engages in facilitating G1–S-phase transition, but is also capable of deregulating other cell cycle checkpoints [15,107,108]. To this end, the first experimental demonstration by Parker et al. showed that EBNA-3C-expressing osteosarcoma cells typically bypass the G2/M cell cycle block when treated with nocodazole, a potent micro-tubule destabilizing drug [107]. It is interesting to point out, that in addition to its role in promoting S-phase entry, cyclin A also plays an important role in controlling entry into mitosis [109,110]. For example, microinjection of cyclin A into Xenopus oocytes stimulates M-phase entry and microinjection of cyclin A-reactive antibodies into human cells initiates G2/M blockage [111,112]. Therefore, it is tempting to speculate that the ability of EBNA-3C to relieve repression of cyclin A/CDK activity [95,96] could also affect cell cycle progression into mitosis in addition to S-phase transition. Moreover, although the cyclin A immune complexes with which EBNA-3C was shown to interact contained CDK2 [96], it is possible that these experiments also isolated cyclin A/CDK1 complexes. The observed increases in CDK activity could, therefore, represent a combined effect on the G1/S kinase CDK2, and the G2/M kinase CDK1. While the mechanism for G2–M progression in cells transformed with EBV has not been well defined, studies have shown that EBNA-3C may also facilitate this stage of the cell cycle. For example, ectopic expression of EBNA-3C in mouse fibroblasts can bypass the mitotic spindle checkpoint activated by a microtubule-destabilizing drug and maintain continuous cell proliferation [24,107,113]. Along these lines, Wade and Allday demonstrated that EBV infection can lead to suppression of the G2/M checkpoint activated by genotoxic drugs [114], although a clear mechanism has not been elucidated. In order to further define a mechanistic pathway by which EBV compromises cell cycle checkpoints, we examined the ability of EBNA-3C to disrupt the genotoxin-induced G2/M checkpoint. This study provided new insights into EBNA-3C-mediated functions in regulating cell cycle activities at the G2/M checkpoint. Our results showed that EBNA-3C directly interacts with Chk2 [113], a downstream modulator of the ATM/ATR signaling pathway. The interaction between these two proteins subsequently led to predominant phosphorylation at S216 of Cdc25c, which triggered its sequestration in the cytoplasm through interaction with 14-3-3 and thereby allows the kinase activation of cylin B/Cdc2 complex and ultimately assists in bypassing the G2/M block [113]. This study is, therefore, the first demonstration of an essential mechanism by which EBNA-3C disrupts the G2/M checkpoint signaling to maintain continuous proliferation of EBV-transformed B cells [113]. In agreement with our finding, a recent study also showed that EBNA-3C expression is absolutely required to attenuate ATM–Chk2-mediated DNA damage responsive signaling for B-cell transformation [108]. A comprehensive scenario of how EBNA-3C manipulates numerous cell cycle components in order to deregulate multiple checkpoints of the mammalian cell cycle is shown in Figure 2.
EBNA-3C: a potent inhibitor of cellular apoptosis
Apoptosis: a major determinant of cellular transformation
Apoptosis, or programmed cell death, is a complex mechanism by which a cell regulates its own destruction to control the process of cell proliferation [72]. Apoptosis occurs normally during development and aging, and as a homeostatic mechanism to maintain cell populations in tissues [72]. Apoptosis also occurs as a defense mechanism, such as in response to DNA-damaging agents or microbial infection, including numerous viruses [115,116]. Although there are a wide variety of stimuli and cellular fate, both physiological and pathological, that can trigger apoptosis, not all cells will necessarily die in response to a specific stimulus. For example, the most salient feature of cancer cells is that they do not normally undergo apoptosis to maintain uncontrolled and continuous cell proliferation [117]. However, certain genotoxic drugs have been identified that can restore the normal apoptotic pathways and, therefore, have the potential for effective treatment of cancer development [73].
The diverse strategies used by viruses to modulate the apoptotic pathway are as varied as the viruses that use them. In addition, for each virus, these strategies are integrated into a wider scheme that manipulates other aspects of host defenses. For example, the persistent infection by tumor viruses can lead to immortalization of the infected cells through disruption of several cell cycle checkpoints coupled with inhibition of cellular apoptosis, which eventually causes oncogenesis [5,116]. In general, this is accomplished by functional inhibition or proteasomal degradation of many important tumor-suppressor proteins that are actively engaged in controlling aberrant cell proliferation or inducing multiple cellular death-signaling pathways such as apoptosis, by tumor virus encoded oncoproteins [5,116]. There is a great deal of evidence that several EBV-encoded essential gene products can efficiently suppress apoptosis [5]. The ability to regulate apoptotic signals appear to be most critical during viral pathogenesis, as apoptosis represents one of the major antiviral responses for removal of the infected cells without affecting the surrounding uninfected cells. EBV has, therefore, evolved multiple mechanisms to ensure the infected cells survive long enough for the virus to establish persistent latent infection and subsequently promote lymphomagenesis. Here, we will focus on reviewing recent studies regarding EBNA-3C-mediated suppression of apoptotic events, which collectively contribute to B-cell transformation (Figure 3).
EBNA-3C inhibits p53-mediated apoptotic activities
As noted in the previous section, tumor-suppressor proteins (e.g., p53) protect cells from malignant transformation by inducing either cell cycle arrest or apoptosis in response to persistent infection of multiple tumor viruses [5]. The p53 gene is mutated or deleted in half of all malignant tumors and the other half of human cancers express wild-type p53 protein, which is competent to induce apoptosis in malignant cells after genotoxic stress, thus offering a potential therapeutic opportunity applicable to a wide range of human tumors expressing wild-type p53 [118]. Not surprisingly, functional p53 or its downstream effectors are inactivated by various viral oncoproteins by releasing cells from cell cycle checkpoints or by protecting cells from p53-dependent apoptosis during oncogenesis [5]. While cell cycle arrest depends on the ability of p53 to induce the transcription of target genes such as the CDK inhibitor p21WAF1/CIP1, apoptosis depends on induction of a distinct class of target genes including bax, puma, perp and many others [119]. Several human tumor virus-encoded oncoproteins, including HBV X protein, HPV E6, Kaposi’s sarcoma-associated herpesvirus LANA and EBV-latent proteins, EBNA-1 and LMP-1, have been shown to inhibit p53 functions via multiple mechanisms [5,120,121]. In harmony with these tumor virus antigens, we have recently demonstrated that EBNA-3C has profound inhibitory effects on p53-mediated activities [98], which certainly provides an additional platform to increase the efficiency of current therapeutic strategies against EBV-associated lymphomas. Using both in vitro and in vivo binding experiments, we showed that EBNA-3C can physically interact with p53 via the same region, amino acid residues 130–190, which has been previously implicated for interaction with several other important cell cycle components, including SCFSkp2, pRb, c-Myc, cyclin A, cyclin E, cyclin D1 and RBP-Jκ [27,31,90,93,95,98]. The inclusion of p53 in this group further emphasizes the importance of this critical domain of EBNA-3C for bypassing the cell cycle checkpoints in EBV-infected cells. Recently, a genetic study using recombinant Bacmid EBV virus expressing conditionally active EBNA-3C, showed that deletion of this particular domain was not able to maintain cell proliferation of EBV-transformed LCLs [27], further indicating the importance of this domain in EBNA-3C. There are three major functional domains that have been identified in p53: an N-terminal transactivation domain (residues 1–80), followed by a central sequence-specific DNA-binding domain (residues 94–293) and a C-terminal oligomerization domain (residues 325–355) [122]. In addition to the oligomerization domain, the C-terminal domain contains two other small regions located at residues 290–325 and 356–393, respectively, implicated for negative regulation of its DNA-binding ability through several post-translational modifications, including phosphorylation and acetylation [122,123]. EBNA-3C was shown to interact with p53 at its two regulatory regions, the core DNA-binding domain and the C-terminal oligomerization domain [98], suggesting that EBNA-3C may regulate p53-mediated transcriptional activity and its DNA-binding ability. Indeed, our results showed that EBNA-3C significantly blocks p53 transcriptional activity by inhibiting its DNA-binding ability [98]. Since previous studies have shown that the C-terminal regulatory domain negatively regulates DNA-binding ability of p53 [123], it is probable that the inactivation of p53 activity by EBNA-3C is owing to the formation of a stable complex between the C-terminal negative regulatory domain of p53 and EBNA-3C.
Several chromatin-modifying enzymes, such as CREB-binding protein CBP/p300 and its associated factor PCAF, have been shown to interact with p53 and increase p53 transcriptional activity through acetylation at multiple lysine residues located at C-terminal domain [123,124]. By contrast, deacetylation of p53 by the HDAC1-containing complex has led to transcriptional repression [124]. Moreover, MDM2, a major negative regulator of p53-mediated activities, has also been shown to recruit HDAC1 activity, which subsequently increases ubiquitin–proteasome-mediated degradation of p53 [125,126]. The previously documented interaction of EBNA-3C with p300 and HDAC1 [40–42] led us to believe that EBNA-3C may manipulate p53 transcriptional activity through regulation of its acetylation status. Our data also indicated that ectopic expression of EBNA-3C was able to abrogate p53-mediated apoptosis in a p53-null cell line, Saos-2, perhaps through regulation of its transcriptional activity [98]. The interaction between EBNA-3C and p53 warrants further investigation as p53 controls cell cycle activities by regulating the genetic integrity, and repression of p53 function by EBNA-3C can result in increased genetic instability contributing to EBV-mediated cellular transformation.
EBNA-3C blocks ING protein-mediated p53 activities
The ING family of gene products, described as type II tumor-suppressor proteins, have been found to be rarely mutated at the genetic level in human cancer. However, at the protein level, their expression is often significantly reduced in various cancers [127]. Interestingly, a number of functional analyses of the ING family of proteins, particularly ING4 and ING5, provided a solid molecular link to p53-mediated regulation of apoptotis [128].
In a recent study, we showed that EBNA-3C can form a p53-independent stable complex with both ING4 and ING5 in EBV-transformed LCLs [99]. However, functional mapping of the binding region of EBNA-3C demonstrated that it associates with both ING4 and ING5 through amino acid residues 129–200 at the amino terminal domain [99], previously shown to interact with p53 [98]. This suggests that ING4 or ING5 may control formation of the complex between EBNA-3C and p53. A possible transient ternary complex formation has also been demonstrated with EBNA-3C, p53 and ING proteins [99]. However, the interaction between EBNA-3C with ING molecules is significantly blocked in the presence of p53 expression, suggesting that p53 hinders the binding of EBNA-3C to ING proteins, possibly through sharing a common binding motif with ING proteins [99].
In fact, competitive binding experiments and domain analysis through deletion mapping demonstrated that both EBNA-3C and p53 are bound to the identical amino acid sequence of both ING proteins [99]. ING4 and ING5 consist of several highly conserved domains, which include a leucine zipper-like motif, two NLS domains (NLS1 and NLS2) and a C-terminal plant homeo-domain (PHD) [129]. The PHD motif is a zinc finger domain that binds to histone H3 in a methylation-sensitive fashion and is a key structural component of any ING proteins [129]. This domain is also essential for ING protein-mediated chromatin remodeling through recruitment of HAT- and HDAC-associated complexes [129]. Similarly to many other ING proteins, ING5 has also been shown to interact with different chromatin remodeling factors, such as MOZ/MORF and HBO1 HAT complexes, via its PHD zinc finger domain [129]. The interaction between the conserved PHD domain of ING5 with both EBNA-3C and p53 not only corroborates the importance of this domain, but also raised the possibility of modulating ING5 functions by EBNA-3C and p53. Unlike ING5, ING4 binds to both EBNA-3C and p53 through the bipartite NLS1 domain, which was previously shown to interact with p53 by a different study group [128]. As observed from our binding experiments, these ING proteins can be deregulated via different mechanisms in response to EBV infection, although they share a high degree of structural and functional similarity. However, one mechanism may be that by blocking the interaction interface between p53 and either ING4 or ING5, EBNA-3C can interfere with both ING4- and ING5-prompted p53-mediated deregulation. Furthermore, we demonstrated that EBNA-3C efficiently rescues both ING4- and ING5-mediated activation of p53 functions, as evident in reporter and apop-tosis assays [99]. Both these ING proteins have been shown to positively regulate p53 activity, perhaps by enhancing acetylation of p53 via recruitiment of p300 HAT activity, although the precise mechanism remains elusive [99]. It is possible that the interaction of EBNA-3C with p300 and HDAC1 [40–42] may result in tempering the acetylation status of both p53 and ING proteins, which eventually define their functions in regulating cell growth. Overall, our findings suggest that these ING proteins are potential targets of EBNA-3C and provide us with further insights into understanding the possible mechanisms by which EBV-mediated transformation is associated with human cancers. Understanding the role of EBNA-3C in ING protein deregulation in EBV-infected cells provides clues into the mode of inactivation by ING proteins, and consequently the possibility that restoration of ING protein expression or functions may have therapeutic value.
EBNA-3C recruits MDM2 E3 ligase activity for enhancing degradation of p53
Ubiquitin–proteasome-dependent proteolysis is one of the fundamental mechanisms for regulating the activity of many cellular proteins involved in cell proliferation and apoptosis, and therefore disruption of this pathway has profound consequences for human cancer development [130]. The targeted degradation of p53 by one of its negative regulators, MDM2, represents a critical circuit in the regulation of p53-mediated tumor-suppressive functions [125]. It has been shown that growth of in vitro EBV-transformed LCLs are sensitive to Nutlin-3a-mediated growth suppression [131]. Nutlin-3a belongs to a small-molecule group of inhibitors that specifically inhibit p53–MDM2 complex formation, and subsequently increases p53 stability and mediated apoptosis in cancer cells carrying wild-type p53 gene [132]. In this line of evidence, we have recently shown that EBNA-3C recruits MDM2 E3-ubiquitin ligase activity for enhancing proteasome dependent proteolysis of p53 [31].
From previous reports, in order to deregulate the mammalian cell cycle, EBNA-3C manipulates the ubiquitin–proteasome machinery [87,92,93]. Noted previously, in contrast to destabilizing proteins, EBNA-3C is also critically involved in stabilizing at least two important cellular oncoproteins, cyclin D1 and c-Myc [90,92]. Our studies demonstrated that in addition to regulating self-ubiquitination, EBNA-3C can also efficiently block ubiquitination of MDM2 [31]. MDM2 protein expression level is overexpressed in many human cancers with wild-type p53, suggesting that it may contribute to tumor progression through inactivation of p53 functions [126,131]. In addition to deregulating p53 activity, MDM2 has also been shown to interact with and modulate the activity, stability and apoptotic function of many important cellular proteins, including pRb and E2F1 [31]. Our results further demonstrated that an essential N-terminal EBNA-3C domain (residues 130–190), previously known to interact with p53, physically interacts with MDM2 via its central acidic domain [31]. Interestingly, studies have shown that this central acidic domain plays an additional role besides the C-terminal ring-finger domain of MDM2 towards its E3 ubiquitin ligase activity [133]. The importance of this acidic domain in the regulation of MDM2-mediated p53 degradation has been extensively studied [133]. For example, binding of p14ARF and pRb to the MDM2 acidic region have been shown to increase p53 stability, while interaction with the chromatin remodeling protein, p300, stimulates MDM2-dependent p53 degradation [133]. Altogether these studies offer a possible mechanistic explanation of EBNA-3C-mediated deregulation of MDM2 stability and, thereby, p53 degradation. Our findings suggest that the multifaceted regulation of p53-MDM2 functions by EBNA-3C serves to enhance the efficiency of EBV-mediated lymphomagenesis [31]. This study is undoubtedly a significant step towards our understanding of the importance of EBNA-3C in development of EBV-associated human lymphomas and to design effective therapies targeting the p53–MDM2 complex.
EBNA-3C stabilizes Gemin3 to inhibit p53-dependent activities
Gemin3, also known as DDX-20 or DP103, was originally discovered from a yeast two-hybrid screen as a binding partner of both EBNA-2 and EBNA-3C [134]. Gemin3 belongs to the DEAD-box family of putative ATP-dependent RNA helicases. However, these proteins appear to be involved in the regulation of a large number of cellular processes including DNA repair, replication, RNA stability, translation initiation and transcription [135]. Soon after the discovery that Gemin3 is an interacting partner of EBV-encoded latent antigens, Gemin3 has subsequently been shown to interact with a number of transcriptional regulators, such as the orphan nuclear receptor SF-1, Egr2, mitogen Ets repressor and SMN [135]. The SMN complex plays an essential role in the synthesis of small nuclear ribonucleoproteins, pre-mRNA splicing, RNA transport, as well as regulation of gene transcription [136]. Interestingly, both EBNA-2 and EBNA-3C have also been shown to associate with the SMN complex [134]. While EBNA-2, in cooperation with Gemin3 and SMN, transcriptionally activates the LMP-1 promoter [134], owing to binding of EBNA-3C to either Gemin3 or SMN, no functional data have been documented. In this context, we have recently shown that EBNA-3C strongly interacts with and subsequently stabilizes Gemin3 expression in EBV-transformed cells [137]. In an attempt to explore the functional consequences as a result of EBNA-3C interaction with Gemin3, our results further demonstrated that Gemin3 forms a direct complex with p53 and this is critically involved in EBNA-3C-mediated inhibition of p53-dependent transcriptional activity and apoptosis [137]. In fact, inhibition of Gemin3 expression using a specific lentivirus construct partly abolished EBNA-3C-mediated inhibition of p53-induced apoptosis [137]. In accordance with our previous studies [31,98,99], this work further appends to the potential mechanism through which EBNA-3C regulates p53 functions.
Gemin3 has previously been depicted as a transcriptional repressor [135], although the mechanism of Gemin3-mediated transcriptional repression is not fully understood. The C-terminal nonconserved region of Gemin3 appears to mediate the interactions between Gemin3 and its target transcription factors, through which Gemin3 represses specific gene transcription [137]. For example, Gemin3 was shown to recruit HDAC activity as well as promote sumoylation, which eventually blocks Ets- and SF-1-mediated transcriptional activation [135]. Since both p53 and EBNA-3C functions have been shown to be extensively affected by these chromatin remodeling factors [5], it is tempting to speculate that the EBNA-3C/Gemin3 complex may also regulate p53-dependent transcriptions and apoptosis through modulation of its acetylation, as well as sumoylation status. A number of recent reports have shown that Gemin3 is a DEAD-box RNA helicase that plays an important role in cancer development. Thus, it would be fascinating to screen helicase inhibitors as a potential therapeutic strategy, possibly through the induction of p53-mediated apoptosis in the context of EBV-associated B-cell lymphomas.
EBNA-3C inhibits DNA damage-induced E2F1-mediated apoptosis
In response to DNA damage signals, E2F1 can induce apoptotic cell death in both a p53-dependent and -independent manner [138,139]. Since both genetic and functional inactivation of p53 has been detected in most human cancers, E2F1-regulated apoptosis certainly offers an additional tumor surveillance mechanism [139]. Thus, DNA damage-induced E2F1-mediated apoptosis can be targeted as a promising therapeutic strategy in controlling several cancers with the mutant p53 gene [140]. As previously discussed, EBNA-3C is critically involved in manipulating the functions of several upstream components of the E2F1 signaling pathway of the G1–S-phase transition of the cell cycle. Of these, the most striking observation was that EBNA-3C can physically interact with pRb and consequently enhance its ubiquitin–proteasome-mediated degradation to facilitate G1–S-phase transition, by relieving the negative regulatory pressure from E2F transcription factors [87]. Furthermore, along these lines, we recently examined whether EBNA-3C can directly regulate E2F1 functions to modulate both cell cycle and apoptotic activities in EBV-transformed B-lymphoma cells. We initiated our study with binding experiments that convincingly, as well as unexpectedly, showed that EBNA-3C forms a pRb-independent complex with E2F1, suggesting that EBNA-3C may regulate E2F1 activity through a different mechanism [97]. Subsequently, we utilized various truncated mutants of both proteins and showed that the N-terminal DNA-binding domain of E2F1 (residues 1–243) responsible for apoptotic induction, binds to two distinct regions of EBNA-3C, located at amino acid residues 100–200 and 621–700 [97]. Interestingly, while the EBNA-3C N-terminal binding sequence shows a direct interaction with E2F1, the interaction between the C-terminal residues of EBNA-3C and E2F1 appears to form an in vivo complex involving other unknown cellular proteins [97]. Importantly, these N-terminal (but not C-terminal) residues of EBNA-3C were shown to be particularly important in maintaining LCLs outgrowth [27], suggesting that EBNA-3C-mediated E2F1 regulation may exert a distinctly different cellular role in its contribution to cell immortalization.
A recent study using a genetically engineered EBV has shown that EBNA-3C attenuates the DNA damage response induced during EBV-mediated B-cell transformation [108]. In agreement with these data, our results in this study showed that EBNA-3C knockout virus is incapable of suppressing E2F1-mediated DNA damage response during the early stages of infection in naive B lymphocytes [97]. Our results persuasively showed that EBNA-3C represses E2F1-mediated transcriptional activity by blocking the E2F1-DNA-binding ability in latent infection using EBNA-3C knockdown LCLs, as confirmed by endogenous chromatin immunoprecipitation experiments [97]. Interestingly, in support of our results, a recent publication from a different group also demonstrated that E2F1 transcript level is upregulated in EBNA-3C knockout virus-infected B-lymphoma cell lines compared with wild-type virus-infected cell lines, using microarray analysis [24]. In the case of E2F3, it was shown to be genetically amplified in certain human cancers, although there was no clear documentation of an oncogenic role for the other ‘activators’ of the E2F family members, E2F1 and E2F2 [141]. In addition to its well-established function in controlling cell proliferation at the G1–S-phase transition, E2F1 is also capable of DNA damage-induced apoptosis through regulation of p73, Apaf-1 and caspase activities [138,140]. Our results demonstrated that, in response to a DNA damage signal, EBNA-3C blocks E2F1-mediated DNA binding and, as a result, its transcriptional activity that finally deactivates E2F1-regulated apoptosis by downregulating certain proapoptotic gene activities, including p73 and Apaf-1 [97].
DNA damage signaling events are clearly involved in the induction of E2F1 and its stabilization through several post-translational modifications [142]. However, the mechanism by which these modifications can lead to E2F1 stabilization remains unclear. E2F1 is regulated through a ubiquitin–proteasome pathway in a cell cycle-dependent manner, which relies upon its dissociation from its negative regulatory protein, pRb, as well as its association with specific E3 ligases, such as SCFSkp2 [97]. As previously described, EBNA-3C recruits SCFSkp2 activity for pRb degradation [87]; therefore, it is tempting to speculate that EBNA-3C may also regulate E2F1 degradation. Our results indeed showed that EBNA-3C enhances E2F1 proteolysis in a ubiquitin–proteasome-dependent manner [97], yet it is unclear whether EBNA-3C recruits SCFSkp2 activity exclusively, as several other ligases were also shown to be actively involved in E2F1 degradation [97]. Altogether, our results portray a plausible model in which EBNA-3C suppresses E2F1 transcriptional activity by both blocking its DNA-binding activity and promoting protein degradation [97]. However, apart from these activities, there could be other potential molecular mechanisms including several post-translational events, phosphorylation and acetylation that may control EBNA-3C-mediated inhibition of E2F1-regulated apoptosis in response to DNA damage signals. In response to DNA damage signals, ATM–Chk2-mediated phosphorylation of E2F1 and, consequently, increasing stability clearly provide us with a molecular link in understanding the critical role of E2F1-mediated cell cycle and apoptotic activities [142]. Importantly, as EBNA-3C was previously shown to bypass the DNA damage-induced ATM–Chk2 signaling cascade in order to establish B-cell transformation through overriding the G2/M block [108,113], it would thus be essential to investigate other plausible mechanisms in regulating E2F1-targeted apoptosis in the context of EBV-associated B-cell lymphomas. It is, as yet, unknown from our studies whether EBNA-3C affects the acetylation status of E2F1 or not, particularly in response to DNA damage. Further studies are required to fully elucidate the combinatorial effects of these different mechanisms by which EBNA-3C exerts a complete inhibition to E2F1-mediated apoptosis. Our data allow us to develop a simple model in which E2F1 can be targeted to promote apoptotic cell death in multiple EBV-linked B-cell lymphomas, regardless of p53 status in the presence of DNA damage signals and, thus, this can serve as a critical therapeutic strategy for targeted drug development.
EBNA-3C manipulates Bim-mediated apoptosis
Two apoptotic cell-death pathways exist in mammals: a cell-intrinsic pathway and a cell-extrinsic pathway. These distinct pathways converge with the activation of caspase cleavage [143]. Initiation of the cell-intrinsic pathway involves the activation of proapoptotic Bcl-2 family members [143]. The Bim tumor-suppressor protein, an essential member of this family, induces apoptosis during lymphocyte maturation or when overexpressed through the regulatory activities of several prosurvival members of the Bcl-2 family, as well as the proapoptotic family member Bax [144]. Bim has also been shown to play an important role in B-cell lymphomagenesis, as deletion of even a single Bim allele can drastically increase B-cell lymphoma development in Eμ-Myc transgenic mice with constitutive c-Myc expression within B cells [145,146]. Deregulation of c-Myc through chromosomal translocations is a characteristic feature of BL [147]. Bim-mediated apoptosis has been shown to be well connected with c-Myc activity, as when c-Myc is mutated or Bim is absent, this results in B-cell lymphomagenesis [146]. Recently, Anderton et al. showed that EBNA-3C along with EBNA-3A repress Bim expression and efficiently alter Bim-mediated proapoptotic signaling in BL cells [148]. This provides a potential mechanistic explanation by which EBV directly contributes to the development of BL, wheras EBNA-3C and/or EBNA-3A block Bim expression in the presence of wild-type c-Myc translocation. The authors noted significant protection from apoptosis in clones expressing the latency III program as compared with cells expressing latency I program [148]. To further analyze the role of the EBNA3 proteins in protection from apoptosis, EBV-negative BL cells were infected with recombinant EBV viruses containing a knockout of either the EBNA-3C or -3A genes. Results confirmed that cells infected with knockout viruses were unable to protect the cells from apoptotic death induced by either a microtubule polymerization inhibitor (nocodazole), a DNA-crosslinking inducer (cisplatin) or a CDK inhibitor (roscovitine) [148]. Moreover, Bim expression was significantly elevated in clones infected with either EBNA-3C or -3A knockout viruses compared with cells infected with wild-type virus or mutant virus lacking expression of other EBNA genes [148]. The regulation of Bim expression was found to be at the level of mRNA synthesis [148]. The authors speculate that both these viral oncoproteins, EBNA-3A and -3C, may allow a permissive B cell environment for c-Myc translocations to occur by preventing apoptosis of damaged cells via suppression of Bim, thus supporting the establishment of BL tumors [148]. Further investigations to delineate the molecular mechanism by which EBV infection can regulate Bim expression has revealed that epigenetic modifications in the regulatory region of Bim play a major role in determining Bim expression in EBV-infected B cells [23]. Epigenetic modifications can efficiently change chromatin structure without varying the DNA sequence and subsequently regulate gene expression in a heritable manner [149]. Two closely associated chromatin modification components have been established, which include hypermethylation of cytosine residues of CpG islands and, second, covalent modifications to the N-terminal tails of histones. These combined effects ultimately result in long-term silencing of many tumor-suppressor genes and provide another hallmark of cancer progression [149]. EBNA-3C has previously been shown to regulate or recruit many chromatin modification enzymes [5,16]. However, the roles of EBNA-3C and EBNA-3A in this process, particularly in modulating methylating status of CpG islands (a common mechanism for silencing transcriptional activation of many tumor-suppressor genes in most cancers), are not yet understood. Nevertheless, the authors propose a model in which EBV infection inhibits Bim transcription via a mechanism involving EBNA-3C- and EBNA-3A-mediated blocking of the efficient assembly of transcription complexes at or around the transcription initiation site, which can expose the large CpG island available for DNA methylation [23].
Future perspective
The broad conclusion that has emerged from a large number of recent studies from our group, as well as others, is that EBNA-3C has equipped itself with a set of sophisticated skills to promote aberrant cell proliferation of EBV-associated B lymphoma cells through inhibition of cellular apoptosis. Over the last decade, our laboratory has been actively involved in elucidating the mechanistic pathways by which EBNA-3C targets these two putative pathways linked to the development of human cancer. However, additional studies are required to develop a comprehensive model for the role of EBNA-3C in B-cell transformation. This will enable us to enhance the efficacy of the current therapeutic regimens against EBV-linked multiple B-cell lymphomas. Currently, large numbers of synthetic and natural compounds have been found that are pharmacologically successful in limiting multiple cancerous growths through induction of cellular apoptosis [150]. Thus these compounds have promise for the development of novel therapies against EBV-associated B-cell lymphomas based on the disruption of the apoptotic process. For now, the basic mechanisms of apoptotic deregulation in these lymphomas have been extensively explored, but its implications for therapeutic purposes must still be determined. A future challenge will be to exploit these mechanistic insights both to gain a better understanding of the biology of EBV infection and to develop novel therapies for the treatment of numerous virus-associated B-cell lymphomas.
Executive summary.
EBV: a ubiquitous lymphotropic virus
EBV is a ubiquitous human lymphotropic γ-herpesvirus. Based on serology, approximately 95% of the world’s adult population has been infected with EBV.
In most cases, EBV establishes a life-long asymptomatic infection without overt serious consequences. However, in some individuals, particularly patients with HIV infection or organ transplantation, the virus is implicated in the development of several malignancies.
EBV primarily infects B lymphocytes and the cells are subsequently growth transformed. These cells are largely nonpermissive for viral replication, but readily express a set of latent viral genes that are implicated in the development of several lymphoproliferative disorders.
From these latent transcripts, only four were shown to be engaged for in vitro B-cell immortalization and naive B cells to leukemic lymphoblasts in vivo.
EBNA-3C: an essential viral oncoprotein
Using a recombinant genetic engineering approach, EBNA-3C was found to be one of the essential latent antigens for B-cell transformation in vitro.
EBNA-3C was initially identified as a viral transcriptional modulator.
Besides regulating transcriptional activation of certain viral genes, EBNA-3C was later shown to be involved in disrupting normal cellular homeostasis by targeting many important signaling pathways.
EBNA-3C modulates multiple cellular machineries
EBNA-3C efficiently deregulates the two most important pathways in cancer development, the cell cycle and apoptosis – apparently mediated by direct protein–protein interaction, transcriptional deregulation and manipulation of protein stability through recruiting the ubiquitin–proteasome pathway.
EBNA-3C interacts with the metastasis suppressor Nm23-H1. The interaction triggers its nuclear localization, which, in turn, regulates its transcriptional activity to modulate the overall metastatic potential of Nm23-H1. EBNA-3C coupled with Nm23-H1 blocks the tumor-suppressor Necdin-mediated transcriptional repression and its antiangiogenic activities.
EBNA-3C accelerates cell cycle activities
EBNA-3C targets retinoblastoma protein and p27 tumor-suppressor proteins for degradation and thereby increases cell cycle activities.
EBNA-3C recruits MRS18-2 to disrupt the retinoblastoma protein–E2F1 complex for increasing cell cycle activities at G1/S phase.
EBNA-3C enhances kinase activities of cyclin/CDK complexes, namely cyclin A/CDK2 and cyclin D1/CDK6, to bypass the G1–S-phase transition.
EBNA-3C blocks cellular apoptosis
EBNA-3C suppresses p53-mediated apoptotic activities through several mechanisms. It involves direct binding as well as blocking the interaction between p53 and its positive regulatory proteins, ING4 and ING5, which thus limits its transcriptional activation. EBNA-3C recruits MDM2 E3 ligase activity to enhance its ubiquitin–proteasome-mediated degradation. In addition, by promoting a complex formation between Gemin3 and p53, EBNA-3C blocks p53 transcriptional as well as apoptotic activities.
In response to DNA-damaging stimuli, EBNA-3C blocks E2F1-mediated apoptotic activities by two distinct mechanisms, which include inhibition of its DNA-binding ability and increasing ubiquitin–proteasome-dependent degradation.
EBNA-3C was also shown to play a crucial role in chromatin remodeling to regulate functional consequences of many cell cycle and apoptotic components, such as Bim-mediated apoptosis.
Acknowledgments
We apologize to all our colleagues whose important work could not be directly cited. We would like to thank our previous laboratory members JS Knight, C Subramanian, N Sharma, M Murakami, T Chaudhuri, R Kaul, B Bajaj, S Verma and Q Cai for their contributions to the EBNA-3C projects. We also express our gratitude to the rest of the laboratory members for their enormous support.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
The research in ES Robertson’s laboratory is supported by Public health service grants CA137894-02 and CA138434-02 from the NCI, USA. A Saha is a Ramanujan fellow, Department of Science & Technology, India. ES Robertson is a scholar of the Leukemia and Lymphoma Society of America. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Saha A, Robertson ES. Epstein–Barr virus-associated B-cell lymphomas: pathogenesis and clinical outcomes. Clin Cancer Res. 2011;17(10):3056–3063. doi: 10.1158/1078-0432.CCR-10-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford DH. Biology and disease associations of Epstein–Barr virus. Philos Trans R Soc Lond B Biol Sci. 2001;356(1408):461–473. doi: 10.1098/rstb.2000.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vereide D, Sugden B. Insights into the evolution of lymphomas induced by Epstein–Barr virus. Adv Cancer Res. 2010;108:1–19. doi: 10.1016/B978-0-12-380888-2.00001-7. [DOI] [PubMed] [Google Scholar]
- 4.Thompson MP, Kurzrock R. Epstein–Barr virus and cancer. Clin Cancer Res. 2004;10(3):803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- 5▪.Saha A, Kaul R, Murakami M, Robertson ES. Tumor viruses and cancer biology: modulating signaling pathways for therapeutic intervention. Cancer Biol Ther. 2010;10(10):961–978. doi: 10.4161/cbt.10.10.13923. Recent comprehensive review of host responses and the unique survival strategies employed by different RNA and DNA tumor viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nat Rev Cancer. 2004;4(10):757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 7.Kutok JL, Wang F. Spectrum of Epstein–Barr virus-associated diseases. Annu Rev Pathol. 2006;1:375–404. doi: 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 8.Klein G, Klein E, Kashuba E. Interaction of Epstein–Barr virus (EBV) with human B-lymphocytes. Biochem Biophys Res Commun. 2010;396(1):67–73. doi: 10.1016/j.bbrc.2010.02.146. [DOI] [PubMed] [Google Scholar]
- 9.Dolcetti RGM, Gloghini A, Carbone A. EBV-associated tumors: pathogenetic insights for improved disease monitoring and treatment. Curr Cancer Ther Rev. 2005;1(1):27–44. [Google Scholar]
- 10.Rezk SAWL. Epstein–Barr virus-associated lymphoproliferative disorders. Hum Pathol. 2007;38(9):1293–1304. doi: 10.1016/j.humpath.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Dolcetti R. B lymphocytes and Epstein–Barr virus: the lesson of post-transplant lymphoproliferative disorders. Autoimmun Rev. 2007;7(2):96–101. doi: 10.1016/j.autrev.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Ma SD, Hegde S, Young KH, et al. A new model of Epstein–Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J Virol. 2010;85(1):165–177. doi: 10.1128/JVI.01512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, Hayward SD. The Ying–Yang of the virus–host interaction: control of the DNA damage response. Future Microbiol. 2011;6(4):379–383. doi: 10.2217/fmb.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hertle ML, Popp C, Petermann S, et al. Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathog. 2009;5(7):e1000506. doi: 10.1371/journal.ppat.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian C, Knight JS, Robertson ES. The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration. Front Biosci. 2002;7:d704–d716. doi: 10.2741/subraman. [DOI] [PubMed] [Google Scholar]
- 16.West MJ. Structure and function of the Epstein–Barr virus transcription factor, EBNA 3C. Curr Protein Pept Sci. 2006;7(2):123–136. doi: 10.2174/138920306776359777. [DOI] [PubMed] [Google Scholar]
- 17▪.Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. Epigenetic repression of p16INK4A by latent Epstein–Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 2010;6(6):e1000951. doi: 10.1371/journal.ppat.1000951. Together with [23,24], demonstrates that EBNA-3C may recruit epigenetic machinery for regulating tumor-suppressor gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yenamandra SP, Sompallae R, Klein G, Kashuba E. Comparative analysis of the Epstein–Barr virus encoded nuclear proteins of EBNA-3 family. Comput Biol Med. 2009;39(11):1036–1042. doi: 10.1016/j.compbiomed.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Zhao B, Dalbies-Tran R, Jiang H, et al. Transcriptional regulatory properties of Epstein–Barr virus nuclear antigen 3C are conserved in simian lymphocryptoviruses. J Virol. 2003;77(10):5639–5648. doi: 10.1128/JVI.77.10.5639-5648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krauer K, Buck M, Flanagan J, Belzer D, Sculley T. Identification of the nuclear localization signals within the Epstein–Barr virus EBNA-6 protein. J Gen Virol. 2004;85(Pt 1):165–172. doi: 10.1099/vir.0.19549-0. [DOI] [PubMed] [Google Scholar]
- 21.Buck M, Burgess A, Stirzaker R, Krauer K, Sculley T. Epstein–Barr virus nuclear antigen 3A contains six nuclear-localization signals. J Gen Virol. 2006;87(Pt 10):2879–2884. doi: 10.1099/vir.0.81927-0. [DOI] [PubMed] [Google Scholar]
- 22.Burgess A, Buck M, Krauer K, Sculley T. Nuclear localization of the Epstein–Barr virus EBNA3B protein. J Gen Virol. 2006;87(Pt 4):789–793. doi: 10.1099/vir.0.81640-0. [DOI] [PubMed] [Google Scholar]
- 23.Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein–Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene. Bim PLoS Pathog. 2009;5(6):e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White RE, Groves IJ, Turro E, Yee J, Kremmer E, Allday MJ. Extensive cooperation between the Epstein–Barr virus EBNA3 proteins in the manipulation of host gene expression and epigenetic chromatin modification. PLoS One. 2010;5(11):e13979. doi: 10.1371/journal.pone.0013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White RE, Ramer PC, Naresh KN, et al. EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J Clin Invest. 2012;122(4):1487–1502. doi: 10.1172/JCI58092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. Epstein–Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci USA. 2011;108(5):1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27▪▪.Maruo S, Wu Y, Ito T, Kanda T, Kieff ED, Takada K. Epstein–Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc Natl Acad Sci USA. 2009;106(11):4419–4424. doi: 10.1073/pnas.0813134106. Demonstrates critical EBNA-3C amino acid residues involved in B-cell transformation through interaction with several important cellular proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein–Barr virus DNA. J Virol. 1995;69(1):231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomkinson B, Robertson E, Kieff E. Epstein–Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67(4):2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30▪.Petti L, Sample J, Wang F, Kieff E. A fifth Epstein–Barr virus nuclear protein (EBNA3C) is expressed in latently infected growth-transformed lymphocytes. J Virol. 1988;62(4):1330–1338. doi: 10.1128/jvi.62.4.1330-1338.1988. First study that evidently shows EBNA-3C expression in virus-transformed B cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31▪▪.Saha A, Murakami M, Kumar P, Bajaj B, Sims K, Robertson ES. Epstein–Barr virus nuclear antigen 3C augments MDM2-mediated p53 ubiquitination and degradation by deubiquitinating MDM2. J Virol. 2009;83(9):4652–4669. doi: 10.1128/JVI.02408-08. Important study demonstrating EBNA-3C may recruit deubiquitinase activity to stabilize MDM2 and subsequently facilitate p53 degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krauer KG, Buck M, Belzer DK, Flanagan J, Chojnowski GM, Sculley TB. The Epstein–Barr virus nuclear antigen-6 protein colocalizes with EBNA-3 and survival of motor neurons protein. Virology. 2004;318(1):280–294. doi: 10.1016/j.virol.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 33▪.Yan X, Mouillet JF, Ou Q, Sadovsky Y. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol Cell Biol. 2003;23(1):414–423. doi: 10.1128/MCB.23.1.414-423.2003. Initial study proposing that EBNA-3C may be involved in RNA biogenesis through interaction with Gemin3 (DP103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34▪▪.Allday MJ, Farrell PJ. Epstein–Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J Virol. 1994;68(6):3491–3498. doi: 10.1128/jvi.68.6.3491-3498.1994. Pioneering work that describes a functional role for EBNA-3C in viral gene transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Mar JC, Maruo S, et al. Epstein–Barr virus nuclear antigen 3C regulated genes in lymphoblastoid cell lines. Proc Natl Acad Sci USA. 2010;108(1):337–342. doi: 10.1073/pnas.1017419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36▪.Radkov SA, Bain M, Farrell PJ, West M, Rowe M, Allday MJ. Epstein–Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol. 1997;71(11):8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. Another initial study that demonstrates EBNA-3C-mediated viral gene transcriptional regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J, Johannsen E, Robertson E, Kieff E. Epstein–Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J Virol. 2002;76(1):232–242. doi: 10.1128/JVI.76.1.232-242.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClellan MJ, Khasnis S, Wood CD, et al. Downregulation of integrin receptor-signaling genes by Epstein–Barr virus EBNA 3C via promoter-proximal and -distal binding elements. J Virol. 2012;86(9):5165–5178. doi: 10.1128/JVI.07161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao B, Sample CE. Epstein–Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein–Barr virus nuclear antigen 2 through sequences encompassing an spi-1/spi-B binding site. J Virol. 2000;74(11):5151–5160. doi: 10.1128/jvi.74.11.5151-5160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian C, Hasan S, Rowe M, Hottiger M, Orre R, Robertson ES. Epstein–Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J Virol. 2002;76(10):4699–4708. doi: 10.1128/JVI.76.10.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41▪.Radkov SA, Touitou R, Brehm A, et al. Epstein–Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73(7):5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. Together with [40,42], demonstrates that EBNA-3C may recruit histone deacetylases and histone acetyltransferase activities for regulating gene transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knight JS, Lan K, Subramanian C, Robertson ES. Epstein–Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J Virol. 2003;77(7):4261–4272. doi: 10.1128/JVI.77.7.4261-4272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bain M, Watson RJ, Farrell PJ, Allday MJ. Epstein–Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol. 1996;70(4):2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuentes-Panana EM, Peng R, Brewer G, Tan J, Ling PD. Regulation of the Epstein–Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J Virol. 2000;74(17):8166–8175. doi: 10.1128/jvi.74.17.8166-8175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimber-Strobl U, Strobl LJ. EBNA2 and Notch signalling in Epstein–Barr virus mediated immortalization of B lymphocytes. Semin Cancer Biol. 2001;11(6):423–434. doi: 10.1006/scbi.2001.0409. [DOI] [PubMed] [Google Scholar]
- 46.Zhou S, Fujimuro M, Hsieh JJ, Chen L, Hayward SD. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J Virol. 2000;74(4):1939–1947. doi: 10.1128/jvi.74.4.1939-1947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S, Sakakibara S, Maruo S, et al. Epstein–Barr virus nuclear protein 3C domains necessary for lymphoblastoid cell growth: interaction with RBP-Jkappa regulates TCL1. J Virol. 2009;83(23):12368–12377. doi: 10.1128/JVI.01403-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashuba E, Kashuba V, Sandalova T, Klein G, Szekely L. Epstein–Barr virus encoded nuclear protein EBNA-3 binds a novel human uridine kinase/uracil phosphoribosyltransferase. BMC Cell Biol. 2002;3:23. doi: 10.1186/1471-2121-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanigaki K, Honjo T. Two opposing roles of RBP-J in Notch signaling. Curr Top Dev Biol. 2010;92:231–252. doi: 10.1016/S0070-2153(10)92007-3. [DOI] [PubMed] [Google Scholar]
- 50▪▪.Robertson ES, Lin J, Kieff E. The amino-terminal domains of Epstein–Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70(5):3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. Pioneering work that describes how EBNA-3C, along with other EBNA-3 proteins, interacts with RBP-Jκ, a downstream modulator of the Notch signaling pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22(2):148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartova E, Krejci J, Harnicarova A, Galiova G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56(8):711–721. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 54.Touitou R, Hickabottom M, Parker G, Crook T, Allday MJ. Physical and functional interactions between the corepressor CtBP and the Epstein–Barr virus nuclear antigen EBNA3C. J Virol. 2001;75(16):7749–7755. doi: 10.1128/JVI.75.16.7749-7755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao LJ, Subramanian T, Vijayalingam S, Chinnadurai G. PLDLS-dependent interaction of E1A with CtBP: regulation of CtBP nuclear localization and transcriptional functions. Oncogene. 2007;26(54):7544–7551. doi: 10.1038/sj.onc.1210569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickabottom M, Parker GA, Freemont P, Crook T, Allday MJ. Two nonconsensus sites in the Epstein–Barr virus oncoprotein EBNA3A cooperate to bind the co-repressor carboxyl-terminal-binding protein (CtBP) J Biol Chem. 2002;277(49):47197–47204. doi: 10.1074/jbc.M208116200. [DOI] [PubMed] [Google Scholar]
- 57▪.Steeg PS, Bevilacqua G, Kopper L, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80(3):200–204. doi: 10.1093/jnci/80.3.200. Describes the discovery of the first metastasis repressor protein, Nm23-H1. [DOI] [PubMed] [Google Scholar]
- 58.Saha A, Robertson ES. Functional modulation of the metastatic suppressor Nm23-H1 by oncogenic viruses. FEBS Lett. 2011;585(20):3174–3184. doi: 10.1016/j.febslet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murakami M, Kaul R, Kumar P, Robertson ES. Nucleoside diphosphate kinase/Nm23 and Epstein–Barr virus. Mol Cell Biochem. 2009;329(1–2):131–139. doi: 10.1007/s11010-009-0123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choudhuri T, Murakami M, Kaul R, et al. Nm23-H1 can induce cell cycle arrest and apoptosis in B cells. Cancer Biol Ther. 2010;9(12):1065–1078. doi: 10.4161/cbt.9.12.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaul R, Murakami M, Choudhuri T, Robertson ES. Epstein–Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J Virol. 2007;81(19):10352–10361. doi: 10.1128/JVI.00886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62▪▪.Subramanian C, Cotter MA, 2nd, Robertson ES. Epstein–Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat Med. 2001;7(3):350–355. doi: 10.1038/85499. Initial study showing that EBNA-3C interacts with Nm23-H1 to regulate its metastatic activity. [DOI] [PubMed] [Google Scholar]
- 63.Kelly GL, Milner AE, Tierney RJ, et al. Epstein–Barr virus nuclear antigen 2 (EBNA2) gene deletion is consistently linked with EBNA3A, -3B, and -3C expression in Burkitt’s lymphoma cells and with increased resistance to apoptosis. J Virol. 2005;79(16):10709–10717. doi: 10.1128/JVI.79.16.10709-10717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64▪.Kaul R, Murakami M, Lan K, Choudhuri T, Robertson ES. EBNA3C can modulate the activities of the transcription factor Necdin in association with metastasis suppressor protein Nm23-H1. J Virol. 2009;83(10):4871–4883. doi: 10.1128/JVI.02286-08. Recent study describing that EBNA-3C, in association with Nm23-H1, can block Necdin-mediated tumor-suppressive activities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67(6):705–712. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 66.Matsumoto K, Taniura H, Uetsuki T, Yoshikawa K. Necdin acts as a transcriptional repressor that interacts with multiple guanosine clusters. Gene. 2001;272(1–2):173–179. doi: 10.1016/s0378-1119(01)00544-3. [DOI] [PubMed] [Google Scholar]
- 67.Harper JV, Brooks G. The mammalian cell cycle: an overview. Methods Mol Biol. 2005;296:113–153. doi: 10.1385/1-59259-857-9:113. [DOI] [PubMed] [Google Scholar]
- 68.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 69.Malumbres M, Harlow E, Hunt T, et al. Cyclin-dependent kinases: a family portrait. Nat Cell Biol. 2009;11(11):1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9(11):910–916. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- 71.Canavese M, Santo L, Raje N. Cyclin dependent kinases in cancer: potential for therapeutic intervention. Cancer Biol Ther. 2012;13(7):451–457. doi: 10.4161/cbt.19589. [DOI] [PubMed] [Google Scholar]
- 72.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5(11):876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- 74.Rizzolio F, Tuccinardi T, Caligiuri I, Lucchetti C, Giordano A. CDK inhibitors: from the bench to clinical trials. Curr Drug Targets. 2010;11(3):279–290. doi: 10.2174/138945010790711978. [DOI] [PubMed] [Google Scholar]
- 75.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24(17):2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 76.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poznic M. Retinoblastoma protein: a central processing unit. J Biosci. 2009;34(2):305–312. doi: 10.1007/s12038-009-0034-2. [DOI] [PubMed] [Google Scholar]
- 78.Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36(3):131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flatt PM, Pietenpol JA. Mechanisms of cell-cycle checkpoints: at the crossroads of carcinogenesis and drug discovery. Drug Metab Rev. 2000;32(3–4):283–305. doi: 10.1081/dmr-100102335. [DOI] [PubMed] [Google Scholar]
- 80.Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotechnol. 2012;2012:170325. doi: 10.1155/2012/170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, Xu ZP, Huang Y, Hamrick HE, Duerksen-Hughes PJ, Yu YN. ATM and ATR: sensing DNA damage. World J Gastroenterol. 2004;10(2):155–160. doi: 10.3748/wjg.v10.i2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6(9):1006–1010. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- 83.Smith J, Tho LM, Xu N, Gillespie DA. The ATM–Chk2 and ATR–Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 84.Yao Y, Dai W. Mitotic checkpoint control and chromatin remodeling. Front Biosci. 2012;17:976–983. doi: 10.2741/3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun SC, Kim NH. Spindle assembly checkpoint and its regulators in meiosis. Hum Reprod Update. 2012;18(1):60–72. doi: 10.1093/humupd/dmr044. [DOI] [PubMed] [Google Scholar]
- 86▪▪.Parker GA, Crook T, Bain M, Sara EA, Farrell PJ, Allday MJ. Epstein–Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13(12):2541–2549. Pioneering study that demonstrates how EBNA-3C is a potent transforming antigen encoded by EBV. [PubMed] [Google Scholar]
- 87▪.Knight JS, Sharma N, Robertson ES. Epstein–Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci USA. 2005;102(51):18562–18566. doi: 10.1073/pnas.0503886102. Evidence that shows EBNA-3C targets major tumor-suppressor protein pRb to override G1–S-phase block. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kashuba E, Yurchenko M, Yenamandra SP, et al. EBV-encoded EBNA-6 binds and targets MRS18-2 to the nucleus, resulting in the disruption of pRb–E2F1 complexes. Proc Natl Acad Sci USA. 2008;105(14):5489–5494. doi: 10.1073/pnas.0801053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. Epstein–Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci USA. 2006;103(51):19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90▪▪.Saha A, Halder S, Upadhyay SK, et al. Epstein–Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1. PLoS Pathog. 2011;7(2):e1001275. doi: 10.1371/journal.ppat.1001275. Recent study that shows EBNA-3C targets cyclin D1 functions to facilitate G1–S-phase transition of the cell cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bashir T, Pagano M. Don’t skip the G1 phase: how APC/CCdh1 keeps SCFSKP2 in check. Cell Cycle. 2004;3(7):850–852. [PubMed] [Google Scholar]
- 92.Bajaj BG, Murakami M, Cai Q, Verma SC, Lan K, Robertson ES. Epstein–Barr virus nuclear antigen 3C interacts with and enhances the stability of the c-Myc oncoprotein. J Virol. 2008;82(8):4082–4090. doi: 10.1128/JVI.02500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93▪▪.Knight JS, Sharma N, Robertson ES. SCFSkp2 complex targeted by Epstein–Barr virus essential nuclear antigen. Mol Cell Biol. 2005;25(5):1749–1763. doi: 10.1128/MCB.25.5.1749-1763.2005. Study demonstrating that EBNA-3C can manipulate the ubiquitin–proteasome pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.von der Lehr N, Johansson S, Larsson LG. Implication of the ubiquitin/proteasome system in Myc-regulated transcription. Cell Cycle. 2003;2(5):403–407. [PubMed] [Google Scholar]
- 95▪.Knight JS, Sharma N, Kalman DE, Robertson ES. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein–Barr virus-infected cells. J Virol. 2004;78(23):12857–12867. doi: 10.1128/JVI.78.23.12857-12867.2004. Shows that EBNA-3C enhances cyclin A activity for regulating S-phase propagation of the cell cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96▪▪.Knight JS, Robertson ES. Epstein–Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J Virol. 2004;78(4):1981–1991. doi: 10.1128/JVI.78.4.1981-1991.2004. Shows that EBNA-3C enhances cyclin A activity for regulating S-phase propagation of the cell cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97▪▪.Saha A, Lu J, Morizur L, Upadhyay SK, Aj MP, Robertson ES. E2F1 mediated apoptosis induced by the DNA damage response is blocked by EBV nuclear antigen 3C in lymphoblastoid cells. PLoS Pathog. 2012;8(3):e1002573. doi: 10.1371/journal.ppat.1002573. Recent study demonstrating that EBNA-3C blocks E2F1-mediated apoptosis in response to DNA-damaging signals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98▪.Yi F, Saha A, Murakami M, et al. Epstein–Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology. 2009;388(2):236–247. doi: 10.1016/j.virol.2009.03.027. Initial study that shows an in vitro as well as an in vivo association between p53 and EBNA-3C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99▪.Saha A, Bamidele A, Murakami M, Robertson ES. EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5. J Virol. 2011;85(5):2079–2088. doi: 10.1128/JVI.02279-10. Study demonstrating that inhibiton of growth family proteins ING4 and ING5 may be involved in regulating cell apoptosis in EBV-infected B cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Spender LC, Cannell EJ, Hollyoake M, et al. Control of cell cycle entry and apoptosis in B lymphocytes infected by Epstein–Barr virus. J Virol. 1999;73(6):4678–4688. doi: 10.1128/jvi.73.6.4678-4688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palmero I, Holder A, Sinclair AJ, Dickson C, Peters G. Cyclins D1 and D2 are differentially expressed in human B-lymphoid cell lines. Oncogene. 1993;8(4):1049–1054. [PubMed] [Google Scholar]
- 102.Kim HR, Jeong JA, Park CH, Lee SK, Lee WK, Jang YS. A role for cell cycle proteins in the serum-starvation resistance of Epstein–Barr virus immortalized B lymphocytes. Biochem Cell Biol. 2002;80(4):407–413. doi: 10.1139/o02-085. [DOI] [PubMed] [Google Scholar]
- 103.Murai Y, Dobashi Y, Okada E, et al. Study on the role of G1 cyclins in Epstein–Barr virus-associated human lymphomas maintained in severe combined immune deficiency (SCID) mice. Int J Cancer. 2001;92(2):232–239. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1171>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 104.Velasco-Velazquez MA, Li Z, Casimiro M, Loro E, Homsi N, Pestell RG. Examining the role of cyclin D1 in breast cancer. Future Oncol. 2011;7(6):753–765. doi: 10.2217/fon.11.56. [DOI] [PubMed] [Google Scholar]
- 105.Lin DI, Barbash O, Kumar KG, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24(3):355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pontano LL, Diehl JA. Speeding through cell cycle roadblocks: Nuclear cyclin D1-dependent kinase and neoplastic transformation. Cell Div. 2008;3:12. doi: 10.1186/1747-1028-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parker GA, Touitou R, Allday MJ. Epstein–Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene. 2000;19(5):700–709. doi: 10.1038/sj.onc.1203327. [DOI] [PubMed] [Google Scholar]
- 108▪▪.Nikitin PA, Yan CM, Forte E, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein–Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8(6):510–522. doi: 10.1016/j.chom.2010.11.004. Recent study that shows EBNA-3C can block DNA damage-induced signaling events to ensure efficient proliferation during B-cell transformation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vardy L, Pesin JA, Orr-Weaver TL. Regulation of Cyclin A protein in meiosis and early embryogenesis. Proc Natl Acad Sci USA. 2009;106(6):1838–1843. doi: 10.1073/pnas.0813237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mateo F, Vidal-Laliena M, Pujol MJ, Bachs O. Acetylation of cyclin A: a new cell cycle regulatory mechanism. Biochem Soc Trans. 2010;38(Pt 1):83–86. doi: 10.1042/BST0380083. [DOI] [PubMed] [Google Scholar]
- 111.Jessus C, Ozon R. How does Xenopus oocyte acquire its competence to undergo meiotic maturation? Biol Cell. 2004;96(3):187–192. doi: 10.1016/j.biolcel.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 112.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11(3):961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113▪.Choudhuri T, Verma SC, Lan K, Murakami M, Robertson ES. The ATM/ATR signaling effector Chk2 is targeted by Epstein–Barr virus nuclear antigen 3C to release the G2/M cell cycle block. J Virol. 2007;81(12):6718–6730. doi: 10.1128/JVI.00053-07. Study that demonstrates EBNA-3C can override G2–M-phase cell cycle blockage by targeting Chk2-mediated activities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wade M, Allday MJ. Epstein–Barr virus suppresses a G(2)/M checkpoint activated by genotoxins. Mol Cell Biol. 2000;20(4):1344–1360. doi: 10.1128/mcb.20.4.1344-1360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Benedict CA, Norris PS, Ware CF. To kill or be killed: viral evasion of apoptosis. Nat Immunol. 2002;3(11):1013–1018. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- 116.White E. Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis. Cell Death Differ. 2006;13(8):1371–1377. doi: 10.1038/sj.cdd.4401941. [DOI] [PubMed] [Google Scholar]
- 117.Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Essmann F, Schulze-Osthoff K. Translational approaches targeting the p53 pathway for anti-cancer therapy. Br J Pharmacol. 2012;165(2):328–344. doi: 10.1111/j.1476-5381.2011.01570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28(3):128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tan S, de Vries EG, van der Zee AG, de Jong S. Anticancer drugs aimed at E6 and E7 activity in HPV-positive cervical cancer. Curr Cancer Drug Targets. 2012;12(2):170–184. doi: 10.2174/156800912799095135. [DOI] [PubMed] [Google Scholar]
- 121.Cai Q, Verma SC, Lu J, Robertson ES. Molecular biology of Kaposi’s sarcoma-associated herpesvirus and related oncogenesis. Adv Virus Res. 2010;78:87–142. doi: 10.1016/B978-0-12-385032-4.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 123.Taira N, Yoshida K. Post-translational modifications of p53 tumor suppressor: determinants of its functional targets. Histol Histopathol. 2012;27(4):437–443. doi: 10.14670/HH-27.437. [DOI] [PubMed] [Google Scholar]
- 124.Gu B, Zhu WG. Surf the post-translational modification network of p53 regulation. Int J Biol Sci. 2012;8(5):672–684. doi: 10.7150/ijbs.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pei D, Zhang Y, Zheng J. Regulation of p53: a collaboration between Mdm2 and Mdmx. Oncotarget. 2012;3(3):228–235. doi: 10.18632/oncotarget.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Di J, Zhang Y, Zheng J. Reactivation of p53 by inhibiting Mdm2 E3 ligase: a novel antitumor approach. Curr Cancer Drug Targets. 2011;11(8):987–994. doi: 10.2174/156800911797264789. [DOI] [PubMed] [Google Scholar]
- 127.Russell M, Berardi P, Gong W, Riabowol K. Grow-ING, age-ING and die-ING: ING proteins link cancer, senescence and apoptosis. Exp Cell Res. 2006;312(7):951–961. doi: 10.1016/j.yexcr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 128.Jafarnejad SM, Li G. Regulation of p53 by ING family members in suppression of tumor initiation and progression. Cancer Metastasis Rev. 2011;31(1–2):55–73. doi: 10.1007/s10555-011-9329-5. [DOI] [PubMed] [Google Scholar]
- 129.Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32(11):509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 130.Dalla Via L, Nardon C, Fregona D. Targeting the ubiquitin–proteasome pathway with inorganic compounds to fight cancer: a challenge for the future. Future Med Chem. 2012;4(4):525–543. doi: 10.4155/fmc.11.187. [DOI] [PubMed] [Google Scholar]
- 131.Forte E, Luftig MA. MDM2-dependent inhibition of p53 is required for Epstein–Barr virus B-cell growth transformation and infected-cell survival. J Virol. 2009;83(6):2491–2499. doi: 10.1128/JVI.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shen H, Maki CG. Pharmacologic activation of p53 by small-molecule MDM2 antagonists. Curr Pharm Des. 2011;17(6):560–568. doi: 10.2174/138161211795222603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23(14):4939–4947. doi: 10.1128/MCB.23.14.4939-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Grundhoff AT, Kremmer E, Tureci O, et al. Characterization of DP103, a novel DEAD box protein that binds to the Epstein–Barr virus nuclear proteins EBNA2 and EBNA3C. J Biol Chem. 1999;274(27):19136–19144. doi: 10.1074/jbc.274.27.19136. [DOI] [PubMed] [Google Scholar]
- 135.Fuller-Pace FV, Jacobs AM, Nicol SM. Modulation of transcriptional activity of the DEAD-box family of RNA helicases, p68 (Ddx5) and DP103 (Ddx20), by SUMO modification. Biochem Soc Trans. 2007;35(Pt 6):1427–1429. doi: 10.1042/BST0351427. [DOI] [PubMed] [Google Scholar]
- 136.Zhou J, Zheng X, Shen H. Targeting RNA-splicing for SMA treatment. Mol Cells. 2012;33(3):223–228. doi: 10.1007/s10059-012-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137▪.Cai Q, Guo Y, Xiao B, et al. Epstein–Barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis. PLoS Pathog. 2011;7(12):e1002418. doi: 10.1371/journal.ppat.1002418. Recent study showing EBNA-3C recruits Gemin3 activity to regulate p53-dependent transcription as well as apoptosis. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 138.Wu Z, Zheng S, Yu Q. The E2F family and the role of E2F1 in apoptosis. Int J Biochem Cell Biol. 2009;41(12):2389–2397. doi: 10.1016/j.biocel.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 139.Polager S, Ginsberg D. p53 and E2F: partners in life and death. Nat Rev Cancer. 2009;9(10):738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 140.Wu Z, Yu Q. E2F1-mediated apoptosis as a target of cancer therapy. Curr Mol Pharmacol. 2009;2(2):149–160. doi: 10.2174/1874467210902020149. [DOI] [PubMed] [Google Scholar]
- 141.Molina-Privado I, Rodriguez-Martinez M, Rebollo P, et al. E2F1 expression is deregulated and plays an oncogenic role in sporadic Burkitt’s lymphoma. Cancer Res. 2009;69(9):4052–4058. doi: 10.1158/0008-5472.CAN-08-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pediconi N, Ianari A, Costanzo A, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5(6):552–558. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 143.Jendrossek V. The intrinsic apoptosis pathways as a target in anticancer therapy. Curr Pharm Biotechnol. 2012;13(8):1426–1438. doi: 10.2174/138920112800784989. [DOI] [PubMed] [Google Scholar]
- 144.Hughes P, Bouillet P, Strasser A. Role of Bim and other Bcl-2 family members in autoimmune and degenerative diseases. Curr Dir Autoimmun. 2006;9:74–94. doi: 10.1159/000090773. [DOI] [PubMed] [Google Scholar]
- 145.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101(16):6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Richter-Larrea JA, Robles EF, Fresquet V, et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116(14):2531–2542. doi: 10.1182/blood-2010-02-268003. [DOI] [PubMed] [Google Scholar]
- 147.Molyneux EM, Rochford R, Griffin B, et al. Burkitt’s lymphoma. Lancet. 2012;379(9822):1234–1244. doi: 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- 148.Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. Two Epstein–Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt’s lymphoma. Oncogene. 2008;27(4):421–433. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- 149.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22(1):50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 150.Ocker M, Hopfner M. Apoptosis-modulating drugs for improved cancer therapy. Eur Surg Res. 2012;48(3):111–120. doi: 10.1159/000336875. [DOI] [PubMed] [Google Scholar]
- 151.Bornkamm GW. Epstein–Barr virus and the pathogenesis of Burkitt’s lymphoma: more questions than answers. Int J Cancer. 2009;124(8):1745–1755. doi: 10.1002/ijc.24223. [DOI] [PubMed] [Google Scholar]
- 152.Schlee M, Schuhmacher M, Holzel M, Laux G, Bornkamm GW. c-Myc impairs immunogenicity of human B cells. Adv Cancer Res. 2007;97:167–188. doi: 10.1016/S0065-230X(06)97007-9. [DOI] [PubMed] [Google Scholar]
- 153.Graham JP, Arcipowski KM, Bishop GA. Differential B-lymphocyte regulation by CD40 and its viral mimic, latent membrane protein 1. Immunol Rev. 2010;237(1):226–248. doi: 10.1111/j.1600-065X.2010.00932.x. [DOI] [PubMed] [Google Scholar]
- 154.Johannsen E, Miller CL, Grossman SR, Kieff E. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJkappa in Epstein–Barr virus-transformed B lymphocytes. J Virol. 1996;70(6):4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garrido JL, Maruo S, Takada K, Rosendorff A. EBNA3C interacts with Gadd34 and counteracts the unfolded protein response. Virol J. 2009;6:231. doi: 10.1186/1743-422X-6-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cotter MA, 2nd, Robertson ES. Modulation of histone acetyltransferase activity through interaction of Epstein–Barr nuclear antigen 3C with prothymosin alpha. Mol Cell Biol. 2000;20(15):5722–5735. doi: 10.1128/mcb.20.15.5722-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Subramanian C, Robertson ES. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J Virol. 2002;76(17):8702–8709. doi: 10.1128/JVI.76.17.8702-8709.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rosendorff A, Illanes D, David G, Lin J, Kieff E, Johannsen E. EBNA3C coactivation with EBNA2 requires a SUMO homology domain. J Virol. 2004;78(1):367–377. doi: 10.1128/JVI.78.1.367-377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]