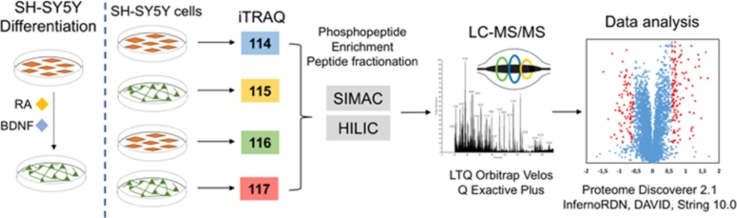
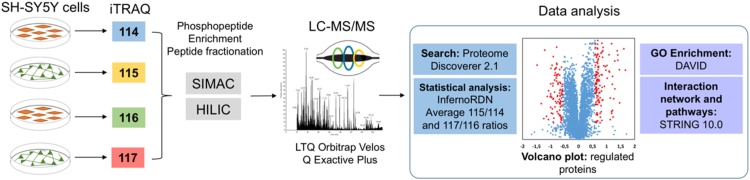
Graphical abstract
Keywords: SH-SY5Y cells, iTRAQ-based proteomics, Neuronal differentiation, Phosphoproteomics
Highlights
-
•
Differentiation analysis of SH-SY5Y cells with iTRAQ strategy is proposed.
-
•
Differentiated SH-SY5Y cells are more appropriated as a neuronal model.
-
•
Upregulated proteins are mainly related to ECM-interaction and apoptosis.
-
•
Proteins to explore as differentiation markers: AGRN, EMILIM-1, AIFM, STMN1.
Abstract
SH-SY5Y neuroblastoma cells are susceptible to differentiation using retinoic acid (RA) and brain-derived neurotrophic factor (BDNF), providing a model of neuronal differentiation. We compared SH-SY5Y cells proteome before and after RA/BDNF treatment using iTRAQ and phosphopeptide enrichment strategies. We identified 5587 proteins, 366 of them with differential abundance. Differentiated cells expressed proteins related to neuronal development, and, undifferentiated cells expressed proteins involved in cell proliferation. Interactive network covered focal adhesion, cytoskeleton dynamics and neurodegenerative diseases processes and regulation of mitogen-activated protein kinase-related signaling pathways; key proteins involved in those processes might be explored as markers for neuronal differentiation.
1. Introduction
In vitro models to study neuron cell biology strongly rely on the cultivation of primary cells from mouse and rat, which have been proven useful to study many aspects of neuron physiology and several disease animal models [1]. Nonetheless, human derived cell are necessary to validate the molecular mechanisms of human diseases, which cannot be completely reproduced in animals, and are more adequate for drug testing and screening [2], [3]. The human derived cell line SH-SY5Y reproduces biochemical and morphological properties of neurons, being often used as in vitro model for human neurons [4]. This lineage was obtained from bone marrow biopsy of a patient with neuroblastoma and can be induced to differentiate into cells with neuron-like phenotype [5], [6], [7], [8].
Induced differentiation of SH-SY5Y cells leads to increase of plasma membrane excitability, appearance of functional synapses, formation and extension of neurites and induction of neurotransmitters and their receptors [4]. During differentiation cells decrease proliferation rate, increase the activity of neuron specific enolase/phosphopyruvate hydratase (NSE), and depending on differentiation protocol, display characteristics of cholinergic, adrenergic or dopaminergic phenotype neurons [4], [9], [10].
Retinoic acid (RA) is used in several protocols to induce neuronal phenotype [4], [11], [12], [13]. RA promotes cell survival activating phosphoinositide 3-kinase/protein kinase B (PI3K/Akt) pathway [13], [14], upregulation of the anti-apoptotic Bcl-2 protein and neurotrophin receptors expression (tyrosine kinase A receptor (TrkA), tyrosine kinase B receptor (TrkB) and neurotrophic factor receptor (RET)) [4]. Also, an increase in neurochemical properties such as the increase of acetyl choline transferase activity, vesicular transport of monoamines, tyrosine hydroxylase (TH) activity and dopamine processing is reported in response to RA treatment [15].
Following RA treatment, neurotrophic factors such as brain-derived neurotrophic factor (BDNF) and nerve growth factor (NGF) have been used to support further differentiation and maintenance of mature neuronal phenotype [16], [17]. This is triggered by BDNF and NGF binding to their respective tropomyosin receptors (Trk) and activation of Ras/Extracellular signal-regulated kinases (Ras/ERK), PI3K/Akt and phospholipase C/calcium (PLCγ/Ca2+) pathways [4]. Combination of RA and BDNF treatment leads to the expression of neuronal markers such as NeuN, synaptophysin, microtubule-associated protein 2 (MAP2), NSE, nestin, among others [9], [16].
Comprehensive analysis of the molecular mechanisms involved in the differentiation of SH-SY5Y was not yet accessed. Published data was obtained from reductionist approaches, e.g. Western blot, quantitative polymerase chain reaction (qPCR) or fluorescence microscopy targeting specific proteins or differentiation markers. A more holistic way of providing high-quality data about presence, abundance, post-translational modifications (PTMs) and interaction of proteins in a specific context [18] can be achieved using Mass spectrometry (MS)-based proteomics. So far, proteomic knowledge on SH-SY5Y comprise few reports which applied two-dimensional difference gel electrophoresis (2D-DIGE) to study differentiation [19], [20] and, more recently, quantitative analysis of nerve growth factor (NGF) driven molecular events [17].
Here, we characterize the proteomic changes of SH-SY5Y cells before and after RA/BDNF induced differentiation using iTRAQ-based quantitative approach via LC–MS/MS and phosphopeptide enrichment for identification of differentially expressed protein profiles in long-term culture of neuron-like differentiation. Our data corroborate previous reports of diverse neuron-related markers and provides additional information about new potential markers for neuron differentiation. We compile a comprehensive resource of pathways and phosphorylation sites, which are altered during neuronal differentiation, they may prove useful to research using in vitro models for human neurons.
2. Material and methods
2.1. Cell culture and differentiation
Human neuroblastoma cell line SH-SY5Y (ATCC − 2266) were cultured in Dulbecco Modified Essential Medium (DMEM)/F12 GlutaMAX™ (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin (Life Technologies) in humidified 5% CO2 and 37 °C incubator. Culture medium was replaced each 3 days until culture reached the appropriate confluency for differentiation or sub-culturing. Differentiation was performed when cells reached 70% confluency following a previously described protocol [16]. At this stage untreated controls were harvested for experiments. Differentiation was induced by treatment with 10 μM retinoic acid (Sigma) added to culture medium (day 0), with medium change at day 2. On day 5 cells were washed three times with DMEM/F12 and incubated with DMEM/F12 GlutaMAX™ (Gibco) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin (Life Technologies) containing 50 ng/mL brain-derived neurotrophic factor (BDNF) (Life Technologies). Medium was replaced on day 8 and 11 and on day 15 cells were used for experiments. Cells were visualized using a Axiovert 135 microscope and documented with AxioVisionLE (Zeiss).
2.2. Western blot
Cells were washed twice with cold phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, homogenized in lysis buffer (2% SDS, 125 mM Tris, pH 6.8, and protease inhibitors cocktail (Roche)), sonicated and boiled for 5 min. Extracted proteins from untreated control cells and differentiated cells were quantified using Qubit® fluorimetric assay (Invitrogen) following manufactures instructions and 20 μg of total protein extracts were resolved in SDS-polyacrylamide gel electrophoresis (PAGE) using 12% acrylamide and transferred to nitrocellulose membrane (Amersham Biosciences). Membrane was blocked with 5% non-fat dry milk in Tris-buffered saline (pH 7.4) and 0.1% Tween-20 and probed against rabbit anti-Tubb3 (1:1000 dilution), mouse anti-Nestin (1:1000 dilution), rabbit anti-MAP2 (1:1000 dilution) antibodies (Millipore). Horseradish peroxidase-conjugated secondary antibodies were used for detection (1:2000 or 1:10,000 dilution) (Cell Signalling Tecnologies) using enhanced chemiluminescence (ECL) kit (Thermo Scientific) in an ImageQuant LAS 4000 camera system (GE Healthcare).
2.3. Protein extraction for proteomics
Cells were washed twice with cold PBS, harvested by scraping them off the plate, collected by centrifugation and stored in −80 °C until sample preparation. 1 × 105 cells were lysed with buffer containing 7 M urea, 2 M thiourea, 50 mM HEPES pH 8, 75 mM NaCl, 1 mM EDTA, 1 mM PMSF and protease/phosphatase inhibitor cocktails (Roche). The lysates were sonicated and centrifuged at 10,000g for 10 min at 4 °C. Total protein content in supernatant was quantified with Qubit® fluorimetric assay (Invitrogen) following manufactures instructions. Two independent biological replicates of each condition were used in the experiments.
2.4. Protein digestion and iTRAQ labeling
Proteins (100 μg of each condition) were incubated with dithiothreitol (DTT) at final concentration of 10 mM for 1 h at 30 °C; afterwards, iodoacetamide (final concentration of 40 mM) was added an incubated for 30 min in the dark at room temperature. Samples were diluted 10-fold with 50 mM HEPES pH 8 and incubated with sequence-grade modified Trypsin (Promega) at 1/50 trypsin/protein ratio for 16 h at 37 °C. The resultant peptides were further labeled using chemical tags for iTRAQ 4-plex reagent kit (AB Sciex) according to the manufacturer’s instructions. Peptides from two biological replicates of each condition were labeled as follows: 114 and 116 isobaric tags for undifferentiated cells (undiff-SH-SY5Y); 115 and 117 isobaric tags for differentiated cells (diff-SH-SY5Y). Labeled peptides were mixed in a unique tube and desalted with C-18 macro spin column (Harvard Apparatus) and then dried in a vacuum centrifuge.
2.5. Phosphopeptide enrichment
Protein extraction, digestion and peptide labeling for phosphoproteome analysis were performed as described in Section 2.4, Poros 20 R2 (C-18) and Oligo R3 (Applied Biosystems) resins were used for desalting before enrichment. Phosphopeptides were enriched using SIMAC (Immobilized Metal Affinity Chromatography – IMAC combined with TiO2) [21]. Briefly, desalted peptides were dissolved in loading buffer (50% acetonitrile (ACN), 0.1% trifluoroacetic acid (TFA)), loaded in PHOS-Select Iron Affinity gel beads (Sigma) and incubated for 30 min at room temperature with end-over-end rotation. The beads were separated by centrifugation and washed twice with loading buffer; the supernatant was transferred to a new tube (flow through fraction). The acid fraction was obtained by adding 20% ACN, 0.1% TFA to the beads and the supernatant was mixed with flow through fraction and vacuum dried. Multiphosphorylated peptides were eluted by adding 1% NH4OH to the beads, cleaned in a microcolumn with Poros Oligo R3 and vacuum dried. The flow through and acid fraction mixture of IMAC were dissolved in 80% ACN, 1 M glycolic acid, 5% TFA and incubated with Titanspheres – TiO2 (GL Sciences) with high shaking for 30 min at room temperature. TiO2 spheres were decanted and washed twice with 80% ACN, 5% TFA; monophosphorylated peptides were eluted by incubating TiO2 with 1% NH4OH for 20 min under shaking. Samples were centrifuged and supernatant was acidified with 1% TFA. Phosphopeptides were cleaned in a microcolumn with Poros Oligo R3 and dried in a vacuum centrifuge.
2.6. Hydrophilic interaction chromatography fractionation and nano-LC–MS/MS analysis
Dried peptides were redissolved in buffer A (90% ACN, 0.1% TFA) and then fractionated using a UPLC System (Shimadzu) connected to a HILIC-TSKGel Amide-80 column (5 cm × 2 mm i.d. × 5 μm) (Supelco). Separation was achieved with a linear gradient of 0 to 30% of buffer B (0.1% TFA) in 45 min at a constant flow rate of 200 μl/min. A total of 26 fractions were collected (those with low absorbance intensity were pooled) and dried in a speed vacuum concentrator. This protocol was applied to peptides for total proteome (with no enrichment) and monophosphorylated peptides obtained from TiO2.
Each fraction or pool of fractions were analyzed in three technical replicates in an Easy-nLC 1000 nano-LC system (Thermo Scientific) coupled to a quadrupole-Orbitrap (Q-Exactive) and/or LTQ Orbitrap Velos mass spectrometers (Thermo Scientific). Fractions were dissolved in 20 μl of 0.1% formic acid (FA), loaded onto a trap column (ReprosilPur C18, 2 cm × 150 μm i. d. × 5 μm) with a flow rate of 5 μl/min and separated on the analytical column (ReprosilPur C18, 30 cm × 75 μm i.d. × 1.7 μm) with a constant flow rate of 300 nL/min and linear gradient of 5–40% of B (95% ACN, 0.1% FA) in 120 min. For electrospray was used 2.0 kV and 200 °C at the inlet of mass spectrometer. The Q-Exactive mass spectrometer was operated in data dependent analysis (DDA) mode with dynamic exclusion of 30 ms and full-scan MS spectra (m/z 375–1800) with resolution of 70,000 (m/z 200), followed by fragmentation of 12 most intense ions with high energy collisional dissociation (HCD), normalized collision energy (NCE) of 30.0 and resolution of 17,000 (m/z 200) in MS/MS scans. The LTQ Orbitrap Velos mass spectrometer was operated in DDA mode with dynamic exclusion of 45 ms and full-scan MS spectra with resolution of 60,000; followed by fragmentation of 5 most intense ions with HCD, NCE of 35 and resolution of 7500 in MS/MS scans. Species with a charge of +1 or greater than +4 were excluded from MS/MS analysis.
2.7. Data analysis
Raw data were processed using Proteome Discoverer 2.1 Software (Thermo Scientific). Peptide identification was performed with Sequest HT algorithm against Homo sapiens database provided by Uniprot (http://www.uniprot.org/downloaded in 26-04-2016). The searches were run with peptide mass tolerance of 10 ppm, MS/MS tolerance of 0.1 Da, tryptic cleavage specificity, 2 maximum missed cleavage sites, fixed modification of carbamidomethyl (Cys) and variable modification of iTRAQ 4-plex (Tyr, Lys and N-terminus), phosphate (Ser, Thr and Tyr) and oxidation (Met); peptides with high confidence based on Xcorr value threshold >2.28 (high-resolution data) and a ΔCn >0.15 were considered for further analysis. The ptmRS algorithm was used for scoring phosphorylation sites considering site probability threshold of 75. False discovery rates (FDR) were obtained using Target Decoy PSM Validator node selecting identifications with a q-value equal or to less than 0.01.
Data for quantification were processed in InfernoRDN (Pacific Northwest National Laboratory, https://omics.pnl.gov/software/infernordn). Only unique peptides were used for quantification. Data from all technical replicates were exported from Proteome Discoverer and normalized using the log2-transformed median. Proteins were considered differentially expressed if the iTRAQ ratios (115/114 and 117/116) were ≥1.5 or ≤0.67 with p-value <0.05 calculated from two-tailed t-test. The Gene Ontology (GO) analysis and pathway enrichment for identified and regulated proteins was performed using the online tool DAVID (https://david.ncifcrf.gov/). The STRING 10.0 tool (http://string-db.org/) and Cytoscape software were used to analyze biological processes and pathways of differentially expressed proteins, the GO terms, signaling pathways and interaction networks were ranked depending on their p-values using the entire data set as background; nods without any interaction were excluded from the analysis. Phosphorylation sequence motifs regulated were identified using the online motif-x algorithm (http://motif-x.med.harvard.edu/).
3. Results and discussion
We present an integrated iTRAQ-based quantification pipeline to characterize proteins modulated during neuroblastoma differentiation. SH-SY5Y cells are widely used as in vitro neuron-like model for studying several aspects of neuronal behavior. In this study we differentiated SH-SY5Y cells with RA and BDNF following previously characterized protocols [4], [16]. Extracted proteins from undifferentiated cells (undiff-SH-SY5Y) and differentiated cells (diff-SH-SY5Y) conditions were digested and subsequently labeled with iTRAQ-4plex and combined in a 1:1:1:1 ratio, labeling was performed for two independent biological experiments and three technical replicates for each condition. Our proteomic analysis also incorporates phosphopeptide enrichment for detecting key phosphorylation sites and phosphoproteins regulated during neuronal differentiation; data from total proteome and phosphoproteome were subjected to statistical analysis based on the averages of 115/114 and 117/116 iTRAQ ratios, GO and KEGG terms enrichment analysis, and interactive network analysis to evaluate key hubs in regulated proteins and their influence in pathways related to neuronal development (Fig. 1). Results presented in this study provide information about differences at proteome level between undiff-SH-SY5Y and diff-SH-SY5Y confirming previous studies and showing additional information about potential markers related to neuroblastoma and neuronal differentiation that might be explored in future research.
Fig. 1.
Experimental pipeline. Undiff/diff-SH-SY5Y cells were subjected to lysis and subsequently proteins were digested using trypsin, the resulting peptides were labeled with iTRAQ-4plex and combined in 1:1:1:1 ratio. Phosphorylated peptides were enriched with SIMAC strategy; multiphosphorylated peptides were analyzed directly by LC–MS/MS while monophosphorylated and nonphosphorylated peptides were prefractionated offline using HILIC. Finally, bioinformatics and statistical analysis were performed.
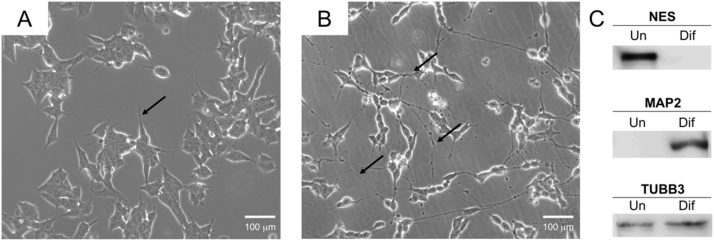
Induced differentiation with RA/BDNF treatment resulted in the appearance of cell projections, which closely resemble neurites, along with strong inhibition of cell proliferation, as previously described [4], [16]. Fig. 2A and B show changes in morphology of diff-SH-SY5Y compared with undiff-SH-SY5Y. Diff-SH-SY5Y present longer projections emerging from the cell body [10] coherent with increased abundancy of MAP2, a neuronal cytoskeletal marker and an indicative of neuronal differentiation (Fig. 2C). Nestin (NES), a marker of neuronal stem cells is present in undiff-SH-SY5Y and is not detected after differentiation. In both undifferentiated and differentiated cells β-III-tubulin is present as a marker of neuronal lineage [9], [22] (Fig. 2C).
Fig. 2.
Cellular morphology of SH-SY5Y cells. (A) Undifferentiated cells, arrow shows short cellular projections. (B) Differentiated cells after sequential incubation with RA/BDNF, arrows show extension and connection of neurites and vesicular transport along them. (C) Protein expression analysis of neuron differentiation markers nestin (NES), microtubule-associated protein 2 (MAP2) and tubulin β-III (TUBB3) using Western blot.
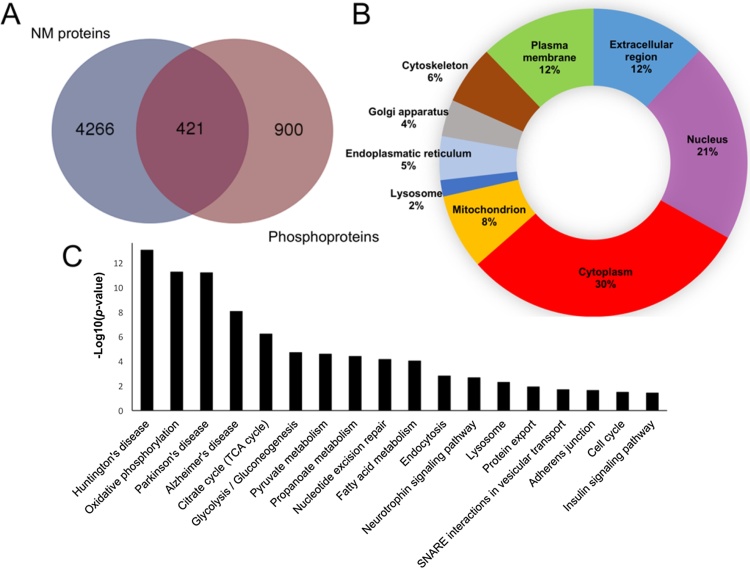
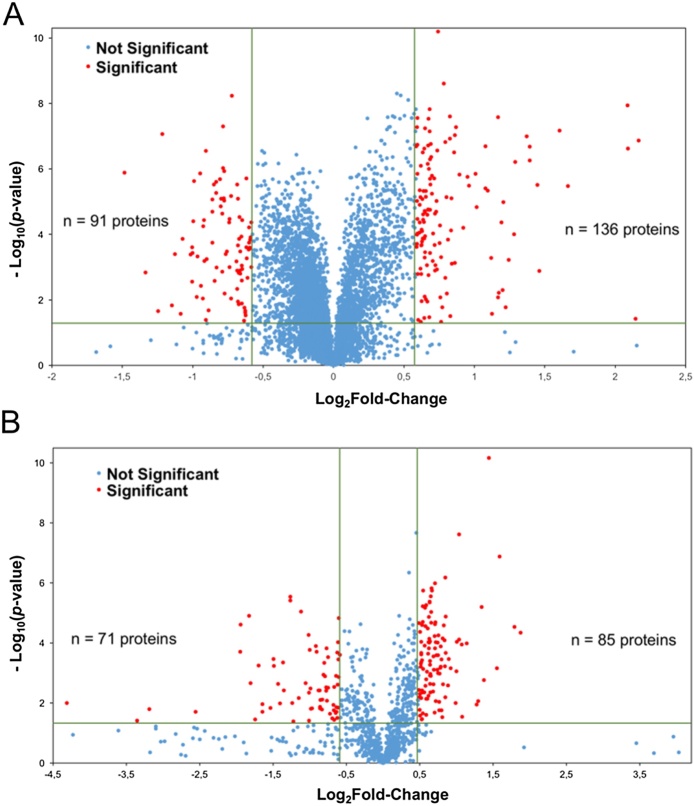
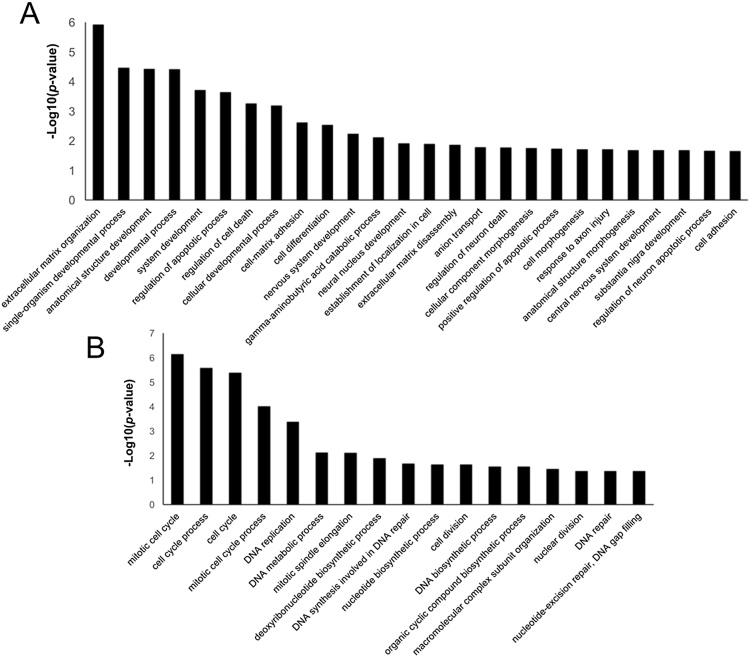
Our analysis identified a total of 5587 protein groups considering a false discovery rate of 1%, 1321 of total protein identified were phosphoproteins with 2683 phosphosites with score >75 assigned by the ptmRS algorithm (Tables 1 and S1–S4, Fig. 3A). Gene ontology was applied to classify identified proteins according to their subcellular localizations. Proteins were widely distributed in different cell compartments demonstrating that extraction protocol was not biased to any cell compartment (Fig. 3B). As summarized in Fig. 3C, the main KEGG pathways enriched in data set were related to some aspects of neuronal phenotype: SNARE interaction in vesicular transport, neurotrophin signaling, metabolic pathways and Huntington, Parkinson and Alzheimer diseases. This proteomic profile corroborates the use of differentiated SH-SY5Y cells as a model to study phenotypic alterations observed in these diseases [23], [24]. To identify the differentially expressed proteins, the main Log2 transformed averages of iTRAQ normalized ratios from each biological and/or technical replicates were plotted in volcano plot (Fig. 4). A total of 366 proteins that presented a mean expression fold change ≥1.5 or ≤0.67 and p-value <0.05 were considered as differentially abundant between two conditions of cell culture. In diff-SH-SY5Y 207 proteins were upregulated; 67 of them with at least one phosphorylation site regulated. On the other hand, 159 proteins were downregulated; 63 of them with at least one phosphorylation site regulated (Tables 1, S5 and S6). According with their GO annotations, top ranked proteins described in Table 2, Table 3 showed regulation of previously reported neuronal markers (nestin, protein Tau (MAPT), microtubule-associated protein 2, gamma enolase or neuron specific enolase (ENO), cellular retinoic acid-binding protein 2 (CRABP2), among others), together with Western blot analysis described in Fig. 2C, these results validate the cell differentiation and quantification method, thereby, they constitute a quality control of presence of regulated proteins. In addition, we describe some potential makers which are not fully explored in SH-SY5Y (Table 2, Table 3). GO annotation analysis showed that upregulated proteins are closely related to neuronal development. Also, attachment to extracellular matrix and regulation of apoptosis are important biological process during differentiation (Figs. 5 A and S1); in contrast, downregulated proteins were related to cell proliferation, DNA synthesis and cell cycle regulation (Figs. 5B and S2). The same GO analysis was done for proteins regulated from phosphopeptide enrichment. Upregulated phosphoproteins showed multiple GO terms strongly associated to neuronal development and behavior as neuron projection, axogenesis, synapse, among others (Fig. S3), downregulated phosphoproteins were involved in RNA splicing, cell cycle and chromosome organization during cell division (Fig. S4). Diverse studies use undifferentiated SH-SY5Y cells as neuronal in vitro model [24], [25], [26], [27], our data suggest that differentiated cells are more adequate for those experiments because they have a differential molecular background and molecular mechanisms compared to undiff-SH-SY5Y which confer a more mature neuron phenotype compassing neuronal pathways only detected in differentiated cells.
Table 1.
Summarized data about proteomics and phosphoproteomics analyses.
| Total Proteins | 5587 |
| Phosphoproteins | 1321 |
| Total Unique Peptides | 29078 |
| Unique Phosphopeptides | 2283 |
| Phosphosites (Score >75) | 2683 |
| Proteins upregulated | 207 |
| Proteins downregulated | 159 |
| Phosphoproteins upregulated | 67 |
| Phosphoproteins downregulated | 63 |
| Ratio S:T:Y | 22:4:1 |
Fig. 3.
Features of the proteomic data set of SH-SY5Y cells differentiation from iTRAQ 4-plex shotgun analysis. (A) Venn diagram of the overlap of non- modified proteins (NM) and phosphoproteins. (B) Subcellular localization of identified proteins. (C) KEGG enriched pathways of proteomic data set obtained from DAVID.
Fig. 4.
Volcano plot representing the protein abundance changes between undiff-SH-SY5Y and diff-SH-SY5Y. Proteins were considered regulated if fold-change values were ≥1.5 or ≤0.67 with p-value < 0.05. (A) Total proteome without phosphopeptide enrichment (227 regulated proteins). (B) Phosphopeptide enrichment (156 regulated proteins). Combination of both data sets showed regulation of 366 proteins.
Table 2.
Top ranked upregulated proteins based on their GO annotations. The phosphorylated peptide and protein ratios are shown as the average ratios between replicates and correspond to the diff-SHSY5Y/undiff-SHSY5Y ratio. The site indicates the phosphorylated residue, its position in the protein and its ratio in parenthesis.
| Uniprot Accesion | Description | Protein Ratio | p-value | Phosphosite Ratio |
|---|---|---|---|---|
| Q96QR8 | Transcriptional activator protein Pur-beta (PURB) | 0.10 | 3.94E-02 | |
| Q16637 | Survival motor neuron protein (SMN1) | 0.30 | 3.58E-02 | Ser28(0.53); Ser31(0.53) |
| P24941 | Cyclin-dependent kinase 2 (CDK2) | 0.31 | 5.64E-04 | Thr14(0.31); Tyr15(0.31) |
| Q15911 | Zinc finger homeobox protein 3 (ZFHX3) | 0.38 | 1.03E-02 | |
| P48681 | Nestin (NES) | 0.42 | 3.89E-06 | Ser471(0.56) |
| Q16512 | Serine/threonine-protein kinase N1 (PKN1) | 0.42 | 1.00E-02 | Ser916(0.42) |
| Q9Y6A5 | Transforming acidic coiled-coil-containing protein 3 (TACC3) | 0.45 | 1.48E-02 | |
| P16949 | Stathmin (STMN1) | 0.46 | 1.51E-03 | Ser16(0.41); Ser38(0.44) |
| O43663 | Protein regulator of cytokinesis 1 (PRC1) | 0.46 | 4.00E-04 | |
| Q13233 | Mitogen-activated protein kinase kinase kinase 1 (MAP3K1) | 0.46 | 9.18E-06 | S923(0.46) |
| Q66K89 | Transcription factor E4F1 (E4F1) | 0.47 | 2.64E-02 | Thr325(0.64) |
| P31350 | Ribonucleoside-diphosphate reductase subunit M2 (RRM2) | 0.48 | 1.38E-04 | |
| Q96AA8 | Janus kinase and microtubule-interacting protein 2 (JAKMIP2) | 0.50 | 4.00E-05 | |
| P17252 | Protein kinase C alpha type (PRKCA) | 0.50 | 2.37E-06 | |
| Q9Y328 | Neuron-specific protein family member 2 (NSG2) | 0.51 | 8.18E-03 | |
| Q9NXR1 | Nuclear distribution protein nudE homolog 1 (NDE1) | 0.51 | 1.52E-02 | Ser282(0.51) |
| Q93045 | Stathmin-2 (STMN2) | 0.51 | 4.65E-04 | |
| P29966 | Myristoylated alanine-rich C-kinase substrate (MARCKS) | 0.53 | 5.68E-05 | Ser26/27(0.44) |
| O43602 | Neuronal migration protein doublecortin (DCX) | 0.53 | 2.80E-07 | Ser415(0.59) |
| P05129 | Protein kinase C gamma type (PRKCG) | 0.55 | 1.51E-02 | Thr514(0.55) |
| Q04724 | Transducin-like enhancer protein 1 (TLE1) | 0.55 | 8.04E-03 | |
| Q9BTT0 | Acidic leucine-rich nuclear phosphoprotein 32 family member E (ANP32E) | 0.56 | 1.68E-02 | |
| P06400 | Retinoblastoma-associated protein (RB1) | 0.57 | 1.16E-03 | Ser249(0.57) |
| Q16566 | Calcium/calmodulin-dependent protein kinase type IV (CAMK4) | 0.57 | 2.06E-06 | |
| Q9NSC5 | Homer protein homolog 3 (HOMER3) | 0.61 | 2.28E-02 | Ser159(0.65) |
| O95343 | Homeobox protein SIX3 (SIX3) | 0.62 | 2.16E-04 | |
| Q71RC2 | La-related protein 4 (LARP4) | 0.63 | 3.70E-02 | Ser583(0.63) |
| Q9NZC4 | ETS homologous factor (EHF) | 0.64 | 1.29E-02 | |
| P20839 | Inosine-5′-monophosphate dehydrogenase 1 (IMPDH1) | 0.64 | 1.13E-03 | |
| Q08211 | ATP-dependent RNA helicase A (DHX9) | 0.64 | 2.16E-02 | Ser87(0.64) |
| P53804 | E3 ubiquitin-protein ligase TTC3 (TTC3) | 0.65 | 2.43E-02 | |
| P0CG34 | Thymosin beta-15A (TMSB15A) | 0.65 | 5.41E-04 |
Table 3.
Top ranked upregulated proteins based on their GO annotations. The phosphorylated peptide and protein ratios are shown as the average ratios between replicates and correspond to the diff-SHSY5Y/undiff-SHSY5Y ratio. The site indicates the phosphorylated residue, its position in the protein and its ratio in parenthesis.
| Uniprot Accesion | Description | Protein Ratio | p-value | Phosphosite ratio |
|---|---|---|---|---|
| Q8N163 | Cell cycle and apoptosis regulator protein 2 (CCAR2) | 1.51 | 5.36E-08 | |
| P27338 | Amine oxidase [flavin-containing] B (MAOB) | 1.52 | 1.62E-03 | |
| Q9UMS6 | Synaptopodin-2 (SYNPO2) | 1.53 | 4.48E-06 | Ser204(1.6); Ser234(1.81); Ser604(1.85) |
| P21796 | Voltage-dependent anion-selective channel protein 1 (VDAC1) | 1.53 | 4.32E-05 | Ser104(1.55) |
| P10636 | Microtubule-associated protein (MAPT) | 1.56 | 2.32E-03 | Thr720(1.56)/Ser721(1.56) |
| O75781 | Paralemmin-1 (PALM) | 1.56 | 5.44E-04 | Ser116(1.54); Ser124(1.51); Ser162(1.60) |
| P27816 | Microtubule-associated protein (MAP4) | 1.56 | 4.46E-03 | Ser297(1.56); Ser2073(1.72) |
| Q96FJ2 | Dynein light chain 2, cytoplasmic (DYNLL2) | 1.57 | 3.49E-04 | |
| O75369 | Filamin-B (FLNB) | 1.58 | 7.00E-06 | Ser856(2.50) |
| Q9BY67 | Cell adhesion molecule 1 (CADM1) | 1.59 | 2.48E-05 | |
| P19320 | Vascular cell adhesion protein 1 (VCAM1) | 1.61 | 1.78E-05 | |
| P07196 | Neurofilament light polypeptide (NEFL) | 1.62 | 1.74E-07 | |
| P05556 | Integrin beta-1 (ITGB1) | 1.62 | 4.26E-07 | Tyr195(1.51) |
| Q8IZJ1 | Netrin receptor UNC5B (UNC5B) | 1.63 | 2.07E-02 | Ser528(1.62) |
| P10415 | Apoptosis regulator Bcl-2 (BCL2) | 1.63 | 6.55E-04 | |
| Q2M2I8 | AP2-associated protein kinase 1 (AAK1) | 1.63 | 2.40E-05 | |
| P46821 | Microtubule-associated protein 1B (MAP1B) | 1.64 | 1.04E-06 | Ser1154(1.68); Ser1265(1.54); Ser1396(1.73) |
| Q8ND76 | Cyclin-Y (CCNY) | 1.64 | 2.04E-05 | Ser326(1.63) |
| Q09666 | Neuroblast differentiation-associated protein AHNAK (AHNAK) | 1.65 | 1.26E-03 | Ser4850(1.50) |
| O15394 | Neural cell adhesion molecule 2 (NCAM2) | 1.66 | 1.18E-03 | |
| P22694 | cAMP-dependent protein kinase catalytic subunit beta (PRKACB) | 1.68 | 2.56E-04 | Ser345(1,68) |
| Q13098 | COP9 signalosome complex subunit 1 (GPS1) | 1.69 | 1.02E-03 | Ser509(1,68) |
| Q12888 | Tumor suppressor p53-binding protein 1 (TP53BP1) | 1.72 | 2.45E-09 | Ser500(1,53) |
| O75396 | Vesicle-trafficking protein SEC22b (TP53BP1) | 1.72 | 1.33E-04 | Ser(1.72) |
| P09104 | Gamma-enolase (ENO2) | 1.73 | 3.98E-05 | |
| P11137 | Microtubule-associated protein 2 (MAP2) | 1.73 | 1.50E-07 | Ser833(2.22); Thr1154(1.52); Ser1782(1.54); Ser1790(1.79) |
| P54826 | Growth arrest-specific protein 1 (GAS1) | 1.75 | 6.37E-03 | |
| P07197 | Neurofilament medium polypeptide (NEFM) | 1.75 | 3.00E-02 | Ser837(1.75) |
| P13591 | Neural cell adhesion molecule 1 (NCAM1) | 1.78 | 7.50E-05 | |
| Q14195 | Dihydropyrimidinase-related protein 3 (DPYSL3) | 1.79 | 7.92E-06 | Ser522(2.00) |
| O60716 | Catenin delta-1 (CTNND1) | 1.80 | 6.05E-04 | Ser857(1.77) |
| O00443 | Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha (DPYSL3) | 1.81 | 2.15E-04 | Ser259(1.81) |
| Q9Y639 | Neuroplastin (NPTN) | 1.86 | 1.49E-06 | |
| P56199 | Integrin alpha-1 (ITGA1) | 2.25 | 2.64E-08 | |
| Q8N111 | Cell cycle exit and neuronal differentiation protein 1 (CEND1) | 2.29 | 4.28E-05 | |
| P49006 | MARCKS-related protein (MARCKSL1) | 2.72 | 6.82E-11 | Thr148(2.93) |
| P29373 | Cellular retinoic acid-binding protein 2 (CRABP2) | 3.17 | 3.29E-06 |
Fig. 5.
Gene Ontology (GO) of the differentially regulated proteins from total proteome data set (without phosphopeptide enrichment). (A) Biological process terms for upregulated proteins. (B) Biological Process terms for downregulated proteins. Only terms with p values less than 0.01 are shown.
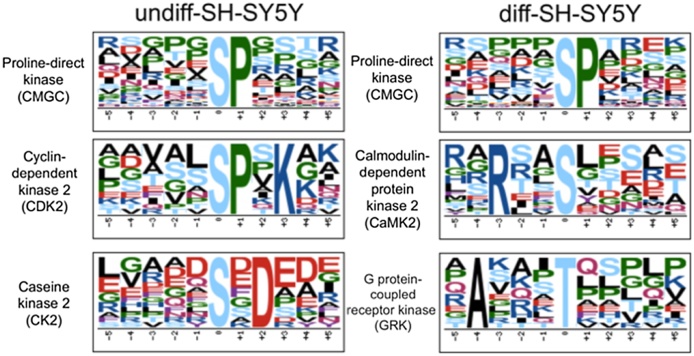
Regulated phosphosites from phosphoproteome data set were analyzed to determine the over-represented sequence motifs using the Motif-x algorithm (Fig. 6). The motif pS-P was found over-represented in both undifferentiated and differentiated cells, 67.2% of regulated phosphopeptides contain this sequence consensus which is a known target for the proline-directed kinase group (CMCG) [28]. The motifs pS-P-X-K (7.8% of total phosphosites) target of cyclin-dependent kinase 2 (CDK2) and surrounded acidic pS-X-D (9.2% of total phosphosites) target of casein kinase 2 (CK2), were the second most relevant over-represented motifs in undiff-SH-SY5Y, which is coherent to a role of these kinases in regulation of cell cycle division [29], [30] and corroborates the enrichment of proliferation related biological processes in GO. Analysis of phosphorylation motifs in differentiated cells revealed that the second most relevant over-represented motifs in these cells were R-X-X-pS (12.4% of total phosphosites) for calmodulin-dependent protein kinase 2 (CaMK2) and a threonine motif belonging to G protein-coupled receptor kinase (GRK) group (7.8% of total phosphosites). CaMK2 participates in neuronal differentiation of NSCs and in synaptic transmission and plasticity in mature neurons [31], [32]. GRKs are involved in dopamine receptor function in striatal neurons and phosphorylation of cytoskeleton proteins, histone deacetylases, among others [33], [34]. These kinases also match the biological processes enriched in GO, which correlate to neuronal function.
Fig. 6.
Over-represented phosphorylation motifs regulated in undiff-SH-SY5Y and diff-SH-SY5Y created by the Motif-x algorithm (significance of 0.0001) and their respective kinases.
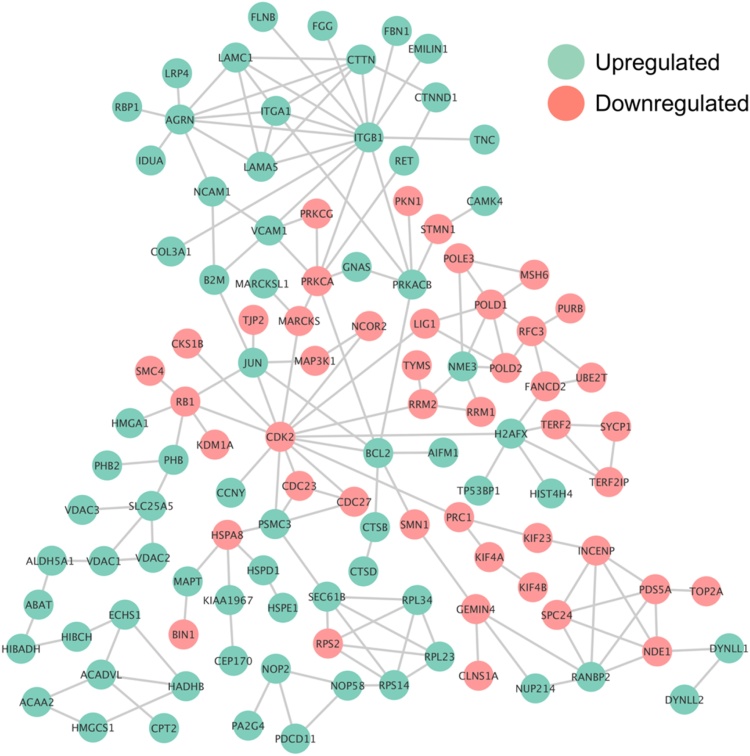
To analyze interaction maps between differentially expressed proteins in diff-SH-SY5Y and undiff-SH-SY5Y, interactive network analysis was performed using STRING 10.0 algorithm plugin in Cytoscape software [35], [36]. Only interactions with high confidence score were considered and proteins without any interaction were excluded, hubs were defined by the presence of six or more interactions (Fig. 7). We observed hubs corresponding to downregulated kinases such as CDK2 and protein kinase Cγ (PRKCG) and upregulated proteins such as BCL2 and integrins (ITGB1/ITGA1). The interactive network also showed alteration of specific signaling pathways and processes related to neuron development (Fig. S5A and B); reflecting the RA/BDNF influence in neuroblastoma differentiation; PI3K-Akt and MAPK pathways were significantly regulated by different hubs in interaction network, most of them more abundant in diff-SH-SY5Y (Fig. S5A). This observation is consistent with GO enrichment analysis and the phenotypic characteristics of undiff/diff-SH-SY5Y cells.
Fig. 7.
Top interactions of proteins presenting variation in abundance between diff-SH-SY5Y and undiff-SH-SY5Y obtained from STRING 10.0 and Cytoscape. The interactive network was constructed with high confidence score data (0.9), proteins without any interaction were excluded from the analysis.
The interactive network presented isoforms of protein kinase C (PKC), MAP3K1, eukaryotic elongation factor 2 kinase (EEFK2) and CDK2 downregulated in diff-SH-SY5Y (Fig. 7). Neuron specific isotype PKCγ (PRKCG) is a kinase localized in soma, dendritic spines, axon and synaptic terminals, playing important role in early development of hippocampal neurons in initial stages of synaptic formation [37]. In addition, PKCγ showed a significant interaction with PKCα isoform which has multiple roles in cellular development. Particularly, it promotes cellular growth activating ERK/MAPK pathway [38]. PKCγ⁄PKCα regulates multiple signaling pathways including oxytocin, Wnt or mTOR (Fig. S5A), which are commonly associated to cell migration, ribosome formation and survival cell proliferation [39], [40]. This is consistent with proliferative and immature neuron properties of undiff-SH-SY5Y mentioned above. One of the targets of PKC is the myristoylated alanine-rich C-kinase substrate (MARCKS) protein that is involved in regulation of cytoskeleton dynamics during embryonic development in brain and in the cells surrounding the neural tube. MARCKS crosslinks actin filaments at the plasma membrane probably to facilitate morphological changes at the membrane and, in addition, has the ability to concentrate signaling molecules within membrane microdomains and interacts with adhesion molecules facilitating synapse, functions that are controlled by its effector domain regulated in phosphorylation dependent manner [41]. Phosphorylation on Ser26, detected in our data set, is specific in neurons but does not affect its association with membranes [42]. Given these characteristics, MARCKS could be explored as marker for SH-SY5Y differentiation.
Phosphorylated protein kinase A (PRKACB) was more expressed in diff-SH-SY5Y (Fig. 7), this kinase is involved in dopamine reception, excitability and control of plasticity and reward-related behavior of spiny neurons [43], characteristics that are correlated with dopaminergic nature described for diff-SH-SY5Y[4]. PRKACB appears as key kinase regulated in different pathways (MAPK, GnRH, among others) (Fig. S-5A) apparently resulting in PRKACB correlation with different neuron phenotypes enriched in the interactive network, such as Parkinson’s disease and GABAergic, serotoninergic, glutamatergic and cholinergic synapse (Fig. S-5B). An interesting interaction partner/target of PRKACB, the phosphorylated stathmin 1 (STMN1) was predominant in undiff-SH-SY5Y. STMN1 binds tubulin in phosphorylation-dependent manner for microtubule dynamics regulation and is highly expressed and regulated in neurons during embryonic development [44]. Unphosphorylated STMN1 is related to plasticity maintenance and neuronal regeneration in hippocampal neurons [44] while temporal phosphorylation in Ser16 and Ser38 is involved in negative regulation of tubulin sequestering, allowing microtubule polymerization. This is important for cell motility, cell proliferation and cell mitosis regulation [44], [45] and our data suggest that STMN1 might be considered as a proliferation-restricted marker in SH-SY5Y regulated by extracellular signaling molecules (Table 2).
The interactive network also exhibited hubs containing apoptosis-related proteins (Fig. 7). Bcl-2 (or BCL2), an anti-apoptotic protein, is strongly involved in neuron survival during neurotrophic factor withdrawal-induced death for control of neuronal population in neuron precursors [46], [47] consistent with the signaling targets of BDNF in diff-SH-SY5Y [4]. The phosphorylated Apoptosis Inducing Factor, Mitochondria Associated 1 (AIFM1) is a flavoprotein essential for nuclear disassembly in apoptotic cells, according to its role in neuronal cells. AIFM1 probably plays a role with BCL2 in diff-SH-SY5Y because its liberation and transport to nucleus from mitochondria depends on BCL2 [48]. Mutations and depletion of AIFM1 result in deficiencies in oxidative phosphorylation and Cowchock syndrome in motor neurons and its regulation contributes to Parkinson’s disease phenotype [48], [49], [50]. These characteristics could be also attributed to diff-SH-SY5Y used in this study. Element survival motor neuron protein 1 (SMN1) interacts and is phosphorylated by BCL2 and was more abundant in undiff-SH-SY5Y. SMN1 is commonly reported as a marker for spinal muscular atrophy. SMN1 interacts with cytoskeleton in growth cones and neurites of primary motor neurons and it is suggested that SMN1 supports motor neuron survival [51], [52], [53]. Decrease in abundance of SMN1 in differentiated cells parallels the decrease in abundance observed during neural development [51], suggesting that more differentiated cells present lesser amounts of this proteins as a general trend. The best-characterized function of SMN1 is the oligomerization and interaction with Gemins 2–8 (Gemin 4 was upregulated in our data set, Table S-6), a complex that assembles small nuclear ribonucleoprotein particles (snRNPs), key components of the spliceosome; this assembly machinery is highly regulated by PKA phosphorylation at Ser28 and Ser31 residues of SMN1 [54], [55] which were regulated in our data set (Table 2). More specific roles for AIFM1 and SMN1 in SH-SY5Y cells have to be established, therefore, because their differential abundance and biological functions, they might be considered as potential differentiation markers during neuroblastoma differentiation.
The adhesiveness and migration are well defined characteristics of SH-SY5Y cells [4] and for this reason regulation of proteins related to ECM interaction was expected because during differentiation they play a key role in neurite development [9], [10]. As fundamental receptor in cell adhesion and migration, integrin β1 (ITGB1) appears as key upregulated protein in diff-SH-SY5Y (Fig. 7, Table 3). Previous reports determinates its role in neuronal polarization, neurite outgrowth and branching by interaction with focal adhesion kinase (FAK) in embryonic mouse dorsal root ganglion (DRG) neurons [56] and neurogenesis, glial differentiation, neuronal migration and formation of neuronal layers in cerebral cortex [57]. Expression of ITGB1 is accompanied by its heterodimeric partner in αβ complex integrin α1 (ITGA1) expression and common integrin ligands as tenascin C (TNC) laminin subunits α and γ (LAMA5 and LAMC1) and collagen (COL3A1). Presence of those receptors is critical in neuronal survival and neurite outgrowth of embryonic hippocampal neurons [58], [59]. Particularly, ITGB1-LAMC1 proteins are relevant in neural tube dynamics but repressed upon differentiation [60] suggesting that expression of these proteins might be restricted to specific type of neurons and/or specific phases of their development. As expected, the integrins and interactive partners are involved in crucial processes of neural differentiation as focal adhesion, ECM-receptor interaction and cell adhesion molecules (Fig. S-5B).
Among the upregulated ITGB1/ITGA1 partners, we found the expression of neuron-secreted proteoglycan agrin (AGRN) upregulated in our data set. AGRN is involved in neuromuscular junction by clustering acetylcholine receptors in synaptogenesis interacting with co-receptor LRP4 [61], also found upregulated. Despite the literature description of agrin as an adherence factor in proliferative undiff-SH-SY5Y cells [62], we suggest that its main role could be more related to synapses of neurites due to the phenotype and GO enrichment of synapses part terms observed in differentiated cells (Figs. 2, 5 and S3). Other upregulated ITGB1 interaction partner closely related with stabilization of ECM interactions is the extracellular glycoprotein EMILIN-1 (Elastin Microfibrils Interface Located proteINs) which promotes extrinsic apoptotic pathways and regulation of Hedgehog and Wnt ligand availability in embryonic fibroblasts. In addition, EMILIN-1 has a antiproliferative response in trophoblasts in synergy with integrins α4/α9β1 [63]. Additionally, the upregulated neural cell adhesion molecule (NCAM), from neuroectodermal origin, appears as downstream response in plasticity changes to the signals of NMDA receptor activity in differentiated SH-SY5Y cells with RA [64]. Similar to agrin, this protein is believed to be a motility factor in neuroblastoma cells when suffers polysialylation and we speculate whether NCAM has low grade of polysialylation in diff-SH-SY5Y that confers more adhesive properties [65]. In addition to substrate adherent regulated proteins, we found the upregulation of δ-catenin (CTNND1), a brain-specific member of the adherens junction. CTNND1 regulates synapses [66] and can be related to the formation of synapses in diff-SH-SY5Y cells.
Overall, the iTRAQ-based quantitative proteomics pipeline, the GO profile, phosphosite analysis and interactive network analysis of regulated proteins addressed here, covered essential features of long-term differentiated SH-SY5Y cells and led us to suggest additional differentiation markers in neuroblastoma cells. During the change of proliferative nature to a neural-like identity, cytoskeleton rearrangements are remarkable. We highlight the relevance of MARCKS, STMN1 and SMN1 in undiff-SH-SY5Y; in the context of neuroblastoma differentiation, their roles could be interpreted as earlier agents responsible for cytoskeleton regulation. On the other hand, interaction with ECM to stimulate neurite outgrowth and control of neuronal population by apoptosis were the main process enriched in diff-SH-SY5Y. Upregulation of interactive partners of ITGB1/ITGA1 such as NCAM, AGRN and CTNND1 reflect changes in the interaction with ECM which correlate to synaptogenesis, plasticity and synapse localization. Apoptosis mechanisms present during differentiation could be explored, starting by upregulated proteins AIFM1 and EMILIN-1. Taken together, our results represent a resource to the field of neuronal differentiation and neurodegenerative diseases, with impact in neuroscience and neuroproteomics research.
4. Conclusions
Here, we applied an integrative iTRAQ-based quantitative proteomics and phosphopeptide enrichment pipelines to study the overall protein expression changes involved in RA/BDNF-based differentiation of SH-SY5Y cells. Compiled information about differentially expressed proteins and regulated phosphosites was closely related to neuronal differentiation aspects such as regulation of proliferation rate, apoptosis, ECM interaction and neurodegenerative associated proteins. Western blot analysis of common neuronal differentiation markers, GO enrichment analysis and regulation of selected pathways in diff-SH-SY5Y reflected a more mature neuron phenotype than undiff-SH-SY5Y used in this work. Together, these analyses suggest some candidates (MARCKS, STMN1, AIFM1, SMN1, AGRN and CTNND1) to be used as potential differentiation markers, which could be applied in a variety of studies using neuroblastoma cells as human in vitro neuronal-like model.
Finally, we would like to emphasize the relevance to use induced differentiated SH-SY5Y cells in human neuronal-like cell line research, specially to cover analysis of pathways involved in diverse neuronal diseases, neuronal development processes, synapses and axon pathfinding.
Conflict of interest
The authors declare no competing financial interest.
Availability of raw files
The RAW files used in this work have been deposited to the Mass Spectrometry Interactive Virtual Environment – MassIVE online repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the data set identifier MSV000080133.
Acknowledgments
The present work was supported by Conselho Nacional de Desenvolvimento Cientifíco e Tecnologico (CNPq) and Fundação Carlos Chagas Filho de Amparo à Pesquisa o Estado do Rio de Janeiro (FAPERJ). The authors also thank Drs. Ana Gisele C. Neves-Ferreira and Richard H. Valente, from the Laboratory of Toxinology/FIOCRUZ, for mass spectrometric analyses using the Q Exactive Plus instrument (acquired through a joint FAPERJ and PIDTS/FIOCRUZ funding) and Laboratório de Apoio ao Desenvolvimento Tecnologico (LADETEC). J. R. M. acknowledges doctoral scholarship support from FAPERJ.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.euprot.2017.06.001.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Gibbons H.M., Dragunow M. Adult human brain cell culture for neuroscience research. Int. J. Biochem. Cell Biol. 2010;42:844–856. doi: 10.1016/j.biocel.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Korecka J.A., Levy S., Isacson O. In vivo modeling of neuronal function, axonal impairment and connectivity in neurodegenerative and neuropsychiatric disorders using induced pluripotent stem cells. Mol. Cell. Neurosci. 2015;73:3–12. doi: 10.1016/j.mcn.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Melo-Braga M.N., Meyer M., Zeng X., Larsen M.R. Characterization of human neural differentiation from pluripotent stem cells using proteomics/PTMomics-current state-of-the-art and challenges. Proteomics. 2015;15:656–674. doi: 10.1002/pmic.201400388. [DOI] [PubMed] [Google Scholar]
- 4.Kovalevich J., Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013;1078:9–21. doi: 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodeur G.M. Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 6.Cheung N.-K.V., a Dyer M. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edsjö A., Holmquist L., Pahlman S. Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin. Cancer Biol. 2007;17:248–256. doi: 10.1016/j.semcancer.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Ross R.A., Spengler B.A. Human neuroblastoma stem cells. Semin. Cancer Biol. 2007;17:241–247. doi: 10.1016/j.semcancer.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Constantinescu R., Constantinescu A.T., Reichmann H., Janetzky B. Neuronal differentiation and long-term culture of the human neuroblastoma line SH-SY5Y. J. Neural Transm. Suppl. 2007:17–28. doi: 10.1007/978-3-211-73574-9_3. http://www.ncbi.nlm.nih.gov/pubmed/17982873 [DOI] [PubMed] [Google Scholar]
- 10.Agholme L., Lindström T., Kgedal K., Marcusson J., Hallbeck M. An in vitro model for neuroscience: differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J. Alzheimer’s Dis. 2010;20:1069–1082. doi: 10.3233/JAD-2010-091363. [DOI] [PubMed] [Google Scholar]
- 11.Cheung Y.T., Lau W.K.W., Yu M.S., Lai C.S.W., Yeung S.C., So K.F., Chang R.C.C. Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology. 2009;30:127–135. doi: 10.1016/j.neuro.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Rhinn M., Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K., Tamiya-Koizumi K., Hagiwara K., Ito H., Takagi A., Kojima T., Suzuki M., Iwaki S., Fujii S., Nakamura M., Banno Y., Kannagi R., Tsurumi T., Kyogashima M., Murate T. Role of down-regulated neutral ceramidase during all-trans retinoic acid-induced neuronal differentiation in SH-SY5Y neuroblastoma cells. J. Biochem. 2012;151:611–620. doi: 10.1093/jb/mvs033. [DOI] [PubMed] [Google Scholar]
- 14.López-Carballo G., Moreno L., Masiá S., Pérez P., Barettino D. Activation of the phosphatidylinositol 3-kinase/Akt signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J. Biol. Chem. 2002;277:25297–25304. doi: 10.1074/jbc.M201869200. [DOI] [PubMed] [Google Scholar]
- 15.Korecka J.A., van Kesteren R.E., Blaas E., Spitzer S.O., Kamstra J.H., Smit A.B., Swaab D.F., Verhaagen J., Bossers K. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Encinas M., Iglesias M., Liu Y., Wang H., Muhaisen A., Ceña V., Gallego C., Comella J.X. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 17.Emdal K.B., Pedersen A.-K., Bekker-Jensen D.B., Tsafou K.P., Horn H., Lindner S., Schulte J.H., Eggert A., Jensen L.J., Francavilla C., Olsen J.V. Temporal proteomics of NGF-TrkA signaling identifies an inhibitory role for the E3 ligase Cbl-b in neuroblastoma cell differentiation. Sci. Signal. 2015;8 doi: 10.1126/scisignal.2005769. ra40–ra40. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Fonslow B.R., Shan B., Baek M.C., Yates J.R. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimmino F., Spano D., Capasso M., Zambrano N., Russo R., Zollo M., Iolascon A. Comparative proteomic expression profile in all-trans retinoic acid differentiated neuroblastoma cell line. J. Proteome Res. 2007;6:2550–2564. doi: 10.1021/pr060701g. [DOI] [PubMed] [Google Scholar]
- 20.Sitek B., Apostolov O., Stühler K., Pfeiffer K., Meyer H.E., Eggert A., Schramm A. Identification of dynamic proteome changes upon ligand activation of Trk-receptors using two-dimensional fluorescence difference gel electrophoresis and mass spectrometry. Mol. Cell. Proteom. 2005;4:291–299. doi: 10.1074/mcp.M400188-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Thingholm T.E., Larsen M.R. Sequential elution from IMAC (SIMAC): An efficient method for enrichment and separation of mono-and multi-phosphorylated peptides. Methods Mol. Biol. 2016;2016:147–160. doi: 10.1007/978-1-4939-3049-4_10. [DOI] [PubMed] [Google Scholar]
- 22.Katsetos C.D., Legido A., Perentes E., Mörk S.J. Class III beta-tubulin isotype: a key cytoskeletal protein at the crossroads of developmental neurobiology and tumor neuropathology. J. Child Neurol. 2003;18:851–866. doi: 10.1177/088307380301801205. discussion 867. [DOI] [PubMed] [Google Scholar]
- 23.Jämsä A., Hasslund K., Cowburn R.F., Bäckström A., Vasänge M. The retinoic acid and brain-derived neurotrophic factor differentiated SH-SY5Y cell line as a model for Alzheimer’s disease-like tau phosphorylation. Biochem. Biophys. Res. Commun. 2004;319:993–1000. doi: 10.1016/j.bbrc.2004.05.075. [DOI] [PubMed] [Google Scholar]
- 24.Krishna A., Biryukov M., Trefois C., Antony P.M.A., Hussong R., Lin J., Heinäniemi M., Glusman G., Köglsberger S., Boyd O., van den Berg B.H.J., Linke D., Huang D., Wang K., Hood L., Tholey A., Schneider R., Galas D.J., Balling R., May P. Systems genomics evaluation of the SH-SY5Y neuroblastoma cell line as a model for Parkinson’s disease. BMC Genom. 2014;15:1154. doi: 10.1186/1471-2164-15-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scifo E., Szwajda A., Debski J., Uusi-Rauva K., Kesti T., Dadlez M., Gingras A.C., Tyynelä J., Baumann M.H., Jalanko A., Lalowski M. Drafting the CLN3 protein interactome in SH-SY5Y human neuroblastoma cells: a label-free quantitative proteomics approach. J. Proteome Res. 2013;12:2101–2115. doi: 10.1021/pr301125k. [DOI] [PubMed] [Google Scholar]
- 26.Yusuf M., Leung K., Morris K.J., Volpi E.V. Comprehensive cytogenomic profile of the in vitro neuronal model SH-SY5Y. Neurogenetics. 2013;14:63–70. doi: 10.1007/s10048-012-0350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Z., Baykal A.T., Gao H., Quezada H.C., Zhang H., Bereczki E., Serhatli M., Baykal B., Acioglu C., Wang S., Ioja E., Ji X., Zhang Y., Guan Z., Winblad B., Pei J.J. MTor is a signaling hub in cell survival: a mass-spectrometry-based proteomics investigation. J. Proteome Res. 2014;13:2433–2444. doi: 10.1021/pr500192g. [DOI] [PubMed] [Google Scholar]
- 28.Adams J.A. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 29.Litchfield D.W. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chohan T.A., Qian H., Pan Y., Chen J. Cyclin-dependent kinase-2 as a target for cancer therapy: progress in the development of CDK2 inhibitors as anti-Cancer agents. Curr. Med. Chem. 2015;22:237–263. doi: 10.2174/0929867321666141106113633. [DOI] [PubMed] [Google Scholar]
- 31.Colbran R.J. Targeting of calcium/calmodulin-dependent protein kinase II. Biochem. J. 2004;378:1–16. doi: 10.1042/BJ20031547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo-Braga M.N., Schulz M., Liu Q., Swistowski A., Palmisano G., Engholm-Keller K., a Jakobsen L., Zeng X., Larsen M.R. Comprehensive quantitative comparison of the membrane proteome, phosphoproteome and sialiome of human embryonic and neural stem cells. Mol. Cell. Proteom. 2013;13:311–328. doi: 10.1074/mcp.M112.026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich E.V., Tesmer J.J.G., Mushegian A., Gurevich V.V. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol. Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurevich E.V., Gainetdinov R.R., Gurevich V.V. G protein-coupled receptor kinases as regulators of dopamine receptor functions. Pharmacol. Res. 2016;111:1–16. doi: 10.1016/j.phrs.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., Von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito N., Shirai Y. Protein kinase C gamma (PKC gamma): function of neuron specific isotype. J. Biochem. 2002;132:683–687. doi: 10.1093/oxfordjournals.jbchem.a003274. [DOI] [PubMed] [Google Scholar]
- 38.Nakashima S. Protein kinase Ca (PKCa): regulation and biological function. J. Biochem. 2002;132:669–675. doi: 10.1093/oxfordjournals.jbchem.a003272. http://www.ncbi.nlm.nih.gov/pubmed/12417014 [DOI] [PubMed] [Google Scholar]
- 39.Komiya Y., Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laplante M., Sabatini D.M. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. 122/20/3589 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brudvig J.J., Weimer J.M. X MARCKS the spot: myristoylated alanine-rich C kinase substrate in neuronal function and disease. Front. Cell. Neurosci. 2015;9:1–10. doi: 10.3389/fncel.2015.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tinoco L.W., Fraga J.L., Anobom C.D., Zolessi F.R., Obal G., Toledo A., Pritsch O., Arruti C. Structural characterization of a neuroblast-specific phosphorylated region of MARCKS. Biochim. Biophys. Acta – Proteins Proteom. 2014;1844:837–849. doi: 10.1016/j.bbapap.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 43.Nagai T., Nakamuta S., Kuroda K., Nakauchi S., Nishioka T., Takano T., Zhang X., Tsuboi D., Funahashi Y., Nakano T., Yoshimoto J., Kobayashi K., Uchigashima M., Watanabe M., Miura M., Nishi A., Kobayashi K., Yamada K., Amano M., Kaibuchi K. Phosphoproteomics of the dopamine pathway enables discovery of rap1 activation as a reward signal in vivo. Neuron. 2016;89:550–565. doi: 10.1016/j.neuron.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Chauvin S., Sobel A. Neuronal stathmins: a family of phosphoproteins cooperating for neuronal development, plasticity and regeneration. Prog. Neurobiol. 2015;126:1–18. doi: 10.1016/j.pneurobio.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- 46.Akhtar R.S., Ness J.M., Roth K.A. Bcl-2 family regulation of neuronal development and neurodegeneration. Biochim. Biophys. Acta – Mol. Cell Res. 2004;1644:189–203. doi: 10.1016/j.bbamcr.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 47.Shacka J.J., Roth K.A. Regulation of neuronal cell death and neurodegeneration by members of the Bcl-2 family: therapeutic implications. Curr Drug Targets CNS Neurol Disord. 2005;4:25–39. doi: 10.2174/1568007053005127. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15723611 [DOI] [PubMed] [Google Scholar]
- 48.Hangen E., De Zio D., Bordi M., Zhu C., Dessen P., Caffin F., Lachkar S., Perfettini J.-L., Lazar V., Benard J., Fimia G.M., Piacentini M., Harper F., Pierron G., Vicencio J.M., Bénit P., de Andrade A., Höglinger G., Culmsee C., Rustin P., Blomgren K., Cecconi F., Kroemer G., Modjtahedi N. A brain-specific isoform of mitochondrial apoptosis-inducing factor: AIF2. Cell Death Differ. 2010;17:1155–1166. doi: 10.1038/cdd.2009.211. [DOI] [PubMed] [Google Scholar]
- 49.Diodato D., Tasca G., Verrigni D., D’Amico A., Rizza T., Tozzi G., Martinelli D., Verardo M., Invernizzi F., Nasca A., Bellacchio E., Ghezzi D., Piemonte F., Dionisi-Vici C., Carrozzo R., Bertini E. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. Eur. J. Hum. Genet. 2016;24:463–466. doi: 10.1038/ejhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sevrioukova I.F. Structure/function relations in AIFM1 variants associated with neurodegenerative disorders. J. Mol. Biol. 2016:1–16. doi: 10.1016/j.jmb.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Giavazzi A., Setola V., Simonati A., Battaglia G. Neuronal-specific roles of the survival motor neuron protein: evidence from survival motor neuron expression patterns in the developing human central nervous system. J. Neuropathol. Exp. Neurol. 2006;65:267–277. doi: 10.1097/01.jnen.0000205144.54457.a3. [DOI] [PubMed] [Google Scholar]
- 52.Kerr D.A., Nery J.P., Traystman R.J., Chau B.N., Hardwick J.M. Survival motor neuron protein modulates neuron-specific apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13312–13317. doi: 10.1073/pnas.230364197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez T.L., Kong L., Wang X., Osborne M.A., Crowder M.E., Van Meerbeke J.P., Xu X., Davis C., Wooley J., Goldhamer D.J., Lutz C.M., Rich M.M., Sumner C.J. Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. J. Neurosci. 2012;32:8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grimmler M., Bauer L., Nousiainen M., Korner R., Meister G., Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burnett B.G., Muñoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. Regulation of SMN protein stability. Mol. Cell. Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ribeiro A., Balasubramanian S., Hughes D., Vargo S., Powell E.M., Leach J.B. b1–Integrin cytoskeletal signaling regulates sensory neuron response to matrix dimensionality. Neuroscience. 2013;248:67–78. doi: 10.1016/j.neuroscience.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid R.S., Anton E.S. Role of integrins in the development of the cerebral cortex 2. Cereb. Cortex. 2003;13:219–224. doi: 10.1093/cercor/13.3.219. [DOI] [PubMed] [Google Scholar]
- 58.Rigato F., Garwood J., Calco V., Heck N., Faivre-Sarrailh C., Faissner A. Tenascin-C promotes neurite outgrowth of embryonic hippocampal neurons through the alternatively spliced fibronectin type III BD domains via activation of the cell adhesion molecule F3/contactin. J. Neurosci. 2002;22:6596–6609. doi: 10.1523/JNEUROSCI.22-15-06596.2002. 20026682 [doi]\r22/15/6596 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Z.-L., a Indyk J., Strickland S. The hippocampal laminin matrix is dynamic and critical for neuronal survival. Mol. Biol. Cell. 2003;14:2665–2676. doi: 10.1091/mbc.E02-12-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao X., Pfaff S.L., Gage F.H. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zong Y., Jin R. Structural mechanisms of the agrin-LRP4-MuSK signaling pathway in neuromuscular junction differentiation. Cell. Mol. Life Sci. 2013;70:3077–3088. doi: 10.1007/s00018-012-1209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolfram T., Spatz J.P., Burgess R.W. Cell adhesion to agrin presented as a nanopatterned substrate is consistent with an interaction with the extracellular matrix and not transmembrane adhesion molecules. BMC Cell Biol. 2008;9:64. doi: 10.1186/1471-2121-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pivetta E., Danussi C., Wassermann B., Modica T.M.E., Del Bel Belluz L., Canzonieri V., Colombatti A., Spessotto P. Neutrophil elastase-dependent cleavage compromises the tumor suppressor role of EMILIN1. Matrix Biol. 2014;34:22–32. doi: 10.1016/j.matbio.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 64.Singh J., Kaur G. Transcriptional regulation of polysialylated neural cell adhesion molecule expression by NMDA receptor activation in retinoic acid-differentiated SH-SY5Y neuroblastoma cultures. Brain Res. 2007;1154:8–21. doi: 10.1016/j.brainres.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 65.Valentiner U., Mühlenhoff M., Lehmann U., Hildebrandt H., Schumacher U. Expression of the neural cell adhesion molecule and polysialic acid in human neuroblastoma cell lines. Int. J. Oncol. 2011;39:417–424. doi: 10.3892/ijo.2011.1038. [DOI] [PubMed] [Google Scholar]
- 66.Matter C., Pribadi M., Liu X., Trachtenberg J.T. δ-Catenin is required for the maintenance of neural structure and function in mature cortex in vivo. Neuron. 2009;64:320–327. doi: 10.1016/j.neuron.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.