We report the first use of near-infrared spectroscopy for rapid, low-cost, noninvasive detection of Zika virus in Aedes aegypti.
Abstract
The accelerating global spread of arboviruses, such as Zika virus (ZIKV), highlights the need for more proactive mosquito surveillance. However, a major challenge during arbovirus outbreaks has been the lack of rapid and affordable tests for pathogen detection in mosquitoes. We show for the first time that near-infrared spectroscopy (NIRS) is a rapid, reagent-free, and cost-effective tool that can be used to noninvasively detect ZIKV in heads and thoraces of intact Aedes aegypti mosquitoes with prediction accuracies of 94.2 to 99.3% relative to quantitative reverse transcription polymerase chain reaction (RT-qPCR). NIRS involves simply shining a beam of light on a mosquito to collect a diagnostic spectrum. We estimated in this study that NIRS is 18 times faster and 110 times cheaper than RT-qPCR. We anticipate that NIRS will be expanded upon for identifying potential arbovirus hotspots to guide the spatial prioritization of vector control.
INTRODUCTION
Zika virus (ZIKV) is a mosquito-borne virus of the family Flaviviridae transmitted to humans primarily by Aedes mosquitoes (1, 2). ZIKV has been of international concern due to its rapid spread in the Americas in 2015–2016 (3) and its association with neurological disorders, such as Guillain-Barré syndrome in adults (4) and microcephaly in infants (5). Timely detection of ZIKV transmission in mosquito populations would be critical for identifying potential hotspots and preventing outbreaks (6). However, adequate mosquito surveillance is missing from most Zika-endemic countries like Brazil, which reported over 1.6 million ZIKV infections between January 2015 and November 2016 (7, 8). Real-time or rapid mapping of arbovirus transmission at the neighborhood or community level could guide the spatial prioritization of vector control (9). To estimate transmission risk, the abundance of mosquito vectors is often used as a proxy for entomological risk and can be determined by trapping adults or using larval measures, such as the Breteau index (10). However, the relationship between vector densities and the prevalence of arbovirus infections in humans is frequently inconsistent across different communities, possibly because vector-human contact also influences infection (11). Viral infection rates among mosquitoes can provide more information about infection risk; however, many arbovirus surveillance programs in low- and middle-income countries do not have the resources to carry out pathogen screening with currently available techniques.
Conventional pathogen screening in mosquito vectors involves time-consuming analysis using molecular techniques, such as quantitative reverse transcription polymerase chain reaction (RT-qPCR), which are often too expensive for the analysis of the large numbers of mosquitoes needed for mosquito surveillance programs (12). For example, when ZIKV was first detected in A. aegypti mosquitoes in Brazil, 550 A. aegypti were grouped into 198 pools, and only three of the pools were ZIKV positive, or a prevalence of ≤0.9% (13). Therefore, routine pathogen surveillance in mosquitoes to identify transmission hotspots would involve testing thousands of mosquitoes each month. The use of honey-baited nucleic acid preservation cards to capture arboviruses from mosquito saliva has been proposed to eliminate the problem of testing individual mosquitoes, but the technique still requires RT-qPCR to test for viral RNA (14). Most of the currently available rapid diagnostic tests (RDTs) for arbovirus detection in mosquitoes rely on the capture of viral antigens through an enzyme-linked immunosorbent assay. A test targeting the dengue nonstructural protein 1 (NS1) has been developed for dengue virus detection in A. aegypti. The NS1 test is sensitive and more affordable than RT-qPCR, and it can be coupled with electronic biosensors for point-of-use diagnosis (15–17). However, the specificity of RDTs using antigen detection is a major limitation, given that cross-reactivity of antigen-based assays among flaviviruses has been reported (17). Similarly, a plaque reduction neutralization test can be used to measure virus-specific neutralizing antibodies, but this test shows cross-reaction with antibodies from other flaviviruses (17). Given that RT-qPCR is expensive and time-consuming and there is still uncertainty about the specificity of many antigen-based tests, arbovirus surveillance programs in endemic countries often rely solely on human case reports (7, 18), a strategy which has been inadequate for containing arbovirus outbreaks.
We sought to understand whether near-infrared spectroscopy (NIRS) can be used as an alternative to molecular techniques to detect ZIKV in A. aegypti mosquitoes. NIRS is a light-based method of chemical analysis that has been used for decades across fields such as agriculture (19), pharmaceuticals (20), and medicine (21). It classifies biological samples based on the type and the concentration of chemical compounds present in the samples. The technique is easy to use, rapid, and noninvasive, and it does not require reagents to operate (19). Previous applications of NIRS to vector-borne diseases include the age determination of biting midges (22) and house flies (23) and the sex separation of tsetse flies (24). More recently, NIRS has been used on major malaria vectors in Africa to noninvasively and rapidly determine their age and species (25, 26) and to predict the age (27, 28) and the presence of endosymbiont Wolbachia (29) in A. aegypti. In addition to insect characterization, NIRS has also been tested in small pilot studies for detecting hepatitis C and human immunodeficiency viruses in human serum and plasma (30, 31) and influenza virus in nasal fluids (32). This study represents the first investigation of the capability of NIRS to noninvasively detect arboviruses in mosquito vectors.
RESULTS
To understand whether NIRS can distinguish ZIKV-infected from uninfected A. aegypti mosquitoes, we reared adult females (cohort 1, n = 275) to 5 to 6 days of age. We fed half with ZIKV-infected blood and the other half with uninfected blood as a control group. Using a near-infrared (NIR) spectrometer, we collected spectra from the head/thorax region of mosquitoes at 4 and 7 days post infection (dpi) and then confirmed the ZIKV loads by RT-qPCR. We found that analysis of NIR spectra by cross-validation and partial least squares (PLS) regression distinguished ZIKV-infected from uninfected mosquitoes with 92.5% accuracy (n = 120). When we applied the ZIKV training model to a subset of samples from cohort 1 that were excluded from the training model (n = 155), we found an overall predictive accuracy of 99.3% (Fig. 1). These results suggest that a randomly selected mosquito from cohort 1 could be predicted as infected with ZIKV or uninfected more than 9 out of 10 times based on the spectra of the other mosquitoes in this cohort. The sensitivity appeared to improve with increasing incubation period (Table 1).
Fig. 1. NIRS differentiation of ZIKV-infected and uninfected A. aegypti mosquitoes using leave-one-out cross-validation analysis.
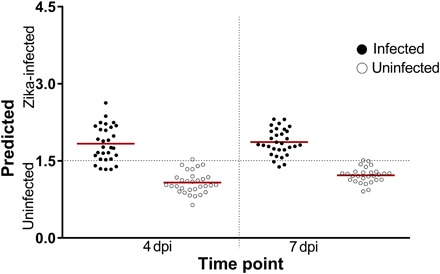
Shown are the predictions of the leave-one-out cross-validation analysis carried out on a subset of mosquitoes in cohort 1 (n = 120) at 4 and 7 dpi. Each circle represents an individual mosquito; infection status confirmed by RT-qPCR is indicated by solid (infected) or empty (uninfected) circles. Red lines indicate the mean prediction value for each group. The vertical axis indicates infection status as predicted by NIRS, with the dotted line indicating the classification cutoff point. Infected mosquitoes shown below the dotted line and uninfected mosquitoes shown above the dotted line were falsely predicted.
Table 1. Sensitivities and specificities of the ZIKV training model.
Predicted sensitivity [true positive rate (TPR)] and specificity (SPC) of NIRS are shown for cohort 1 samples used in the training set at 4 dpi (n = 61) and 7 dpi (n = 59) and cohort 1 samples that were excluded from the training set (prevalidation) at 4 dpi (n = 65) and 7 dpi (n = 90). Predicted sensitivity and specificity of NIRS are shown for heads/thoraces of cohort 2 samples at 4 dpi (n = 136), 7 dpi (n = 138), and 10 dpi (n = 138) and for abdomens at 4 dpi (n = 136), 7 dpi (n = 138), and 10 dpi (n = 138).
| Experiment set | 4 dpi | 7 dpi | 10 dpi | |||
| %TPR | %SPC | %TPR | %SPC | %TPR | %SPC | |
| Cohort 1. Training set | 83.3 | 96.8 | 93.5 | 96.4 | — | — |
| Cohort 1. Prevalidation set | 100.0 | 94.1 | 100.0 | 100.0 | — | — |
| Cohort 2. Test set (heads/thoraces) |
98.7 | 98.3 | 100.0 | 98.3 | 100.0 | 86.7 |
| Cohort 2. Test set (abdomens) | 98.7 | 85.0 | 96.2 | 80.0 | 97.4 | 68.3 |
To determine whether the ZIKV training model developed from mosquitoes in cohort 1 was robust enough to detect ZIKV infections in a set of independent mosquitoes (that is, unknown to the model), we reared a second cohort (cohort 2, n = 412) of female A. aegypti mosquitoes and infected half with ZIKV. In addition to determining the overall predictive accuracy of the ZIKV training model, we determined the accuracy of the model on the spectra collected from both heads/thoraces and abdomens and at a longer incubation period. Accordingly, the spectra were collected from the heads/thoraces and abdomens of ZIKV-infected and control mosquitoes in cohorts 2 at 4, 7, and 10 dpi. Using the ZIKV training model developed from cohort 1, we predicted the mosquitoes in cohort 2 as ZIKV-infected or uninfected with an overall predictive accuracy of 97.3% for heads/thoraces (n = 412) and 88.8% (n = 412) for abdomens (Fig. 2). Table 1 shows the sensitivity and specificity at various time points for cohort 1 and cohort 2 mosquito samples for heads/thoraces and abdomens as predicted by NIRS.
Fig. 2. NIRS predictions of ZIKV infection in heads/thoraces and abdomens of A. aegypti mosquitoes on a blind data set.
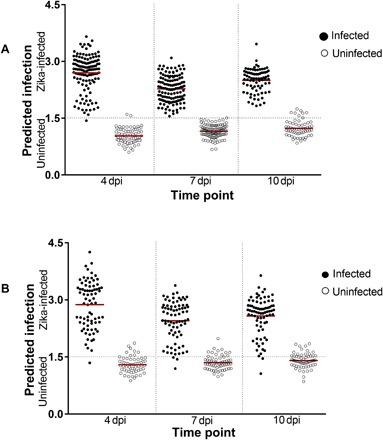
(A) Predicted infections at 4, 7, and 10 dpi in heads/thoraces for the subset of cohort 1 (n = 155) that were excluded from the model and all cohort 2 samples (n = 412). (B) Predictions for abdomens from cohort 2 samples (n = 412). Each circle represents an individual mosquito; infection status confirmed by RT-qPCR is indicated by solid (infected) or empty (uninfected) circles. Red lines indicate the mean prediction value for each group. The vertical axis indicates infection status as predicted by NIRS, with the dotted line indicating the classification cutoff point. Infected mosquitoes shown below the dotted line and uninfected mosquitoes shown above the dotted line were falsely predicted.
The highest accuracy (227 of 228 mosquitoes correctly predicted) was obtained when heads/thoraces were scanned at 7 dpi, suggesting that this could be the time point when the effects of ZIKV infection on the chemistry of the mosquito are most pronounced. Although prediction accuracy was lower for 10 dpi than for the first two time points, the sensitivity of this assay was still remarkably high considering that this time point was not accounted for in the training model. The observed higher prediction accuracies of spectra from heads/thoraces compared to spectra from abdomens could be due to the fact that abdomens were not included in the training model. Alternatively, the higher accuracy of head/thorax spectra could suggest that ZIKA infection generates a more marked chemical change in the head/thorax than in the abdomen. Given that the Brazilian strain of ZIKV is only detectable in the heads/thoraces of A. aegypti beginning at 4 to 7 dpi (33), there may be an enhanced mosquito immune response to the virus at this time point. Nevertheless, accuracies of 84.8 to 92.6% in the abdomens indicate that the model is still of value for Zika detection in these tissues, particularly when the head/thorax is unavailable for analysis. Together, these results support the external validity of the NIRS-based model for the identification of ZIKV-infected A. aegypti mosquitoes.
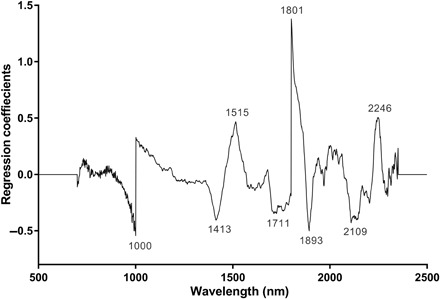
To further understand the spectral changes influencing the differentiation of ZIKV-infected and uninfected mosquitoes by NIRS, we analyzed the raw spectra (Fig. 3) using PLS regression. There were noticeable differences between the shapes of the average spectra of ZIKV-infected (red) and uninfected mosquitoes (blue), indicating differences in their absorbance characteristics. Important wavelengths identified in the PLS regression coefficients plot (Fig. 4) were observed at 1000, 1413, 1515, 1711, 1801, 1893, 2109, and 2246 nm. Peaks at 1000 and 1801 nm should be considered with caution, as they may be an artifact of the spectrometer used. The spectrometer used in this study contains three sensors: a silicon sensor (350 to 1000 nm) and two InGaAs sensors (1001 to 1800 nm and 1801 to 2500 nm). The transition points between these sensors are 1000 and 1801 nm. However, it is also possible that the peaks at 1000 and 1801 nm truly correspond to molecular components of ZIKV. Further studies using improvements to this instrument or using different equipment are needed to eliminate this source of error. A stronger light source, more efficient fibers, or improved sensors in the current spectrometer could improve the signal-to-noise ratio and minimize or eliminate this noise. The spectral region encompassing peaks at 1413 and 1515 nm is associated with the O–H and N–H overtones (19), which might correspond to glycoproteins in the viral envelope (34) or to antimicrobial peptides produced by the mosquito immune system as part of the Toll pathway (35). Given that NIR spectroscopy has only recently been introduced to the fields of virology (36) and medical entomology (22, 24–27, 29), there is limited information on the NIR bands corresponding to specific physical and chemical changes associated with viral infections in mosquitoes. Future studies should use more informative mid-infrared sensors to elucidate the chemical changes influencing the observed differences.
Fig. 3. Average NIR spectra in the 350- to 2500-nm region from heads/thoraces of ZIKV-infected (red) and uninfected (blue) A. aegypti mosquitoes.
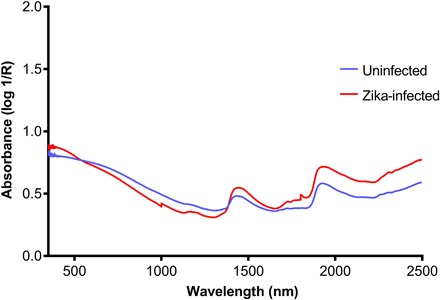
Fig. 4. Regression coefficients using eight factors in the PLS model based on the NIR spectra in the 700- to 2350-nm region for differentiating ZIKV-infected from uninfected A. aegypti.
Here, we calculated that ZIKV detection by NIRS was 18 times faster and 110 times cheaper than RT-qPCR. Given the technique’s rapid, high-throughput, and reagent-free nature, hundreds of samples can be processed in a day by unskilled technicians, enabling rapid predictions of potential disease transmission, which in turn could facilitate a rapid action plan to stop major disease outbreaks.
DISCUSSION
The value of NIR spectroscopy as a research tool in vector-borne diseases is only beginning to be realized. In this novel application of NIR spectroscopy to detect ZIKV in mosquitoes, we found NIR prediction accuracies of 94.2 to 99.3% in differentiating ZIKV-infected and uninfected A. aegypti when heads/thoraces are analyzed. Because our aim was to detect ZIKV in vivo, we were unable to distinguish the contributions of the virus itself from those of the mosquito immune response when analyzing the chemical and physical changes associated with changes in the NIR spectra. Future studies of ZIKV in serum and a variety of media using mid-infrared sensors can clarify which chemical compounds are responsible for the differences that we observed between infected and uninfected mosquitoes.
At present, NIR detection of ZIKV is not “field ready,” as the heterogeneity of field-collected mosquitoes must be incorporated into NIR models to achieve robust validity for field applications. Field samples are much more likely to be desiccated and damaged than laboratory mosquitoes, and they represent a variety of ages and feeding statuses. Therefore, the next steps in the development of the technique should include testing it on field populations of A. aegypti to determine its sensitivity and specificity for arbovirus detection relative to RT-qPCR. The technique must also be evaluated against different arboviruses in a variety of media to determine the effects of background and to test for any cross-reactivity.
One major drawback of NIRS is the need to purchase a spectrometer with high enough sensitivity to identify the virus. The estimated cost of commercially available benchtop spectrometers that are sensitive enough (defined by a high signal-to-noise ratio) to collect spectra from mosquitoes is $60,000. Because of the budget restrictions often faced by the public health sector in many disease-endemic countries, it may be necessary to establish NIRS processing centers to scan mosquito samples sent in from entire regions or countries. Because there is no need for reagents, the cost of acquiring such a spectrometer would be recovered after scanning approximately 10,000 mosquitoes. More affordable microspectrometers are commercially available for less than $500; however, the sensitivity of these spectrometers needs to be tested because of their lower resolution, inherent noise, and more limited range of wavelengths. As the technology evolves and the market for spectrometers grows, the cost and size of spectrometers are expected to decrease significantly. Another major drawback of NIRS is the need to develop a separate data library for each arbovirus or pathogen of interest. However, once a robust data library for ZIKV detection in field mosquitoes is established for a given country, the data library could then be packaged and used by mosquito control programs throughout the country to screen for ZIKV infections in unknown field samples without the need for building new models. If NIRS is as accurate in detecting ZIKV in field mosquitoes as it is in the laboratory, then the speed, cost effectiveness, and noninvasiveness of the technique would make it an attractive technology for the surveillance of mosquito-borne viruses in the future.
Here, we calculated that ZIKV detection by NIRS was 18 times faster and 110 times cheaper than RT-qPCR. However, the user often becomes faster with repeated use of the spectrometer. If used as a prescreening tool for RT-qPCR or other molecular techniques, NIRS can save time and money by quickly separating infected from uninfected mosquitoes. Because there is no need to dissect or manipulate mosquitoes for NIRS, it is also a safer tool than those assays that require handling potentially infected mosquito tissues. NIRS data can be analyzed by a number of commercially available software packages, as well as free, open-source programs built in R. The data can also be saved in a variety of formats that could be incorporated into decision support systems based on multicriteria system analysis, similar to the system proposed for Ebola control (37). Information on pathogen circulation has been the key element missing from predictive models of arboviruses thus far. Therefore, we believe that rapid arbovirus detection in mosquitoes by NIRS has the potential to improve the quality and timeliness of surveillance data, thereby accelerating responses to arbovirus outbreaks.
MATERIALS AND METHODS
Mosquito rearing
All A. aegypti mosquitoes were reared in the insectary of the Laboratório de Mosquitos Transmissores de Hematozoários, Pavilhão Carlos Chagas, Instituto Oswaldo Cruz, Rio de Janeiro, Brazil, under identical conditions: 27°C, 70% humidity, and 12:12 hours light/dark. Larvae were fed on TetraMin tropical flakes (Tetra Melle) until pupation. Pupae were transferred into cages (40 cm × 40 cm × 30 cm) for emergence. Adults were allowed to mate for 3 to 4 days and were given 10% sugar solution ad libitum up until 36 hours before infection. Two separate cohorts of mosquitoes reared at different time points were used in this experiment: One was used to train the model (cohort 1), and the second was used to test the accuracy of the model (cohort 2).
Experimental ZIKV infection
We used the currently circulating strain of Brazilian ZIKV [BRPE243/2015 (BRPE)] (38), which was isolated from a Zika-infected patient in late 2015 and maintained in cell culture. Viral titers were quantified via plaque-forming assay before experimental infection. In two separate experiments, 1 ml of ZIKV-infected supernatant was harvested from culture, mixed with 2 ml of rabbit blood and 0.5 mM adenosine 5′-triphosphate (as a phagostimulant), and was used to orally infect female A. aegypti mosquitoes at 5 to 6 days post-emergence. The viral titer used for mosquitoes in both cohorts was 1.9 × 106 PFU (plaque-forming units)/ml. For each cohort and at each time point, there was a control group of mosquitoes fed with blood and virus-free culture medium.
Scanning of mosquitoes
Mosquitoes from cohort 1 were collected for scanning at 4 and 7 dpi. For cohort 2, mosquitoes were collected for scanning at 4, 7, and 10 dpi. Mosquitoes were killed just before scanning by placing them in a closed jar with an acetate-soaked cotton ball for 1 min. Whole mosquitoes were then arranged on their sides on a Spectralon diffuse reflectance stage for scanning, as previously described (26). The heads/thoraces of mosquitoes in cohort 1 and the heads/thoraces and abdomens (collecting one spectrum per body part) of mosquitoes in cohort 2 were positioned in the center of the light from a fiber-optic probe. Spectra were collected using a LabSpec 4 i NIR spectrometer (Malvern Panalytical) with an internal 18.6-W light source and a 3.2-mm-diameter fiber-optic probe (Model 135325 Rev B, ASD Inc.), according to established protocols (26). The average raw spectra collected from the heads/thoraces of ZIKV-infected and uninfected mosquitoes are shown in Fig. 3.
Confirmation of ZIKV infection by RT-qPCR
Viral RNA was extracted from sampled mosquitoes with a QIAamp Viral RNA Mini kit (Qiagen). Viral RNA detection and quantification in each individual were performed through RT-qPCR with the SuperScript III Platinum One-Step qRT-PCR Kit (Invitrogen) in QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) using previously published primers and amplification conditions (13). Virus copy numbers were calculated through absolute quantification in each run using a standard curve of a seven-point dilution series (102 to 108 copies) of in vitro transcribed ZIKV RNA (39). Virus titers for mosquitoes at each time point are shown in Fig. 5.
Fig. 5. ZIKV load in A. aegypti cohorts at 4, 7, and 10 dpi with a Brazilian ZIKV isolate (BRPE243/2015) provided at a titer of 1.9 × 106 PFU/ml.
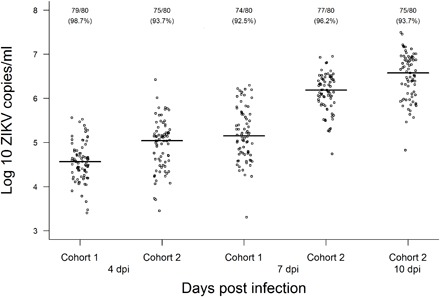
Viral copies were determined by RT-qPCR in individual mosquito homogenates with a standard curve of a seven-point dilution series (102 to 108 copies/ml) of in vitro transcribed ZIKV RNA. The number of infected mosquitoes followed by the infection rate (in parentheses) is shown for each condition.
Data analysis
We trained and tested the ZIKV detection model according to previously published methods (26). Spectra from 120 (n = 61 infected, n = 59 uninfected) mosquitoes in cohort 1 were included in the analysis, having excluded the spectra of those samples that failed to amplify ZIKV RNA in RT-qPCR. To minimize noise, only the wavelengths between 700 and 2350 nm were analyzed, and spectra were mean-centered before applying the PLS regression method in GRAMS Plus/IQ software (Thermo Galactic). The number of PLS factors was chosen on the basis of the predicted residual error sum of squares and the regression coefficients plot. In the PLS model, a value of “1” was assigned to the spectra of uninfected mosquitoes, and a value of “2” was assigned to the spectra of ZIKV-infected mosquitoes. A model-predicted value of 1.5 was considered the threshold for correct classification. The resulting eight-factor model was then applied to a prevalidation set of mosquitoes from cohort 1 that were not included in the model (n = 155 spectra from heads/thoraces) and independent prediction sets (n = 412 spectra from heads/thoraces and n = 412 spectra from abdomens of mosquitoes in cohort 2, having excluded the spectra of those samples that failed to amplify in RT-qPCR). The accuracy of the model to detect the presence or absence of ZIKV was tested on mosquitoes at 4, 7, and 10 dpi. The regression coefficient plot used to differentiate ZIKV-infected from uninfected female A. aegypti is shown in Fig. 4.
Cost and time effectiveness calculation
It took approximately 30 s to position one mosquito on its side on a Spectralon diffuse reflectance stage and to collect a spectrum from its head/thorax using NIRS. Of this time, collecting the spectrum took approximately 15 s. Using NIRS, a total of 50 min was required to scan 100 mosquitoes. Comparatively, it took 2 full days (7.5 hours per day × 2 days = 900 min) to prepare mosquitoes, extract DNA, and run RT-qPCR to detect ZIKA infection in 100 mosquitoes. This makes NIRS 18 times faster than RT-qPCR (900 min/50 min = 18). We disregarded the delivery time for PCR reagents, which in some countries can be in the order of months.
For cost analysis, we excluded the cost of a NIR spectrometer and a real-time PCR thermal cycler. After the initial purchase of a NIR spectrometer, no reagents were required; therefore, operational running costs for NIRS were only related to labor. For this study, we used a research assistant for $10 per hour to carry out NIRS experiments for 50 min and scan 100 samples. Comparatively, RT-qPCR costs ~$10 per sample processed ($1000 for 100 samples). Labor costs for RT-qPCR were estimated at $10 per person per hour working for 900 min amounting to $160 for running RT-qPCR on 100 samples for 2 days. As per our calculations, NIRS in this study was 116 times cheaper than PCR ($1160/$10 = 116).
Acknowledgments
We thank M. C. Bonaldo for providing quantified RNA of ZIKV. In addition, we thank S. Ciocchetta for collecting pilot data before this study. We also thank M. T. Petersen and I. Dias da Silveira for technical support. Lastly, we thank the two reviewers for the feedback on the initial draft of this manuscript. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). USDA is an equal opportunity provider and employer. Funding: This work was supported by the United States Agency for International Development Combating Zika and Future Threats (award no. AID-OAA-F-16-00100), the Brazilian Research Councils MCTIC/FNDCT-CNPq/MEC-CAPES/MS-Decit E14/2016 (award no. 440929/2016-4), and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro E18/2015. Grand Challenges Canada Stars for Global Health funded by the Government of Canada provided funding for pilot data collection (award no. 0781-06). The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript. The corresponding author had full access to all the data in the study and had the final responsibility for the decision to submit for publication. Author contributions: J.N.F., R.M.-d.-F., and M.T.S.-L. conceived the study; J.N.F., L.M.B.d.S., G.A.G., M.R.D., F.E.D., R.M.-d.-F., and M.T.S.-L. designed the experiments; L.M.B.d.S., T.C.-C., G.A.G., and M.R.D. performed the experiments; M.T.S.-L. carried out the analysis; and J.N.F. wrote the manuscript. All authors reviewed the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
REFERENCES AND NOTES
- 1.Musso D., Gubler D. J., Zika virus. Clin. Microbiol. Rev. 29, 487–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chouin-Carneiro T., Vega-Rua A., Vazeille M., Yebakima A., Girod R., Goindin D., Dupont-Rouzeyrol M., Lourenço-de-Oliveira R., Failloux A. B., Differential susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika virus. PLOS Negl. Trop. Dis. 10, e0004543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization, Mosquito (Vector) Control Emergency Response and Preparedness for Zika Virus (World Health Organization, 2016). [Google Scholar]
- 4.Oehler E., Watrin L., Larre P., Leparc-Goffart I., Lastère S., Valour F., Baudouin L., Mallet H., Musso D., Ghawche F., Zika virus infection complicated by Guillain-Barré syndrome—Case report, French Polynesia, December 2013. Euro Surveill. 19, 20720 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Cordeiro M. T., Pena L. J., Brito C. A., Gil L. H., Marques E. T., Positive IgM for Zika virus in the cerebrospinal fluid of 30 neonates with microcephaly in Brazil. Lancet 387, 1811–1812 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ayllón T., Campos R. M., Brasil P., Morone F. C., Câmara D. C. P., Meira G. L. S., Tannich E., Yamamoto K. A., Carvalho M. S., Pedro R. S., Schmidt-Chanasit J., Cadar D., Ferreira D. F., Honório N. A., Early evidence for Zika virus circulation among Aedes aegypti mosquitoes, Rio de Janeiro, Brazil. Emerg. Infect. Dis. 23, 1411–1412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Oliveira W. K., de França G. V. A., Carmo E. H., Duncan B. B., de Souza Kuchenbecker R., Schmidt M. I., Infection-related microcephaly after the 2015 and 2016 Zika virus outbreaks in Brazil: A surveillance-based analysis. Lancet 390, 861–870 (2017). [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization, Global Vector Control Response 2017–2030 (World Health Organization, 2017). [Google Scholar]
- 9.Pepin K. M., Leach C. B., Marques-Toledo C., Laass K. H., Paixao K. S., Luis A. D., Hayman D. T., Johnson N. G., Buhnerkempe M. G., Carver S., Grear D. A., Tsao K., Eiras A. E., Webb C. T., Utility of mosquito surveillance data for spatial prioritization of vector control against dengue viruses in three Brazilian cities. Parasit. Vectors 8, 98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diallo M., Dia I., Diallo D., Diagne C. T., Ba Y., Yactayo S., Perspectives and challenges in entomological risk assessment and vector control of chikungunya. J. Infect. Dis. 214, S459–S465 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Honório N. A., Nogueira R. M., Codeço C. T., Carvalho M. S., Cruz O. G., Magalhães M. A. F. M., de Araújo J. M., de Araújo E. S., Gomes M. Q., Pinheiro L. S., da Silva Pinel C., Lourenço-de-Oliveira R., Spatial evaluation and modeling of dengue seroprevalence and vector density in Rio de Janeiro, Brazil. PLOS Negl. Trop. Dis. 3, e545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu W., Novak R. J., Detection probability of arbovirus infection in mosquito populations. Am. J. Trop. Med. Hyg. 71, 636–638 (2004). [PubMed] [Google Scholar]
- 13.Ferreira-de-Brito A., Ribeiro I. P., Miranda R. M., Fernandes R. S., Campos S. S., Silva K. A., Castro M. G., Bonaldo M. C., Brasil P., Lourenço-de-Oliveira R., First detection of natural infection of Aedes aegypti with Zika virus in Brazil and throughout South America. Mem. Inst. Oswaldo Cruz 111, 655–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson B. J., Kerlin T., Hall-Mendelin S., van den Hurk A. F., Cortis G., Doggett S. L., Toi C., Fall K., McMahon J. L., Townsend M., Ritchie S. A., Development and field evaluation of the sentinel mosquito arbovirus capture kit (SMACK). Parasit. Vectors 8, 509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasik D. A., Mulchandani A., Yates M. V., Point-of-use nanobiosensor for detection of dengue virus NS1 antigen in adult Aedes aegypti: A potential tool for improved dengue surveillance. Anal. Chem. 90, 679–684 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Sylvestre G., Gandini M., de Araújo J. M., Kubelka C. F., Lourenço-de-Oliveira R., Maciel-de-Freitas R., Preliminary evaluation on the efficiency of the kit Platelia Dengue NS1 Ag-ELISA to detect dengue virus in dried Aedes aegypti: A potential tool to improve dengue surveillance. Parasit. Vectors 7, 155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aziz H., Zia A., Anwer A., Aziz M., Fatima S., Faheem M., Zika virus: Global health challenge, threat and current situation. J. Med. Virol. 89, 943–951 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Achee N. L., Gould F., Perkins T. A., Reiner R. C. Jr, Morrison A. C., Ritchie S. A., Gubler D. J., Teyssou R., Scott T. W., A critical assessment of vector control for dengue prevention. PLOS Negl. Trop. Dis. 9, e0003655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D. A. Burns, E. W. Ciurczak, Handbook of Near-Infrared Analysis (CRC press, 2008). [Google Scholar]
- 20.Roggo Y., Chalus P., Maurer L., Lema-Martinez C., Edmond A., Jent N., A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J. Pharm. Biomed. Anal. 44, 683–700 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Bale G., Elwell C. E., Tachtsidis I., From Jöbsis to the present day: A review of clinical near-infrared spectroscopy measurements of cerebral cytochrome-c-oxidase. J. Biomed. Opt. 21, 091307 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Reeves W. K., Peiris K. H. S., Scholte E.-J., Wirtz R. A., Dowell F. E., Age-grading the biting midge Culicoides sonorensis using near-infrared spectroscopy. Med. Vet. Entomol. 24, 32–37 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Perez-Mendoza J., Dowell F. E., Broce A. B., Throne J. E., Wirtz R. A., Xie F., Fabrick J. A., Baker J. E., Chronological age-grading of house flies by using near-infrared spectroscopy. J. Med. Entomol. 39, 499–508 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Dowell F., Parker A. G., Benedict M. Q., Robinson A. S., Broce A. B., Wirtz R. A., Sex separation of tsetse fly pupae using near-infrared spectroscopy. Bull. Entomol. Res. 95, 249–257 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Sikulu M., Killeen G. F., Hugo L. E., Ryan P. A., Dowell K. M., Wirtz R. A., Moore S. J., Dowell F. E., Near-infrared spectroscopy as a complementary age grading and species identification tool for African malaria vectors. Parasit. Vectors 3, 49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayagaya V. S., Michel K., Benedict M. Q., Killeen G. F., Wirtz R. A., Ferguson H. M., Dowell F. E., Non-destructive determination of age and species of Anopheles gambiae s.l. using near-infrared spectroscopy. Am. J. Trop. Med. Hyg. 81, 622–630 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Sikulu-Lord M. T., Milali M. P., Henry M., Wirtz R. A., Hugo L. E., Dowell F. E., Devine G. J., Near-infrared spectroscopy, a rapid method for predicting the age of male and female wild-type and Wolbachia infected Aedes aegypti. PLOS Negl. Trop. Dis. 10, e0005040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebman K., Swamidoss I., Vizcaino L., Lenhart A., Dowell F., Wirtz R., The influence of diet on the use of near-infrared spectroscopy to determine the age of female Aedes aegypti mosquitoes. Am. J. Trop. Med. Hyg. 92, 1070–1075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikulu-Lord M. T., Maia M. F., Milali M. P., Henry M., Mkandawile G., Kho E. A., Wirtz R. A., Hugo L. E., Dowell F. E., Devine G. J., Rapid and non-destructive detection and identification of two strains of Wolbachia in Aedes aegypti by near-infrared spectroscopy. PLOS Negl. Trop. Dis. 10, e0004759 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saade J., Pacheco M. T. T., Rodrigues M. R., Silveira L. Jr, Identification of hepatitis C in human blood serum by near-infrared Raman spectroscopy. Spectroscopy 22, 387–395 (2008). [Google Scholar]
- 31.Sakudo A., Tsenkova R., Onozuka T., Morita K., Li S., Warachit J., Iwabu Y., Li G., Onodera T., Ikuta K., A novel diagnostic method for human immunodeficiency virus type-1 in plasma by near-infrared spectroscopy. Microbiol. Immunol. 49, 695–701 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Sakudo A., Baba K., Ikuta K., Discrimination of influenza virus-infected nasal fluids by Vis-NIR spectroscopy. Clin. Chim. Acta 414, 130–134 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Costa-da-Silva A. L., Ioshino R. S., Araújo H. R., Kojin B. B., Zanotto P. M., Oliveira D. B., Melo S. R., Durigon E. L., Capurro M. L., Laboratory strains of Aedes aegypti are competent to Brazilian Zika virus. PLOS ONE 12, e0171951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai L., Song J., Lu X., Deng Y. Q., Musyoki A. M., Cheng H., Zhang Y., Yuan Y., Song H., Haywood J., Xiao H., Yan J., Shi Y., Qin C. F., Qi J., Gao G. F., Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Xi Z., Ramirez J. L., Dimopoulos G., The Aedes aegypti toll pathway controls dengue virus infection. PLOS Pathog. 4, e1000098 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakudo A., Suganuma Y., Kobayashi T., Onodera T., Ikuta K., Near-infrared spectroscopy: Promising diagnostic tool for viral infections. Biochem. Biophys. Res. Commun. 341, 279–284 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phelps C., Madhavan G., Rappuoli R., Colwell R., Fineberg H., Beyond cost-effectiveness: Using systems analysis for infectious disease preparedness. Vaccine 35, A46–A49 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Faria N. R., Azevedo R. D. S. D. S., Kraemer M. U. G., Souza R., Cunha M. S., Hill S. C., Thézé J., Bonsall M. B., Bowden T. A., Rissanen I., Rocco I. M., Nogueira J. S., Maeda A. Y., Vasami F. G. D. S., Macedo F. L. L., Suzuki A., Rodrigues S. G., Cruz A. C. R., Nunes B. T., Medeiros D. B. A., Rodrigues D. S. G., Queiroz A. L. N., da Silva E. V. P., Henriques D. F., da Rosa E. S. T., de Oliveira C. S., Martins L. C., Vasconcelos H. B., Casseb L. M. N., de Brito Simith D., Messina J. P., Abade L., Lourenço J., Alcantara L. C. J., de Lima M. M., Giovanetti M., Hay S. I., de Oliveira R. S., Lemos P. D. S., de Oliveira L. F., de Lima C. P. S., da Silva S. P., de Vasconcelos J. M., Franco L., Cardoso J. F., Vianez-Júnior J. L. D. S. G., Mir D., Bello G., Delatorre E., Khan K., Creatore M., Coelho G. E., de Oliveira W. K., Tesh R., Pybus O. G., Nunes M. R. T., Vasconcelos P. F. C., Zika virus in the Americas: Early epidemiological and genetic findings. Science 352, 345–349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonaldo M. C., Ribeiro I. P., Lima N. S., Dos Santos A. A. C., Menezes L. S. R., da Cruz S. O. D., de Mello I. S., Furtado N. D., de Moura E. E., Damasceno L., da Silva K. A., de Castro M. G., Gerber A. L., de Almeida L. G. P., Lourenço-de-Oliveira R., Vasconcelos A. T., Brasil P., Isolation of infective Zika virus from urine and saliva of patients in Brazil. PLOS Negl. Trop. Dis. 10, e0004816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]


