Abstract
Ethologists discovered over 100 years ago that some lifelong behavioural patterns were acquired exclusively during restricted developmental phases called critical periods (CPs). Developmental song learning in zebra finches is one of the most striking examples of a CP for complex learned behaviour. After post-hatch day 65, whether or not a juvenile male can memorize the song of a ‘tutor' depends on his experiences in the month prior. If he experienced a tutor, he can no longer learn, but if he has been isolated from hearing a tutor the learning period is extended. We aimed to identify how tutor experience alters the brain and controls the ability to learn. Epigenetic landscapes are modulated by experience and are able to regulate the transcription of sets of genes, thereby affecting cellular function. Thus, we hypothesized that tutor experiences determine the epigenetic landscape in the auditory forebrain, a region required for tutor song memorization. Using ChIPseq, RNAseq and molecular biology, we provide evidence that naturalistic experiences associated with the ability to learn can induce epigenetic changes, and propose transcriptional plasticity as a mediator of CP learning potential.
Keywords: songbird, epigenetics, plasticity, gene regulation, critical period
1. Background
Some behaviours can only be acquired during critical periods (CPs), phases in development when experience remodels receptive neural circuits in large and persistent ways [1,2]. In brain regions with CPs, sufficient relevant experience transforms them from receptive, to closed. Closing prevents further experience-dependent plasticity, resulting in a stable behavioural pattern. CPs therefore provide a rich substrate for discovering mechanisms that control the potential for behavioural learning.
Juvenile male zebra finches learn to sing from an adult ‘tutor’ male; females cannot sing. Tutor song memorization is the sensory foundation for learned song [3]. Birds hear song all day every day, but only memorize tutor song they hear from post-hatch day 30 (P30) to P65 (figure 1) [4–8]. The CP for tutor song memorization is considered to be cognitive, rather than sensory, because it is closed exclusively by tutor song experience and not by other types of auditory signals including vocalizations such as calls. Tutor song therefore (i) provides the juvenile with a model for a meaningful song structure and (ii) prevents future sensory song learning. Importantly, the developing motor circuit appears largely unaffected by a lack of tutor song input [9]. Therefore, there is compelling evidence that the basis for the shift in the ability to acquire tutor song is the modulation of a brain area required for tutor song memorization, the auditory forebrain [10–12] (but see [13]).
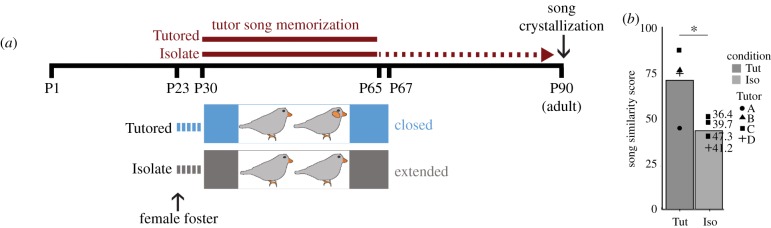
Figure 1.
Experimental design and song behaviour. (a) CP for tutor song memorization in the Tutored and Isolate condition, and the conditions used to close and extend the CP, superimposed on a developmental timeline. Red dashed line indicates Isolates have CP for tutor song memorization beyond P65. (b) Graph of song similarity scores showing higher levels of tutor song similarity in Tutored birds compared to Isolate birds. Isolate bird songs were compared to all adult tutors; the highest similarity score is plotted. Symbols reflect which tutor adult was the comparator, numbers next to Isolate data points indicate the difference in song copying scores between that bird and the Tutored bird who lived with the relevant tutor. Asterisk indicates p < 0.05. (Online version in colour.)
CP plasticity is probably driven by sets of genes, whose regulation must be coordinated to properly shape multiple cell and circuit properties that control the ability to learn. Epigenetic mechanisms such as post-translational modifications (PTMs) of histone tails directly influence gene expression without altering genomic DNA sequence. Histone PTMs work in a combinatorial fashion but specific PTMs indicate the potential for transcriptional activity [14]. For example, tri-methylation on lysine 4 of histone H3 (H3K4me3) and Polymerase 2 (Pol2) are found near transcription start sites and indicate that the corresponding gene is expressed, while H3K27me3 is found across repressed genes, and H3K9me3 is characteristic of transcriptionally inactive heterochromatin [15–18]. Histone PTMs are altered by experience and associated with learning and memory, thus epigenetic mechanisms could coordinate gene sets that regulate cognitive CPs [19–22].
Our aim was to identify differences in the auditory forebrain that permit or prevent the ability to memorize tutor song. Because epigenetic mechanisms regulate gene expression patterns in response to the environment, we hypothesized that tutor experience closes the CP by changing histone PTM profiles. We raised males under conditions consistent with those that close or extend the CP and, using ChIPseq, RNAseq and molecular biology, we consistently identified transcriptional regulation as a mechanism differentially affected by tutor experience. We found these distinctions despite assaying the entire auditory forebrain, which includes multiple cell types and several subregions, indicating that mechanisms associated with the ability for tutor song memorization are robust. Our results are among the first to provide insight that epigenetic mechanisms may regulate neural plasticity in brain areas required to learn complex natural behaviour.
2. Material and methods
(a). Birds
All juveniles were hatched in laboratory breeding aviaries. We raised two independent, replicate sets of males to P67 in controlled environments designed to disambiguate the epigenomic contributions of experience and maturation (figure 1). To standardize rearing conditions and ensure that birds in the ‘Isolate’ condition were unable to memorize tutor song, all males were removed from their home cages at P23 and placed with one to two other juvenile males that were within 3 days of age of each other, plus two adult foster females, in a cage housed within a sound attenuating chamber (figure 1) [4–8,10,12]. At P30, the first age at which tutor experience results in song copying, individual males were moved to live with either one adult (tutor) male and one adult female (‘Tutored’), or two adult females (‘Isolate’) [8]. Each triad lived in a cage within its own chamber that prevents song copying from other chambers [10]. Housing with two females standardizes the social complexity between groups and exposes juveniles to conspecific vocalizations; females call but do not sing, and their calls are not learned. Between 6.5 and 9 h post-lights-on (early to mid-afternoon, when singing behaviour lulls; no juveniles sang within 30 min of sacrifice) on P67, juveniles were sacrificed, and the auditory forebrain was rapidly bilaterally dissected and flash frozen within 2 min. Auditory forebrain samples for the P67 RNAseq experiment were collected following the same procedures. We also raised a group of males that were left in the breeding aviary in which they hatched until P32; P32 auditory forebrains were collected similarly as those for the P67 males. All tissue was stored at −80°C until further processing.
(b). P67 song analysis
To confirm that Tutored and Isolate birds were producing distinct types of songs, we placed a webcam into each chamber to record song from P65 to P67. We quantitatively compared each tutor's song to the appropriate Tutored bird's song. We also compared each tutor to each Isolate bird's song (electronic supplementary material, Methods). The adult song served as the template for the P67 songs' similarity metrics in Sound Analysis 2011 (electronic supplementary material, Methods) [23]. A t-test using song similarity scores at α < 0.05 was used to assess that the Tutored songs were more similar to their tutor's song than the Isolates were.
(c). ChIPseq
Chromatin immunoprecipitation followed by DNA sequencing (ChIPseq) for H3K9me3 (Active Motive cat# 39161; Carlsbad, CA), H3K27me3 (Millipore cat# 07-449), H3K4me3 (Active Motive cat# 39159) and Pol2 (Active Motive cat# 39097) was performed on two independent pools of Tutored and Isolate biological replicates (electronic supplementary material, Methods). Input control sequencing was performed on a combined pool of P67 Isolate and Tutored chromatin in equal proportions, or P32 chromatin. To obtain enough chromatin for all four ChIPs plus the Input controls, both hemispheres from three to four individuals from each condition were pooled. Chromatin isolation, ChIP and DNA sequencing were performed at Active Motif (Services and Low Cell ChIPseq kit cat# 53084; electronic supplementary material, Methods).
Reads were mapped to the zebra finch genome assembly (Ensembl genebuild taeGut3.2.4) with BWA [24]. We called non-differential and differential peaks with SICER v1.1 using peak and gap settings appropriate for each ChIP at a 10 × 10–5 threshold [25,26]. Resulting exclusive differential peaks were employed in downstream analysis (electronic supplementary material, Methods). We used custom algorithms for the zebra finch genome with the Ensembl gene set v84 as the universe to run Gene Ontology (GO; www.ark-genomics.org/tools/GOfinch), KEGG (www.ark-genomics.org/tools/KEGGfinch) and transcription factor binding site analysis [27,28]. Panther Protein Classes were obtained from www.geneontology.org. Our data were compared with an online database to identify cell-type specific genes (web.stanford.edu/group/barres_lab/brain_rnaseq.html; electronic supplementary material, Methods) [29]. We calculated the Z-score for two population proportions with α < 0.05. We processed the biological replicates of Tutored and Isolate ChIPseq samples independently to confirm distribution of called peaks, proportion of genes differentially associated with each PTM and Pol2, and significant GO categories. Replicate 1 data are presented here, Replicate 2 data showed similar results and are therefore in the electronic supplementary material, Methods, figure S3 and table S1.
(d). RNAseq
Total RNA was extracted from an independent set of P67 Isolate or Tutored auditory forebrains (bilateral, n = 8 per condition) with TRI Reagent (Life Technologies), treated with RNase free DNase I (Turbo DNase; Thermo Fisher) and cleaned with MinElute columns (Qiagen). All samples had RIN ≥ 7.7. Sequencing reads were checked for quality, trimmed and aligned to the genome assembly (taeGut3.2.4), assigned to gene models and statistically tested for differential abundance in Tutored and Isolate samples (α < 0.05; electronic supplementary material, Methods).
(e). Immunohistochemistry and in situ hybridization
We raised another set of Tutored (n = 5) and Isolate (n = 7) males. At P67, birds were transcardially perfused with 4% paraformaldehyde in 0.025 M phosphate-buffered saline (PBS) and whole brains were dissected. A cryostat was used to section brains (20 µm) in series for molecular analysis of four genes/proteins across the approximately 1 mm lateral extent of the auditory forebrain. To label oligodendrocytes, we hybridized one series of thaw-mounted sections with a Digoxigenin-labelled myelin proteolipid protein (PLP) antisense riboprobe in vitro transcribed from linearized zebra finch EST FE722130 in an in situ hybridization protocol described previously [30,31]. To label mature neurons, astrocytes and endothelial cells, we performed immunohistochemistry for NeuN, glutamine synthetase (GluL) and zona occludens-1 (ZO1), respectively. We used primary antibodies: mouse anti-NeuN (EMD Millipore #MAB377), mouse anti-GLUL (Atlas Antibodies # AMAb91103), mouse anti-ZO-1 (Thermo Fisher #ZO1-1A12). Sections were incubated in primary antibody prepared in PBS-Tween 20 containing 1% normal serum either overnight (NeuN, GluL) or for 48 h (ZO1; electronic supplementary material, Methods). Images were captured with a light microscope, analysed with FIJI, and t-tests with α < 0.05 were used to assess statistically significant differences between Tutored and Isolate measures [32] (electronic supplementary material, Methods).
3. Results
(a). Behavioural confirmation of tutor effects on song structure
Each Tutored juvenile's song was more similar to his tutor's song than any Isolate bird's were to that same tutor (t-test p = 0.03; figure 1, replicate 2; electronic supplementary material, figure S3). Because the Isolate birds had no opportunity to memorize tutor song, the difference in Tutored-tutor and Isolate-tutor similarity scores indicates that the Tutored birds learned song. Based on prior studies demonstrating that tutor experience is required for closing of the CP, we interpret these data to be consistent with age-matched males with either closed (Tutored) or extended (Isolate) CPs [4,6,33].
(b). ChIPseq peaks for PTMs and Pol2 map to expected distributions across the genome
For each ChIPseq, we generated at least 30 million reads, calling more than 17 000 peaks (H3K9me3: Tutored = 19 002, Isolate = 19 518; H3K27me3: Tutored = 24 321, Isolate = 23 375; H3K4me3: Tutored = 17 061, Isolate = 17 686; Pol2: Tutored = 18 311, Isolate = 19 656; ChIPseq data can be accessed at GEO, accession number GSE91399). Consistent with published profiles, called peaks for H3K9me3 and H3K27me3 spread throughout intergenic regions, whereas H3K4me3 and Pol2 localized to more proximal promoter regions (electronic supplementary material, figure S1) [15,34–38]. The distribution of peaks relative to functional genomic features is similar between Tutored and Isolate datasets (figure 2a). As expected, read density within called peaks for H3K4me3 and Pol2 are more closely correlated with each other than with H3K9me3 and H3K27me3 (electronic supplementary material, figure S2). The read counts per called peaks between the two biological replicates are also highly correlated (Pearson correlation coefficients ≥ 0.95; electronic supplementary material, figure S3).
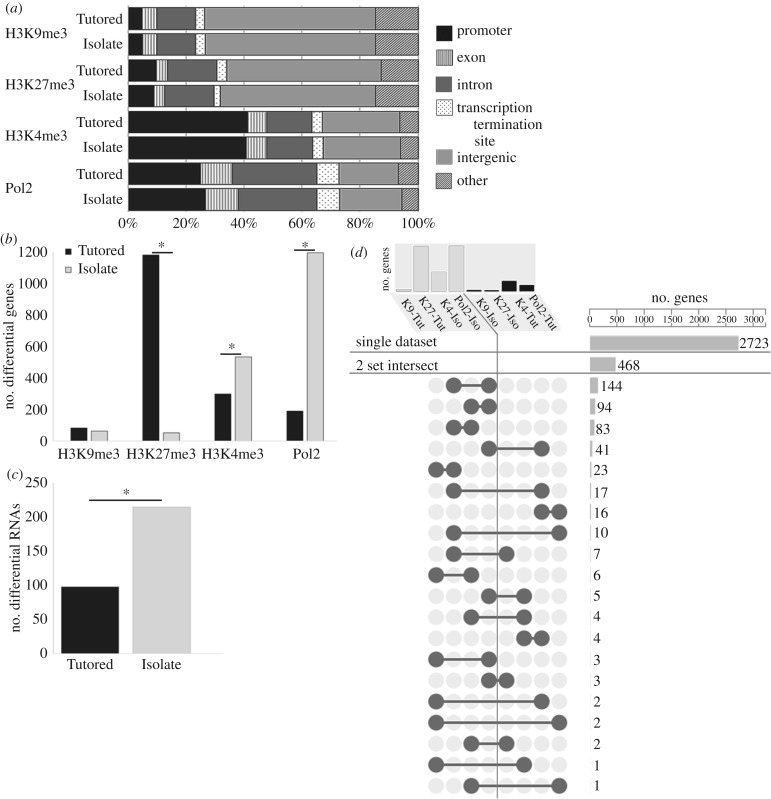
Figure 2.
ChIPseq and RNAseq demonstrate more transcriptional regulation in the Isolate condition. (a) ChIPseq read alignment demonstrates expected genomic distribution of PTM and Pol2 peaks, with similar distributions in Tutored and Isolate samples. (b) The number of Isolate and Tutored genes differentially associated with H3K9me3 (Tutored: 83, Isolate: 62), H3K27me3 (Tutored: 1181, Isolate: 51), H3K4me3 (Tutored: 298, Isolate: 533), and Pol2 (Tutored: 189, Isolate: 1196). (c) RNAseq showed a greater number of relatively more abundant RNAs in Isolate compared to Tutored auditory forebrain. (b,c) Asterisk designates significantly different proportions at p < 0.05. (d) ChIPseq datasets are ordered to display the number of genes (top) in Isolate-on (light grey) and Tutored-on (black) genesets. Lower panel illustrates specificity; 2723 of 3228 genes are unique to one dataset, 468 more genes were represented in two datasets. Barbell graphics show which two datasets share genes, with the number of genes on the right. Adapted from UpSet [39].
(c). Isolate auditory forebrain exhibits more transcription than Tutored auditory forebrain
We then identified 18 477 peaks that were differentially associated with PTMs or Pol2 between Tutored and Isolate auditory forebrains. We assigned these peaks to specific genes to further understand the epigenetic effect of tutor experience, and that found differentially modified genes were more likely to be associated with active H3K4me3 and Pol2 in the Isolate auditory forebrain (H3K4me3: Tutored = 298, Isolate = 533; Pol2: Tutored = 189, Isolate = 1196) and more likely to be associated with inactive H3K9me3 and H3K27me3 in the Tutored (H3K9me3: Tutored = 82, Isolate = 62; H3K27me3: Tutored = 1181, Isolate = 51), indicating greater probability for transcription in Isolate auditory forebrain (figure 2b). These differences were significant when considered as proportions of the entire gene set for H3K27me3, H3K4me3 and Pol2 (H3K27me3: Z-score = 33.30, p < 0.0001; H3K4me3: Z-score = 8.34, p < 0.0001; Pol2: Z-score = 44.33, p < 0.0001, H3K9me3: Z-score = 1.66, p = 0.09). The same pattern was obtained in the second replicate (electronic supplementary material, figure S3).
RNAseq on an independent set of birds confirmed significantly greater transcription in Isolate compared to Tutored auditory forebrain; 215 transcripts were more abundant in Isolate and 98 transcripts were more abundant in Tutored (Z-score = 6.64, p < 0.001; figure 2c; electronic supplementary material, figure S5; RNAseq data can be accessed at GEO, accession number GSE91399). We did not see a significant correlation between Pol2 ChIPseq and RNAseq genesets. This not entirely surprising given that RNAseq reflects steady-state RNA dynamics, not active transcription like Pol2 binding, and there is precedence for a disconnect between these two measures [40]. While GO category analysis did not reveal any significantly over-represented functional categories at adjusted thresholds, possibly due to relatively low numbers of differentially expressed genes, the Isolate set did have three times the number of genes involved in nucleic acid binding than the Tutored data (Isolate: 32 genes, Tutored: 9 genes). Of the top categories represented, only the Isolate dataset had genes related to DNA function (electronic supplementary material, table S6).
(d). Differential tutor experience alters complexity of transcriptional regulation
We expected that categorical analyses of genes differentially associated with PTMs or Pol2 would reveal distinct cellular functions in Isolate and Tutored conditions. Instead, we saw that similar processes were represented in both conditions (electronic supplementary material, table S2). This was not because the differential gene sets lacked specificity: of the 3228 total unique Human Gene Nomenclature Committee gene names present in any of the eight datasets, 84% (2723 genes) were found exclusively in one dataset (figure 2d). Another 14% (468 genes) were present in only two datasets. Further, of those 468 genes, 75% were present in datasets consistent with higher transcription in Isolate than Tutored auditory forebrain (i.e. H3K4me3/Pol2 in Isolate, or H3K9me3/H3K27me3 in Tutored; figure 2d) [39].
The high level of exclusivity and consistency suggested that what functionally distinguished Tutored and Isolate auditory forebrains was the direction in which a key set of cellular processes were regulated, rather than which processes were regulated. We therefore created composite lists coherent for the genes that would be more highly transcribed (on) in either condition (‘Tutored-on’: genes associated with H3K4me3, Pol2 in Tutored and H3K9me3, H3K27me3 in Isolate datasets; ‘Isolate-on’: genes associated with H3K4me3, Pol2 in Isolate and H3K9me3, H3K27me3 in Tutored datasets) and verified this pattern of differential regulation with ingenuity pathway analysis (electronic supplementary material, figure S4 and table S3).
GO analysis with the Tutored-on and Isolate-on genesets provided a more cohesive view of the cellular processes that may be modulated depending on tutor experience. We used the entire Ensembl gene set because there was no a priori expectation of what genes may be differentially associated with modified histones and Pol2. Our results suggested a type of regulatory loop wherein the chromatin landscape influences the transcriptional probability of genes that function to regulate transcriptional processes. For example, in the Isolate-on geneset, the GO biological process represented by the most genes was ‘regulation of transcription, DNA-dependent’ and ‘nucleus’ was the most abundant cellular component (table 1). Furthermore, all but one of the GO categories significantly over-represented in Replicate 1 Isolate-on was also significantly over-represented in Replicate 2 Isolate-on data (electronic supplementary material, table S1). In total, 350 genes in Replicate 1 are implicated in transcriptional regulation, including transcription factors, chromatin binding proteins and histone deacetylases. Further, of the 216 genes with GO annotation for chromatin present across all Replicate 1 datasets; 85% were in the Isolate-on geneset, including histone demethylases (e.g. KDM5A). Thirty-nine microRNA genes were also differentially associated with PTMs and Pol2, of which 34 (87%) were in the Isolate-on geneset (electronic supplementary material, table S7). Notably, genes annotated for nucleic acid binding were also the most abundant subset of genes more highly expressed in the Isolate than Tutored auditory forebrain in the RNAseq data.
Table 1.
GO analysis. Isolate-on genes are significantly over-represented in GO categories related to transcriptional regulation based on Fisher's test of adjusted p-values (α < 0.05). No categories were significantly over- or under-represented in the Tutored-on geneset.
| GO ID | GO description | expected | observed | adj. Fisher |
|---|---|---|---|---|
| Isolate-on | ||||
| 0003700 | sequence-specific DNA binding transcription factor activity | 46 | 82 | 0.00057 |
| 0005634 | nucleus | 201 | 265 | 0.00057 |
| 0043565 | sequence-specific DNA binding | 38 | 69 | 0.00096 |
| 0003677 | DNA binding | 81 | 123 | 0.001 |
| 0006355 | regulation of transcription, DNA-dependent | 60 | 95 | 0.0014 |
| 0004984 | olfactory receptor activity | 15 | 1 | 0.0014a |
| 0050911 | detection of chemical stimulus involved in sensory perception of smell | 15 | 1 | 0.0014a |
| 0007275 | multicellular organismal development | 13 | 29 | 0.0053 |
| 0005737 | cytoplasm | 165 | 210 | 0.026 |
| 0005515 | protein binding | 494 | 557 | 0.026 |
| 0000122 | negative regulation of transcription from RNA polymerase II promoter | 22 | 40 | 0.036 |
| 0045944 | positive regulation of transcription from RNA polymerase II promoter | 35 | 57 | 0.045 |
aCategories significantly under-represented.
(e). Predicted binding sites for canonical learning and memory transcription factors were over-represented in Isolate-on genes
Epigenetic PTMs regulate and coordinate transcription in part by controlling accessibility of regulatory regions to transcription factors. We asked if specific transcription factor binding sites were over-represented in the Tutored-on and Isolate-on genesets (electronic supplementary material, table S8). Interestingly, binding sites for transcription factors such as CREB, Fos and AP1, which orchestrate learning and memory events, as well as MEF2A, which influences activity-dependent excitatory synaptic stability, were over-represented in the Isolate-on geneset only [41–43]. Further, KEGG analysis showed that the Extracellular signal Regulated Kinase (ERK) signalling pathway (gga04010) is significantly over-represented for factors in the Isolate-on geneset (adjusted Fisher p = 0.0029). We confirmed that RNAs for these transcription factors were represented in the RNAseq data, indicating that they were expressed.
(f). Tutor experience does not significantly affect major cell-type abundance
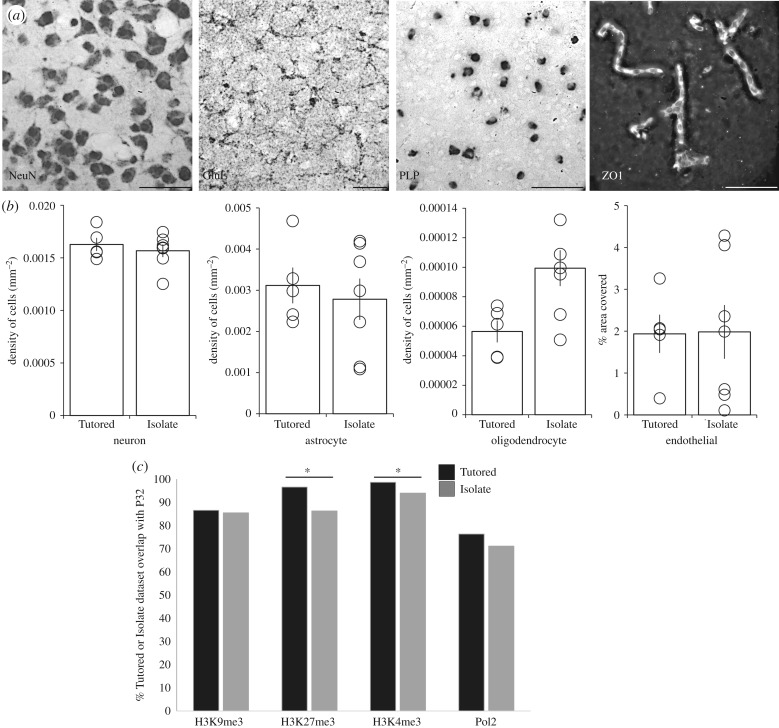
It is possible that different cell compositions could lead to distinct chromatin profiles, but we did not detect any significant differences in the relative abundance of major cell populations in Tutored and Isolate auditory forebrain (figure 3a,b; NeuN: t10 = 0.01, p = 0.99 ; GluL: t10 = 0.48, p = 0.65; PLP: t9 = 1.54, p = 0.16; ZO1: t10 = 0.05, p = 0.96). Further, cross-referencing Tutored and Isolate genesets with a database of RNAs enriched in distinct cell types revealed that few (less than 5%; range: 6–116 genes; electronic supplementary material, figure S6) of our genes were cell-type-specific [29].
Figure 3.
Distinct epigenetic landscapes are not explained by differences in major cell types or delayed epigenetic maturation. (a,b) There was no difference in cell number normalized for area (neurons, astrocytes and microglia), or in the percentage of the area covered with cells (endothelial) between Tutored and Isolate auditory forebrain. (a) Representative high magnification images. Scale bars, 100 µm. (b) Graphs of quantification. O denotes individual data points; bars are s.e.m. (c) Proportionally more Tutored than Isolate genes are represented in the P32 datasets; significantly different for H3K27me3 (Z-score = 3.77, p < 0.001) and H3K4me3 (Z-score = 3.17, p = 0.002), denoted by asterisks, but not for H3K9me3 (Z-score = 0.19, p = 0.85) or Pol2 (Z-score = 1.47, p = 0.14).
(g). Tutor song isolation does not extend earlier epigenetic profile
We next tested the possibility that without tutor experience, the Isolate auditory forebrain would preserve the epigenetic profile that was present at the beginning of the CP. We identified thousands of genes associated with each PTM and Pol2 at P32 (H3K9me3: 6936, H3K27me3: 5907, H3K4me3: 7985, Pol2: 6277). To determine the extent to which the Tutored and Isolate datasets overlapped with the P32 datasets, we cross-referenced the gene names and found that 71–93% of the Isolate genes were present in the equivalent P32 dataset, whereas 76–98%, of the Tutored genes overlapped with the P32 datasets. Thus, the Isolate epigenetic landscape was not more similar to P32 than the Tutored condition (figure 3c).
4. Discussion
Tutored and Isolate birds differed only in their experience with a singing male for 37 days of their development; both sets of birds had social and vocal interactions with two adults. Despite this subtle manipulation, we found that naturalistic experiences like those previously shown to determine CP closing also alter the epigenetic profiles [4–6,33]. It is possible that additional distinctions would emerge from examination of restricted anatomical divisions of the auditory forebrain, but, importantly, we found reliable epigenetic signatures of tutor experience without micro-dissecting its subregions or cell types, as is often advocated (e.g. [22]).
Our results consistently implicated regulation of transcriptional processes as a defining distinction between Tutored and Isolate auditory forebrains. Transcription and translation are essential to direct the cellular changes that support learning and long-term memory formation [44]. Tutor experience may therefore restrict transcriptional plasticity and minimize the ability to learn via epigenetic mechanisms [43,45,46]. This process may explain why basal gene expression is higher in the pre-CP juvenile than in the adult auditory forebrain [30,31]. Collectively, the evidence supports the idea that over the course of the CP, tutoring sufficiently represses chromatin such that the cellular plasticity required for subsequent tutor song learning is prevented, and the CP closes.
This proposal is compatible with the idea that ‘epigenetic priming’ within adult neural circuits prepares downstream nodes to receive information, as well as findings that increased histone acetylation, which increases the probability of transcription, improves learning in adults [19,47,48]. Interestingly, genes associated with the extended CP have binding sites for transcription factors well known to modulate learning and memory [44]. For example, during tutor experience, ERK signalling is required in the auditory forebrain for tutor song copying [10] and we found that transcription factors of the ERK pathway are over-represented in the set of genes predicted to be more highly expressed in the Isolate birds. This raises the possibility that the genes that are important for tutor song memorization, and those involved in preventing future sensory song learning, are coordinately regulated. Future experiments to test the causal relationships between chromatin profiles, learning and memory, and the extended ability to learn will advance understanding of all of these fields in developing and mature animals.
Our results contribute to understanding CP mechanisms more generally. For example, we were surprised that the P32 chromatin profile overlaps more with that of the Tutored compared to Isolate auditory forebrain; our expectation was that because both P32 and Isolate males are able to memorize tutor song, they would have more similar chromatin landscapes. Our data therefore suggest that in the auditory forebrain, CP extension is an active process with unique chromatin regulation. There is also support for epigenetic regulation in the mammalian visual system CP [49]. This perceptual CP in the primary visual cortex is controlled by sensory deprivation [50,51]. In comparison, the CP for tutor song memorization probably occurs within higher-order processing areas and is specifically controlled by song exposure [52]. These findings taken together indicate that continued investigation of epigenetic mechanisms will likely be fruitful for elucidating specialized and generalizable features of CPs.
5. Conclusion
We applied emergent epigenetic technologies to a learning CP to address a long-standing question of how early life experience influences brain and behaviour. Results support a model in which extended CP learning is characterized by epigenetically mediated ‘transcriptional plasticity’ that is required for the cellular plasticity which underlies learning and memory. These findings therefore stimulate new avenues of investigation into mechanisms that promote and limit the ability to learn.
Supplementary Material
Acknowledgements
We thank Elisa Gores for technical help with the immunohistochemistry, Dr Paul Labhart for ChIPseq analysis, and Dr Denise Garcia and Dr Chris Balakrishnan for helpful comments on the manuscript.
Ethics
Procedures were in accordance with the National Institute of Health guidelines for the care and use of animals for experimental procedures and approved by the University of Chicago Institutional Animal Care and Use Committee (ACUP #72220).
Data accessibility
Sequence data are accessible on GEO, record GSE91399.
Authors' contributions
Conceptualization, methodology, writing, funding acquisition, resources was performed by T.K.K. and S.E.L. Investigation was done by T.K.K., S.A., A.B. and S.E.L.
Competing interests
We declare we have no competing interests.
Funding
Funding was received from University of Chicago and Whitehall Foundation.
References
- 1.Knudsen EI. 2004. Sensitive periods in the development of the brain and behavior. J. Cogn. Neurosci. 16, 1412–1425. ( 10.1162/0898929042304796) [DOI] [PubMed] [Google Scholar]
- 2.Hess EH. 1959. Imprinting. An effect of early experience, imprinting determines later social behavior in animals. Science 130, 133–141. ( 10.1126/science.130.3368.133) [DOI] [PubMed] [Google Scholar]
- 3.Immelmann K. 1969. Song development in the zebra finch and other estrilid finches. In Bird vocalizations (ed. Hinde R.), pp. 61–74. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 4.Eales LA. 1985. Song learning in zebra finches—some effects of song model availability on what is learnt and when. Anim. Behav. 33, 1293–1300. ( 10.1016/S0003-3472(85)80189-5) [DOI] [Google Scholar]
- 5.Eales LA. 1987. Song learning in female-raised zebra finches—another look at the sensitive phase. Anim. Behav. 35, 1356–1365. ( 10.1016/S0003-3472(87)80008-8) [DOI] [Google Scholar]
- 6.Morrison RG, Nottebohm F. 1993. Role of a telencephalic nucleus in the delayed song learning of socially isolated zebra finches. J. Neurobiol. 24, 1045–1064. ( 10.1002/Neu.480240805) [DOI] [PubMed] [Google Scholar]
- 7.Price PH. 1979. Developmental determinants of structure in zebra finch song. J. Comp. Physiol. Psychol. 93, 260–277. ( 10.1037/h0077553) [DOI] [Google Scholar]
- 8.Roper A, Zann R. 2006. The onset of song learning and song tutor selection in fledgling zebra finches. Ethology 112, 458–470. ( 10.1111/J.1439-0310.2005.01169.X) [DOI] [Google Scholar]
- 9.Mori C, Wada K. 2015. Audition-independent vocal crystallization associated with intrinsic developmental gene expression dynamics. J. Neurosci. 35, 878–889. ( 10.1523/JNEUROSCI.1804-14.2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.London SE, Clayton DF. 2008. Functional identification of sensory mechanisms required for developmental song learning. Nat. Neurosci. 11, 579–586. ( 10.1038/nn.2103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yanagihara S, Yazaki-Sugiyama Y. 2016. Auditory experience-dependent cortical circuit shaping for memory formation in bird song learning. Nat. Commun. 7, 11946 ( 10.1038/ncomms11946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmadiantehrani S, London SE. 2017. Bidirectional manipulation of mTOR signaling disrupts socially mediated vocal learning in juvenile songbirds. Proc. Natl Acad. Sci. USA 114, 9463–9468. ( 10.1073/pnas.1701829114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canopoli A, Zai AT, Hahnloser RH. 2016. Lesions of a higher auditory brain area during a sensorimotor period do not impair birdsong learning. Matters ( 10.19185/matters.201603000018) [DOI] [Google Scholar]
- 14.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128, 693–705. ( 10.1016/j.cell.2007.02.005) [DOI] [PubMed] [Google Scholar]
- 15.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. ( 10.1016/j.cell.2007.05.009) [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903. ( 10.1038/ng.154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. 2010. Histone modification levels are predictive for gene expression. Proc. Natl Acad. Sci. USA 107, 2926–2931. ( 10.1073/pnas.0909344107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18. ( 10.1038/nrg2905) [DOI] [PubMed] [Google Scholar]
- 19.Lattal KM, Wood MA. 2013. Epigenetics and persistent memory: implications for reconsolidation and silent extinction beyond the zero. Nat. Neurosci. 16, 124–129. ( 10.1038/nn.3302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarome TJ, Lubin FD. 2014. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol. Learn. Mem. 115, 116–127. ( 10.1016/j.nlm.2014.08.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roidl D, Hacker C. 2014. Histone methylation during neural development. Cell Tissue Res. 356, 539–552. ( 10.1007/s00441-014-1842-8) [DOI] [PubMed] [Google Scholar]
- 22.Halder R, et al. 2016. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci. 19, 102–110. ( 10.1038/nn.4194) [DOI] [PubMed] [Google Scholar]
- 23.Tchernichovski O, Nottebohm F, Ho CE, Pesaran B, Mitra PP. 2000. A procedure for an automated measurement of song similarity. Anim. Behav. 59, 1167–1176. ( 10.1006/anbe.1999.1416) [DOI] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S, Grullon S, Ge K, Peng W. 2014. Spatial clustering for identification of ChIP-Enriched Regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol. 1150, 97–111. ( 10.1007/978-1-4939-0512-6_5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pepke S, Wold B, Mortazavi A. 2009. Computation for ChIP-seq and RNA-seq studies. Nat. Meth. 6, S22–S32. ( 10.1038/nmeth.1371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Watson M. 2009. CORNA: testing gene lists for regulation by microRNAs. Bioinformatics (Oxford, England) 25, 832–833. ( 10.1093/bioinformatics/btp059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blatti C, Sinha S. 2014. Motif enrichment tool. Nucleic Acids Res. 42, W20–W25. ( 10.1093/nar/gku456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11 929–11 947. ( 10.1523/jneurosci.1860-14.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin H, Clayton DF. 1997. Localized changes in immediate-early gene regulation during sensory and motor learning in zebra finches. Neuron 19, 1049–1059. ( 10.1016/S0896-6273(00)80396-7) [DOI] [PubMed] [Google Scholar]
- 31.London SE, Dong S, Replogle K, Clayton DF. 2009. Developmental shifts in gene expression in the auditory forebrain during the sensitive period for song learning. Dev. Neurobiol. 69, 437–450. ( 10.1002/dneu.20719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. ( 10.1038/nmeth.2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clayton NS. 1987. Song learning in cross-fostered zebra finches: a re-examination of the sensitive phase. Behaviour 102, 67–81. ( 10.1163/156853986X00054) [DOI] [Google Scholar]
- 34.Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. 2005. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 6, R64 ( 10.1186/gb-2005-6-8-r64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. 2005. A high-resolution map of active promoters in the human genome. Nature 436, 876–880. ( 10.1038/nature03877) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88. ( 10.1016/j.cell.2007.05.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heintzman ND, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318. ( 10.1038/ng1966) [DOI] [PubMed] [Google Scholar]
- 38.Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotech. 28, 817–825. ( 10.1038/nbt.1662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. 2014. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph 20, 1983–1992. ( 10.1109/TVCG.2014.2346248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mokry M, Hatzis P, Schuijers J, Lansu N, Ruzius F-P, Clevers H, Cuppen E. 2012. Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes. Nucleic Acids Res. 40, 148–158. ( 10.1093/nar/gkr720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nonaka M, Kim R, Sharry S, Matsushima A, Takemoto-Kimura S, Bito H. 2014. Towards a better understanding of cognitive behaviors regulated by gene expression downstream of activity-dependent transcription factors. Neurobiol. Learn. Mem. 115, 21–29. ( 10.1016/j.nlm.2014.08.010) [DOI] [PubMed] [Google Scholar]
- 42.Rashid AJ, Cole CJ, Josselyn SA. 2014. Emerging roles for MEF2 transcription factors in memory. Genes, Brain Behav. 13, 118–125. ( 10.1111/gbb.12058) [DOI] [PubMed] [Google Scholar]
- 43.Alberini CM. 2009. Transcription factors in synaptic plasticity and learning and memory A2—squire. In Encyclopedia of neuroscience (ed. Larry R.), pp. 1081–1092. Oxford, UK: Academic Press. [Google Scholar]
- 44.Kandel ER. 2012. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 5, 14 ( 10.1186/1756-6606-5-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis HP, Squire LR. 1984. Protein synthesis and memory: a review. Psychol. Bull. 96, 518–559. ( 10.1037/0033-2909.96.3.518) [DOI] [PubMed] [Google Scholar]
- 46.Hernandez PJ, Abel T. 2008. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol. Learn. Mem. 89, 293–311. ( 10.1016/j.nlm.2007.09.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gervain J, Vines BW, Chen LM, Seo RJ, Hensch TK, Werker JF, Young AH. 2013. Valproate reopens critical-period learning of absolute pitch. Front. Syst. Neurosci. 7, 102 ( 10.3389/fnsys.2013.00102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graff J, Tsai L-H. 2013. Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 14, 97–111. ( 10.1038/nrn3427) [DOI] [PubMed] [Google Scholar]
- 49.Baroncelli L, Scali M, Sansevero G, Olimpico F, Manno I, Costa M, Sale A. 2016. Experience affects critical period plasticity in the visual cortex through an epigenetic regulation of histone post-translational modifications. J. Neurosci. 36, 3430–3440. ( 10.1523/jneurosci.1787-15.2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hensch TK. 2005. Critical period plasticity in local cortical circuits. Nat. Rev. Neurosci. 6, 877–888. ( 10.1038/nrn1787) [DOI] [PubMed] [Google Scholar]
- 51.Takesian AE, Hensch TK. 2013. Balancing plasticity/stability across brain development. In Progress in brain research (eds MM Merzenich, M Nahum, TM van Vleet), pp. 3–34. Oxford, UK: Elsevier. [Google Scholar]
- 52.London SE. In press Developmental song learning as a model to understand neural mechanisms that limit and promote the ability to learn. Behav. Processes. ( 10.1016/j.beproc.2017.11.008) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are accessible on GEO, record GSE91399.