Abstract
Myotube formation by fusion of myoblasts and subsequent elongation of the syncytia is essential for skeletal muscle formation. However, molecules that regulate myotube formation remain elusive. Here we identify PIEZO1, a mechanosensitive Ca2+ channel, as a key regulator of myotube formation. During myotube formation, phosphatidylserine, a phospholipid that resides in the inner leaflet of the plasma membrane, is transiently exposed to cell surface and promotes myoblast fusion. We show that cell surface phosphatidylserine inhibits PIEZO1 and that the inward translocation of phosphatidylserine, which is driven by the phospholipid flippase complex of ATP11A and CDC50A, is required for PIEZO1 activation. PIEZO1-mediated Ca2+ influx promotes RhoA/ROCK-mediated actomyosin assemblies at the lateral cortex of myotubes, thus preventing uncontrolled fusion of myotubes and leading to polarized elongation during myotube formation. These results suggest that cell surface flip-flop of phosphatidylserine acts as a molecular switch for PIEZO1 activation that governs proper morphogenesis during myotube formation.
Myotube formation by fusion of myoblasts is essential for skeletal muscle formation, but which molecules regulate this process remains elusive. Here authors identify the mechanosensitive PIEZO1 channel as a key element, whose activity is regulated by phosphatidylserine during myotube formation.
Introduction
Transbilayer relocation of phospholipids at the plasma membrane is critical for various cellular processes such as cell division, signal transduction, and vesicular transport1–4. Phosphatidylserine (PS), a negatively charged phospholipid, normally resides in the inner leaflet of the plasma membrane5. Controlled cell surface exposure of PS acts as a potent promoter of blood coagulation, apoptotic cell engulfment, and myogenesis6–9. Mammalian skeletal muscles are formed by the fusion of mononucleated precursor cells (myoblasts) into unusually elongated multinucleated cells called myotubes, whose formation relies on orchestrated cell-to-cell fusion and elongation of multinucleated syncytia10, 11. During myotube formation, PS transiently translocates to the outer leaflet of the plasma membrane and recognition of cell surface-exposed PS by PS receptors induces contact-dependent signaling to promote fusion with neighboring myoblasts9, 12–15. However, it remains unclear how the transbilayer relocation of PS at the plasma membrane is controlled during myotube formation.
Several members of the type IV subfamily of P-type adenosine triphosphatases (P4-ATPases) that are complexed with an auxiliary CDC50 subunit act as a phospholipid flippase that translocates the cell surface-exposed PS to the inner leaflet of the plasma membrane3, 4, 8, 16–18. In mammals, at least 14 members of P4-ATPases, designated ATP8A1 through ATP11C, and three CDC50 family proteins (CDC50A, CDC50B, and CDC50C) have been identified3, 4, 17, 18. ATP8A1, ATP8A2, ATP8B1, ATP8B2, ATP8B4, ATP10A, ATP10D, ATP11A, and ATP11C are localized to the plasma membrane, whereas ATP9A, ATP9B, ATP10B, and ATP11B are distributed to intracellular membranes3, 4, 8, 16–18. Among the cell surface-localized P4-ATPases, ATP8A1, ATPA2, ATP8B1, ATP11A, and ATP11C have been shown to catalyze the inward translocation of PS at the plasma membrane3, 4, 8, 16–18. As first reported in yeast4, 19, complex association with CDC50 family proteins is required for transport of these P4-ATPases from endoplasmic reticulum to the plasma membrane, where they play a dominant role in maintaining the asymmetric distribution of PS in the bilayer leaflet3, 4, 8, 16–18, 20.
Although little is known about the physiological functions of mammalian P4-ATPases, deficiencies of at least three P4-ATPases, ATP8A2, ATP8B1, and ATP11C, can cause severe human disease3, 17, 18, 21, 22. Mutations identified in ATP8A2, which is highly expressed in the brain, testes, and retina, are associated with cerebellar ataxia, mental retardation and disequilibrium syndrome17, 18. Mutations in ATP8B1 cause liver disorders such as progressive familial intrahepatic cholestasis type 1 (PFIC1) and benign recurrent intrahepatic cholestasis type 1 (BRIC1)17. A mutation in ATP11C is liked to a congenital hemolytic anemia22. In vivo studies in mouse models have also contributed to our understanding of the physiological functions of mammalian P4-ATPases: Atp8a1-deficient mice exhibit delayed hippocampus-dependent learning, Atp8a2-mutant mice display neurological abnormalities such as ataxia derived from axonal degeneration and loss of visual/auditory functions, and Atp11c-mutant mice show arrested B cell development17, 21. ATP11A is ubiquitously expressed in various tissues16 and deletion of Atp11a results in lethality during embryogenesis23. The function of P4-ATPases in skeletal muscle, however, remains to be elucidated.
Here we identify the phospholipid flippase complex of ATP11A and CDC50A as a critical regulator for activation of the mechanosensitive Ca2+ channel PIEZO124, 25 during myotube formation. We show that the phospholipid flippase-mediated translocation of cell surface-exposed PS is a prerequisite for activation of PIEZO1 and that PIEZO1-mediated Ca2+ influx promotes RhoA/ROCK-dependent actomyosin assemblies26, thus leading to controlled cell fusion and the polarized elongation of multinucleated myotubes. The inhibitory effect of cell surface-exposed PS on PIEZO1 is strictly dependent on the headgroup structure of PS, and is controlled by manipulating the amount of PS present on the cell surface. Furthermore, the myoblast-specific disruption of Atp11a results in the formation of abnormal myofibres that fuse with each other during muscle regeneration after injury.
Results
ATP11A/CDC50A is required for myotube formation
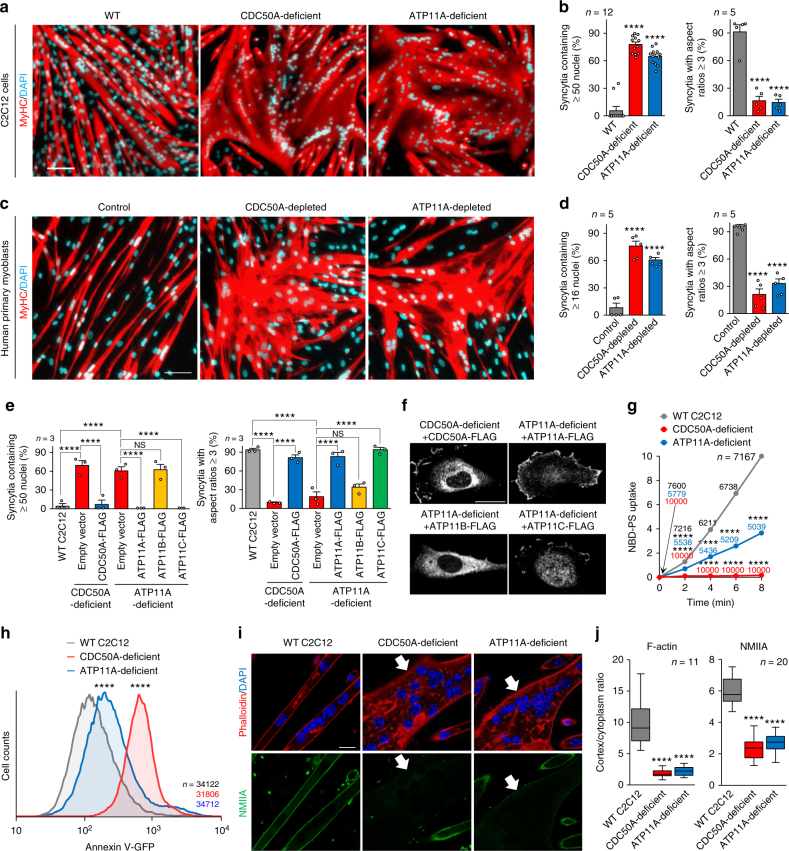
In this study, we detected the expression of seven genes encoding P4-ATPases and a single gene for a CDC50 family protein, CDC50A, in mouse C2C12 myoblasts12, 27 (Supplementary Figure 1a–c). Under myogenic differentiation conditions as indicated by the expression of muscle-type myosin heavy chain (MyHC), C2C12 myoblasts changed cell shape to a bipolar form and fused to generate elongated multinucleated myotubes, thereby providing a quantifiable in vitro model of myogenesis (Fig. 1a). CDC50A-deficient C2C12 cells produced by the CRISPR/Cas9 system28 (Supplementary Figure 1d) differentiated and fused with neighboring cells, but formed aberrantly enlarged syncytia with an undefined cell shape (Fig. 1a). Nuclei number and aspect ratios of the syncytia indicated excessive cell fusion and defects in polarized elongation in the CDC50A-deficient cells (Fig. 1b and Supplementary Figure 1e). Among the P4-ATPases expressed in C2C12 cells, ATP8B1, ATP8B2, ATP11A, ATP11B, and ATP11C were found to associate with CDC50A (Supplementary Figure 1f). We then established a series of P4-ATPase-deficient cell lines (Supplementary Figure 1d) and demonstrated that ATP11A-deficient cells formed enlarged syncytia with morphological features similar to those of the CDC50A-deficient cells (Fig. 1a, b and Supplementary Figure 1e). Live-cell imaging showed that neighboring C2C12 myoblasts fused with each other, and the resulting syncytia elongated longitudinally to form mature myotubes (Supplementary Figure 1g and Supplementary Movie 1). In contrast, ATP11A-deficient and CDC50A-deficient cells excessively fused with neighboring myoblasts, and the resulting syncytia spread without polarized elongation, forming an enlarged sheet-like morphology (Supplementary Figure 1g, Supplementary Movies 2 and 3). These observations were further confirmed using human primary myoblasts in which the phospholipid flippase complex of ATP11A and CDC50A was also robustly expressed (Supplementary Figure 2a). siRNA-mediated depletion of either CDC50A or ATP11A caused similar morphological abnormalities in syncytia without affecting differentiation (Fig. 1c, d and Supplementary Figure 2b, c). Retrovirus-mediated gene transfer was conducted to confirm the role of the phospohlipid flippase complex in myotube formation. Morphological abnormalities in the CDC50A-deficient or ATP11A-deficient C2C12 syncytia were rescued by re-expression of FLAG-tagged CDC50A or ATP11A, respectively (Fig. 1e, f and Supplementary Figure 3a–c). Importantly, ATP11C (a P4-ATPase localized to the plasma membrane like ATP11A) prevented the morphological abnormalities seen in the ATP11A-deficient C2C12 syncytia, but ATP11B residing in intracellular membranes did not (Fig. 1e, f and Supplementary Figure 3a–c).
Fig. 1.
Defective myotube formation by CDC50A-deficient and ATP11A-deficient myoblasts. a–d Aberrant morphologies of PS flippase-deficient myotubes. a Syncytia formed by WT and CDC50A-deficient or ATP11A-deficient C2C12 myoblasts were visualized by immunofluorescent staining with anti-MyHC antibody (differentiated cells, red) and DAPI (nuclei, cyan). b Left: cell fusion evaluated as percentages of syncytia containing ≥50 nuclei in a. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3 in a. c Syncytia formed by human primary myoblasts transfected with control, CDC50A or ATP11A siRNA were visualized by immunofluorescent staining with anti-MyHC antibody and DAPI. d Left: cell fusion evaluated as percentages of syncytia containing ≥16 nuclei in c. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3 in c. e, f Rescue of morphologies in PS flippase-deficient myotubes by overexpression of PS flippase complex components. e Syncytia formed by WT, CDC50A-deficient or ATP11A-deficient C2C12 myoblasts expressing FLAG-tagged CDC50A, ATP11A, ATP11B or ATP11C were visualized by immunofluorescent staining with anti-MyHC antibody and DAPI in Supplementary Figure 3b. Left: cell fusion evaluated as percentages of syncytia containing ≥50 nuclei. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3. f Localization of FLAG-tagged proteins expressed in CDC50A-deficient or ATP11A-deficient C2C12 myoblasts. g, h PS exposure on PS flippase-deficient myoblasts. g Flow cytometry analysis of inward translocation (flip) activity of fluorescence-labelled PS (NBD-PS) at the plasma membrane of WT, CDC50A-deficient and ATP11A-deficient C2C12 myoblasts. h Flow cytometry histogram of WT, CDC50A- and ATP11A-deficient C2C12 myoblasts labelled with annexin V-GFP. i, j Mislocalization of cortical actomyosin in PS flippase-deficient myotubes. i Localization of F-actin (phalloidin, red) and NMIIA (anti-NMIIA antibody, green) in WT, CDC50A-deficient and ATP11A-deficient C2C12 syncytia. Arrows indicate syncytia with diminished peripheral NMIIA accumulation. j Cortex vs. cytoplasm ratio of F-actin and NMIIA signals in i. ****P < 0.0001 (Student’s t-test). NS not significant, n sample number. Bar graphs represent mean ± S.E.M. Box and whiskers graph-line: median, box: upper and lower quartiles, whiskers: maxima and minima. Scale bars: 100 μm (a, c), 20 μm (f, i)
We analyzed the inward translocation activity of PS at the plasma membrane using the fluorescence-labelled PS analogue NBD-PS8, 20. After incubation of cells for various amounts of time with NBD-PS, we removed the fluorescent lipid remaining in the outer leaflet by washing with lipid-free bovine serum albumin (BSA). Flow cytometric analysis of cells incorporating NBD-PS into the inner leaflet showed that PS translocation was significantly reduced in both CDC50A-deficient and ATP11A-deficient C2C12 cells (Fig. 1g). Flow cytometric analysis of PS-exposing cells labelled with the PS-binding protein annexin V8 (Fig. 1h), fluorescent spectroscopic analysis using the surface charge-sensitive probe F2N12S29 (Supplementary Figure 1h) and quantification of PS contents by thin-layer chromatography (Supplementary Figure 1i) demonstrated that PS was exposed on the cell surface in CDC50A-deficient and ATP11A-deficient cells, without changes in PS content. Although previous studies9, 12–15 have shown that PS is transiently exposed on the cell surface during myotube formation, the molecular mechanisms underlying PS exposure remain unclear. In relation to this event, caspases are transiently activated in the early phase of myotube formation30, 31. Although caspase-mediated degradation of ATP11A has been shown to be responsible for PS exposure during apoptosis16, it remains unclear whether ATP11A degradation is involved in PS exposure during myotube formation. Here we found that ATP11A expression was significantly reduced in the early phase of C2C12 cell differentiation and restored by the presence of a caspase inhibitor, suggesting that the caspase-mediated transient cleavage of ATP11A is responsible for PS exposure on the cell surface (Supplementary Figure 1j). Taken together, these results suggest that the clearance of cell surface-exposed PS by the phospholipid flippase complex of ATP11A and CDC50A (PS flippase) plays a crucial role in the myotube formation. During myotube formation, assemblies of F-actin and non-muscle myosin IIA (NMIIA) create actomyosin fibers underneath the plasma membrane, which prevents uncontrolled fusion of adjacent myotubes and generates a lateral compression force to support polarized elongation10, 32. Both F-actin and NMIIA were enriched at the lateral cortex of wild-type (WT) C2C12 myotubes, but neither significant accumulation of F-actin nor NMIIA to the cell periphery was observed in the sheet-like syncytium of the CDC50A-deficient and ATP11A-deficient cells (Fig. 1i). Analysis of actomyosin localization by quantifying cortex/cytoplasm ratios33 clearly demonstrated the suppressed assembly of cortical F-actin and NMIIA in the sheet-like syncytium, which accumulated in unfused mononuclear cells (Fig. 1i, j).
PIEZO1 is required for myotube formation
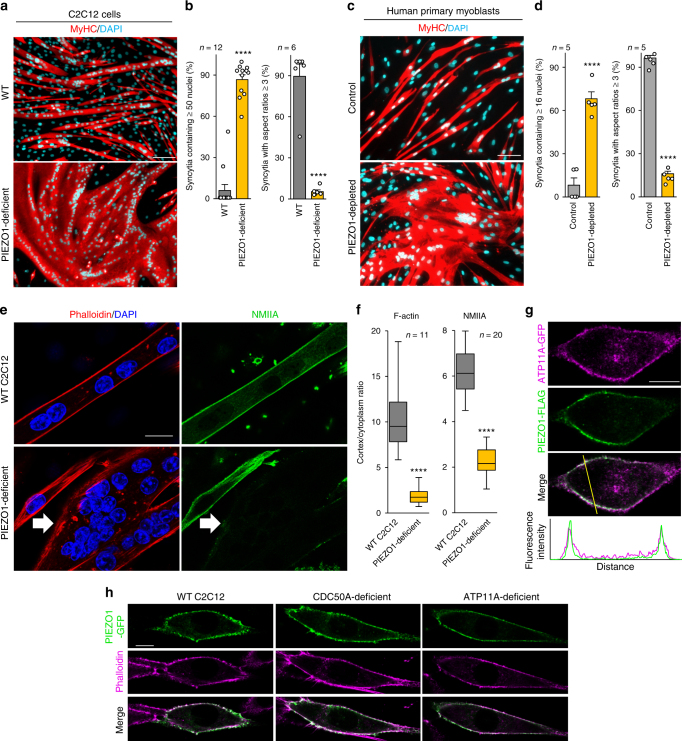
Myotube formation and the accompanying cortical actomyosin assembly are dependent on the influx of Ca2+ across the plasma membrane11. Because mechanical tension is crucial for myoblast fusion events34, we examined the effect of the depletion of mechanosensitive Ca2+-permeable channels expressed in C2C12 myoblasts on myotube formation (Supplementary Figure 4a). Of the Ca2+ channels examined, siRNA-mediated depletion of PIEZO124, 25, a mechanosensitive Ca2+ channel predominantly expressed during myotube formation, resulted in the formation of sheet-like syncytia (Supplementary Figure 4a, b), showing a morphological phenotype quite similar to that observed in the ATP11A-deficient and CDC50A-deficient cells (Fig. 1a). Thus, we established PIEZO1-deficient C2C12 cells using the CRISPR/Cas9 system28 (Supplementary Figure 4c), in which the mutations completely abolished the function of PIEZO1 as a Ca2+ channel (Supplementary Figure 4d–f). Quantitative analyses of syncytium morphology (Fig. 2a, b and Supplementary Figure 4g, h) and time-lapse observations (Supplementary Movie 4) confirmed excessive cell fusion and the cell-elongation defect in PIEZO1-deficient C2C12 cells. siRNA-mediated depletion of PIEZO1 in human primary myoblasts also caused morphological abnormalities in myotube formation similar to those observed in PS flippase-deficient cells (Fig. 2c, d and Supplementary Figure 2a–c). Moreover, there was no significant accumulation of cortical F-actin or NMIIA in the syncytium formed by the PIEZO1-deficient C2C12 cells (Fig. 2e, f). These results suggest that both PS flippase and PIEZO1 are involved in the cortical actomyosin assembly, as well as in myotube formation.
Fig. 2.
Defective myotube formation by PIEZO1-deficient myoblasts. a–d Aberrant morphologies of PIEZO1-deficient myotubes. a Syncytia formed by WT and PIEZO1-deficient C2C12 myoblasts were visualized by immunofluorescent staining with anti-MyHC antibody (differentiated cells, red) and DAPI (nuclei, cyan). b Left: cell fusion evaluated as percentages of syncytia containing ≥50 nuclei in a. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3 in a. c Syncytia formed by human primary myoblasts transfected with control or PIEZO1 siRNA were visualized by immunofluorescent staining with anti-MyHC antibody (differentiated cells, red) and DAPI (nuclei, cyan). d Left: cell fusion evaluated as percentages of syncytia containing ≥16 nuclei in c. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3 in c. e, f Mislocalization of cortical actomyosin in PIEZO1-deficient myotubes. e Localization of F-actin (phalloidin, red) and NMIIA (anti-NMIIA antibody, green) at the cell periphery of WT and PIEZO1-deficient C2C12 syncytia. Arrows indicate PIEZO1-deficient syncytia with diminished peripheral accumulation of NMIIA. f Cortex vs. cytoplasm ratio of F-actin and NMIIA signals in e. g, h Normal cell surface expression of PIEZO1 in PS flippase-deficient myoblasts. g Co-localization of GFP-tagged ATP11A (magenta) and FLAG-tagged PIEZO1 (green) in WT C2C12 myoblasts. Merged images and signal intensities are shown in the bottom panels. h Co-localization of GFP-tagged PIEZO1 (anti-GFP antibody, green) and F-actin (phalloidin, magenta) at the cell periphery of WT, CDC50A-deficient and ATP11A-deficient C2C12 myoblasts. Merged images are shown in the bottom panel. ****P < 0.0001 (Student’s t-test). n sample number. Bar graphs represent mean ± S.E.M. Box and whiskers graph-line: median, box: upper and lower quartiles, whiskers: maxima and minima. Scale bars: 100 μm (a, c), 20 μm (e), 10 μm (g, h)
PS flippase is required for PIEZO1 activation
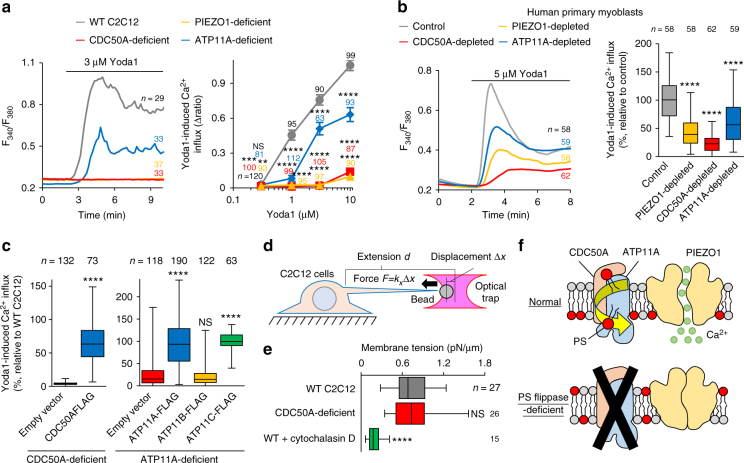
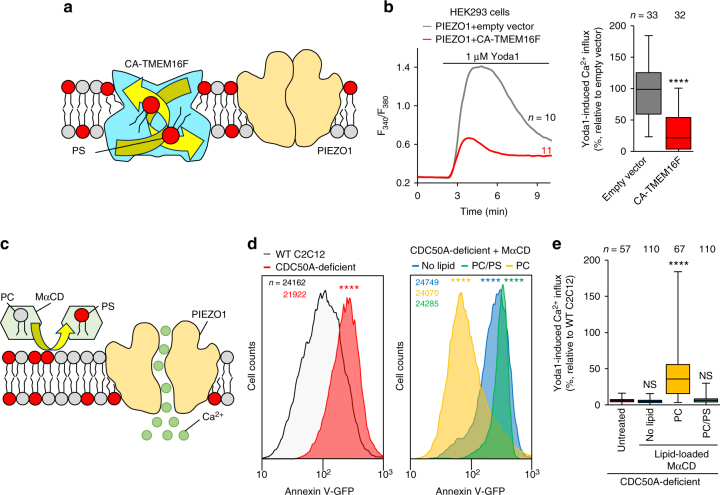
To eliminate the possible effect of defective PS flippase expression on translocation of PIEZO1 to the cell surface, we examined the cellular localization of PIEZO1 in flippase-deficient C2C12 cells. Both ATP11A and PIEZO1 were predominantly localized on the plasma membrane (Fig. 2g), and deficiency of CDC50A or ATP11A had no significant effect on the cell surface localization of PIEZO1 (Fig. 2h). Next, PIEZO1-mediated Ca2+ influx was monitored by Fura2 ratiometric imaging using the PIEZO1 activator Yoda1, which mimics mechanical activation35, 36. Ca2+ influx in response to Yoda1 was completely abolished in the CDC50A-deficient C2C12 cells and significantly reduced in the ATP11A-deficient cells (Fig. 3a). The inhibitory effect on PIEZO1 activation was also observed in human primary myoblasts transfected with siRNA against CDC50A or ATP11A (Fig. 3b). PIEZO1-mediated Ca2+ influx in the CDC50A-deficient or ATP11A-deficient C2C12 cells was recovered by exogenous expression of CDC50A or one of the PS flippases (i.e., ATP11A or ATP11C8), respectively (Fig. 3c and Supplementary Figure 5a). PIEZO1 overexpression restored the Yoda1-elicited Ca2+ influx in the PIEZO1-deficient C2C12 cells, but not in the CDC50A-deficient cells (Supplementary Figure 5b).
Fig. 3.
Impaired PIEZO1 activation in PS flippase-deficient myoblasts. a, b Suppression of agonist-induced PIEZO1 activation by PS flippase deficiency. a Left: Fura2 ratiometric measurements (F340/F380) of Yoda1-induced Ca2+ influx in WT, PIEZO1-deficient, CDC50A-deficient, and ATP11A-deficient C2C12 myoblasts. Right: quantification of Yoda1-induced Ca2+ influx as the maximal increment of F340/F380 (∆ ratio) in left. b Left: Fura2 ratiometric measurements of Yoda1-induced Ca2+ influx in human primary myoblasts transfected with control, PIEZO1, CDC50A or ATP11A siRNA. Right: quantification of Yoda1-induced Ca2+ influx in left. c Rescue of agonist-induced PIEZO1 activation in PS flippase-deficient myoblasts by overexpression of PS flippase complex components. Quantification of Yoda1-induced Ca2+ influx in CDC50A-deficient (left) or ATP11A-deficient (right) C2C12 myoblasts expressing FLAG-tagged CDC50A, ATP11A, ATP11B or ATP11C. d, e Normal plasma membrane tension in PS flippase-deficient myoblasts. d Schematic model of membrane tension measurement using an optical trap. e Quantification of membrane tension in WT, CDC50A-deficient and cytochalasin D-treated WT C2C12 cells. f Schematic model showing PS flippase-mediated inward translocation of cell surface-exposed PS as a prerequisite for PIEZO1 activation. **P < 0.01, ***P < 0.001, and ****P < 0.0001 (Student’s t-test). NS not significant, n sample number. Box and whiskers graph―line: median, box: upper and lower quartiles, whiskers: maxima and minima
Because lateral membrane tension is the physical stimulus that activates PIEZO137, we next examined whether depletion of PS flippase might affect plasma membrane tension. Membrane tension was measured by pulling a thin tube of membrane from the cell surface with an adhesive polystyrene bead in an optical trap38 (Fig. 3d). No significant change was observed between WT and CDC50A-deficient C2C12 cells (Fig. 3e and Supplementary Figure 5c). In contrast, inhibition of actin polymerization by treatment with cytochalasin D significantly reduced membrane tension, as previously reported38 (Fig. 3e and Supplementary Figure 5c). These results raise the intriguing possibility that the diminished response of PIEZO1 in PS flippase-deficient cells is due to the change in the transbilayer distribution of PS at the plasma membrane (Fig. 3f).
Cell surface flip-flop of PS regulates PIEZO1 activity
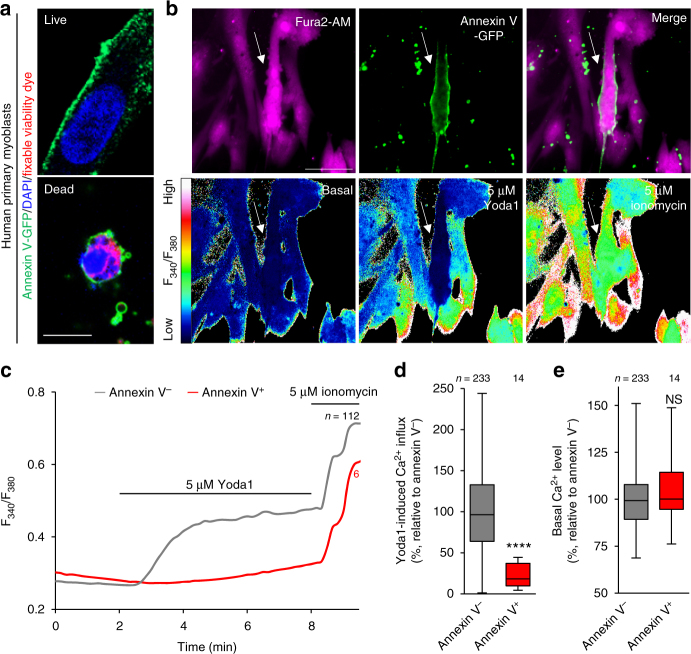
Next, we investigated the possible inhibitory effect of cell-surface-exposed PS on PIEZO1 activation in the course of myotube formation. Consistent with the results obtained from mouse differentiating myoblasts9, 12–15, cell surface exposure of PS was clearly observed on viable human primary myoblasts, accounting for about 20% of differentiating myoblasts (Fig. 4a). In PS-exposing myoblasts, PIEZO1-mediated Ca2+ influx was strikingly impaired (Fig. 4b–d), whereas both the basal Ca2+ concentration and ionomycin-induced Ca2+ influx were normal (Fig. 4c, e). To assess whether inward translocation of PS at the plasma membrane is a prerequisite for PIEZO1 activation, the transbilayer distribution of PS was manipulated by co-expression of PIEZO1 with a constitutively active form of TMEM16F (CA-TMEM16F), a phospholipid scramblase that induces cell-surface exposure of PS by bidirectional transport of phospholipids between the bilayer leaflets7, 39 (Fig. 5a). Ca2+ influx via PIEZO1 was significantly suppressed by the expression of CA-TMEM16F, compared to that of empty vector-transfected HEK293 cells (Fig. 5b). In addition, the inhibitory effect of cell-surface PS was further confirmed by manipulating the cell-surface contents of PS using the methyl-α-cyclodextrin (MαCD)-catalyzed phospholipid exchange method40 (Fig. 5c, d). PIEZO1-mediated Ca2+ influx in CDC50-deficient C2C12 cells was significantly recovered when cell surface-exposed PS was exchanged with exogenous phosphatidylcholine (PC), but not with a mixture containing PS (PC/PS) (Fig. 5e). These results indicate that PS flippase-mediated inward translocation of cell surface PS, which was exposed during myotube formation9, 12–15, is essential for PIEZO1 activation.
Fig. 4.
Impaired PIEZO1 activation in PS-exposing human primary myoblasts during myotube formation. a Detection of cell surface-exposed PS on differentiating human primary myoblasts by annexin V-GFP (PS, green), DAPI (nuclei, blue) and fixable viability dye (dead cells, red). b Fura2 imaging of Ca2+ influx in annexin V-GFP-labelled human primary myoblasts (arrow) upon addition of Yoda1 and ionomycin. Representative traces (c), quantification of Yoda1-induced Ca2+ influx (d) and basal Ca2+ level (e) in b. ****P < 0.0001 (Student’s t-test). NS not significant, n sample number. Box and whiskers graph―line: median, box: upper and lower quartiles, whiskers: maxima and minima. Scale bars: 10 μm (a), 20 μm (b)
Fig. 5.
Suppression of PIEZO1 activation by cell surface-exposed PS. a, b Suppression of agonist-induced PIEZO1 activation by overexpression of phospholipid scramblases. a Schematic illustration showing inhibition of PIEZO1 activation by CA-TMEM16-mediated bidirectional translocation of PS in the plasma membrane. b Representative traces (left) and quantification (right) of Yoda1-induced Ca2+ influx in HEK293 cells co-expressing PIEZO1 and CA-TMEM16F. c–e Rescue of agonist-induced PIEZO1 activation in PS flippase-deficient myoblasts by MαCD-catalyzed replacement of cell surface-exposed PS with exogenous PC. c Schematic illustration showing restoration of PIEZO1 activation by phospholipid exchange between PC-loaded MαCD and PS-exposing myoblasts. d, e Flow cytometry histograms of annexin V-GFP labelling (d) and quantification of Yoda1-induced Ca2+ influx (e) of CDC50A-deficient C2C12 myoblasts pre-incubated with no lipid-, PC/PS mixture (3:1)- or PC-loaded MαCD. ****P < 0.0001 (Student’s t-test). NS not significant, n sample number. Box and whiskers graph-line: median, box: upper and lower quartiles, whiskers: maxima and minima
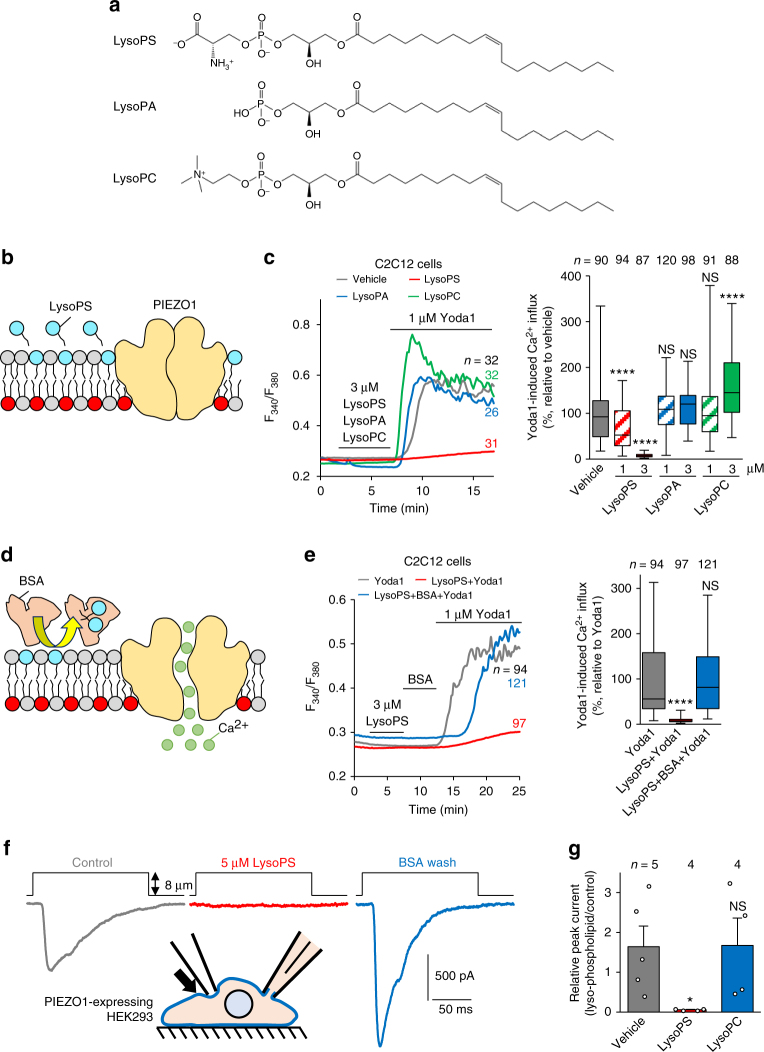
The direct effect of cell surface-exposed PS was further examined by treatment with a deacylated form of PS, lyso-phosphatidylserine (LysoPS), which readily inserts into the outer leaflet of the plasma membrane41 (Fig. 6a, b). Ca2+ influx via PIEZO1 was dose-dependently suppressed by the incorporation of LysoPS into the plasma membrane of C2C12 cells, while the incorporation of a zwitterionic phospholipid, lyso-phosphatidylcholine (LysoPC), or an anionic phospholipid, lyso-phosphatidic acid (LysoPA), showed no significant inhibitory effect on Ca2+ influx (Fig. 6a, c). Furthermore, slight modification of the serine moiety of LysoPS abolished its ability to inhibit PIEZO1 activity, suggesting that the stereospecific inhibition of PIEZO1 is mediated by the serine headgroup of PS (Supplementary Figure 5d). Ca2+ influx via PIEZO1 was restored when LysoPS on the cell surface was removed by washing with lipid-free BSA8 (Fig. 6d, e). To confirm the inhibitory effect of LysoPS on membrane tension-induced PIEZO1 activation37, we conducted electrophysiological measurements of the PIEZO1 current by applying mechanical force to the cell surface using an electrically-driven glass probe24. In PIEZO1-expressing HEK293 cells, indentation of the cell surface by 8 μm evoked a transient inward current, consistent with that observed in a previous study24 (Fig. 6f). The addition of LysoPS significantly blunted the mechanically gated currents, while removal of cell surface-inserted LysoPS with BSA completely restored the responsiveness of PIEZO1 to mechanical stimuli (Fig. 6f). No significant change in the current was observed with the addition of LysoPC (Fig. 6g). In addition, application of negative pressure (−20 mmHg) to the cell surface evoked a transient inward current in PIEZO1-expressing cells, and the addition of LysoPS in the recording pipettes significantly blunted the PIEZO1 currents (Supplementary Figure 5e). These data suggest that the phospholipids with phosphoserine headgroups present on the outer leaflet of the plasma membrane are responsible for inhibition of PIEZO1 activation.
Fig. 6.
Suppression of PIEZO1 activation by cell surface-inserted LysoPS. a Chemical structures of LysoPS, LysoPA and LysoPC. b–d Suppression of agonist-induced PIEZO1 activation by cell-surface LysoPS. b Schematic model showing suppression of PIEZO1 activation by insertion of LysoPS to the cell surface. c Representative traces (left) and quantification (right) of Yoda1-induced Ca2+ influx in WT C2C12 myoblasts treated with vehicle, LysoPS, LysoPA or LysoPC. d Schematic model showing restoration of PIEZO1 activation by removal of cell surface-inserted LysoPS with lipid-free BSA. e Representative traces (left) and quantification (right) of Yoda1-induced Ca2+ influx in WT C2C12 myoblasts treated with LysoPS and washed with lipid-free BSA. f, g Impairment of mechanical stimulation-induced PIEZO1 activation by cell-surface LysoPS. f Representative traces of the mechanically-activated current evoked by indentation (8 μm) using a glass probe in PIEZO1-expressing HEK293 cells before (grey, control) and during administration of 5 μM LysoPS (red), followed by removal of LysoPS with lipid-free BSA (blue). g Relative peak currents induced by mechanical stimulation before (control) and during administration of vehicle (grey), 5 μM LysoPS (red), and 5 μM LysoPC (blue) in f. *P < 0.05 and ****P < 0.0001 (Student’s t-test). NS not significant, n sample number. Bar graphs represent mean ± S.E.M. Box and whiskers graph―line: median, box: upper and lower quartiles, whiskers: maxima and minima
PS flippase and PIEZO1 regulate cortical actomyosin assembly
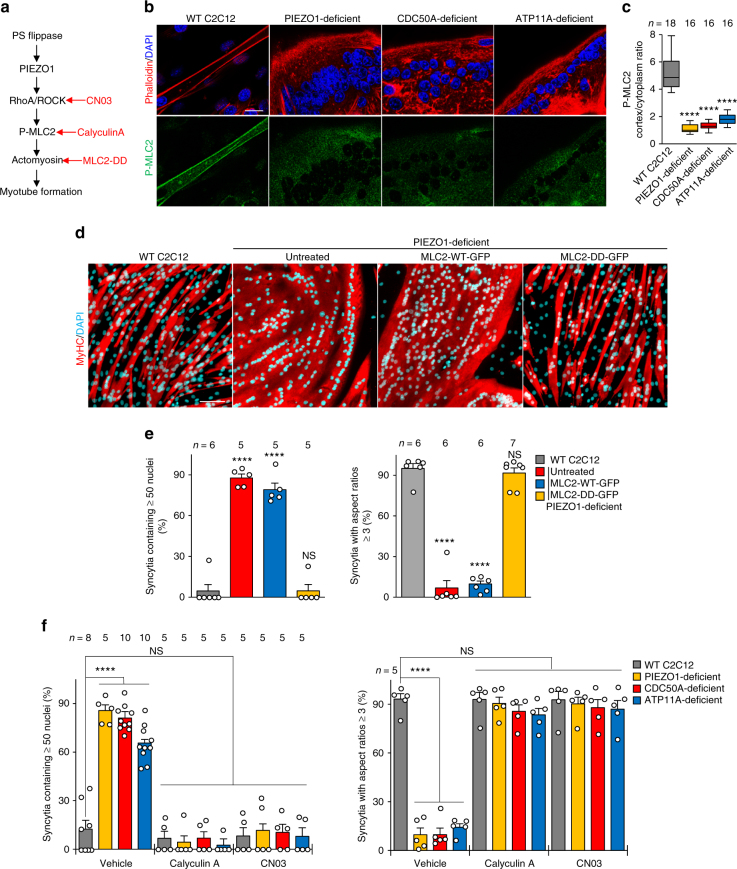
Rho-associated, coiled-coil-containing protein kinase (ROCK) is a primary effector of RhoA GTPase and induces phosphorylation of myosin light chain 2 (MLC2) at the Thr-18 and Ser-19 residues, resulting in NMII activation and subsequent actomyosin assembly26. The active forms of RhoA and ROCK were co-localized with F-actin, NMIIA, and MLC2, and partially with PIEZO1, at the lateral cortex of bipolar C2C12 myoblasts (Supplementary Figure 6a). These observations, together with a previous report that the RhoA/ROCK pathway is activated by PIEZO142, suggest that the RhoA/ROCK/actomyosin pathway plays a dominant role in PS flippase-mediated and PIEZO1-mediated myotube formation (Fig. 7a). We found that a phosphorylated form of MLC2 (P-MLC2) had clearly accumulated at the lateral cortex of WT myotubes, while no significant localization was observed in the PIEZO1-deficient, CDC50A-deficient or ATP11A-deficient C2C12 cells (Fig. 7b, c). Stable expression of a phospho-mimetic form of MLC2 (MLC2-DD: T18D, S19D) totally prevented aberrant myotube formation in PIEZO1-deficient C2C12 cells (Fig. 7d, e and Supplementary Figure 6b), indicating that MLC2 phosphorylation is a downstream event of PIEZO1 that leads to actomyosin assembly. Furthermore, both CN03, a selective RhoA activator, and calyculin A, a myosin II activator that inhibits dephosphorylation of MLC2, prevented the myotube formation defects observed with CDC50A-deficient, ATP11A-deficient, and PIEZO1-deficient C2C12 cells (Fig. 7f and Supplementary Figure 6c, d). These results collectively indicate that flippase-mediated PS translocation at the plasma membrane regulates PIEZO1 activation, which promotes RhoA/ROCK-mediated phosphorylation of MLC2 and subsequent assembly of cortical actomyosin fibers, thereby controlling fusion and polarized elongation during myotube formation.
Fig. 7.
RhoA/ROCK-mediated actomyosin formation via the PS flippase/PIEZO1 pathway required for myotube morphology. a Schematic diagram of the PS flippase/PIEZO1 axis (black) and activators for the RhoA/ROCK/actomyosin pathway (red). b, c Suppressed cortical accumulation of phosphorylated MLC2 (P-MLC2) in PS flippase-deficient and PIEZO1-deficient myotubes. b Localization of P-MLC2 (anti-P-MLC2 antibody, green), F-actin (phalloidin, red), and nuclei (DAPI, cyan) in WT, PIEZO1-deficient, CDC50A-deficient, and ATP11A-deficient C2C12 syncytia. Intense co-localization of P-MLC2 and F-actin was observed on the cell periphery of WT myotubes. c Cortex vs. cytoplasm ratio of P-MLC2 signals in b. d, e Rescue of myotube formation by stable expression of a phospho-mimetic form of MLC2 in PIEZO1-deficient C2C12 syncytia. d Syncytia formed by WT and PIEZO1-deficient C2C12 myoblasts stably expressing WT MLC2 (MLC2-WT-GFP) or phospho-mimetic MLC2 (MLC2-DD-GFP) were visualized by immunofluorescent staining with anti-MyHC antibody (differentiated cells, red) and DAPI (nuclei, cyan). e Left: cell fusion evaluated as percentages of syncytia containing ≥50 nuclei in d. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3 in d. f Rescue of myotube formation by activation of the RhoA/ROCK/actomyosin pathway in PS flippase- or PIEZO1-deficient C2C12 syncytia. Syncytia formed by WT, PIEZO1-deficient, CDC50A-deficient, and ATP11A-deficient C2C12 syncytia after treatment with calyculin A (a myosin II activator) or CN03 (a selective RhoA activator) were visualized by immunofluorescent staining with anti-MyHC antibody and DAPI. Morphologies of the syncytia are shown in Supplementary Figure 6c. Left: cell fusion evaluated as percentages of syncytia containing ≥50 nuclei. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3. ****P < 0.0001 (Student’s t-test). NS not significant, n sample number. Bar graphs represent mean ± S.E.M. Box and whiskers graph-line: median, box: upper and lower quartiles, whiskers: maxima and minima. Scale bars: 20 μm (b), 100 μm (d)
A role of ATP11A in muscle regeneration
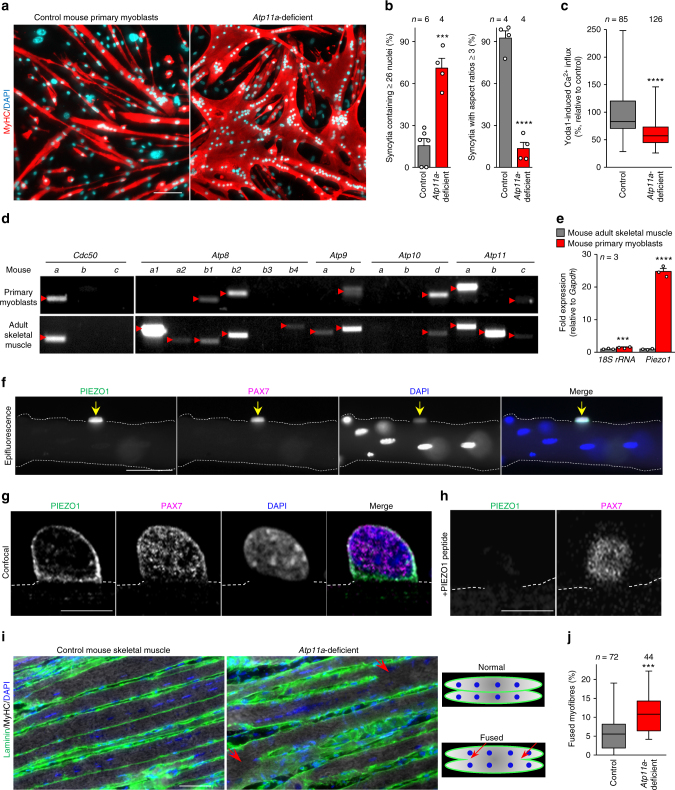
Since systemic knockout of Atp11a results in lethality during embryogenesis23, we generated myoblast-specific conditional Atp11a-deficient mice utilizing Myf5-cre transgenic mice that expressed Cre-recombinase in myoblasts43 (Supplementary Figure 7a) and examined the muscle phenotype. Primary myoblasts isolated from the myoblast-specific Atp11a-deficient mice formed aberrantly enlarged syncytia upon differentiation (Fig. 8a, b and Supplementary Figure 7b), showing the morphological abnormalities observed with ATP11A-deficient myoblasts (Fig. 1a–d). PIEZO1-mediated Ca2+ influx was also attenuated in the Atp11a-deficient primary myoblasts (Fig. 8c). However, no obvious morphological or behavioural abnormalities were observed in Atp11a-deficient mice by either hematoxylin-eosin staining of cross/longitudinal muscle sections or functional analyses such as grip strength and treadmill running tests. To further evaluate the physiological function of ATP11A and PIEZO1 in myogenesis, we performed detailed analyses on the stage-specific and tissue-specific expression of ATP11A and PIEZO1 in mouse muscle tissues. The expression profile of mRNA encoding P4-ATPases in primary myoblasts isolated from adult skeletal muscle was quite different from that of adult skeletal muscle: among P4-ATPases expressed on the plasma membrane16, Atp11a was a major flippase in primary myoblasts, while strong expression of Atp8A1, Atp8b2, and Atp11a was observed in adult skeletal muscle (Fig. 8d). In developing muscle27, a variety of cell surface P4-ATPases including Atp8a1, Atp8a2, Atp8b1, Atp8b2, Atp9a, Atp10d, Atp11a, and Atp11c were expressed16 (Supplementary Figure 7c). These results suggest that these P4-ATPases may functionally compensate for the defective expression of ATP11A during developmental myogenesis. Furthermore, no significant expression of PIEZO1 was detected in developing or adult muscle (Fig. 8e and Supplementary Figure 7c). However, we did find robust expression of PIEZO1 in primary myoblasts as well as in Pax7-positive satellite cells, a population of myogenic progenitor cells in adult muscle44 (Fig. 8f, g). The fluorescent signal of the PIEZO1 antibody in satellite cells was significantly diminished in the presence of the epitope peptide, confirming the specificity of the antibody (Fig. 8h). In silico analysis of adult regenerating myofibres after muscle injury45 showed that expression levels of Piezo1 as well as Myog (a myogenic marker) were significantly upregulated after induction of muscle injury (Supplementary Figure 7d). These observations prompted us to examine the physiological function of ATP11A in muscle regeneration.
Fig. 8.
A role of ATP11A in morphogenesis during myofibre regeneration. (a, b) a Aberrant morphologies of Atp11a-deficient primary myotubes. Syncytia formed by control or Atp11a-deficient mouse primary myoblasts were visualized by immunofluorescent staining with anti-MyHC antibody (differentiated cells, red) and DAPI (nuclei, cyan). b Left: cell fusion evaluated as percentages of syncytia containing ≥26 nuclei in a. Right: polarized elongation evaluated as percentages of syncytia with aspect ratios ≥3 in a. c Suppression of agonist-induced PIEZO1 activation in Atp11a-deficient mouse primary myoblasts. Quantification of Yoda1-induced Ca2+ influx in control or Atp11a-deficient mouse primary myoblasts. d Semi-quantitative RT-PCR analysis of CDC50 family members and P4-ATPases in mouse primary myoblasts (upper panels) and adult skeletal muscle (lower panels). Arrowheads denote specific bands. e Expression levels of Piezo1 in mouse primary myoblasts and adult skeletal muscle, evaluated by quantitative RT-PCR. Gapdh and 18S rRNA were used as internal controls. f–h Detection of endogenous PIEZO1 protein in satellite cells. Epifluorescence (f) and confocal (g, h) images of mouse myofibres visualized by immunofluorescent staining with anti-PIEZO1, anti-PAX7 antibodies and DAPI (nuclei). Specificity of the anti-PIEZO1 antibody was confirmed by the decreased fluorescence signal in the sample treated with the epitope peptide (h). Arrows indicate Pax7-positive satellite cells. Dashed lines indicate the cell periphery of myofibres. i, j Aberrant morphologies of Atp11a-deficient regenerated myofibres after cardiotoxin-induced muscle degeneration. i Longitudinal sections prepared from control (left) and Atp11a-deficient adult muscle (right) were probed with anti-laminin antibody (green), anti-MyHC antibody (grey), and DAPI (nuclei, blue). Arrows indicate apparent fusion sites with neighboring myofibres. j Evaluation of fused myofibres in i. ***P < 0.001 and ****P < 0.0001 (Student’s t-test). NS not significant, n sample number. Bar graphs represent mean ± S.E.M. Box and whiskers graph-line: median, box: upper and lower quartiles, whiskers: maxima and minima. Scale bars: 100 μm (a), 50 μm (f, i), 10 μm (g, h)
Adult skeletal muscle has the ability to efficiently regenerate after different types of injury. Muscle regeneration is mediated by satellite cells residing beneath the basal lamina of muscle fibers, which are activated after injury and undergo myogenic commitment to become fusion-competent myoblasts44. The resulting myoblasts fuse with each other to generate nascent syncytia that mature into functional myofibres10. In this study, we evaluated the morphology of regenerating myofibres in Atp11a-deficient tibialis anterior (TA) muscle. Cardiotoxin, a myotoxic agent that causes degeneration and concomitant regeneration of myofibres46, was injected into TA muscle of Atp11a-deficient mice. The regenerating muscle tissues were harvested at 2 weeks post-cardiotoxin injection, then morphological analysis was conducted by staining longitudinal sections from the cardiotoxin-injected muscles with anti-laminin antibody (for the extracellular matrix), anti-MyHC antibody (for myofibres), and DAPI (for nuclei). Our immunohistological analyses demonstrated that, upon cardiotoxin administration, abnormal myofibres that fused with each other were evident in cardiotoxin-injected Atp11a-deficient TA muscle (Fig. 8i, j), as observed in regenerating muscles of mutant mice that display the in vitro hyperfusion phenotype47. These results suggest that ATP11A-mediated PIEZO1 activation plays a crucial role in proper morphogenesis during myofibre regeneration.
Discussion
The present study provides evidence that the transbilayer redistribution of PS at the plasma membrane, which is mediated by the phospholipid flippase complex of ATP11A and CDC50A, plays a critical role in regulating the Ca2+ channel function of the integral membrane protein PIEZO1. Recent studies of structural determination of membrane proteins and molecular simulations of lipid–protein interactions have demonstrated that the structure, function, and dynamics of membrane proteins are significantly affected by associated lipid molecules48, 49. However, it remains unclear how lipids act and regulate the function of membrane proteins in biological membranes, because these structural and biophysical studies have been conducted in model systems where membrane proteins are purified and reconstituted in artificial membranes. There is a key difference between artificial and biological membranes; in biological membranes, phospholipids are asymmetrically distributed between the two leaflets of the bilayer, and are actively transported across the bilayer leaflets by the actions of a family of lipid-transport machineries such as phospholipid flippase and scramblase.
Phospholipid flippase catalyzes selective translocation of specific phospholipid species from the outer to the inner leaflet of cellular membranes, thereby generating the asymmetric transbilayer lipid distribution3, 4, 17, 18. A locally unbalanced increase in the phospholipid number on one side of the membrane drives bending of the membrane to induce membrane curvature, which supports recruitment of membrane curvature-sensing proteins in vesicle budding within Golgi and endosomal membranes3, 4. In addition to affecting the curved structure of membranes, phospholipid flippase is thought to regulate the electrostatic properties of the inner leaflet of the plasma membrane3. Phospholipid flippase-mediated local enrichment of PS in the inner leaflet strengthens the electrostatic interaction between the basic domain of cytosolic proteins and the membrane, thereby playing a critical role in the recruitment and activation of various signaling molecules such as small GTPases and those with pleckstrin homology domain50, 51. Recent studies have also shown that lateral reorganization of PS to nanocluster formation drives K-Ras assembly on the inner leaflet via interactions between Ras basic residues and negatively charged PS50.
Electrostatic interactions of proteins with anionic phospholipids such as PS, phosphatidic acid, and phosphoinositides in the inner leaflet of the plasma membrane50, 52 have been shown to play a crucial role in controlling the function of membrane-associated proteins by the so-called ‘electrostatic switch' mechanism53–57. Based on the results described here, we propose a lipid-mediated mechanism, namely the ‘flip-flop switch’ mechanism, for tempo-spatial activation of PIEZO1, in which the regulated change in the transbilayer distribution of particular lipids, such as PS, regulates the function of the ion channel.
Methods
Reagents
siRNAs and antibodies are summarized in Supplementary Tables 1 and 2, respectively. 1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoserine (18:1-06:0, NBD-PS), 1-oleoyl-2-hydroxy-sn-glycero-3-phospho-L-serine (18:1, LysoPS), 1-oleoyl-2-hydroxy-sn-glycero-3-phosphocholine (18:1, LysoPC), 1-oleoyl-2-hydroxy-sn-glycero-3-phosphate (18:1, LysoPA), 1,2-dioleoyl-sn-glycero-3-phosphocholine (18:1, DOPC) and 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (18:1, DOPS) were purchased from Avanti. LysoPS (18:1) analogues were provided by J.A. and T.O.58, 59.
Plasmids
cDNA clone of human PIEZO1 was purchased from Promega. MmCdc50a cDNA was obtained from pCMV-Tag4A-CDC50A20. The other mouse cDNA clones were obtained from mouse (C57BL/6J, Japan SLC Inc.) brain or C2C12 myoblasts (ATCC). pcDNA3.1-Clover-mRuby2 (#49089), mPIEZO1-IRES-eGFP (#80925), pX260 (#42229), and pX330 (#42230) were purchased from Addgene. pGFP-AHD was generated according to previous literature60.
pX330-PGKpuro was generated by inserting a PGK promoter and a puromycin-resistant gene (from pX260) into pX330. Target sequences were determined using CRISPRdirect61 (Supplementary Figures 1d and 4c) and introduced into pX330-PGKpuro. pmClover-N1 and pmClover-C1 were generated by replacing EGFP in pEGFP-N1 and pEGFP-C1 (Clontech) with mClover (monomeric Clover with A206K mutation), respectively. pATP11A-mClover and pmClover-NMIIA were generated by subcloning of ATP11A and NMIIA into pmClover-N1 and pmClover-C1, respectively. pHsPIEZO1-FLAG was made by subcloning HsPIEZO1 into pEGFP-N1, followed by replacing EGFP with a FLAG-tag. pHsPIEZO1-IRES-EGFP was made by inserting HsPIEZO1 into pEGFP-N1, followed by replacing EGFP with the IRES-EGFP sequence obtained from pMmPIEZO1-IRES-EGFP. pHsPIEZO1-Clover was generated by subcloning HsPIEZO1 into pcDNA3.1 (Invitrogen) and inserting Clover at amino acid position 1591 in PIEZO137. pMmPIEZO1Δ838-849-IRES-EGFP was generated by switching from the WT to the deleted sequence between the NotI and EcoRI sites of pMmPIEZO1-IRES-EGFP. Plasmids for full-length ATP11A, truncated ATP11A (1 bp insertion) and truncated PIEZO1 (13 bp deletion) were generated by subcloning the corresponding cDNA fragments with a C-terminal FLAG tag into pIRES2-Venus. pMLC2-WT-mClover-P2A-puro was constructed by subcloning MLC2 into pEGFP-N1, followed by replacing EGFP with the gene cassette composed of mClover, a self-cleavage P2A sequence and a puromycin-resistant gene (from pX260). pMLC2-DD-mClover-P2A-puro was constructed by introducing phospho-mimetic mutations (T18D/S19D) into pMLC2-WT-mClover-P2A-puro. pCDC50A and pCDC50A-FLAG were generated by subcloning CDC50A into pCMV-Tag4A (Agilent Technologies) with and without a stop codon, respectively. pCA-TMEM16F-mRuby2 was generated by subcloning the cDNA encoding CA-TMEM16F39 into pEGFP-N1 and replacing EGFP with mRuby2. pEGFP-P4-ATPases and pEGFP-AP3D1 were generated by subcloning the corresponding full-length into pEGFP-C1. Plasmids of mechanosensitive channels (pMSCs) were generated by inserting the qPCR-targeting fragment into pMD-20 (Takara). Retroviral vectors expressing P4-ATPases or CDC50A were generated by subcloning the corresponding full-length with FLAG- or mClover-tag at the C-terminus into pMXs-puro (Dr. Toshio Kitamura, the University of Tokyo). Plasmids were confirmed by sequencing.
Cells
C2C12, HEK293 cells, and human primary myoblasts were purchased from ATCC, RIKEN BRC and Thermo, respectively. Plat-E cells were a gift from Dr. Toshio Kitamura. Cells were grown in 10–20% FBS/DMEM in a 37 °C incubator with 5% CO2.
For CRISPR/Cas9-mediated gene editing28, C2C12 cells were transfected with pX330-PGKpuro using ViaFect (Promega) for 1 day and cultured with puromycin (1–3 μg/mL) for 3–5 days. Survived cells were cloned by limited dilution and genotyped (Supplementary Figures 1d and 4c). Data about CDC50A-deficient, ATP11A-deficient, and PIEZO1-deficient C2C12 cells were obtained from the corresponding clone #1 unless otherwise noted.
For stable lines expressing MLC2, C2C12 cells were transfected with pMLC2-WT-mClover-P2A-puro or pMLC2-DD-mClover-P2A-puro using CUY21EDITII electroporator (BEX) and cultured with puromycin. For stable lines expressing P4-ATPases or CDC50A, Plat-E cells were transfected with the pMXs-puro vectors using Viafect. Resultant retroviruses were used to infect C2C12 cells. Two days after infection, the cells were cultured with puromycin.
Myotube formation
C2C12 cells or primary myoblasts confluently grown in 20% FBS/DMEM were maintained in differentiation media [DMEM containing 2% horse serum (Gibco)] for the indicated periods. For RNA interference, cells were transfected twice with siRNA using Lipofectamine RNAiMAX (Thermo) and maintained in differentiation medium for 3 days. In pharmacological experiments, cells cultured in differentiation media for 2 days were incubated with calyculin A (1 nM, Wako) or CN03 (2 μg/mL, Cytoskeleton) for an additional 2 days. Z-VAD-FMK (50 µM, Peptide institute) was added to cells 1 day before switching to differentiation medium and maintained for 4 days.
Four days after differentiation, myotubes were stained with anti-MyHC antibody (14-6503-82, dilution 1:500, eBioscience) and DAPI (Dojindo), and visualized with an epifluorescence microscope (Axio-Observer.Z1, Zeiss) with a 10× objective lens. The number of nuclei, cell area and best-fit ellipse aspect ratio were calculated using ImageJ software. Cell fusion was evaluated as the percentage of nuclei in multinucleate MyHC+ cells (containing ≥16 nuclei) versus the total number of nuclei in MyHC+ cells in each microscopic field27. Polarized elongation was evaluated as percentages of the sum area occupied by MyHC+ cells with ≥3 aspect ratios versus the total area of MyHC+ cells in the field62, 63. Differentiation level was evaluated as percentages of nuclei in MyHC+ cells versus the total number of nuclei in each microscopic field27.
RT-PCR
Total RNA was isolated using ISOGEN II (Nippon Gene) and NucleoSpin RNA (Macherey-Nagel). cDNA was generated with PrimeScript II RTase (Takara) or ReverTra Ace qPCR RT Master Mix (Toyobo). Semi-qPCR was performed with EmeraldAmp MAX PCR Master Mix (Takara). qPCR was performed with PowerUp SYBR Green Master Mix (Thermo) using the StepOne system (Thermo). Copy numbers were determined using standard curves from pEGFP-P4-ATPases, pMSCs and pEGFP-AP3D1, and compared to Ap3d164. Relative expression was calculated using the 2−ΔΔCt method. Primers are listed in Supplementary Tables 3 and 4.
Co-immunoprecipitation and immunoblotting
For co-immunoprecipitation, HEK293 cells were transfected with pEGFP-P4-ATPases and pCDC50A-FLAG using X-tremeGENE HP DNA (Roche). Two days after transfection, the cells were lysed in cold buffer (150 mM NaCl, and 50 mM Tris at pH 7.4 containing 1% Triton X-100 and protease inhibitors). After removal of insoluble fractions by centrifugation, the supernatants were incubated with anti-FLAG M2 affinity gel (Sigma) for 30 min on ice. After washing with 0.1% Triton X-100/TBS, immunoprecipitates were eluted with FLAG peptides (500 μg/mL, Sigma) for 30 min on ice. Resulting supernatants were incubated in SDS sample buffer at 50 °C for 5 min.
For immunoblotting, proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. The membrane was blocked in TBS containing 5% skim milk and 0.1% Tween 20, stained with antibodies and visualized using an enhanced chemiluminescent reagent. Uncropped immunoblots are shown in Supplementary Figure 8.
Immunofluorescent analyses
Cells were fixed with 4% paraformaldehyde (PFA)/PBS, permeabilized in 0.1% Triton X-100/PBS, blocked in 1–2% BSA/PBS and probed with antibodies. An anti-goat tertiary antibody (A-11055, dilution 1:500, Thermo) was used to detect P-MLC2. Optical sections in the middle of cells were obtained using a confocal microscope (LSM710, Zeiss) with a 63× objective lens. Fluorescence profiles were analyzed using Zen software (Zeiss).
To determine PIEZO1 localization, either pHsPIEZO1-Clover or pHsPIEZO1-FLAG alone, pHsPIEZO1-FLAG together with pGFP-AHD, pMLC2-WT-mClover-P2A-puro or pmClover-NMIIA (1:1), or pHsPIEZO1-FLAG/pATP11A-mClover/pCDC50A (1:1:1) were introduced into C2C12 myoblasts by electroporation. Cells were cultured on μ-Dishes ibiTreat for 1 day and probed with phalloidin-TRITC (Sigma), anti-FLAG (F1804, dilution 1:500, Sigma), anti-GFP (598, dilution 1:1000, MBL), and anti-ROCK1 (GTX113266, dilution 1:100, GeneTex) antibodies.
To analyze NMIIA or P-MLC2 localization33, C2C12 cells on μ-Dishes ibiTreat were placed in differentiation medium for 3 days and stained with phalloidin-TRITC, DAPI, anti-NMIIA (M8064, dilution 1:50, Sigma), and anti-P-MLC2 (3671, dilution 1:50, CST) antibodies. Cell edges were divided into multiple rectangles orthogonal to the cell periphery. Within the fluorescence profile in each rectangle, the peak intensity between 0 and 5 μm from the cell edge and the mean intensity between 2 and 7 μm from the peak position were calculated. The cortex/cytoplasm ratio was determined as the average of the peak versus the mean intensity of all rectangles.
Time-lapse imaging
C2C12 cells on μ-Dishes ibiTreat were placed in differentiation medium for 2 days and visualized with an LCV110 incubator microscope equipped with a 20× objective lens (Olympus). DIC images were captured every 20 min for 2 days.
NBD-PS flipping assay
C2C12 myoblasts were detached by Cell Dissociation Solution Non-enzymatic 1× (Sigma), harvested by centrifugation and washed with HBSS (Thermo). After being placed on ice for at least 10 min, the assay was initiated by incubation with 0.1 μM NBD-PS for the indicated periods at 15 °C. The reaction was terminated by resuspension of the cells in HBSS containing fatty-acid-free BSA (3 mg/mL, Sigma). Cells (0.5–1 × 104) were analyzed with a flow cytometer (Cytomics FC 500, Beckman). Median fluorescence intensities of cell populations were determined using FlowJo software (Tree Star).
Phospholipid exchange
C2C12 cells trypsinized for annexin V labelling or grown on coverslips for Ca2+ measurements were incubated in PBS or DMEM containing 4 mM MαCD (2,6-Di-O-methyl-α-cyclodextrin, Wako) and 0.3 mM DOPC or 3:1 (mol:mol) DOPC/DOPS for 20 min at 37 °C40.
Annexin V labelling
For flow cytometry, C2C12 cells were detached, re-suspended in binding buffer (140 mM NaCl, 2.5 mM CaCl2 and 10 mM HEPES at pH 7.4) and incubated with annexin V-EGFP (Promokine) at room temperature (RT) for 5 min. Cells (2–4 × 104) were analyzed with a flow cytometer (Cytomics FC 500, Beckman). Histograms were drawn using FlowJo. Gating strategies are shown in Supplementary Figure 8.
For confocal microscopy, human primary myoblasts were differentiated on μ-Dishes ibiTreat for 2 days. After washing with binding buffer, the cells were incubated with Fixable Viability Dye eFluor 660 (Thermo) at RT for 15 min. Cells were treated with annexin V-EGFP at RT for 5 min, fixed with 4% PFA/PBS at RT for 10 min and stained with DAPI.
For Ca2+ measurements, human primary myoblasts were differentiated on Cell Desk LF1 (Sumitomo Bakelite) for 2 days, incubated with Fura2-AM (15 μM, Dojindo) at RT for 30 min, washed with binding buffer and incubated with annexin V-EGFP at RT for 5 min.
F2N12S measurements
C2C12 cells were detached, suspended in HBSS and placed on ice for at least 10 min. Then, the cells (0.1−1 × 107 cells/mL) were incubated with F2N12S (0.1 μM, Thermo) for 5 min at 15 °C. Emission spectra were recorded at an excitation wavelength of 400 nm using an LS55 Fluorescence Spectrometer (Perkin Elmer) at RT. Spectra were averaged over three replicates and corrected for lamp intensity variations and blanks.
Ca2+ measurements
Ca2+ measurements were conducted according to previous literature65. HEK293 cells were transfected with pMmPIEZO1-IRES-EGFP and pCA-TMEM16F-mRuby2 using ViaFect. The cells were then seeded onto poly-L-lysine-coated coverslips at one day post transfection and cultured overnight. C2C12 cells on poly-L-lysine-coated coverslips were transfected with plasmids expressing PIEZO1 or ATP11A and cultured overnight. Human primary myoblasts on poly-L-lysine-coated coverslips were transfected with siRNA and cultured for 2 days. Cells were loaded with Fura2-AM (5 μM) in 10% FBS/DMEM at 37 °C for 40 min and washed with HEPES-buffered saline (HBS)65. The coverslip was placed in a perfusion chamber mounted on a microscope (Axio-observer Z1) at RT. Time-lapse images were recorded every 10 s. Transfected cells were identified by fluorescent proteins. Cells were perfused with HBS for 2 min, then the perfusion solution was replaced with HBS containing Yoda1 (0.3–10 μM, Maybridge or Tocris) for 8 min. To analyze the effect of lyso-lipids, C2C12 cells were perfused with HBS containing 1–3 μM lyso-lipid for 5 min, then the perfusion solution was changed to 1 μM Yoda1/HBS for 8 min. After treatment with 3 μM LysoPS for 5 min, the perfusion solution was changed to HBS containing fatty-acid-free BSA (5 mg/mL) for 5 min, followed by 1 μM Yoda1/HBS for 8 min. To analyze the effect of LysoPS analogues, cells were incubated with 3 μM LysoPS analogues for 5 min, perfused with HBS for 2 min, then the perfusion solution was changed to 1 μM Yoda1/HBS for 10 min. Ratiometric images (F340/F380) were analyzed with Physiology software (Zeiss). Yoda1-induced Ca2+ influx was quantified as the difference in the Fura2 ratio between its maximum value and that at 1 min from imaging initiation.
Electrophysiology
HEK293T cells were transfected with pMmPIEZO1-IRES-EGFP using Lipofectamine 2000 (Thermo) and reseeded onto coverslips. Whole-cell recordings on HEK293T expressing MmPIEZO1 at 24–36 h post-transfection were conducted according to previous literature24. The internal solution contained (in mM): 140 CsCl, 5 EGTA, 10 HEPES, pH = 7.4 adjusted with CsOH, and the standard bath solution contained (in mM): 140 NaCl, 5 KCl, 2 CaCl2, 2 MgCl2, 10 glucose, and 10 HEPES, pH = 7.4 adjusted with NaOH). The folding potential was −60 mV and currents were recorded at a whole-cell configuration using Axopatch 200B amplifier (Molecular Devices), filtered at 5 kHz with a low-pass filter and digitized with Digidata 1440 A (Axon Instruments). Data were acquired with pCLAMP 10 (Axon Instruments). Glass pipettes (8250; King Precision Glass Inc.) had a resistance of 2–4 MΩ. Mechanical stimulation was provided by membrane indentation (8 μm) with a glass pipette for 150 ms. Currents were recorded before (control) and after administration of 5 μM lyso-phospholipid for 1 min, and after washing with fatty-acid-free BSA for 1 min.
Cell-attached recordings were carried out according to previous literature66. Electrophysiological recordings on MmPIEZO1-expressing HEK293 cells at 36–60 h post-transfection were conducted using an EPC10 amplifier and Patchmaster software (HEKA Elektronik). Data were acquired at 5 kHz and filtered at 2.9 kHz. Glass pipettes (1.5 OD, 0.85 ID; Sutter Instrument Company) had a resistance of 1.5–4 MΩ. The internal and bath solutions were prepared according to previous literature66. Pressure was administrated using a syringe while monitoring the pressure with a manometer. Patches were held at −60 mV and stimulated with negative pressure for 2 s. LysoPS (3 μM) was added to the internal solution.
Membrane tension measurements
An optical trap system (MMS-1064–200–2L/2E/2S, Sigma-koki) with a 1064-nm laser was used to trap cell-attached beads. A 100× oil objective (UPLSAPO100XO, NA = 1.4, Olympus) mounted on an inverted microscope (IX71, Olympus) was used to trap beads and visualize cells and beads in bright field. A motorized stage (BIOS-225T, Sigma-koki) controlled by a custom LabView program (National Instruments) was used to pull cells away from the stationary laser trap.
Trap stiffness calibration was performed by measuring the position variance of a trapped bead undergoing Brownian motion—a passive stiffness calibration based on equipartition theorem, assuming the thermal energy for each degree of freedom to be kBT/2 (where kB is the Boltzmann constant and T the absolute temperature). The particle is trapped in a harmonic potential in the x-direction,
| 1 |
kx is the trap stiffness and 〈x2〉 the position variance of the trapped particle. Using the information of the trapped particle coordinates:
| 2 |
The same applies for fluctuation in the y-axis direction.
Images of a trapped bead were captured at 1000 frames/s for 10 s, with the exposure time of the CCD camera (Zyla 5.5 cMOS, Andor) set to 1 ms. The center of mass coordinates of the trapped bead was obtained using ImageJ software. Variance in the x-axis direction 〈x2〉 was obtained by a Gaussian fit to the probability distribution of x. This resulted in a spring constant kx = 7.29 ± 0.50 [pN/μm]. We also confirmed that ky was similar to that of kx.
For tether pulling experiments, C2C12 cells on poly-L-lysine-coated glass bottom dishes (Matsunami) were incubated in 10% FBS/DMEM containing streptavidin-coated polystyrene beads (1.76 μm diameter, Spherotech) conjugated with biotinylated concanavalin A (J-OIL MILLS) for 1–2 h at 37 °C. For actin depolymerization, cells were incubated with cytochalasin D (20 μM, Sigma) for 2 h and washed with 10% FBS/DMEM. Cell-attached beads were trapped and pulled by moving the stage at 1 μm/s. Images were captured with a CCD camera (Zyla 5.5 cMOS, Andor) every 10 ms.
To obtain membrane tension, force-extension curves were acquired by calculating the force F exerted on the trapped bead as
| 3 |
where Δx is the deviation of the trapped bead from its original position. A linear fit was applied to the first linear part of the force-extension curves.
PS content
PS was separated from total lipid by two-dimensional thin-layer chromatography with the first solvent system of chloroform/methanol/acetic acid (65:25:10, v/v/v) and the second solvent system of chloroform/methanol/formic acid (65:25:10, v/v/v) on a silica plate and assessed by inorganic phosphate quantification67.
Analysis of Atp11a-deficient mice
Animal care, ethical usage and procedures were approved by the Animal Care Use and Review Committee of the Department of Engineering of Kyoto University. A transgenic mouse strain (Atp11atm1a(KOMP)Wtsi) harboring the ‘knockout-first’ conditional cassette68 in the Atp11a gene was purchased from EMMA. Atp11atm1a(KOMP)Wtsi was crossed with B6-Tg(CAG-FLPe)36 mice69 to generate Atp11atm1c(KOMP)Wtsi mice. The resultant mice were further mated with Myf5-cre transgenic mice on a C57BL/6 background43, to generate myoblast-specific Atp11a-deficient mice.
Myoblasts were isolated from extensor digitorum longus (EDL) muscles according to previous literature70. EDL myofibres were fixed with 4% PFA/PBS, permeabilized in 0.5% Triton X-100/PBS and blocked in 3% BSA/PBS. Samples were stained with anti-Pax7 and anti-PIEZO1 antibodies in the presence or absence of PIEZO1 peptide (5 μg/mL, Novus Biologicals) and visualized with a confocal microscope (LSM-800, Zeiss) or an epifluorescence microscope (Axio-observer Z1).
Cardiotoxin experiments were carried out according to previous literature46. Fifty microliters of 10 μM cardiotoxin (Sigma or Latoxan) was injected into tibialis anterior muscle of 8- to 12-week-old mice. The muscle was harvested at 2 weeks post-injection, and snap-frozen in isopentane cooled with liquid nitrogen. Longitudinal cryosections (thickness, 7 µm) obtained from the muscle were stained with anti-MyHC and anti-laminin antibodies and visualized with a microscope (Axio-observer Z1). In silico analysis on regenerating muscle was conducted by using NCBI dataset (#GDS4924)45.
Statistics and reproducibility
Representative figures are shown as data from a single experiment of multiple experiments or average data from pools across multiple experiments. Samples in quantitative data indicate microscopic fields (morphological analysis), single cells (flow cytometry, localization analysis, Ca2+ imaging, membrane tension measurement, electrophysiology), cell cultures (RT-PCR, PS quantification, F2N12S measurement) and muscles (in silico analysis). Quantitative data of single-cell-based or microscopic analyses were derived from pools across multiple experiments. All tests were repeated at least three times. Statistical significance was determined with the paired Student’s t-test. Bar and box-whiskers graphs, sample size (n) and P values are specified in the figure legends. No statistical methods to pre-specify sample size were used. No randomization was used. No blinding was done.
Data availability
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request.
Electronic supplementary material
Description of Additional Supplementary Information
Acknowledgements
We thank S. Sawada, K. Akiyoshi, T. Tamura, and I. Hamachi (Kyoto University) for valuable advice and technical assistance. We thank Kyoto University Live Imaging Center for time-lapse observations. pMXs-puro vector and a retrovirus packaging cell line Plat-E were kind gifts from T. Kitamura (the University of Tokyo). We thank the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP) and its funders for providing the mutant mouse line (Atp11atm1a(KOMP)Wtsi). Funding information may be found at www.sanger.ac.uk/mouseportal and associated primary phenotypic information at www.mousephenotype.org. This study was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas (15H05930 to M.U.); Fund for the Promotion of Joint International Research (15K21744 to M.U.); Grants-in-Aid for Scientific Research (KAKENHI) (25293012, 17H03805 to M.U.; 15H04846 to Y.H.) from Japan Society for the Promotion of Science (JSPS) and Ministry of Education, Culture, Sports, Science and Technology (MEXT); AMED-PRIME from Japan Agency for Medical Research and Development (JP17gm5810016 to Y.H.); intramural research grants (26-8, 29-4) for neurological and psychiatric disorders of NCNP (to Y.H.); Grant-in-Aid for JSPS Research Fellow (16J11409 to M.Ts.); and grants from the Takeda science and Ono medical research foundations (to Y.H.).
Author contributions
M.U. conceived the project. M.U. and Y.H. designed the project. M.Ts., Y.H., and M.U. interpreted data and wrote the paper. M.Ts. performed most experiments. K.I. performed Ca2+ measurements. R.N. established and characterized cell lines. M.O. produced and characterized Atp11a-deficient mice. A.S. and K.N. performed flow cytometry. M.M. and Y.M. supervised electrophysiological measurements. J.I. provided GFP-AHD construct. R.S. and M.Ta. supervised membrane tension measurements. T.O. and J.A. provided LysoPS analogues. M.K. and T.T. contributed to generate conditional KO mice. Y.N. and R.M. supervised the assay for myotube formation and isolation of primary myoblasts. Y.T. and M.To. supervised PIEZO1 analysis.
Competing interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-04436-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuji Hara, Email: hara@sbchem.kyoto-u.ac.jp.
Masato Umeda, Email: umeda@sbchem.kyoto-u.ac.jp.
References
- 1.Emoto K, Umeda M. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 2000;149:1215–1224. doi: 10.1083/jcb.149.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panatala R, Hennrich H, Holthuis JC. Inner workings and biological impact of phospholipid flippases. J. Cell Sci. 2015;128:2021–2032. doi: 10.1242/jcs.102715. [DOI] [PubMed] [Google Scholar]
- 4.Sebastian TT, Baldridge RD, Xu P, Graham TR. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta. 2012;1821:1068–1077. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murate M, et al. Transbilayer distribution of lipids at nano scale. J. Cell Sci. 2015;128:1627–1638. doi: 10.1242/jcs.163105. [DOI] [PubMed] [Google Scholar]
- 6.Hankins HM, Baldridge RD, Xu P, Graham TR. Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic. 2015;16:35–47. doi: 10.1111/tra.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki J, Umeda M, Sims PJ, Nagata S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 2010;468:834–838. doi: 10.1038/nature09583. [DOI] [PubMed] [Google Scholar]
- 8.Segawa K, et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344:1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- 9.van den Eijnde SM, et al. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 2001;114:3631–3642. doi: 10.1242/jcs.114.20.3631. [DOI] [PubMed] [Google Scholar]
- 10.Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hindi SM, Tajrishi MM, Kumar A. Signaling mechanisms in mammalian myoblast fusion. Sci. Signal. 2013;6:re2. doi: 10.1126/scisignal.2003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochreiter-Hufford AE, et al. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;497:263–267. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SY, et al. Stabilin-2 modulates the efficiency of myoblast fusion during myogenic differentiation and muscle regeneration. Nat. Commun. 2016;7:10871. doi: 10.1038/ncomms10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leikina E, et al. Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J. Cell Biol. 2013;200:109–123. doi: 10.1083/jcb.201207012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong J, Conboy IM. Phosphatidylserine directly and positively regulates fusion of myoblasts into myotubes. Biochem. Biophys. Res. Commun. 2011;414:9–13. doi: 10.1016/j.bbrc.2011.08.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segawa K, Kurata S, Nagata S. Human type IV P-type ATPases that work as plasma membrane phospholipid flippases and their regulation by caspase and calcium. J. Biol. Chem. 2016;291:762–772. doi: 10.1074/jbc.M115.690727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Mark VA, Elferink RP, Paulusma CC. P4 ATPases: flippases in health and disease. Int. J. Mol. Sci. 2013;14:7897–7922. doi: 10.3390/ijms14047897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Marques RL, Theorin L, Palmgren MG, Pomorski TG. P4-ATPases: lipid flippases in cell membranes. Pflug. Arch. 2014;466:1227–1240. doi: 10.1007/s00424-013-1363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato U, et al. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:37855–37862. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- 20.Kato U, et al. Role for phospholipid flippase complex of ATP8A1 and CDC50A proteins in cell migration. J. Biol. Chem. 2013;288:4922–4934. doi: 10.1074/jbc.M112.402701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen JP, et al. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front. Physiol. 2016;7:275. doi: 10.3389/fphys.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arashiki N, et al. ATP11C is a major flippase in human erythrocytes and its defect causes congenital hemolytic anemia. Haematologica. 2016;101:559–565. doi: 10.3324/haematol.2016.142273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The phenotype summary of systemic Atp11a-deficient mice in the website of IMPC (international mouse phenotyping consortium) http://www.mousephenotype.org/data/genes/MGI:1354735
- 24.Coste B, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Lewis AH, Grandl J. Touch, tension, and transduction—the function and regulation of piezo ion channels. Trends Biochem. Sci. 2016;42:57–71. doi: 10.1016/j.tibs.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millay DP, et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shynkar VV, et al. Fluorescent biomembrane probe for ratiometric detection of apoptosis. J. Am. Chem. Soc. 2007;129:2187–2193. doi: 10.1021/ja068008h. [DOI] [PubMed] [Google Scholar]
- 30.Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc. Natl Acad. Sci. USA. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray TV, et al. A non-apoptotic role for caspase-9 in muscle differentiation. J. Cell Sci. 2008;121:3786–3793. doi: 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- 32.Mukai A, Hashimoto N. Localized cyclic AMP-dependent protein kinase activity is required for myogenic cell fusion. Exp. Cell Res. 2008;314:387–397. doi: 10.1016/j.yexcr.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Ramanathan SP, et al. Cdk1-dependent mitotic enrichment of cortical myosin II promotes cell rounding against confinement. Nat. Cell Biol. 2015;17:148–159. doi: 10.1038/ncb3098. [DOI] [PubMed] [Google Scholar]
- 34.Kim JH, et al. Mechanical tension drives cell membrane fusion. Dev. Cell. 2015;32:561–573. doi: 10.1016/j.devcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syeda R, et al. Chemical activation of the mechanotransduction channel Piezo1. eLife. 2015;4:e07369. doi: 10.7554/eLife.07369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, et al. Endothelial cation channel PIEZO1 controls blood pressure by mediating flow-induced ATP release. J. Clin. Invest. 2016;126:4527–4536. doi: 10.1172/JCI87343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox CD, et al. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat. Commun. 2016;7:10366. doi: 10.1038/ncomms10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieber AD, Yehudai-Resheff S, Barnhart EL, Theriot JA, Keren K. Membrane tension in rapidly moving cells is determined by cytoskeletal forces. Curr. Biol. 2013;23:1409–1417. doi: 10.1016/j.cub.2013.05.063. [DOI] [PubMed] [Google Scholar]
- 39.Segawa K, Suzuki J, Nagata S. Constitutive exposure of phosphatidylserine on viable cells. Proc. Natl Acad. Sci. USA. 2011;108:19246–19251. doi: 10.1073/pnas.1114799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G, et al. Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids. Proc. Natl Acad. Sci. USA. 2016;113:14025–14030. doi: 10.1073/pnas.1610705113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daleke DL, Huestis WH. Incorporation and translocation of aminophospholipids in human erythrocytes. Biochemistry. 1985;24:5406–5416. doi: 10.1021/bi00341a019. [DOI] [PubMed] [Google Scholar]
- 42.Eisenhoffer GT, et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tallquist MD, Weismann KE, Hellström M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 44.Wang YX, Rudnicki MA. Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol. 2011;13:127–133. doi: 10.1038/nrm3265. [DOI] [PubMed] [Google Scholar]
- 45.Lukjanenko L, Brachat S, Pierrel E, Lach-Trifilieff E, Feige JN. Genomic profiling reveals that transient adipogenic activation is a hallmark of mouse models of skeletal muscle regeneration. PLoS ONE. 2013;8:e71084. doi: 10.1371/journal.pone.0071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohn RD, et al. Disruption of Dag1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell. 2002;110:639–648. doi: 10.1016/S0092-8674(02)00907-8. [DOI] [PubMed] [Google Scholar]
- 47.Charrin S, et al. Normal muscle regeneration requires tight control of muscle cell fusion by tetraspanins CD9 and CD81. Nat. Commun. 2013;4:1674. doi: 10.1038/ncomms2675. [DOI] [PubMed] [Google Scholar]
- 48.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whorton MR, MacKinnon R. X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature. 2013;498:190–197. doi: 10.1038/nature12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, et al. SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science. 2015;349:873–876. doi: 10.1126/science.aaa5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S, et al. Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 2015;34:669–688. doi: 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 53.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem. Sci. 1995;20:272–276. doi: 10.1016/S0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 54.Fairn GD, et al. An electrostatic switch displaces phosphatidylinositol phosphate kinases from the membrane during phagocytosis. J. Cell Biol. 2009;187:701–714. doi: 10.1083/jcb.200909025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alexander RT, et al. Membrane surface charge dictates the structure and function of the epithelial Na+/H+ exchanger. EMBO J. 2011;30:679–691. doi: 10.1038/emboj.2010.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeffries O, Tian L, McClafferty H, Shipston MJ. An electrostatic switch controls palmitoylation of the large conductance voltage- and calcium-activated potassium (BK) channel. J. Biol. Chem. 2012;287:1468–1477. doi: 10.1074/jbc.M111.224840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sung PJ, et al. Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc. Natl Acad. Sci. USA. 2013;110:20593–20598. doi: 10.1073/pnas.1306431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwashita M, et al. Synthesis and evaluation of lysophosphatidylserine analogues as inducers of mast cell degranulation. Potent activities of lysophosphatidylthreonine and its 2-deoxy derivative. J. Med. Chem. 2009;52:5837–5863. doi: 10.1021/jm900598m. [DOI] [PubMed] [Google Scholar]
- 59.Uwamizu A, et al. Lysophosphatidylserine analogues differentially activate three LysoPS receptors. J. Biochem. 2015;157:151–160. doi: 10.1093/jb/mvu060. [DOI] [PubMed] [Google Scholar]
- 60.Aoki K, et al. A RhoA and Rnd3 cycle regulates actin reassembly during membrane blebbing. Proc. Natl Acad. Sci. USA. 2016;113:E1863–E1871. doi: 10.1073/pnas.1600968113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren K, Crouzier T, Roy C, Picart C. Polyelectrolyte multilayer films of controlled stiffness modulate myoblast cells differentiation. Adv. Funct. Mater. 2008;18:1378–1389. doi: 10.1002/adfm.200701297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prager-Khoutorsky M, et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 64.Hildyard, J. C. & Wells, D. J. Identification and validation of quantitative PCR reference genes suitable for normalizing expression in normal and dystrophic cell culture models of myogenesis. PLoS Curr. 6 (2014). 10.1371/currents.md.faafdde4bea8df4aa7d06cd5553119a6. [DOI] [PMC free article] [PubMed]
- 65.Endo Y, et al. Dominant mutations in ORAI1 cause tubular aggregate myopathy with hypocalcemia via constitutive activation of store-operated Ca²+ channels. Hum. Mol. Genet. 2015;24:637–648. doi: 10.1093/hmg/ddu477. [DOI] [PubMed] [Google Scholar]
- 66.Lewis AH, Grandl J. Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. Elife. 2015;4:e12088. doi: 10.7554/eLife.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rouser G, Siakotos AN, Fleischer S. Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids. 1966;1:85–86. doi: 10.1007/BF02668129. [DOI] [PubMed] [Google Scholar]
- 68.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanki H, Suzuki H, Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp. Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- 70.Nagata Y, Partridge TA, Matsuda R, Zammit PS. Entry of muscle satellite cells into the cell cycle requires sphingolipid signaling. J. Cell Biol. 2006;174:245–253. doi: 10.1083/jcb.200605028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
Data supporting the findings of this manuscript are available from the corresponding authors upon reasonable request.