Abstract
Aim:
International collaboration is needed to enable large-scale pharmacogenomics studies in childhood asthma. Here, we describe the design of the Pharmacogenomics in Childhood Asthma (PiCA) consortium.
Materials & methods:
Investigators of each study participating in PiCA provided data on the study characteristics by answering an online questionnaire.
Results:
A total of 21 studies, including 14,227 children/young persons (58% male), from 12 different countries are currently enrolled in the PiCA consortium. Fifty six percent of the patients are Caucasians. In total, 7619 were inhaled corticosteroid users. Among patients from 13 studies with available data on asthma exacerbations, a third reported exacerbations despite inhaled corticosteroid use. In the future pharmacogenomics studies within the consortium, the pharmacogenomics analyses will be performed separately in each center and the results will be meta-analyzed.
Conclusion:
PiCA is a valuable platform to perform pharmacogenetics studies within a multiethnic pediatric asthma population.
Keywords: : asthma, children, consortium, genetics, pharmacogenomics, treatment
Asthma is the most common chronic disease in childhood. Although it cannot be cured, effective treatments are available to decrease the symptoms, maintain lung function and prevent future exacerbations [1]. Standard treatment regimens for persistent asthma include regular use of inhaled corticosteroids (ICS) combined with long-acting β2 agonists (LABA) and short-acting β2 agonists (SABA) as needed [2]. There is heterogeneity in response to treatment; approximately 30–40% of the patients receiving ICS, do not show an improvement in lung function and remain uncontrolled [3–6]. Uncontrolled asthma is associated with low quality of life for patients and can be life threatening [7,8]. Furthermore, unscheduled physician visits and hospital admissions due to exacerbations are responsible for almost half of the costs of asthma management [9,10].
Poor adherence to medication, ongoing environmental exposures, disease severity and misdiagnosis influence response to treatment in asthmatic patients. In addition, it has been shown that genetic variation contributes to the heterogeneity in treatment response [11]. To date, a large number of candidate gene studies and several genome-wide association studies (GWAS) have been conducted to study the pharmacogenomics of childhood asthma [12,13]. However, one of the main unmet needs for pediatric asthma management is the lack of clinically available biomarkers (e.g., pharmacogenetic markers) to guide asthma treatment. Genetic associations have been reported with three commonly used outcome measures (i.e., asthma exacerbations, asthma symptoms and lung function) [14,15]. Different outcomes might reflect different aspects of asthma control and the heterogeneity in the outcome measures complicates the comparison of study results. In addition, most studies have been performed in relatively small study populations. There is a need for international collaboration in the field of pharmacogenomics of asthma to obtain large sample sizes of well-phenotyped asthmatic children to perform large scale meta-analyses to assess the clinical value of genetic markers for asthma management and identify markers that can guide asthma treatment [16,17]. There have been successful efforts to establish consensus on diagnosis and management of asthma [18,19]. The Pharmacogenomics in Childhood Asthma (PiCA) consortium was initiated in December 2013 and brings together asthma studies that have genetic data and treatment outcome measures. The main goals of the PiCA consortium are to create a platform to identify new pharmacogenomic markers in asthma by conducting GWAS meta-analyses. To replicate these new and also previously identified loci that are associated with treatment response and finally, to develop pharmacogenetics-guided algorithms to guide asthma therapy to improve symptoms and reduce/prevent future exacerbations. This is the first consortium that focuses on pharmacogenomics in childhood asthma. In this study, we describe the characteristics of the study populations currently included in the PiCA consortium, assess the outcome measures that can be used to study treatment response within the consortium and describe the design of the pharmacogenomics studies that will be performed within PiCA
Methods
PiCA consortium
The PiCA consortium was established in December 2013 by the pharmacogenomics research group of AH Maitland van der Zee (Utrecht University, The Netherlands) by expanding existing and new collaborations. Studies were identified from the literature, at conferences and by references of other PiCA collaborators. Studies were eligible to participate in the PiCA consortium if:
Data on asthmatic children or young persons were collected;
DNA samples were collected or could be collected;
Data were collected on asthma drug use;
Data were collected on treatment outcome.
PiCA is a growing consortium and new studies can join the consortium if they meet the inclusion criteria [20].
Data collection
An online questionnaire (created using SurveyMonkey [ 21]) was sent to the investigators of each cohort to collect information about the patients and design of the studies.
Characteristics of the studies & study populations
Information was collected on the following characteristics of the studies: study design (i.e., asthma cohort, clinical trial and [high risk] birth cohort), country where the study was conducted and location of patient enrollment (type of healthcare center: primary, secondary or tertiary care). Per study, the following data were collected on the study populations: the age range (in years.), number of male asthmatics and the number of patients in distinct ethnic groups (i.e., Caucasians, African–Americans, Hispanics and Asians). In order to assess the potential of PiCA to perform pharmacogenomics studies, the numbers of patients with a reported use of asthma medication (ICS, SABA, LABA, leukotriene modifiers, anti-IgE and oral corticosteroids [OCS]) were collected per study. It was also ascertained whether data regarding environmental exposures and atopy were collected. The source for the DNA collection (i.e., blood and saliva) and availability of whole-genome genotyping data were assessed.
Outcome measures & treatment response
The presence of information on exacerbations, asthma symptoms and lung function was assessed for each study. A severe exacerbation was considered as a short course (3–5 days) OCS use or a hospitalization/emergency room (ER) visit according to the American Thoracic Society/European Respiratory Society (ATS/ERS) 2009 statement [22]. The presence of information on unscheduled general practitioner visits or asthma-related absences from school was also assessed. The two outcomes have been used as indicators of exacerbations in several pharmacogenomics studies. For asthma symptoms, presence of information on validated asthma symptom questionnaires (asthma control questionnaire [ACQ] or asthma control test [ACT]) was assessed within the studies. The comparability of the results of these two questionnaires has been shown previously [23]. Patients with ACQ scores ≥0.75 and ACT scores <20 were considered to have poor asthma control. In addition, availability of information on asthma symptoms based on guidelines (i.e., Global Initiative for Asthma [GINA] and ATS/ERS) was also assessed. According to the availability of data in each study, the number of patients with exacerbations despite regular use of ICS was collected. For observational studies, the presence of any of these outcomes in the preceding 6 or 12 months was gathered. Asthma diagnosis is difficult in infants and preschool children. Hence, from birth cohorts within the PiCA consortium, we collected outcomes of children ≥6 years of age with physician-diagnosed asthma. Cohen's Kappa statistic was calculated per study, to show the overlap between patients experiencing exacerbations and asthma symptoms [24]. This was calculated for those studies in which both outcomes were available. The analysis was performed in R (package ‘irr’) [25].
Furthermore, since lung function measures are widely used as a response outcome in asthma, it was ascertained whether data regarding lung function measurements, especially changes in Forced expiratory volume in the first second (FEV1) from baseline over time (before and after treatment) and changes in FEV1 after SABA use were also collected within the studies included in the consortium.
Results
Baseline characteristics of the studies & patients
Currently, 21 asthma studies from 12 different countries are enrolled in the PiCA consortium. PiCA includes 15 asthma cohorts, 3 birth cohorts, 2 high-risk birth cohorts (inclusion of infants based on allergic history of the mother) and 1 clinical trial (Table 1).
Table 1. . Pharmacogenomics in Childhood Asthma characteristics: study design and patient characteristics.
| Study | Country | Study design | Recruiting centers | Asthmatic patients (n) | Age (range, years) | Male n, (%) | Mean (SD) FEV1% predicted baseline | Medication | DNA source | Ref. | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICS | LABA | SABA | LTRA | ||||||||||
| BAMSE | Sweden | General birth cohort | Primary care | 420 | 0–16 | 242 (57.6) | 103 (11.0) | 226 | 57 | 218 | – | Peripheral blood§ | [28] |
| BREATHE | UK | Asthma cohort | Primary and secondary care | 1570 | 3–22 | 1017 (64) | 96.6 (15.5) | 959 | 62 | 1505 | 210 | Saliva§ | [27] |
| British Columbia Childhood Asthma Cohort | Canada | Asthma cohort | Tertiary/quaternary referral center | 343 | 1–18 | 223 (65) | – | 343 | 54 | 343 | 79 | Buccal cell and saliva | |
| CAMP | USA | RCT | Tertiary care | 1041 | 5–12 | 621 (59) | 95.6 ± 18 | 311 | – | 418 | – | Peripheral blood§ | [29] |
| COPSAC2000 | Denmark | High-risk birth cohort | Written invitation | 43 | 0–7 | 22 (51) | 94.4 (12.1) | 43 | † | 43 | † | Peripheral blood§ | [30] |
| COPSAC2010 | Denmark | General birth cohort | Written invitation | 90 | 0–5 | 52 (57) | 97.1 (12.1) | 90 | 0 | 90 | † | Peripheral blood§ | |
| COPSACSevere† | Denmark | Asthma cohort | Registry based | 1173 | 0–25 | 791 (67) | – | † | † | † | † | Peripheral blood | |
| DUCHA | Greece | Asthma cohort | Tertiary care | 193 | 5–14 | 179 (92) | 101.2 (12.8) | 193 | 56 | 18 | 25 | Peripheral blood | |
| ESTATe | The Netherlands | Case–control | Primary care | 111 | 4–19 | 67 (60) | – | 110 | 42 | 111 | 2 | Saliva§ | |
| followMAGICS | Germany/Austria | Asthma cohort | Secondary and tertiary care | 313 | 7–25 | 194 (62) | – | 150 | 104 | 107 | 27 | Peripheral blood§ | [31] |
| GALA II‡ | USA | Case–control | Secondary care, community- and clinic-based recruitment | 2377 | 8–21 | 1288 (54) | 90.8 (16.2) | 1174 | 368 | 1900 | 610 | Peripheral blood and saliva§ | [32] |
| Generation R¶ | The Netherlands | Population-based birth cohort | Primary, secondary and tertiary care | 399 | Fetal – ongoing | 249 (62.4) | 100 (12.8) | 200 | 50 | 280 | 10 | Umbilical cord blood§ | [33] |
| GOASC‡ | Spain | Asthma cohort | Secondary and tertiary care | 125 | 2–18 | 76 (60) | 94.6 (15.2) | 125 | 78 | 14 | 107 | Peripheral blood and saliva | [34] |
| PACMAN | The Netherlands | Asthma cohort | Primary care | 995 | 4–12 | 616 (61) | – | 844 | 229 | 819 | 87 | Saliva§ | [35] |
| PAGES | UK | Asthma cohort | Primary, secondary and tertiary care | 701 | 2–18 | 519 (74) | 94 (16) | 648 | 347 | 696 | 286 | Saliva | [36] |
| PASS | UK | Asthma cohort | Tertiary care | 525 | 5–18 | 307 (58) | – | 525 | 395 | 525 | 369 | Peripheral blood and saliva§ | [37] |
| PIAMA | The Netherlands | General birth cohort/high-risk birth cohort | Primary care | 428 | 8 | 254 (59.3) | 105.4 (12.2) | 208 | 28 | 210 | 5 | [38] | |
| SAGE II‡ | USA | Case–control | Secondary care, community- and clinic-based recruitment | 987 | 8–21 | 503 (51) | 98.7 (14.1) | 670 | 171 | 822 | 96 | Peripheral blood and saliva§ | [32] |
| Singapore Cross Sectional Genetic Epidemiology Study‡ | Singapore | Asthma cohort | Tertiary care | 1450 | 18–25 | 600 (41) | 76.9 (12.8) | 394† | † | † | † | † | [39] |
| Slovenia | Slovenia | Asthma cohort | Tertiary care | 350 | 5–19 | 162 (46) | 89.9 (14.85) | 193 | † | † | 86 | Peripheral blood | [40] |
| Study of asthma in Puerto Rican children (HPR) | USA | Case–control | Tertiary care and population-based probabilistic sampling design | 593 | 6–14 | 320 (53.9) | 88.5 (16.5) | 213 | 9 | 452 | 133 | Peripheral blood§ | [60] |
| Total: 21 studies | 12 countries | 14,227 | 7619 | 2050 | 8571 | 2132 | |||||||
†Data collection ongoing.
‡Patient inclusion ongoing.
§Studies with GWAS data available.
¶Patient follow-up ongoing, numbers based on participation until 1 April 2015, aged 9 years.
BAMSE: Swedish abbreviation for Children, Allergy, Milieu, Stockholm, Epidemiology; CAMP: Childhood asthma management program; DUCHA: Democritus University Child Hospital;ESTATe: Effectiveness and safety of treatment with asthma therapy in children; GALA II: Genes-environment and admixture in Latino Americans; GOASC: Genetics of asthma in Spanish children; ICS: Inhaled corticosteroid; LABA: Long-acting β2 agonist; LTRA: Leukotriene receptor antagonist; PACMAN: Pharmacogenetics of asthma medication in children: medication with anti-inflammatory effect; PASS: Pharmacogenetics of adrenal suppression; PIAMA: The Prevention and incidence of asthma and mite allergy; RCT: Randomized controlled trial; SABA: Short-acting β2 agonist.
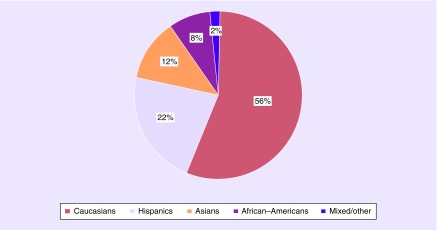
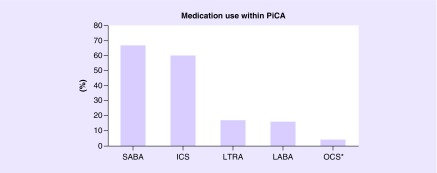
In total, PiCA includes data of 14,227 asthmatic patients up to 25 years of age. In 17 studies (80%), asthma was based on physician diagnosis and/or hospital records. For three studies, asthma diagnosis was based on parental-reported asthma diagnosis. PACMAN included children with a regular use of asthma medication. Analysis of PiCA children showed that 58% are male. From almost all patients within PiCA (97%), information was available on ethnic background. The majority of the asthmatic patients in PiCA are Caucasians (56%), 12% are Asians, 22% are Hispanic and 8% have an African/African–American background and the remaining (2%) have mixed/other ethnic backgrounds (Figure 1). In the PiCA consortium studies, data on medication use were collected based on parental/patient reports (17 studies), pharmacy records (nine studies) and physician's prescriptions (five studies). Medication data were available for 12,736 patients. Most of the patients in the studies were treated with ICS (n = 7619) and SABA (n = 8571). Furthermore, 2050 patients received LABA and 2132 used LTRA. OCS as a maintenance medication was used in 568 patients (Figure 2). In line with clinical asthma guidelines, most patients were treated with a combination of different asthma medications.
Figure 1. . Ethnic backgrounds of the asthmatic patients.
Figure 2. . Medication use within PiCA.
OCS* considered as long-term therapy.
Outcome measures & treatment response
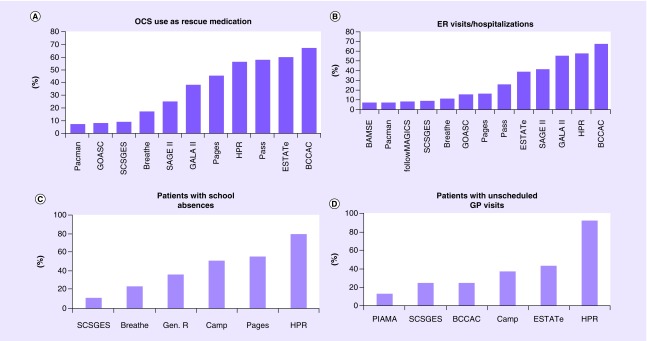
Thirteen studies had information on exacerbations and approximately a third of the patients had severe exacerbations despite ICS treatment. In 11 studies (including 5769 patients), data were available on OCS use as rescue medication despite ICS treatment. The prevalence of OCS use ranged between 7 and 67% in different studies, and in total, 1929 (33%) PiCA patients on ICS had received rescue OCS in the preceding 6–12 months of the study visit. Thirteen studies had data available on asthma-related ER visits or hospitalizations despite ICS (n = 6095). The prevalence of ER visits/hospitalizations ranged between 7 and 67%. In total, 1806 (29%) patients reported asthma-related ER visits or hospitalizations. Data on asthma-related school absences despite ICS use were available for 2587 patients in six studies. Furthermore, data on unscheduled general practitioner visits were available for 1479 patients in six studies (Figure 3). The total number of patients experiencing exacerbations in each study is shown in Supplementary Table 1.
Figure 3. . Exacerbations despite regular use of ICS in the preceiding 6 months or year.
A) Percentage of OCS users as a rescue medication in 11 PiCA studies. B) Percentage of patients with ER visit/hospitalization in 13 PiCA studies. C) Percentage of patients with asthma-related school absences in 6 PiCA studies. D) Percentage of patients with unscheduled GP visits in 6 studies.
BCCAC; British Columbia Childhood Asthma Cohort, Gen.R; Generation R, SCSGES; Singapore Cross Sectional Genetic Epidemiology Study.
In PASS and BREATHE exacerbation data were available in the preceding 6 months.
Validated scaled questionnaires to assess current asthma symptoms (ACQ and ACT) were used in five studies (DUCHA, ESTATe, PACMAN, PAGES and Singapore Cross Sectional Genetic Epidemiology Study; in a total of 2070 patients). In this population, 37% (n = 766) of the patients had ACQ scores ≥0.75 or ACT scores <20 indicating poor asthma control. Furthermore, a modified version of the 1978 ATS–Division of Lung Diseases Epidemiology Questionnaire [41] was used to assess current asthma control in GALA II and SAGE II in 1725 patients; 41% had uncontrolled asthma symptoms based on this questionnaire. In addition to these scaled questionnaires, several other categorical measures of symptoms were used in studies. Modified GINA definition for long-term asthma control was used in BAMSE (n = 226), with 34% of the patients having poor asthma symptoms. In the PIAMA study (n = 110), 43% of the patients using inhaled steroids had uncontrolled asthma at 8 years of age. Guidelines of the Dutch Pediatric Society (NVK), which follow the GINA guidelines, were used to define uncontrolled asthma [42].
Regarding lung function measurements, changes in FEV1 after bronchodilator were measured in seven studies and changes in FEV1 from baseline were measured in four studies.
Information on asthma severity was available for 5608 PiCA patients. The number of severe asthmatics according to ATS/ERS, GINA and British Thoracic Society/Scottish Intercollegiate Guidelines Network (step four or higher) guidelines was 838.
Overlap between exacerbations & asthma symptoms
In three studies (GALA II, PACMAN and SAGE II), we could assess the overlap between exacerbations (defined by OCS use) and patients with asthma symptoms. In all three patient populations, there was only a slight-to-fair agreement between these two outcomes (κ: 0.03–00.21); 46–72% of the patients with reported OCS use as a rescue medication also had uncontrolled asthma symptoms according to the asthma questionnaire. The overlap between patients with ER visits/hospitalizations in the past 6–12 months and uncontrolled asthma symptoms in four studies (BAMSE, GALA II, PACMAN, and SAGE II) was also poor (κ: 0.03–0.22); 41–55% of the patients with ER visits/hospitalizations had uncontrolled asthma symptoms (Supplementary Table 2).
Pharmacogenomic studies in PiCA
DNA samples have been collected in 20 studies, and for 1 study the DNA collection is still ongoing. The source of DNA per study is shown in Table 1.
A protocol written by the research center interested in a specific research question will be sent to the principal investigators of the consortium for review. Next, the protocol will be sent to all PiCA studies. Centers that are willing to participate will perform the association analysis and the results will be sent to research center that initially initiated the research proposal. In case, individual study lacks resources or expertise to perform the analyses, other PiCA collaborators will help to perform the analysis.
GWAS in PiCA
Currently, GWAS data are available for 13 studies (n = 6743; Supplementary Table 2). In addition, 1967 DNA samples from five studies will be genotyped; BAMSE (n = 400), BREATHE (n = 92), PAGES (n = 514), GoShare (n = 561) and SLOVENIA (n = 400).
In the discovery phase of the GWAS, genotyped samples will be imputed with the Michigan imputation server (available at: https://imputationserver.sph.umich.edu). After imputation and quality check, association analysis will be performed with efficient and parallelizable association container toolbox (EPACTS; available at: http://genome.sph.umich.edu/wiki/EPACTS). Principal component analysis and adjustment for gender and age will be performed when necessary. GWAS meta-analysis will be performed by METASOFT [43]. In the replication phase, association analysis will be performed for the top hits identified in the discovery phase.
Candidate gene approach in PiCA
Candidate gene studies will be conducted for newly identified SNPs from GWAS meta-analyses and for previously identified SNPs in GWAS of childhood asthma onset and pharmacogenomics of asthma and SNPs that might associate with treatment response based on biological pathways.
Association analysis will be performed in the studies that have genotype or imputed data with high quality. The results of the association analysis will be meta-analyzed.
Discussion
The PiCA consortium is a unique initiative that brings together data from 14,227 asthmatic children/young adults from 12 different countries worldwide. In genetic association studies, replication of the results across populations with different ethnic backgrounds is of high importance in order to support the findings of the pharmacogenomics analysis [44]. The PiCA consortium is a novel platform to study the pharmacogenomics of uncontrolled childhood asthma despite asthma treatment.
It is important to study pharmacogenomics of childhood asthma in addition to adult asthma, since asthma phenotypes differ between children and adults and findings in adult studies cannot be translated directly to the pediatric asthma population [45]. For example, a genetic variant influencing FBXL7 expression has been found by the CAMP group to associate with improvement in asthma symptoms in response to ICS in two pediatric populations, but it failed to replicate in adults [13]. Several GWAS of response to asthma medication have been published by the CAMP study group [46–48] and they can be found in the National Human Genome Research Institute and the European Bioinformatics Institute (NHGRI-EBI) GWAS catalog [49]. In addition, variation in the ADRB2 gene has been associated with altered LABA response, but mainly in pediatric populations [27,50–52]. Hence, it is important to study treatment response in asthmatic children. However, assessing treatment response in asthmatic children remains a challenging subject, as symptoms may vary over time. Different measures of uncontrolled asthma (i.e., exacerbations, symptoms or lung function) might reflect distinct dimensions of the disease. It has been previously shown that demographic characteristics and biomarker profiles of children with severe exacerbations were different from children with persistent symptoms [15], and children without asthma symptoms can be prone to severe exacerbations [53]. Furthermore, It has been shown that the definition of treatment response influences the genetic risk profile associated with drug response [46,54,55]. Calculated κ values showed only minimal-to-moderate agreements between asthma symptoms and exacerbations. Since different dimensions of uncontrolled asthma include different patient populations and overlap only partly, distinct outcome measures need to be studied separately. An important strength of PiCA is the collection of well-defined asthma outcomes in >14,000 individuals for future pharmacogenomics studies within the PiCA consortium, we will perform analyses using distinct measures of poor treatment response that reflect different dimensions of asthma.
Within the PiCA consortium, we included study designs such as observational asthma cohorts and (high risk) birth cohorts. An observational study (cohort or case control) is a common approach to assess pharmacogenomics and should not be undervalued. Observational studies can provide valuable evidence for clinically relevant pharmacogenomics markers. Once identified, the next step would be further replication and developing a prognostic biomarker test with additional replication for generalizability and investigating the functional biology to interrogate the mechanistic aspect of the replicated findings.
Major strengths of the design of the PiCA consortium are: inclusion of patients from mild-to-severe asthmatics with thoroughly investigated outcome and phenotype data (i.e., exacerbations and asthma symptoms); and the coverage of the broad spectrum of pediatric asthmatic medication users, which will make it possible to assess the value of pharmacogenetics for subgroups of patients. Study heterogeneity makes it possible to assess the generalizability of findings across multiple designs and/or multiple ethnicities. Sensitivity analyses can be used to assess for which group a certain marker might have the highest clinical value.
In addition to large-scale pharmacogenomics studies, which are the main goal of this consortium, PiCA also has potential to study other factors influencing treatment outcomes, such as continued exposure to allergens or epigenomics. However, obtaining additional biological samples or data might be complicated for some PiCA studies; this might only be possible in part of the PiCA population. Several potential limitations of this consortium should be acknowledged. One of the limitations of PiCA could be population stratification. However, this heterogeneity will help us to identify different genetic markers associated with the treatment response in patients with different ethnicities. Furthermore, it will help us to discover pharmacogenomics markers that are associated with the treatment response in asthmatics regardless of the ethnic background of the patients. In genome-wide association analyses, we will adjust the results of each cohort by principal components when necessary. In candidate gene studies, the analyses will be performed separately for each study and the results will be meta-analyzed. Furthermore, we will also perform sensitivity analysis by conducing separate analysis for patients with different ethnic backgrounds. The results of these analyses will be compared and in the presence of a significant difference, they will be reported. Another limitation could be the wide age range of the patients included in PiCA, although this does reflect the general asthma population in clinical practice, infant onset asthma might be a different phenotype from asthma in teenagers [56]. In addition, asthma diagnosis is complicated at a young age, and infants and preschool children can suffer from symptoms (such as wheezing) similar to those caused by asthma. In PiCA, we will only include children who were still experiencing asthma symptoms at ≥6 years of age. In the majority of the PiCA studies (17 out of 21), asthma was based on physician diagnosis and/or hospital records. Although criteria for physician diagnosis might differ between countries, this difference reflects current clinical practice.
This is the first large effort to unite childhood asthma studies with a common interest in pharmacogenetics. Various studies within PiCA have collected detailed information on asthmatic children and followed children prospectively, making PiCA a unique platform for collaboration and validation. Several other studies (Asthma Genetics in Hungary, EUROPA from The Netherlands, GoShare from the UK and the Canadian asthma cohort) are still in the stage of recruiting patients, data and genotyping DNA samples, and will participate in the future projects of the PiCA consortium. In other fields, such as in cardiovascular pharmacogenomics, large research consortia have delivered key discoveries [26,57–59]. PiCA is a growing consortium and it provides the opportunity to study pharmacogenetics on a large scale, paving the way for precision medicine in asthma.
Summary points.
PiCA consortium
The Pharmacogenomics in Childhood Asthma (PiCA) consortium is a newly established pharmacogenomics consortium that brings pediatric asthma studies worldwide together to enable large-scale pharmacogenetics and pharmacogenomics studies.
Baseline characteristics of the patients
PiCA currently includes data of 14,227 asthmatic children/young adults up to 25 years of age and is multi-ethnic. The PiCA population is 56% Caucasian, 22% Hispanic, 12% Asian, 8% African/African–American and 2% mixed/other.
Medication use within PiCA
From 12,736 patients with available data on medication use in the PiCA consortium; 7619 patients received ICS, 8571 received SABA, 2050 received LABA and 2132 used LTRA. OCS as a maintenance medication was used by 568 patients.
Outcome measures & treatment response
There are different measures to study treatment outcomes, e.g., OCS use as rescue medication, asthma-related ER visits or hospitalizations, lung function measures or reported asthma symptoms.
Data on asthma-related Emergency Room/hospitalizations despite ICS treatment were recorded in 13 studies in the PiCA consortium, 11 studies had data available on OCS use as rescue medication despite ICS use. Changes in forced expiratory volume in 1 second (FEV1) after bronchodilator were measured in seven studies and changes in FEV1 from baseline were measured in four studies. Validated scaled questionnaires to assess current asthma symptoms (ACQ and ACT) were used in five studies.
Pharmacogenomic studies in PiCA
PiCA is a collaborative platform to initiate novel pharmacogenetic/omic studies for discovery or replication. PiCA researchers can propose studies and each PiCA study interested to participate, can perform their own data-analysis using pre-approved study protocols.
Conclusion
PiCA is a growing consortium and it provides the opportunity to study pharmacogenetics/omics of pediatric asthma on a large scale using different measures of uncontrolled asthma (i.e., exacerbations, symptoms, or lung function) paving the way for precision medicine.
Supplementary Material
Footnotes
Financial & competing interests disclosure
This study was supported by a grant from Stichting Astma Bestrijding (SAB grant 2014/063) and ERA-Net ERACoSysMed (SysPharmPedia 99). Maria Pino-Yanes was supported by the grant AC15/00015 by Instituto de Salud Carlos III within the ERACoSysMed 1st Joint Transnational Call from the European Union (SysPharmPedia 99), under the Horizon 2020. The Hartford-Puerto Rico study was supported by grants HL079966 and HL117191 from the US NIH. The PASS study was funded by the UK Department of Heath Chair of Pharmacogenetics which was awarded to Professor Munir Pirmohamed. BAMSE was supported by The Swedish Research Council, The Swedish Heart-Lung Foundation, Stockholm County Council (ALF), and the Strategic Research Programme (SFO) in Epidemiology at Karolinska Institutet. The ESTATe study was supported by a Zon MW research grant (113201006). Dr AH Maitland-van der Zee and Dr. Susanne Vijverberg received an unrestricted grant for research on Pharmacogenomics in pediatric asthma from GSK (PACMAN), Dr. AH Maitland-van der Zee was part of an advisory board meeting on benralizumab from AstraZeneca. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Global Initiative for Asthma. Global strategy for asthma management and prevention. 2016. www.ginasthma.org
- 2.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. 2004;59(5):469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 3.Szefler SJ, Martin RJ, King TS, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. 2002;109(3):410–418. doi: 10.1067/mai.2002.122635. [DOI] [PubMed] [Google Scholar]
- 4.Szefler SJ, Phillips BR, Martinez FD, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. 2005;115(2):233–242. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Malmstrom K, Rodriguez-Gomez G, Guerra J, et al. Oral montelukast, inhaled beclomethasone, and placebo for chronic asthma. A randomized, controlled trial. Montelukast/Beclomethasone Study Group. 1999;130(6):487–495. doi: 10.7326/0003-4819-130-6-199903160-00005. [DOI] [PubMed] [Google Scholar]
- 6.Langmack EL, Martin RJ. Heterogeneity of response to asthma controller therapy: clinical implications. 2010;16(1):13–18. doi: 10.1097/MCP.0b013e328333af9c. [DOI] [PubMed] [Google Scholar]
- 7.Bahadori K, Doyle-Waters MM, Marra C, et al. Economic burden of asthma: a systematic review. 2009;9:24. doi: 10.1186/1471-2466-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman KR, Boulet LP, Rea RM, Franssen E. Suboptimal asthma control: prevalence, detection and consequences in general practice. 2008;31(2):320–325. doi: 10.1183/09031936.00039707. [DOI] [PubMed] [Google Scholar]
- 9.Williams AE, Lloyd AC, Watson L, Rabe KF. Cost of scheduled and unscheduled asthma management in seven European Union countries. 2006;15(98):4–9. [Google Scholar]
- 10.Barnett SBL, Nurmagambetov TA. Costs of asthma in the United States: 2002–2007. 2011;127(1):145–152. doi: 10.1016/j.jaci.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Fleming L, Wilson N, Bush A. Difficult to control asthma in children. 2007;7(2):190–195. doi: 10.1097/ACI.0b013e3280895d0c. [DOI] [PubMed] [Google Scholar]
- 12.Pijnenburg MW, Szefler S. Personalized medicine in children with asthma. 2015;16(2):101–107. doi: 10.1016/j.prrv.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Park H-W, Dahlin A, Tse S, et al. Genetic predictors associated with improvement of asthma symptoms in response to inhaled corticosteroids. 2014;133(3):664.e5–669.e5. doi: 10.1016/j.jaci.2013.12.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers AJ, Tantisira KG, Fuhlbrigge AL, et al. Predictors of poor response during asthma therapy differ with definition of outcome. 2009;10(8):1231–1242. doi: 10.2217/PGS.09.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu AC, Tantisira K, Li L, Schuemann B, Weiss ST, Fuhlbrigge AL. Predictors of symptoms are different from predictors of severe exacerbations from asthma in children. 2011;140(1):100–107. doi: 10.1378/chest.10-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans WE, Relling MV. Moving towards individualized medicine with pharmacogenomics. 2004;429(6990):464–468. doi: 10.1038/nature02626. [DOI] [PubMed] [Google Scholar]
- 17.Yip VLM, Hawcutt DB, Pirmohamed M. Pharmacogenetic markers of drug efficacy and toxicity. 2015;98(1):61–70. doi: 10.1002/cpt.135. [DOI] [PubMed] [Google Scholar]
- 18.Custovic A, Ainsworth J, Arshad H, et al. The Study Team for Early Life Asthma Research (STELAR) consortium “Asthma e-lab”: team science bringing data, methods and investigators together. 2015;70(8):799–801. doi: 10.1136/thoraxjnl-2015-206781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bacharier LB, Boner A, Carlsen K-H, et al. Diagnosis and treatment of asthma in childhood: a PRACTALL consensus report. 2008;63(1):5–34. doi: 10.1111/j.1398-9995.2007.01586.x. [DOI] [PubMed] [Google Scholar]
- 20.Pica consortium. www.pica-consortium.org
- 21.SurveyMonkey. www.surveymonkey.com/
- 22.Reddel HK, Taylor DR, Bateman ED, et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. 2009;180(1):59–99. doi: 10.1164/rccm.200801-060ST. [DOI] [PubMed] [Google Scholar]
- 23.Khalili B, Boggs PB, Shi R, Bahna SL. Discrepancy between clinical asthma control assessment tools and fractional exhaled nitric oxide. 2008;101(2):124–129. doi: 10.1016/S1081-1206(10)60199-8. [DOI] [PubMed] [Google Scholar]
- 24.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. 2005;37(5):360–363. [PubMed] [Google Scholar]
- 25.R Development Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2008. R: a language and environment for statistical computing.www.R-project.org [Google Scholar]
- 26.Ferreira MAR, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. 2011;378(9795):1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer CNA, Lipworth BJ, Lee S, Ismail T, Macgregor DF, Mukhopadhyay S. Arginine-16 beta2 adrenoceptor genotype predisposes to exacerbations in young asthmatics taking regular salmeterol. 2006;61(11):940–944. doi: 10.1136/thx.2006.059386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickman M, Melén E, Berglind N, et al. Strategies for preventing wheezing and asthma in small children. 2003;58(8):742–747. doi: 10.1034/j.1398-9995.2003.00078.x. [DOI] [PubMed] [Google Scholar]
- 29.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. 1999;20(1):91–120. [PubMed] [Google Scholar]
- 30.Bisgaard H. The Copenhagen Prospective Study on Asthma in Childhood (COPSAC): design, rationale, and baseline data from a longitudinal birth cohort study. 2004;93(4):381–389. doi: 10.1016/S1081-1206(10)61398-1. [DOI] [PubMed] [Google Scholar]
- 31.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 32.Nishimura KK, Galanter JM, Roth LA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. 2013;188(3):309–318. doi: 10.1164/rccm.201302-0264OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnenschein-van der Voort AMM, Jaddoe VW, Raat H, et al. Fetal and infant growth and asthma symptoms in preschool children: the Generation R Study. 2012;185(7):731–737. doi: 10.1164/rccm.201107-1266OC. [DOI] [PubMed] [Google Scholar]
- 34.Pino-Yanes M, Sánchez-Machín I, Cumplido J, et al. IL-1 receptor-associated kinase 3 gene (IRAK3) variants associate with asthma in a replication study in the Spanish population. 2012;129(2):573–575. 575.e1–575.e10. doi: 10.1016/j.jaci.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Koster ES, Raaijmakers JAM, Koppelman GH, et al. Pharmacogenetics of anti-inflammatory treatment in children with asthma: rationale and design of the PACMAN cohort. 2009;10(8):1351–1361. doi: 10.2217/pgs.09.79. [DOI] [PubMed] [Google Scholar]
- 36.Turner SW, Ayres JG, Macfarlane TV, et al. A methodology to establish a database to study gene environment interactions for childhood asthma. 2010;10:107. doi: 10.1186/1471-2288-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawcutt DB, Jorgensen AL, Wallin N, et al. Adrenal responses to a low-dose short synacthen test in children with asthma. 2016;84(5):793. doi: 10.1111/cen.13069. [DOI] [PubMed] [Google Scholar]
- 38.Zuidgeest MGP, Koster ES, Maitland-van der Zee A-H, et al. Asthma therapy during the first 8 years of life: a PIAMA cohort study. 2010;47(2):209–213. doi: 10.3109/02770900903483790. [DOI] [PubMed] [Google Scholar]
- 39.Andiappan AK, Sio YY, Lee B, et al. Functional variants of 17q12–21 are associated with allergic asthma but not allergic rhinitis. 2016;137(3):758.e3–766.e3. doi: 10.1016/j.jaci.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 40.Berce V, Kozmus CEP, Potočnik U. Association among ORMDL3 gene expression, 17q21 polymorphism and response to treatment with inhaled corticosteroids in children with asthma. 2013;13(6):523–529. doi: 10.1038/tpj.2012.36. [DOI] [PubMed] [Google Scholar]
- 41.Ferris BG. Epidemiology Standardization Project (American Thoracic Society) 1978;118(6 Pt 2):1–120. [PubMed] [Google Scholar]
- 42.de Jongste J, Vrijlandt EJLE. Astma bijkinderen: samenvatting van de herziene rich-tlijnen van de Sectie Kinderlongziekten vande NVK. 2007. [Guideline “Asthma in Children” for pediatric pulmonologists]. Hilversum: SKL.
- 43.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. 2011;88(5):586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall IP, Blakey JD. Genetic association studies in thorax. 2005;60(5):357–359. doi: 10.1136/thx.2005.040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleming L, Murray C, Bansal AT, et al. The burden of severe asthma in childhood and adolescence: results from the paediatric U-BIOPRED cohorts. 2015;46(5):1322–1333. doi: 10.1183/13993003.00780-2015. [DOI] [PubMed] [Google Scholar]
- 46.Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. 2011;365(13):1173–1183. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park HW, Dahlin A, Tse S, et al. Genetic predictors associated with improvement of asthma symptoms in response to inhaled corticosteroids. 2014;133(3):664–669.e5. doi: 10.1016/j.jaci.2013.12.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tantisira KG, Damask A, Szefler SJ, et al. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. 2012;185(12):1286–1291. doi: 10.1164/rccm.201111-2061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hindorff L, Parkinson H, Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuurhout MJL, Vijverberg SJH, Raaijmakers JAM, et al. Arg16 ADRB2 genotype increases the risk of asthma exacerbation in children with a reported use of long-acting β2-agonists: results of the PACMAN cohort. 2013;14(16):1965–1971. doi: 10.2217/pgs.13.200. [DOI] [PubMed] [Google Scholar]
- 51.Wechsler ME, Kunselman SJ, Chinchilli VM, et al. Effect of beta2-adrenergic receptor polymorphism on response to longacting beta2 agonist in asthma (LARGE trial): a genotype-stratified, randomised, placebo-controlled, crossover trial. 2009;374(9703):1754–1764. doi: 10.1016/S0140-6736(09)61492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner S, Francis B, Vijverberg S, et al. Childhood asthma exacerbations and the Arg16 β2-receptor polymorphism: a meta-analysis stratified by treatment. 2016;138(1):107–113. doi: 10.1016/j.jaci.2015.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carroll CL, Schramm CM, Zucker AR. Severe exacerbations in children with mild asthma: characterizing a pediatric phenotype. 2008;45(6):513–517. doi: 10.1080/02770900802017751. [DOI] [PubMed] [Google Scholar]
- 54.Leusink M, Vijverberg SJH, Koenderman L, et al. Genetic variation in uncontrolled childhood asthma despite ICS treatment. 2015;16(2):158–168. doi: 10.1038/tpj.2015.36. [DOI] [PubMed] [Google Scholar]
- 55.Vijverberg SJH, Tavendale R, Leusink M, et al. Pharmacogenetic analysis of GLCCI1 in three north European pediatric asthma populations with a reported use of inhaled corticosteroids. 2014;15(6):799–806. doi: 10.2217/pgs.14.37. [DOI] [PubMed] [Google Scholar]
- 56.Depner M, Fuchs O, Genuneit J, et al. Clinical and epidemiologic phenotypes of childhood asthma. 2013;189(2):129–138. doi: 10.1164/rccm.201307-1198OC. [DOI] [PubMed] [Google Scholar]
- 57.Owen RP, Altman RB, Klein TE. PharmGKB and the international warfarin pharmacogenetics consortium: the changing role for pharmacogenomic databases and single-drug pharmacogenetics. 2008;29(4):456–460. doi: 10.1002/humu.20731. [DOI] [PubMed] [Google Scholar]
- 58.Paternoster L, Standl M, Chen CM, et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. 2012;44(2):187–192. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forno E, Sordillo J, Brehm J, et al. Genome-wide interaction study of dust mite allergen on lung function in children with asthma. 2016;(17):30154–30159. doi: 10.1016/j.jaci.2016.12.967. pii: S0091–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.