Abstract
Mitochondria within a cell exist as a population in a dynamic morphological continuum. The balance of mitochondrial fusion and fission dictates a spectrum of shapes from interconnected networks to fragmented individual units. This plasticity bestows the adaptive flexibility needed to adjust to changing cellular stresses and metabolic demands. The mechanisms that regulate mitochondrial dynamics, their importance in normal cell biology, and the roles they play in disease conditions are only beginning to be understood. Dysfunction of mitochondrial dynamics has been identified as a possible disease mechanism in Parkinson’s disease. This chapter will introduce the budding field of mitochondrial dynamics and explore unique characteristics of affected neurons in Parkinson’s disease that increase susceptibility to disruptions in mitochondrial dynamics.
2.1 Introduction
Mitochondrial Dynamics refers to the observation that mitochondria within the individual cell go through fusion and fission events. Visually, this results in a morphological spectrum with contrasting degrees of elongation and fragmentation. Plasticity bestows the adaptive flexibility needed to adjust to changing cellular stresses and metabolic demands. Constant network remodeling also establishes a mechanism for quality control of the mitochondrial population with important ramifications for long-term function and health.
While our understanding of mitochondrial dynamics is just beginning, descriptions of morphological transitions by mitochondria can be traced to reports dating back nearly a century. In 1914, Lewis and Lewis elegantly describe witnessing fusion and fission events along with an incredible range of structures exhibited by mitochondria in cultured cells (Lewis and Lewis 1914). The body of knowledge surrounding mitochondrial dynamics has expanded greatly from these early studies and this chapter provides a brief introduction into this exciting field.
2.2 Mitochondrial Dynamics Proteins
The state of balance between four dynamin-related proteins essentially controls mitochondrial fusion and fission. From yeast to humans, these highly conserved enzymes share homologous GTPase and transmembrane regions that form complexes and alter the curvature of the mitochondrial membranes. It is the relative activities of oppositional forces that together determine mitochondrial morphology. Complete network fragmentation can result from increased expression or activation of fission proteins. However the mitochondrial network will also fragment if fusion activity is inhibited. Similarly, elongated tubular networks occur with enhanced fusion activity as well as through blockage of fission. These extremes are reminders that antagonism between counteracting enzymatic forces sets the shape of mitochondria and therefore must always be considered simultaneously when deciphering network morphology.
2.3 Fusion
Fusion is categorized into two forms and three mitochondrial localized GTPases control the difference between transient and complete fusion events. Transient fusion involves only outer membranes while complete fusion requires merging of both inner and outer membranes. A complete fusion event occurs with a rapid diffusion of soluble mitochondrial components followed by a more gradual mixing of membrane elements (Twig et al. 2006; Partikian et al. 1998; Karbowski et al. 2004a; Jakobs et al. 2003; Jakobs 2006; Arimura et al. 2004; Busch et al. 2006). This process is believed to bestow complementation between units and increased homogeneity over the network. Complementation is a key mechanism by which mitochondria can rescue a damaged unit within the network. The effect of loss of fusion has been assessed in several model systems. Network fragmentation and susceptibility to apoptosis, decreased mitochondrial membrane potential and oxygen consumption, and increased ROS production are seen with blocking fusion and underscore its importance in maintaining mitochondrial integrity.
Two homologous proteins known as mitofusin 1 and mitofusin 2 (Mfn1 and Mfn2) function together to merge the outer membranes of mitochondria. Both proteins share relevant functional domains and connect adjacent membranes through coiled-coil antiparallel homotypic (Mfn1–Mfn1) and heterotypic (Mfn1–Mfn2) dimers. The GTPase activity of Mfn1 is higher compared to Mfn2, thus the relative proportion of dimer combinations has important functional consequences for fusion rates within the cell (Chen et al. 2003; Koshiba et al. 2004). Turnover occurs in part by polyubiq-uitination-mediated recruitment of chaperone proteins, such as p97, which mediate retrotranslocation of mitofusins and promote their proteasomal degradation (Tanaka et al. 2010a ). Curiously, Mfn2 appears to have other crucial functions in the cell beyond mitochondrial fusion. One such function is tethering mitochondria and endoplasmic reticulum during calcium exchange between the organelles (de Brito and Scorrano 2008). In neurons, Mfn2 has been shown to play a role in motility by connecting mitochondria to the Miro/Milton transport complex (Misko et al. 2010).
Inner mitochondrial membranes are joined via the protein encoded by Optic Atrophy type 1 gene (OPA1) (Song et al. 2009). Expression of OPA1 is highly regulated at the transcriptional level with eight possible isoforms available through alternative splicing (Song et al. 2007). Imported OPA1 protein localizes to the intermembrane space and is further processed by several proteases to produce five additional isoform variations (Ehses et al. 2009). Functional differences between isoforms are not entirely understood but it is known that both long and short forms of OPA1 are needed to maintain fusion capacity (Song et al. 2007; Duvezin-Caubet et al. 2006). In both soluble and membrane associated forms, OPA1 exists in a complex with mitofusins (Cipolat et al. 2004). This interaction is crucial for complete fusion as cleavage of OPA1 disrupts the complex and limits mitochondria to only transient fusion events. In this way, proteolytic removal of long isoforms provides a mechanism for creating network fragmentation in response to stress (Griparic et al. 2007). The specific molecular signals that trigger processing of OPA1 remain largely a mystery but clearly both induction of apoptosis and dissipation of mitochondrial membrane potential induce OPA1 cleavage (Gottlieb 2006; Guillery et al. 2008; Lee et al. 2004; Olichon et al. 2007). This effect may represent a stopgap attempt to limit spread of damaged material within the mitochondrial network by isolating units that pose a risk or have been selected for mitophagy.
2.4 Fission
Fission is crucially involved in numerous important cell pathways including mitochondrial inheritance by daughter cells during cellular division, differentiation of post-mitotic cells such as neurons and cardiomyocytes, mitophagy, and forms of cell death (Lee et al. 2004, 2011a; Yu et al. 2005; Gomes and Scorrano 2008; Mendl et al. 2011; Grohm et al. 2010; Karbowski 2010; Jourdain et al. 2009; Wilkerson and Sankar 2011; Choudhary et al. 2011; Kane and Youle 2010; Shroff et al. 2009; Ishihara et al. 2009; Frank et al. 2001). Loss of fission results in increased mitochondrial connectivity, loss of mtDNA, bioenergetic deficiency, and alterations in apoptosis (Landes and Martinou 2011; Westermann 2010; Sheridan and Martin 2010; Parone et al. 2008). One cytosolic GTPase performs the division of fused mitochondria. Dynamin-related protein 1 (Drp1) translocates to mitochondrial scission sites and polymerizes into structures that surround the perimeter of the organelle (Fukushima et al. 2001). Polymerization activates the GTPase domain of Drp1, which literally causes constriction and pinching of a single unit into two individual daughters (Legesse-Miller et al. 2003). Sub-cellular localization and activity of Drp1 is regulated by several post-translational modifications, such as phosphorylation, ubiquitination, nitrosylation, and sumoylation (Figueroa-Romero et al. 2009; Cho et al. 2009; Wang et al. 2011a; Santel and Frank 2008; Braschi et al. 2009; Taguchi et al. 2007).
Network fragmentation occurs in response to various factors including intracellular calcium levels, mitochondrial membrane potential, and ATP availability (Yoon et al. 2003; Kong et al. 2005). For example, the calcium-sensitive phosphatase calcineurin promotes fission by dephosphorylating cytosolic Drp1, which causes translocation to mitochondria (Scorrano 2005). Two additional proteins that reside on the outer mitochondrial membrane act together as a receptor for organizing Drp1 to sites of fission. Mitochondrial fission 1 protein (hFis1) is a transmembrane protein that marks sites of division (Yu et al. 2005; Koch et al. 2005; Serasinghe and Yoon 2008; Otera et al. 2010; James et al. 2003). Mitochondrial Fission Factor (Mff) interacts with hFis1 and serves as an adaptor that recruits Drp1 to promote polymerization (Otera et al. 2010). Recent studies have demonstrated that Mff is required for fission but hFis1 is dispensible (Otera et al. 2010; Huang et al. 2011a). This surprising finding suggests the existence of other proteins that can supersede hFis1 and act as alternative receptors for Mff and Drp1.
In summary, there are three key steps for mitochondrial fragmentation. First is the localization of fission adaptor proteins, such as hFis1 and Mff, to fission sites. Second Drp1 must be recruited from the cytosol and polymerize at fission sites. Finally, there must be an accompanying inhibition of fusion through cleavage of long Opa1 isoforms within mitochondria. These are the three minimal steps that are required for a continuous network of fused mitochondria to transition towards fragmentation.
2.5 Approaches for Measurement of Mitochondrial Dynamics
While early observational studies describe mitochondrial fusion and fission, direct experimental proof was first obtained using polyethylene glycol (PEG)-mediated cell fusion assays (Legros et al. 2002; Neuspiel et al. 2005). In this method, two separate cultures of cells have their mitochondria labeled with different molecular probes, such as green and red fluorescent proteins (GFP, RFP). Combining the two cell populations in the presence of PEG detergent promotes fusion of plasma membranes and subsequent mixing of mitochondrial populations. Fluorescence heterogeneity is detected in the resultant pool, with cells containing mitochondria purely expressing GFP or RFP while other units display a mixture of both colors. These studies provide direct proof of fusion between individual mitochondria but significant practical limitations of the methodology left ample room for improvement. Recently, more physiologically relevant studies have employed an expanded arsenal of probes in the form of dyes and proteins that allow precise, detailed data collection on a wide range of parameters.
The tracking of dyes that accumulate within mitochondria in a membrane potential dependant manner, such as Tetramethylrhodamine ethyl ester perchlorate (TMRE), was a significant advancement in the study of mitochondrial dynamics. Confocal imaging studies of cells labeled with TMRE reveal stable mitochondrial membrane potentials maintained for a period of 40–80 s followed by a sudden drop of more than 15 mV (Loew et al. 1993). These studies were pioneering in our understanding of mitochondrial biology but limited by an inability to assure that the detected mitochondrion did not fuse and/or divide during the recording time. For example, fission can occur without movement of the two daughter mitochondria or involve only the inner (but not the outer) mitochondrial membrane (Twig et al. 2006; Malka et al. 2005). Therefore fission cannot be reliably identified by observation of separation of a mitochondrion into two segments. Similarly, the repositioning of a mitochondrion to become juxtaposed to another mitochondrion is not an indication that a fusion event occurred (Twig et al. 2006). The use of photoactivatable proteins was a breakthrough because it overcame these technical difficulties of imaging individual organelles that move and change morphology within a complex architecture (Betzig et al. 2006; Patterson and Lippincott-Schwartz 2002).
The creation of tools that allow laser-mediated photoactivation of mitochondrial matrix-targeted GFP (mtPA-GFP) facilitates improved biophysical and morphologic measurements as it is nontoxic for the cells and can therefore be used to make observations over an extended period of time (Karbowski et al. 2004a; Arimura et al. 2004; Busch et al. 2006). Overall fusion rates for a cell can be quantified by activating the mtPA-GFP in a subset of the mitochondrial population and then tracking diffusion of the fluorescence signal over time, as the mtPA-GFP spreads to non-activated fusion partners. While fusion rates vary across cell types and conditions, several studies have shown that when 10–20% of population is activated, the mtPA-GFP equilibrates across the entire network in approximately 45 min (Karbowski et al. 2004a, b; Twig et al. 2008a). This is predicted to result in homogeneity in protein content and function across the mitochondrial population. In addition to analyzing the properties of the entire mitochondrial network within a cell, mtPA-GFP can be used to assess attributes of an individual mitochondrion. These observable characteristics include the size, shape, membrane potential, motility, and temporal properties of fusion. For example, photoactivation of a selected mitochondrion enables real time tracking of that individual. Long-term monitoring of single mitochondrial units with activated mtPA-GFP in INS1 and COS7 cells has allowed for direct quantification of fusion rates. These studies revealed the frequency of fusion to be once every 5–20 min per mitochondrion (Twig et al. 2008a). The duration of fusion events is typically brief, lasting ~100 s and followed by fission (Arimura et al. 2004; Twig et al. 2008a). Thus mitochondria spend most of their time as individual solitary units. These studies provided the groundwork for a concept of the mitochondrial life cycle consisting of two stages, the pre-fusion period (solitary period) and the post-fusion period when mitochondria are connected together (networked period).
The combination of both TMRE and mtPA-GFP has two significant additional benefits in measuring biophysical properties of mitochondria (Twig et al. 2006; Molina and Shirihai 2009). First, it provides means for accurate determination of organelle boundaries that can be easily followed despite movement within a dense mitochondrial network. It is also beneficial because it allows comparison of the fluorescence intensities of the two probes to get a ratiometric value. This offers a tool for quantification of changes in membrane potential that are independent of exact focal plane. By avoiding the need to perform repeated imaging through the entire z-axis, monitoring can be done with greatly reduced phototoxicity. The combination approach with TMRE and mtPA-GFP extends the permissible recording periods for tracking mitochondria within a cell from minutes to hours. This advancement helped reveal that mitochondria maintain stable membrane potential during their solitary period for up to 2 h (Twig et al. 2008a; Wikstrom et al. 2007).
One major limitation of direct user-based microscopy studies is they tend to be labor intensive and therefore not amenable to high-throughput screening. Recent description of an innovative cell-free fusion assay addresses this shortcoming with a luciferase-based approach that will allow large-scale screens of modifiers of mitochondrial dynamics (Schauss et al. 2010). Specifically, the assay is based on a bimolecular complementation approach using both mitochondrial targeted yellow fluorescent protein (YFP) and luciferase constructs separated by a leucine zipper. The two split proteins are expressed separately in large cultures of cells from which mitochondrial populations are isolated and purified. During the assay the two populations of mitochondria are mixed and through fusion the split proteins are able to combine to form functional molecules. This new system holds great promise as it provides multiple highly quantifiable readouts. Linking luciferase activity to fusion events provides a much-needed tool for rapid, large-scale screening of conditions that affect mitochondrial dynamics. The methods described here provided important insights into mitochondrial biology yet they likely represent a mere beginning, as creative new approaches expand boundaries in this emerging field.
2.6 Benefits from Mitochondrial Dynamics
Responsive mitochondrial dynamics is an essential part of an array of cellular processes including mitosis, fuel sensing, ATP production, mitophagy, and apoptosis (Arimura et al. 2004; Twig et al. 2008a; Nakada et al. 2001a, b; Skulachev 2001; Liesa et al. 2008; Molina et al. 2009). In some situations, entire network fragmentation is necessary to facilitate autophagic clearance of mitochondria (mitophagy), such as during erythrocyte maturation, sperm mitochondria in oocyte fertilization and apoptosis dependant on PTP opening (Takano-Ohmuro et al. 2000; Shitara et al. 2000; Elmore et al. 2001). On a local level, fusion allows mixing and complementation between two units. In a fused state, exchange of components such as solutes, metabolites, and soluble proteins occurs rapidly (Partikian et al. 1998; Arimura et al. 2004; Chen et al. 2003, 2005; Chen and Chan 2005; Shaw and Nunnari 2002; Griffin et al. 2006) while membrane embedded proteins and mitochondrial DNA spread more slowly (Twig et al. 2006; Legros et al. 2004; Gilkerson et al. 2008; Wikstrom et al. 2009). The ability of mitochondria to fuse together reduces content heterogeneity and thus is a first line of defence against dysfunction (Legros et al. 2002; Chen et al. 2005, 2011; Legros et al. 2004; Hori et al. 2011; Chan 2006; Ono et al. 2001; Mazzoni and Falcone 2011).
Maintaining quality control through mitochondrial dynamics simultaneously optimizes bioenergetic efficiency and reduces risks associated with oxidative phos-phorylation by removal of damaged material. For example, inhibition of mitochondrial fission leads to an increase in oxidized proteins along with decreased maximal oxygen consumption rates during uncoupled respiration. These findings suggest the accumulation of oxidized material is due to a loss of clearance rather than increased production of ROS. Failure to properly remove damaged components impairs mitochondrial function and limits reserve capacity. These outcomes are particularly important for long-lived cells with high metabolic demands.
The dependence of quality control on fission may stem from the ability to generate unequal daughter units. Most fission events produce heterogeneous daughters with opposite membrane potential “de flections,” usually greater than 5 mV. Oxidized and damaged material is also inequitably distributed and this ability to regularly create uneven fission events suggests a selective mechanism of intra-mitochondrion segregation and separation. The net effect of numerous cycles of asymmetric divisions and selective isolation is the ability to concentrate undesirable material within a minimal number of units. Damage laden mitochondria ultimately get isolated and prevented from fusing with the rest of the network through reduction in fusion proteins (Twig et al. 2008a, b). In various cell types, loss of membrane potential leads to the polyubiquitination and proteasomal degradation of proteins associated with the mitochondrial outer membrane, such as Miro and mitofusins. In addition to degradation of individual proteins, ubiquitination serves to recruit autophagy-related scaffold adaptors, such as p62 and HDAC6 (Huang et al. 2011b; Lee et al. 2011b ). These scaffold proteins bind to polyubiquitin chains and serve essentially as receptors for autophagosomes to facilitate lysosomal degradation of the mitochondrial unit through mitophagy. Blocking autophagy is sufficient to cause a buildup of damaged material including mitochondria, particularly in energy intensive tissues such as brain, heart, liver, kidney, and pancreatic beta cells (Twig et al. 2008a ; Jung and Lee 2009 ; Taneike et al. 2010; Kimura et al. 2011 ). It is worth noting in these cases that dysfunctional mitochondria accumulate without requiring any additional toxins or mitochondrial stressors. Mitochondrial turnover is a major proportion of the basal autophagic processing within cells, especially those with elevated metabolic demands. Long-lived post-mitotic cells with chronic high levels of turnover are inherently vulnerable to disruptions in the quality control pathway (Terman et al. 2010).
2.7 Regulation of Mitochondrial Dynamics
Multiple levels of cell signaling are involved in regulating mitochondrial dynamics. Despite its complexity, the system can be broken down into two simple categories, global and local regulation (Hyde et al. 2010). Figure 2.1 illustrates regulatory elements during the mitochondrial lifecycle and Table 2.1 lists examples of global and local control during fusion, fission, and the solitary period. Control derived from the cellular macroenvironment that affects the entire mitochondrial network is categorized as global regulation of mitochondrial dynamics, such as during the cell cycle (Lee et al. 2004; Taguchi et al. 2007; Scarpulla 2002a; Arakaki et al. 2006 ). At different stages of mitotic cell division there is transcriptional control of dynamics proteins that lead to opposite extremes in network morphology. A concert of transcription factors mediates increases in mitochondrial mass, respiratory capacity, and energy production that are required during S phase (Scarpulla 2002a, b). Accordingly, there is hyperfusion of the network during G1-S phase while hyper-fragmentation occurs in late S and M phases.
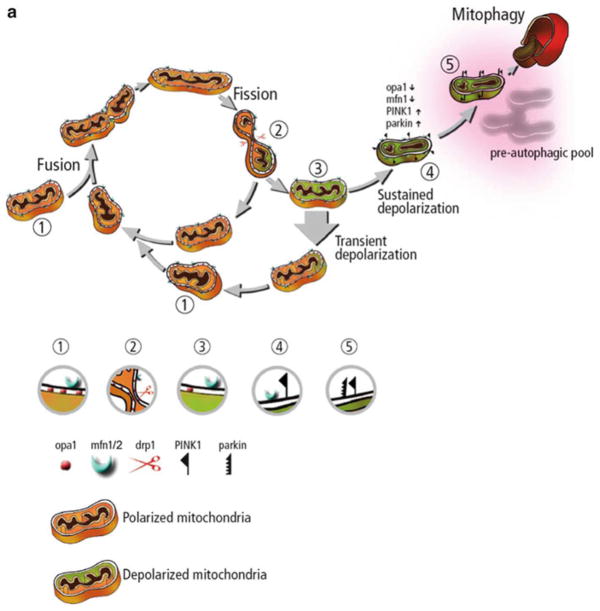
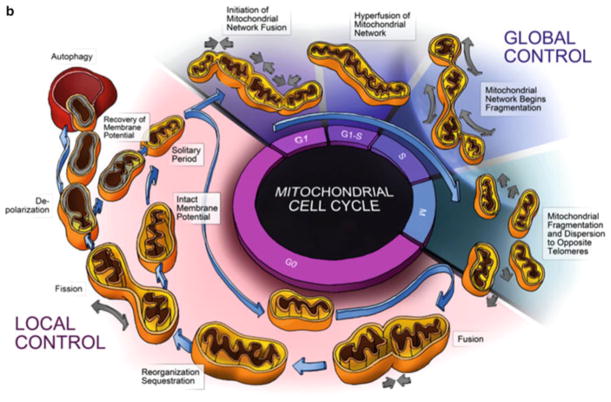
Fig. 2.1.
Organellar and cellular controls of the mitochondrial life cycle. The mitochondria life cycle. (a) The mitochondria life cycle. Mitochondria go through continuous cycles of fusion and fission. Each cycle last 5–20 min. Fusion is brief (1) and triggers fission events (2). A daughter mitochondrion may maintain intact membrane potential (orange ) or depolarize (3, green). When depolarized a subsequent fusion event is unlikely to occur, unless the mitochondrial re-polarizes. As a result, depolarized daughter mitochondria remain solitary. Depolarized and solitary mitochondria (4) remain for 1–4 h in a pre-autophagic pool before being consumed by the autophagic machinery. (b) The interaction of the mitochondria life cycle with the cell cycle—this diagram depicts the normal life cycle of an individual mitochondrion during the G0 phase of the cell cycle. The mitochondrion undergoes fusion, fission, depolarization, and degradation by autophagy. This process is depicted as one of local control whereby mitochondrial events are largely dictated by the local energetic status and associated local signals. During the cell cycle global signals cause concerted changes in the mitochondrial population, as noted by hyperfusion in the G1-S and fragmentation during the M phase. These global population effects are governed by the cellular demand for energy required by cell division and the need for homogenization and sequestration of cellular components during met-phase. The cell cycle serves as an elegant example of the parities of local and global control
Table 2.1.
Local (organellar) versus global (cellular) controls of mitochondrial dynamics
| Control mechanism | Outcome | References |
|---|---|---|
| Fission | ||
| Global cellular control | ||
| Recruitment of Drp1 to mitochondria is calcium dependent and regulated by calcineurin | Elevated cytosolic calcium levels activate calcineurin to dephosphorylate Drp1 | Yoon et al. (2003), Kong et al. (2005), Kaddour-Djebbar et al. (2010), Hom et al. (2010), Cribbs and Strack (2007), Cereghetti et al. (2008, 2010), Tan et al. (2011), Wang et al. (2011c) |
| Prolonged exercise increases transcription and expression of Fis1 while decreasing mitofusins | Acute increases in metabolic demands of skeletal muscle stimulate fission | Ding et al. (2010b) |
| BH3 only proteins and Bax/Bak induce fission, Bax/Bak in healthy cells control fusion through MFN2 | Interact at mitochondrial scission sites to promote fission | Karbowski (2010), Shroff et al. (2009), Karbowski et al. (2002), Wu et al. (2011), Sheridan et al. (2008) |
| Sumoylation of Drp1 occurs by multiple enzymes and is present at fission sites | Protects Drp1 from degradation and increases fission activity | Figueroa-Romero et al. (2009), Braschi et al. (2009), Harder et al. (2004), Dimmer and Scorrano (2006) |
| High levels of oxidative stress causes fragmentation of the mitochondrial network | Drp1 phosphorylation and Bid translocation increase fission activity | Grohm et al. (2010), Qi et al. (2011) |
| Amino acid and other nutrient deprivation causes hyperfusion by downregulation of Drp1 | Fused mitochondrial network evades autophagic degradation during starvation | Rambold et al. (2011a, b) |
| Drp1 expression is transcriptionally activated by p53 protein in response to apoptotic stimuli | The miR-30 family of micro-RNA limit fission by suppressing p53 expression and Drp1 activation | Li et al. (2010) |
| Inhibition of histone deacetylases induces mitochondrial elongation | Fused networks occur due to decreased Fis1 expression and reduced Drp1 translocation | Lee et al. (2012) |
| Phosphorylation of Drp1 has opposing effects on fission depending on the kinase | Example: phosphorylation of Drp1 by cAMP kinase increases yet PKC delta decreases fission | Cribbs and Strack (2007), Qi et al. (2011), Kim et al. (2011) |
| Local organelle control | ||
| Knockdown of Mff promotes elongation of the network and overexpression of Mff promotes fission | Mff recruits Drp-1 to fission sites on the outer membrane independently of hFis1 | Otera et al. (2010), Gandre-Babbe and van der Bliek (2008) |
| Blocking the mitochondrial Na+/Ca2+ exchanger increases interaction between Drp1 and Fis1 | Elevated levels of mitochondrial calcium increases fission activity | Kaddour-Djebbar et al. (2010) |
| Treatment with cysteine-alkylators inhibits fission and fast mitochondrial movement | Loss of movement coincided with microtubule- dependent thin mitochondrial extensions | Bowes and Gupta (2005, 2008) |
| Conformation specificities and self-interaction dictate the ability of Fis1 to recruit fission machinery | Fis1 activity is regulated by two interaction interfaces and its ability to oligomerize | Serasinghe and Yoon (2008), Zhang and Chan (2007) |
| Fusion | ||
| Global cellular control | ||
| Activation of PGC1a/PGC-1b/ERRa induces MFN2 mRNA. PGC-1b induces mitochondrial fusion by Mfn2 | Increased fusion activity accompanies mitochondrial biogenesis | Liesa et al. (2008), Soriano et al. (2006) |
| Mitochondrial tubularization and network fusion at G1-S of cell cycle | G1-S stimulates global mitochondrial fusion, mitosis stimulates fission | Lee et al. (2004), Taguchi et al. (2007), Arakaki et al. (2006), Kashatus et al. (2011) |
| Variation in isoform expression of OPA1 occurs via alternative gene splicing | Expression of the eight splice variants differs across tissues and alter fusion and apoptotic activity | Song et al. (2007), Olichon et al. (2007), Frezza et al. (2006) |
| The promoter region of MFN2 is a target of the tumor suppressor protein, p53 | Mfn2 mRNA and protein levels are up-regulated in a p53-dependant manner | Wang et al. (2010) |
| Local organelle control | ||
| Functional interaction of OPA1 with MFN1 and physical interaction between mitofusins and OPA1 | Protein complex spans the two mitochondrial membranes and permits fusion activity | Cipolat et al. (2004), Guillery et al. (2008) |
| OPA1 processing by metalloproteases can block fusion activity and alter cristae structure | Provides a mechanism for fusion inhibition at the local level by protease activity | Song et al. (2007, 2009), Ehses et al. (2009), Duvezin-Caubet et al. (2006), Griparic et al. (2007), Guillery et al. (2008), Baricault et al. (2007), Kieper et al. (2010) |
| Low levels of local GTP induce outer membrane tethering while complete fusion events require high intra-mitochondrial GTP levels | Initial fusion is promoted in energy deficient environments yet complete fusion is regulated by energetic status of the organelles | Meeusen et al. (2006) |
| G-protein beta2 is enriched in the mitochondrial membrane and interacts with Mfn1 to regulate fusion | Gbeta2 regulates the mobility of Mfn1 within the outer membrane and promotes fusion | Zhang et al. (2010) |
| Bcl-x(L) increases rates of fusion and fission with an observed overall network elongation | Bcl-x(L) increases mitochondrial mass concurrent with elevated dynamics cycling | Berman et al. (2009) |
| Solitary period | ||
| Global cellular control | ||
| Global ADP levels increase mitochondrial movement to synapses | ADP signals mitochondrial motility | Mironov (2009) |
| G-protein coupled receptor, Ga12, is expressed in mitochondria and regulates motility | GPCRs are sensitive to GDP/GTP levels and can regulate mitochondrial motility | Andreeva et al. (2008) |
| Bnip3 expression induces Drp1 mediated fission and parkin translocation in adult myocytes | Increased fission activity and parkin translocation enhanced mitophagy | Lee et al. (2011a) |
| Local organelle control | ||
| Mitochondrial movement along microtubules occurs in an energy-dependent manner | Individual mitochondria move at different rates along microtubules based on ATP levels | Yi et al. (2004), Miller and Sheetz (2004), Guo et al. (2005) |
| Local redox status of mitochondria impacts membrane potential and velocity of movement | Elevated oxidation leads to depolarization and to increased motility | Gerencser and Nicholls (2008) |
| PINK1 and Parkin target Miro, mitofusins and other outer membrane proteins for proteasomal degradation and promote mitophagy | Proteasomal degradation of Miro and mitofusins isolate and immobilize mitochondria which increases the pre-autophagic pool | Tanaka et al. (2010a), Wang et al. (2011b), Weihofen et al. (2009), Ziviani et al. (2010), Ziviani and Whitworth (2010), Poole et al. (2008, 2010), Yang et al. (2008), Glauser et al. (2011), Rakovic et al. (2011), Gegg et al. (2010) |
Local regulation occurs at the level of the microenvironment of an individual mitochondrion. Fission events are controlled checkpoints for generation of polarized and depolarized mitochondria; therefore changes in membrane potential distribution are a mechanistic example of local regulation (Twig et al. 2008a; Wikstrom et al. 2007). Loss of membrane potential and ATP production causes cleavage and degradation of fusion proteins by mitochondrial proteases and the proteasome (Song et al. 2007; Chan and Chan 2011; Chan et al. 2011). Decreased fusion capacity results in increased time spent in the solitary phase, and if membrane potential is not recovered, the mitochondrion enters into the pre-autophagic pool (Cipolat et al. 2004, 2006; Baricault et al. 2007; Twig and Shirihai 2011; Rambold et al. 2011a).
Another crucial local regulator of mitochondrial dynamics is the degree of movement of an individual mitochondrion (Twig and Shirihai 2011). Movement greatly increases the chances of fusion perhaps in part because microtubule transport aligns mitochondria (Twig et al. 2010). Alignment facilitates pole interaction between mitochondria and thus increases the likelihood of tethering between mitofusins. Motility depends on the calcium-sensitive mitochondrial Rho GTPase (Miro), which connects mitochondria to the ATP-dependant motor enzymes, dynein and kinesin (Boldogh and Pon 2007; Fehrenbacher et al. 2004; Saotome et al. 2008). Milton is a protein that complexes with Miro and is also required for transport of mitochondria (Glater et al. 2006; Rice and Gelfand 2006). Calcium binding to Miro inhibits motility by causing detachment of the motor protein complex from microtubules (Wang and Schwarz 2009a, b; Wang et al. 2011b). Calcium exchangers, serving a role in calcium buffering, are dependant on ATP, resulting in a dependency of the buffering capacity on mitochondrial ATP synthesis. This is leading to the detachment of mitochondria in calcium rich spots, thus creating a localized mechanism for selective delivery of mitochondria to cellular regions with unmet ATP needs (Yi et al. 2004). Elevated levels of cytosolic calcium inhibit motility by binding to Miro and decreased supply of ATP lowers the activity of ATPase-driven motor proteins. Kinesins themselves are unaffected by calcium levels, so by coupling both ATP availability and local calcium concentrations this local regulation specifically impacts mitochondrial movement and not general microtubule transport.
In neurons, proper distribution of mitochondria is of the upmost importance. The degree of dendritic arborization correlates with mitochondrial content and is dependant on Miro activity (Macaskill et al. 2009; Russo et al. 2009). Mitochondria with high membrane potential and elevated ATP production travel anterogradely to synaptic regions where there is a very high demand for energy (Miller and Sheetz 2004). Appropriately, global elevation of ADP levels in neurons increases delivery of mitochondria to synapses (Mironov 2009). On the contrary, there is fast retrograde transport of depolarized mitochondria with low membrane potential back to the soma to facilitate lysosomal degradation (Boldogh and Pon 2007; Gerencser and Nicholls 2008; Hollenbeck and Saxton 2005). In addition to supplying ATP, mitochondria fulfill a crucial role by buffering cytosolic calcium. The abundant neurotransmitter glutamate activates ionotropic NMDA receptors resulting in local increases in calcium influx. This relationship establishes an important regulatory mechanism for local inhibition of mitochondrial transport by Miro at active synaptic sites (Saotome et al. 2008; Wang and Schwarz 2009b; Macaskill et al. 2009).
The transcriptional and post-translational regulation of OPA1 serves as an excellent final example of both global and local control of mitochondrial dynamics. Through global signaling pathways, alterations in gene transcription can create eight isoforms of OPA1 (Landes et al. 2010). The distinct functions of the different forms of OPA1 are not well understood but clearly they can perform unique activities such as stabilizing cristae and protecting mtDNA (Semenzato et al. 2011; Merkwirth et al. 2008; Frezza et al. 2006; Elachouri et al. 2011; Yu-Wai-Man et al. 2010). A shift in isoform production affects all mitochondria undergoing protein import and therefore represents a form of global regulation. On the other hand, imported OPA1 is cleaved to produce variants of different lengths by several mitochondrial proteases, including MPP, OMA1, PARL, and Yme1L (Song et al. 2007, 2009; Ehses et al. 2009; Cipolat et al. 2004, 2006; Griparic et al. 2007; Guillery et al. 2008; Ishihara et al. 2004). This proteolytic processing is dependant on membrane potential, metal ion levels, and ATP availability. Both long and short isoforms are required for proper inner membrane fusion and so represents a means of local regulation. For example, stress-induced cleavage of OPA1 long isoforms by OMA1 can disrupt interaction with Mfn1 and thereby block complete fusion (Ehses et al. 2009; Cipolat et al. 2004, 2006; Guillery et al. 2008). In this way, OPA1 is regulated both at the global and local levels to control fusion of mitochondria.
2.8 Mitochondrial Dynamics and Pathology
Alterations to mitochondrial fusion and fission have been demonstrated in several pathological conditions including neurodegeneration, obesity, and type II diabetes. Mutations in genes that encode for fusion proteins provide the clearest connection between mitochondrial dynamics and disease. Charcot-Marie-Tooth (CMT) disease Type 2A is caused by mutations in MFN2 and results in peripheral nervous system dysfunction (Ching et al. 2010; Casasnovas et al. 2010; Ouvrier and Grew 2010; Feely et al. 2011). The most common form of hereditary optic neuropathy is caused by mutations in OPA1 (Ferre et al. 2009; Nochez et al. 2009; Yu-Wai-Man et al. 2011a, b). These diseases confirm the importance of mitochondrial fusion in cell survival and also illustrate the existence of selective susceptibility amongst neuronal subtypes.
Neurons in general are vulnerable to mitochondrial dysfunction due to extreme energy demands coupled with complex, polarized cell structures. Degeneration in Parkinson’s disease (PD) occurs selectively in neurons that exemplify these combined susceptibilities and genetic studies strongly implicate defects in mitochondrial dynamics and quality control (Wang et al. 2011b; Braak and Del Tredici 2008; Braak et al. 2004; Narendra and Youle 2011; Dagda and Chu 2009; Whitworth and Pallanck 2009). Specific cellular morphological characteristics create inherent challenges to the networking of mitochondrial populations within PD-sensitive neurons. The A9 dopaminergic (A9-DA) neurons of the substantia nigra elegantly illustrate this principle (Braak and Del Tredici 2008; Braak et al. 2004; Ferrer et al. 2011).
The nigral A9-DA neurons are so polarized and branched that their somas account for less than 1% of total cell volume (Sulzer 2007). Massive neuritic arborization occurs in both axonal and dendritic compartments such that each A9-DA neuron may contain more than 300,000 synapses in its axonal field alone (Arbuthnott and Wickens 2007; Matsuda et al. 2009; Surmeier et al. 2010a, b). Extreme cellular morphology creates a major logistical hurdle and heightens susceptibility to disruptions in mitochondrial transport. Dispersion also limits protective mechanisms of complementation and quality control by decreasing the likelihood of fusion events. Synapses are the most energy-demanding region of the neuron as well as being sites of voltage-gated calcium influx. Proper mitochondrial distribution is therefore critical not only to provide ATP but also to buffer calcium levels (Oliveira 2010; MacAskill et al. 2010).
To reach the pre-synaptic compartment, mitochondria must travel along long, thin, and poorly myelinated axons in A9-DA neurons (Braak et al. 2004). Each of these characteristics increases both metabolic demand and the parallel risk of oxidative stress. Length correlates with surface area and longer axons have increased requirements for ATPase activity by the sodium potassium exchanger (Na+/K+ ATPase). Greater distances also increase energy expenditures and travel time for motor proteins bringing cargo back and forth from soma to synapse. Longer retrograde transit times for damaged mitochondria autophagocytosed at synapses likely increases the risk and extent of oxidative damage en route back to lysosomes (Terman et al. 2010; Yue 2007; Yue et al. 2009). Like lanes on a road, width also impacts axonal transport. Thin caliber axons are spatially restrained and this reduces capacity for delivery of both mitochondria and autophagosomes. Thin A9-DA axons have higher surface area to volume ratios and therefore elevated energy demands due to higher basal Na+/K+ ATPase activity. Similarly, myelin limits the amount of surface area involved in ion exchange and this insulation dramatically impacts energy demands for maintaining ionic gradients required axonal conductance. Creating specialized sub-domains of the axon permits clustering of mitochondria in energy intensive micro-regions and that may promote mitochondrial fusion (Ohno et al. 2011). Poorly myelinated A9-DA neurons require high Na+/K+ ATPase activity over the entire length of the axon are thereby denied the potential benefits of mitochondrial clustering. Collectively, these characteristics of A9-DA neurons likely synergize to heighten sensitivity to disruptions in mitochondrial dynamics, motility, and mitophagy.
Studies of genetic mutations that cause recessive forms of familial PD support these predictions of heightened susceptibility of disruptions in mitochondrial dynamics and quality control in A9-DA neurons. Mutations in DJ-1, PINK1, and Parkin cause parkinsonism and originally these genes were thought to have disparate functions but recently their cellular roles were unified around mitochondrial dynamics and quality control (Narendra and Youle 2011; Dagda and Chu 2009; Whitworth and Pallanck 2009; Chu 2010a; Irrcher et al. 2010; Thomas et al. 2011). Together these genes provide protection against the extremes of mitochondrial membrane potential and ROS production. In this thermostat analogy DJ-1 targets mitochondria that produce excess ROS with normal to high membrane potential. The other extreme is handled by PINK1, which identifies mitochondria with little ROS production due to depolarized membrane potential. Parkin acts as a downstream effecter of both DJ-1 and PINK1 pathways to facilitate selective autophagic clearance of targeted mitochondria.
DJ-1 is a cytosolic chaperone protein that translocates to mitochondria in response to oxidative stress (Canet-Aviles et al. 2004; Moore et al. 2005; Xiong et al. 2009). Loss of DJ-1 function leads to aberrant mitochondrial morphology and function (Irrcher et al. 2010; Thomas et al. 2011; Goldberg et al. 2005; Krebiehl et al. 2010) as well as increased sensitivity to mitochondrial toxins and ROS (Canet-Aviles et al. 2004; Kim et al. 2004, 2005; Ved et al. 2005; Zhang et al. 2005; Paterna et al. 2007; Taira et al. 2004 ; Menzies et al. 2005 ; Meulener et al. 2005). The protective functions of DJ-1 are dependant on a specific cysteine residue located at site 106 and this single amino acid allows DJ-1 to function as a cytoplasmic sensor of mitochondrial oxidative stress (Irrcher et al. 2010; Canet-Aviles et al. 2004; Blackinton et al. 2005, 2009). In support of this role, mitochondria isolated from mice lacking DJ-1 display greater ROS generation and phenotypes associated with loss of DJ-1 are reversed with anti-oxidants (Irrcher et al. 2010; Thomas et al. 2011). Replacing wild-type protein rescues the phenotypes of DJ-1 knockout cells but not when the replacement protein is mutated at site 106. Exactly how DJ-1 regulates mitochondrial ROS production is not fully clear but two plausible mechanisms include modulation of complex I activity and regulation of uncoupling protein expression (Hayashi et al. 2009; Guzman et al. 2010). Additionally, oxidation of DJ-1 results in binding with the cytosolic E3 ubiquitin ligase Parkin (Moore et al. 2005). DJ-1 functions upstream in this pathway as Parkin overexpression can rescue phenotypes associated with loss of DJ-1 (Irrcher et al. 2010; Thomas and Cookson 2009) but not the reverse (Dodson and Guo 2007).
Maintenance of homeostasis within the mitochondrial population is not complete with only protection from excess ROS by DJ-1. Production of ROS is connected to membrane potential and when a mitochondrion becomes depolarized it decreases ROS output. In this situation the dysfunctional mitochondrion would be undetected by the DJ-1-mediated quality control mechanism. An additional surveillance mechanism is needed to guard against this other potential extreme situation. PTEN-induced kinase 1 (PINK1) is a protein kinase with a mitochondrial targeting signal and a putative transmembrane domain (Chu 2010a, b; Mills et al. 2008). PINK1 is continuously imported into the intermembrane space where it is immediately targeted for degradation by several mitochondrial proteases. This normal turnover is interrupted when mitochondria depolarize and PINK1 accumulates, allowing for kinase signaling to bring about selective mitochondrial removal (Jin et al. 2010; Narendra et al. 2010a). In this way PINK1 provides a mechanism for monitoring mitochondrial function that is not based on ROS production. Loss of PINK1 function leads to a buildup of damaged mitochondrial material along with decreased membrane potential and ATP synthesis (Dagda and Chu 2009 ; Exner et al. 2007 ; Liu et al. 2009, 2011; Grunewald et al. 2009; Marongiu et al. 2009; Dagda et al. 2009a, b; Wood-Kaczmar et al. 2008; Gandhi et al. 2009). Mitochondrial dysfunction occurs prior to the onset of any neu-rodegeneration in mice lacking the PINK1 gene (Gispert et al. 2009; Narendra et al. 2008, 2009, 2010). One downstream event of PINK1 stabilization is the binding and phosphorylation of Miro/Milton mitochondrial transport complexes (Wang et al. 2011b; Weihofen et al. 2009). This PINK1 effect may isolate depolarized mitochondria by limiting their transport and help in the clearance by mitophagy.
Another consequence of depolarization-induced PINK1 accumulation related to mitophagy is the mitochondrial recruitment of Parkin. Cytosolic Parkin is selectively recruited to depolarized mitochondria, which facilitates autophagic elimination of the dysfunctional units (Narendra et al. 2008, 2009, 2010a) and PINK1 is required for this process (Geisler et al. 2010a, b; Vives-Bauza et al. 2010a, b, c; Vives-Bauza and Przedborski 2010). By attaching lysine 48 (K48) linked polyubiquitin chains, Parkin promotes proteasomal degradation of mitochondrial outer membrane proteins such as Miro, mitofusins, and several transporters (Tanaka et al. 2010a, b; Chan and Chan 2011; Chan et al. 2011; Ziviani et al. 2010; Ziviani and Whitworth 2010; Poole et al. 2008, 2010 ; Tanaka 2010 ). In this way K48-mediated proteasomal turnover of outer membrane proteins immobilizes and isolates damaged mitochondria to increase the likelihood of autophagic clearance. In addition, Parkin also creates recruitment signals for mitophagy via lysine 63-linked (K63) ubiquitin chains. Scaffold proteins, such as HDAC6 and p62/SQSTM1, bind to K63-ubiquitin and facilitate localization to the aggresome and clearance by mitophagy (Huang et al. 2011b; Geisler et al. 2010a; Lee et al. 2010; Okatsu et al. 2010; Narendra et al. 2010b; Ding et al. 2010a).
Quality control of mitochondrial performance occurs through two pathways in which either DJ-1 or PINK1 utilize Parkin as a downstream effector, summarized in Fig. 2.2. Age of onset studies from PD patients support this conceptual hierarchy. Mutations in DJ-1 and PINK1 cause an early onset of Parkinson symptoms relative to the sporadic disease (~30–50 compared to ~60–80 years of age) (Abou-Sleiman et al. 2004; Mizuno et al. 2006). In accordance with being downstream in both pathways, Parkin mutations tend to have very early disease onset with a large number of juvenile cases occurring before the age of 30 (Kitada et al. 1998; Nisipeanu et al. 1999, 2001; Oliveri et al. 2001; Lucking et al. 2000).
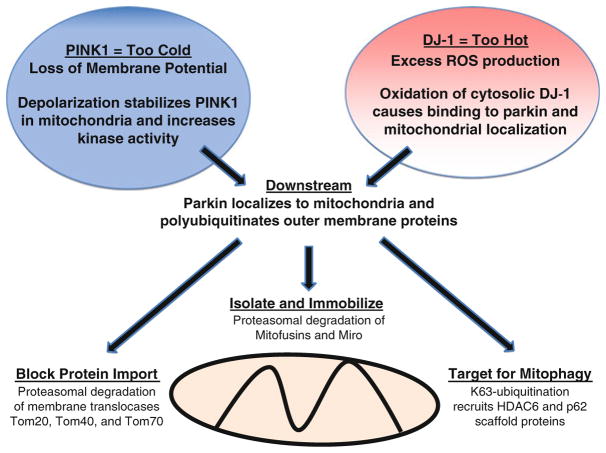
Fig. 2.2.
The thermostat model of mitochondrial quality control. Separate pathways maintain mitochondrial homeostasis by safeguarding against functional extremes. In the cold extreme, PINK1 is stabilized within mitochondria upon membrane depolarization leading to increased kinase signaling. Alternatively, oxidative activation of cytosolic DJ-1 occurs in response to the hot extreme of excess ROS production. Downstream of both PINK1 and DJ-1 pathways is the recruitment of the E3 ligase Parkin and attachment of K63 and K48 polyubiquitin chains to mitochondrial outer membrane proteins. Damaged mitochondria are isolated and immobilized by proteasomal degradation of K48-tagged proteins, such as mitofusins and Miro. Proteasomal clearance of mitochondrial translocases prevents repopulation of the outer membrane with newly synthesized replacement proteins. Finally, K63 polyubiquitin chains selectively identifies mitochondria for autophagic clearance by recruitment of scaffold proteins, such as HDAC6 and p62
2.9 Open Questions and Controversies
Studies have shown that Mff is required for fission and hFis1 is not (Otera et al. 2010). This raises the potential for the existence of other mitochondrial outer membrane proteins that can bind and anchor Mff for Drp1 recruitment. The identity of these additional proteins is not known nor is it known how they differ functionally from hFis1. Finally, hFis1 is definitely involved in mitochondrial fission but its function is not required. Could hFis1 be important for a specific form or aspect of fission, such as in the mechanism behind the generation of unequal division?
One crucial area of future study is understanding of the regulatory mechanisms for mitochondrial dynamics and how they connect to quality control. These are of particular interest as therapeutic targets since strategies aimed at maximizing upkeep of mitochondrial performance would have broad application across health. Can enhancement of mitochondrial dynamics, quality control, and turnover be a viable therapeutic strategy for treatment of chronic diseases such as diabetes and neurodegeneration?
A large portion of publications connecting the DJ-1-PINK1-Parkin pathways were performed with cell lines in nonphysiologic conditions. The degree to which these pathways exist in neurons, surprisingly, remains a matter of debate (Van Laar et al. 2011). It is also worth acknowledging that most patients with Parkin mutations lack Lewy Bodies, the intracellular neuropathological hallmarks of PD (Ahlskog 2009). Assuming the role of Parkin in neurons is to execute mitophagy downstream of DJ-1 and PINK1 signaling, the question of what connects Parkin-mediated mitophagy to Lewy Body formation is another tantalizing question facing the field of PD research.
Contributor Information
Andrew Ferree, Department of Pharmacology, and Department of Neuroscience, Boston University School of Medicine, Boston, MA, USA.
Orian Shirihai, Department of Medicine, Boston University School of Medicine, 650 Albany Street, Boston, MA 02118, USA.
References
- Abou-Sleiman PM, Healy DG, Wood NW. Causes of Parkinson’s disease: genetics of DJ-1. Cell Tissue Res. 2004;318(1):185–188. doi: 10.1007/s00441-004-0922-6. [DOI] [PubMed] [Google Scholar]
- Ahlskog JE. Parkin and PINK1 parkinsonism may represent nigral mitochondrial cytopathies distinct from Lewy body Parkinson’s disease. Parkinsonism Relat Disord. 2009;15(10):721–727. doi: 10.1016/j.parkreldis.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA, Voyno-Yasenetskaya TA. G alpha12 is targeted to the mito-chondria and affects mitochondrial morphology and motility. FASEB J. 2008;22(8):2821–2831. doi: 10.1096/fj.07-104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki N, et al. Dynamics of mitochondria during the cell cycle. Biol Pharm Bull. 2006;29(9):1962–1965. doi: 10.1248/bpb.29.1962. [DOI] [PubMed] [Google Scholar]
- Arbuthnott GW, Wickens J. Space, time and dopamine. Trends Neurosci. 2007;30(2):62–69. doi: 10.1016/j.tins.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Arimura S, et al. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc Natl Acad Sci USA. 2004;101(20):7805–7808. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baricault L, et al. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res. 2007;313(17):3800–3808. doi: 10.1016/j.yexcr.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Berman SB, et al. Bcl-x L increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol. 2009;184(5):707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Blackinton J, et al. Effects of DJ-1 mutations and polymorphisms on protein stability and subcellular localization. Brain Res Mol Brain Res. 2005;134(1):76–83. doi: 10.1016/j.molbrainres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Blackinton J, et al. Post-transcriptional regulation of mRNA associated with DJ-1 in sporadic Parkinson disease. Neurosci Lett. 2009;452(1):8–11. doi: 10.1016/j.neulet.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17(10):502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Bowes TJ, Gupta RS. Induction of mitochondrial fusion by cysteine-alkylators ethacrynic acid and N-ethylmaleimide. J Cell Physiol. 2005;202(3):796–804. doi: 10.1002/jcp.20178. [DOI] [PubMed] [Google Scholar]
- Bowes T, Gupta RS. Novel mitochondrial extensions provide evidence for a link between microtubule-directed movement and mitochondrial fission. Biochem Biophys Res Commun. 2008;376(1):40–45. doi: 10.1016/j.bbrc.2008.08.120. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. Invited Article: Nervous system pathology in sporadic Parkinson disease. Neurology. 2008;70(20):1916–1925. doi: 10.1212/01.wnl.0000312279.49272.9f. [DOI] [PubMed] [Google Scholar]
- Braak H, et al. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004;318(1):121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10(7):748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KB, et al. Mitochondrial dynamics generate equal distribution but patchwork localization of respiratory Complex I. Mol Membr Biol. 2006;23(6):509–520. doi: 10.1080/09687860600877292. [DOI] [PubMed] [Google Scholar]
- Canet-Aviles RM, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA. 2004;101(24):9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasnovas C, et al. Phenotypic spectrum of MFN2 mutations in the Spanish population. J Med Genet. 2010;47(4):249–256. doi: 10.1136/jmg.2009.072488. [DOI] [PubMed] [Google Scholar]
- Cereghetti GM, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105(41):15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti GM, Costa V, Scorrano L. Inhibition of Drp1-dependent mitochondrial fragmentation and apoptosis by a polypeptide antagonist of calcineurin. Cell Death Differ. 2010;17(11):1785–1794. doi: 10.1038/cdd.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chan NC, Chan DC. Parkin uses the UPS to ship off dysfunctional mitochondria. Autophagy. 2011;7(7):771–772. doi: 10.4161/auto.7.7.15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20(9):1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14(Spec No 2):R283–R289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160(2):189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280(28):26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu Y, Dorn GW., 2nd Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109(12):1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching CK, et al. A novel mitofusin 2 gene mutation causing Charcot-Marie-Tooth type 2A disease in a Chinese family. Chin Med J (Engl) 2010;123(11):1466–1469. [PubMed] [Google Scholar]
- Cho DH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324(5923):102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V, et al. Novel role of androgens in mitochondrial fission and apoptosis. Mol Cancer Res. 2011;9(8):1067–1077. doi: 10.1158/1541-7786.MCR-10-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Hum Mol Genet. 2010a;19(R1):R28–R37. doi: 10.1093/hmg/ddq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT. Tickled PINK1: mitochondrial homeostasis and autophagy in recessive Parkinsonism. Biochim Biophys Acta. 2010b;1802(1):20–28. doi: 10.1016/j.bbadis.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S, et al. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci USA. 2004;101(45):15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S, et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126(1):163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8(10):939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Chu CT. Mitochondrial quality control: insights on how Parkinson’s disease related genes PINK1, parkin, and Omi/HtrA2 interact to maintain mitochondrial homeostasis. J Bioenerg Biomembr. 2009;41(6):473–479. doi: 10.1007/s10863-009-9255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009a;284(20):13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Zhu J, Chu CT. Mitochondrial kinases in Parkinson’s disease: converging insights from neurotoxin and genetic models. Mitochondrion. 2009b;9(5):289–298. doi: 10.1016/j.mito.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Dimmer KS, Scorrano L. (De)constructing mitochondria: what for? Physiology (Bethesda) 2006;21:233–241. doi: 10.1152/physiol.00010.2006. [DOI] [PubMed] [Google Scholar]
- Ding WX, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010a;285(36):27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, et al. Response of mitochondrial fusion and fission protein gene expression to exercise in rat skeletal muscle. Biochim Biophys Acta. 2010b;1800(3):250–256. doi: 10.1016/j.bbagen.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr Opin Neurobiol. 2007;17(3):331–337. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S, et al. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J Biol Chem. 2006;281(49):37972–37979. doi: 10.1074/jbc.M606059200. [DOI] [PubMed] [Google Scholar]
- Ehses S, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187(7):1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elachouri G, et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21(1):12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore SP, et al. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15(12):2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Exner N, et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27(45):12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feely SM, et al. MFN2 mutations cause severe phenotypes in most patients with CMT2A. Neurology. 2011;76(20):1690–1696. doi: 10.1212/WNL.0b013e31821a441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher KL, et al. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr Biol. 2004;14(22):1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Ferre M, et al. Molecular screening of 980 cases of suspected hereditary optic neuropathy with a report on 77 novel OPA1 mutations. Hum Mutat. 2009;30(7):E692–E705. doi: 10.1002/humu.21025. [DOI] [PubMed] [Google Scholar]
- Ferrer I, et al. Neuropathology of sporadic Parkinson disease before the appearance of par-kinsonism: preclinical Parkinson disease. J Neural Transm. 2011;118(5):821–839. doi: 10.1007/s00702-010-0482-8. [DOI] [PubMed] [Google Scholar]
- Figueroa-Romero C, et al. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009;23(11):3917–3927. doi: 10.1096/fj.09-136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1(4):515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Frezza C, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126(1):177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Fukushima NH, et al. The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission. Mol Biol Cell. 2001;12(9):2756–2766. doi: 10.1091/mbc.12.9.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S, et al. PINK1-associated Parkinson’s disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell. 2009;33(5):627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19(6):2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19(24):4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010a;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Geisler S, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010b;6(7):871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- Gerencser AA, Nicholls DG. Measurement of instantaneous velocity vectors of organelle transport: mitochondrial transport and bioenergetics in hippocampal neurons. Biophys J. 2008;95(6):3079–3099. doi: 10.1529/biophysj.108.135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW, et al. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J Cell Biol. 2008;181(7):1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gispert S, et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS One. 2009;4(6):e5777. doi: 10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, et al. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173(4):545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser L, et al. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem. 2011;118(4):636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, et al. Nigrostriatal dopaminergic deficits and hypokinesia caused by inactivation of the familial Parkinsonism-linked gene DJ-1. Neuron. 2005;45(4):489–496. doi: 10.1016/j.neuron.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Scorrano L. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta. 2008;1777(7–8):860–866. doi: 10.1016/j.bbabio.2008.05.442. [DOI] [PubMed] [Google Scholar]
- Gottlieb E. OPA1 and PARL keep a lid on apoptosis. Cell. 2006;126(1):27–29. doi: 10.1016/j.cell.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Griffin EE, Detmer SA, Chan DC. Molecular mechanism of mitochondrial membrane fusion. Biochim Biophys Acta. 2006;1763(5–6):482–489. doi: 10.1016/j.bbamcr.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Griparic L, Kanazawa T, van der Bliek AM. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol. 2007;178(5):757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohm J, Plesnila N, Culmsee C. Bid mediates fission, membrane permeabilization and peri-nuclear accumulation of mitochondria as a prerequisite for oxidative neuronal cell death. Brain Behav Immun. 2010;24(5):831–838. doi: 10.1016/j.bbi.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Grunewald A, et al. Differential effects of PINK1 nonsense and missense mutations on mitochondrial function and morphology. Exp Neurol. 2009;219(1):266–273. doi: 10.1016/j.expneurol.2009.05.027. [DOI] [PubMed] [Google Scholar]
- Guillery O, et al. Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential. Biol Cell. 2008;100(5):315–325. doi: 10.1042/BC20070110. [DOI] [PubMed] [Google Scholar]
- Guo X, et al. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47(3):379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Guzman JN, et al. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468(7324):696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14(4):340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hayashi T, et al. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochem Biophys Res Commun. 2009;390(3):667–672. doi: 10.1016/j.bbrc.2009.10.025. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118(Pt 23):5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom J, et al. Regulation of mitochondrial fission by intracellular Ca2+ in rat ventricular myocytes. Biochim Biophys Acta. 2010;1797(6–7):913–921. doi: 10.1016/j.bbabio.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori A, Yoshida M, Ling F. Mitochondrial fusion increases the mitochondrial DNA copy number in budding yeast. Genes Cells. 2011;16(5):527–544. doi: 10.1111/j.1365-2443.2011.01504.x. [DOI] [PubMed] [Google Scholar]
- Huang P, Galloway CA, Yoon Y. Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins. PLoS One. 2011a;6(5):e20655. doi: 10.1371/journal.pone.0020655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, et al. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011b;6(6):e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde BB, Twig G, Shirihai OS. Organellar vs cellular control of mitochondrial dynamics. Semin Cell Dev Biol. 2010;21(6):575–581. doi: 10.1016/j.semcdb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrcher I, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum Mol Genet. 2010;19(19):3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117(Pt 26):6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11(8):958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Jakobs S. High resolution imaging of live mitochondria. Biochim Biophys Acta. 2006;1763(5–6):561–575. doi: 10.1016/j.bbamcr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Jakobs S, Schauss AC, Hell SW. Photoconversion of matrix targeted GFP enables analysis of continuity and intermixing of the mitochondrial lumen. FEBS Lett. 2003;554(1–2):194–200. doi: 10.1016/s0014-5793(03)01170-0. [DOI] [PubMed] [Google Scholar]
- James DI, et al. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278(38):36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Jin SM, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191(5):933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain I, Gachet Y, Hyams JS. The dynamin related protein Dnm1 fragments mitochon-dria in a microtubule-dependent manner during the fission yeast cell cycle. Cell Motil Cytoskeleton. 2009;66(8):509–523. doi: 10.1002/cm.20351. [DOI] [PubMed] [Google Scholar]
- Jung HS, Lee MS. Macroautophagy in homeostasis of pancreatic beta-cell. Autophagy. 2009;5(2):241–243. doi: 10.4161/auto.5.2.7518. [DOI] [PubMed] [Google Scholar]
- Kaddour-Djebbar I, et al. Specific mitochondrial calcium overload induces mitochondrial fission in prostate cancer cells. Int J Oncol. 2010;36(6):1437–1444. doi: 10.3892/ijo_00000629. [DOI] [PubMed] [Google Scholar]
- Kane LA, Youle RJ. Mitochondrial fission and fusion and their roles in the heart. J Mol Med (Berl) 2010;88(10):971–979. doi: 10.1007/s00109-010-0674-6. [DOI] [PubMed] [Google Scholar]
- Karbowski M. Mitochondria on guard: role of mitochondrial fusion and fission in the regulation of apoptosis. Adv Exp Med Biol. 2010;687:131–142. doi: 10.1007/978-1-4419-6706-0_8. [DOI] [PubMed] [Google Scholar]
- Karbowski M, et al. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159(6):931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, et al. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol. 2004a;164(4):493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004b;166(7):1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus DF, et al. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13(9):1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieper N, et al. Modulation of mitochondrial function and morphology by interaction of Omi/HtrA2 with the mitochondrial fusion factor OPA1. Exp Cell Res. 2010;316(7):1213–1224. doi: 10.1016/j.yexcr.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AJ, Lee CS, Schlessinger D. Bex3 associates with replicating mitochondria and is involved in possible growth control of F9 teratocarcinoma cells. Gene. 2004;343(1):79–89. doi: 10.1016/j.gene.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Kim RH, et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3, 6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci USA. 2005;102(14):5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, et al. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44(4):532–544. doi: 10.1016/j.molcel.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol. 2011;22(5):902–913. doi: 10.1681/ASN.2010070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Koch A, et al. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16(11):5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, et al. Regulation of Ca2+-induced permeability transition by Bcl-2 is antagonized by Drpl and hFis1. Mol Cell Biochem. 2005;272(1–2):187–199. doi: 10.1007/s11010-005-7323-3. [DOI] [PubMed] [Google Scholar]
- Koshiba T, et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305(5685):858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Krebiehl G, et al. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One. 2010;5(2):e9367. doi: 10.1371/journal.pone.0009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landes T, Martinou JC. Mitochondrial outer membrane permeabilization during apoptosis: the role of mitochondrial fission. Biochim Biophys Acta. 2011;1813(4):540–545. doi: 10.1016/j.bbamcr.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Landes T, et al. OPA1 (dys)functions. Semin Cell Dev Biol. 2010;21(6):593–598. doi: 10.1016/j.semcdb.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Lee YJ, et al. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15(11):5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, et al. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189(4):671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, et al. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2011a;301(5):H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, et al. Autophagy negatively regulates keratinocyte inflammatory responses via scaffolding protein p62/SQSTM1. J Immunol. 2011b;186(2):1248–1258. doi: 10.4049/jimmunol.1001954. [DOI] [PubMed] [Google Scholar]
- Lee JS, et al. Histone deacetylase inhibitors induce mitochondrial elongation. J Cell Physiol. 2012;227(7):2856–2869. doi: 10.1002/jcp.23027. [DOI] [PubMed] [Google Scholar]
- Legesse-Miller A, Massol RH, Kirchhausen T. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol Biol Cell. 2003;14(5):1953–1963. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, et al. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13(12):4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, et al. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117(13):2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- Lewis MR, Lewis WH. Mitochondria in tissue culture. Science. 1914;39(1000):330–333. doi: 10.1126/science.39.1000.330. [DOI] [PubMed] [Google Scholar]
- Li J, et al. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6(1):e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, et al. Mitochondrial fusion is increased by the nuclear coactivator PGC-1beta. PLoS One. 2008;3(10):e3613. doi: 10.1371/journal.pone.0003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. PINK1 defect causes mitochondrial dysfunction, proteasomal deficit and alpha-synuclein aggregation in cell culture models of Parkinson’s disease. PLoS One. 2009;4(2):e4597. doi: 10.1371/journal.pone.0004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc Natl Acad Sci USA. 2011;108(31):12920–12924. doi: 10.1073/pnas.1107332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew LM, et al. Imaging in five dimensions: time-dependent membrane potentials in individual mitochondria. Biophys J. 1993;65(6):2396–2407. doi: 10.1016/S0006-3495(93)81318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucking CB, et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. N Engl J Med. 2000;342(21):1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- Macaskill AF, et al. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009;61(4):541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill AF, Atkin TA, Kittler JT. Mitochondrial trafficking and the provision of energy and calcium buffering at excitatory synapses. Eur J Neurosci. 2010;32(2):231–240. doi: 10.1111/j.1460-9568.2010.07345.x. [DOI] [PubMed] [Google Scholar]
- Malka F, et al. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep. 2005;6(9):853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marongiu R, et al. Mutant Pink1 induces mitochondrial dysfunction in a neuronal cell model of Parkinson’s disease by disturbing calcium flux. J Neurochem. 2009;108(6):1561–1574. doi: 10.1111/j.1471-4159.2009.05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda W, et al. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J Neurosci. 2009;29(2):444–453. doi: 10.1523/JNEUROSCI.4029-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni C, Falcone C. The importance of mitochondrial fusion in aging. Cell Cycle. 2011;10(21):3631. doi: 10.4161/cc.10.21.18181. [DOI] [PubMed] [Google Scholar]
- Meeusen S, et al. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127(2):383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Mendl N, et al. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci. 2011;124(Pt 8):1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- Menzies FM, Yenisetti SC, Min KT. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr Biol. 2005;15(17):1578–1582. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- Merkwirth C, et al. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22(4):476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulener M, et al. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr Biol. 2005;15(17):1572–1577. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- Miller KE, Sheetz MP. Axonal mitochondrial transport and potential are correlated. J Cell Sci. 2004;117(Pt 13):2791–2804. doi: 10.1242/jcs.01130. [DOI] [PubMed] [Google Scholar]
- Mills RD, et al. Biochemical aspects of the neuroprotective mechanism of PTEN-induced kinase-1 (PINK1) J Neurochem. 2008;105(1):18–33. doi: 10.1111/j.1471-4159.2008.05249.x. [DOI] [PubMed] [Google Scholar]
- Mironov SL. Complexity of mitochondrial dynamics in neurons and its control by ADP produced during synaptic activity. Int J Biochem Cell Biol. 2009;41(10):2005–2014. doi: 10.1016/j.biocel.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Misko A, et al. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci. 2010;30(12):4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno Y, et al. Progress in familial Parkinson’s disease. J Neural Transm Suppl. 2006;70:191–204. doi: 10.1007/978-3-211-45295-0_30. [DOI] [PubMed] [Google Scholar]
- Molina AJ, Shirihai OS. Monitoring mitochondrial dynamics with photoactivatable [corrected] green fluorescent protein. Methods Enzymol. 2009;457:289–304. doi: 10.1016/S0076-6879(09)05016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina AJ, et al. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes. 2009;58(10):2303–2315. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, et al. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum Mol Genet. 2005;14(1):71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- Nakada K, Inoue K, Hayashi J. Interaction theory of mammalian mitochondria. Biochem Biophys Res Commun. 2001a;288(4):743–746. doi: 10.1006/bbrc.2001.5838. [DOI] [PubMed] [Google Scholar]
- Nakada K, et al. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat Med. 2001b;7(8):934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- Narender T, et al. An unprecedented biogenetic-type chemical synthesis of 1(15–>11) abeotaxanes from normal taxanes. J Nat Prod. 2010;73(4):747–750. doi: 10.1021/np900722j. [DOI] [PubMed] [Google Scholar]
- Narendra DP, Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox Signal. 2011;14(10):1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, et al. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 2009;5(5):706–708. doi: 10.4161/auto.5.5.8505. [DOI] [PubMed] [Google Scholar]
- Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010a;8(1):e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, et al. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010b;6(8):1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M, et al. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J Biol Chem. 2005;280(26):25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- Nisipeanu P, et al. Autosomal-recessive juvenile parkinsonism in a Jewish Yemenite kindred: mutation of Parkin gene. Neurology. 1999;53(7):1602–1604. doi: 10.1212/wnl.53.7.1602. [DOI] [PubMed] [Google Scholar]
- Nisipeanu P, et al. Parkin gene causing benign autosomal recessive juvenile parkinsonism. Neurology. 2001;56(11):1573–1575. doi: 10.1212/wnl.56.11.1573. [DOI] [PubMed] [Google Scholar]
- Nochez Y, et al. Acute and late-onset optic atrophy due to a novel OPA1 mutation leading to a mitochondrial coupling defect. Mol Vis. 2009;15:598–608. [PMC free article] [PubMed] [Google Scholar]
- Ohno N, et al. Myelination and axonal electrical activity modulate the distribution and motility of mitochondria at CNS nodes of Ranvier. J Neurosci. 2011;31(20):7249–7258. doi: 10.1523/JNEUROSCI.0095-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15(8):887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A, et al. OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 2007;14(4):682–692. doi: 10.1038/sj.cdd.4402048. [DOI] [PubMed] [Google Scholar]
- Oliveira JM. Mitochondrial bioenergetics and dynamics in Huntington’s disease: tripartite synapses and selective striatal degeneration. J Bioenerg Biomembr. 2010;42(3):227–234. doi: 10.1007/s10863-010-9287-6. [DOI] [PubMed] [Google Scholar]
- Oliveri RL, et al. The parkin gene is not involved in late-onset Parkinson’s disease. Neurology. 2001;57(2):359–362. doi: 10.1212/wnl.57.2.359. [DOI] [PubMed] [Google Scholar]
- Ono T, et al. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28(3):272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191(6):1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouvrier R, Grew S. Mechanisms of disease and clinical features of mutations of the gene for mitofusin 2: an important cause of hereditary peripheral neuropathy with striking clinical variability in children and adults. Dev Med Child Neurol. 2010;52(4):328–330. doi: 10.1111/j.1469-8749.2010.03613.x. [DOI] [PubMed] [Google Scholar]
- Parone PA, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3(9):e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partikian A, et al. Rapid diffusion of green fluorescent protein in the mitochondrial matrix. J Cell Biol. 1998;140(4):821–829. doi: 10.1083/jcb.140.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterna JC, et al. DJ-1 and Parkin modulate dopamine-dependent behavior and inhibit MPTP-induced nigral dopamine neuron loss in mice. Mol Ther. 2007;15(4):698–704. doi: 10.1038/sj.mt.6300067. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297(5588):1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105(5):1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, et al. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5(4):e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, et al. Aberrant mitochondrial fission in neurons induced by protein kinase C{delta} under oxidative stress conditions in vivo. Mol Biol Cell. 2011;22(2):256–265. doi: 10.1091/mbc.E10-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A, et al. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS One. 2011;6(3):e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Lippincott-Schwartz J. Together we are stronger: fusion protects mitochondria from autophagosomal degradation. Autophagy. 2011;7(12):1568–1569. doi: 10.4161/auto.7.12.17992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, et al. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci USA. 2011b;108(25):10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice SE, Gelfand VI. Paradigm lost: milton connects kinesin heavy chain to miro on mito-chondria. J Cell Biol. 2006;173(4):459–461. doi: 10.1083/jcb.200604071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo GJ, et al. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J Neurosci. 2009;29(17):5443–5455. doi: 10.1523/JNEUROSCI.5417-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Frank S. Shaping mitochondria: the complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60(7):448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- Saotome M, et al. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci USA. 2008;105(52):20728–20733. doi: 10.1073/pnas.0808953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002a;1576(1–2):1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002b;286(1):81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- Schauss AC, et al. A novel cell-free mitochondrial fusion assay amenable for high-throughput screenings of fusion modulators. BMC Biol. 2010;8:100. doi: 10.1186/1741-7007-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L. Proteins that fuse and fragment mitochondria in apoptosis: confissing a deadly con-fusion? J Bioenerg Biomembr. 2005;37(3):165–170. doi: 10.1007/s10863-005-6572-x. [DOI] [PubMed] [Google Scholar]
- Semenzato M, Cogliati S, Scorrano L. Prohibitin(g) cancer: aurilide and killing by Opa1-dependent cristae remodeling. Chem Biol. 2011;18(1):8–9. doi: 10.1016/j.chembiol.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Serasinghe MN, Yoon Y. The mitochondrial outer membrane protein hFis1 regulates mitochondrial morphology and fission through self-interaction. Exp Cell Res. 2008;314(19):3494–3507. doi: 10.1016/j.yexcr.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JM, Nunnari J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002;12(4):178–184. doi: 10.1016/s0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, Martin SJ. Mitochondrial fission/fusion dynamics and apoptosis. Mitochondrion. 2010;10(6):640–648. doi: 10.1016/j.mito.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Sheridan C, et al. Bax- or Bak-induced mitochondrial fission can be uncoupled from cytochrome C release. Mol Cell. 2008;31(4):570–585. doi: 10.1016/j.molcel.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Shitara H, et al. Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics. 2000;156(3):1277–1284. doi: 10.1093/genetics/156.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff EH, et al. BH3 peptides induce mitochondrial fission and cell death independent of BAX/BAK. PLoS One. 2009;4(5):e5646. doi: 10.1371/journal.pone.0005646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev VP. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem Sci. 2001;26(1):23–29. doi: 10.1016/s0968-0004(00)01735-7. [DOI] [PubMed] [Google Scholar]
- Song Z, et al. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178(5):749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, et al. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20(15):3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FX, et al. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes. 2006;55(6):1783–1791. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- Sulzer D. Multiple hit hypotheses for dopamine neuron loss in Parkinson’s disease. Trends Neurosci. 2007;30(5):244–250. doi: 10.1016/j.tins.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Surmeier DJ, Guzman JN, Sanchez-Padilla J. Calcium, cellular aging, and selective neuronal vulnerability in Parkinson’s disease. Cell Calcium. 2010a;47(2):175–182. doi: 10.1016/j.ceca.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, et al. What causes the death of dopaminergic neurons in Parkinson’s disease? Prog Brain Res. 2010b;183:59–77. doi: 10.1016/S0079-6123(10)83004-3. [DOI] [PubMed] [Google Scholar]
- Taguchi N, et al. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282(15):11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Taira T, et al. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5(2):213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Ohmuro H, et al. Autophagy in embryonic erythroid cells: its role in maturation. Eur J Cell Biol. 2000;79(10):759–764. doi: 10.1078/0171-9335-00096. [DOI] [PubMed] [Google Scholar]
- Tan AR, et al. Elevated intracellular calcium causes distinct mitochondrial remodelling and calcineurin-dependent fission in astrocytes. Cell Calcium. 2011;49(2):108–114. doi: 10.1016/j.ceca.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Tanaka A. Parkin-mediated selective mitochondrial autophagy, mitophagy: Parkin purges damaged organelles from the vital mitochondrial network. FEBS Lett. 2010;584(7):1386–1392. doi: 10.1016/j.febslet.2010.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010a;191(7):1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Matsuda N, Okatsu K. Mechanisms underlying the cause of Parkinson’s disease: the functions of Parkin/PINK1. Rinsho Shinkeigaku. 2010b;50(11):867. doi: 10.5692/clinicalneurol.50.867. [DOI] [PubMed] [Google Scholar]
- Taneike M, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6(5):600–606. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- Terman A, et al. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid Redox Signal. 2010;12(4):503–535. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KJ, Cookson MR. The role of PTEN-induced kinase 1 in mitochondrial dysfunction and dynamics. Int J Biochem Cell Biol. 2009;41(10):2025–2035. doi: 10.1016/j.biocel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KJ, et al. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20(1):40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14(10):1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, et al. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am J Physiol Cell Physiol. 2006;291(1):C176–C184. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008a;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008b;1777(9):1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, et al. Biophysical properties of mitochondrial fusion events in pancreatic beta-cells and cardiac cells unravel potential control mechanisms of its selectivity. Am J Physiol Cell Physiol. 2010;299(2):C477–C487. doi: 10.1152/ajpcell.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laar VS, et al. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20(5):927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ved R, et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J Biol Chem. 2005;280(52):42655–42668. doi: 10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Przedborski S. PINK1 points Parkin to mitochondria. Autophagy. 2010;6(5):674–675. doi: 10.4161/auto.6.5.12068. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, et al. PINK1/Parkin direct mitochondria to autophagy. Autophagy. 2010a;6(2):315–316. doi: 10.4161/auto.6.2.11199. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, et al. Control of mitochondrial integrity in Parkinson’s disease. Prog Brain Res. 2010b;183:99–113. doi: 10.1016/S0079-6123(10)83006-7. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010c;107(1):378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. Imaging axonal transport of mitochondria. Methods Enzymol. 2009a;457:319–333. doi: 10.1016/S0076-6879(09)05018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009b;136(1):163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, et al. Mitofusin-2 is a novel direct target of p53. Biochem Biophys Res Commun. 2010;400(4):587–592. doi: 10.1016/j.bbrc.2010.08.108. [DOI] [PubMed] [Google Scholar]
- Wang H, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011a;286(13):11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011b;147(4):893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, et al. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011c;17(1):71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- Weihofen A, et al. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking. Biochemistry. 2009;48(9):2045–2052. doi: 10.1021/bi8019178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11(12):872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? J Bioenerg Biomembr. 2009;41(6):499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- Wikstrom JD, et al. beta-Cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes. 2007;56(10):2569–2578. doi: 10.2337/db06-0757. [DOI] [PubMed] [Google Scholar]
- Wikstrom JD, Twig G, Shirihai OS. What can mitochondrial heterogeneity tell us about mitochondrial dynamics and autophagy? Int J Biochem Cell Biol. 2009;41(10):1914–1927. doi: 10.1016/j.biocel.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Wilkerson DC, Sankar U. Mitochondria: a sulfhydryl oxidase and fission GTPase connect mitochondrial dynamics with pluripotency in embryonic stem cells. Int J Biochem Cell Biol. 2011;43(9):1252–1256. doi: 10.1016/j.biocel.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Wood-Kaczmar A, et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS One. 2008;3(6):e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, et al. Bax is essential for Drp1-mediated mitochondrial fission but not for mitochondrial outer membrane permeabilization caused by photodynamic therapy. J Cell Physiol. 2011;226(2):530–541. doi: 10.1002/jcp.22362. [DOI] [PubMed] [Google Scholar]
- Xiong H, et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119(3):650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci USA. 2008;105(19):7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Weaver D, Hajnoczky G. Control of mitochondrial motility and distribution by the calcium signal: a homeostatic circuit. J Cell Biol. 2004;167(4):661–672. doi: 10.1083/jcb.200406038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, et al. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23(15):5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, et al. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J Cell Sci. 2005;118(Pt 18):4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- Yue Z. Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 2007;3(2):139–141. doi: 10.4161/auto.3602. [DOI] [PubMed] [Google Scholar]
- Yue Z, et al. The cellular pathways of neuronal autophagy and their implication in neurode-generative diseases. Biochim Biophys Acta. 2009;1793(9):1496–1507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Wai-Man P, et al. OPA1 mutations cause cytochrome c oxidase deficiency due to loss of wild-type mtDNA molecules. Hum Mol Genet. 2010;19(15):3043–3052. doi: 10.1093/hmg/ddq209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies – disease mechanisms and therapeutic strategies. Prog Retin Eye Res. 2011a;30(2):81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Wai-Man P, et al. Genetic screening for OPA1 and OPA3 mutations in patients with suspected inherited optic neuropathies. Ophthalmology. 2011b;118(3):558–563. doi: 10.1016/j.ophtha.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci USA. 2007;104(47):18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, et al. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: implications for pathogenesis. Hum Mol Genet. 2005;14(14):2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. G-protein beta2 subunit interacts with mitofusin 1 to regulate mitochondrial fusion. Nat Commun. 2010;1:101. doi: 10.1038/ncomms1099. [DOI] [PubMed] [Google Scholar]
- Ziviani E, Whitworth AJ. How could Parkin-mediated ubiquitination of mitofusin promote mitophagy? Autophagy. 2010;6(5):660–662. doi: 10.4161/auto.6.5.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci USA. 2010;107(11):5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]