Abstract
Background
Bombax ceiba L. (the red silk cotton tree) is a large deciduous tree that is distributed in tropical and sub-tropical Asia as well as northern Australia. It has great economic and ecological importance, with several applications in industry and traditional medicine in many Asian countries. To facilitate further utilization of this plant resource, we present here the draft genome sequence for B. ceiba.
Findings
We assembled a relatively intact genome of B. ceiba by using PacBio single-molecule sequencing and BioNano optical mapping technologies. The final draft genome is approximately 895 Mb long, with contig and scaffold N50 sizes of 1.0 Mb and 2.06 Mb, respectively.
Conclusions
The high-quality draft genome assembly of B. ceiba will be a valuable resource enabling further genetic improvement and more effective use of this tree species.
Keywords: Bombax ceiba, genome assembly, annotation, evolution
Data Description
Introduction
Bombax ceiba Linn. (Malvaceae), commonly known as the cotton tree or red silk cotton tree, is a spectacular flowering tree with a height of up to 40 meters (Fig. 1a) that is found in tropical and sub-tropical Asia as well as northern Australia [1]. It has been chosen as the “city flower” of the cities of Kaohsiung and Guangzhou for its large, showy flowers with thick, waxy, red petals that densely clothe leafless branch tips in late winter and early spring (Fig. 1b, c). B. ceiba is a source of food, fodder, fiber, fuel, medicine, and many other valuable goods for natives of many Asian countries [2]. For example, its fruits are good sources of silk-cotton for making mattresses, cushions, pillows, and quilts [3], while its timbers are widely used in matches, boxes, and splints [4]. Moreover, studies on the cotton tree have shown that it produces many novel secondary metabolites and have explored its traditional medicinal usage by various tribal communities [1, 2, 5, 6]. In addition to its economic and medicinal value, B. ceiba is an ecologically important plant: it is a reforestation pioneer that survives easily in low-rainfall and well-drained conditions [7] and has been identified as a plant species suitable for municipal greening because of its capacity to counteract the detrimental effects of air pollution [8, 9].
Figure 1:
Example of the red silk cotton tree (B. ceiba). A) Natural habitat of B. ceiba (image from Guanglong Ou). B) B. ceiba used as municipal greening trees (image from Jianmei Wu). C) The flower of B. ceiba (image from Renbin Zhu).
Despite the considerable economic and ecological importance of B. ceiba, the genomic information available for this species is limited, which has hindered its utilization. Here we report a draft genome sequence for B. ceiba that is expected to facilitate and expand its use.
Sampling and sequencing
All samples were collected from Yuanmou, Yunnan Province, China (25°40′50.06″ N, 101°53′27.76″ E). Genomic DNA was extracted from leaves of a single tree using the Plant Genomic DNA kit (Tiangen, Beijing, China). A SMRTbell DNA library was then prepared and sequenced using P6, C4 chemistry according to the manufacturer's protocols (Pacific Biosciences, CA, USA), and a 20-kb SMRTbell library was generated using a BluePippin DNA size selection instrument (Sage Science, MA, USA) with a lower size limit of 10 kb. Single-molecule real-time sequencing of long reads was conducted on a PacBio Sequel platform with 19 SMRT cells. A total of 86.0 Gb of genomic data with an average read length of 8.4 kbwas generated after quality filtering (Supplementary Table S1). In addition, a separate paired-end (PE) DNA library with an insert size of 400 bp (amplification by eight PCR cycles) was constructed and sequenced using the Illumina platform (PE 150) to enable a genome survey. The NGS sequencing produced 36.1 Gb of raw data, of which 20.0 Gb remained after filtering.
Total RNA was extracted from the bud, root, bark, flower, and fruit tissues of one B. ceiba individual using the QIAGEN RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). RNA-seq libraries were then prepared using the TruSeq RNA Library Preparation Kit (Illumina, CA, USA), and paired-end sequencing with a read length of 150 bp was conducted on the HiSeq 2000 platform, yielding 44.41 Gb of clean data (30,816,034−51,191,192 reads per sample) (Supplementary Table S2).
Genome size and heterozygosity estimation
The genome size of B. ceiba was estimated by the K-mer method [10] using sequencing data from the Illumina DNA library. Quality-filtered reads were subjected to 17-mer frequency distribution analysis using the Jellyfish program [10]. The count distribution of 17-mers followed a Poisson distribution, with the highest peak occurring at a depth of 22 (Supplementary Table S3 and Fig. S1). The estimated genome size was approximately 809,166,127 bp, and the heterozygosity rate of the B. ceiba genome was approximately 0.88%.
Genome assembly
Genome assembly was performed on full PacBio long reads using FALCON v0.3.0 [11]. Error correction and pre-assembly were carried out with the FALCON pipeline after evaluating the outcomes of using different parameters in FALCON during the pre-assembly process. Based on the contig N50 results, a length_cutoff of 11kb and a length_cutoff_pr of 11.5kb for the assembly step were ultimately chosen. The draft assembly was polished using Arrow [12], which mapped the PacBio reads to the assembled genome with the Blasr pipline [13]. The preliminary genome assembly was approximately 852 Mb in size, with a contig N50 size of 727 kb. A GC depth analysis was conducted to assess the potential contamination during sequencing and the coverage of the assembly, revealing that the genome had an average GC content of 33.3% and a unimodal GC content distribution (Supplementary Fig. S2). The GC depth as well as the sequencing depth of the genome assembly suggested that there was no contamination from other species (Supplementary Fig. S3). To further assess contaminations, we searched all sequences of the genome assembly against the NCBI non-redundant nucleotide database (Nt) with BLASTN [14] (E-value ≤ 1e−5). In total, 2,494 significant hits were achieved. The top-hit species were Theobroma cacao and Gossypium species, comprising more than 69% ( 1,733 hits) of the hits (Supplementary Table S4). Only five hits from four non-plant species (Psyllidae sp., Trioza eugeniae, Diptacus sp., and Dichorragia nesimachus) were detected (Supplementary Table S4), suggesting there was no potential contamination from non-plant species in the genome of Bombax ceiba.
Scaffolding with BioNano optical mapping
The purified genomic DNA of B. ceiba was embedded in an agarose layer, digested with Nt. BspQI enzyme, and labeled. The molecules were counterstained using the protocol provided with the IrysPrep Reagent Kit (BioNano Genomics, San Diego, USA). Samples were then loaded into IrysChips and imaged on an Irys imaging instrument (BioNano Genomics, San Diego, USA). After filtering using a molecule length cutoff of <150kb, a molecule SNR of <2.75, a label SNR of <2.75, and a label intensity of >0.8, 160.0 Gb of BioNano clean data were obtained, with the N50 size of the labeled single molecules being 269.9 kb (Supplementary Table S5).
A molecular quality report was generated by aligning the BioNano library sequences to the initial PacBio genome assembly, yielding a map rate of 34.2%. Using the PacBio genome assembly data as a reference, a reference genome assembly was conducted based on the clean BioNano data. A genome map consisting of 2,023 consensus maps was assembled, yielding a genome size of 1.09 Gb with an N50 size of 0.7 Mb. The average molecule coverage depth of the genome map was about 27 folds. To obtain a longer scaffold, the de novo assembly of PacBio reads was then mapped to the BioNano single-molecule genomic map. After scaffolding, the contig assembly contained 3,105 scaffolds with a scaffold N50 of 1.5 Mb.
To fill the gaps in the scaffolds, the Blasr pipline [13] was used to map the PacBio long reads to the draft genome assembly scaffolding with BioNano optical mapping. The draft was polished using PBJelly 2 (PBJelly, RRID:SCR_012091) [15] over three iterations. Reads from the Illumina DNA library (400bp) were then aligned against the genome assembly using the BWA software (BWA, RRID:SCR_010910) to fill the gaps and correct potential sequencing errors of the assembly, and a mapping rate of 99.2% was achieved [16]. The final assembly was polished using Pilon [17], yielding a final draft genome of approximately 895 Mb, with contig and scaffold N50 sizes of 1.0 Mb and 2.06 Mb, respectively (SupplementaryTable S6).
Evaluation of the completeness of the genome assembly gene space
To evaluate the coverage of the assembly, we aligned all the RNA-seq reads against the B. ceiba genome assembly using HISAT [18] with default parameters. The percentage of aligned reads ranged from 84.78% to 91.08% (Supplementary Table S2). We then used Benchmarking Universal Single-Copy Orthologs (BUSCO, RRID:SCR_015008) [19] to search the annotated genes in the assembly for the 1,440 single-copy genes conserved among all embryophytes. About 94.4% of the complete BUSCOs were found in the assembly (Supplementary Table S7). These results suggested that the genome assembly was complete and robust.
Genome annotation
The repeat sequences in the genome consisted of simple sequence repeats (SSRs), moderately repetitive sequences, and highly repetitive sequences. The MISA tool [20] was used to search for SSR motifs in the B. ceiba genome, with default parameters. A total of 454,435 SSRs were identified in this way: 310,369, 105,004, 30,925, 6,448, 1,165,and 524 mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide repeats, respectively (Supplementary Table S8).
To identify known transposable elements (TEs) in the B. ceiba genome, RepeatMasker (RepeatMasker, RRID:SCR_012954) [21] was used to screen the assembled genome against the Repbase (v. 22.11) [22] and Mips-REdat libraries [23]. In addition, de novo evolved annotation was performed using RepeatModeler v. 1.0.11 (RepeatModeler, RRID:SCR_015027) [21]. The combined results of the homology-based and de novo predictions indicated that repeated sequences account for 60.30% of the B. ceiba genome assembly (Supplementary Table S9), with long terminal repeats accounting for the greatest proportion (47.86%) (Supplementary Table S9).
Homology-based ncRNA annotation was performed by mapping plant rRNA, miRNA, and snRNA genes from the Rfam database (release 13.0) [24] to the B. ceiba genome using BLASTN [14] (E-value ≤ 1e−5). tRNAscan-SE v1.3.1 (tRNAscan-SE, RRID:SCR_010835) [25] was used (with default parameters for eukaryotes) for tRNA annotation. RNAmmer v1.2 [26] was used to predict rRNAs and their subunits. These analyses identified 496 miRNAs, 894 tRNAs, 6,772 rRNAs, and 727 snRNAs (Supplementary Table S10).
The homology-based and de novo predictions were also used to annotate protein coding genes. For homology-based predictions, protein sequences from four species (Arabidopsis thaliana, Carica papaya, Gossypium arboretum, and T. cacao) (Supplementary Table S11) were mapped onto the B. ceiba genome; the aligned sequences and the corresponding query proteins were then filtered and passed to GeneWise v2.4.1 (GeneWise, RRID:SCR_015054) [27] to search for accurately spliced alignments. For the de novo predictions, we first randomly selected 1,000 full-length genes from the homology-based predictions to train model parameters for Augustus v3.0 (Augustus: Gene Prediction, RRID:SCR_008417) [28], GeneID v1.4.4 [29], GlimmerHMM (GlimmerHMM, RRID:SCR_002654) [30], and SNAP [31]. Augustus v3.0 [28], GeneID v1.4.4 [29], GlimmerHMM [30], and SNAP [31] were then used to predict genes based on the training set. Finally, EVidenceModeler v1.1.1 [32] was used to integrate the predicted genes and generate a consensus gene set (Supplementary Table S11). Genes with TEs were discarded using the TransposonPSI [33] package. Low-quality genes consisting of fewer than 50 amino acids and/or exhibiting premature termination were also removed from the gene set, yielding a final set of 52,705 genes. The final set's average transcript length, average CDS length, and exon number per gene were 2,418.37 bp, 1,019.38 bp, and 4.57, respectively (Supplementary Table S12 and Fig. S4).
The annotations of the predicted genes of B. ceiba were screened for homology against the Uniprot (release 2017_10) and KEGG (release 84.0) databases using Blastall [14] and KAAS [34]. Then, the InterProScan (InterProScan, RRID:SCR_005829) [35] package was used to annotate the predicted genes using the InterPro (5.21–60.0) database. In total, 47,105 of the total 52,705 genes (89.37 %) were annotated with potential functions (Supplementary Table S13).
Phylogenetic tree construction and divergence time estimation
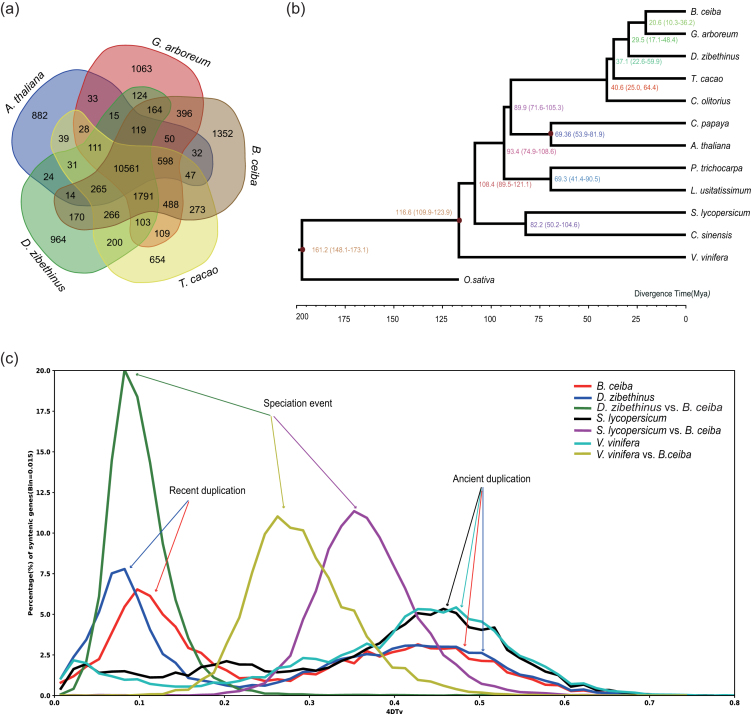
To investigate the evolutionary position of B. ceiba, we compared its genome to the genome sequences of 12 other plants. These included four plants in the Malvales order (Gossypium arboreum, Durio zibethinus, Corchorus olitorius, and T. cacao), seven plants from different orders in the same Eudicots clade (Arabidopsis thaliana, Carica papaya, Linum usitatissimum, Populus trichocarpa, Camellia sinensis, Solanum lycopersicum, and Vitis vinifera), and Oryza sativa as an outgroup. Genome sequences from A. thaliana, T. cacao, C. papaya, L. usitatissimum, P. trichocarpa, C. sinensis, S. lycopersicum, V. vinifera, and O. sativa were downloaded from Phytozome v. 12.0 [36]. Gene sequences of G. arboreum, C. olitorius, and D. zibethinus were downloaded from the NCBI Database (PRJNA335838, PRJNA215141 and PRJNA400310). We used the OrthoMCL (v2.0.9) pipeline (OrthoMCL DB: Ortholog Groups of Protein Sequences, RRID:SCR_007839) [37] (BLASTP E-value ≤ 1e−5) to identify potentially orthologous gene families within these genomes. Gene family clustering identified 16,586 gene families containing 37,736 genes in B. ceiba (Fig. 2a). Of these, 906 gene families were unique to B. ceiba (Supplementary Table S14). B. ceiba and other Malvales plants had the largest number of shared gene families among the studied plants.
Figure 2:
Phylogenetic relationships and genomic comparisons between B. ceiba and other plants. A) A Venn diagram of shared gene families between B. ceiba and three other Malvales plants, with A. thaliana as an outgroup. Each number represents a gene family number. B) Inferred phylogenetic tree across 13 plant species. The estimated divergence time (Mya) is shown at each node. C) Whole-genome duplication (WGD) events of four plants (B. ceiba, D. zibethinus, S. lycopersicum, and V. vinifera) inferred by four-fold synonymous third-codon transversion (4DTv) estimations. Peaks corresponding to speciation, recent, and ancient WGDs are indicated by arrows.
Phylogenetic analysis was performed using 172 single-copy orthologous genes from common gene families found by OrthoMCL [37] (Supplementary Fig. S5). We codon-aligned each gene family using MUSCLE (MUSCLE, RRID:SCR_011812) [38] and curated the alignments with Gblocks v0.91b [39]. Phylogeny analysis was performed using RAxML (RAxML, RRID:SCR_006086) v 8.2.11 [40] with the GTRGAMMA model and 100 bootstrap replicates. We then used MCMCTREE as implemented in PAML v4.9e (PAML, RRID:SCR_014932) [41] to estimate the divergence times of B. ceiba from the other plants. The parameter settings of MCMCTREE were as follows: clock = 2, RootAge ≤ 1.73, model = 7, BDparas = 110, kappa_gamma = 62, alpha_gamma = 11, rgene_gamma = 23.18, and sigma2_gamma = 14.5. In addition, the divergence times of O. sativa (148–173 Mya), V. vinifera (110–124 Mya),and A. thaliana (53–82 Mya) were used for fossil calibration. The phylogenetic analysis showed that B. ceiba is more closely related to G. arboraum than to D. zibethinus (Supplementary Fig. S6), which supports the well-established hypothesis of a close relationship between Bombacaceae and Malvaceae [42, 43]. Recent phylogenetic studies have suggested that the group traditionally referred to as Bombacaceae (which includes the tribe Durioneae) is not actually monophyletic, and that the genera of the tribe Durioneae should be excluded from Bombacaceae. Most members of the erstwhile family Bombacaceae have been transferred to the subfamily Bombacoideae within the family Malvaceae [43]. This phylogenetic ordering was supported by our phylogenetic analysis of the complete chloroplast genomes of Marvel plants [44]. The estimated divergence time of B. ceiba and D. zibethinus was 29.5 Mya, while that of B. ceiba and G. arboretum was about 20.6 Mya (Fig. 2b).
Genes under positive selection
B. ceiba is an ecologically important plant that could survive in extreme climate conditions, such as hot-dry valleys [7]. According to the neutral theory of molecular evolution [45], the ratio of nonsynonymous substitution rate (Ka) and synonymous substitution rate (Ks) of protein coding genes can be used to identify genes that show signatures of natural selection. We calculated average Ka/Ks values and conducted the branch-site likelihood ratio test using Codeml implemented in the PAML package [41] to identify positively selected genes in the B. ceiba lineage. These genes might contribute to the adaption to unfavorable environments. Thirty-six genes with signatures of positive selection were identified (P ≤ 0.05), of which 32 genes could be annotated with potential functions in the Swissport database (Supplementary Table S15). One gene is homologous to a desiccation protectant protein coding gene (Lea14). There is a strong association of LEA proteins with abiotic stress tolerance, particularly during dehydration and cold stress [46]. This gene could potentially contribute to the adaption of B. ceiba to the dry valley environment. Another gene showing signs of positive selection is homologous to the gene coding for Kelch domain-containing protein 4. The Kelch domain-containing proteins are involved in regulating major processes such as growth, development, and biotic and abiotic stress responses in plants [47, 48]. Some researchers suggested that the E3 ubiquitin-protein ligase (RFWD3) has potential roles in plant stress responses [49, 50]. Twenty-one positively selected sites were identified in the CACTIN protein coding gene. The CACTIN protein was characterized as a negative regulator of many different developmental processes, such as embryogenesis [51]. While literature reports are rare, other identified genes might also be associated with the ecological adaption of B. ceiba. It should be noted that this is just a preliminary analysis of the functions of these genes; further studies would be needed to clarify their roles.
Whole-genome duplication and gene family expansion analysis
We used 4DTv estimation to detect WGD events in the B. ceiba genome. To this end, paralogous sequences of B. ceiba, T. cacao, V. vinfira, S. lycopersicum, and D. zibethinus was identified with OrthoMCL [37]. Then, protein sequences for each of these plants were aligned against each other with Blastp [14] (using an E-value threshold of ≤1e−5) to identify conserved paralogs in each species. Finally, potential WGD events in each genome were evaluated based on their 4DTv distribution. The WGD analysis suggested that B. ceiba experienced the same WGD events as other Dicotyledons, and that B. ceiba and D. zibethinus went through their WGD events before diverging from their common ancestor (Fig. 2c).
The OrthoMCL gene family analysis results were analyzed further by using Computational Analysis of Gene Family Evolution, v3.0 [52] to detect expanded gene families. This approach revealed 5,612 expanded gene families and 1,902 contracted gene families in the B. ceiba lineage (Supplementary Fig. S7).
Conclusion
This paper reports the sequencing, assembly, and annotation of the B. ceiba genome along with details of its evolutionary history. The genomic data generated in this work will be a valuable resource for further genetic improvement and effective use of the red silk cotton tree.
Availability of supporting data
The raw data from our genome project was deposited in the SRA (Sequence Read Archive) database of National Center for Biotechnology Information with Bioproject ID PRJNA429932. The assembly and annotation of the B. ceiba genome and other supporting data, including BUSCO results, are available in the GigaScience database, GigaDB [53]. Versions and main parameters of the software used in this study are provided in Supplementary Table S16.
Additional files
Figure S1. Frequency distribution of the 17-mer graph analysis used to estimate the size of the B. ceiba genome.
Figure S2. GC content distribution of the B. ceiba genome. The GC content was established using 500-bp slidingwindows.
Figure S3. The GC depth distribution of the B. ceiba genome.
Figure S4. Comparison of gene structure characteristics in B. ceiba to that in other plants. a, CDS length; b, exon length; c, exon number; d, gene length; e, intron length.
Figure S5. Gene orthology determined by comparing genomes using the OrthoMCL software.
Figure S6. The maximum-likelihood phylogeny of B. ceiba and 13 other plants.
Figure S7. Gene family expansions and contractions in B. ceiba and 13 other plants.
Table S1. Sequencing statistics from the PacBio platform.
Table S2. Summary of the transcriptomes and their mapping rates on the genome assembly.
Table S3. Estimation of genome size based on 17-mer statistics.
Table S4. Blast results of B. ceiba genome against the NCBI Nt database.
Table S5. Summary of the BioNano optical mapping data.
Table S6. Summary of the final genome assembly.
Table S7. Summary of BUSCO analysis results.
Table S8. Summary of the SSR search results.
Table S9. Repeat annotation of the B. ceiba genome assembly.
Table S10. Summary of non-protein-coding gene annotations in the B. ceiba genome assembly.
Table S11. Gene annotation statistics of the B. ceiba genome assembly.
Table S12. Comparative gene statistics.
Table S13. Functional annotation of predicted genes of B. ceiba.
Table S14. Summary statistics of gene families in 13 plant species.
Table S15. Candidate positively selected genes in the B. ceiba lineage.
Table S16. Versions and main parameters of the software used in this study.
Abbreviations
SMRT: single molecule real time; 4DTv: four-fold synonymous third-codon transversion.
Competing interests
S.S. is an employee of Nextomics Bioscences. Other authors declare that they have no competing interests.
Authors’ contributions
L.T. and B.T. designed the project; H.W., C.L., and H.C. collected samples and extracted the DNA and RNA samples; Y.G., S.S., H.,W. and C.L. worked on sequencing and data analyzing; Y.G. wrote the manuscript; L.T., B.T., and D.D. revised the manuscript; Y. G., H. W., C. L., H. C., D. D., S. S., L. Y., L. H., Y. F., B. T. and L. T. read and approved the final version of the manuscript.
Supplementary Material
2/23/2018 Reviewed
2/27/2018 Reviewed
3/19/2018 Reviewed
4/8/2018 Reviewed
Acknowledgements
We thank Guanglong Ou, Jianmei Wu, and Renbin Zhu for offering photos of B. ceiba. This study was financially supported by the National Science Foundation of China (grants 31460561, 31760103, 31460179, and 31660680), the Key Laboratory of Forest Resources Conservation and Utilization in the Southwest Mountains of China (Southwest Forestry University), Ministry of Education, and the Yunnan Applied Basic Research Project (grant 2017FD145).
References
- 1. Barwick M. Tropical and Subtropical Trees. Portland: Timber Press, 2004. [Google Scholar]
- 2. Jain V, Verma SK. Pharmacology of Bombax Ceiba Linn. Berlin Heidelberg: Springer, 2012. [Google Scholar]
- 3. Chand S, Singh AK. In vitro propagation of Bombax ceiba L. (Silkcotton). Silvae Genetica. 1999;48(6):313–7. [Google Scholar]
- 4. Nair GS, Bai Y. Ethnobotanical value of dry, fallen ovaries of Bombax ceiba L. (Bombacaceae: Malvales). J Threatened Taxa. 2012;4(15):3443–6. [Google Scholar]
- 5. Ngwuluka NC. Are Bombax buonopozense and Bombax Malabaricum possible nutraceuticals for age management?. Prev Med. 2012;54(S3):64–70. [DOI] [PubMed] [Google Scholar]
- 6. Pankaj HC, Somshekhar SK. Bombax ceiba Linn.: pharmacognosy, ethnobotany and phyto-pharmacology. Pharmacogn Commun. 2012;2(3):2–9. [Google Scholar]
- 7. Zhou Z, Ma H, Lin K et al. RNA-seq reveals complicated transcriptomic responses to drought stress in a nonmodel tropic plant, Bombax ceiba L. Evol Bioinform. 2015;11(S1):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng C, Wen D, Sun Z et al. Response of some plants for municipal greening to air pollutants. J Trop Subtrop Bot. 2002;10(4):321–7. [Google Scholar]
- 9. Elhagrassi AM, Ali MM, Osman AF et al. Phytochemical investigation and biological studies of Bombax Malabaricum flowers. Nat Prod Res. 2011;25(2):141–51. [DOI] [PubMed] [Google Scholar]
- 10. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of K-mers. Bioinformatics. 2011;27(6):764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. PacBio Falcon GitHub: https://github.com/PacificBiosciences/falcon, Accessed 18 Mar 2018. [Google Scholar]
- 12. PacBio Genomic Consensus GitHub:https://github.com/PacificBiosciences/GenomicConsensus. Accessed 18 Mar 2018. [Google Scholar]
- 13. Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (Blasr): application and theory. BMC Bioinformatics. 2012;13(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Camacho C, Coulouris G, Avagyan V et al. Blast+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Worley KC, English AC, Richards S et al. Improving genomes using long reads and Pbjelly 2. In: International Plant and Animal Genome Conference Xxii. San Diego, CA, 10-15 January 2014. [Google Scholar]
- 16. Li H, Durbin R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Oxford University Press, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker BJ, Abeel T, Shea T et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim D, Langmead B, Salzberg SL. Hisat: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simão FA, Waterhouse RM, Ioannidis P et al. Busco: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. [DOI] [PubMed] [Google Scholar]
- 20. Thiel T, Michalek W, Varshney RK et al. Exploiting est databases for the development and characterization of gene-derived Ssr-markers in barley (Hordeum Vulgare L.). Theor Appl Genet. 2003;106(3):411–22. [DOI] [PubMed] [Google Scholar]
- 21. Tarailograovac M, Chen N. Using repeatmasker to identify repetitive elements in genomic sequences. Current Protocols in Bioinformatics2009;3:4–14. [DOI] [PubMed] [Google Scholar]
- 22. Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015;6(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas N, Martis MM, Roessner SK et al. Mips plantsdb: a database framework for comparative plant genome research. Nucleic Acids Res. 2013;41:1144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalvari I, Argasinska J, Quinones-Olvera N et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2017, 46, D335–D42.;10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lowe TM, Eddy SR. Trnascan-Se: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lagesen K, Hallin P, Rødland EA et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Birney E, Durbin R. Using genewise in the drosophila annotation experiment. Genome Res. 2000;10(4):547–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stanke M, Steinkamp R, Waack S et al. Augustus: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:309–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blanco E, Parra G, Guigó R. Using geneid to identify genes. Curr Protocol Bioinformatics. 2007;4(3):1–28. [DOI] [PubMed] [Google Scholar]
- 30. Majoros WH, Pertea M, Salzberg SL. Tigrscan and Glimmerhmm: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–9. [DOI] [PubMed] [Google Scholar]
- 31. Bromberg Y, Rost B. Snap: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35(11):3823–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haas BJ, Salzberg SL, Wei Z et al. Automated eukaryotic gene structure annotation using evidencemodeler and the program to assemble spliced alignments. Genome Biol. 2008;9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tansposon PSI:http://transposonpsi.sourceforge.net/. Accessed 18 Mar 2018. [Google Scholar]
- 34. Moriya Y, Itoh M, Okuda S et al. Kaas: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quevillon E, Silventoinen V, Pillai S et al. Interproscan: protein domains identifier. Nucleic Acids Res. 2005;33:116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. M GD, Shengqiang S, Russell H et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(Database issue):D1178–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Stoeckert CJ, Roos DS. Orthomcl: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–77. [DOI] [PubMed] [Google Scholar]
- 40. Stamatakis A. Raxml Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. [DOI] [PubMed] [Google Scholar]
- 42. Baum DA, Smith DW, Yen A et al. Phylogenetic relationships of malvatheca (Bombacoideae and Malvoideae; Malvaceae Sensu Lato) as inferred from plastid DNA sequences. Am J Bot. 2004;91(11):1863–71. [DOI] [PubMed] [Google Scholar]
- 43. Heywood VH, Brummitt RK, Culham A et al. Flowering Plant Families of the World. Richmond, Surrey: Royal Botanic Gardens, 2007. [Google Scholar]
- 44. Gao Y, Wang H, Liu C et al. Complete chloroplast genome sequence of the red silk cotton tree (Bombax ceiba). Mitochondr DNA B. 2018;3(1):315–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press, 1983. [Google Scholar]
- 46. Shinde S, Nurul IM, Ng CK. Dehydration stress-induced oscillations in LEA protein transcripts involves abscisic acid in the moss, physcomitrella patens. New Phytol. 2012;195(2):321–8. [DOI] [PubMed] [Google Scholar]
- 47. Feder A, Burger J, Gao S et al. Focus on metabolism: a Kelch domain-containing F-box coding gene negatively regulates flavonoid accumulation in muskmelon. Plant Physiol. 2015;169(3):1714–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X, Gou M, Guo C et al. Down-regulation of Kelch domain-containing F-box protein in arabidopsis enhances the production of (Poly)phenols and tolerance to ultraviolet radiation. Plant Physiol. 2015;167(2):337–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Serrano I, Campos L, Rivas S. Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Front Plant Sci. 2018;9(139, 1–7.):10.3389/fpls.2018.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Duplan V, Rivas S. E3 Ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci. 2014;5(42, 1–6.):doi:10.3389/fpls.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baldwin KL, Dinh EM, Hart BM et al. CACTIN is an essential nuclear protein in Arabidopsis and may be associated with the eukaryotic spliceosome. FEBS Lett. 2013;587(7):873–9. [DOI] [PubMed] [Google Scholar]
- 52. De Bie T, Cristianini N, Demuth JP et al. Cafe: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–71. [DOI] [PubMed] [Google Scholar]
- 53. Gao Y, Wang H, Liu C et al. Supporting data for “De novo genome assembly of the red silk cotton tree (Bombax ceiba)”. GigaScience Database. http://dx.doi.org/10.5524/100445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2/23/2018 Reviewed
2/27/2018 Reviewed
3/19/2018 Reviewed
4/8/2018 Reviewed