Abstract
Gene presence–absence polymorphisms segregating within species are a significant source of genetic variation but have been little investigated to date in natural populations. In plant pathogens, the gain or loss of genes encoding proteins interacting directly with the host, such as secreted proteins, probably plays an important role in coevolution and local adaptation. We investigated gene presence–absence polymorphism in populations of two closely related species of castrating anther-smut fungi, Microbotryum lychnidis-dioicae (MvSl) and M. silenes-dioicae (MvSd), from across Europe, on the basis of Illumina genome sequencing data and high-quality genome references. We observed presence–absence polymorphism for 186 autosomal genes (2% of all genes) in MvSl, and only 51 autosomal genes in MvSd. Distinct genes displayed presence–absence polymorphism in the two species. Genes displaying presence–absence polymorphism were frequently located in subtelomeric and centromeric regions and close to repetitive elements, and comparison with outgroups indicated that most were present in a single species, being recently acquired through duplications in multiple-gene families. Gene presence–absence polymorphism in MvSl showed a phylogeographic structure corresponding to clusters detected based on SNPs. In addition, gene absence alleles were rare within species and skewed toward low-frequency variants. These findings are consistent with a deleterious or neutral effect for most gene presence–absence polymorphism. Some of the observed gene loss and gain events may however be adaptive, as suggested by the putative functions of the corresponding encoded proteins (e.g., secreted proteins) or their localization within previously identified selective sweeps. The adaptive roles in plant and anther-smut fungi interactions of candidate genes however need to be experimentally tested in future studies.
Keywords: copy number variation, pathogen, population structure, adaptive variation
Introduction
Gene presence–absence polymorphisms segregating within species are probably a widespread and important source of genetic variation (Conrad et al. 2006, 2010). Several gene copy number changes have been shown to be adaptive, due to their effects on gene expression and phenotypic diversity (Henrichsen et al. 2009; Orozco et al. 2009). However, the proportion and type of genes affected by presence–absence polymorphism in natural populations have been little explored, and such studies are only now being facilitated by new sequencing technologies (Schrider and Hahn 2010; Albalat and Cañestro 2016).
Several fungal plant pathogen species have small, compact genomes (Möller and Stukenbrock 2017), rendering them highly suitable for the fine mapping of intraspecific differences in gene content, as low repeat content and high-quality genome assemblies facilitate structural variant discovery (Guan and Sung 2016). Furthermore, some of the best-documented examples of adaptive evolution through gene loss or gene gain have been identified in fungal pathogen–host plant interactions (Fouché et al. 2018). During host infection, fungal pathogens secrete proteins (i.e., effectors) that may be recognized by the host in a gene-for-gene relationship and trigger plant defense responses (Jones and Dangl 2006). Losses or gains of genes encoding proteins interacting with the host defense system have a strong impact on fitness (Presti et al. 2015). In agricultural ecosystems, the loss of entire effector genes, leading to the emergence of virulence, has been documented in several crop pathogens (Orbach et al. 2000; Schürch et al. 2004; Gout et al. 2006, 2007; Hartmann et al. 2017). The acquisition of new effector genes through horizontal gene transfer or duplication events is also associated with the emergence of virulence (Friesen et al. 2006; Khang et al. 2008; Jonge et al. 2012). The widespread use of new resistant host genotypes carrying specific resistance genes probably imposes a strong selective pressure on fungal pathogen populations in agricultural crops, promoting loss of the cognate effector gene or the retention of recently gained genes with potentially beneficial effects in terms of virulence, according to the “arms race” model of evolution (Brown and Tellier 2011). Comparisons of multiple genomes within agricultural pathogen species have shown that gene content variation can affect hundreds of genes, including a high proportion of effector genes and genes located in rapidly evolving genomic regions, in particular regions rich in repetitive elements (Gout et al. 2006; Yoshida et al. 2009, 2016; Plissonneau et al. 2016; Hartmann and Croll 2017; Plissonneau et al. 2018).
Despite its importance for coevolution, we know very little about the extent of gene presence–absence polymorphism in fungal plant pathogens in natural ecosystems. Several factors may drive specific dynamics of gene content change in natural populations. For example, wild plant parasites undergo frequent population extinction–recolonization events, with major consequences for infection success and population genetic structure (Tack and Laine 2014). High environmental heterogeneity and seed or spore banks also affect pathogen population structure and the process of coevolution in natural conditions (Laine and Tellier 2008; Koopmann et al. 2017). Host ecotype and environmental factors also have a strong impact on local pathogen populations, driving local adaptation (Laine et al. 2014; Stam et al. 2017). In natural conditions, the coevolutionary dynamics of plant–parasite interactions appear to follow essentially a “trench warfare” model, favoring balanced polymorphism at loci involved in host–parasite coevolution (Brown and Tellier 2011). In addition, the virulence of pathogen populations in wild pathosystems is usually a quantitative trait (Alexander and Antonovics 1995; Thrall et al. 2001; Koskela et al. 2002; Laine et al. 2011), suggesting that presence–absence variations of effector gene may be a less frequent mechanism of coevolution than in crops, in which virulence is more often a binary trait, with resistant varieties completely impeding infection (Möller and Stukenbrock 2017).
Microbotryum lychnidis-dioicae (MvSl) and M. silenes-dioicae (MvSd) are two closely related species of castrating anther-smut fungi. These highly specialized plant pathogens parasitize the white campion Silene latifolia and the red campion S. dioica, respectively (Le Gac et al. 2007; Refrégier et al. 2008). Virulence is mostly a quantitative trait, and no gene-for-gene relationship has been identified in anther-smut fungi (Alexander et al. 1993; Alexander and Antonovics 1995; Biere and Antonovics 1996; Chung et al. 2012). Populations of MvSl are genetically more diverse than those of MvSd, and form distinct genetic clusters, corresponding to the footprints of southern glacial refugia. By contrast, MvSd displays little population differentiation across Europe (Vercken et al. 2010; Badouin et al. 2017). The strong costructure between the anther-smut pathogen MvSl and its host, together with cross-inoculation experiments, are consistent with plant local adaptation (Kaltz et al. 1999; Feurtey et al. 2016). MvSl has undergone numerous selective sweeps, consistent with rapid coevolution, affecting almost 17% of the genome, whereas only 2% of the genome is affected in MvSd (Badouin et al. 2017). These two fungal species have total haploid genome size of ∼30 Mb; hyphae in plants are dikaryotic and teliospores are diploid. Both species have dimorphic mating-type chromosomes containing large regions without recombination, but with extensive rearrangements and gene losses due to permanent heterozygosity (Fontanillas et al. 2014; Badouin 2015; Branco et al. 2017). By contrast, the autosomes display low levels of heterozygosity, due to high rates of selfing and the rarity of outcrossing events (Giraud et al. 2008; Vercken et al. 2010; Badouin et al. 2017).
We investigated homozygous gene presence–absence polymorphisms on autosomes of the two sister anther-smut species MvSl and MvSd, using available genome sequence data and high-quality reference genome assemblies. The genes displaying presence–absence polymorphism were mostly recently acquired, in a single species, through duplications in multiple-gene families and their absence alleles segregated at low frequencies. The phylogeographic structure of presence–absence polymorphism in MvSl corresponded to the previously identified genetic clusters based on SNPs. Altogether, these findings suggest that most gene presence–absence polymorphism is neutral. Nevertheless, the putative functions of some genes affected by presence–absence polymorphism (e.g., secreted proteins) or their localization within previously identified selective sweeps suggest that some gene loss or gain events may be adaptive. However, functional validation in future studies will be needed to test adaptive roles in plant and anther-smut fungi interactions.
Materials and Methods
Genome Data Used for Gene Presence–Absence Polymorphism Calling
We analyzed the genome sequences of 39 MvSl isolates and 19 MvSd isolates collected from across Western Europe, previously obtained with Illumina paired-end sequencing technology, with a mean coverage of 100× (Whittle et al. 2015; Badouin et al. 2017). In total, 14 strains were sequenced in these previous studies as haploids of the a1 or a2 mating type, whereas the remaining 47 strains were sequenced as diploids. We downloaded raw data publicly available from the NCBI Short Read Archive (SRA) under the BioProject IDs PRJNA295022 and PRJNA269361 (see summary in supplementary table S1, Supplementary Material online).
We studied gene presence–absence polymorphism in MvSl and MvSd, using the high-quality reference genomes of the MvSl-1064 strain and the MvSd-1303 strain, previously obtained and annotated for gene models (see summary in supplementary table S2, Supplementary Material online; Branco et al. 2017). We focused on autosomes, which are highly homozygous in these species (Badouin 2015). We therefore used only one of the two available haploid genomes for each strain for further analyses (the haploid genomes of the a1 mating type). We used the MvSl-1064-A1-R4 assembly from the European Nucleotide Archive (ENA), accession number ERS1013679, and the MvSd-1303-D assembly, accession number ERS1436592 (Branco et al. 2017).
Read Mapping
We mapped Illumina reads against the reference genomes of each species. We trimmed Illumina raw reads of the MvSl and MvSd genomes for sequence quality and removed adapter sequences with the Cutadapt v1.12 software (Martin 2011). We used the options: -q 10, 10; –minimum-length 50; -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC; -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATT. We aligned MvSl trimmed reads against the MvSl-1064-A1-R4 genome and MvSd trimmed reads against the MvSd-1303-D a1 genome. We used the short-read aligner bowtie2 v2.1.0 (Langmead et al. 2009) for the mapping of trimmed reads with the following software options: –very-sensitive-local –phred33 -X 1000. We removed PCR duplicates with the MarkDuplicates tool of Picard tools version 1.88 (http://broadinstitute.github.io/picard). We improved alignment accuracy in indel regions, by locally realigning the mapped reads with the RealignerTargetCreator and IndelRealigner tools of the Genome Analysis Toolkit (GATK) version 3.7 (McKenna et al. 2010). Mean genome-wide mapping coverage of MvSl strains ranged from 54× to 350× and 72× to 156× for the MvSl-1064-A1-R4 and MvSd-1303-D reference genomes, respectively. As differences in coverage between samples may lead to a bias in the specificity and sensitivity of copy number variation detection (Guan and Sung 2016), we normalized the coverage of all isolates to a single value (30×), by sampling reads at random from alignment files with the samtools v1.3.1 software (Li et al. 2009).
Gene Presence–Absence Polymorphism Calling
We used a combination of read depth-based and split read-based methods to call homozygous missing fragments in the genomes analyzed, relative to the reference genomes of the MvSl-1064 and MvSd-1303 strains. Mating-type chromosomes contain an extensive region without recombination that is permanently sheltered in a heterozygous state, which led to genomic decay and in particular the loss of hundreds of genes (Fontanillas et al. 2014; Badouin 2015). We therefore focused here exclusively on autosomes. We first used the algorithm implemented in CNVnator v0.3, which uses all mapped reads and performs a statistical analysis of read coverage in bins along the genome sequence (Abyzov et al. 2011). We set the bin size at 100 bp and retained only deletion calls, that is, missing fragments, fulfilling the following criteria: P value < 0.05, length > 500 bp, q0 < 0.4 and normalized read coverage < 0.3. As a second method, we used the Pindel v0.5.7 software, which uses information about paired reads for which only one end can be mapped. Based on the anchor point of the mapped read, the insert size and the direction of the unmapped read, Pindel predicts missing fragments by breaking the unmapped read into fragments and mapping them separately (Ye et al. 2009). We converted .bam alignment files into Pindel input format with the sam2pindel tool of Pindel. We ran Pindel with default options and used the Pindel pindel2vcf tool to convert output files into variant calling format. We retained only homozygous deletion calls of >500 bp, supported by at least 15 reads (allele depth >15). The calling of missing fragments is more challenging in short contigs, due to frequent poor-quality assembly and high repeat content. We therefore restricted our analyses to the largest contigs (minimum size of 147 kb), which summed covered 90% of the length of the autosomal genome of the MvSl-1064 and MvSd-1303 strains. To detect genes present in the reference genomes that were absent from the Illumina genomes, we retained the missing fragments called with each method that covered at least one gene, over >90% of its length. We assessed the overlaps between missing fragments and gene models masked for repeats, with the bedtools “intersect” command (Quinlan and Hall 2010). We considered gene absence events identified by both the read depth-based method and the split read-based method as single gene absence events. We determined the total length of the genomic region affected by missing fragments for each strain by summing the lengths of missing fragments detected by the two methods. To avoid counting twice the lengths of overlapping missing fragments detected by the two methods, we took into account the reunion of overlapping missing fragments.
Quality Control of Detected Gene Presence–Absence Polymorphism
To assess the overall quality of gene presence–absence polymorphism calls, we performed several quality control steps. First, we checked the gene absence events called with our pipeline using Illumina sequencing of the very same strain (Illumina-resequenced MvSl-1064 strain; Badouin et al. 2017) against the reference high-quality genome of the MvSl-1064 strain (Branco et al. 2017). Second, we used a comparison of high-quality genome assemblies of two MvSl strains, MvSl-1064 (used as a reference for gene presence–absence calling) and a newly sequenced strain, MvSl-1318 (see section “Analyses of strain-specific genes using de novo genome assemblies” below). We compared the number of gene absence events called with our pipeline by using the Illumina-resequenced MvSl-1064 strain (Badouin et al. 2017) against the MvSl-1318 reference genome with the number of genes found to be specific to the MvSl-1318 reference genome (i.e., lacking from the MvSl-1064 reference genome) in the comparison of the two high-quality genome assemblies. We studied global synteny between genome assemblies using the nucmer command from MUMmer package v3.1 (Kurtz et al. 2004). To identify gene sequences of the MvSl-1318 strain that were absent in the MvSl-1064 strain, we performed BLASTn analyses in the genome of the MvSl-1064 strain using the sequences of predicted genes of the MvSl-1318 strain as a query. Genes were considered as absent if no BLAST hit was found on the orthologous contig with a minimum identity of 90%, a bit score value of 100 and a length of at least 90% of the gene length. We used the Integrative Genomics Viewer (IGV) browser to visually inspect read mapping in genomic regions with detected gene absence events (Robinson et al. 2011; Thorvaldsdóttir et al. 2013). Finally, we validated the presence–absence polymorphism inferred in silico for two genes using polymerase chain reactions (PCR) on DNAs from the same strains whose genomes were studied. Primers were designed in the gene coding sequences using Primer 3.0 (Rozen and Skaletsky 2000). For the gene MvSl-1064-A1-R4_MC01-1g01717 in the MvSl-1064 reference genome, we performed PCR in nine MvSl strains using 5′ GATGTCGATGCGCTCTTTGT 3′ and 3′ CGTCATCAGTGTGCCCTTTT 5′ as forward and reverse primers, respectively. For the gene MvSdioicae_1303_FR02_D_N206_PbcR_C014g07422 in the MvSd-1303 reference genome, we performed PCR in nine MvSd strains using 5′ GACATCAGGCACCACTCACA 3′ and 3′ ATCCACCCGTCAAATTCGCA 5′ as forward and reverse primers, respectively. PCR reactions were conducted in a 30 µl volume containing 5–10 ng genomic DNA, 0.4 mM each of forward and backward primers, 0.25 mM dNTP, 0.75 U Taq polymerase (DreamTaq, Thermo Fisher, Inc.), PCR buffer. PCR products were amplified for 30 cycles. The resulting amplicons were examined on 1.5% agarose gels. We also tested for significant differences in gene absence calls between haploid and diploid strains within MvSl and MvSd populations.
Gene Ontology Analysis and Functional Annotation
We assigned genes to functional categories using InterproScan v5.24-63.0 (Zdobnov and Apweiler 2001) that provided information for protein family (Pfam), gene ontology (GO), and pathways. We used SignalP v4.1 (Petersen et al. 2011) and TMHMM v2.0 (Krogh et al. 2001) to predict putative secreted proteins. We defined secreted proteins as proteins with a predicted signal peptide but no predicted transmembrane helices. Putative small secreted proteins were defined as secreted proteins with a size inferior to 300 amino acids and unknown functional domain. We filtered out from the set of genes affected by presence–absence polymorphism all genes with a transposable element activity related domain, that is, reverse transcriptase (PF00078, PF07727), integrase (PF00665, PF13976), Helitron helicase-like domain (PF14214), and GAG-polypeptide of long terminal repeat (LTR) copia-type (PF14223). GO enrichment analysis of genes affected by presence–absence polymorphism was performed with the hypergeometric test implemented in the R package GOstats. GO enrichment P values were adjusted for multiple testing with the Bonferroni correction and Benjamini and Hochberg (1995) correction using the p.adjust() R function.
Annotation of Repetitive Elements
Annotation of repetitive elements was performed using a set of repeat sequences (available upon request) detected de novo in a large set of 41 genomes of the Microbotryum genus. Briefly, repeat motifs were detected using a combination of two tools, LTRharvest (Ellinghaus et al. 2008) that performs high-quality annotation of LTR retrotransposons and RepeatModeler (Smit and Hubley 2008) that uses results from three other programs, RECON, RepeatScout, and Tandem Repeats Finder. Detected repeat motifs were clustered based on sequence similarities (min. 90% of similarity) and the best representative sequences per cluster, that is, the centroid sequences, were kept. A set of 3599 repeat sequences was built after removing redundant sequences among centroid sequences, and keeping only transposable element sequences present in at least seven genomes. We annotated genome assemblies using the set of 3599 repeat sequences as library in RepeatMasker v4.0.3 (Smit et al. 2013). We masked gene models for transposable elements in all the genomes. Telomeric and centromeric repeats were identified by performing a BLASTn search of telomeric and centromeric motifs previously identified in the MvSl-1064 genome (Badouin 2015). Subtelomeres were defined as regions within 100 kb of the contig extremities containing telomeric repeats. Centromeres were defined as regions within 100 kb of the contig extremities containing centromeric repeats.
Expression Data
To assess expression levels of genes affected by presence–absence polymorphisms, we used RNAseq data collected using the MvSl-1064 strain (i.e., the same strain as the MvSl reference genome) both using in vitro cultures and in vivo late infection (Perlin et al. 2015). We trimmed raw reads using cutadapt v1.12 (Martin 2011) and mapped trimmed reads to the MvSl-1064 genome using TopHat v2.1.1 (Trapnell et al. 2009). We used the featureCounts program from the Subread v1.5.3 package (Liao et al. 2014) to calculate absolute transcript abundance (i.e., number of mapped reads for each transcript). For each tested condition, we computed reads per kilobase of transcript per million mapped reads (RPKM) values using a customized R script.
Comparative Genomics Analyses
For comparative genomics analyses, we used three additional genomes from previously sequenced Microbotryum species (Branco et al. 2017): M. violaceum s. str. collected on S. nutans (MvSn ERS1013671), M. lagerheimii collected on S. vulgaris (MvSv ERS1013677), and M. intermedium collected on Salvia pratensis (Mint ERS1324257). To investigate gene orthology relationships between species, we compared the predicted protein sequences of the genome assemblies of MvSl-1064, MvSd-1303, MvSn, MvSv, and Mint, with BLASTp+ v2.30. The output was processed in orthAgogue (Ekseth et al. 2014) that uses Markov clustering to obtain orthologous groups. To study the synteny between the MvSl-1064 and MvSd-1303 genomes, we used the nucmer command from MUMmer package v3.1 (Kurtz et al. 2004) and performed BLASTn+ v2.30 analysis on repeat masked genomic sequences. BLAST hits were stringently filtered for a minimum alignment length of 500 bp, a minimum identity of 80%, and a bit score value of 80.
Population Structure and Statistical Analyses
Principal component analyses were computed in the R package {ADEGENET} (dudi.pca function; Jombart 2008; Jombart and Ahmed 2011). Neighbor-joining trees were performed with the R package {Ape} (Paradis et al. 2004). These analyses were performed based on unique events of gene presence–absence polymorphism, that is, using only a single gene per missing fragments, to be conservative regarding the number of gene gain or loss events. To compare the population structure based on gene presence–absence polymorphism with the population structure based on SNPs, we called genome-wide SNPs for MvSl strains against the MvSl-1064-A1-R4 genome and for MvSd strains against the MvSd-1303-D a1 genome. For SNP calling, we used GATK version 3.7 (McKenna et al. 2010). We ran HaplotypeCaller on each strain individually using a diploid mode. We performed joint variant calls using GenotypeGVCFs on a merged gvcf variant file. SNP call filtering for quality was performed using VariantFiltration and following the GATK Good Practices for hard-filtering of variants (QUAL < 250; QD < 2; MQ < 30.0; −10.5 > MQRankSum > 10.5; −5 > ReadPosRankSum > 5; FS > 60; SOR > 3). We kept only biallelic SNPs with a high genotyping rate (> 90%) on autosomes. In total, we detected 216, 878 biallelic SNPs in MvSl and 33, 694 biallelic SNPs in MvSd. The variant call format files are available upon request. Pairwise FST were calculated for SNPs and gene presence–absence polymorphisms between genetic clusters using the R package {hierfstats} (Goudet 2005) implementing Yang’s algorithm (Yang 1998). To test the significance of the FST value obtained between the MvSd populations that we found differentiated based on gene presence–absence polymorphism, we performed a permutation test with 1000 bootstraps, assigning randomly cluster identity to each strain, keeping the cluster sizes constant, and computing mean FST values for each permutation. We performed Kruskal–Wallis rank sum test and Pearson’s chi-squared test in the R software v3.3.3. Plots were performed using the software Circos v0.67-7 (Krzywinski et al. 2009) and the R package {ggplot2} (Wickham 2009).
Analyses of Strain-Specific Genes Using De Novo Genome Assemblies
For identifying genes absent from the reference genome but present in the genomes of other MvSl strains, we performed de novo genome assemblies. As MvSl showed a population structure with four genetic clusters (Badouin et al. 2017), we performed de novo assemblies for three MvSl strains belonging to the genetic clusters different from that of the reference MvSl-1064 genome. We assembled the Illumina genomes of the MvSl-1005 and the MvSl-140-01 strains that belonged to the MvSl Northwestern and MvSl Southwester genetic clusters, respectively (Badouin et al. 2017), and that showed high read depth (233× and 89×, respectively). The assemblies of the MvSl-1005 and the MvSl-140-01 Illumina genomes were performed in SpAdes v3.11.0 (Bankevich et al. 2012) with the following options: –only-assembler –careful -cov-cutoff auto using the whole set of trimmed Illumina reads of each genome. KmerGenie (Chikhi and Medvedev 2014) was used to estimate the best k-mer length for genome de novo assembly. The best predicted k values were 61 and 87 for the MvSl-1005 genome and the MvSl-140-01 genome, respectively. These assemblies had lower quality than the MvSl-1064 genome assembly (see summary in supplementary table S2, Supplementary Material online) and they are available upon request. In order to obtain a high-quality genome assembly of a second MvSl strain, we sequenced separately the two haploid genomes (corresponding to the a1 and a2 mating types) of the diploid MvSl-1318 strain (collected on S. latifolia, Olomouc, Czech Republic, Coord. GPS: 49.569172 and 17.287057, previously shown to belong to the MvSl Eastern genetic cluster), with P6/C4 Pacific Biosciences SMRT technology. The sequencing and assembly of the MvSl-1318 genomes were generated as described in (Branco et al. 2017). Briefly, we assembled the genome using the wgs-8.2 version of the PBcR assembler and polished the assembly with the quiver software (https://github.com/PacificBiosciences/GenomicConsensus). The gene models were predicted with EuGene using a training data set as described in (Branco et al. 2017). The assembly of the haploid genome of a1 mating type (MvSl-1318-D) and the assembly of the haploid genome of a2 mating type (MvSl-1318-T) are available at GenBank under the BioProject accession number PRJNA437556 (BioSample IDs SAMN08667584 and SAMN08667587, respectively). The raw reads are available at NCBI SRA under the BioProject accession number PRJNA437556 (BioSample IDs SRR6825078 and SRR6825079 for MvSl-1318-D and MvSl-1318-T, respectively). We only used the a1 haploid genome for our analyses. Gene models were masked for repeats.
To detect strain-specific genes in the assembled genomes of the MvSl-1318, MvSl-140-01, and MvSl-1005 strains, we applied the same procedure as the one used to detect genes present in the reference genomes and absent from the Illumina genomes, for consistency. We mapped Illumina sequencing data of the MvSl-1064 strain (Badouin et al. 2017) against the three genome assemblies and called gene presence–absence polymorphism with our gene presence–absence detection pipeline. We restricted our analyses to the largest contigs, which covered 90% of the autosomal genome length in the three genomes (minimum size of 317,619, 11,561, and 11,985 bp in the MvSl-1318 strain, MvSl-140-01 strain, and MvSl-1005 strain, respectively). We identified autosomal contigs in the three genome assemblies by global synteny analyses with the genome assemblies of the MvSl-1064 strain using the nucmer command from MUMmer package (Kurtz et al. 2004). As the MvSl-140-01 and MvSl-1005 strains were sequenced as diploid, we excluded any contigs with large synteny with the mating-type chromosomes a1 and a2. The MvSl-1064-A2-R4 assembly (the haploid genomes of a2 mating type) was accessed from ENA at accession number ERS459551. To analyze population structure of MvSl-1318-specific genes, we called gene presence–absence polymorphism with our pipeline in all MvSl strains against the MvSl-1318 assembly and selected missing fragments affecting the MvSl-1318-specific genes. Mean genome-wide mapping coverage of MvSl strains ranged from 53× to 345× on the MvSl-1318-D reference genome.
Results
Gene Presence–Absence Event Calling and Quality Control
We called homozygous gene presence–absence polymorphisms by comparing the available high-coverage Illumina genomes of 38 MvSl strains and 19 MvSd strains sampled across Europe (supplementary table S1, Supplementary Material online; Whittle et al. 2015; Badouin et al. 2017) with high-quality reference genomes for these two species: MvSl-1064 and MvSd-1303 (supplementary table S2, Supplementary Material online; Badouin 2015; Branco et al. 2017). Read mapping identified genes present in the reference genomes that were absent from the Illumina genomes. We called missing fragments of >500 bp by two different methods, to increase the sensitivity of copy number variation calls. The read depth-based method involved scanning the genome for local variation in read depth and calls missing fragments in genomic regions with low read-depth values (Abyzov et al. 2011). The split read-based method is based on the identification of read pairs for which only one read is mapped onto the genome (supplementary fig. S1, Supplementary Material online; Ye et al. 2009; Lin et al. 2015). We focused on missing fragments affecting >90% of gene lengths on autosomes, after masking gene models for repeated elements. Merging the missing fragments detected by the two methods resulted in the calling of 810 missing fragments in MvSl and 99 in MvSd. Most missing fragments were small (median length of 2.0 kb in MvSl and 3.1 kb in MvSd; supplementary fig. S2A, Supplementary Material online). In both MvSl and MvSd, there was little overlap between methods concerning the missing fragments detected (18.7% and 12.5%, respectively). This was expected, as the two methods were based on different read mapping approaches. The overlap between missing fragments detected by the two methods covered a mean of 68 kb per individual in MvSl, and 19 kb in MvSd (supplementary fig. S2B, Supplementary Material online). Most missing fragments affected a single gene (supplementary fig. S2C, Supplementary Material online). In MvSl, 473 missing fragments affected a single gene and 38 affected groups of at least five genes. In MvSd, 48 missing fragments affected a single gene and one affected a group of five genes. In total, we detected 953 gene absence events in MvSl, and 161 in MvSd. The number of missing genes detected per strain ranged from 2 to 50, with a median value of 24 genes in MvSl, and 1 to 15 with a median value of eight genes in MvSd (supplementary fig. S2D, Supplementary Material online).
Several quality control checks indicated that the inferences for the missing fragments were reliable. First, no missing fragments were detected following genome resequencing of the MvSl-1064 strain (Badouin et al. 2017), the source of the reference genome sequence. Second, we found a large overlap between the missing fragments detected with our pipelines and those detected by comparing high-quality genome assemblies for two MvSl strains. Out of 15 genes detected to be missing in one strain with our pipeline, four genes were detected present using BLAST analyses on de novo assemblies (supplementary text S1 and supplementary fig. S3, Supplementary Material online); however, BLAST analyses can match paralogs. The visual inspection of read mapping in genomic regions with detected gene absence events using a genome browser suggested a rate of false positive of ∼15% for all analyzed genomes. Finally, PCR tests validated in silico inferences for two genes affected by presence–absence polymorphism. The gene MvSl-1064-A1-R4_MC01-1g01717 could not be amplified in five MvSl strains that were predicted to lack the gene in silico. The gene MvSdioicae_1303_FR02_D_N206_PbcR_C014g07422 could not be amplified in three MvSd strains that were predicted to lack the gene in silico.
Contrasting Gene Presence–Absence Polymorphism Patterns between Species
We found that different genes and different numbers of genes displayed presence–absence polymorphism in MvSl and MvSd. We identified 186 autosomal genes displaying presence–absence polymorphism in the set of 38 MvSl strains studied, corresponding to 2% of the total gene content of MvSl-1064 autosomes (fig. 1, right panel; supplementary table S3, Supplementary Material online). We identified 51 autosomal genes displaying presence–absence polymorphism in the set of 19 MvSd strains, corresponding to 0.6% of the total gene content of MvSd-1303 autosomes (fig. 1, left panel; supplementary table S3, Supplementary Material online). Fewer genes displayed presence–absence polymorphism in MvSd than in MvSl, consistent with the lower SNP and microsatellite diversity previously reported in MvSd (Vercken et al. 2010; Badouin et al. 2017). In agreement with this diversity difference, we identified 216, 878 biallelic SNPs in MvSl and 33, 694 biallelic SNPs in MvSd by mapping MvSl and MvSd Illumina genomes on their respective reference genome. Gene absence alleles segregated at low frequency in both species, with a median frequency of 5% in MvSl and 10% in MvSd (fig. 1E;supplementary table S3, Supplementary Material online).
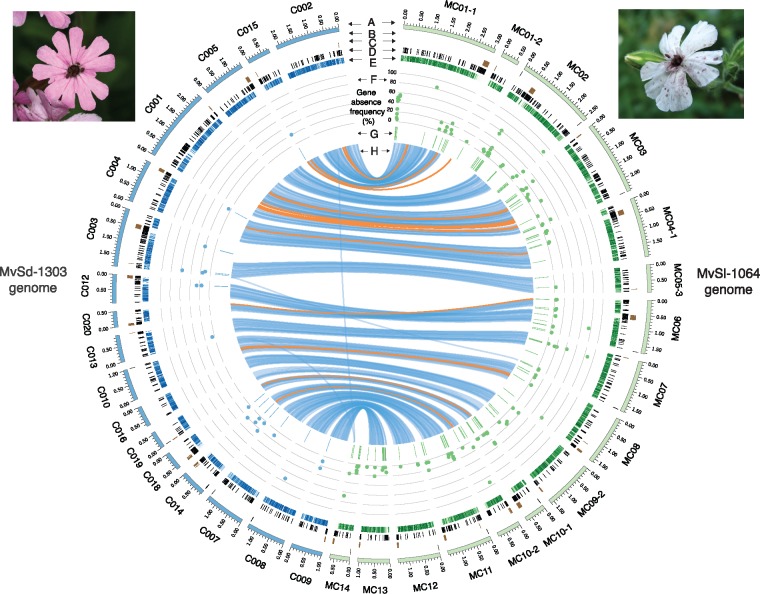
Fig. 1.
—Genome-wide location of gene presence–absence polymorphism events and their frequencies in Microbotryum lychnidis-dioicae (MvSl) and Microbotryum silenes-dioicae (MvSd) strains. The tracks represent the following features: (A) Contigs > 500 kb in length from the MvSd-1303 (left, blue tracks) and MvSl-1064 (right, green tracks) reference genomes; (B) Location of subtelomeric repeats; (C) Location of centromeric repeats; (D) Location of transposable elements; (E) Gene density in 10-kb nonoverlapping windows; the color gradient shows gene density differences from 0% (light) to 100% (dark); (F) Location of genes displaying presence–absence polymorphism (x axis) and frequency of the gene absence allele (y axis, from 0% to 100%) in MvSd (left, blue dots) and MvSl strains (right, green dots); (G) Location of fragments identified as missing in MvSd (left, blue track) and MvSl strains (right, green track); (H) Links representing collinearity of genomic regions >10 kb between the MvSl-1064 and MvSd-1303 reference genomes, with the orange links corresponding to inversions. The images show spores from MvSd in the anthers of Silene dioica (left) and spores from MvSl in the anthers of S. latifolia (right).
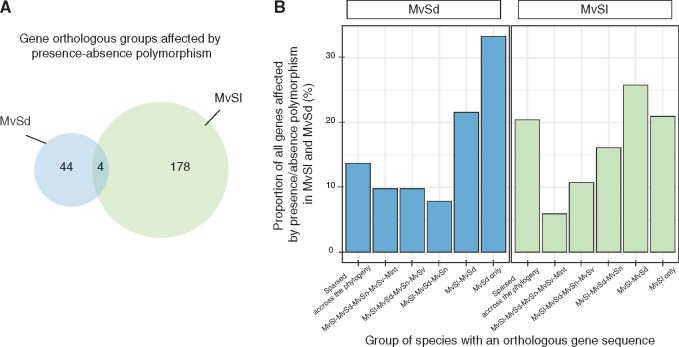
We searched for orthology relationships between the genes identified as displaying presence–absence polymorphism on MvSl-1064 and MvSd-1303 autosomes. The two closely related species shared 7, 869 groups of orthologous autosomal genes, containing 83.3% and 78.2% of the total gene content of MvSl-1064 and MvSd-1303 autosomes, respectively. In total, 59% of the genes displaying presence–absence polymorphism in MvSl and 49% of those in MvSd belonged to orthologous groups present in both species (fig. 2B). However, only four orthologous groups were affected by presence–absence polymorphism in both species (fig. 2A). Gene presence–absence polymorphism affected different genomic regions in the two species (fig. 1G).
Fig. 2.
—Evolutionary history of genes displaying presence–absence polymorphism. (A) Venn diagram of shared and specific orthologous gene group numbers displaying presence–absence polymorphism in Microbotryum lychnidis-dioicae (MvSl) and Microbotryum silenes-dioicae (MvSd). (B) Groups of Microbotryum species with orthologous gene sequences, for the genes displaying presence–absence polymorphism in MvSl and MvSd. We searched for orthologous groups in the sister species MvSl and MvSd and three closely related Microbotryum species: M. violaceum s. str. (MvSn), M. lagerheimii (MvSv), and M. intermedium (Mint).
Functional Characteristics, Evolutionary History, and Genomic Environment of the Genes Displaying Presence–Absence Polymorphism
Most of the genes displaying presence–absence polymorphism encoded proteins of unknown function, to a greater extent than genome average. Indeed, a total of 86% and 88% of the genes displaying presence–absence polymorphism encoded proteins of unknown function compared to 44% and 45% for all autosomal genes in MvSl and MvSd, respectively. Based on the available expression data for MvSl-1064 in three types of conditions, poor and rich in vitro medium and in planta (Perlin et al, 2015), we showed that the genes displaying presence–absence polymorphism in MvSl had lower levels of expression than other genes (comparisons of RPKM values; Kruskal–Wallis rank sum tests P value < 2.2e-16 in all conditions tested). The genes displaying presence–absence polymorphism in MvSl included a high proportion of genes not expressed in planta (RPKM < 1; 37%, vs 10% for other genes; one-tailed Fisher’s exact test P value < 2.2e-16). In both species, the genes displaying presence–absence polymorphism were also significantly shorter than other genes (difference in median gene length in MvSl strains= 928 bp; Kruskal–Wallis rank sum test P value < 2.2e-16; difference in median gene length in MvSd strains = 852 bp; Kruskal–Wallis rank sum test P value = 1.5e-07). Finally, the genes displaying presence–absence polymorphism included a higher proportion of genes from multiple-copy gene families (24% and 25% of all genes displaying presence–absence polymorphism in MvSl and MvSd, respectively) than other genes (7% and 9% in MvSl and MvSd, respectively).
The genes displaying presence–absence polymorphism and encoding proteins with predicted functions included a large proportion of genes associated with biological processes relating to DNA repair mechanisms and the cellular response to stress in MvSl and to glycoprotein metabolism and glycosylation in MvSd. Corresponding gene ontology terms were not significantly enriched after Bonferroni multiple testing correction, but were significantly enriched after Benjamini and Hochberg multiple testing correction (supplementary table S4, Supplementary Material online). We found no enrichment in any specific cellular compartment. In particular, genes encoding secreted proteins, which frequently play a major role in the interactions of fungal pathogens with their hosts (Fouché et al. 2018), were not overrepresented among the genes displaying presence–absence polymorphism. Three genes encoding small secreted proteins were nevertheless absent from some MvSl and MvSd strains. In MvSl, genes encoding a secreted peptidase and a histidine phosphatase, both potentially involved in pathogenicity (Monod et al. 2002; Albataineh and Kadosh 2016; Vincent et al. 2016), also displayed presence–absence polymorphism (supplementary table S3, Supplementary Material online). Five genes encoding proteins involved in RNA interference activity (Piwi domain, Argonaute domain) were absent in one MvSd strain (supplementary table S3, Supplementary Material online). Some plant pathogens use the RNA interference machinery to prevent the expression of host immunity genes (Weiberg and Jin 2015).
We investigated the evolutionary history of genes displaying presence–absence polymorphism by searching for orthologs in three Microbotryum species closely related to the sister species MvSl and MvSd, using the available genomes of M. violaceum s. str. (MvSn), M. lagerheimii (MvSv), and M. intermedium (Mint) (Branco et al. 2017). Most of the genes displaying presence–absence polymorphism (46.7% in MvSl and 54.8% in MvSd, fig. 2B) were recently acquired, that is, were present only in one or both of the sister species MvSl and MvSd and not in the other Microbotryum species. Only 5.9% of the genes displaying presence–absence polymorphism in MvSl and 9.8% of those displaying presence–absence polymorphism in MvSd were present in all five Microbotryum species. This finding indicates that presence–absence polymorphism is rare in ancient genes, that were present in the common ancestor of the five Microbotryum species. Most of the genes displaying presence–absence polymorphism in MvSl and MvSd instead corresponded to species-specific genes, that is, genes recently gained in a single species, after their divergence from other species (fig. 2B).
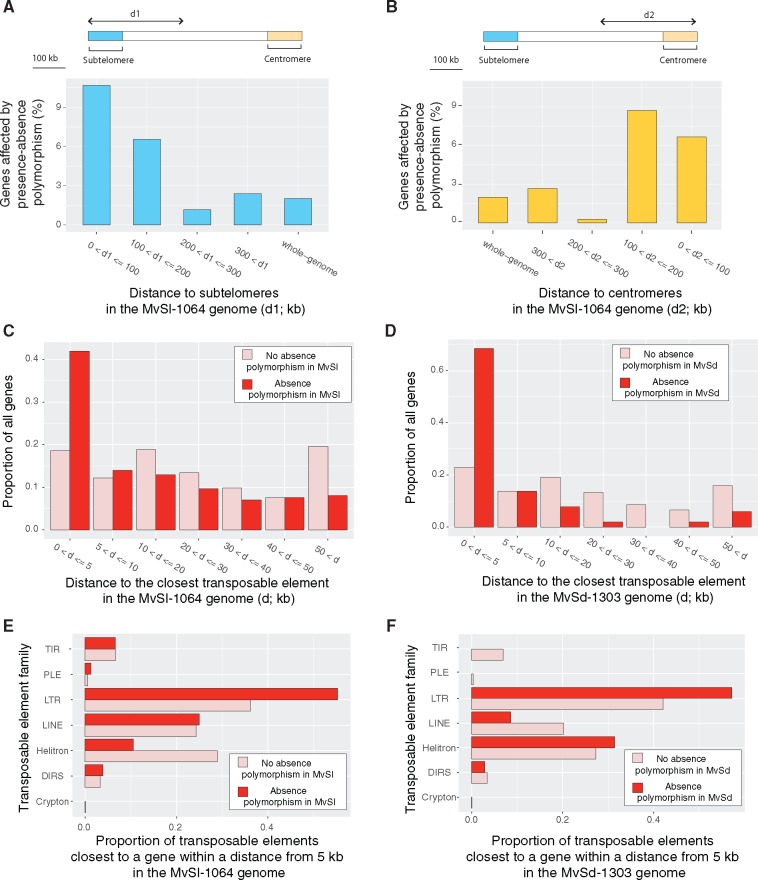
The genes displaying presence–absence polymorphism in MvSl were found in subtelomeric regions (defined as regions within 100 kb of the contig extremities and containing telomeric repeats) more frequently than would be expected by chance (fig. 3A). A total of 10.7% of genes in subtelomeric regions were affected by presence–absence polymorphism compared to 2% in the whole genome. We checked by visual inspection that the gene absence events called in subtelomeric regions were not artifacts due to low mapping quality. The small number of subtelomeric repeats in the MvSd-1303 genome precluded testing for an enrichment in gene presence–absence polymorphism in subtelomeric regions in MvSd, but gene absence events were frequently found close to the ends of the contigs (i.e., within 100 kb of the contig extremities). The genes displaying presence–absence polymorphism in MvSl were also found closer to centromeric regions (defined as regions within 100 kb of the contig extremities and containing centromeric repeats) than would be expected by chance (fig. 3B). The genes displaying presence–absence polymorphism were also significantly closer to transposable elements than other genes. In MvSl, 42% of the genes displaying presence–absence polymorphism were located within 5 kb of the nearest transposable element, against only 19% of other genes (fig. 3C). In MvSd, 69% of genes displaying presence–absence polymorphism were located within 5 kb of the nearest transposable element, against only 23% of other genes (fig. 3D). We found significant differences in the abundances of the transposable element families closest to the genes displaying presence–absence polymorphism and those closest to other genes in MvSl, but not in MvSd (fig. 3E and F). LTR transposons, the most frequent family of repeats in the MvSl-1064 reference genome (Hood 2005; Perlin et al. 2015), were significantly more frequently found closest to genes displaying presence–absence polymorphism than to other genes (one-tailed Fisher’s exact test P value = 0.0007345 in MvSl; not significant in MvSd). Conversely, members of the Helitron family were less frequently found closest to genes displaying presence–absence polymorphism than to other genes in MvSl (one-tailed Fisher’s exact test P value = 0.0001393; not significant in MvSd).
Fig. 3.
—Genome features associated with gene presence–absence polymorphism in Microbotryum lychnidis-dioicae (MvSl) and Microbotryum silenes-dioicae (MvSd). (A and B) Proportion of genes displaying presence–absence polymorphism according to the distance to subtelomeres (A) and centromeres (B) in MvSl. Regions within 100 kb of contig extremities and containing subtelomeric repeats were considered to be subtelomeric. Regions within 100 kb of contig regions containing centromeric repeats were considered to be centromeric. Only contigs of >500 kb in length and containing subtelomeric motifs and centromeric repeats were included in the analysis (MC02, MC03, MC06, MC07, MC10-1, MC11, MC12, MC14). (C and D) Distance (kb) to the closest transposable element for genes displaying presence–absence polymorphism and genes not displaying presence–absence polymorphism in MvSl (C) and MvSd (D). Distances were classified into seven categories. (E and F) Family of the closest transposable element for genes displaying presence–absence polymorphism and genes not displaying presence–absence polymorphism in MvSl (E) and MvSd (F). Only transposable elements located <5 kb away from a gene were considered.
Population Structure of Gene Presence–Absence Events
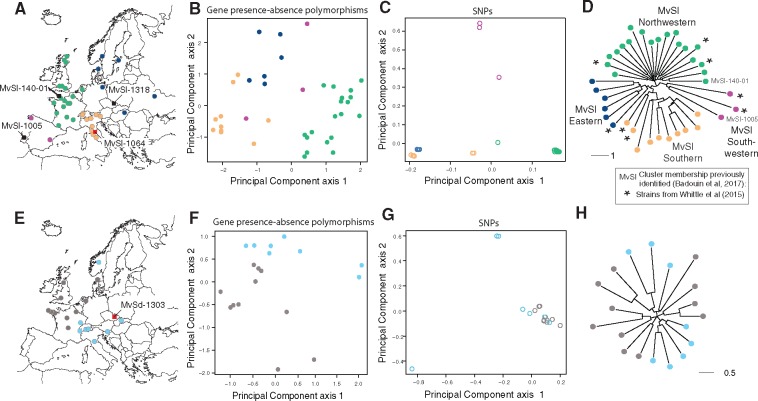
We investigated the population structure of gene presence–absence polymorphism in each species. For MvSl, we identified four genetic clusters based on the three first components of a principal component analysis and a dendrogram (fig. 4A–D). These clusters were consistent with the population subdivision previously described based on genome-wide SNPs (fig. 4C) and corresponding to the phylogeographic footprints of glacial refugia (Badouin et al. 2017). The congruence of the genetic subdivisions based on SNPs and gene presence–absence polymorphism in MvSl resulted from the large proportion of cluster-specific presence or absence among the genes affected by presence–absence polymorphisms (64%; fig. 5A). Within MvSl, we found higher mean pairwise FST for SNPs (mean value = 0.32 for pairwise comparisons) than for gene presence–absence polymorphisms (mean value = 0.15). Within MvSd, we observed low levels of population structure (fig. 4E–H), consistent with previous findings for SNPs (fig. 4G) and microsatellites (Vercken et al. 2010; Badouin et al. 2017). However, a principal component analysis of gene presence–absence polymorphism suggested here the existence of genetic differentiation between strains across one east–west longitudinal gradient (fig. 4E and F). When calculating mean pairwise FST between the eight MvSd strains sampled in Eastern Europe and the eleven MvSd strains sampled in Western Europe (fig. 4E), we found higher differentiation for gene presence–absence (mean pairwise FST value = 0.12) than for SNPs (mean pairwise FST value = 0.08). The observed mean FST value calculated between these two groups for gene presence–absence polymorphisms was higher than expected by chance, lying within the 1% extreme values in 1000 permutations.
Fig. 4.
—Population structure of gene presence–absence polymorphism in Microbotryum lychnidis-dioicae (MvSl) and Microbotryum silenes-dioicae (MvSd). (A) Geographic origin of the MvSl strains. Sampling location is indicated by circles for the MvSl strains sequenced with Illumina technology. The colors correspond to the genetic clusters identified by the dendrogram (D). The sampling locations for the reference genomes used for mapping are indicated by squares in red. The three MvSl strains that were de novo assembled (i.e., the MvSl-1318 strain originating from the MvSl Eastern cluster, the MvSl-140-1 strain originating from the MvSl Northwestern cluster, and the MvSl-1005 strain originating from the MvSl Southwestern cluster) are indicated by squares in black. (B) Principal component analysis of gene presence–absence polymorphism for MvSl, based on 124 unique gene presence–absence polymorphisms, that is, using only a single gene per missing fragment. Points correspond to strains and are colored according to the genetic clusters identified on the dendrogram (D). (C) Principal component analysis of 216, 878 genome-wide SNPs for MvSl. Points correspond to strains and are colored according to the genetic clusters identified on the dendrogram (D). (D) Neighbor-joining tree representing the genetic distance between the 38 MvSl strains on the basis of gene presence–absence polymorphism, based on 124 unique gene presence–absence polymorphisms. Points correspond to strains and are colored according to the genetic clusters identified in the dendrogram. The four genetic clusters (Northwestern, Southwestern, Eastern, and Southern) described by (Badouin et al. 2017) on the basis of single-nucleotide polymorphisms (SNPs) are indicated. The strains sequenced by (Whittle et al. 2015) are indicated by stars. The MvSl-140-1 strain originating from the MvSl Northwestern cluster and the MvSl-1005 strain originating from the MvSl Southwestern cluster that were de novo assembled are indicated. The scale bar indicates the number of differences between individuals. (E) Geographic origin of the MvSd strains. The sampling locations for MvSd strains sequenced with Illumina technology are indicated by circles. Colors indicate position on a geographic east–west gradient. The sampling location for the reference genome used for mapping is indicated by a square. (F) Principal component analysis of gene presence–absence polymorphism for MvSd, based on 35 unique gene presence–absence polymorphisms. Points correspond to strains, and are colored according to a geographic east–west gradient, as shown on the map (H). (G) Principal component analysis of genome-wide 33, 694 SNPs for MvSd. Points correspond to strains and are colored according to the genetic clusters identified on the dendrogram (D). (H) Neighbor-joining tree representing the genetic distance between the 19 MvSd strains on the basis of gene presence–absence polymorphism, based on 35 unique gene presence–absence polymorphisms. The scale bar indicates the number of differences between individuals. Points correspond to strains and are colored according to position on a geographic east–west gradient, as shown on the map.
Fig. 5.
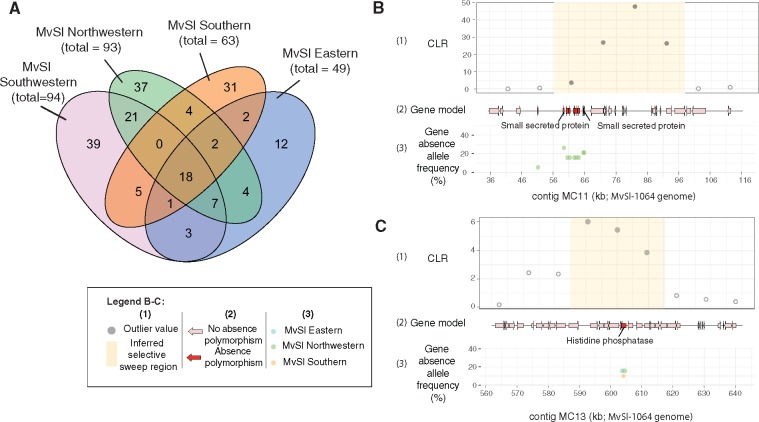
—Gene presence–absence polymorphism within individual genetic clusters in Microbotryum lychnidis-dioicae (MvSl) and localization within previously identified selective sweep regions. (A) Venn diagram for shared and unique gene presence–absence polymorphism events in the four MvSl genetic clusters. Genetic clusters were defined on the basis of dendrograms (fig. 4D). (B) Localization of a gene presence–absence polymorphism within a selective sweep previously identified by (Badouin et al. 2017) in the Northwestern genetic cluster for MvSl. The region with coordinates 36–116 kb on the MvSl-1064 MC11 contig is represented, corresponding to the MvSlA1A2r3c_S01: 2964344-3005424 selective sweep region described by (Badouin et al. 2017); (C) Localization of a gene presence–absence polymorphism within a selective sweep previously identified by (Badouin et al. 2017) in the Northwestern genetic cluster for MvSl. The region with coordinates 560–640 kb on the MvSl-1064 MC13 contig is represented, corresponding to the MvSlA1A2r3c_S11: 445169-475073 selective sweep region described by (Badouin et al. 2017). For (B) and (C), the following are shown in each panel, from top to bottom: (1) The composite likelihood ratio (CLR) from a SweeD analysis with outlier values in light blue and the inferred selective sweep region in yellow. All data were retrieved from (Badouin et al. 2017); (2) Gene models for the MvSl-1064 strain, for genes displaying presence–absence polymorphism in the Northwestern cluster, shown in red; (3) Frequency of gene absence in the Northwestern genetic cluster (green), Eastern genetic cluster (blue), and Southern genetic cluster (orange) for MvS1.
We investigated the possible presence of gene presence–absence polymorphisms in regions previously identified to have been affected by a recent selective sweep in the MvSl Northwestern genetic cluster (Badouin et al. 2017). The MvSl Northwestern genetic cluster displayed no enrichment of gene presence–absence polymorphisms in selective sweep regions (Pearson’s chi-squared test χ2 = 0.24379, P = 0.6215). Nevertheless, 21 of the 93 genes absent from at least one MvSl Northwestern strain were located within a selective sweep region, including three genes encoding secreted proteins (fig. 5B and C;supplementary table S3, Supplementary Material online). In selective sweep regions, seven gene absence were rare (frequency < 5%) and three gene absence events were almost fixed (frequency > 70%), as expected for selective sweeps (supplementary fig. S4, Supplementary Material online). Some of the gene presence–absence events, particularly those concerning genes encoding secreted proteins, may thus be adaptive, although functional studies are needed to confirm this.
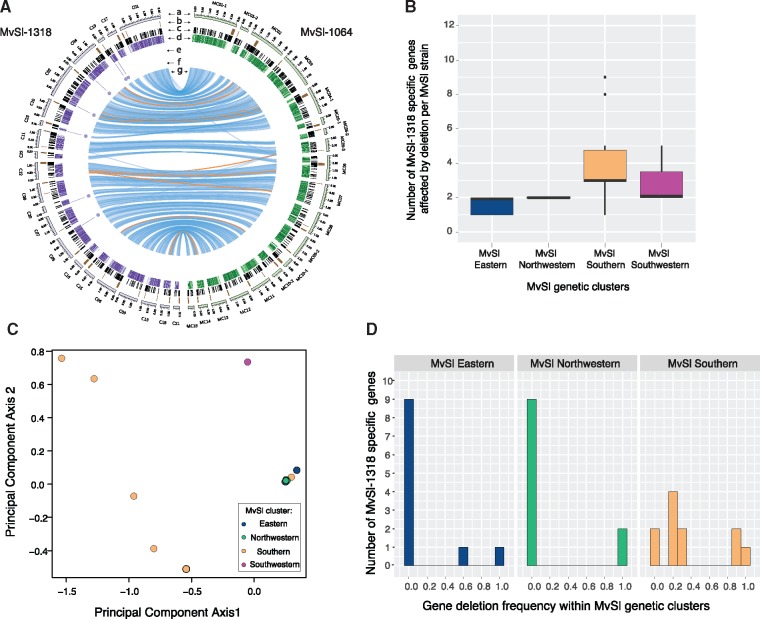
For identifying genes absent from the MvSl-1064 genome reference but present in the genomes of other MvSl strains, we generated a high-quality genome assembly for the MvSl-1318 strain from the MvSl Eastern cluster and we assembled de novo the Illumina genomes of the MvSl-140-01 strain from the MvSl Northwestern cluster, and of the MvSl-1005 strain from the MvSl Southwestern cluster (fig. 4A;supplementary text S2, Supplementary Material online). Using our gene presence–absence detection pipeline, we showed that the MvSl-1318 genome contained 11 genes absent from the MvSl-1064 genome (fig. 6A). We identified nine genes in the MvSl-140-01 genome and 15 in the MvSl-1005 genome that were absent from the MvSl-1064 genome. The number of strain-specific genes identified in the three de novo assembled genomes (i.e., absent from the MvSl-1064 genome) was consistent with the number of genes present in the MvSl-1064 genome and absent from other MvSl strains (supplementary fig. S2D, Supplementary Material online). The population structure of the presence–absence polymorphism of strain-specific genes in MvSl confirmed the MvSl phylogeographic structure corresponding to the plant local adaptation (fig. 6C and D).
Fig. 6.
—Identification of strain-specific genes in three Microbotryum lychnidis-dioicae (MvSl) de novo assembled genomes and effect of population structure on gene presence–absence polymorphism. (A) Comparison between high-quality genome assemblies of the MvSl strains MvSl-1064 and MvSl-1318. The different tracks are: (a) Contigs of the MvSl-1318 (left, purple tracks) and MvSl-1064 reference genomes (right, green tracks). Only contigs >320 kb were represented; (b) Location of centromeric repeats; (c) Location of transposable elements; (d) Gene density in 10-kb nonoverlapping windows (the gradient shows differences from 0% to 100%); (e) Location of detected missing fragments in MvSl-1064; (f) Location of missing genes in MvSl-1064, that is, MvSl-1318-specific genes; (g) Links representing collinearity of genomic regions >10 kb between the MvSl-1064 and MvSl-1318 reference genomes, with the orange links corresponding to inversions. (B) Distribution of the number of MvSl-1318-specific genes predicted to be absent per MvSl strain and per genetic cluster. (C) Principal component analysis of gene presence–absence polymorphism of MvSl-1318-specific genes in MvSl strains based on six unique gene presence–absence polymorphisms, that is, using only a single gene per missing fragments. Points correspond to strains and are colored according to the genetic clusters identified by the dendrogram (fig. 4D). (D) Distribution of gene absence frequency of MvSl-1318-specific genes in the four MvSl genetic clusters.
Discussion
Gene Content Variation Affects Recently Gained Genes in Anther-Smut Fungi
Using a robust gene presence–absence calling procedure based on two different read mapping methods, we characterized the degree of gene content variation on autosomes of the fungal anther-smut species MvSl and MvSd. Autosomal chromosomes are highly homozygous and have a total assembly size of ∼30 Mb in both species (Badouin 2015; Branco et al. 2017). The read depth-based and split read-based methods were complementary, as they were based on different read mapping approaches and detected different gene presence–absence polymorphism events. We detected two to 50 missing genes per strain relative to the reference genome for the species concerned. We performed de novo genome assemblies for three additional MvSl strains to validate the degree of intraspecific gene content variation between two strains, as we detected about a dozen genes that were absent from the MvSl reference genome but present in other strains. Our results supported the view that one single reference genome underestimates the gene space of fungal species (Peter et al. 2018; Plissonneau et al. 2018). The level of gene presence–absence polymorphism in anther-smut pathogens and the sizes of the missing DNA fragments were consistent with findings for yeasts. In Saccharomyces cerevisiae, gene content variation concerns a median of 31 genes per strain (Bergström et al. 2014). We found that gene presence–absence polymorphism contributed to the genetic variation of anther-smut fungi, consistent with the view that gene gains and losses are important sources of fungal genetic variation (Taylor et al. 2017; Fouché et al. 2018).
The functional characteristics of the genes displaying presence–absence polymorphism were similar in MvSl and MvSd: mostly genes of unknown function, with low expression levels, small sizes and enriched in genes from multiple-copy gene families. These functional characteristics suggest that the genes displaying presence–absence polymorphism do not have housekeeping functions and have been gained recently. Our finding that a large proportion of the genes displaying presence–absence polymorphism belong to multiple-gene families suggests that these genes originate from recent duplication events in multiple-gene families. Following a duplication event, a gene is gained and one of the copies can diverge extensively and acquires a new function (Tautz and Domazet-Lošo 2011). Short length and low level of expression are also common features of genes that have recently arisen de novo, as reported for Zymoseptoria tritici, a fungal pathogen of wheat (Plissonneau et al. 2016; Hartmann and Croll 2017). A search for orthologs in outgroup species also suggested that most of the presence–absence polymorphisms observed probably resulted from recent gene gains in MvSl or MvSd. New genes may arise from noncoding DNA through the spontaneous evolution of an open reading frame and the gain of cis-regulatory elements (Carvunis et al. 2012; McLysaght and Guerzoni 2015), or through horizontal gene transfer (Marcet-Houben and Gabaldón 2010; Slot and Rokas 2011; Ropars et al. 2015), We found that gene presence–absence polymorphism was particularly prevalent in subtelomeric regions and close to repeats, as previously reported for yeasts and for the plant pathogens M. oryzae and Z. tritici (Gallone et al. 2016; Plissonneau et al. 2016; Steenwyk et al. 2016; Yoshida et al. 2016), consistent with the view that duplication and excision events may be driven by repeated elements. The high content of repeats found in subtelomeres and centromeres (fig. 1B–D) might at least partly explain the enrichment of gene presence–absence polymorphism events in these genomic regions.
Gene Presence–Absence Polymorphism and Host–Pathogen Coevolution in Natural Environments
Most of the gene presence–absence polymorphism detected is likely neutral. Indeed, previously identified selective sweep regions were not enriched in gene presence–absence events. In addition, gene absence alleles were skewed toward low-frequency variants in the two anther-smut fungal species. Neutral evolution for gene presence–absence polymorphism is also consistent with previous findings in other plant fungal pathogens. Whole-genome sequence comparisons between Magnaporthe oryzae strains specific to rice, millet, wheat, or oat revealed for instance that most of the genes displaying presence–absence polymorphism were genes whose functional domains were present multiple times in the genome, that is, genes likely displaying high levels of functional redundancy (Yoshida et al. 2016). In the wheat pathogen Z. tritici, mainly weak negative selection and neutrality have been shown to act on presence–absence polymorphism of recently gained genes, but also divergent selection in some cases (Hartmann and Croll 2017). In the anther-smut pathogens MvSl and MvSd, we also found that a few of the recently gained genes may be adaptive, with predicted functions potentially involved in host–pathogen interactions, such as secreted proteins, and/or being located within regions subject to recent selective sweeps. The enrichment in functions linked to cellular stress responses of genes displaying presence–absence polymorphism may also reflect adaptation to specific environments. The loss of genes encoding proteins involved in RNA interference activity might also be adaptive, as some plant pathogens use the RNA interference machinery to prevent the expression of host immunity genes (Weiberg and Jin 2015), but this has not been investigated in anther-smut fungi. We found that different genomic regions displayed gene presence–absence polymorphism in MvSl and MvSd specialized on different host plants, with differences in the affected secreted protein-encoding genes, in particular. The genes involved in the coevolution of anther-smut fungi with their host plants are unknown (Badouin et al. 2017), and the genes displaying presence–absence polymorphism within MvSl and MvSd populations and corresponding to secreted proteins and/or located within selective sweeps represent potentially interesting candidates for future exploration. Functional validation assays using gene transformation tools recently established for anther-smut fungi (Toh et al. 2016) should be performed in future studies to validate an adaptive role of the gene presence–absence events with relevant functions of genomic regions.
We identified only five genes encoding secreted proteins displaying presence–absence polymorphism in MvSl, and three in MvSd. These numbers correspond to lower percentages of total gene content than reported for several fungal crop pathogens, such as the rice BLAST pathogen M. oryzae, the wheat pathogens Z. tritici and Stagonospora nodorum, and the generalist crop pathogens Verticillium dahliae and Fusarium oxysporum (Yoshida et al. 2009; Ma et al. 2010; Jonge et al. 2013; Syme et al. 2013; Plissonneau et al. 2016). Many gene-for-gene relationships have been documented in crop-pathogen systems, several of which corresponded to recent losses or gains of genes in the fungus, to prevent host recognition by the plant defense system (Orbach et al. 2000; Gout et al. 2006, 2007; Jonge et al. 2012; Hartmann et al. 2017; Fouché et al. 2018). By contrast, no gene-for-gene relationship has been reported for anther-smut fungi, despite extensive studies on genetic variation in host resistance and fungal pathogenicity. Instead, the probability of infection is a quantitative trait (Alexander et al. 1993; Alexander and Antonovics 1995; Biere and Antonovics 1996; Chung et al. 2012), which likely corresponds to a control of host recognition by other genetic changes than gene loss or gain. Differences in coevolutionary dynamics between anthropized and natural pathosystems, such as the “arms race” and “trench warfare” evolution models (Brown and Tellier 2011) may also result in different mechanisms of adaptation to the host. More studies of this type are required, on noncrop pathogens in particular, to test hypotheses concerning the evolutionary causes of structural variation frequencies affecting genes involved in coevolutionary processes.
Diversity and Phylogeographic Structure
Gene presence–absence polymorphisms were more frequent in MvSl than in MvSd (2% vs 0.5% of all autosomal genes). The degree of gene presence–absence polymorphism in MvSd may be slightly underestimated, due to the lower reference genome quality for MvSd than for MvSl and the lower number of sequenced strains (supplementary table S2, Supplementary Material online). However, this is unlikely to account for the magnitude of the difference observed, which was consistent with those reported for SNPs and microsatellites (Vercken et al. 2010; Badouin et al. 2017).
The analysis of gene presence–absence polymorphism in MvSl revealed a population structure highly similar to that reported on the basis of SNPs and microsatellite data, with the same four genetic clusters corresponding to footprints of glacial refugia (Vercken et al. 2010; Badouin et al. 2017). The addition of MvSl strains (Whittle et al. 2015) compared to previous phylogeographic studies revealed very high levels of genetic diversity among the strains of the Southwestern cluster, as expected for strains derived from southern glacial refugia, which have been little explored to date (Vercken et al. 2010; Badouin et al. 2017). The correspondence between the structures revealed by analyses of neutral markers and those revealed by analyses of gene presence–absence polymorphism further reinforce the view that most of such polymorphism is neutral. In particular, gene gain or loss within gene families may be neutral due to functional redundancy (Albalat and Cañestro 2016). However, the host plant S. latifolia also displays the same genetic subdivision in Europe and local adaptation between clusters: plants from a given cluster are more resistant to MvSl strains from the same cluster (Feurtey et al. 2016). Some of the gene presence–absence polymorphism events that are congruent with these clusters may, therefore, also correspond to adaptive polymorphism resulting from coevolution with the host plant. Interestingly, this analysis of gene presence–absence polymorphism suggested the existence of a geographical population structure in MvSd, across an east–west gradient in Europe. Although this population structure had not been detected before, our analysis of genome-wide SNP data called against an MvSd reference genome partly recovered a similar population structure. However, PCAs and FST values indicated a stronger structure in gene presence–absence polymorphism than in SNPs in MvSd, which may reflect distinct population genetic properties of genetic markers in populations, such as mutation rate, or may result from an effect of selection on some of the gene gains/losses. The population structure based on gene presence–absence polymorphism in MvSd may thus reflect, at least partly, adaptive events, possibly in response to a population subdivision of the host in terms of resistance genes. Indeed, a population subdivision of the host plant species S. dioica separates the Western and Eastern populations (Hathaway et al. 2009; Rautenberg et al. 2010). However, here again, functional validation studies are needed to test this hypothesis.
In conclusion, our findings show that gene presence–absence polymorphism contributes to intraspecific genetic variation in anther-smut fungi, although to a lower extent in terms of total gene content than previously reported for fungal crop pathogens. Few genes encoding secreted proteins showed presence–absence polymorphisms, as expected given the lack of gene-for-gene relationships in anther-smut fungi. Gene presence–absence polymorphism mostly affected recently gained genes, found in a single species, which were likely mostly neutral, with a few cases perhaps corresponding to innovations allowing adaptation to the host or interaction with the environment, which should be validated using functional studies in the near future.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
PacBio sequencing was conducted at the IGM Genomics Center, University of California, San Diego, La Jolla, CA. This work was supported by the European Research Council (ERC) (starting grant GenomeFun 309403 to TG); the French National Research Agency (ANR) (Gandalf ANR Grant ANR-12-ADAP-0009 to TG); and a Marie Curie European grant (PRESTIGE-2016-4-0013 to FEH). FEH received the Young Biological Researcher Prize from the Fondation des Treilles, created by Anne Gruner Schlumberger, which supports research in Science and Art (http://www.les-treilles.com; last accessed February 12, 2018). JTB was supported by an IDEEV funding.
Literature cited
- Abyzov A, Urban AE, Snyder M, Gerstein M.. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 216:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R, Cañestro C.. 2016. Evolution by gene loss. Nat Rev Genet. 177:379–391. [DOI] [PubMed] [Google Scholar]
- Albataineh MT, Kadosh D.. 2016. Regulatory roles of phosphorylation in model and pathogenic fungi. Med Mycol. 544:333–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander HM, Antonovics J.. 1995. Spread of anther-smut disease (Ustilago violacea) and character correlations in a genetically variable experimental population of Silene alba. J Ecol. 835:783–794. [Google Scholar]
- Alexander HM, Antonovics J, Kelly AW.. 1993. Genotypic variation in plant disease resistance–physiological resistance in relation to field disease transmission. J Ecol. 812:325–333. [Google Scholar]
- Badouin H. 2015. Chaos of rearrangements in the mating-type chromosomes of the anther-smut fungus Microbotryum lychnidis-dioicae. Genetics 2004:1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouin H, et al. , . 2017. Widespread selective sweeps throughout the genome of model plant pathogenic fungi and identification of effector candidates. Mol Ecol. 267:2041–2062. [DOI] [PubMed] [Google Scholar]
- Bankevich A, et al. , . 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 195:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 57:289–300. [Google Scholar]
- Bergström A, et al. , . 2014. A high-definition view of functional genetic variation from natural yeast genomes. Mol Biol Evol. 314:872–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biere A, Antonovics J.. 1996. Sex-specific costs of resistance to the fungal pathogen Ustilago violacea (Microbotryum violaceum) in Silene alba. Evolution 503:1098–1110. [DOI] [PubMed] [Google Scholar]
- Branco S, et al. , . 2017. Evolutionary strata on young mating-type chromosomes despite the lack of sexual antagonism. Proc Natl Acad Sci U S A. 11427:7067–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JKM, Tellier A.. 2011. Plant-parasite coevolution: bridging the gap between genetics and ecology. Annu Rev Phytopathol. 491:345–367. [DOI] [PubMed] [Google Scholar]
- Carvunis A-R, et al. , . 2012. Proto-genes and de novo gene birth. Nature 4877407:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R, Medvedev P.. 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 301:31–37. [DOI] [PubMed] [Google Scholar]
- Chung E, Petit E, Antonovics J, Pedersen AB, Hood ME.. 2012. Variation in resistance to multiple pathogen species: anther smuts of Silene uniflora. Ecol Evol. 29:2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK.. 2006. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 381:75–81. [DOI] [PubMed] [Google Scholar]
- Conrad DF, et al. , . 2010. Origins and functional impact of copy number variation in the human genome. Nature 4647289:704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekseth OK, Kuiper M, Mironov V.. 2014. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics 305:734–736. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 91:18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtey A, et al. , . 2016. Strong phylogeographic co-structure between the anther-smut fungus and its white campion host. New Phytol. 2123:668–679. [DOI] [PubMed] [Google Scholar]
- Fontanillas E, et al. , . 2014. Degeneration of the non-recombining regions in the mating-type chromosomes of the anther-smut fungi. Mol Biol Evol. 324:928–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouché S, Plissonneau C, Croll D.. 2018. The birth and death of effectors in rapidly evolving filamentous pathogen genomes. Curr Opin Microbiol. 46:34–42. [DOI] [PubMed] [Google Scholar]
- Friesen TL, et al. , . 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet. 388:953–956. [DOI] [PubMed] [Google Scholar]
- Gallone B, et al. , . 2016. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 1666:1397–1410.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Yockteng R, Lopez-Villavicencio M, Refregier G, Hood ME.. 2008. Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism. Eukaryot Cell 75:765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. 2005. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 51:184–186. [Google Scholar]
- Gout L, et al. , . 2006. Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol. 601:67–80. [DOI] [PubMed] [Google Scholar]
- Gout L, et al. , . 2007. Genome structure impacts molecular evolution at the AvrLm1 avirulence locus of the plant pathogen Leptosphaeria maculans. Environ Microbiol. 912:2978–2992. [DOI] [PubMed] [Google Scholar]
- Guan P, Sung W-K.. 2016. Structural variation detection using next-generation sequencing data: a comparative technical review. Methods 102:36–49. [DOI] [PubMed] [Google Scholar]
- Hartmann FE, Croll D.. 2017. Distinct trajectories of massive recent gene gains and losses in populations of a microbial eukaryotic pathogen. Mol Biol Evol. 3411:2808–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FE, Sánchez-Vallet A, McDonald BA, Croll D.. 2017. A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements. ISME J. 115:1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway L, Malm JU, Prentice HC.. 2009. Geographically congruent large-scale patterns of plastid haplotype variation in the European herbs Silene dioica and S. latifolia (Caryophyllaceae). Bot J Linn Soc. 1612:153–170. [Google Scholar]
- Henrichsen CN, Chaignat E, Reymond A.. 2009. Copy number variants, diseases and gene expression. Hum Mol Genet. 18(R1):R1–R8. [DOI] [PubMed] [Google Scholar]
- Hood ME. 2005. Repetitive DNA in the automictic fungus Microbotryum violaceum. Genetica 1241:1–10. [DOI] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 2411:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Ahmed I.. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 2721:3070–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 4447117:323–329. [DOI] [PubMed] [Google Scholar]
- Jonge R, et al. , . 2012. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci U S A. 10913:5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonge R, et al. , . 2013. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 238:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltz O, Gandon S, Michalakis Y, Shykoff JA.. 1999. Local maladaptation in the anther-smut fungus Microbotryum violaceum to its host plant Silene latifolia: evidence from a cross-inoculation experiment. Evol Int J Org Evol. 532:395–407. [DOI] [PubMed] [Google Scholar]
- Khang CH, Park S-Y, Lee Y-H, Valent B, Kang S.. 2008. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol Plant Microbe Interact. 215:658–670. [DOI] [PubMed] [Google Scholar]
- Koopmann B, Müller J, Tellier A, Živković D.. 2017. Fisher–Wright model with deterministic seed bank and selection. Theor Popul Biol. 114:29–39. [DOI] [PubMed] [Google Scholar]
- Koskela T, Puustinen S, Salonen V, Mutikainen P.. 2002. Resistance and tolerance in a host plant-holoparasitic plant interaction: genetic variation and costs. Evol Int J Org Evol. 56:899–908. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 3053:567–580. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. , . 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 199:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. , . 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L, Burdon JJ, Dodds PN, Thrall PH.. 2011. Spatial variation in disease resistance: from molecules to metapopulations. J Ecol. 991:96–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L, Burdon JJ, Nemri A, Thrall PH.. 2014. Host ecotype generates evolutionary and epidemiological divergence across a pathogen metapopulation. Proc R Soc B 2811787:20140522.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L, Tellier A.. 2008. Heterogeneous selection promotes maintenance of polymorphism in host–parasite interactions. Oikos 1179:1281–1288. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 103:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gac M, Hood ME, Fournier E, Giraud T.. 2007. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evol Int J Org Evol. 611:15–26. [DOI] [PubMed] [Google Scholar]
- Li H, et al. , . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 307:923–930. [DOI] [PubMed] [Google Scholar]
- Lin K, Smit S, Bonnema G, Sanchez-Perez G, de Ridder D.. 2015. Making the difference: integrating structural variation detection tools. Brief Bioinform. 165:852–864. [DOI] [PubMed] [Google Scholar]
- Ma L-J, et al. , . 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 4647287:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet-Houben M, Gabaldón T.. 2010. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 261:5–8. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 171:10–12. [Google Scholar]
- McKenna A, et al. , . 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 209:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Guerzoni D.. 2015. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc B 3701678:20140332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller M, Stukenbrock EH.. 2017. Evolution and genome architecture in fungal plant pathogens. Nat Rev Microbiol. 1512:756–771. [DOI] [PubMed] [Google Scholar]
- Monod M, et al. , . 2002. Secreted proteases from pathogenic fungi. Int J Med Microbiol. 292(5-6):405–419. [DOI] [PubMed] [Google Scholar]
- Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B.. 2000. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 1211:2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, et al. , . 2009. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet. 1821:4118–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 202:289–290. [DOI] [PubMed] [Google Scholar]
- Perlin MH, et al. , . 2015. Sex and parasites: genomic and transcriptomic analysis of Microbotryum lychnidis-dioicae, the biotrophic and plant-castrating anther smut fungus. BMC Genomics 161:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, et al. , . 2018. Genome evolution across 1, 011 Saccharomyces cerevisiae isolates. Nature 556:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 810:785–786. [DOI] [PubMed] [Google Scholar]
- Plissonneau C, Hartmann FE, Croll D.. 2018. Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biol. 161:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plissonneau C, Stürchler A, Croll D.. 2016. The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio 75:e01231-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti LL, et al. , . 2015. Fungal effectors and plant susceptibility. Annu Rev Plant Biol. 661:513–545. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 266:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautenberg A, Hathaway L, Oxelman B, Prentice HC.. 2010. Geographic and phylogenetic patterns in Silene section Melandrium (Caryophyllaceae) as inferred from chloroplast and nuclear DNA sequences. Mol Phylogenet Evol. 573:978–991. [DOI] [PubMed] [Google Scholar]
- Refrégier G, et al. , . 2008. Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation. BMC Evol Biol. 81:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. , . 2011. Integrative genomics viewer. Nat Biotechnol. 291:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J, et al. , . 2015. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Curr Biol. 2519:2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H.. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 132:365–386. [DOI] [PubMed] [Google Scholar]
- Schrider DR, Hahn MW.. 2010. Gene copy-number polymorphism in nature. Proc R Soc Lond B Biol Sci. 2771698:3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch S, Linde CC, Knogge W, Jackson LF, McDonald BA.. 2004. Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol Plant Microbe Interact. 1710:1114–1125. [DOI] [PubMed] [Google Scholar]
- Slot JC, Rokas A.. 2011. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr Biol. 212:134–139. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R.. 2008. RepeatModeler Open-1.0 [cited 2017 Nov 30]. Available from: http://www.repeatmasker.org; last accessed November 30, 2017.
- Smit AFA, Hubley R, Green P.. 2013. RepeatMasker Open-4.0 [cited 2017 Nov 30]. Available from: http://www.repeatmasker.org; last accessed November 30, 2017.
- Stam R, Scheikl D, Tellier A.. 2017. The wild tomato species Solanum chilense shows variation in pathogen resistance between geographically distinct populations. PeerJ 5:e2910.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Soghigian JS, Perfect JR, Gibbons JG.. 2016. Copy number variation contributes to cryptic genetic variation in outbreak lineages of Cryptococcus gattii from the North American Pacific Northwest. BMC Genomics 171:700.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syme RA, Hane JK, Friesen TL, Oliver RP.. 2013. Resequencing and comparative genomics of Stagonospora nodorum: sectional gene absence and effector discovery. G3 (Bethesda) 3:959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack AJM, Laine A-L.. 2014. Ecological and evolutionary implications of spatial heterogeneity during the off-season for a wild plant pathogen. New Phytol. 2021:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 1210:692.. [DOI] [PubMed] [Google Scholar]
- Taylor JW, et al. , . 2017. Sources of fungal genetic variation and associating It with phenotypic diversity. Microbiol Spectr 55: doi: 10.1128/microbiolspec.FUNK-0057-2016. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 142:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall PH, Burdon JJ, Young A.. 2001. Variation in resistance and virulence among demes of a plant host–pathogen metapopulation. J Ecol. 895:736–748. [Google Scholar]
- Toh SS, Treves DS, Barati MT, Perlin MH.. 2016. Reliable transformation system for Microbotryum lychnidis dioicae informed by genome and transcriptome project. Arch Microbiol. 1988:813–825. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 259:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercken E, et al. , . 2010. Glacial refugia in pathogens: european genetic structure of anther smut pathogens on Silene latifolia and Silene dioica. PLoS Pathog. 612:e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent D, et al. , . 2016. Editorial: how can secretomics help unravel the secrets of plant-microbe interactions? Front Plant Sci. 7:1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A, Jin H.. 2015. Small RNAs—the secret agents in the plant–pathogen interactions. Curr Opin Plant Biol. 26:87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CA, Votintseva A, Ridout K, Filatov DA.. 2015. Recent and massive expansion of the mating-type-specific region in the smut fungus Microbotryum. Genetics 1993:809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York, NY: Springer Science & Business Media. [Google Scholar]
- Yang R-C. 1998. Estimating hierarchical F-Statistics. Evolution 524:950–956. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z.. 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2521:2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, et al. , . 2009. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 215:1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, et al. , . 2016. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics 171:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R.. 2001. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics 179:847–848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.