Wheat miR9678, specifically expressed in scutellum of developing and germinating wheat seeds, affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling
Abstract
Seed germination is important for grain yield and quality and rapid, near-simultaneous germination helps in cultivation; however, cultivars that germinate too readily can undergo preharvest sprouting (PHS), which causes substantial losses in areas that tend to get rain around harvest time. Moreover, our knowledge of mechanisms regulating seed germination in wheat (Triticum aestivum) remains limited. In this study, we analyzed function of a wheat-specific microRNA 9678 (miR9678), which is specifically expressed in the scutellum of developing and germinating seeds. Overexpression of miR9678 delayed germination and improved resistance to PHS in wheat through reducing bioactive gibberellin (GA) levels; miR9678 silencing enhanced germination rates. We provide evidence that miR9678 targets a long noncoding RNA (WSGAR) and triggers the generation of phased small interfering RNAs that play a role in the delay of seed germination. Finally, we found that abscisic acid (ABA) signaling proteins bind the promoter of miR9678 precursor and activate its expression, indicating that miR9678 affects germination by modulating the GA/ABA signaling.
INTRODUCTION
Regulation of seed germination and dormancy allow plants to adapt to various environmental conditions. Primary dormancy inhibits the germination of newly produced seeds, thus preventing germination during long periods of unsuitable environmental conditions (Koornneef et al., 2002; Shu et al., 2016; Penfield, 2017). Therefore, seed germination is a well-timed checkpoint that can help plants avoid detrimental effects of unfavorable environmental conditions on plant establishment and reproductive growth (Finch-Savage and Leubner-Metzger, 2006). Accordingly, time of seed germination is associated with seedling survival rate and subsequent vegetative plant growth, ultimately affecting crop yield and quality. Many crops exhibit weak dormancy as a result of selection for rapid and uniform germination at sowing. This has led to seeds prone to preharvest sprouting (PHS) following wet and cool conditions, causing substantial losses in yield and quality (Barrero et al., 2015). Hence, for improving agronomic performance, it is critical to apply strategies that concomitantly produce uniform and rapid germination at sowing and to prevent PHS (Bewley and Black, 1994). This requires an improvement of our knowledge about mechanisms regulating seed germination.
The widely cultivated crop hexaploid wheat, Triticum aestivum (2n=6x=42; genomes BBAADD), provides 20% of the calories consumed by humans and is valued for its high yield, nutritional value, and processing qualities. To optimize agronomic production, wheat cultivars with rapid and uniform seed germination at planting are required, which is capable of producing great vegetative growth (TeKrony and Egli, 1991). However, selection for these traits can yield cultivars with weak dormancy, leading to PHS in the rainy season that tends to overlap with the harvest season in various geographical areas (Bewley, 1997; Liu et al., 2013). During PHS, seeds germinate in the spike before harvest when prolonged rainfall occurs; this results in reduced yield and quality (Liu et al., 2015). Understanding the underlying mechanisms that control wheat seed germination and dormancy is expected to provide key information for approaches to regulate germination and thus prevent PHS to obtain uniform and rapid germination (Mares and Mrva, 2014).
Genetic approaches have identified several quantitative trait loci and genes that affect seed dormancy and germination in wheat. For example, quantitative trait loci on chromosomes 2B (Munkvold et al., 2009; Somyong et al., 2014), 3A (Liu et al., 2008), and 4A (Ogbonnaya et al., 2008) have been shown to affect PHS resistance. TaMFT-3A, a wheat homolog of Arabidopsis (Arabidopsis thaliana) MOTHER OF FLOWERING LOCUS T AND TERMINAL FLOWER 1-LIKE, and its homolog TaPHS1 are associated with seed dormancy and PHS (Nakamura et al., 2011; Liu et al., 2013). Two adjacent candidate genes, PM19-A1 and A2, located on chromosome 4A, also act as positive regulators of seed dormancy (Barrero et al., 2015). However, these studies are focused on PHS and regulation of seed dormancy and do not provide information on seed germination. Recently, an integrative transcriptome and proteome analysis of wheat embryos and endosperm during seed germination identified a large number of transcripts and proteins that were differentially expressed in the embryo and endosperm, thus providing information about genes involved in germination (He et al., 2015; Yu et al., 2016). Nevertheless, addressing PHS while retaining high germination rates still requires more knowledge about the genes and mechanisms regulating wheat seed germination.
The plant hormones abscisic acid (ABA) and gibberellin (GA) are the primary hormones that antagonistically regulate seed dormancy and germination (Gubler et al., 2005; Finkelstein et al., 2008; Penfield, 2017). ABA, which is synthesized during seed maturation, can establish and maintain dormancy and inhibit germination; ABA levels decrease before the onset of germination (Kanno et al., 2010). In contrast to ABA, increased GA levels promote germination by causing the secretion of hydrolytic enzymes that weaken the structure of the seed testa; moreover, GA also stimulates embryo growth potential (Holdsworth et al., 2008; Yano et al., 2009). In Arabidopsis, mutants that are severely defective in GA biosynthesis, such as ga1-3 (mutated in GA1), show strong dormancy and inhibited seed germination (Sun et al., 1992; Olszewski et al., 2002). GA signaling is also required for efficient seed germination. For example, mutation of the GA signaling gene SLEEPY1 leads to increased seed dormancy (Ariizumi and Steber, 2007). Mutants defective in GA 2-OXIDASE (GA2ox), which deactivates bioactive GAs, show enhanced seed germination under dark conditions (Yamauchi et al., 2007). Although these studies show that ABA and GA play key roles in seed germination, knowledge about whether and how these hormones control seed germination in wheat is still very limited.
In wheat seeds, GAs are synthesized in the scutellum, which is a modified cotyledon and highly adapted as an absorptive or haustorial structure (Payne and Walker-Smith, 1987). GAs then diffuse from scutellum to the starchy endosperm (Appleford and Lenton, 1997). Besides producing GAs, the scutellum in the germinated wheat seed functions in the transfer of sugars and amino acids to the growing seedling (Aoki et al., 2006; Domínguez et al., 2012). Therefore, the scutellum and the genes specifically expressed in scutellum such as TaMFT play important roles in seed germination (Nakamura et al., 2011).
Studies in Arabidopsis show that microRNAs (miRNAs) act as both activators and repressors of seed germination and dormancy by interacting with ABA and GA signaling (Martin et al., 2010; Das et al., 2015). Specifically, miR159 regulates mRNA encoding GAMYB transcription factors, which affect GA-mediated programmed cell death in the aleurone layer, a developmental process that is important for seed germination (Reyes and Chua, 2007; Alonso-Peral et al., 2010). Similarly, miR160 mediates downregulation of AUXIN RESPONSE FACTOR10, which plays a role in seed germination and postembryonic development (Liu et al., 2007; Nonogaki, 2008). In addition, DELAY OF GERMINATION1 regulates seed dormancy and flowering times in lettuce (Lactuca sativa) and Arabidopsis by controlling levels of miR156 and miR172 (Huo et al., 2016). The biogenesis of phased small interfering RNAs (phasiRNAs), another type of small RNAs (sRNAs) in plants, depends upon an initial targeting event by a sRNA (miRNA or phasiRNA) on a primary transcript (Axtell, 2017). These phasiRNAs can, in turn, regulate other genes in cis or trans (tasiRNA) (Allen et al., 2005; Chen et al., 2007; Allen and Howell, 2010; Axtell, 2013). TasiRNAs are involved in the control of plant development, including hormone signaling (Das et al., 2015). Although tasiRNAs have not yet directly been implicated in seed germination, their crosstalk with miRNAs and hormone signaling in feedback loops (Martin et al., 2010), as well as role in seed development (Zhang et al., 2013), indicate their potential function in seed maturation and germination.
In this study, we provide experimental evidence that miR9678, a miRNA specifically expressed in the scutellum, affects wheat seed germination. This miRNA (previously named miR021b) had been found to be specifically expressed in the embryos of developing and germinating seeds (Sun et al., 2014). Here, we showed that wheat-specific miR9678 triggered the generation of phasiRNAs by targeting a long noncoding RNA and that, in turn, this negatively affected seed germination. Overexpression of miR9678 delayed germination and improved resistance to PHS in wheat and reduced bioactive GA levels. In addition, components of the ABA signaling pathway bound the promoter of the miR9678 precursor and activated the expression of this miRNA gene. Overall, our findings provide indications of a novel regulatory mechanism for wheat seed germination and PHS. In this mechanism, miR9678 modulates abscisic acid/gibberellin signaling to generate phased siRNAs, thus negatively affecting seed germination.
RESULTS
miR9678 Is Specifically Expressed in the Scutellum of Wheat Seeds
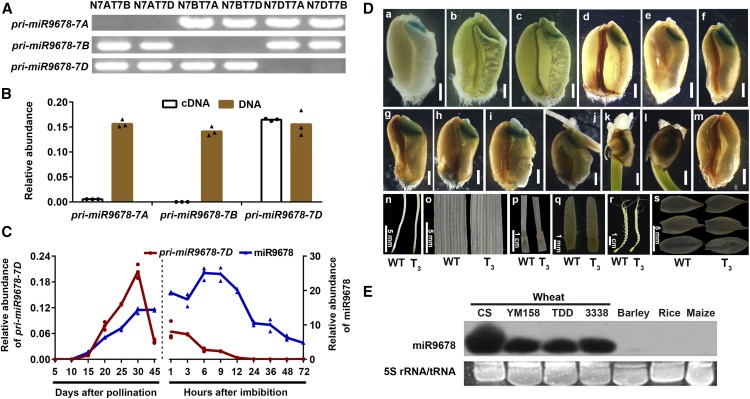
In a previous genome-wide analysis, we identified a 22-nucleotide miRNA, miR9678 (previously named miR021b), which was specifically expressed in the embryos of developing and germinating seeds, suggesting that it could be involved in regulating germination (Han et al., 2014; Sun et al., 2014). As a first step in the functional characterization of miR9678, we aimed to identify its precursor primary-miR9678 (pri-miR9678) loci. A BLAST search using the mature miR9678 sequence against the draft wheat genome sequence (http://plants.ensembl.org/Triticum_aestivum/Info/Index) identified three precursor sequences located on chromosomes 7A, 7B, and 7D. These loci were verified by amplifying copy-specific fragments from the genomic DNA of the wheat cultivar Chinese Spring (CS) nullisomic-tetrasomic lines (Figure 1A), which lack one pair of chromosomes but have extra homoeologous ones in compensation. The amplification fragments disappeared when the corresponding chromosome was deleted from CS lines. Therefore, these three pri-miR9678 loci were considered to be homoeologs and were designated pri-miR9678-7A, pri-miR9678-7B, and pri-miR9678-7D. The level of these precursors in germinating seeds was determined by RT-qPCR and the results showed that the amount of pri-miR9678-7D RNA was much higher than pri-miR9678-7A and pri-miR9678-7B RNAs (Figure 1B). We also observed that pri-miR9678-7D transcript started to accumulate at 15 d after pollination (DAP), with a peak around 30 DAP; subsequently, it decreased at seed maturity and it kept decreasing during germination, when miR9678 was around its maximum level (Figure 1C). Interestingly, miR9678 rapidly decreased to a very low level in a period from 9 to 24 h after imbibition (HAI). These results indicated that mature miR9678 was stored during seed maturation and in dry seeds and disappeared after the onset of germination, implying that miR9678 levels might be negatively associated with seed germination.
Figure 1.
Expression Analysis of the Wheat-Specific miR9678.
(A) PCR amplification of genomic DNA extracted from nullisomic-tetrasomic series of wheat CS cultivar using specific primers. The lines tested are indicated, where N7AT7B represents nullisomic7A-tetrasomic7B, for example.
(B) Relative abundance of three miR9678 precursors in 28-DAP embryos. DNA of the Chinese Spring cultivar was used to analyze the amplification efficiency of three pairs of primers. The RT-qPCR data were normalized to the wheat ACTIN gene, and each bar in the graph corresponds to the mean value of three data points, which are shown as circles (cDNA) and triangles (DNA).
(C) Expression of pri-miR9678-7D precursor transcript (red) and mature miR9678 (blue) during seed maturation and germination in wheat embryos. miR9678 and pri-miR9678-7DRNA levels were measured with RT-qPCR and normalized to the wheat U6 and wheat ACTIN genes, respectively. Data are presented as a line connecting the average of three data points (reported as circles and triangles).
(D) Pri-miR9678-7D:GUS expression in T3 generation transgenic wheat seeds, roots, leaves, stems, spikes, and glumes. A total of 10 independent transformants were analyzed; one of these transformants is shown as a representative example. a to l, transgenic wheat seeds; a,15 DAP; b, 20 DAP; c, 25 DAP; d, dry seed; e, 3 HAI; f, 6 HAI; g, 9 HAI; h, 12 HAI; i, 24 HAI; j, 36 HAI; k, 48 HAI; l, 72 HAI. m, Wild-type seeds at 6 HAI. Bar = 1 mm for a to m. n, roots; o, leaves; p, stems; q, young spikes with 5 mm in length; r, young spikes with 5 cm in length; s, glumes.
(E) RNA gel blot analysis to detect miR9678 in 12-HAI embryos of four different wheat varieties (Chinese Spring [CS], Yangmai158 [YM158], TDD and 3338) and of barley, rice, and maize. Ethidium bromide-stained polyacrylamide gel with 5S rRNA and tRNA are shown as loading control.
To analyze the expression pattern of pri-miR9678-7D, we generated a pri-miR9678-7D promoter-GUS construct by fusing the 2-kb putative promoter fragment directly upstream of the GUS reporter gene. We then transformed this construct into wheat and stained for GUS activity, finding that the pri-miR9678-7D drove expression only in the scutellum of seeds, but not in other tissues including roots, leaves, stems, spikes, and glumes (Figure 1D). During seed maturation, we detected GUS activity in the scutellum at 15, 20, and 25 DAP (Figure 1D, a to c). GUS activity was also detected in the scutellum from the point of imbibition to 72 HAI, but this persistence of GUS exceed that of its transcript, very likely due to long half-life of GUS protein (Li et al., 1999) (Figure 1D, d to l).
To test whether miR9678 was also produced in other species, we searched for miR9678 homologs in the genome of all plant species in NCBI, but we failed to find any sequences that form the hairpin foldback structures required for miRNA generation. To further corroborate that miR9678 was specific to wheat, we examined its expression abundance in embryos of germinated seeds from four wheat varieties (CS, Yangmai158, TDD, and 3338) and from other cereals like barley (Hordeum vulgare), rice (Oryza sativa), and maize (Zea mays). We observed high levels of miR9678 in all wheat cultivars, whereas no expression was detected in barley, maize, and rice (Figure 1E). Together, our findings showed that miR9678 was a wheat-specific miRNA that was expressed only in the grain scutellum with an expression pattern negatively correlated with seed germination.
miR9678 Targets a Long Noncoding RNA
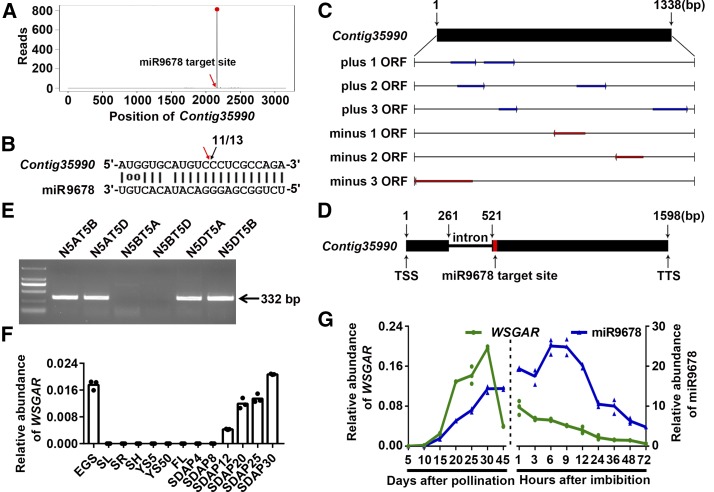
To find the target genes of miR9678, we interrogated wheat ESTs, antisense RNAs, and genome sequence draft databases. This approach identified 15 candidate target genes of miR9678 (Supplemental Table 1). As a first approach for the experimental validation of these targets, two degradome libraries from germinating seeds mixed from 1, 3, and 6 HAI and from 9, 12, and 24 HAI were constructed and sequenced. Among the 15 candidate targets, the cleavage of only Contig35990 was supported by the degradome sequencing. As shown in Figure 2A, the most highly abundant 5′ ends of RNA degradation products of Contig35990 were located in the miR9678 reverse complementary binding site, indicating that Contig35990 was cleaved at the miR9678 binding site. Modified RNA ligase-mediated 5′-rapid amplification of cDNA ends (RLM-5′-RACE) further confirmed that cleavage of Contig35990 occurred at the 10th nucleotide from the 5′ end of miR9678 (Figure 2B).
Figure 2.
miR9678 Targets the Long Noncoding RNA WSGAR.
(A) The degradome sequencing of putative target Contig35990 (we name this sequence WSGAR) is presented as target plots (t-plots). The red dot indicates the signature abundance of cleavage. The red arrow in the t-plots indicates the miR9678 target site (x axis).
(B) miR9678 and Contig35990/WSGAR mRNA alignment along with the cleavage frequencies detected using RLM-5′-RACE. The red arrow represents the predicted cleavage site by miR9678. Black arrow indicates the 5′ ends of the miR9678-guided cleavage products, and numbers (11/13) are the ratios of the cleaved products with respect to total sequenced fragments.
(C) Open reading frame (ORF) prediction of the full-length transcript of Contig35990/WSGAR.
(D) Genome structure of Contig35990/WSGAR. The red box indicates the miR9678 binding site. The black boxes and line indicate exons and an intron, respectively. TSS and TTS indicate the transcription start site and transcription termination site, respectively.
(E) PCR amplification results of the nullisomic-tetrasomic series of the CS wheat cultivar using specific primers, where the lines are indicated such that N5AT5B represents the nullisomic5A-tetrasomic5B line, for example.
(F) RNA level of WSGAR in various wheat tissues. EGS, embryos in germinating seeds; SL, seedling leaves; SR, seedling roots; SH, shoots; YS5, 2- to 5-mm young spikes; YS50, 20 to 50-mm young spikes; FL, flag leaves; SDAP4 to SDAP30 represent whole seeds at 4, 8, 12, 20, 25, and 30 DAP, respectively. The expression levels were normalized to that of ACTIN. Each bar in the graph corresponds to the mean value of three data points, which are shown by circles.
(G) Level of mature miR9678 (blue) and WSGAR RNA (green) in embryos during seed maturation and germination in wheat. The miR9678 and WSGAR expression levels were normalized to U6 and ACTIN, respectively. Data are presented as a line connecting the average of three data points (reported as triangles and circles).
To test whether Contig35990 transcribes a protein-coding RNA, its 1338-bp full-length cDNA was identified by 5′- and 3′-RACE. All open reading frames in this cDNA were shorter than 100 amino acids, with no significant similarity to any protein in the NCBI database (Figure 2C). Therefore, we concluded that it represented a long noncoding RNA. Using the Contig35990 sequence as a query against the wheat genome sequence database, we identified a single-copy sequence, located on chromosome 5B and containing a 260-bp-long intron (Figure 2D). The location of this sequence was confirmed using PCR with Contig35990-specific primers and CS nullisomic-tetrasomic lines (Figure 2E). Accumulation of Contig35990 RNA was analyzed in various organs, showing that it was expressed at high levels in embryos of germinating seeds (EGS) and whole developing seeds after pollination (Figure 2F); accordingly, the noncoding gene was named Wheat Seed Germination Associated RNA (WSGAR) (Figure 2F). Comparison of miR9678 and WSGAR RNA levels in embryos from seed maturation to germination indicated that WSGAR transcript accumulated from 15 to 30 DAP, decreased at maturation, and rapidly disappeared at 1 to 12 HAI, when miR9678 was most abundant (Figure 2G). Overall, our results show that miR9678 targets the long noncoding RNA WSGAR in vivo.
miR9678 Triggers Production of PhasiRNAs from the WSGAR Transcript
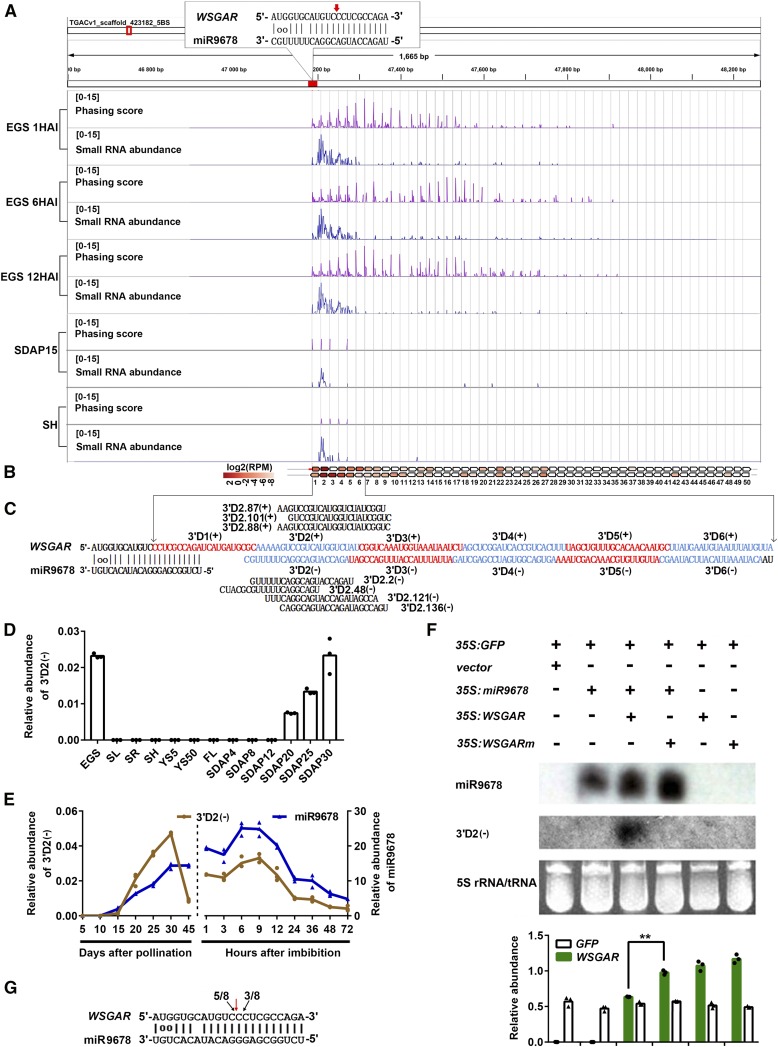
We observed that miR9678 had a U at the 5′ position and that all three precursors exhibited asymmetry in their foldback structures in the form of a nonpaired nucleotide within their miRNA sequences (Supplemental Figure 1). These characteristics were consistent with those of classical 22-nucleotide miRNA triggering the biogenesis of phasiRNAs in plants (Chen et al., 2010; Cuperus et al., 2010). Accordingly, we searched for the presence of phasiRNAs putatively arising from WSGAR transcript in the small RNA sequencing data sets from embryos of germinating seeds at 1, 6, and 12 HAI, shoots (SH) and developing seeds at 15 DAP (SDAP15). Detailed examination of the mapping profile revealed that the miR9678 cleavage site set the phase of the phasiRNA production and that this occurred especially in EGS compared with SH and SDAP15 (Figure 3A). The cleavage site of miR9678 set the main phase of phasiRNAs of the first several cycles (D1–D11), although out-of-phase siRNA signal was detected (especially after the D11 cycle), a commonly observed phenomenon possibly caused by the in cis action of generated phasiRNAs (Xia et al., 2013). Among all the siRNA isoforms produced from WSGAR, the ones located immediately after the site of miR9678-mediated cleavage showed highest abundance (Figure 3B) and also had some 20 to 22-nucleotide variants (Figure 3C). When we analyzed the level of one phasiRNA with high abundance, designated 3′D2(-) according to its location and antisense polarity, we observed that it was highly detected in EGS and developing whole seeds (Figure 3D). Furthermore, analysis of phasiRNA 3′D2(-) during seed maturation and germination confirmed that the expression profile of this phasiRNA overlapped with that of miR9678 (Figure 3E).
Figure 3.
miR9678 Triggers PhasiRNA Biogenesis at the WSGAR Locus.
(A) Phasing score and read abundance distribution along WSGAR locus in embryos of germinating seeds (EGS) at 1, 6, and 12 HAI, and shoots (SH) and developing seeds at 15 DAP (SDAP15). Phasing score was calculated based on the mapping results of 21-nucleotide sRNAs following Xia’s method (Xia et al., 2013). Paring of miR9678 and target site is denoted in the top-left corner with the cleavage site marked with a red arrow. Gray gridlines show the 21-nucleotide phasing pattern set up by the miR9678 cleavage site.
(B) The position and expression level of WSGAR-derived phasiRNAs from the first phasing register. Each box represents one phasiRNA generated from the indicated position within WSGAR locus.
(C) Phased pattern of 21-nt siRNAs along WSGAR sequence after the cleavage guided by miR9678. The sense phasiRNAs are designated as 3′D1(+), 3′D2(+), 3′D3(+), etc., while the antisense phasiRNAs are designated in the same way, but with a minus symbol. Some phasiRNA variants produced from the WSGAR plus and minus strands are shown.
(D) Level of phasiRNA 3′D2(-) in various wheat tissues, as in Figure 2D, normalized to the U6 gene. Each bar in the graph corresponds to the mean value of three data points, which are shown by circles.
(E) Comparison of mature miR9678 (blue) and phasiRNA 3′D2(-) (yellow) level in embryos during seed maturation and germination. Data were normalized to the wheat U6 gene. Data are presented as a line connecting the average of three data points (reported as circles and triangles).
(F) Accumulation of miR9678, phasiRNA 3′D2(-), and WSGAR RNA determined in N. benthamiana leaves after transient transformation. The expression levels of miR9678 and phasiRNA 3′D2(-) were measured with RNA gel blotting. The 5S rRNA/tRNA bands were visualized via ethidium bromide staining of polyacrylamide gels, which served as a loading control. The expression level of WSGAR and GFP transcripts measured with RT-qPCR were normalized to the N. benthamiana ACTIN gene. Each bar in the graph corresponds to the mean value of three data points, which are shown as circles or triangles. The double asterisks represent significant differences determined by the Student’s t test at P < 0.01.
(G) Mapping of WSGAR cleavage sites by miR9678 using RLM-5′-RACE in N. benthamiana leaves agroinfiltrated with 35S:miR9678 and 35S:WSGAR. The red arrow represents the predicted cleavage site by miR9678. The black arrows indicate the 5′ ends of the miR9678-guided cleavage products, and numbers (5/8 and 3/8) are the ratios of the cleaved products with respect to total sequenced fragments.
Further experimental support for the finding that miR9678 induced production of 21-nucleotide phasiRNAs from WSGAR locus was achieved by transiently coexpressing the miR9678 precursor and WSGAR transcript in Nicotiana benthamiana leaf tissue (Supplemental Figure 2; Figure 3F). The results showed that mature miR9678 can be efficiently produced in N. benthamiana leaves and WSGAR-derived 3′D2(-) phasiRNA was only detected when vectors carrying WSGAR and miR9678 precursor were coexpressed (Figure 3F). Conversely, formation of 3′D2(-) was not detected when the assay used for WSGAR with a mutation resistant to miR9678-mediated cleavage (35S:WSGARm) (Supplemental Figure 2; Figure 3F). In addition, the level of WSGAR RNA was significantly lower when the miR9678 precursor and WSGAR were coexpressed compared with when the WSGAR transcript was expressed alone or when miR9678 cleavage-resistant WSGAR mutant was coexpressed with the miR9678 precursor (Figure 3F). Finally, RLM-5′-RACE assays showed that miR9678 precursor led to WSGAR RNA cleavage at the 10th or 11th nucleotide in N. benthamiana leaves coexpressing WSGAR transcript, which was consistent with cleavage in wheat germinating seeds (compare Figures 2B and 3G). These data suggested that miR9678 directed the degradation of WSGAR transcript and triggered secondary phasiRNA biogenesis.
The Level of miR9678 Is Negatively Associated with Seed Germination
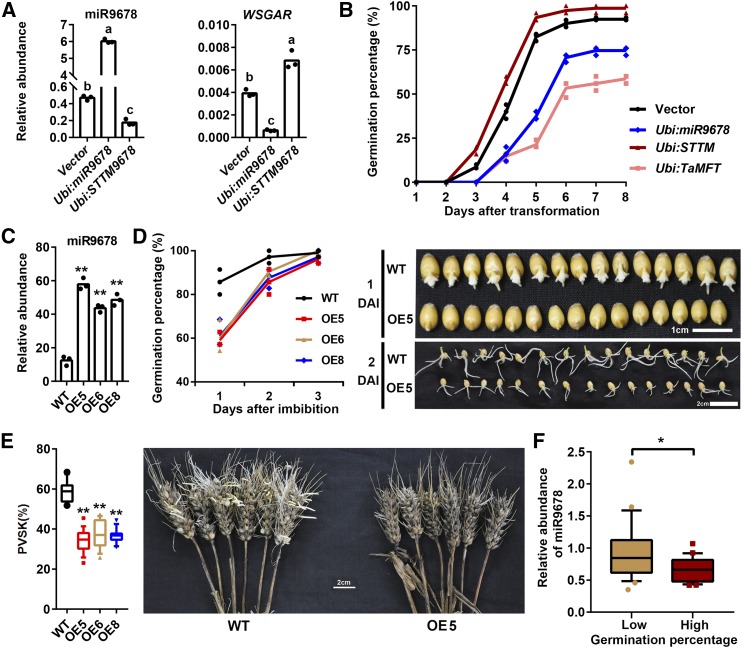
Based on its expression pattern (Figure 1), we postulated that miR9678 could be negatively associated with seed germination. To provide experimental evidence for this function, we transiently overexpressed or silenced miR9678 in cultured immature embryos and examined the effects on germination (Supplemental Figure 3). The TaMFT gene, which represses immature embryo germination when overexpressed (Nakamura et al., 2011), was used as positive control, and empty vector was used as a negative control. Overexpression of miR9678 (Ubi:miR9678) was associated with a decrease in WSGAR RNA and delayed germination of immature embryo (Figures 4A to 4C). Conversely, miR9678 silencing (Ubi:STTM9678) was associated with an increase in WSGAR RNA and increase in germination percentage. These results were corroborated by stable transformation of wheat plants overexpressing the miR9678 precursor pri-miR9678-7D under the control of the maize ubiquitin promoter (OE lines). A total of eight independent transgenic lines were obtained and three lines (OE5, OE6, and OE8) were selected for further analyses because they showed (1) higher miR9678 levels, (2) a decrease in WSGAR levels, and (3) an increase of phasiRNA 3′D2(-) levels (Figure 4C; Supplemental Figure 4). The results indicated that OE5, OE6, and OE8 plants exhibited a statistically significant reduction of the germination percentage at 1 and 2 d after imbibition (DAI) compared with the wild type (Figure 4D). Subsequently, we measured PHS of these lines as the percentage of visible sprouted kernels (PVSK) in a spike. The miR9678 OE lines exhibited a PVSK of 34.3 to 37.3%, while the wild type reached a PVSK of 58.5% (Figure 4E). Hence, miR9678 overexpression strongly improved resistance to PHS. In addition, we found that ectopic overexpression of miR9678 also affected developmental phenotypes, like plant height (Supplemental Figure 5A) and kernel size and weight (Supplemental Figure 5B), which decreased compared with the wild type. Since miR9678 was specifically expressed in the scutellum, we compared the scutellum structure between miR9678 OE5 line and the wild type, but no alteration was found (Supplemental Figure 6).
Figure 4.
miR9678 Is Negatively Associated with Seed Germination.
(A) Levels of miR9678 and WSGAR mRNA after transient transformation of wheat immature embryos with constructs overexpressing (Ubi:miR9678) or silencing (Ubi:STTM9678) miR9678. The RNA was extracted from immature embryos two days after transformation. Each bar in the graph corresponds to the mean value of three data points, which are shown by circles. Letters above the bar represent significant difference according to a least significant difference or Dunnett’s T3 post-hoc test (where equal variances were not assumed) (P < 0.05).
(B) Time course of the germination percentage after transformation with empty vector (black line), Ubi:miR9678 (blue line), Ubi:STTM9678 (red line), or Ubi:TaMFT (pink line) constructs. Data are represented as a line connecting the average of three data points (reported using different symbols for different samples: circle, triangles, etc.).
(C) Levels of miR9678 in the wild type and three miR9678 OE transgenic wheat lines were tested by RT-qPCR using embryos from germinating seeds at 6 HAI. Each bar in the graph corresponds to the mean value of three data points, which are shown by circles. The double asterisks represent statistically significant differences as determined by the Student’s t test at P < 0.01.
(D) Germination percentage of the wild type and three miR9678 OE transgenic wheat lines. Data are represented as a line connecting the average of three data points (reported using different symbols for different samples: circle, triangles, etc.). Representative pictures of seeds of the wild type and miR9678 OE5 at 1 and 2 DAI are also shown.
(E) Box plots summarize the percentage of visible sprouted kernels (PVSK) from 18 replicates of wild-type and miR9678 OE5, OE6, and OE8 plants. Lines in the box plots indicate the median. The 10th/90th percentiles of outliers are shown. The double asterisks represent significant differences determined by the Student’s t test at P < 0.01. Representative pictures of spikes from the wild type and OE5 are shown. Bar = 2 cm.
(F) Comparison of miR9678 abundance between wheat cultivars with high (≥80%) (n = 22) and low (≤30%) (n = 21) germination percentage at 1 DAI. Lines in the box plots indicate the median. The 10th/90th percentiles of outliers are shown. The single asterisk represents significant differences as determined by Student’s t test at P < 0.05. The expression of miR9678 was analyzed using embryos at 6 HAI and was normalized to the U6 gene.
As a further validation of our results, 22 wheat cultivars with high germination percentages (≥80%) and 21 cultivars with low germination percentages (≤30%) at 1 DAI were used to determine the association between miR9678 expression and seed germination. The results showed that miR9678 abundance at 6 HAI was higher in cultivars with low germination percentage compared with those with high germination percentage (Figure 4F). Altogether, our findings provided evidence that the level of miR9678 was associated with a delay in seed germination and improvement of PHS resistance.
The Level of miR9678-Triggered PhasiRNAs Is Negatively Associated with Seed Germination
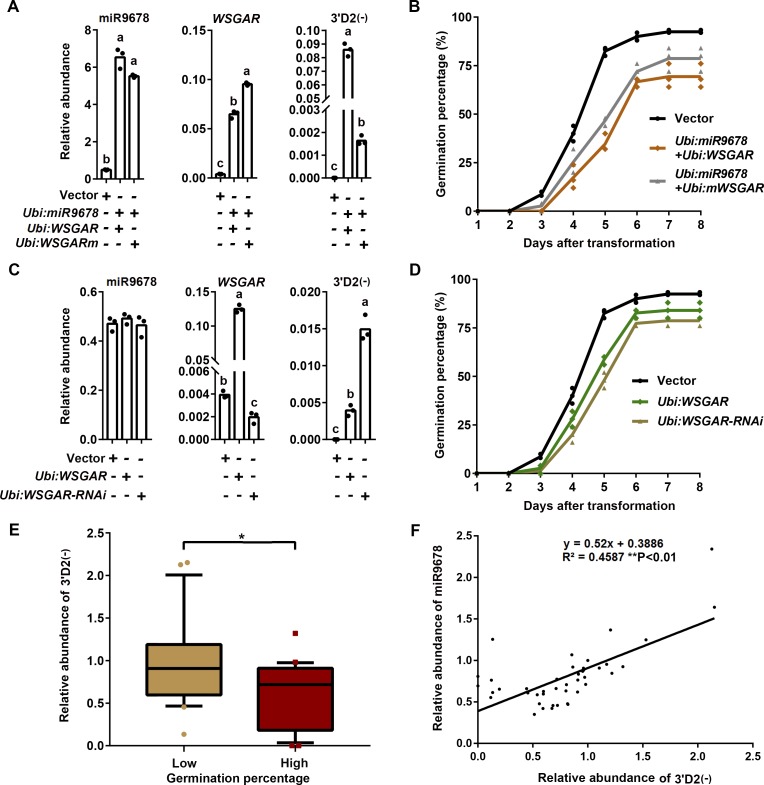
To test whether miR9678 negatively regulates seed germination through the production of WSGAR-derived phasiRNAs, we transiently transformed cultured wheat immature embryos using vectors that coexpressed miR9678 with WSGAR or with WSGARm resistant to miR9678-mediated cleavage (Supplemental Figure 3A). The accumulation of phasiRNA 3′D2(-), due to coexpression of miR9678 and WSGAR mRNA, induced a slight decrease of germination percentage compared with coexpression of miR9678 and WSGARm (Figures 5A and 5B). Furthermore, we observed that both transient overexpression of WSGAR transcript and its RNAi-directed silencing (Supplemental Figure 3) led to accumulation of phasiRNA 3′D2(-) (Figure 5C) because siRNAs from RNAi vectors were also located around miRNA cleavage site. Again, accumulation of phasiRNA 3′D2(-) was associated with a lower germination percentage (Figure 5D). Finally, the association of phasiRNA 3′D2(-) with germination was assessed in wheat cultivars with high and low germination percentage (see above). The phasiRNA 3′D2(-) level at 6 HAI was more abundant in seeds of cultivars with low germination percentage compared with those with high germination percentage (Figure 5E). In addition, the phasiRNA 3′D2(-) level was positively correlated with abundance of miR9678 (Figure 5F). Our findings indicated that the levels of miR9678-mediated phasiRNAs were negatively associated with seed germination.
Figure 5.
PhasiRNAs Are Negatively Associated with Seed Germination.
(A) Abundance of miR9678, WSGAR, and 3′D2(-) after Ubi:miR9678 transformation of immature embryos in combination with Ubi:WSGAR or Ubi:WSGARm. The RNA was extracted from immature embryos 2 d after transformation to perform RT-qPCR. Each bar in the graph corresponds to the mean value of three data points, which are shown by circles. Letters above the bar represent significant difference according to a least significant difference or Dunnett’s T3 post-hoc test (where equal variances were not assumed) (P < 0.05).
(B) Time course of the germination percentage after transformation with empty vector (black line), Ubi:miR9678 combined withUbi:WSGAR (orange line), or Ubi:miR9678 combined with Ubi:WSGARm (gray line) constructs. Data are represented as a line connecting the average of three data points (reported using different symbols for different samples: circle, triangles, etc.).
(C) Level of miR9678, WSGAR, and 3′D2(-) after Ubi:WSGAR or Ubi:WSGAR-RNAi transformation of immature embryos. RT-qPCR data are presented as the mean value of three data points, which are shown by circles. Letters above the bar represent significant difference according to a least significant difference or Dunnett’s T3 post-hoc test (where equal variances were not assumed) (P < 0.05).
(D) Time course of changes in percentage germination after transformation with empty vector (black line), Ubi:WSGAR (green line), or Ubi:WSGAR-RNAi (brown line). Data are presented as in (B).
(E) Comparison of 3′D2(-) expression levels between the wheat cultivars with high (≥80%) (n = 22) and low germination percentage (≤30%) (n = 21) used in Figure 4F. The abundance of 3′D2(-) was normalized to the wheat U6 gene. Lines in the box plots indicate the median. The 10th/90th percentiles of outliers are shown. The single asterisk represents significant differences as determined by Student’s t test at P < 0.05.
(F) Correlation of expression levels of miR9678 and 3′D2(-). The expression levels of miR9678 and 3′D2(-) were analyzed using embryos at 6 HAI in 43 wheat cultivarsand were normalized to the wheat U6 gene.
miR9678 Affects Expression of Genes Involved in Germination and GA Homeostasis
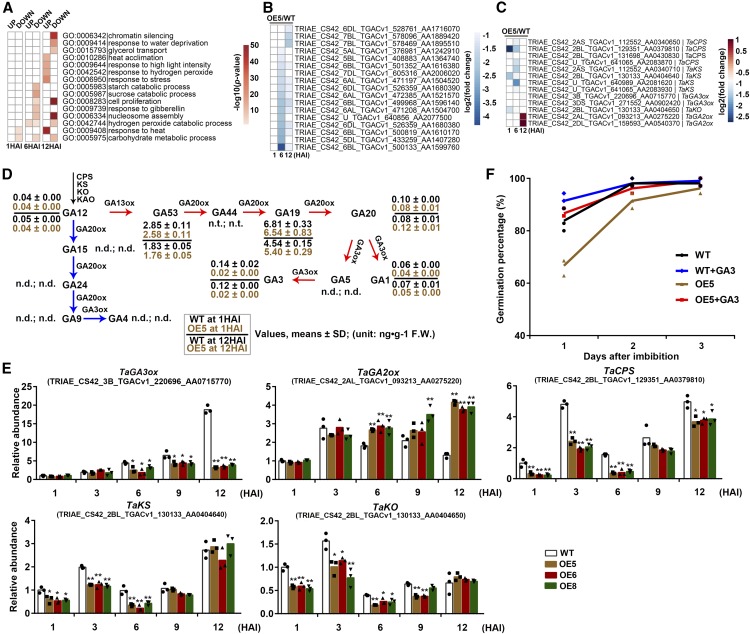
To determine the downstream targets regulated by miR9678, high-throughput RNA sequencing (RNA-seq) was performed to identify differences in transcript level between wild-type and OE5 seeds. This analysis was done using RNA extracted from embryos of germinating seeds at 1, 6, and 12 HAI. We found that a total of 820, 868, and 1290 genes were downregulated in 1, 6, and 12 HAI OE5 compared with wild-type seeds (|log2(fold change)| ≥ 0.8; P < 0.01), respectively (Supplemental Data Set 1). A total of 488, 425, and 682 genes were upregulated in the same samples. Gene Ontology (GO) functional annotation analysis revealed that genes downregulated in 6 HAI OE5 seeds were mainly enriched for participation in GA response and starch and sucrose catabolism, while in 12 HAI OE5 seeds were highly enriched for participation in cell proliferation and nucleosome assembly (Figure 6A). Genes downregulated in the OE5 line included 17 sequences encoding α-amylase (Figure 6B), consistent with the delayed germination of OE5 seeds. Indeed, the expression of α-amylase is triggered by active GAs and the enzymes are secreted from the aleurone layer into the endosperm to catalyze the hydration reaction of stored starch, thus providing a carbohydrate source used during germination (Huang and Varriano-Marston, 1980). In OE5 seeds, 11 genes encoding key enzymes of the GA biosynthesis pathway were also downregulated. These genes include four ENT-COPALYL DIPHOSPHATE SYNTHASE (TaCPS) genes, four ENT-KAURENESYNTHASE (TaKS) genes, ENT-KAURENE OXIDASE (TaKO), and two GA3-OXIDASE (TaGA3ox) genes (Figure 6C). Finally, we noticed that the expression of two GA2-oxidase genes, which encode the TaGA2ox enzyme catalyzing GA deactivation reaction, were upregulated in 12 HAI OE5 seeds (Figure 6C). Therefore, we speculated that miR9678 delayed seed germination through inhibiting GA biosynthesis and increasing GA degradation. Overall, the results from RNA-seq analysis indicated that miR9678 overexpression affected the expression of genes involved in germination.
Figure 6.
miR9678 Affects Expression of Genes Encoding α-Amylase and Enzymes Involved in Bioactive GA Biogenesis.
(A) GO analysis of genes differentially expressed in 1, 6, and 12 HAI seeds of the wild type and OE5. The color in each cell indicates log10 (P values) of the GO enrichment and a blank cell indicates no significant difference.
(B) Differential expression of α-amylase genes at 1, 6, and 12 HAI in wild-type and OE5 seeds was identified using RNA-seq analysis and is illustrated in the heat map.
(C) Differential expression of GA biogenesis and deactivation genes at 1, 6, and 12 HAI in wild-type and OE5 seeds was identified using RNA-seq analysis and is illustrated in the heat map.
(D) Levels of each of the indicated molecules from the GA biosynthesis pathway in seeds from the wild type and the miR9678 OE5, at 1 and 12 HAI. FW, fresh weight; n.d., not detected; n.t., not tested. The blue arrows indicate GAs from the non-13-hydroxylation pathway, which were below the detection limit under the tested experimental conditions. The red arrows indicated the early 13-hydroxylationpathway (GA53, GA19, GA20, GA1, and GA3). The values shown are averages from three independent replicates and the sd is indicated (n = 3).
(E) RT-qPCR quantification of RNA levels of TaGA3ox, TaGA2ox, TaCPS, TaKS, and TaKO in seeds of the wild type and OE5, OE6, and OE8 at 1, 3, 6, 9, and 12 HAI. The abundance was normalized to the ACTIN gene, and the abundance of the wild type at 1 HAI was set as 1. Each bar in the graph corresponds to the mean value of three data points, which are shown with different symbols for different samples. The single and double asterisks represent significant differences as determined by the Student’s t test at P < 0.05 and P < 0.01, respectively.
(F) Germination percentage of wild-type and OE5 seeds treated with exogenous GA3. Dry seeds were submerged in GA3 (60 mg/L) for 3 d, and germination percentages were calculated at daily intervals. Data are represented as a line connecting the average of three data points (reported using different symbols for different samples: circle, triangles, etc.).
Since the genes involved in GA biosynthesis were downregulated by miR9678 overexpression (Figure 6C) and seed germination is predominantly regulated by the balance of GA and ABA levels (Shu et al., 2016), we measured ABA and GA contents in seeds of the wild type and the OE5 line. No differences in ABA content between wild-type and OE5 seeds were detected (Supplemental Figure 7A). Conversely, we did detect changes in the levels of bioactive GAs (Figure 6D). The levels of major endogenous GA metabolites were assessed in seeds at 1 and 12 HAI when seeds were undergoing imbibition and protrusion, respectively. At these stages, the levels of all GAs of the non-13-hydroxylation pathway (GA15, GA24, GA9, and GA4) were below the detection limit. However, we could detect and analyze levels of GAs from the early 13-hydroxylationpathway (GA53, GA19, GA20, GA1, and GA3) (Figure 6D). The levels of bioactive GA1 and GA3 decreased in the OE5 line compared with the wild type. In agreement with this, the abundance of TaGA3ox, which encodes an enzyme catalyzing the biosynthesis of GA3 and GA1 from GA19 and GA20 (Yamaguchi, 2008), was decreased in three OE lines including OE5, OE6, and OE8, compared with the wild type (Figure 6E). Moreover, the abundance of TaGA2ox, which encodes the TaGA2ox enzyme catalyzing the GA deactivation reaction, was upregulated in OE5, OE6, and OE8 at 6 and 12 HAI, compared with the wild type (Figure 6E). The content of GA12, the common precursor of all GAs (Graeber et al., 2014), also decreased in 12 HAI seeds of the OE5 line (Figure 6D). This reduction was consistent with the lower expression of TaCPS, TaKS, and TaKO in all three OE lines (Figure 6E). TaCPS is involved in the conversion of geranylgeranyl diphosphate to the tetracyclic hydrocarbon intermediate ent-kaurene, while TaKO and TaKS catalyze the biosynthesis of GA12 (Yamaguchi, 2008; Hedden and Thomas, 2012). The level of GA53, the precursor of the early 13-hydroxylation pathway (Yamaguchi, 2008), was decreased in OE5 seeds only at 1 HAI (Figure 6D). In addition, in 12 HAI seeds, the levels of GA19 and GA20, precursors of GA1 and GA3 (Yamaguchi, 2008), were higher in OE5 than in the wild type. These results suggested that miR9678 overexpression caused seeds to require a high GA levels for germination. To confirm this result, OE5, OE6, and OE8 seeds were treated with exogenous GA3 and we observed that their delayed germination was restored to the wild-type percentage (Figure 6F; Supplemental Figures 7B and 7C).
miR9678 Overexpression Does Not Affect Transcriptome Changes through Transcript Cleavage Mediated by WSGAR-Derived PhasiRNAs
The RNA-seq expression analysis showed that miR9678 overexpression was associated with transcriptome changes of various genes related to seed germination. To shed light on the mechanism of miR9678 activity, considering that phasiRNAs in Arabidopsis act in trans (tasiRNAs) to mediate cleavage of their target transcripts (Vazquez et al., 2004), we assessed whether some of transcriptome changes observed in the miR9678 OE5 line were due to the RNA cleavage mediated by WSGAR-derived phasiRNAs. To this end, we sequenced two degradome libraries, produced from a mix of wild-type germinating seeds collected at 1, 3, and 6 HAI or at 9, 12, and 24 HAI [i.e., when the level of both miR9678 and phasiRNA 3′D2(-) were higher, Figures 2E and 3E]. Although two α-amylase genes and TaGA3ox, exhibiting downregulation in OE5 versus the wild type, contained binding sites for the WSGAR-derived phasiRNAs, none had cleavage verified by high confidence degradome sequencing data (Supplemental Data Set 2 and Supplemental Figure 8) or RLM-5′-RACE (Supplemental Figure 8). These data indicate that, although WSGAR-derived phasiRNAs were negatively associated with seed germination, the transcriptome changes in miR9678 OE lines appeared not to be caused by direct RNA cleavage mediated by WSGAR-derived phasiRNA. Hence, similar to other phasiRNAs (Fan et al., 2016), WSGAR-derived phasiRNAs possibly act through other unknown mechanisms.
Proteins Responsive to ABA Signaling Regulate miR9678 Expression
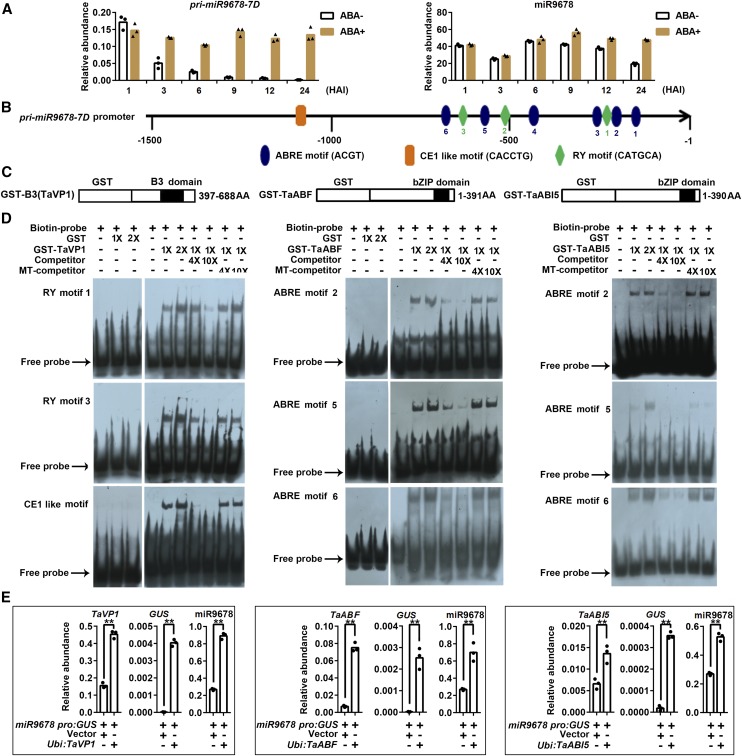
To detect if ABA affects miR9768 expression, we treated germinating wheat seeds with ABA. Both pri-miR9678-7D and mature miR9678 were upregulated after ABA treatment (Figure 7A), indicating that miR9678 responded to the ABA signaling pathway. Analysis of the pri-miR9678-7D promoter sequence identified multiple sequence motifs potentially bound by components of the ABA signaling pathway. These motifs included ABA response elements (ABREs), Sph (RY) elements, and Coupling Element 1 (CE1)-like (Figure 7B; Supplemental Figure 9). In Arabidopsis, ABSCISIC ACID INSENSITIVE5 (ABI5) binds ABREs and ABI3 binds RY and CE1-like motifs, thus activating the response to the ABA signaling pathway (Suzuki et al., 2005). Accordingly, we used electrophoretic mobility shift assays (EMSAs) and GST-fused recombinant proteins to test whether wheat homologs of these proteins could also bind the pri-miR9678-7D promoter (Figure 7C). Various combinations of labeled and unlabeled probes were used in these experiments (Figure 7D). GST-fused TaVP1, a wheat homolog of maize viviparous 1 (VP1) and of Arabidopsis ABI3, bound two of three RY elements and the unique CE1-like elements (Figure 7D; Supplemental Figure 10). The specificity of the binding was corroborated by observation that a molar excess of unlabeled probe prevented the band shift in the EMSA, but mutated variants did not. Similarly, three out of the six ABREs located in the pri-miR9678-7D promoter were specifically bound by GST-TaABF and GST-TaABI5 (Figure 7D; Supplemental Figure 10). The binding of the three components of the wheat ABA signaling pathway and its effect on pri-miR9678-7D promoter expression was further analyzed by transient transformation assays of wheat immature embryos. The pri-miR9678-7D promoter-GUS construct cotransformed with vectors overexpressing TaVP1, TaABF, or TaABI5 induced a statistically significant increase of GUS and endogenous miR9678 levels (Figure 7E). Overall, our findings indicated that the pri-miR9678-7D promoter was bound and activated by components of the ABA signaling pathway, suggesting that miR9678 mediated the crosstalk between ABA and GA hormones to affect seed germination in wheat.
Figure 7.
TaVP1, TaABI5, and TaABF Bind to the Promoter of the miR9678 Precursor.
(A) Analysis of miR9678 and pri-miR9678-7D transcript accumulation in germinating seeds at 1, 3, 6, 9, and 12 HAI with and without ABA (70 mg/L) treatment. miR9678 and pri-miR9678-7D abundance was normalized to that of U6 and ACTIN, respectively. Each bar in the graph corresponds to the mean value of three data points, which are shown as circles or triangles.
(B) Six ABREs, ACGT (blue ovals); three Sph (RY) elements, CATGCA (green squares); and one Coupling Element 1 (CE1) like, CACCTG (orange rectangle), were located in the promoter of pri-miR9678-7D.
(C) Schematic diagrams of the GST fusion constructs used in the EMSA assays.
(D) EMSAs of the recombinant proteins fused to GST, with nine biotin-labeled probes, competitor probes (biotin-unlabeled), and mutant competitor probes (MT-competitor) derived from the miR9678 promoters. The sequences of the probes and mutant competitors are shown in Supplemental Figure 9. “+” and “−” indicate the presence and absence of corresponding probes, proteins or competitors. “1×” and “2×” indicate the amount of proteins in the reaction. “4×” and “10×” indicate 4- and 10-fold molar excess of competitor probes relative to biotin-labeled probes.
(E) TaVP1, TaABF, and TaABI5 induced miR9678 expression. The pri-miR9678-7D promoter-GUS (miR9678pro:GUS) construct was transiently coexpressed with Ubi:TaVP1, Ubi:TaABF, or Ubi:TaABI5 in immature embryos. Levels of TaVP1, TaABI5, TaABF, and GUS transcript were normalized to that of the wheat ACTIN gene, while miR9678 was normalized to that of U6. The RNA was extracted from immature embryos 2 d after transformation. Each bar in the graph corresponds to the mean value of three data points, which are shown by circles. The double asterisks represent significant differences determined by the Student’s t test at P < 0.01.
DISCUSSION
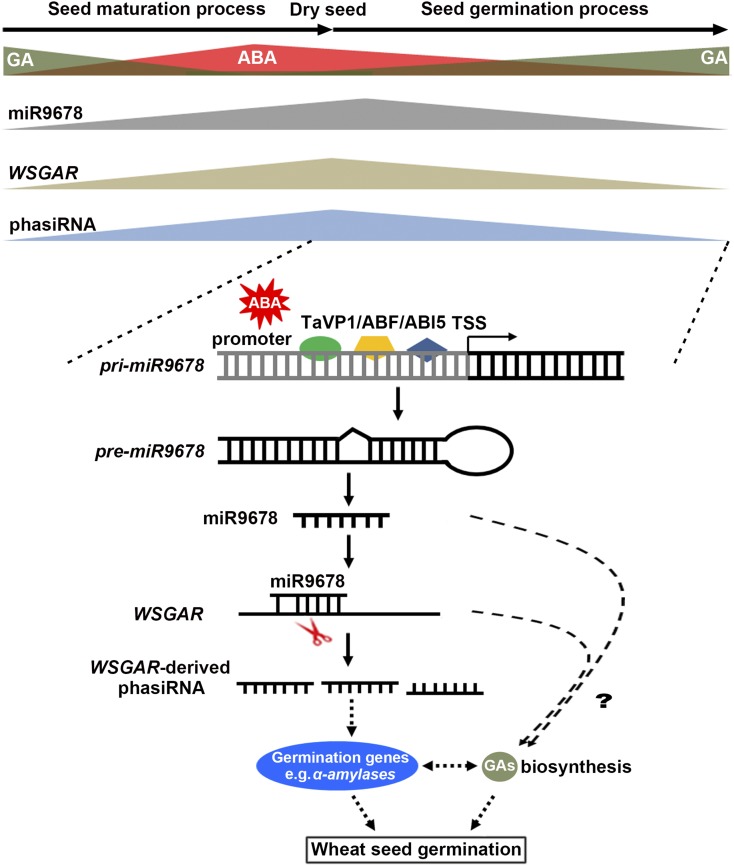
In this work, we provide evidence that 22-nucleotide wheat miR9678 is specifically expressed in the scutellum of germinating seeds and that its accumulation negatively correlates with germination. In addition, miR9678 induces the production of phasiRNAs from a long noncoding RNA WSGAR, and phasiRNA production is also negatively associated with germination. Furthermore, we show that miR9678 accumulation in transgenic miR9678 overexpressing lines (OE lines) reduces the bioactive GA content and affects the expression of genes that are associated with proper germination. Based on these results, we infer that this wheat-specific miR9678-WSGAR-phasiRNA pathway plays a role in regulating seed germination through influencing GA signaling (Figure 8). In this pathway, ABA is also involved in the regulation of miR9678 production. ABA promotes the expression of miR9678, and the production of miR9678 negatively affects GA biosynthesis and the expression of genes involved in seed germination, probably through its experimentally proven target gene WSGAR and subsequently generated phasiRNAs (Figure 8). In general, the regulatory circuit of miR9678-WSGAR-phasiRNA plays an important role in the regulation of seed germination in wheat by mediating the ABA-GA balance. However, a few questions remain to be addressed to fully understand how this pathway affects seed germination.
Figure 8.
Speculative Model for the Mechanism of miR9678-Mediated Regulation of Germination via Modulation of ABA-GA Crosstalk.
A timeline describing accumulation of ABA, GAs, miR9678, WSGAR RNA, and WSGAR-derived phasiRNAs during various stages, including seed maturation, dry seed, and germination is depicted at the top. During seed maturation, the miR9678 promoter is bound by major regulators of ABA signaling (i.e., TaVP1/TaABF/TaABI5). Subsequently, miR9678 negatively affects expression of genes involved in GA biosynthesis and germination through WSGAR-derived phasiRNAs or other unknown routes, which leads to a reduction in bioactive GAs and α-amylase levels, and thus is negatively associated with seed germination. The question mark and dashed lines indicate routes and aspects of model that still require experimental support. See Discussion for further details.
One major outstanding question is how the phasiRNAs derived from WSGAR cause the effect on seed germination, i.e., what the target genes of these phasiRNAs are. So far, although a couple of phasiRNAs or tasiRNAs are known to function in trans to target certain genes (Vazquez et al., 2004), the majority are functionally uncharacterized. We set out to search for the downstream target genes of WSGAR-derived phasiRNA using genome-wide target prediction in combination with degradome sequencing and RLM-5′-RACE. The candidate target genes obtained did not belong to a certain gene family or functional pathway, nor did they group as tasiRNAs usually do. Moreover, no clear degradome or RLM-5′-RACE results were found to support the cleavage of predicted target genes. Recently, a similar case was reported in rice, in which PMS1, a long noncoding RNA targeted by miR2118, was found to be associated with photoperiod-sensitive male sterility (Fan et al., 2016). PMS1T generates phasiRNAs, and although its results suggested an association between the abundance of the PMS1T-phasiRNA and male sterility under long-day conditions, this study also failed to uncover the downstream target genes of PMS1T-derived phaiRNAs, using techniques similar to ours (Fan et al., 2016). Therefore, it is likely that phasiRNAs may function through other unknown mechanisms, rather than guiding mRNA cleavage. One possibility is translational inhibition, a way that sRNAs commonly function in other species (Lanet et al., 2009; Yu and Wang, 2010; Ma et al., 2013). WSGAR-derived phasiRNAs could inhibit the translation of genes critical for GA homeostasis, thus decreasing seed germination. Indeed a few genes involved in GA-biogenesis have potential target sites for WSGAR-derived phasiRNAs (Supplemental Figure 8). We speculate that the decreased expression of GA homeostasis-related genes is likely a feedback effect of the reduction of mRNA translation efficiency by WSGAR-derived phasiRNAs, as mRNA degradation can be modulated by alterations in translation (Huch and Nissan, 2014). Reduced translation can drive mRNAs toward degradation, due to the assembly of decay factors that replace the translation initiation machinery, reinforce the initial repression, and enhance decay rates (Huch and Nissan, 2014). The second mechanism of WSGAR-derived phasiRNAs is action in cis, i.e., phasiRNAs can target their parental genes where they originate. We detected moderate signal for out-of-phase siRNAs produced from WSGAR (Figure 3A), indicating that the cis-function of phasiRNAs is indeed relevant for WSGAR. However, the cis-function is just an intermediate step of this regulatory circuit; the subsequent steps that connect cis-function to GA homeostasis remain a mystery.
Our data provide evidence that miR9678 affects seed germination depending on WSGAR-derived phasiRNAs, but we cannot exclude the possibility that other mechanisms besides miR9678-mediated cleavage could affect the abundance of WSGAR and 3′D2(-) or miR9678 might target genes other than WSGAR due to the availability of limited wheat genomic resources (e.g., incomplete genome sequences), and in this case the effects on GA homeostasis and eventually seed germination might result from a combination of effects on the function of different miR9678 target genes, not solely caused by WSGAR-derived phasiRNAs. In fact, in OE5 plants, we detected the downregulation of α-amylase genes and genes required for the production of specific GAs (i.e., TaCPS, TaKO, and TaKS) that were not among the putative targets of the WSGAR-derived phasiRNAs or miR9678. In addition, we must also consider that miR9678 overexpression was achieved using a constitutive promoter and that this might have led to pleiotropic effects. Indeed, OE5 lines exhibit additional developmental defects, along with the delay in germination. The constitutive miR9678 overexpression may also account for a fraction of the transcriptome changes we found in OE5 line. However, we validated the change in mRNA level of genes known to be involved in GA biosynthesis and in germination, as well as germination phenotypes, in three independent miR9678-overexpressing lines. Hence, we believe that our findings provide sufficient support to conclude the involvement of miR9678-WSGAR-phasiRNAs in affecting seed germination. Further work will be required to fully elucidate the underlying mechanisms, including the possible use of a scutellum-specific promoter and the production of miR9678-null mutants, along with the development of new techniques for the characterization of phasiRNA target genes.
We found evidence supporting a model in which miR9678 acts as a “mediator” of the crosstalk between ABA and GA in regulating seed germination in wheat (Figure 8). miR9678 promoter is bound and activated by TaVP1/TaABF/TaABI5, which have been reported as major regulators of ABA signaling. In addition, the expressions of pri-miR9678-7D and mature miR9678 are upregulated after ABA treatment. These findings suggest that miR9678 affects bioactive GA content and germination is responsive to ABA signal. This observation is in agreement with previous findings that some genes/proteins affect the ABA and/or the GA pathways to regulate seed germination. For example, the Arabidopsis FUSCA3 B3 transcription factor is essential for seed maturation and negatively regulates GA levels (Curaba et al., 2004; Gazzarrini et al., 2004). Maize VP1 can mediate the repression of genes downstream of GAs, such as α-amylase in aleurone cells (Hoecker et al., 1999). The sorghum (Sorghum bicolor) ABA signaling components SbABI4 and SbABI5 interact in vitro with a fragment of the SbGA2-OXIDASE3 promoter containing an ABA-responsive complex (Cantoro et al., 2013). In wheat, TaMFT was exclusively expressed in the scutellum and coleorhiza and plays an important role in the regulation of germination through suppression of GA synthesis (Nakamura et al., 2011). However, all these previous studies provide only partial information about the possible mechanisms involved in the mediation of the ABA/GA balance during germination. Here, we provide more direct evidence about the mechanism that links ABA-miR9678-GAs ultimately affecting germination.
PHS causes harvest losses and, more importantly, a reduction in end-product quality (Mares and Mrva, 2014). Direct annual losses caused by PHS approach $1 billion worldwide (Black et al., 2006). Therefore, avoidance of PHS with a concomitant increase of germination rate remains an important goal for wheat breeding. In this context, our study provides important information about potential targets for breeding programs. For example, we showed that expression of miR9678 and phasiRNA 3′D2(-) has a negative relationship with germination rate in 43 modern wheat cultivars; therefore, alleles associated with desirable levels of expression of miR9678 and phasiRNA 3′D2(-) could be used in marker-assisted selection to fine-tune germination. Furthermore, the observation that miR9678 overexpression reduces germination and improves resistance to PHS suggests that artificial regulation of this miRNA could be used to modulate the germination rate in wheat. However, miR9678 overexpression induces pleiotropic effects, negatively affecting traits important for yield and quality, such as kernel size and weight. Therefore, this study represents the starting point for additional investigations to address the extent of miR9678 involvement in seed germination and to optimize strategies to improve PHS resistance.
METHODS
Plant Materials and Phenotypic Analysis
The name and geographical origins of all wheat (Triticum aestivum) cultivars used in this research are listed in Supplemental Data Set 3. Plant materials used for molecular analysis were grown in a growth chamber at a relative humidity of 75% and 26/20°C day/night temperatures, with a light intensity of 3000 lux (Master GreenPower CG T 400W E40; Philips). Embryos from developing seeds at 5, 10, 15, 20, 25, 30, and 45 DAP were dissected. Seeds harvested at 45 DAP were dried and placed at room temperature for 38 d and then were soaked in a Petri dish for germination and the embryos from germinating seeds that had soaked for 1, 3, 6, 9, 12, 24, 36, 48, and 72 h were dissected. Shoots were harvested from 5-d-old seedlings, when the leaves extended to the tip of the coleoptiles. Seedling leaves and roots were collected from seedlings when the third leaf had emerged by at least 50%. Young spikes (2–5 mm and 20–50 mm) were separated from the top of the node. Flag leaves were cut when spikes were at the beginning of flowering, and seeds of 4, 8, and 12 DAP were collected. For each sample, tissues from three different planting pots or Petri dishes were harvested as three groups of biological replicates. All samples were immediately frozen in liquid nitrogen and stored at −80°C until further use.
Wheat cultivars used for seed reproduction and for phenotypic analysis were grown in the field from 2013 to 2015 at three different locations including the experimental field of China Agricultural University in Beijing, Shijiazhuang in the Hebei province, and Sanyuan in the Shannxi province. A total of 15 individual plants were used for phenotypic analysis.
Small RNA Gel Blot and RT-qPCR Analysis
Small RNA gel blot analysis was performed as previously described (Sun et al., 2014). TaqMan small assay probes were ordered from Applied Biosystems to quantify U6 (probe ID:CS70LBK), miR9678 (probe ID:CSI1LU5), and 3′D2(-) (probe ID: CSWR1RG) levels, following the manufacturer’s instructions. Ten nanograms of total RNA was used to synthesize cDNA from small RNA with 1.5 μL of 10× reverse transcription buffer, 0.15 μL of 100 mM deoxynucleotide triphosphates, 0.19 μL of an RNase inhibitor (20 units/μL), 1μL of MultiScrib Reverse Transcriptase (50 units/μL), and 3 μL of a stem-loop reverse transcription primer (Applied Biosystems) in a 15-μL reaction mixture. The real-time PCR assay was performed in a 20-μL reaction mixture containing 1.33 μL of product from the reverse transcription reaction, 10 μL of TaqMan 2× Universal PCR master mix, and 1 μL of 20× TaqMan assay mix, including miRNA-specific primers and the small RNA-specific TaqMan MGBprobe. The reaction was incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Reaction products were normalized to the expression level of the U6 gene (GenBank: X63066.1). A Mir-XmiRNA first-strand synthesis kit and SYBR Advantage qPCR Premix (TaKaRa) were used for the miR169 (positive control) and WSGAR-derived phasiRNAs reverse transcription reactions and RT-PCR according to the manufacturer’s instructions. PCR conditions were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 58°C for 15 s, 72°C for 20 s, and then 72°C for 5 min. The relative expression of miR169 was normalized to the wheat ACTIN gene in each sample. Differences in the relative transcript amounts were calculated using the 2−ΔCT method. For each sample, the PCR amplification was repeated three times, and the average values of 2−ΔCT were used to determine the differences of gene expression by Student’s t test. Three biological replications were performed with similar results and one replication was shown.
All primers are listed in Supplemental Data Set 4.
GUS Staining
The 2-kb sequence from the pri-miR9678-7D promoter was cloned into the plant GUS reporter expression vector pBGWSF7 (Invitrogen) and used for transformation in wheat cultivar Kenong199. The seeds, roots, leaves, stems, spikes, and glumes from wheat transgenic lines were cut, vacuum-infiltrated with staining buffer (2 mM potassium ferricyanide, 10 mM phosphate buffer, 8 mM EDTA, 0.5% Triton X-100, and 1 mg mL−1 X-Gluc), and subsequently incubated overnight at 37°C. The tissue was then incubated in ethanol:acetic acid (1:1) for 6 h and washed in 80% ethanol. A total of 50 T0 generation transgenic lines were stained and 10 with high GUS expression were selected for T3 generation reproduction. For homozygous lines, 20 seeds from each plant were stained. The staining results from T3 plants were shown in the text. The samples were observed using light microscopy (Olympus SEX16).
Degradome Sequencing
Prediction of miR9678 and phasiRNAs targets were performed using psRNATarget (http://plantgrn.noble.org/psRNATarget/). The parameters were set as follows: maximum expectation of 3 for phasiRNAs and 5 for miR9678; length for complementarity scoring of 20; target accessibility: (1) allowed maximum energy to unpair the target site as 30.0; (2) flanking length around target site for target accessibility analysis, 17 bp upstream and 13 bp downstream; (3) range of central mismatch leading to translational inhibition: 9 to 11 nucleotides. These predicted targets were further analyzed by degradome sequencing.
Two degradome libraries were constructed using pools of equal amounts of RNA from the embryo samples either at 1, 3, and 6 HAI or at 9, 12, and 24 HAI. Briefly, the library was constructed as follows: Approximately 200 ng poly(A)-enriched RNA was annealed with biotinylated random primers. After RNA fragments were streptavidin captured using biotinylated random primers, the RNAs with monophosphate at 5ʹ end were ligated to a 5ʹ adaptor. First-strand cDNA was generated from the ligated sequence after reverse transcription using random hexamer 3′ primer. A number of DNA products were produced by PCR amplification. The library was single-end sequenced using an Illumina HiSeq 2500 platform at LC-BIO following the vendor's recommended protocol. About 40 M high-quality single-end reads 50 bp in length were generated from each library.
For the degradome sequencing data, the reads with low quality and matched to other noncoding RNA such as rRNA, tRNA, and small nuclear RNA were removed using Sickle (https://github.com/najoshi/sickle) with parameters: se mode; -t sanger –q 20 –l 18. The remained reads were aligned to the wheat genome (TGACv1), allowing no more than one mismatch. Targets of miR9678 were identified using CleaveLand (version 4.3) (Addo-Quaye et al., 2009; Brousse et al., 2014). The target plot (t-plot) was generated by the CleaveLand software with default parameters. The x axis represented the nucleotide position on the target transcript. The y axis represented the intensity (number of degradome reads) of the corresponding positions based on the degradome data. The predicted cleavage sites with number of associated reads are denoted as red dots in the t-plots. (Supplemental Figure 8)
Modified 5′- and 3′-RACE
To acquire the full-length WSGAR cDNA sequence, we performed 5′-RACE and 3′-RACE experiments following the manufacturer's instructions accompanying the GeneRacer kit (Invitrogen). To map the cleavage sites in the WSGAR transcript, we performed modified RLM-5′-RACE following the manufacturer's instructions of the GeneRacer kit (Invitrogen). The cDNA templates were amplified through two rounds of PCR with the universal sense or antisense primers provided in the kit and two gene-specific primers (Supplemental Data Set 4). The PCR products were cloned into the pEASY vector (TransGen) and sequenced.
Infiltration of Nicotiana benthamiana with Agrobacterium tumefaciens
The sequences of the miR9678 precursor pri-miR9678-7D, WSGAR, WSGARm, and GFP were cloned into the pSuper1300 vector and placed under the control of the CaMV 35S promoter. The miR9678 binding site mutation in WSGARm was generated using PCR with mutated primers, and all primers used in these assays are listed in Supplemental Data Set 4. All vectors were introduced into Agrobacterium strain GV3101, and the bacteria were injected into N. benthamiana leaves with a syringe. For coinjection with two or three different constructs, the bacteria were resuspended in infiltration medium (10 mM MgCl2 and 10 mM MES containing 150 mM acetosyringone) at OD600 = 1 and incubated for 3 h at room temperature. The leaf zones of infiltration were harvested for RNA isolation 3 d postinjection. The 35S:GFP construct was used as a control for co-agroinfiltration.
Transient Expression Assay in Immature Embryos
The sequences of the miR9678 precursor pri-miR9678-7D, WSGAR, WSGARm, STTM9678, and TaMFT were cloned into the pWMB003 vector, under the control of the maize ubiquitin promoter (provided by Xingguo Ye, Chinese Academy of Agricultural Science). STTM9678 was designed and constructed as described by Yan et al. (2012). The partial sense and antisense sequences of WSGAR were ligated into the pWMB006 vector to generate the Ubi:WSGAR-RNAi construct. The wheat cultivar CB037 (provided by Xingguo Ye, Chinese Academy of Agricultural Science) was used for transformation. The detailed procedure for the transient gene expression assay in immature embryos was described by Nakamura et al. (2011).
For cotransformation of TaVP1, TaABF, or TaABI5 with promoter of pri-miR9678-7D, the cDNAs were cloned into the pWMB003 vector downstream of ubiquitin promoter, while pri-miR9678-7D promoter was cloned upstream GUS gene in the pBGWSF7 vector (Invitrogen). The vectors were mixed at a ratio of 1:1 and used for cotransformation.
Wheat Transformation
The pri-miR9678-7D genomic sequence was cloned into the vector pAHC25 (Taylor et al., 1993), allowing its ectopic overexpression under the control of the maize ubiquitin promoter. The plasmid was transformed into immature embryos of wheat cultivar Jimai5265 using particle bombardment. Since pAHC25 vector contained the phosphimothricin acetyl transferase gene (bar), transformants were selected using Basta herbicide. Genomic integration of the transgene in each generation was confirmed by PCR screening for either bar or the transformed genes. The T3 and T4 transgenic generation lines were used for experiments.
Germination Tests and PHS Measurement
To evaluate germination and sprouting rates, wheat seeds and spikes were harvested at physiological maturity as characterized by the loss of green color on the spike. Then, the air-dried seeds were stored in a freezer at −20°C to maintain dormancy and the air-dried spikes were immediately used to test sprouting rates. All germination tests were performed in three independent replicates. For seed germination, 35 seeds were sown in 3.5-cm Petri dishes containing 4 mL distilled water. The dishes were incubated in the dark for 1 to 3 days at 22°C. Seeds were considered to be germinated when radicle protrusion was visible and germination percentages were calculated at daily intervals. In the sprouting rate test, spikes were immersed in distilled water for 3 h and then bundled with wet gauze and placed in a plastic bag before they were incubated in a moist chamber at 22°C. After 7 d of incubation, geminated spikes were dried at 100°C for 2 h and then at 80°C for 12 h in an oven. Dried spikes were opened by hand, and germinated and nongerminated seeds in each spike were counted.
Quantification of Endogenous GA
For GA measurements, the seeds of the wild type and miR9678 OE5 (T4 generation transgenic plants) were placed in 9-cm Petri dishes containing 15 mL distilled water in the dark at 22°C. Embryos were harvested at 1 and 12 HAI. Quantification of endogenous GAs was performed as previously described (Chen et al., 2012). Each experiment was performed in three replications.
RNA-Seq
Total RNA was isolated from the embryos of the wild type and the miR9678 OE5 line (OE5) at 1, 6, and 12 HAI using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For each sample, 35 seeds were harvested and divided into three groups for three biological repeats. Bar-coded cDNA libraries were constructed using Illumina Poly-A Purification TruSeq library reagents and sequenced on Illumina HiSeq 2500 platform. About 10 Gb of high-quality 125-bp paired-end reads were generated from each library. The overall sequencing quality of the reads in each sample was evaluated by FastQC software (http://www.bioinformatics.babraham.ac.uk). Poor-quality bases were removed using Sickle (Joshi and Fass, 2011) in its paired end model with parameters: -t sanger –q 20 –l 50. The remaining reads were aligned to the wheat reference genome sequence (TGACv1; http://plants.ensembl.org/Triticum_aestivum/Info/Index) (Clavijo et al., 2017) using the default parameters for TopHat v2.09 (Kim et al., 2013), and only the uniquely mapped reads were retained for the following analysis. MultiBamCov subfunction in BEDtools package (Quinlan and Hall, 2010) was used to report the number of reads mapped to respective high-confidence genes in each sample. The Bioconductor package “edgeR” (Robinson et al., 2010) was used to perform differential expression analysis between OE5 and the wild type at each time point, and only the genes with an absolute value of log2 (fold change) ≥ 0.8 and P value < 0.01 were considered as differentially expressed genes.
For GO analysis, cDNAs of high-confidence genes were aligned to the protein sequences of rice (Oryza sativa) and Arabidopsis thaliana using BLASTX with an e-value cutoff of 1e-05. Only the best alignment with identity ≥50% and aligned length ≥30 amino acids were kept. We obtained the GO annotations based on the orthologous genes with the priority: rice > Arabidopsis. GO enrichment analysis of differentially expressed genes was implemented using the GOseq R package, which is based on the Wallenius noncentral hypergeometric distribution and can adjust for gene length bias (Young et al., 2010). GO terms with FDR < 0.05 were considered significantly enriched.
RT-qPCR Analysis
The RT-qPCR method used for expression analysis of all transcripts, except for small RNAs, was performed as described previously (Yang et al., 2016). For each sample, the PCR amplification was repeated three times, and the average values of 2−ΔCT were used to determine the differences of gene expression by a Student's t test. Three biological replications, with tissues harvested from three different planting pots or Petri dishes, were performed with similar results and one replication was shown.
EMSA
The full-length sequences of TaABF and TaABI5 and the partial sequence of TaVP1 containing the B3 domain were cloned into the pGEX6P-1 vector as a translational fusion to GST (primers are listed in Supplemental Data Set 4). Expression of recombinant proteins in Transetta (DE3) Escherichia coli (TransGen) was induced with 0.2 mM IPTG in Luria Bertani buffer overnight at 16°C. The cells were subsequently harvested, washed, and resuspended in 30 mL of PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) containing 1 mM PMSF (Sigma-Aldrich) and half a tablet of protease inhibitor cocktail (Roche), and cells were sonicated for 1 h and centrifuged at 13,000g for 45 min. The supernatant was filtered through a 0.22-μm membrane into a 50-mLtube. The supernatant was mixed with 1 mL of GST MAG Agarose Beads (Novagen) and shaken overnight at 4°C. The GST beads were washed with 5 mL PBS, four times, and the fusion proteins were eluted from the beads by incubation at 4°C for more than 4 h with 50 mM Tris-HCl (pH 8.0) supplemented with 10 mM reduced glutathione. Protein concentrations were determined using a Nano Drop 2000 spectrophotometer (Thermo Scientific).
The biotin probe was 5′ end labeled with biotin. The double-stranded oligonucleotides used in the assays were annealed by cooling from 100°C to room temperature in annealing buffer. The DNA binding reactions were performed in 20 μL with 1× binding buffer (100 mM Tris, 500 mM KCl, and 10 mM DTT, pH 7.5), 10% glycerol, 0.5 mM EDTA, 7.5 mM MgCl2, 14 mM 2-mercaptoethanol, 0.05% (v/v) Nonidet P-40, and 50 ng μL−1poly(dI-dC). Competition analysis with an unlabeled DNA fragment (same sequence as for the labeled probe) can be used to test the specificity of the complex formation to the DNA sequence. A 4- and 10- fold molar excess of an unlabeled DNA fragment was added to the binding reaction, 5 min prior to the labeled probe. After incubation at room temperature for 30 min, samples were loaded onto a 6% native polyacrylamide gel. Electrophoretic transfer to a nylon membrane and detection of the biotin-labeled DNA was performed according to the manufacturer’s instructions (Light Shift Chemiluminescent EMSA kit; Thermo Scientific).
Statistical Analysis
The statistical analysis, including Student's t test, LSD, Dunnett's T3 post-hoc test, and correlation, were performed by SPSS software version 21 (SPSS/IBM). All bar charts, line charts and box plot results were graphically presented using Graph Pad Prism software (version 7.0).
Primers for RT-qPCR and Vector Construction
All primers used in this research are listed in Supplemental Data Set 4.
Accession Numbers
Gene ID numbers for all sequences described in this research can be found in Supplemental Data Set 4. RNA-seq data and degradome sequencing data are available at the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP093334 and SRP040143.
Supplemental Data
Supplemental Figure 1. miRNA sequences triggering phased siRNA in different plant species and foldback structure of their precursors.
Supplemental Figure 2. Constructs used for transient transformation of N. benthamiana.
Supplemental Figure 3. Constructs used for transient transformation of immature wheat embryos and observed phenotype of immature embryo germination.
Supplemental Figure 4. Levels of miR9678, WSGAR, and phasiRNA 3′D2(-) in wild-type and miR9678 overexpression wheat transgenic lines.
Supplemental Figure 5. Phenotype of miR9678 OE5, OE6, and OE8 transgenic lines.
Supplemental Figure 6. The scutellum structure in wild-type and miR9678 overexpressing line OE5.
Supplemental Figure 7. miR9678 does not affect ABA levels and delayed germination phenotype of OE lines can be rescued by GA3.
Supplemental Figure 8. Putative targets of phasiRNAs identified by RLM-5′-RACE and degradome sequencing.
Supplemental Figure 9. pri-miR9678-7D promoter sequence cis-element analysis and the positions and sequences of probes and their mutant competitors corresponding to ABA-responsive elements.
Supplemental Figure 10. Results of EMSA performed using TaVP1, TaABF, and TaABI5 recombinant proteins fused to GST and 10 biotin-labeled probes.
Supplemental Table 1. Predicted miR9678 target genes.
Supplemental Data Set 1. Differentially expressed transcripts in the miR9678 overexpression line and wild type at 1, 6, and 12 HAI.
Supplemental Data Set 2. Putative WSGAR-derived phasiRNAs targets, based on degradome sequencing.
Supplemental Data Set 3. Names and geographical origins of all wheat cultivars used in this research.
Supplemental Data Set 4. Primers and gene IDs for sequences used in this study.
Supplemental File 1. Statistical analysis.
Acknowledgments
We thank Aiju Zhao (Hebei Academy of Agriculture and Forestry Science) and Caixia Gao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for assistance in wheat genetic transformation, and Xiaoyu Zhang (University of Georgia) for suggestions about experimental design. We thank Yuqi Feng (Wuhan University) for assistance with GA quantification. This work was supported by the Major Program of the National Natural Science Foundation of China (31290210), the National Key Research and Development Program of China (2016YFD0101004), and the National Natural Science Foundation of China (31322041).
AUTHOR CONTRIBUTIONS
G.G., Z.N., Q.S., and Y.Y. conceived this project and designed all experiments. G.G., F.S., J.C., M.X., J.D., Z.H., and H.P. performed experiments. N.H. and B.W. contributed to wheat transformation. Y.Y., V.R., and R.X. contributed to manuscript writing and revision. X.L. and R.X. analyzed data.
Footnotes
Articles can be viewed without a subscription.
References
- Addo-Quaye C., Miller W., Axtell M.J. (2009). CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Howell M.D. (2010). miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin. Cell Dev. Biol. 21: 798–804. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Alonso-Peral M.M., Li J., Li Y., Allen R.S., Schnippenkoetter W., Ohms S., White R.G., Millar A.A. (2010). The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 154: 757–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N., Scofield G.N., Wang X.D., Offler C.E., Patrick J.W., Furbank R.T. (2006). Pathway of sugar transport in germinating wheat seeds. Plant Physiol. 141: 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleford N.E.J., Lenton J.R. (1997). Hormonal regulation of α-amylase gene expression in germinating wheat (Triticum aestivum) grains. Physiol. Plant. 100: 534–542. [Google Scholar]
- Ariizumi T., Steber C.M. (2007). Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19: 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Axtell M.J. (2017). Lost in translation? microRNAs at the rough ER. Trends Plant Sci. 22: 273–274. [DOI] [PubMed] [Google Scholar]
- Barrero J.M., et al. (2015). Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 16: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D. (1997). Seed germination and dormancy. Plant Cell 9: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D., Black M. (1994). Seeds: Physiology of Development and Germination. (Boston: Springer; ). [Google Scholar]
- Black M., Bewley J.D., Halmer P. (2006). The Encyclopedia of Seeds Science, Technology and Uses. (Oxfordshire, UK: CABI Publishing; ). [Google Scholar]
- Brousse C., Liu Q., Beauclair L., Deremetz A., Axtell M.J., Bouché N. (2014). A non-canonical plant microRNA target site. Nucleic Acids Res. 42: 5270–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoro R., Crocco C.D., Benech-Arnold R.L., Rodríguez M.V. (2013). In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 64: 5721–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Li Y.H., Wu S.H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.L., Fu X.M., Liu J.Q., Ye T.T., Hou S.Y., Huang Y.Q., Yuan B.F., Wu Y., Feng Y.Q. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905: 67–74. [DOI] [PubMed] [Google Scholar]
- Clavijo B.J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J., Moritz T., Blervaque R., Parcy F., Raz V., Herzog M., Vachon G. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136: 3660–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.S., Karmakar P., Nandi A.K., Sanan-Mishra N. (2015). Small RNA mediated regulation of seed germination. Front. Plant Sci. 6: 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez F., Moreno J., Cejudo F.J. (2012). The scutellum of germinated wheat grains undergoes programmed cell death: identification of an acidic nuclease involved in nucleus dismantling. J. Exp. Bot. 63: 5475–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., et al. (2016). PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. USA 113: 15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage W.E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171: 501–523. [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7: 373–385. [DOI] [PubMed] [Google Scholar]
- Graeber K., et al. (2014). DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. USA 111: E3571–E3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Millar A.A., Jacobsen J.V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8: 183–187. [DOI] [PubMed] [Google Scholar]
- Han R., Jian C., Lv J., Yan Y., Chi Q., Li Z., Wang Q., Zhang J., Liu X., Zhao H. (2014). Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.). BMC Genomics 15: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Zhu C., Dong K., Zhang T., Cheng Z., Li J., Yan Y. (2015). Comparative proteome analysis of embryo and endosperm reveals central differential expression proteins involved in wheat seed germination. BMC Plant Biol. 15: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Hoecker U., Vasil I.K., McCarty D.R. (1999). Signaling from the embryo conditions Vp1-mediated repression of alpha-amylase genes in the aleurone of developing maize seeds. Plant J. 19: 371–377. [DOI] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54. [DOI] [PubMed] [Google Scholar]
- Huang G., Varriano-Marston E. (1980). alpha-Amylase activity and preharvest sprouting damage in Kansas hard white wheat. J. Agric. Food Chem. 28: 509–512. [DOI] [PubMed] [Google Scholar]
- Huch S., Nissan T. (2014). Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. RNA 5: 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo H., Wei S., Bradford K.J. (2016). DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 113: E2199–E2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N., Fass J. (2011). Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. https://github.com/najoshi/sickle.
- Kanno Y., Jikumaru Y., Hanada A., Nambara E., Abrams S.R., Kamiya Y., Seo M. (2010). Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 51: 1988–2001. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Bentsink L., Hilhorst H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5: 33–36. [DOI] [PubMed] [Google Scholar]
- Lanet E., Delannoy E., Sormani R., Floris M., Brodersen P., Crété P., Voinnet O., Robaglia C. (2009). Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21: 1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu Y.H., Hagen G., Guilfoyle T. (1999). Expression of the auxin-inducible GH3 promoter/GUS fusion gene as a useful molecular marker for auxin physiology. Plant Cell Physiol. 40: 675–682. [Google Scholar]
- Liu P.P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52: 133–146. [DOI] [PubMed] [Google Scholar]
- Liu S., Cai S., Graybosch R., Chen C., Bai G. (2008). Quantitative trait loci for resistance to pre-harvest sprouting in US hard white winter wheat Rio Blanco. Theor. Appl. Genet. 117: 691–699. [DOI] [PubMed] [Google Scholar]
- Liu S., Sehgal S.K., Li J., Lin M., Trick H.N., Yu J., Gill B.S., Bai G. (2013). Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics 195: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Sehgal S.K., Lin M., Li J., Trick H.N., Gill B.S., Bai G. (2015). Independent mis-splicing mutations in TaPHS1 causing loss of preharvest sprouting (PHS) resistance during wheat domestication. New Phytol. 208: 928–935. [DOI] [PubMed] [Google Scholar]
- Ma X., Kim E.J., Kook I., Ma F., Voshall A., Moriyama E., Cerutti H. (2013). Small interfering RNA-mediated translation repression alters ribosome sensitivity to inhibition by cycloheximide in Chlamydomonas reinhardtii. Plant Cell 25: 985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares D.J., Mrva K. (2014). Wheat grain preharvest sprouting and late maturity alpha-amylase. Planta 240: 1167–1178. [DOI] [PubMed] [Google Scholar]
- Martin R.C., Liu P.P., Goloviznina N.A., Nonogaki H. (2010). MicroRNA, seeds, and Darwin?: diverse function of miRNA in seed biology and plant responses to stress. J. Exp. Bot. 61: 2229–2234. [DOI] [PubMed] [Google Scholar]
- Munkvold J.D., Tanaka J., Benscher D., Sorrells M.E. (2009). Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor. Appl. Genet. 119: 1223–1235. [DOI] [PubMed] [Google Scholar]
- Nakamura S., et al. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23: 3215–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H. (2008). Repression of transcription factors by microRNA during seed germination and postgerminaiton: Another level of molecular repression in seeds. Plant Signal. Behav. 3: 65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbonnaya F.C., Imtiaz M., Ye G., Hearnden P.R., Hernandez E., Eastwood R.F., van Ginkel M., Shorter S.C., Winchester J.M. (2008). Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor. Appl. Genet. 116: 891–902. [DOI] [PubMed] [Google Scholar]
- Olszewski N., Sun T.P., Gubler F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.): S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne J.W., Walker-Smith D.J. (1987). Isolation and identification of proteins from the peptide-transport carrier in the scutellum of germinating barley (Hordeum vulgare L.) embryos. Planta 170: 263–271. [DOI] [PubMed] [Google Scholar]
- Penfield S. (2017). Seed dormancy and germination. Curr. Biol. 27: R874–R878. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J.L., Chua N.H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49: 592–606. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K., Liu X.D., Xie Q., He Z.H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9: 34–45. [DOI] [PubMed] [Google Scholar]
- Somyong S., Ishikawa G., Munkvold J.D., Tanaka J., Benscher D., Cho Y.G., Sorrells M.E. (2014). Fine mapping of a preharvest sprouting QTL interval on chromosome 2B in white wheat. Theor. Appl. Genet. 127: 1843–1855. [DOI] [PubMed] [Google Scholar]
- Sun F., Guo G., Du J., Guo W., Peng H., Ni Z., Sun Q., Yao Y. (2014). Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol. 14: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Goodman H.M., Ausubel F.M. (1992). Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Ketterling M.G., McCarty D.R. (2005). Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis. Plant Physiol. 139: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.G., Vasil V., Vasil I.K. (1993). Enhanced GUS gene expression in cereal/grass cell suspensions and immature embryos using the maize uhiquitin-based plasmid pAHC25. Plant Cell Rep. 12: 491–495. [DOI] [PubMed] [Google Scholar]
- TeKrony M.D., Egli B.D. (1991). Relationship of seed vigor to crop yield: A Review. Crop Sci. 31: 816–822 [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79. [DOI] [PubMed] [Google Scholar]
- Xia R., Meyers B.C., Liu Z., Beers E.P., Ye S., Liu Z. (2013). MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell 25: 1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Takeda-Kamiya N., Hanada A., Ogawa M., Kuwahara A., Seo M., Kamiya Y., Yamaguchi S. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48: 555–561. [DOI] [PubMed] [Google Scholar]
- Yan J., Gu Y., Jia X., Kang W., Pan S., Tang X., Chen X., Tang G. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu X., Xin M., Du J., Hu Z., Peng H., Rossi V., Sun Q., Ni Z., Yao Y. (2016). Genome-wide mapping of targets of maize histone deacetylase HDA101 reveals its function and regulatory mechanism during seed development. Plant Cell 28: 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R., Kanno Y., Jikumaru Y., Nakabayashi K., Kamiya Y., Nambara E. (2009). CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 151: 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Wang H. (2010). Translational inhibition by microRNAs in plants. Prog. Mol. Subcell. Biol. 50: 41–57. [DOI] [PubMed] [Google Scholar]
- Yu Y., Zhen S., Wang S., Wang Y., Cao H., Zhang Y., Li J., Yan Y. (2016). Comparative transcriptome analysis of wheat embryo and endosperm responses to ABA and H2O2 stresses during seed germination. BMC Genomics 17: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang S., Han S., Li X., Tong Z., Qi L. (2013). Deciphering small noncoding RNAs during the transition from dormant embryo to germinated embryo in Larches (Larix leptolepis). PLoS One 8: e81452. [DOI] [PMC free article] [PubMed] [Google Scholar]