Abstract
Background
The calcineurin B-like protein (CBL)-interacting protein kinase (CIPK) signaling pathway responds to various abiotic stresses in plants.
Results
Wheat CIPK23, isolated from wheat drought transcriptome data set, was induced by multiple abiotic stresses, including drought, salt, and abscisic acid (ABA). Compared with wild-type plants, TaCIPK23-overexpression wheat and Arabidopsis showed an higher survival rate under drought conditions with enhanced germination rate, developed root system, increased accumulation of osmolytes, and reduced water loss rate. Over-expression of TaCIPK23 rendered transgenic plants ABA sensitivity, as evidenced by delayed seed germination and the induction of stomatal closure. Consistent with the ABA-sensitive phenotype, the expression level of drought- and ABA-responsive genes were increased under drought conditions in the transgenic plants. In addition, using yeast two-hybrid system, pull-down and bimolecular fluorescence complementation (BiFc) assays, TaCIPK23 was found to interact with TaCBL1 on the plasma membrane.
Conclusions
These results suggest that TaCIPK23 plays important roles in ABA and drought stress responses, and mediates crosstalk between the ABA signaling pathway and drought stress responses in wheat.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1306-5) contains supplementary material, which is available to authorized users.
Keywords: ABA, CIPK, CBL, Drought tolerance, Induced mechanism, Wheat
Background
Plants have developed a broad range of defense strategies and a complex network of signal transduction pathways [1, 2]. Calcium (Ca2+) serves as a ubiquitous secondary messenger that is involved in multiple physiological and developmental processes in plants [3, 4]. Unfavorable environmental conditions, such as drought, salt, extreme temperatures, and pathogen infection are all known to trigger discreet spatial and temporal changes in the concentration of [Ca2+]cyt in plant cells, leading to specific cellular responses [5]. Ca2+ sensor proteins, including Ca2+-dependent protein kinases (CDPKs), calmodulins (CAMs), and calcineurin B-like proteins (CBLs), decode these specific signatures and interact with targeted proteins to relay signals [6–8]. Plants have evolved complex Ca2+-decoding mechanisms. The CBL-CIPK network is an example of a significantly diverged Ca2+-decoding system in plants [9]. CBLs show significant similarity with both the regulatory β subunit of calcineurin (CNB) and the neuronal calcium sensors (NCS) of animals [10]. CBLs harbor four elongation factor (EF) hand motifs as the structural basis for Ca2+ binding. These EF hands specifically target a group of CDPKs designated as the CBL-interacting protein kinases (CIPKs) [9]. CIPKs consist of an N-terminal kinase catalytic domain and a C-terminal regulatory domain. The N-terminal kinase catalytic domain is related to sucrose non-fermenting kinase (SNF1) and AMP-activated protein kinase (AMPK) [10, 11]. The C-terminal regulatory domain contains a conserved NAF (Asn-Ala-Phe)/FISL (Phe-Ile-Ser-Leu) motif, consisting of 24 unique amino acid residues, that is essential and sufficient for interaction of CIPK proteins with CBLs [6, 12]. The protein phosphatase interaction (PPI) domain, containing 37 unique amino acid residues, is conserved in Arabidopsis protein kinase S (PKS) and in the DNA damage repair and replication block checkpoint kinase, Chk1, from various organisms including humans [13, 14]. The PPI motif, adjacent to the NAF/FISL motif, is necessary for interaction with abscisic acid-insensitive (ABI) protein phosphatases [14].
Comparative genomic analysis of CBL-CIPK genes in plants provides details about the functions, complexity, and conservation of the CBL/CIPK family and the evolution of the CBL-CIPK signaling network [15]. To date, a total of 10 CBLs and 26 CIPKs in Arabidopsis and 10 CBLs and 31 CIPKs in rice have been identified [16–18]. Although the specificity of the interactions of most of the CBLs and CIPKs has been confirmed using yeast two-hybrid assay experiments in Arabidopsis, the function of CBLs and CIPKs remains elusive [19, 20]. CBL4 interacts with CIPK24 to form specific complexes that function in activating plasma membrane-localized Na+/H+ antiporters and vacuolar H+-ATPases to promote salt tolerance [11]. CIPK21 is involved in the regulation of osmotic stress response in Arabidopsis through interaction with the vacuolar Ca2+ sensors CBL2 and CBL3 under salt stress conditions [21]. CBL1 and/or CBL9 interact with CIPK23, and control activation of the inward K+ channel AKT1, thereby regulating K+ uptake under low-K+ conditions [22]. The CBL-CIPK signaling pathway plays important roles in plant responses to environmental stresses [23]. To date, investigations of the CBL-CIPK network have mainly concentrated on how ion channels are involved in the influx or efflux of various ions. However, how the CBL-CIPK network participates in drought stress responses in plants have not been extensively reported [22, 24].
Bread wheat (Triticum avestivum L.) is one of the global staple crops and is mainly grown in arid and semi-arid regions. Serious water scarcity can cause dramatic yield losses in wheat production systems. Although extensive studies have been made that elucidate the role of the CBL-CIPK signaling pathway in Arabidopsis, wheat CIPKs remain poorly investigated; likely owing to the polyploid nature of the bread wheat genome and absence of a complete genome sequence [24, 25]. In the present study, a drought-responsive gene, TaCIPK23, was isolated based on a wheat drought de novo transcriptome sequencing experiment [26]. Over-expression of TaCIPK23 conferred drought tolerance in transgenic wheat and Arabidopsis. Further, the TaCIPK23 transgenic lines were more sensitive to ABA than the wild-type plants. TaCIPK23 enhanced the expression of a group of drought- and ABA-responsive genes under drought stress conditions. These results reveal a positive role for TaCIPK23 in conferring drought tolerance and regulating ABA signaling in plants.
Results
Identification of drought-responsive CIPK genes in wheat
Previously, the wheat genome was predicted to contain 71 TaCIPK genes [27]. In order to isolate drought-inducible CIPK genes in wheat, we analyzed the expression pattern of TaCIPK genes in wheat drought transcriptome database (http://www.ncbi.nlm.nih.gov/sra) [26]. A total of 21 CIPKs were found to responsive to drought stress at the transcriptional level (Table 1). Among the drought-inducible TaCIPKs, TaCIPK23 (Genbank No. KD502068) had the most pronounced induction of expression. To explore relationships among these drought-responsive TaCIPKs and previously-reported plant CIPKs, a phylogenetic tree of drought-responsive TaCIPKs and their orthologs from rice, soybean, rapeseed, Aegilops tauschii, Triticum urartu, maize, sorghum and Arabidopsis was constructed. The tree was divided into 7 subgroups. The 21 drought-responsive wheat CIPKs were distributed across 6 subgroups (I, III, IV, V, VI, and VIII; not present in subgroups II,). TaCIPK23 was included in subgroup I (Additional file 1: Figure S1).
Table 1.
Wheat CIPKs responsive to drought stress
| Gene | Gene ID | CK | Drought | Fold change | Up/Down | P-value |
|---|---|---|---|---|---|---|
| TaCIPK2-D | Unigene19129_All | 20.8224 | 61.194 | 1.5553 | Up | 4.72E-239 |
| TaCIPK4-B | CL16864.Contig2_All | 8.4632 | 0.7856 | −3.4293 | Down | 5.97E-45 |
| TaCIPK5-A | Unigene2976_All | 1.8326 | 4.2486 | 1.2131 | Up | 8.71E-05 |
| TaCIPK10-D | Unigene16768_All | 63.5323 | 6.9792 | −3.1864 | Down | 0 |
| TaCIPK14-A | CL11984.Contig1_All | 63.795 | 9.5433 | −2.7409 | Down | 0 |
| TaCIPK16-B | CL3500.Contig2_All | 1.4303 | 10.6707 | 2.8993 | Up | 2.03E-85 |
| TaCIPK16-D | CL3500.Contig1_All | 1.2472 | 7.7425 | 2.6341 | Up | 2.03E-55 |
| TaCIPK17-A | CL3945.Contig6_All | 3.7199 | 7.7386 | 1.0568 | Up | 7.44E-05 |
| TaCIPK17-B | CL3945.Contig5_All | 2.4916 | 10.8935 | 2.1283 | Up | 2.14E-08 |
| TaCIPK17-D | CL3945.Contig3_All | 5.4597 | 19.2913 | 1.8211 | Up | 2.15E-32 |
| TaCIPK19-A | CL6163.Contig1_All | 3.5214 | 0.845 | −2.0591 | Down | 2.16E-06 |
| TaCIPK19-B | CL10082.Contig2_All | 4.1148 | 26.2689 | 2.6745 | Up | 2.37E-167 |
| TaCIPK21-B | CL13954.Contig1_All | 6.1266 | 28.492 | 2.2174 | Up | 4.03E-169 |
| TaCIPK21-D | CL13954.Contig3_All | 0.2096 | 2.1735 | 3.3743 | Up | 3.97E-07 |
| TaCIPK23-A | CL5365.Contig4_All | 0.9841 | 23.129 | 4.5548 | Up | 3.01E-04 |
| TaCIPK23-B | CL5365.Contig1_All | 1.892 | 8.3534 | 2.1425 | Up | 2.97E-47 |
| TaCIPK23-D | CL5365.Contig2_All | 0.4497 | 2.2338 | 2.3125 | Up | 4.80E-14 |
| TaCIPK24-D | CL7296.Contig3_All | 6.8562 | 16.3986 | 1.2581 | Up | 3.57E-50 |
| TaCIPK26-D | CL6292.Contig1_All | 0.9064 | 2.5742 | 1.5059 | Up | 3.60E-12 |
| TaCIPK28-A | Unigene7442_All | 1.3571 | 5.1916 | 1.9357 | Up | 1.40E-07 |
| TaCIPK29-B | CL10071.Contig2_All | 2.8721 | 47.0435 | 4.0338 | Up | 0 |
TaCIPK23 is induced by multiple abiotic stresses
To investigate if TaCIPK23 expression is responsive to diverse stress factors, we conducted quantitative real-time reverse transcription (qRT)-PCR experiments. The expression of TaCIPK23 was remarkably induced by drought, reaching a peak at 1 h (~ 5-fold) (Fig. 1a). Expression of TaCIPK23 increased after treatment with 10% PEG6000 and reached a peak (~ 3.5-fold) at 0.5 h (Fig. 1b). Similarly, the expression of TaCIPK23 was enhanced by salt, reaching peak values (~ 4-fold) at 1 h after treatment (Fig. 1c). TaCIPK23 expression was also strongly induced by exogenous ABA, reaching its highest level at 1 h (~ 8-fold) (Fig. 1d). The results suggest that TaCIPK23 is responsive to various abiotic stresses and might function at the intersection of different signaling pathways.
Fig. 1.
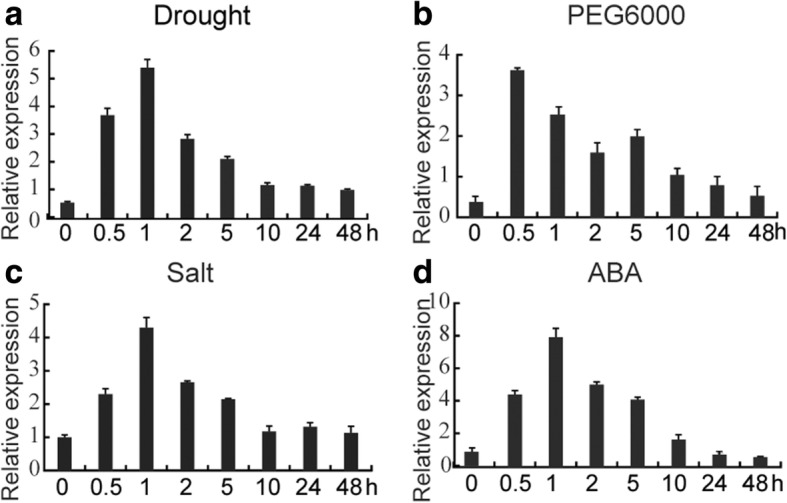
Expression patterns of TaCIPK23 under various abiotic stresses and subcellular localization of TaCIPK23 in wheat protoplasts. Expression profiles of TaCIPK23 after treatment with a Drought, b PEG6000, c NaCl, and d ABA for 0, 0.5, 1, 2, 5, 10, 24, and 48 h. Transcript levels were quantified by qRT-PCR assays. Expression of Actin was analyzed as a loading control. Ten wheat seedlings were combine as a single sample. Each data point is the mean (±SE) of three experiments
TaCIPK23 confers drought resistance in Arabidopsis and wheat
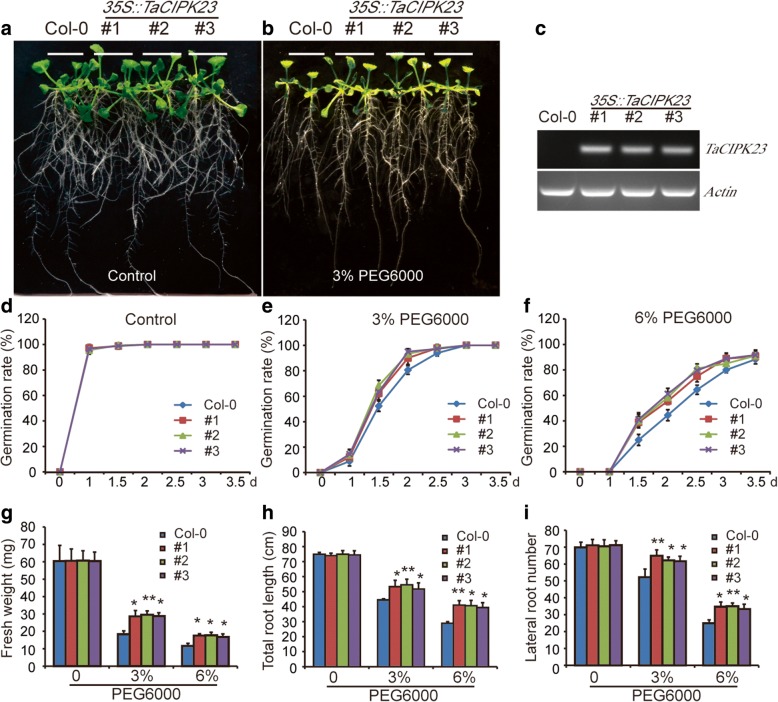
To estimate the capacity of the TaCIPK23-overexpression lines to withstand drought, PEG6000 was used to simulate drought stress. Germination percentages were based on radicle emergence after PEG6000 treatment. In the absence of PEG6000, both TaCIPK23-overexpression lines and wild-type materials had similar germination (Fig. 2d). However, in the presence of PEG6000, the germination of TaCIPK23-overexpression seeds was inhibited less dramatically than that of wild-type seeds (Fig. 2e, f). The TaCIPK23-overexpression lines and wild-type seedlings were grown on medium supplemented with PEG6000. As the concentration of PEG6000 (0, 3, and 6%) increased, the growth of wild-type plants was significantly impaired relative to the growth of TaCIPK23-overexpression lines (Fig. 2b). Compared with the wild-type plants, the TaCIPK23-overexpression lines displayed significantly greater fresh weights, longer root lengths, and a larger number of lateral roots under PEG6000-induced drought stress conditions (Fig. 2g–i). Furthermore, after 3-week-old transgenic and the wild-type seedlings were deprived of water for 2-weeks, the survival rate of TaCIPK23-overexpression lines is significantly higher than that of wild-type plants (Fig. 3a, b).
Fig. 2.
Over-expression of TaCIPK23 in Arabidopsis increased seeds germination rate and root growth under drought treatment. a-b Five-day-old seedlings grown on 1/2 MS were transferred to 1/2 MS medium containing various concentrations of PEG6000 (0 and 3%). c RT-PCR analysis of TaCIPK23 gene expression from TaCIPK23-overexpression and wild-type (Col-0) plants. Expression of Actin was analyzed as a loading control. d-f Time-course of germination percentages of transgenic lines and wild-type seeds in response to PEG6000 treatment. Each data point is the mean (±SE) of three experiments (each with 100 seeds for each line). g Measurements of fresh weight. h Measurements of total root length. i Measurement of lateral roots number. Each data point is the mean (±SE) of three experiments (30 seedlings per experiment). Significant differences from the WT are denoted by two asterisks corresponding to P < 0.01 by Student’s t-tests
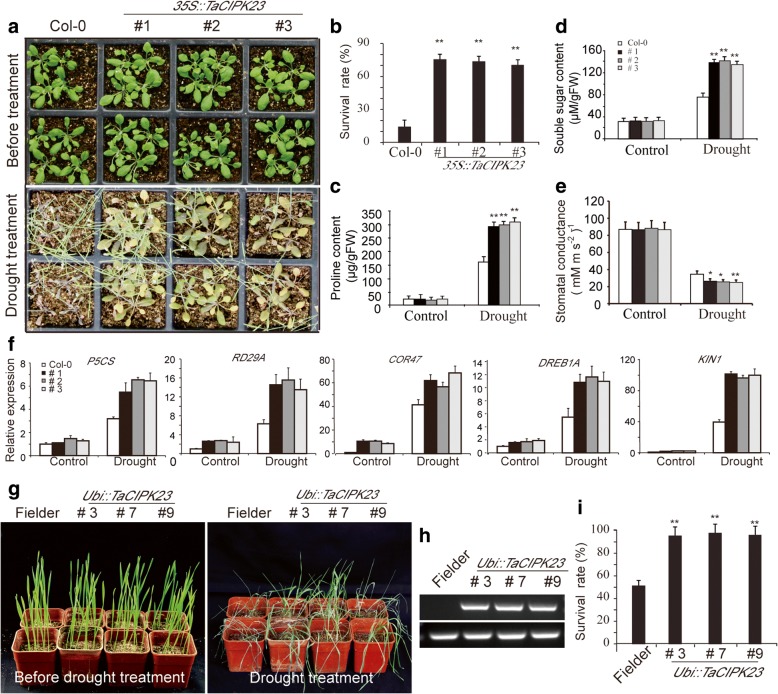
Fig. 3.
Over-expression of TaCIPK23 enhanced drought tolerance in transgenic Arabidopsis and wheat. a Drought tolerance phenotypes of TaCIPK23-overexpression lines under water-deficit conditions. 7-day-old TaCIPK23-overexpression and wild-type plants were grown for 2 weeks in pots under normal conditions. The plants were subject to another 2-week drought treatment. b Survival rate of the water-stressed plants was monitored 5 days after rewatering. c Pro content measurement. d Souble sugar content measurement. e Measurement of stomatal conductance. Each data point is the mean (±SE) of three experiments (10 seedlings per experiment). Significant differences from the WT are denoted by two asterisks corresponding to P < 0.01 by Student’s t-tests. f The expression level of drought-responsive genes was altered in TaCIPK23- overexpressing plants under drought conditions. Two-week-old TaCIPK23-overexpressing and wild-type seedlings with 6% PEG6000 treatment were used for RNA isolation. Transcript level was quantified by qRT-PCR assays. Expression of Actin was analyzed as a control. Each data point is the mean (±SE) of three experiments (10 seedlings per experiment) Data are means (±SE) of three independent experiments. g Drought tolerance phenotypes of TaCIPK23-overexpression wheat seedlings under drought conditions. 10-day-old TaCIPK23-overexpression and wild-type (Fielder) wheat seedlings were without irrigation for 16 days. h RT-PCR analysis of TaCIPK23 gene expression from Ubi-TaCIPK23 transgenic and control plants. Expression of Actin was analyzed as a loading control. i Survival rate of the water-stressed wheat seedlings was monitored 5 days after rewatering
In addition, 10-day-old transgenic and the control wheat seedlings were deprived of water for 16 days. Contrast to the exhibited severe wilting symptoms of wild-type plants, most of the TaCIPK23-overexpression lines remained green and wilting slightly (Fig. 3g). Survival rate was monitored 5 days after rewatering. The survival rate of TaCIPK23 transgenic wheat is significantly higher than that of wild-type plants (Fig. 3i).
To investigate the potential physiological mechanism for the improved drought tolerance of TaCIPK23-overexpression lines, the proline and soluble sugar content in wild-type and TaCIPK23-overexpression plants were measured under both normal growth and drought conditions. Under normal growth conditions, the proline and soluble sugar content of TaCIPK23-overexpression lines and wild-type were similar (Fig. 3c, d). Proline accumulation in the transgenic lines was significantly higher than in wild-type plants under drought conditions (Fig. 3c). TaCIPK23-overexpression plants also accumulated higher soluble sugar content than wild-type plants (Fig. 3d). Furthermore, stomatal conductance in the transgenic lines was significantly lower than in wild-type plants under drought conditions (Fig. 3e). These results indicated that the TaCIPK23-overexpression lines have improved drought tolerance, likely resulting from accelerated root growth, increased accumulation of osmoprotectants and reduced water loss rate.
To investigate the possible molecular mechanisms of TaCIPK23 in drought responses, the expression of several drought- and ABA-responsive markers was investigated, including ABI1 [28], P5CS [29, 30], RD29A [31], RD29B [31], DREB1A [32], COR47 [33], KIN1 [33], and ZAT12 [34]. A 2-fold change in expression was arbitrarily considered to be an induction of expression. qRT-PCR analyses revealed that there were no significant differences in the transcript levels of P5CS, RD29A and DREB1A between TaCIPK23-overexpression and wild-type plants under normal conditions. However, under drought stress conditions, the expression of these genes was significantly enhanced in TaCIPK23-overexpression lines as compared with the wild-type plants. Compared with the wild-type plants, under normal and drought conditions, the expression of COR47, and KIN1 were was much stronger in the TaCIPK23-overexpression lines (Fig. 3f). ABI1, ZAT12, and RD29B were not significantly differentially expressed in the TaCIPK23-overexpression lines in either normal or drought conditions (Additional file 1: Figure S2).
Increased ABA sensitivity in TaCIPK23 transgenic Arabidopsis
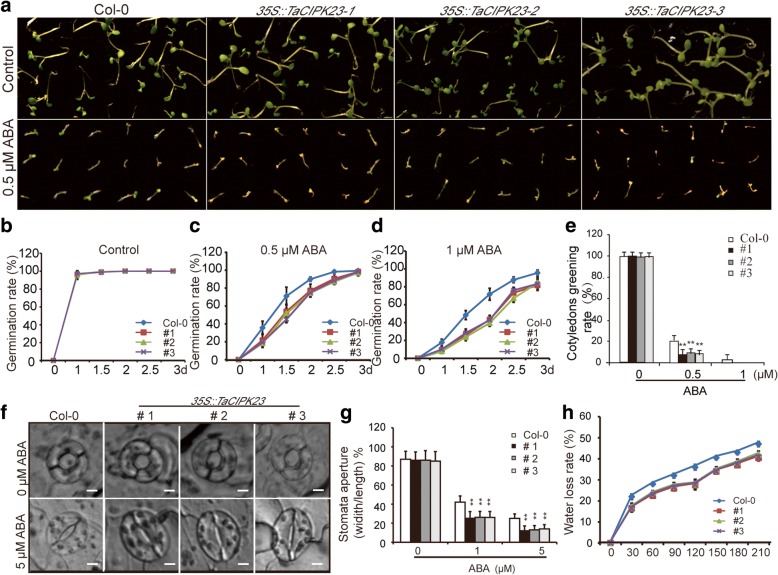
To examine the role of TaCIPK23 in plant ABA responses, the seeds from the transgenic lines and wild-type plants were germinated in the presence or absence of ABA (0, 0.5, or 1 μM) and germination percentages (both radicle emergence and cotyledon greening) were calculated from observation data. Germination percentages were based on radicle emergence after ABA treatment. Photographs were taken 5 days after germination. In the absence of ABA, both TaCIPK23-overexpression lines and wild-type materials had similar germination (Fig. 4b). However, in the presence of ABA, the seeds of lines TaCIPK23-overexpression lines germinated much more slowly than did wild-type seeds (Fig. 4c, d). Differences between TaCIPK23-overexpression lines and wild-type seedlings were even more apparent in cotyledon greening (Fig. 4e). These results indicated that germinating TaCIPK23-overexpression seeds were more sensitive to ABA than wild-type seeds.
Fig. 4.
Over-expression of TaCIPK23 in Arabidopsis increased ABA sensitivity. a TaCIPK23-overexpression and wild-type seeds were germinated on 1/2 MS medium supplemented with different concentrations ABA (0 and 0.5 μM). b-d Time-course of germination percentages of TaCIPK23-overexpression lines and wild-type seeds in response to ABA treatment. e Quantitative evaluation of leaf opening and greening rates. Each data point is the mean (±SE) of three experiments (each with 35 seeds for each line). f ABA-mediated stomatal closure. 3-week-old rosette leaves of TaCIPK23-overexpression lines and wild-type plants grown under continuous light were immersed in stomatal opening solution for 2.5 h and in ABA solution (0, and 5 μM) for an additional 2.5 h. Bars = 10 μm. g Measurement of stomatal apertures. h Measurement of leaf water loss. Each data point is the mean (±SE) of three experiments (30 stomata per experiment). Significant differences from the WT are denoted by one or two asterisks corresponding to P < 0.05 and P < 0.01, respectively, by Student’s t-tests. Significant differences from the WT are denoted by one or two asterisks corresponding to P < 0.05 and P < 0.01, respectively, by Student’s t-tests
Based on the ABA sensitivity in the germination assay, we tested whether ABA-sensitive stomatal movement in adult plants was altered in the transgenic lines. 3-week-old mature leaves were treated with different concentrations of ABA for 2.5 h, and the length and width of stomata were measured. Stomatal movement profiles were measured as the ratio of width to length [22, 35]. In the absence of ABA treatment, TaCIPK23-overexpression and wild-type plants showed comparable average stomatal apertures (Fig. 4f, g). However, after treatment with increasing concentrations of ABA (0, 1, and 5 μM) for 2.5 h, the average stomatal apertures were reduced more dramatically in the TaCIPK23-overexpression plants than in wild-type plants (Fig. 4f, g). The water loss rates of detached leaves of the TaCIPK23-overexpression lines leaves were lower than those of detached wild-type leaves (Fig. 4h). Plants lose water mainly through stomata. These results suggest that over-expression of TaCIPK23 can improve ABA sensitivity of plants and can regulate stomatal movement to reduce water loss rate.
TaCIPK23 interacts with TaCBL1 in vivo and in vitro
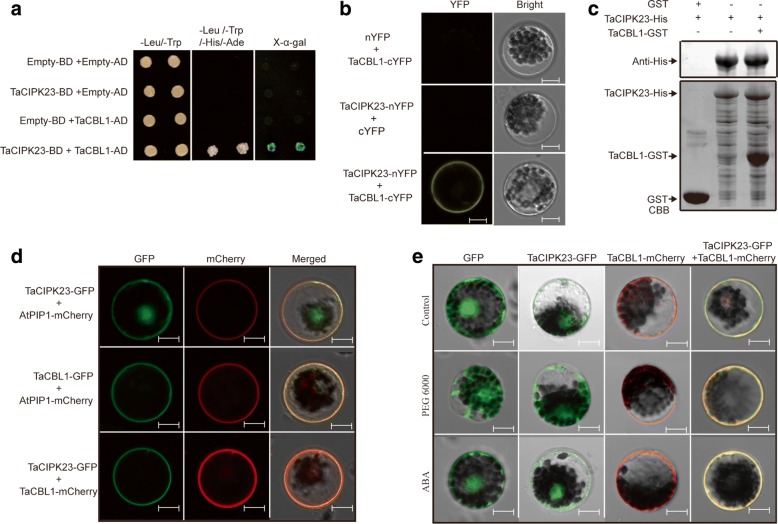
To explore the cellular mechanism(s) through which TaCIPK23 participates in drought stress responses, TaCIPK23 was used as bait protein to screen a wheat cDNA library in yeast two-hybrid assays. One interacting candidate, named TaCBL1, was obtained in this experiment (Fig. 5a).
Fig. 5.
TaCIPK23 interacts with TaCBL1 on the plasma membrane. a Yeast two-hybrid analysis of TaCIPK23 interaction with TaCBL1. b BiFC assay of the interaction of TaCIPK23 with TaCBL1. Fluorescence and bright field images of wheat protoplast co-transfected with constructs encoding fusion proteins TaCIPK23-YFPN and TaCBL1-YFPC. Bar = 12 μm. c In vitro pull-down assays were performed by incubating His-TaCIPK23 with GST-TaCBL1 or GST. GST-TaCBL1 was able to pull down His-TaCIPK23 fusion proteins (upper middle panel), but not GST (upper right panel) as detected by Western blot analysis with His antibodies. Coomassie Brilliant Blue (CBB) staining was used to visualize His-TaCIPK23 fusion proteins, GST and the GST-TaCBL1 fusion proteins used in each assay. d Localization of TaCIPK23 and TaCBL1 proteins in wheat protoplasts. Images were observed under a laser scanning confocal microscope. Bar = 12 μm. AtPIP1-mCherry protein was used as membrane localization marker. e Volume change of wheat protoplast transfected with GFP alone, TaCBL1-mCherry alone, TaCIPK23-GFP alone, and TaCBL1-mCherry/TaCIPK23-GFP constructs, respectively, with PEG6000 and ABA treatments. Images were observed under a laser scanning confocal microscope. Bar = 12 μm
Pull-down experiment was carried out to corroborate the interaction of TaCIPK23 and TaCBL1 in vitro. His-TaCIPK23 and TaCBL1 fused to GST (glutathione-S-transferase) were expressed in E. coli (BL21). Purified TaCBL1-GST and GST protein were immobilized to glutathione beads, respectively. His-TaCIPK23 protein was incubated with the beads in a pull-down assay. In the analysis, the His-TaCIPK23 protein co-purified with TaCBL1-GST (Fig. 5c; top right panel) but not with GST (Fig. 5c; top left panel), indicating a specific interaction between TaCIPK23 and TaCBL1.
To further corroborate the interaction between TaCIPK23 and TaCBL1 in wheat cells, the TaCIPK23-YFPN (N-terminal fragment of yellow fluorescent protein) and TaCBL1-YFPC (C-terminal fragment of yellow fluorescent protein) recombination vectors were transformed into wheat protoplasts; fluorescence signals were mainly observed on the plasma membrane. In contrast, no positive signals were observed when TaCIPK23-YFPN and YFPC or YFPN and TaCBL1-YFPC constructs were co-transformed into wheat protoplasts (Fig. 5b). In addition, subcellular localization assays show that TaCIPK23 was localized to cytoplasm, nucleus, and plasma membrane and TaCBL1 is a membrane-localized protein (Fig. 5d). The AtPIP2-mCherry fusion protein was used as plasma membrane marker [36]. When TaCIPK23-GFP and TaCBL1-mCherry co-expressed in wheat protoplasts, TaCBL1 attached TaCIPK23 to the membrane (Fig. 5d).
To investigate the significance of the direct interaction between TaCIPK23 and TaCBL1, we test volume change of wheat protoplast transfected with GFP alone, TaCBL1-mCherry alone, TaCIPK23-GFP alone, and TaCBL1-mCherry/TaCIPK23-GFP constructs, respectively, with PEG6000 and ABA treatments. As is shown in Fig. 5e, the interaction between TaCBL1 and TaCIPK23 was not affected by PEG6000 or ABA treatments. The protoplast volume was similar under normal and ABA conditions. After PEG6000 treatment, protoplasts transfected with GFP constructs was severely damaged, and the volume of these protoplasts was significantly smaller than those protoplasts transfected with TaCBL1-mCherry alone, TaCIPK23-GFP alone, and both TaCBL1-mCherry and TaCIPK23-GFP constructs. Compared with protoplasts transfected with TaCBL1-mCherry and TaCIPK23-GFP, respectively, the lower injury of volume was found in protoplasts co-transfected with TaCBL1-mCherry and TaCIPK23-GFP constructs. These results suggest that TaCIPK23 maybe interact with TaCBL1 to form a CBL/CIPK complex on the plasma membrane.
Discussion
Many studies have suggested that the CBL-CIPK signaling pathway plays important roles in abiotic stress responses, including drought stress [37, 38]. Little is known about the diverse functions of wheat CIPK genes in drought response. Understanding the mechanism of wheat CIPKs mediated drought stress response has a great practical benefit for the development of stress-tolerant wheat. In the present study, a CIPK gene, named TaCIPK23, was isolated from wheat drought transcriptome databases (Table 1). Biochemical and transgenic studies further supported TaCIPK23 contributed to plant ABA signaling and drought tolerance.
Transgenic Arabidopsis was wildly selected to investigation wheat genes function [39–41]. Although the amino acid sequence of plant CIPKs is highly conserved, their functions are distinctive. Wild-type Arabidopsis is selected for transformation to investigate the function of TaCIPK23 in abiotic stress responses, but not the mutant. In this study, TaCIPK23-overexpression plants has a more developed roots system (Fig. 2h, i) and accumulated significantly higher amounts of proline and soluble sugars than did wild-type plants under drought conditions (Fig. 3c, d), a result consistent with the higher survival rate of the TaCIPK23-overexpression lines in response to drought stress (Fig. 3b). Root length and lateral root number are phenotype traits for assessing adaptability to environmental stresses [42, 43]. A deep taproot system associated with a moderate number of lateral roots enables plants to absorb enough water and minerals for sustaining the viability of plants [43]. Over-expression of constitutively activated mutant CaCIPK6 in transgenic tobacco enhanced root growth, which in turn improved its drought and salt tolerance [20]. Under water-deficit conditions, plants accumulate compatible osmolytes such as proline and soluble sugars to protect their subcellular structures from damage [44, 45]. Proline and soluble sugars function in lowering the cellular osmotic potential and restoring intracellular solute concentrations, which prevent water loss from cells [46, 47]. Previous studies reported that the accumulation of Pro and soluble sugars in OsCIPK03- and OsCIPK12-overexpression plants was significantly higher than that in wild-type plants under abiotic stress conditions, which contributed to the improved tolerance of OsCIPK03- and OsCIPK12-overexpressing plants to cold and drought stresses, respectively [47]. These results indicated that over-expression of TaCIPK23 in plant exhibited developed root system and increased the content of osmoprotectants that conferred drought tolerance in transgenic plants. Furthermore, this result was consequently confirmed in wheat (Fig. 3i). To further investigate the role of TaCIPK23 in abiotic stress responses, we also attempt to knock-down the gene in wheat via RNA interference, but the transcript abundance of TaCIPK23 was little effect due to the complicated wheat genome. In the future, it is necessary to generate CRISPR-mediated mutants knocking out the three TaCIPK23 homologs in wheat.
ABA plays critical roles in regulating root growth, seed germination, stomatal movement, vegetative growth, and stress responses [7, 48, 49]. Under drought conditions, an elevated level of ABA induces stomatal closure to reduce water loss [46]. ABA-induced stomatal closure represents a major mechanism for plant adaptation to drought [22, 47]. Previous studies reported that cipk3 loss-of-function mutants were hypersensitive to ABA [7]. BnCIPK6M over-expression in transgenic Arabidopsis caused hypersensitivity to ABA, whereas silencing of its homologous gene AtCIPK6 conferred plant ABA insensitive growth phenotypes [50]. In this study, the expression of TaCIPK23 was induced by exogenous ABA (Fig. 1d). Detailed phenotypic analyses revealed that the TaCIPK23-overexpression lines were hypersensitive to ABA with delayed seed germination and small stomatal aperture after exogenous ABA treatment (Fig. 4). Furthermore, the water loss rate of the TaCIPK23-overexpression lines was lower than that of wild-type plants (Figs. 3e and 4h). Plants lose water mainly through stomata. The lower stomatal conductance in TaCIPK23-overexpression lines causes the reduced water loss rate under drought stress conditions, which contributes the drought tolerance of TaCIPK23-overexpression plants. TaCIPK23 positively regulated several drought- and ABA-responsive genes expression under drought conditions (Fig. 3f). These results indicate that TaCIPK23 positively modulates plant drought tolerance through ABA-dependent and -independent pathways.
Rapid responses to diverse growth conditions are crucial for plants survival and flourish [51]. Stress perception and signal amplification are involved in stress responses and adaptation [51]. Previous studies indicated that CBL proteins interact with, and are phosphorylated by specific functional interacting CIPKs [51, 52]. Interaction with CBLs is essential for the activation of CIPKs in vivo [53]. Phosphorylation of CBLs by their functional interacting CIPKs enhances complex stability [52]. In vitro kinase activity assays showed that the substrate phosphorylation activity of CIPK24 was negligible in the absence of CBL4, but the CBL4/CIPK24 complex had a basal level of activity for substrate phosphorylation even in the absence of Ca2+ [54]. Interaction of SOS2 and SCaBP8, enhanced by SOS2 phosphorylation of SCaBP8 and not requiring Ca2+, stabilizes the SCaBP8-SOS2 interaction, which in turn activates plasma membrane Na+/H+ exchange to improve salt tolerance [55]. Phosphorylation of SCaBP1 by SOS2-LIKE PROTEIN KINASE5 (PKS5) activates their interaction and negatively regulates the activity of AHA2 [56]. In this study, as is illustrated in Fig. 5, TaCIPK23 interacted with TaCBL1 on the plasma membrane. Compared with protoplasts transfected with TaCIPK23-GFP vector alone, the lower injury of cellular structure was found in protoplasts co-transfected with TaCBL1-mCherry and TaCIPK23-GFP constructs. The interaction of TaCIPK23 and TaCBL1 may be able to form a CBL/CIPK complex and enhance the activity of TaCIPK23, contributing to drought stress tolerance.
Methods
Plant materials and stress treatment
Arabidopsis ecotypes Col-0 was used in this study. Seeds were germinated on 1/2 MS medium (Duchefa) with 2% sucrose and were subsequently transferred to soil. The plants were grown in a greenhouse at 22 °C under long-day conditions (16 h light/8 h dark photoperiod) at a light intensity of around 100 μΜ.m− 2 s− 2. To generate TaBZR2D-overexpressing plants, the coding region of TaBZR2D was introduced into the plant transformation vector pBI121 under the control of the CaMV 35S promoter. The constructs were confirmed by sequencing and then transformed into wild-type plants (Col-0) by the vacuum infiltration method [57]. Seeds of wild-type and TaCIPK23-overpression (independent transgenic lines 1, 2, and 3) plants were sterilized with 30% bleach. After 3 days of stratification at 4 °C, the plates were transferred to a growth chamber. For the germination assay, the sterilized seeds of wild-type and TaCIPK23-overpression plants were sown on 1/2 MS growth medium with various concentrations of PEG6000 (0, 3%, or 6%, Merck, USA) or ABA (0, 0.5, or 1.0 μM, Sigma-Aldrich, USA). For the root growth assay, wild-type and transgenic Arabidopsis seeds were germinated on MS agar medium for 7 days, followed by transfer to MS growth medium containing various concentrations PEG6000 (0, 3%, or 6%, Merck, USA). Photographs were taken after 7 days of growth and root lengths were evaluated using Epson Expression 11000XL root system scanning analyzer (Epson, Japan). At least 20 seedlings were measured for each genotype. To test drought tolerance at later developmental stages, 14-day-old seedlings were withheld from watering for 14 days, by which time the plants growth were severely affected, and survival rates were calculated for each group of plants. Three independent measurements of 30 seedlings were averaged.
Wheat seedlings (T. aestivum L. cultivar Xiaobaimai) were grown in 1/2 Hoagland liquid medium at 22 °C under long-day conditions (16 h light/8 h dark photoperiod) at a light intensity of around 100 μΜ.m− 2 s− 2 for 2 weeks. For the drought treatment, seedlings were transferred onto filter paper, and dried at 25 °C under 60% humidity conditions. For PEG6000, salt, and ABA treatments, seedling roots were immersed in half-strength Hoagland solution containing 10% PEG6000, 200 mM NaCl, or 100 μM ABA and sampled at 0, 0.5, 1, 2, 5, 10, 24, and 48 h. Harvested seedlings were immediately frozen in liquid nitrogen and stored at − 80 °C prior to RNA extraction. Wheat cultivar fielder was employed as the receptor material to generate the transgenic plants. The ORF of TaCIPK23 was introduced into the pWMB110 plant transformation vector. The construct was confirmed by sequencing and then transformed into wild-type plants by Agrobacterium-mediated wheat transformation system. For the drought tolerance assay, 10-day-old wheat seedlings were deprived of water for 16 d.
Sequence analysis
The wheat drought de novo transcriptome data was available in the Sequence Read Achive (SRA) accession number SRP071191 [26]. The full-length cDNA of TaCIPK23 was obtained using gene-specific primers. The PCR products were cloned into the pEASY-T1 vector (TransGene, China) and sequenced. All CIPK sequences in the rice, soybean, rapeseed, Aegilops tauschii, Triticum urartu, maize, sorghum and Arabidopsis genome were identified by using BLASTP, TBLASTN, and the “motif” algorithms to retrieve GenBank database (http://www.ncbi.nlm.nih.gov) [58]. Sequences were aligned with ClustalW using the MEGA5.1 program [59]. The alignment was then used to create a phylogenetic tree with the MEGA5.1 program based on a neighbor-joining method; the confidence level of monophyletic groups was estimated using a bootstrap analysis of 10,00 replicates [59]. The complete amino acid sequence of TaCIPK23 was analyzed with the protein structure prediction tool available at http://www.sbg.bio.ic.ac.uk/phyre2.
Plasmid construction for localization analysis
The ORF of TaCIPK23 and TaCBL1 was cloned into the 16318hGFP vector, respectively and fused with the GFP reporter gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The recombinant plasmids were transformed into wheat mesophyll protoplasts with a PEG-mediated method. Expression of fusion proteins was monitored after 12 h of incubation in darkness, and images were captured under a laser scanning confocal microscope (Zeiss LSM700, Germany) [42]. The expressed AtPIP2-mCherry fusion protein was used as plasma membrane marker [36].
Yeast two-hybrid screening
TaCIPK23 was cloned into the pGBKT7 plasmid. TaCBL1 was cloned into the pGADT7 plasmid. Yeast strain AH109 expressing pGADT7-TaCBL1 (prey) was transformed with pGBKT7-TaCIPK23 (baits). Transformed yeast cells were selected on synthetic complete medium lacking SD/ -Trp/ -Leu following the lithium acetate method (according to TRANFOR protocol). Interaction was determined on synthetic complete medium SD/−Leu/−Trp/-His/−Ade (Sigma-Aldrich, USA) supplemented with 5-Bromo-4-chloro-3-indoxyl-α-D-galactopyranoside (X-α-Gal) (Sigma-Aldrich, USA), at 30 °C for 3 d. Empty prey or bait vectors were transformed with TaCIPK23 or TaCBL1 plasmids as negative controls. All bait proteins were tested for self-activation; none were found to activate the reporter genes LacZ [21].
Protein purification and pull-down assays
TaCIPK23 was inserted into the prokaryotic expression vector pcold TF DNA (TaKaRa, Japan). TaCBL1 was inserted into the pGEX-4 T-1 plasmid. His-TaCIPK23 and GST-TaCBL1 were expressed in E. coli and purified by standard procedures using, respectively, Ni and glutathione agarose beads (GE Healthcare, USA). Briefly, 100 ml of BL21 cells grown overnight and expressing the desired constructs were transferred into 500 ml of LB and grown at 37 °C for 3 h. Isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM) was then added to the media and incubated overnight at 16 °C to induce protein expression. The bacterial cells were sonicated in phosphate-buffered saline (PBS) with 1% Triton X-100 and centrifuged at 10, 000 g for 10 min to remove insoluble cell debris. The supernatant was incubated with PBS pre-equilibrated with Ni or lutathione agarose beads and rotated at 4 °C for 2 h. After washing five times with PBS, His-tagged proteins was eluted using 5 mM glutathione and 125 mM imidazole.
For pull-down assays, His-TaCIPK23 was incubated with GST-TaCBL1 or GST in binding buffer [25 mM HEPES (pH 7.6), 12.5 mM MgCl2, 150 mM KCl, 0.1% NP-40, and 20% glycerol] buffer for 2 h at 4 °C, respectively. After six washes with buffer [100 mM NaCl, 1 Mm EDTA, 0.5% NP-40, and 20 mM TRIS (pH 8.0)], the proteins were analysed on SDS-polyacrylamide gels followed by immunoblotting using anti-His monoclonal antibodies (NEB, USA) at a 1:1000 dilution. IRDye 800CW anti-mouse IG (H + L) at a 1:15000 dilution (LI-COR, USA) was used as the second antibody. The western blots were developed with Odyssey CLx Infrared Imaging Systems (LI-COR, USA) [42, 60, 61].
Bimolecular fluorescence complementation (BiFC) assay
TaCIPK23 and TaCBL1 were cloned into the pSPYCE and pSPYNE plasmids, respectively. For the BiFC assays, the TaCIPK23-pSPYNE and TaCBL1-pSPYCE reconstruction vectors were transformed into common wheat mesophyll protoplasts by a PEG-mediated method. Expression of fusion proteins was monitored after 12 h of incubation in darkness, and images were captured under a laser scanning confocal microscope (Zeiss LSM700, Germany). Yellow fluorescent protein (YFP) fluorescence signals were collected in the 500–570 nm wavelength range. For chloroplast autofluorescence, the wavelength range monitored was 630–700 nm [42, 62].
qRT-PCR
Total RNA from Arabidopsis and wheat seedlings was extracted using an RNAprep plant kit (TIANGEN, China). First-strand cDNA was synthesized using a PrimeScript First-Strand cDNA Synthesis kit (TaKaRa, Japan). The qRT-PCR reactions were performed using an ABI Prism 7500 real-time PCR system (ThermoFisher Scientific, USA) using SYBR Green Master Mix (TIANGEN, China) in a total volume of 25 μl and was performed with three technical replications for each sample. A quantitative analysis was performed using the 2-ΔΔCT method [63].
Measurements of proline content, soluble sugar content, and stomatal conductance
Seven-day-old Arabidopsis seedlings were grown on identical plates filled with a 1:1 mixture of vermiculite and humus. After an additional 3 weeks growth, the seedlings were treated with 10% (w/v) PEG6000 for 7 additional days. Pro concentration was determined as described [64]; Soluble sugars contents were assayed as described [47]. All the measurements were repeated three times, and the Student’s t-test was used for statistical analysis. For stomatal conductance assay, 3-week-old Arabidopsis seedlings were deprive of water for 8 additional days, stomatal conductance was determined as described [65].
Measurement of water loss rate and stomatal aperture
For the water loss assays, 3-week-old Arabidopsis rosette leaves were detached and weighed immediately on a piece of weighing paper and then placed on a laboratory bench and weighed at designated times (0, 30, 60, 120, 180, 210, and 240 min). The percentage loss of fresh weight was calculated on the basis of the initial weight of the detached leaves. The percentage of water loss was calculated as previously described [64]. The experiment was repeated at least three times. Each repetition of the experiment included three replicates for each sampled material.
For the stomatal closure assays, four rosette leaves from 3-week-old plants (grown under 8 h light/16 h dark at 22 °C; 70% relative humidity) were floated in opening buffer (10 mM KCl, 7.5 mM iminodiacetic acid, and 10 mM MES-Tris, pH 6.15) under light [100 μmol/(m2. s)] for 4 h, as described previously [35, 51], with some modifications. After the stomata are fully opened, leaves were transferred to ABA-containing buffer (0, 1, and 5μΜ ABA) for 2.5 h for stomatal closing response analysis. The adaxial side of the leaf epidermis was peeled off using tape and observed with a laser scanning confocal microscope (Zeiss LSM700, Germany). Somatal images were photographed with a laser scanning confocal microscope (Zeiss LSM700, Germany), and analyzed using Photoshop CS5 software (Adobe Systems, USA). Stomatal apertures of wild-type and 35S:TaCIPK23 leaves were measured as the ratio of width to length after ABA treatment. Thirty stomata used for analysis were from the central region of the leaves from 3 individual plants.
Additional file
Figure S1. Phylogenetic tree of CIPK proteins from rice, soybean, rapeseed, Aegilops tauschii, Triticum urartu, maize, sorghum and Arabidopsis . The phylogenetic tree was constructed based on the sequence alignments. The phylogenetic tree of CIPK proteins was constructed with MEGA5.1 program with the neighbor-joining method. The numbers beside the branches represent bootstrap values based on 1000 replications. Figure S2. The expression level of drought-responsive genes was altered in TaCIPK23- overexpressing plants under drought conditions. Two-week-old TaCIPK23-overexpressing and wild-type seedlings with 6% PEG6000 treatment were used for RNA isolation. Transcript level was quantified by qRT-PCR assays. Expression of Actin was analyzed as a control. Each data point is the mean (±SE) of three experiments (10 seedlings per experiment). Table S1. Primers used for qRT-PCR assays. (PDF 496 kb)
Acknowledgments
We are grateful to Drs. Rui-Lian Jing and Yong-Fu Fu (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing wheat seeds and for the BiFC system, respectively. We also thank Dr. Dongying Gao (Department of Plant Sciences, University of Georgia, USA) for suggestions on the manuscript.
Funding
This research was financially supported by the National Transgenic Key Project of the Ministry of Agriculture of China (2018ZX08009100 and 2016ZX08002–002), the Funding Project for Beijing Advanced Innovation Center for Food Nutrition and Human Health and the Open Research Fund Program of Beijing Key Lab of Plant Resource Research and Development, Beijing Technology and Business University.
Availability of data and materials
The transcriptome data was available in the Sequence Read Achive (SRA) under accession number SRP071191. All the supporting data are included as Additional files.
Abbreviations
- [Ca2+]cyt
cytosolic Ca2+
- ABA
Abscisic acid
- AMPK
AMP-activated protein kinase
- BiFC
Bimolecular fluorescence complementation
- Ca2+
Calcium
- CAMs
Calmodulins
- CBL
Calcineurin B-like protein
- CIPK
CBL interacting protein kinase
- CNB
Regulatory β subunit of calcineurin
- NCS
Neuronal calcium sensors
- PPI
Protein phosphatase interaction
- SNF1
Sucrose non-fermenting kinase
Authors’ contributions
ZSX coordinated the project, conceived and designed experiments, and edited the manuscript; XYC performed experiments and wrote the first draft; YTD conducted the bioinformatic work and performed experiments; JDF and TFY generated and analyzed data; CTW and MC contributed with editing the manuscript. JC provided analytical tools and managed reagents; YZM coordinated the project and edited the manuscript. All authors have read and approved the final manuscript.
Competing interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This research is not applicable to the ethics approval and consent.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Xiao-Yu Cui and Yong-Tao Du contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1306-5) contains supplementary material, which is available to authorized users.
Contributor Information
Xiao-Yu Cui, Email: cuixiaoyu763@163.com.
Yong-Tao Du, Email: yongtaodu1994@126.com.
Jin-dong Fu, Email: fujindong@caas.cn.
Tai-Fei Yu, Email: yutaifei824@163.com.
Chang-Tao Wang, Email: wangct@btbu.edu.cn.
Ming Chen, Email: chenming02@caas.cn.
Jun Chen, Email: chenjun01@caas.cn.
You-Zhi Ma, Email: mayouzhi@caas.cn.
Zhao-Shi Xu, Email: xuzhaoshi@caas.cn.
References
- 1.Wang QY, Guan YC, Wu YR, Chen HL, Chen F, Chu CC. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol. 2008;67(6):589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- 2.Xu ZS, Chen M, Li LC, Ma YZ. Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol. 2011;53(7):570–585. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- 3.Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22(3):541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002;14:S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight MR, Campbell AK, Smith SM, Trewavas AJ. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 1991;352(6335):524–526. doi: 10.1038/352524a0. [DOI] [PubMed] [Google Scholar]
- 6.Albrecht V, Ritz O, Linder S, Harter K, Kudla J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+−regulated kinases. EMBO J. 2001;20(5):1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell. 2003;15(2):411–423. doi: 10.1105/tpc.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004;16(7):1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luan S. Signalling drought in guard cells. Plant Cell Environ. 2002;25(2):229–237. doi: 10.1046/j.1365-3040.2002.00758.x. [DOI] [PubMed] [Google Scholar]
- 10.Kudla J, Xu Q, Harter K, Gruissem W, Luan S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci U S A. 1999;96(8):4718–4723. doi: 10.1073/pnas.96.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci U S A. 2000;97(12):6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez-Barrena MJ, Fujii H, Angulo I, Martinez-Ripoll M, Zhu JK, Albert A. The structure of the C-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol Cell. 2007;26(3):427–435. doi: 10.1016/j.molcel.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13(6):1383–1399. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohta M, Guo Y, Halfter U, Zhu JK. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci U S A. 2003;100(20):11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Q, An L, Li W. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014;33(2):203–214. doi: 10.1007/s00299-013-1507-1. [DOI] [PubMed] [Google Scholar]
- 16.Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134(1):43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyzenga WJ, Liu HX, Schofield A, Muise-Hennessey A, Stone SL. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J Exp Bot. 2013;64(10):2779–2791. doi: 10.1093/jxb/ert123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piao HL, Xuan YH, Park SH, Je BI, Park SJ, Park SH, Kim CM, Huang J, Wang GK, Kim MJ, et al. OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol Cells. 2010;30(1):19–27. doi: 10.1007/s10059-010-0084-1. [DOI] [PubMed] [Google Scholar]
- 19.Kim KN, Cheong YH, Gupta R, Luan S. Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 2000;124(4):1844–1853. doi: 10.1104/pp.124.4.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009;58(5):778–790. doi: 10.1111/j.1365-313X.2009.03812.x. [DOI] [PubMed] [Google Scholar]
- 21.Pandey GK, Kanwar P, Singh A, Steinhorst L, Pandey A, Yadav AK, Tokas I, Sanyal SK, Kim BG, Lee SC, et al. Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 2015;169(1):780–792. doi: 10.1104/pp.15.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52(2):223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- 23.Mahajan S, Pandey GK, Tuteja N. Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch Biochem Biophys. 2008;471(2):146–158. doi: 10.1016/j.abb.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Yang WQ, Kong ZS, Omo-Ikerodah E, Xu WY, Li Q, Xue YB. Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.) J Genet Genomics. 2008;35(9):531–543. doi: 10.1016/S1673-8527(08)60073-9. [DOI] [PubMed] [Google Scholar]
- 25.Deng XM, Zhou SY, Hu W, Feng JL, Zhang F, Chen LH, Huang C, Luo QC, He YZ, Yang GX, et al. Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol Plantarum. 2013;149(3):367–377. doi: 10.1111/ppl.12046. [DOI] [PubMed] [Google Scholar]
- 26.He GH, Xu JY, Wang YX, Liu JM, Li PS, Chen M, Ma YZ, Xu ZS. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016;16(1):116. doi: 10.1186/s12870-016-0806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun T, Wang Y, Wang M, Li TT, Zhou Y, Wang XT, Wei SY, He GY, Yang GX. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015;15 [DOI] [PMC free article] [PubMed]
- 28.Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9(5):759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggiero B, Koiwa H, Manabe Y, Quist TM, Inan G, Saccardo F, Joly RJ, Hasegawa PM, Bressan RA, Maggio A. Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in arabidopsis. Plant Physiol. 2004;136(2):3134–3147. doi: 10.1104/pp.104.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchishinozaki K, Wada K, Harada Y, Shinozaki K. Correlation between the induction of a gene for Delta(1)-Pyrroline-5-carboxylate Synthetase and the accumulation of proline in Arabidopsis-Thaliana under osmotic-stress. Plant J. 1995;7(5):751–760. doi: 10.1046/j.1365-313X.1995.07050751.x. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi-Shinozaki K, Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet. 1993;236(2–3):331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- 32.Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17(3):287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 33.Kurkela S, Borg-Franck M. Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol. 1992;19(4):689–692. doi: 10.1007/BF00026794. [DOI] [PubMed] [Google Scholar]
- 34.Davletova S, Schlauch K, Coutu J, Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139(2):847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha CV, Leyva-Gonzalez MA, Osakabe Y, Tran UT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Yamaguchi S, Dong NV, et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci U S A. 2014;111(2):851–856. doi: 10.1073/pnas.1322135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandersson E, Danielson JAH, Rade J, Moparthi VK, Fontes M, Kjellbom P, Johanson U. Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J. 2010;61(4):650–660. doi: 10.1111/j.1365-313X.2009.04087.x. [DOI] [PubMed] [Google Scholar]
- 37.Weinl S, Kudla J. The CBL-CIPK Ca2+−decoding signaling network: function and perspectives. New Phytol. 2009;184(3):517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- 38.Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14(1):37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Zhang N, Yin Y, Liu X, Tong S, Xing J, Zhang Y, Pudake RN, Izquierdo EM, Peng H, Xin M, et al. The E3 ligase TaSAP5 alters drought stress responses by promoting the degradation of DRIP proteins. Plant Physiol. 2017;175(4):1878–1892. doi: 10.1104/pp.17.01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu P, Xu ZS, Lu PP, Hu D, Chen M, Li LC, Ma YZ. A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J Exp Bot. 2013;64(10):2915–2927. doi: 10.1093/jxb/ert133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma BA, Lin Q, Zhang ZB, Zhang JS, et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35(6):1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- 42.Liu P, Xu ZS, Pan-Pan L, Hu D, Chen M, Li LC, Ma YZ. A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J Exp Bot. 2013;64(10):2915–2927. doi: 10.1093/jxb/ert133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manavalan LP, Guttikonda SK, Tran LSP, Nguyen HT. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009;50(7):1260–1276. doi: 10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- 44.Armengaud P, Thiery L, Buhot N, Grenier-de March G, Savoure A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol Plantarum. 2004;120(3):442–450. doi: 10.1111/j.0031-9317.2004.00251.x. [DOI] [PubMed] [Google Scholar]
- 45.Xiang Y, Huang Y, Xiong L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007;144(3):1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farooq M, Wahid A, Lee DJ, Ito O, Siddique KHM. Advances in drought resistance of Rice. Crit Rev Plant Sci. 2009;28(4):199–217. doi: 10.1080/07352680902952173. [DOI] [Google Scholar]
- 47.Zhang HY, Mao XG, Jing RL, Chang XP, Xie HM. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J Exp Bot. 2011;62(3):975–988. doi: 10.1093/jxb/erq328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bu Q, Lv T, Shen H, Luong P, Wang J, Wang Z, Huang Z, Xiao L, Engineer C, Kim TH, et al. Regulation of drought tolerance by the F-box protein MAX2 in Arabidopsis. Plant Physiol. 2014;164(1):424–439. doi: 10.1104/pp.113.226837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo PJ, Xiang FN, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151(1):275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Ren F, Zhou L, Wang QQ, Zhong H, Li XB. The Brassica napus Calcineurin B-like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J Exp Bot. 2012;63(17):6211–6222. doi: 10.1093/jxb/ers273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Kim WT. The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 2013;162(3):1733–1749. doi: 10.1104/pp.113.220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du WM, Lin HX, Chen S, Wu YS, Zhang J, Fuglsang AT, Palmgren MG, Wu WH, Guo Y. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 2011;156(4):2235–2243. doi: 10.1104/pp.111.173377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang CQ, Schumaker KS, Zhu JK. Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell. 2004;16(2):435–449. doi: 10.1105/tpc.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halfter U, Ishitani M, Zhu JK. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. P Natl Acad Sci USA. 2000;97(7):3735–3740. doi: 10.1073/pnas.97.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin HX, Yang YQ, Quan RD, Mendoza I, Wu YS, Du WM, Zhao SS, Schumaker KS, Pardo JM, Guo Y. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 PROTEIN kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell. 2009;21(5):1607–1619. doi: 10.1105/tpc.109.066217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuglsang AT, Guo Y, Cuin TA, Qiu QS, Song CP, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell. 2007;19(5):1617–1634. doi: 10.1105/tpc.105.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 58.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quan RD, Lin HX, Mendoza I, Zhang YG, Cao WH, Yang YQ, Shang M, Chen SY, Pardo JM, Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19(4):1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye HX, Li L, Guo HQ, Yin YH. MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109(49):20142–20147. doi: 10.1073/pnas.1205232109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh MH, Wang XF, Kota U, Goshe MB, Clouse SD, Huber SC. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. P Natl Acad Sci USA. 2009;106(2):658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011;18(1):17–29. doi: 10.1093/dnares/dsq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bu QY, Lv TX, Shen H, Luong P, Wang J, Wang ZY, Huang ZG, Xiao LT, Engineer C, Kim TH, et al. Regulation of Drought Tolerance by the F-Box Protein MAX2 in Arabidopsis(1[C][W][OPEN]) Plant Physiol. 2014;164(1):424–439. doi: 10.1104/pp.113.226837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang WH, Chen J, Liu TW, Chen J, Han AD, Simon M, Dong XJ, He JX, Zheng HL. Regulation of the calcium-sensing receptor in both stomatal movement and photosynthetic electron transport is crucial for water use efficiency and drought tolerance in Arabidopsis. J Exp Bot. 2014;65(1):223–234. doi: 10.1093/jxb/ert362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic tree of CIPK proteins from rice, soybean, rapeseed, Aegilops tauschii, Triticum urartu, maize, sorghum and Arabidopsis . The phylogenetic tree was constructed based on the sequence alignments. The phylogenetic tree of CIPK proteins was constructed with MEGA5.1 program with the neighbor-joining method. The numbers beside the branches represent bootstrap values based on 1000 replications. Figure S2. The expression level of drought-responsive genes was altered in TaCIPK23- overexpressing plants under drought conditions. Two-week-old TaCIPK23-overexpressing and wild-type seedlings with 6% PEG6000 treatment were used for RNA isolation. Transcript level was quantified by qRT-PCR assays. Expression of Actin was analyzed as a control. Each data point is the mean (±SE) of three experiments (10 seedlings per experiment). Table S1. Primers used for qRT-PCR assays. (PDF 496 kb)
Data Availability Statement
The transcriptome data was available in the Sequence Read Achive (SRA) under accession number SRP071191. All the supporting data are included as Additional files.