Abstract
The antibacterial effects of aminoglycosides are based on their association with the A-site of bacterial rRNA and interference with the translational process in the bacterial cell, causing cell death. The clinical use of aminoglycosides is complicated by resistance and side effects, some of which arise from their interactions with the human mitochondrial 12S rRNA and its deafness-associated mutations, C1494U and A1555G. We report a rapid assay that allows screening of aminoglycoside compounds to these classes of rRNAs. These screening tools are important to find antibiotics that selectively bind to the bacterial A-site rather than human, mitochondrial A-sites and its mutant homologues. Herein, we report our preliminary work on the optimization of this screen using 12 anthraquinone–neomycin (AMA–NEO) conjugates against molecular constructs representing five A-site homologues, Escherichia coli, human cytosolic, mitochondrial, C1494U, and A1555G, using a fluorescent displacement screening assay. These conjugates were also tested for inhibition of protein synthesis, antibacterial activity against 14 clinically relevant bacterial strains, and the effect on enzymes that inactivate aminoglycosides. The AMA–NEO conjugates demonstrated significantly improved resistance against aminoglycoside-modifying enzymes (AMEs), as compared with NEO. Several compounds exhibited significantly greater inhibition of prokaryotic protein synthesis as compared to NEO and were extremely poor inhibitors of eukaryotic translation. There was significant variation in antibacterial activity and MIC of selected compounds between bacterial strains, with Escherichia coli, Enteroccocus faecalis, Citrobacter freundii, Shigella flexneri, Serratia marcescens, Proteus mirabilis, Enterobacter cloacae, Staphylococcus epidermidis, and Listeria monocytogenes exhibiting moderate to high sensitivity (50–100% growth inhibition) whereas Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiellla pneumoniae, and MRSA strains expressed low sensitivity, as compared to the parent aminoglycoside NEO.
Keywords: translation inhibition, aminoglycoside modifying enzymes, antibacterial activity
Graphical abstract
Aminoglycosides and their derivatives are widely used clinical antibiotics.1,2 Aminoglycosides bind primarily to the decoding region of the ribosome and subsequently interfere with incorporation of amino acids into the growing peptide chain, sterically hindering the extrusion of the nascent peptide from the ribosome and/or interfering with tRNA selection at the mRNA codon site, respectively.3 Upon binding to the decoding site of bacterial ribosomes, the primary antibacterial mechanism of action involves binding to the A-site of the 16S rRNA, thus inducing template misreadings during the translation process, thereby leading to the synthesis of faulty proteins and ultimately cell death.4,5 The ability of aminoglycosides to bind different RNA structures enormously increases their potential use as therapeutic agents to fight other human diseases.2 For example, their propensity to bind micro (mi) RNAs makes them attractive agents for the treatment of multiple cancer types.6–9 Propagation of viruses was found to be inhibited by aminoglycosides, which bind to viral RNA.10,11 Additionally, it has been recently reported that rationally designed new compounds can selectively bind to the eukaryotic ribosome, resulting in a much higher ability to read through premature false stop codons present in mutant mRNAs that are hallmarks of certain disease types.12 However, undesirable side effects caused by aminoglycosides call for a new generation of aminoglycoside derivatives with reduced toxicity, selective binding to the target RNA, and inhibition of bacterial translation.
One of the approaches to modulate the function of an aminoglycoside toward the goal of developing novel antibacterials is to conjugate it with small nucleic acid binders using various linkers.13,14 It has been demonstrated that the flexibility and length of the linker used in the conjugation plays a critical role in the RNA binding affinity and the resulting antibacterial properties of the compounds.14 Our choice for a conjugate moiety was anthraquinone, a flat planar aromatic ring system, the derivatives of which bind nucleic acids and are known for their various health benefits and antibacterial properties.15,16 We have previously reported the synthesis of anthraquinone–NEO conjugates and their interaction with a variety of nucleic acids.17 Anthraquinone conjugates effectively stabilize nucleic acid structures using a dual binding mode involving intercalation by the anthraquinone moiety and NEO binding within the nucleic acid major groove.17,18
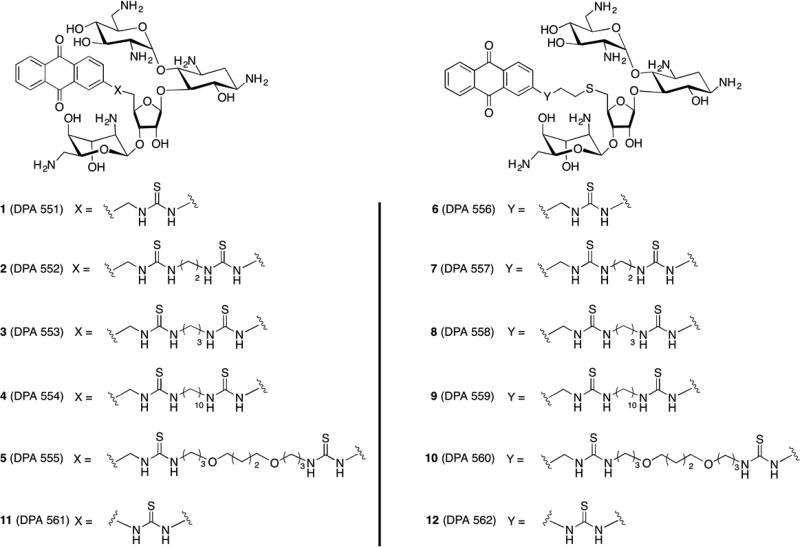
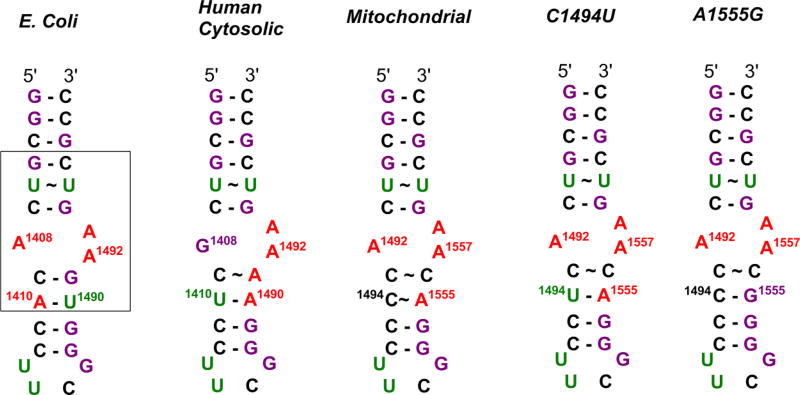
Resistance to aminoglycoside antibiotics is conferred via a class of enzymes called aminoglycoside-modifying enzymes (AMEs),19–22 which includes aminoglycoside acetyltransferases (AACs), nucleotidyltransferases (ANTs), and phosphotransferases (APHs), that catalyze the modification at aminoglycoside hydroxyl or amine functionalities, rendering the drugs unable to bind their ribosomal target.23 We have previously demonstrated that conjugation of aminoglycosides to small molecules renders them poor substrates for AMEs.13,14,24,25 Here, we examine 12 new anthraquinone–NEO conjugates (AMA–NEO) (1–12) with thiourea linkers for their antibacterial properties, translation inhibition, resistance to AMEs, and binding selectivity for five different 27-nucleotide RNA hairpin constructs representing A-site homologues (Figures 1 and 2). The Escherichia coli A-site is a highly conserved region for aminoglycoside binding in the bacterial ribosome. The mitochondrial A-site differs from the bacterial A-site in the identity of two noncanonical base pairs at positions 1493–1554 and 1494–1555. The C1494U and A1555G sequences are derived from mutated mitochondrial 12S rRNAs that carry one-base mutations at positions 1494 and 1555, respectively, and are associated with aggravated ototoxicity due to increased drug binding.22,26,27 Hypersensitivity of A1555G and C1494U mutations is most likely due to similarity between the secondary structures of bacterial and mitochondrial mutant A-sites due to the presence of canonical base pairs in position 1494–1555.28,29 Altogether, these findings challenge researchers to develop antibiotics that will bind preferentially to the bacterial A-site, rather than mitochondrial or deaf mutation A-sites. The human cytosolic A-site, or the eukaryotic homologue, stands out from other A-sites due to the guanine substitution for adenine at position 1408 (E. coli numbering). Guanine reduces the affinity of an A-site for many aminoglycosides by causing a steric hindrance at the preferred binding site, leaving bacterial and mitochondrial ribosomes as primary binding targets for aminoglycosides.30
Figure 1.
Structures of AMA–NEO conjugates used in this study. Compound purity was verified by RP-HPLC and HPLC purity profiles and has been reported previously.31
Figure 2.
Secondary structures of A-site models used in this study. Bases are colored as follows: adenines, red; cytosines, black; guanines, purple; uridines, green. Box indicates the A-site sequences of interest in this study.
RESULTS AND DISCUSSION
Screening Studies against A-Site Analogues
We have shown previously that fluorescent NEO conjugates bind to E. coli and human cytosolic A-sites at a 1:1 stoichiometric ratio.32 Here, we apply binding studies of AMA–NEO conjugates with different linkers to mitochondrial A-site and its two mutant homologues, C1494U and A1555G.13 The synthesis of these compounds has been reported recently.31
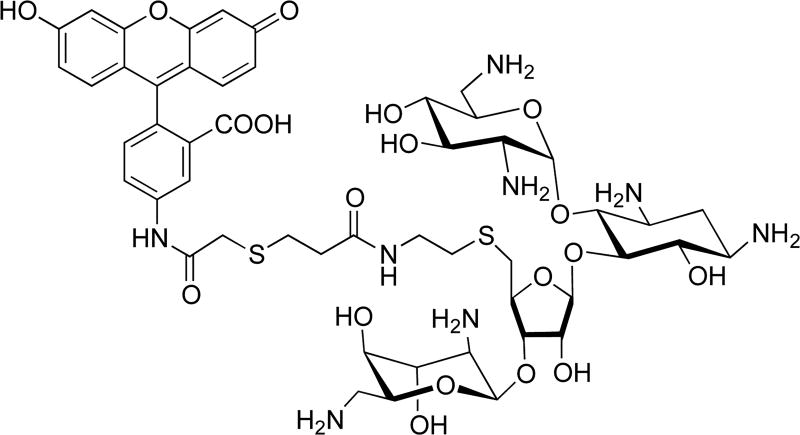
Binding selectivity of AMA–NEO conjugates 1–12 to A-sites was assessed by fluorescent displacement assay using F–NEO as a reporter.33 F–NEO is a conjugate of NEO and fluorescein that binds to an A-site at 1:1 ratio like NEO, as was demonstrated by binding studies (Figure 3).33 F–NEO emission is reduced in the bound state and is enhanced upon displacement. Dissociation constants (Kd) for F–NEO and A-sites are listed in Table 1. F–NEO Kd values for E. coli, mitochondrial, and A1555G A-sites are in the 4–6 nM range, but values for the human cytosolic and C1494U A-sites are roughly 5 times higher.
Figure 3.
Structure of F–NEO, a molecular reporter of compound binding.
Table 1.
Kd for F–NEO and IC50 and SF Values for NEO and Five A-Sites
| A-site | Kd (F–NEO), nM | IC50 (NEO), nM | SF (NEO) |
|---|---|---|---|
| E. coli | 4.1 ± 0.8 | 87.3 ± 5.9 | 1.0 |
| human cytosolic | 23.3 ± 3.1 | 76.5 ± 13.2 | 0.2 |
| mitochondrial | 6.0 ± 1.4 | 87.1 ± 8.3 | 0.7 |
| C1494U | 21.5 ± 4.0 | 67.9 ± 2.0 | 0.2 |
| A1555G | 5.4 ± 1.4 | 119.2 ± 3.7 | 0.5 |
To assess the binding selectivity of NEO, a reference compound, its IC50 was measured for each A-site RNA hairpin. Here, IC50 is the drug concentration at which the F–NEO emission is 50% of the maximum. IC50 values for NEO are essentially the same, within experimental error, for E. coli, mitochondrial, C1494U, and human cytosolic A-sites, but the direct comparison of selectivities is not possible without taking into account their affinity for F–NEO (Table 1). Therefore, we introduced a selectivity factor (SF) parameter, which demonstrates the binding preference of a compound for E. coli A-site over the other A-sites. The SF for E. coli A-site is 1. An SF value below 1 for a particular compound is indicative of a less preferable binding for a target A-site, as compared with the E. coli A-site RNA. Calculated SF values for NEO and target A-sites follow the following relationship: E. coli ~ mitochondrial > A1555G > C1494U ~ human cytosolic. Aminoglycosides preferably bind to mitochondrial mutant A-site homologues over the human and Mycobacterium smegmatis bacterial A-site.29,34 However, the E. coli homologue used in our study has a different primary sequence resulting in a 1410A–1490U base pair instead of a 1410G–1490C pair, which is found in the A-site homologue from M. smegmatis used in the aforementioned studies.29,34,35 These studies demonstrate the importance of base-pair identity and structural geometry surrounding the aminoglycoside binding pocket.29
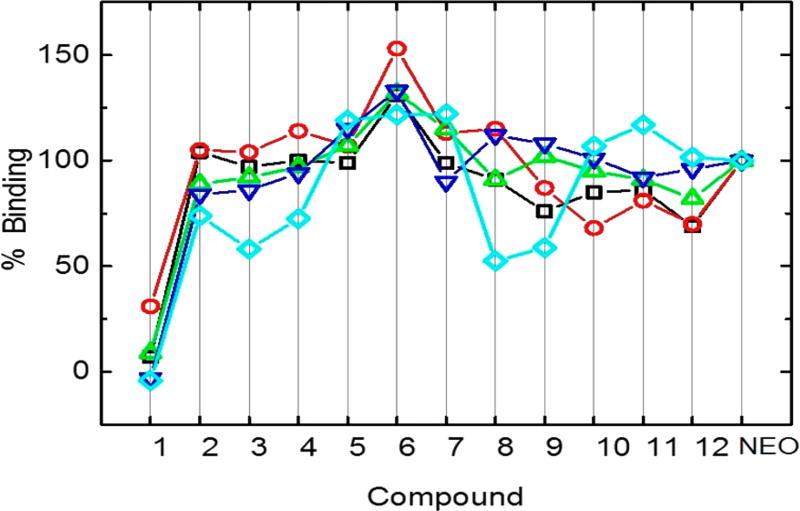
To assess the preference of AMA–NEO conjugates 1–12 for a particular A-site RNA, compounds were initially screened at a single concentration of drug. Emission intensities of displaced F–NEO were converted into percent binding and plotted for each A-site (Figure 4). In general, screening results demonstrate that the AMA–NEO conjugates’ binding affinity to model A-sites is within 50% from NEO affinity with the exception of conjugate 1, the weakest binder. IC50 values measured for compounds 2, 5, and 6 (Table 2) are approximately 1–2 times higher than analogous NEO values. Their binding selectivity factors are similar to those found for NEO, within error.
Figure 4.
Percent binding relative to NEO for compounds 1–12. Screening of the compounds was performed with the following model A-sites: E. coli (black squares), human (red circles), mitochondrial (green triangles), C1494U (blue inverted triangles), and A1555G (cyan rhombuses).
Table 2.
IC50 and Selectivity Factors for Compounds 2, 5, and 6
| 2 | 5 | 6 | ||||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| A-site | IC50, nM | SF | IC50, nM | SF | IC50, nM | SF |
| E. coli | 172.5 ± 1.2 | 1.0 | 151.4 ± 9.8 | 1.0 | 161.9 ± 13.2 | 1.0 |
| human cytosolic | 146.3 ± 10.1 | 0.2 | 140.3 ± 10.1 | 0.2 | 129.2 ± 1.2 | 0.2 |
| mitochondrial | 167.7 ± 1.2 | 0.7 | 121.9 ± 5.0 | 0.8 | 95.7 ± 2.1 | 1.1 |
| C1494U | 98.6 ± 2.1 | 0.3 | 98.6 ± 2.1 | 0.3 | 66.3 ± 4.9 | 0.4 |
| A1555G | 269.5 ± 10.2 | 0.5 | 249.1 ± 46.7 | 0.5 | 196.2 ± 4.2 | 0.6 |
Screening for Antibacterial Activity
The method of screening compounds with potential antibiotic activity at a single-point concentration against bacterial strains was developed by De La Fuente and co-workers. This allows for high-throughput screening as utilized in our laboratory to screen conjugate libraries13,36 and is a quick, reliable, and efficient means for preliminary selection of antimicrobial compounds. AMA–NEO conjugates 1–12 were screened with a single-point concentration at 6.3 µM against five Gram-positive strains, Enterococcus faecalis, Staphylococcus epidermidis, Listeria monocytogenes, and two methicillin-resistant Staphylococcus aureus (MRSA) strains; nine Gram-negative strains, Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Shigella flexneri, Pseudomonas aeruginosa, Serratia marscescens, Klebsiella pneumoniae, and Proteus mirabilis; and Acinetobacter baumannii. All of these strains are recognized as clinically important opportunistic pathogens, which include the ESKAPE pathogens (Enterobacter spp., Staphylococcus spp., Klebsiella spp., Acinetobacter spp., Pseudomonas spp., Entercoccus spp.). The percent inhibition of bacterial growth was determined for each compound relative to NEO and 2-aminomethylanthraquinone (AMA) (Table 3). No significant activity against NEO-resistant MRSA strains, P. aeruginosa, A. baumanni, or K. pneumoniae, was observed. In contrast, significant growth inhibition was observed (80–100%) with most compounds against E. coli, S. marcescens, P. mirabilis, E. cloacae, C. freundii, S. epidermidis, and L. monocytogenes. The anthraquinone derivative imparted some antibacterial activity on E. faecalis, P. mirabilis, and L. monocytogenes. Anthraquinones are known to stack or intercalate between nucleic acid bases, and some AMA derivatives found in nature have been found to have antibacterial activity.37 It is likely that the anthraquinone derivatives used here affect the antibacterial activity of some of the conjugates by modulating the rRNA binding and the bacterial translation machinery, and these properties will be examined in the following section. For MIC studies we chose five representative compounds (2, 5, 6, 10, and 12) that had significant antimicrobial activity (>80%) in the single-point screen. Compound 5 demonstrated a significantly lower MIC value in the range of 50–100 µM for MRSA 33591 than for NEO with an MIC of 400 µM. In contrast, no significant inhibition was observed with compounds 1–12 for the MRSA A960649 strain, which possesses a 4′-aminoglycoside nucleotidyltransferase gene, ant(4′), and a double-functioning 2″-aminoglycoside phosphotransferase/6′-aminoglycoside acetyltransferase gene aac(6′)/aph(2″), according to supplier specifications. Compound 6 inhibited MRSA A960649 at an MIC > 100 µM. Additionally, three of the tested compounds, 2, 5, and 6, demonstrated moderate inhibition of E. coli and P. mirabilis growth, with MIC values of 12.5 µM (Table 4), which is consistent with single-point screening data from Table 3. Moderate inhibition activity was also observed in C. freundii, E. cloacae, S. flexneri, and S. marcescens for compounds 2, 5, 10, and 12, consistent with the single-point screen for these strains. The MIC for compound 6 was not determined because little to no inhibition was observed with the single-point screen for most of strains, with the exceptions of C. freundii, L. monocytogenes, and S. epidermidis. Low MIC values, consistent with screening results, were observed for compounds 2, 5, 10, and 12 in S. epidermidis and L. monocytogenes, most notably with compounds 2, 5, and 10 with an MIC value of 1.6 µM that matches the MIC range for the NEO control. To identify the possibility of membrane interactions with our compounds, the conjugates were assayed for hemolysis of rabbit erythrocytes (Table 5). There was little measurable hemolysis at a compound concentration of 100 µM, which was within the concentration range for MIC values determined for the selected bacterial strains that had significant inhibition identified in our single-point concentration screen of 6.3 µM. This result indicates that the mechanism of bactericidal activity of the compounds was not likely due to lysis of the bacterial cell membrane. Anthraquinone and its derivatives can stack or intercalate between RNA bases. It is hypothesized that the combination of anthraquinone with NEO, with an appropriate linker, can induce increased binding to the target site. The role of the various linkers serves to identify what structural modifications are necessary to both inhibition of enzymatic attack by bacterial resistance enzymes along with strategic positioning of the anthraquinone moiety within the rRNA and its subsequent effects on the translation machinery.
Table 3.
Inhibition of Bacterial Growth (%) by Compounds 1–12 and Commercial Aminoglycosides at 6.3 µMa
| compound | E. coli | E. faecalis | K. pneumoniae | P. mirabilis | MRSA 33591 | MRSA A960649 | A. baumannii |
|---|---|---|---|---|---|---|---|
| 1 | 7 | 11 | 0 | 0 | 7 | 2 | 0 |
| 2 | 84 | 39 | 0 | 87 | 6 | 3 | 0 |
| 3 | 53 | 3 | 0 | 14 | 6 | 3 | 0 |
| 4 | 73 | ND | 0 | 80 | 15 | 2 | ND |
| 5 | 74 | 11 | 0 | 80 | 15 | 0 | 0 |
| 6 | 87 | 29 | 0 | 52 | 0 | 12 | 0 |
| 7 | 71 | 0 | 2 | 23 | 7 | 3 | 0 |
| 8 | 67 | 46 | 0 | 31 | 0 | 1 | 0 |
| 9 | 38 | 9 | 0 | 2 | 5 | 0 | 0 |
| 10 | 62 | 0 | 0 | 4 | 2 | 0 | 0 |
| 11 | 36 | 12 | 0 | 12 ± 5 | 2 | 0 | 0 |
| 12 | 52 | 28 | 0 | 8 | 0 | 2 | 0 |
| AMA | 2 | 27 | 0 | 19 | 0 | 0 | 0 |
| NEO | 100 | 39 | 0 | 96 | 0 | 0 | 91 |
| compound | C. freundii | E. cloacae | L. monocytogenes | P. aeruginosa | S. epidermidis | S. flexneri | S. marcescens |
|---|---|---|---|---|---|---|---|
| 1 | 54 ± 11 | 24 | 100 | 10 | 98 | 0 | 0 |
| 2 | 97 | 100 | 100 | 19 | 97 | 43 | 81 ± 11 |
| 3 | 65 ± 9 | 27 | 100 | 9 | 100 | 16 | 0 |
| 4 | ND | ND | ND | ND | ND | ND | ND |
| 5 | 97 | 76 | 100 | 16 | 100 | 78 | 93 |
| 6 | 95 | 52 | 100 | 18 | 100 | 48 ± 6 | 0 |
| 7 | 86 | 28 | 100 | 0 | 100 | 23 ± 6 | 0 |
| 8 | 96 | 54 | 100 | 16 | 100 | 54 ± 10 | 67 |
| 9 | 96 | 33 | 100 | 0 | 100 | 53 ± 6 | 8 |
| 10 | 96 | 27 | 100 | 0 | 100 | 67 | 18 ± 6 |
| 11 | 94 | 18 | 100 | 16 | 100 | 36 ± 5 | 63 |
| 12 | 95 | 38 ± 8 | 100 | 4 | 100 | 38 ± 7 | 68 ± 5 |
| AMA | 4 | 0 | 37 | 5 | 0 | 9 | 0 |
| NEO | 97 | 100 | 100 | 11 | 95 | 100 | 100 |
Standard error (±) is given for replicate assays when the error was >5%.
Table 4.
MIC Values (µM) for Compounds 2, 5, 6, 10, and 12
| compound | E. coli | P. mirabilis | MRSA 33591 | MRSA A960649 | C. freundii |
|---|---|---|---|---|---|
| 2 | 12.5 | 12.5 | ND | ND | 3.2 |
| 5 | 12.5 | 12.5 | 50–100 | ND | 6.3 |
| 6 | 12.5 | 50–100 | ND | >100 | ND |
| 10 | ND | ND | ND | ND | 6.3 |
| 12 | ND | ND | ND | ND | 6.3 |
| NEO | 1.56 | 6.3 | 400 | 400 | 0.78 |
| compound | E. cloacae | L. monocytogenes | S. epidermidis | S. flexneri | S. marcescens |
|---|---|---|---|---|---|
| 2 | 12.5 | 1.6 | 3.2 | 12.5 | 12.5 |
| 5 | 12.5 | 1.6 | 1.6 | 6.3 | 12.5 |
| 6 | ND | ND | ND | ND | ND |
| 10 | 12.5–25 | 1.6 | 1.6 | 12.5 | >25 |
| 12 | >25 | 3.2 | 3.2 | 12.5 | 12.5–25 |
| NEO | 3.25 | 1.6 | 1.6 | 6.3 | 3.2 |
Table 5.
Evaluation of Hemolysis of Rabbit Erythrocytes (%) in the Presence of AMA–NEO Conjugates (100 µM)a
| compound | % hemolysis |
|---|---|
| Triton X-100 | 100.0 |
| NEO | 7.0 ± 0.4 |
| DMSO | 4.0 ± 0.1 |
| AMA | 10.0 ± 4.6 |
| 1 | 4.0 ± 0.1 |
| 2 | 10.0 ± 0.2 |
| 3 | 4.0 ± 0.8 |
| 4 | 4.0 ± 0.1 |
| 5 | 5.0 ± 1.2 |
| 6 | 4.0 ± 0.9 |
| 7 | 5.0 ± 0.4 |
| 8 | 5.0 ± 1.7 |
| 9 | 4.0 ± 0.3 |
| 10 | 4.0 ± 0.6 |
| 11 | 5.0 ± 0.8 |
| 12 | 5.0 ± 0.2 |
Red blood cells were incubated with each test compound at 37 °C for 1 h. Experiments were performed in triplicate, and the results are expressed as the average percent hemolysis relative to the positive control (Triton X-100).
Although most of the compounds have reduced antibacterial activity compared to NEO, the most antibacterially active compounds, 2, 4, and 5, have a single methylene group connecting NEO and a linker, but, interestingly, substitution of the methylene group for the longer ethyl thioether linkage led to the loss of antibacterial activity (compounds 7, 9, and 10) in most of the strains with the exceptions of C. freundii, L. monocytogenes, and S. epidermidis, which were sensitive to most or all compounds. On the contrary, substitution of CH2 in compound 1, the worst A-site binder, for the ethyl thioether linkage significantly improved binding and antibacterial properties of its counterpart, compound 6, even though there was no clear correlation between affinity to the E. coli A-site and antibacterial properties of the compounds.
In Vitro Inhibition of Translation
All compounds tested for antibacterial activity were assayed in a cell-free translation system for prokaryotes (Figure S4). Many of the compounds were stronger inhibitors of protein synthesis at nanomolar concentrations as compared to the parent aminoglycoside (NEO). This was most notable for compounds 2, 4, 5, 10, and 11 with IC50 values of 50, 40, 35, 71, and 63 nM, respectively, compared with NEO (IC50 = 139 nM), whereas compound 1 did not inhibit translation, was the weakest binder in NEO displacement assays, and exhibited the weakest antibacterial activity. These results are consistent with that reported by others where NEO and NEO-conjugates exhibit selectivity for prokaryotic translation.38 The IC50 values for compounds 2, 5, 10, and 11 correlate well with the moderate to high antibacterial activities observed with C. freundii, L. monocytogenes, and S. epidermidis, with compound 2 exhibiting the broadest spectrum of activity against bacterial strains (Table 3). Although compound binding to the rRNA A-site is similar between the A-site constructs (Figure 4), the overall pattern of weak and strong association is consistent with data from the antimicrobial assays and in vitro translation assays. For these compounds, the appropriate linker allows both anthraquinone and aminoglycoside binding to inhibit translation in vivo. The control AMA did not inhibit in vitro translation of luxAB indicating AMA does not interfere with the translation components. In addition, specificity of inhibition of prokaryotic translation by these compounds was also tested using select compounds. Results for the in vitro translation assay for eukaryotes clearly show that inhibition of translation is not observed for compounds 1, 5, 11, and the NEO control (Figure S5).
AME Studies
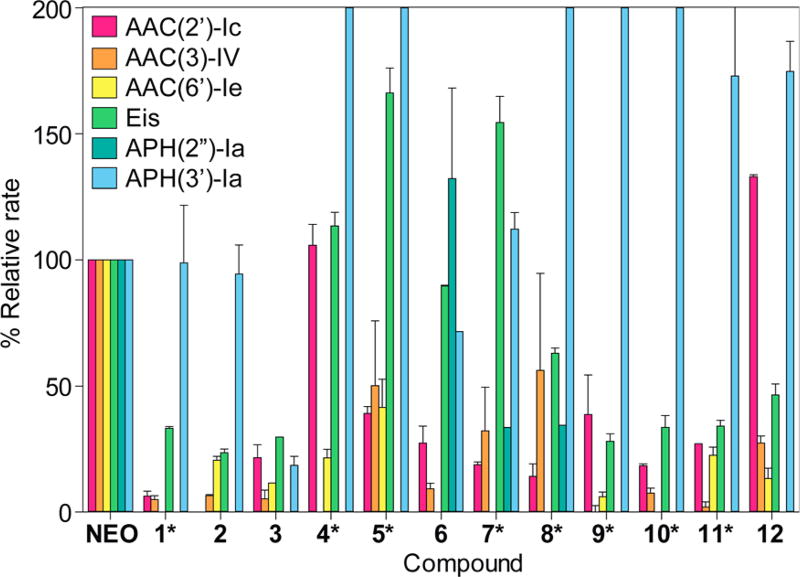
To determine if our novel AMA–NEO conjugates could resist the action of AMEs, we tested them against a panel of six AMEs: AAC(6′)-Ie, AAC(3)-IV, AAC(2′)-Ic, Eis, APH(2″)-Ia, and APH(3′)-Ia. In general, all AMEs tested showed reduced reaction rates with conjugates 1–12, with the exceptions of APH(3′)-Ia and, to some extent, Eis (Figure 5). The observed increases in reaction rates for APH(3′)-Ia could be caused by an increased affinity for this particular conjugation, or even just altered kinetics of the substrate with this particular phosphotransferase. It is also fairly difficult to design aminoglycosides that completely avoid modification by Eis, as this particular AAC modifies multiple amines on aminoglycosides.39 All regiospecific AACs showed a reduction in reaction rate with conjugates 1–12 when compared to the rate of the parent NEO. Some exceptions exist (e.g., AAC(2′)-Ic with compounds 4 and 12); however, given the complexity of AMEs and aminoglycoside structures, it is not surprising that a single modification does not result in avoiding modifications by all AMEs. Previous studies have shown that certain modifications work better for different enzymes. For example, dimerization of NEO with various linkers does not affect the rate of AAC(2′)-Ic,14 whereas the conjugation of NEO does not impede the rate of AAC(3)-IV’s reaction;40 conversely, the addition of short peptide chains seems to slow the rates of all the AMEs tested.32 This observation does not apply to just NEO; other modified aminoglycosides have also shown enzyme-selective rate impediments.41–43 For many of the compounds, there is corroboration from binding assays, AME studies, in vitro translation inhibition assays, and antibacterial activity (Figures 4 and 5; Tables 3 and 4). Most notable was compound 2, which expresses increased resistance to AME AAC(2′)-Ic attack, a higher value in the NEO displacement assay, a stronger inhibition of translation as compared to NEO, and the broadest spectrum of antimicrobial activity. That this compound is resistant to attack by common bacterial antibiotic resistance enzymes is significant. Because many of the conjugates were poorer substrates for most of the tested AMEs as compared with NEO, this knowledge can lead to a better understanding for designing drugs resistant to bacterial enzyme modification.
Figure 5.
Bar graph showing the relative rates of AME activity in the presence of AMA–NEO conjugates normalized to the parent aminoglycoside (NEO). The scale of the y axis was capped at 200% to highlight the usefulness of the compounds that were reacted more slowly than NEO, rather than leaving the graph at full size and highlight the compounds that reacted more quickly than NEO. For the compounds under modification by APH(3′)-Ia that went over 200% activity, the actual values follow: 4, 347 ± 94; 5, 254 ± 46; 8, 249 ± 45; 9, 292 ± 40; and 10, 278 ± 78.
CONCLUSION
A rapid assay that allows us to screen rRNA compounds has been expanded to include numerous rRNA targets of biological relevance to aminoglycoside function in the clinic. With the exception of compound 1, all studied conjugates demonstrated binding that was comparable to NEO for model A-sites, but their antibacterial properties and abilities to act as substrates for AMEs varied significantly. This variation was also mirrored in the prokaryotic vitro translation inhibition assay. A clear specificity of the AMA–NEO conjugates for inhibition of prokaryotic translation was confirmed, as these compounds do not inhibit eukaryotic translation even at much higher concentrations. Overall, the conjugates were poorer substrates for the tested AMEs as compared with NEO, allowing a better understanding of developing antibacterial compounds resistant to bacterial enzymatic attack. Most of the bacterial strains used in this study represent genera that are normal inhabitants of the human body and are responsible for community and nosocomial acquired infections and are under selective pressure for developing resistance to antimicrobial drugs. It is notable that there is significant variation in the antibacterial properties of these 12 compounds within and between strains ranging from insignificant to high inhibition. E. cloacae was moderately inhibited by two of the compounds, whereas S. epidermidis and L. monocytogenes were inhibited by all compounds tested. Bacterial inhibition by modified constructs requires a multitude of factors that must act in unison. For example, a balancing act between RNA binding, inhibition of translation, AME activity, and uptake and efflux mechanisms for these synthetic ligands needs to be achieved for the development of novel antimicrobials. Our data here show that by adding the anthraquinone unit, we can modulate the RNA binding, translation inhibition, and AME activity, but significant improvements in bacterial inhibition were not seen. The uptake of the modified conjugates likely affects their antibacterial property, and further studies with permeable strains and AME mutant strains will be necessary to further investigate the role of modifications made. Useful information can be derived to better understand bacterial physiologic responses to different chemical constructs, as bacteria are sensitive indicators of change in the environment. This variation in response can be attributed to the vast differences in metabolism and physiological characteristics between species and strains of bacteria. For example, capsule production and biofilm formation by K. pneumoniae can impede uptake of compounds and support antibiotic tolerance. C. freundii was sensitive to most of the compounds tested. It has been reported that C. freundii is responsible for a third of all opportunistic infections of the respiratory, urinary, and blood systems in the clinical environment.44 The normal habitat for L. monocytogenes of soil and water is responsible for foodborne illness and can cause bacteremia and meningitis. This organism was sensitive to all compounds. Significant inhibition by some compounds in S. epidermidis and L. monocytogenes was comparable to the MIC values found for NEO. S. epidermidis, a ubiquitous inhabitant of human skin, is responsible for many opportunistic infections via catheters and other medical implants. With heavy usage of topical antibiotics S. epidermidis serves as a natural reservoir for antibiotic resistance genes. Development of novel variations on currently used antibiotics can help limit the spread of opportunistic pathogens in the clinical environment.
METHODS
Materials
Aminoglycosides and buffer components were purchased from Fisher Scientific (Pittsburgh, PA, USA) or Sigma-Aldrich (St. Louis, MO, USA) and used without further purification. MH II broth and agar were purchased from BD (Sparks, MD, USA). Tryptic soy (TS) broth and agar were purchased from MP Biomedicals (Solon, OH, USA).
Microbial Studies
A. baumannii ATCC 19606, E. cloacae ATCC 13047, E. faecalis ATCC 29212, E. coli ATCC 25922, L. monocytogenes ATCC 19115, P. aeruginosa ATCC 27853, S. epidermidis ATCC 12384, S. marcescens ATCC 13880, and MRSA ATCC 33591 were purchased from the American Type Culture Collection (Manassas, VA, USA). C. freundii 4747 CFAA, S. flexneri 2457 NR-517, P. mirabilis HM-752, MRSA A960649 NR-45914, and K. pneumoniae NR-151410 were obtained from BEI Resources (Manassas, VA, USA). K. pneumoniae was cultured in TS broth. MH II broth was used for culturing of all other strains. Lysed horse blood (5%) was amended to MH II for culturing L. monocytogenes. Microbial cultures were grown in broth overnight at 37 °C to OD595 ~ 0.4–0.9. The cell concentration was measured spectrophotometrically at 595 nm and diluted with broth to a final concentration of 2.5 × 105 CFU/mL.
Compounds 1–12 were screened against bacterial strains at a single 6.3 µM concentration using 96-well plates. In each well, 90 µL of bacterial culture was mixed with either 10 µL of H2O (positive control), test compound, NEO (negative control), 2-aminomethylanthraquinone (AMA-derivative control), or DMSO (vehicle control) solution. Each plate contained 90 µL of broth with 10 µL of sterile water as a broth sterility control. All compounds and controls were tested at least in duplicate. Plates were incubated for 20–24 h at 37 °C, with the exception of a 48 h incubation for L. monocytogenes, which grows more slowly. The bacterial concentration was determined by absorbance at 595 nm by using a TECAN M1000Pro plate reader. The percent inhibition and percent growth were calculated using the formula
where A is the average absorbance of the well with added compound, Ab is the broth absorbance, and Ac is the absorbance of the positive control (normal growth of bacteria without treatment).
The double-dilution method was used to determine MIC values for selected compounds that yielded significant inhibition in the single-point assay. Compound concentrations ranged from 0.39 to 100 µM. All measurements were performed in triplicate. The MIC values were determined as an average minimal concentration of the compound at which percent growth was ≤5% as compared to the positive control without antibiotic.
Hemolysis Assay
The hemolysis assay was performed as previously described.45 Rabbit erythrocytes (2% w/w) were incubated for 1 h at 37 °C with each of the test compounds starting with a concentration of 100 µM. Controls included phosphate-buffered saline (negative control), 0.5% DMSO (vehicle control), AMA (derivative control), NEO (aminoglycoside control), and Triton X-100 (1% w/v) as the positive control for 100% hemolysis. After incubation, assay tubes were centrifuged at 10,000 rpm for 10 min at room temperature. The supernatant was removed and transferred to a 96-well clear plate, and absorbance was measured at 540 nm using a microplate reader (Infinite M1000Pro, TECAN). The results were expressed as the average percent hemoglobin released as compared with the Triton X-100 control in triplicate assays.
Screening against A-Sites
The method for the fluorescence-based ribosome binding assay used here has been previously described.46 RNA for A-site analogues was ordered from IDT (Coralville, IA, USA) in the desalted form and used without further purification. Samples were dissolved in DEPC-treated H2O, diluted with buffer to the desired concentrations, heated to 90 °C, and quenched on ice to favor hairpin formation. The following sequences for the 27-base A-site models were used in the study:
E. coli, 5′-GGCGUCACACCUUCGGGUGAAGUCGCC-3′;
human cytosolic, 5′-GGCGUCGCUCCUUCGGGAAAAGUCGCC-3′;
mitochondrial, 5′-GGCGUCACCCCUUCGGGACAAGUCGCC-3′;
C1494U, 5′-GGCGUCACUCCUUCGGGACAAGUCGCC-3;
A1555G, 5′-GGCGUCACCCCUUCGGGGCAAGUCGCC-3′.
All fluorescence experiments were performed in duplicate in 10 mM HEPES, 50 mM NaCl, and 0.4 mM EDTA (pH 7) in black solid 96-well plates. Plates were scanned at λex = 485 nM/λem = 525 nM using a TECAN M1000Pro plate reader. Single-point emission data for the emission maxima were collected in triplicate for duplicate samples. To determine IC50 values, fluorescence emission data were averaged and plotted as a function of log concentration of compound (Figures S1, S2, and S3).
A fluorescent displacement assay developed in-house was utilized for compound screening against A-site RNAs.33 F–NEO was mixed with an A-site at a 1:1 ratio to form a complex at 0.1 µM. NEO or its AMA conjugates were added to the complex at 0.3 µM. Controls included the F–NEO complex without added drug and negative control with test compound without F–NEO. Plates were incubated for 10 min at room temperature before the emission scan. Percent binding relative to NEO was calculated according to the formula
where I is the intensity of the complex with added drug, Ic is the intensity of the control without a drug, and INEO is the intensity of the complex with added NEO.
Selectivity factors for A-sites relative to the E. coli A-site were calculated using the following formula:14
Determination of Selectivity Factors
This calculation is intended for a quick and convenient estimation of binding preferences of a compound (cmpd) to one A-site versus another. We conducted an exchange reaction between A-site–F–NEO complex and a compound with binding constant KE. coli for the E. coli A-site and Khuman for the human A-site:
The following relationship is true:
KdF–NEO(E. coli) and KdF–NEO(human) are dissociation constants between F–NEO and E. coli and human A-sites, respectively, and Kdcmpd(E. coli) and Kdcmpd(human) are dissociation constants between a compound and E. coli and human A-sites, respectively.
After regrouping:
Measurement of KE. coli and Khuman is not possible under our experimental conditions (for example, utilized concentrations, signal detection, etc.); therefore, we substitute K with conveniently measured IC50, a total concentration of compound or NEO, at which the emission intensity at half the maximum value is observed (see the Supporting Information). A higher IC50 value corresponds to a lower binding affinity when IC50 values for the same A-site are considered. After substitution of the ratio KE. coli/Khuman by IC50 (E. coli)/IC50 (human), we can calculate the selectivity factor representing the approximation of the binding preference of a compound to E. coli over the human A-site:
Cell-free Translation Assays for Prokaryotic and Eukaryotic Systems
Cell-free in vitro translation assays for prokaryote and eukaryote systems were used employing the luxAB reporter gene. IC50 values were determined as the total concentration of compound or NEO at which the luminescence intensity is half the maximum value observed. For determination of IC50 of test compounds in prokaryotic systems, the E. coli S30 Extract System for Circular DNA (Promega, L1020) was used. A Master Mix (MM) was created by combining 180 µL of S30 premix, 135 µL of S30 extract, 95 µL of nanopure water, and 40 µL of amino acid mixture for a total volume of 450 µL. Each test compound was assayed at 10 concentration points serially diluted in DMSO for final concentrations of 1.25 µM–2.4 nM. The pBESTluc provided in the kit was diluted from 10 to 54.4 µL with 1× TE buffer. A volume of 12.5 µL of MM was aliquoted to a 1.5 mL centrifuge tube, followed by 0.5 µL of test compound. The tubes were then mixed and centrifuged briefly. The tubes were held at room temperature for 20 min, after which 0.4 µL of pBESTluc was added. After brief mixing and centrifugation, the tubes were incubated at 37 °C for 60 min. The tubes were put on ice for a 5 min inactivation period. After gentle mixing by pipet, 5 µL was aliquoted to a white half-volume 96-well plate (supplied by Greiner). Thirty-five microliters of a 1 mM luciferin solution (Promega) was added to each well, and the plate was read for luminescence after a 30 s shaking period. Luminescence was normalized to DMSO vehicle controls. Data were processed with Graphpad Prism 6.
For determination of the IC50 for eukaryotic systems, rabbit reticulocyte lysate kits were used (Promega). Cell lysate, RNasin, amino acids, and RNA were thawed on ice. Master Mix (MM) was created by combining 300 µL of cell lysate, 5 µL of RNasin, 137 µL of nanopure water, and 8 µL of amino acid mix for a total volume of 450 µL. Compounds were serially diluted in DMSO so that the final concentrations ranged from 4.2 µM to 5 nM. MM (12.5 µL) was aliquoted to a 1.5 mL centrifuge tube, followed by 0.5 µL of compound. The tubes were then mixed and centrifuged briefly. The tubes were held at room temperature for 20 min after which 0.4 µL of RNA was added. After brief mixing and centrifugation, the tubes were incubated at 30 °C for 90 min. The tubes were put on ice for a 5 min inactivation period. After gentle mixing by pipet, 5 µL was aliquoted to a white half-volume 96-well plate (supplied by Greiner). Thirty-five microliters of 1 mM luciferin solution (Promega) was added to each well, and the plate was read for luminescence after a 30 s shaking period. Luminescence was normalized to DMSO controls. Data were processed with Graphpad Prism 6.
Testing of AME Activities on Conjugates 1–12
To determine if various AMEs would have the power to inactivate our AMA–NEO conjugates, we used standard assays to visualize the modification of compounds 1–12. We used NEO as a control. All AME enzymes, AAC(6′)-Ie,47 AAC(3)-IV,47 AAC(2′)-Ic,48 Eis,48 APH(2″)-Ia,25 and APH(3′)-Ia,24 were purified and tested as previously described. All reactions were monitored at 25 °C (with the exception of AAC(6′)-Ie and APH(2″)-Ia, which were monitored at 37 °C) on a SpectraMax M5 microplate reader and performed in duplicate. All rates were normalized to NEO.
Acetylation
The activity of the acetyltransferases was monitored using Ellman’s method, coupling the release of the product (CoASH) with DTNB and monitored at 412 nm (ε 14,150 cm−1 M−1). Briefly, reactions (200 µL) containing NEO conjugate (100 µM) and AcCoA (500 µM for Eis and 150 µM for all other acetyltransferases) were incubated with the enzymes (0.125 µM for AAC(3)-IV and AAC(2′)-Ic; 0.5 µM for all remaining acetyltransferases) in the presence of DTNB (2 mM) and the appropriate buffer (50 mM MES, pH 6.6, for AAC(6′)-Ie and AAC(3)-IV, 50 mM Tris-HCl, pH 7.5, for AAC(6′)-Ib, 100 mM sodium phosphate, pH 7.4, for AAC(2′)-Ic, and 50 mM Tris-HCl, pH 8.0, for Eis). Using kinetic measurements, reading every 30 s for 30 min, initial rates of the reactions were calculated using the first 2 min of the reaction.
Phosphorylation
The phosphorylation activity of APH(2″)-Ia and APH(3′)-Ia was monitored at 340 nM through the consumption of NADH in the well-established enzyme-coupled response to the production of ADP. Reactions (200 µL) containing AMA–NEO conjugate (100 µM), Tris-HCl (50 mM, pH 8.0), MgCl2 (10 mM), KCl (40 mM), NADH (0.5 mg/mL), PEP (2.5 mM), GTP (2 mM), and PK/LDH (4 µL) were initiated by the addition of the phosphotransferase (1 µM). Reactions were monitored kinetically, taking measurements every 30 s for 30 min. Initial rates were determined using the first 5 min of the reaction.
Supplementary Material
Acknowledgments
This research was supported by NIH Grants GM097917 and AI114114 to D.P.A., NIH Grant I090048 to S.G.-T., and the Vasser-Woolley fellowship to A.K.O. We thank Dr. Souvik Sur for building the computer model.
ABBREVIATIONS
- AAC
aminoglycoside acetyltransferase
- AME
aminoglycoside-modifying enzyme
- AMK
amikacin
- ANT
aminoglycoside nucleotidyltransferase
- APH
aminoglycoside phosphotransferase
- Eis
enhanced intracellular survival
- NEO
neomycin
- MIC
minimum inhibitory concentration
- MH II
Mueller Hinton II
- TS
tryptic soy
- MRSA
methicillin-resistant S. aureus
- DEPC
diethylpyrocarbonate
- AMA
2-aminomethylanthraquinone
Footnotes
ASSOCIATED CONTENT
- Titration plots of A-site constructs; inhibition of in vitro translation for prokaryotic and eukaryotic systems (PDF)
The authors declare the following competing financial interest(s): DPA has ownership interest in NUBAD LLC.
References
- 1.Arya DP. In: Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery. Wang B, editor. Wiley; 2007. p. 319. [Google Scholar]
- 2.Willis B, Arya DP. An expanding view of aminoglycoside-nucleic acid recognition. Adv. Carbohydr. Chem. Biochem. 2006;60:251–302. doi: 10.1016/S0065-2318(06)60006-1. [DOI] [PubMed] [Google Scholar]
- 3.Davies J, Davis BD. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J. Biol. Chem. 1968;243:3312–3316. [PubMed] [Google Scholar]
- 4.Davies J, Benveniste R, Kvitek K, Ozanne B, Yamada T. Aminoglycosides: biologic effects of molecular manipulation. J. Infect. Dis. 1969;119:351–354. doi: 10.1093/infdis/119.4-5.351. [DOI] [PubMed] [Google Scholar]
- 5.Davies J, Gilbert W, Gorini L. Streptomycin, suppression, and the code. Proc. Natl. Acad. Sci. U. S. A. 1964;51:883–890. doi: 10.1073/pnas.51.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran TP, Vo DD, Di Giorgio A, Duca M. Ribosome-targeting antibiotics as inhibitors of oncogenic microRNAs biogenesis: old scaffolds for new perspectives in RNA targeting. Bioorg. Med. Chem. 2015;23:5334–5344. doi: 10.1016/j.bmc.2015.07.062. [DOI] [PubMed] [Google Scholar]
- 7.Vo DD, Tran TP, Staedel C, Benhida R, Darfeuille F, Di Giorgio A, Duca M. Oncogenic MicroRNAs Biogenesis as a Drug Target: Structure-Activity Relationship Studies on New Aminoglycoside Conjugates. Chem.–Eur. J. 2016;22:5350–5362. doi: 10.1002/chem.201505094. [DOI] [PubMed] [Google Scholar]
- 8.Watkins D, Jiang L, Arya DP. A pH sensitive high throughput assay for miRNA binding of a peptide-aminoglycoside (PA) library. PLoS One. 2015;10:1–23. doi: 10.1371/journal.pone.0144251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson MD, Lund AH. MicroRNA and cancer. Mol. Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariza-Mateos A, Diaz-Toledano R, Block TM, Prieto-Vega S, Birk A, Gomez J. Geneticin Stabilizes the Open Conformation of the 5′ Region of Hepatitis C Virus RNA and Inhibits Viral Replication. Antimicrob. Agents Chemother. 2016;60:925–935. doi: 10.1128/AAC.02511-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranjan N, Kumar S, Watkins D, Wang D, Appella DH, Arya DP. Recognition of HIV-TAR RNA using neomycin–benzimidazole conjugates. Bioorg. Med. Chem. Lett. 2013;23:5689–5693. doi: 10.1016/j.bmcl.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabbvarapu N, Degani Y, Shavit M, Smolkin B, Belakhov V, Baasov T. Design of novel aminoglycoside derivatives with enhanced suppression of diseases-causing nonsense mutations. ACS Med. Chem. Lett. 2016;7:418–423. doi: 10.1021/acsmedchemlett.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, Watkins D, Jin Y, Gong C, King A, Washington AZ, Green KD, Garneau-Tsodikova S, Oyelere AK, Arya DP. Rapid Synthesis, RNA Binding, and Antibacterial Screening of a Peptidic-Aminosugar (PA) Library. ACS Chem. Biol. 2015;10:1278–1289. doi: 10.1021/cb5010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y, Watkins D, Degtyareva NN, Green KD, Spano MN, Garneau-Tsodikova S, Arya DP. Arginine-linked neomycin B dimers: synthesis, rRNA binding, and resistance enzyme activity. MedChemComm. 2016;7:164–169. doi: 10.1039/C5MD00427F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cock IE. The Genus Aloe: Phytochemistry and Therapeutic Uses Including Treatments for Gastrointestinal Conditions and Chronic Inflammation. Prog. Drug Res. 2015;70:179–235. doi: 10.1007/978-3-0348-0927-6_6. [DOI] [PubMed] [Google Scholar]
- 16.Vasas A, Orban-Gyapai O, Hohmann J. The Genus Rumex: Review of traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2015;175:198–228. doi: 10.1016/j.jep.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Xue L, Xi H, Kumar S, Gray D, Davis E, Hamilton P, Skriba M, Arya DP. Probing the recognition surface of a DNA triplex: binding studies with intercalator-neomycin conjugates. Biochemistry. 2010;49:5540–5552. doi: 10.1021/bi100071j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranjan N, Davis E, Xue L, Arya DP. Dual recognition of the human telomeric G-quadruplex by a neomycin-anthraquinone conjugate. Chem. Commun. 2013;49:5796–5798. doi: 10.1039/c3cc42721h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies J, Wright GD. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 20.Wright GD, Berghuis AM, Mobashery S. Aminoglycoside antibiotics: structures, functions, and resistance. Adv. Exp. Med. Biol. 1998;456:27–69. [PubMed] [Google Scholar]
- 21.Thompson PR, Hughes DW, Wright GD. Regiospecificity of Aminoglycoside Phosphotransferase from Enterococci and Staphylococci (APH(3′)-IIIa. Biochemistry. 1996;35:8686–8695. doi: 10.1021/bi960389w. [DOI] [PubMed] [Google Scholar]
- 22.Serpersu EH, Ozen C, Wright E. Studies of enzymes that cause resistance to aminoglycosides antibiotics. Methods Mol. Med. 2008;142:261–271. doi: 10.1007/978-1-59745-246-5_20. [DOI] [PubMed] [Google Scholar]
- 23.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist. Updates. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watkins D, Kumar S, Green KD, Arya DP, Garneau-Tsodikova S. Influence of linker length and composition on enzymatic activity and ribosomal binding of neomycin dimers. Antimicrob. Agents Chemother. 2015;59:3899–3905. doi: 10.1128/AAC.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green KD, Chen W, Garneau-Tsodikova S. Effects of altering aminoglycoside structures on bacterial resistance enzyme activities. Antimicrob. Agents Chemother. 2011;55:3207–3213. doi: 10.1128/AAC.00312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am. J. Hum. Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Estivill X, Govea N, Barcelo E, Badenas C, Romero E, Moral L, Scozzri R, D’Urbano L, Zeviani M, Torroni A. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am. J. Hum. Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobbie SN, Pfister P, Bruell C, Westhof E, Bottger EC. Analysis of the Contribution of Individual Substituents in 4,6-Aminoglycoside-Ribosome Interaction. Antimicrob. Agents Chemother. 2005;49:5112–5118. doi: 10.1128/AAC.49.12.5112-5118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbie SN, Akshay S, Kalapala SK, Bruell CM, Shcherbakov D, Bottger EC. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20888–20893. doi: 10.1073/pnas.0811258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch SR, Puglisi JD. Structural Origins of Aminoglycoside Specificity for Prokaryotic Ribosomes. J. Mol. Biol. 2001;306:1037–1058. doi: 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]
- 31.Watkins D, Gong C, Kellish P, Arya DP. Probing A-form DNA: a fluorescent aminosugar probe and dual recognition by anthraquinone-neomycin conjugates. Bioorg. Med. Chem. 2016 doi: 10.1016/j.bmc.2016.11.003. in press. [DOI] [PubMed] [Google Scholar]
- 32.Jiang L, Watkins D, Jin Y, Gong C, King A, Washington AZ, Green KD, Garneau-Tsodikova S, Oyelere AK, Arya DP. Rapid Synthesis, RNA Binding, and Antibacterial Screening of a Peptidic-Aminosugar (PA) Library. ACS Chem. Biol. 2015;10:1278–1289. doi: 10.1021/cb5010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watkins D, Norris FA, Kumar S, Arya DP. A fluorescence-based screen for ribosome binding antibiotics. Anal. Biochem. 2013;434:300–307. doi: 10.1016/j.ab.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamasaki K, Rando RR. Specific binding of aminoglycosides to a human rRNA construct based on a DNA polymorphism which causes aminoglycoside-induced deafness. Biochemistry. 1997;36:12323–12328. doi: 10.1021/bi970962r. [DOI] [PubMed] [Google Scholar]
- 35.Recht MI, Douthwaite S, Puglisi JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 1999;18:3133–3138. doi: 10.1093/emboj/18.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De La Fuente R, D Sonawane N, Arumainayagam D, Verkman AS. Small molecules with antimicrobial activity against E. coli and P. aeruginosa identified by high-throughput screening. Br. J. Pharmacol. 2006;149:551–559. doi: 10.1038/sj.bjp.0706873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Zhao H, Kong W, Jin CZY, Qu Y, Xiao X. Microcalorimetric assay on the antimicrobial property of five hydroxyanthraquinone derivatives in rhubarb (Rheum palmatum L.) to Bifidobacterium adolescentis. Phytomedicine. 2010;17:684–689. doi: 10.1016/j.phymed.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Carriere M, Vijayabaskar V, Applefield D, Harvey I, Garneau P, Lorsch J, Lapidot A, Pelletier J. Inhibition of protein synthesis by aminoglycoside-arginine conjugates. RNA. 2002;8:1267–1279. doi: 10.1017/s1355838202029059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houghton JL, Biswas T, Chen W, Tsodikov OV, Garneau-Tsodikova S. Chemical and structural insights into the regioversatility of the aminoglycoside acetyltransferase Eis. ChemBioChem. 2013;14:2127–2135. doi: 10.1002/cbic.201300359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watkins D, Kumar S, Green KD, Arya DP, Garneau-Tsodikova S. Influence of linker length and composition on enzymatic activity and ribosomal binding of neomycin dimers. Antimicrob. Agents Chemother. 2015;59:3899–3905. doi: 10.1128/AAC.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkov-Zrihen Y, Green KD, Labby KJ, Feldman M, Garneau-Tsodikova S, Fridman M. Synthesis and evaluation of hetero- and homodimers of ribosome-targeting antibiotics: antimicrobial activity, in vitro inhibition of translation, and drug resistance. J. Med. Chem. 2013;56:5613–5625. doi: 10.1021/jm400707f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herzog IM, Green KD, Berkov-Zrihen Y, Feldman M, Vidavski RR, Eldar-Boock A, Satchi-Fainaro R, Eldar A, Garneau-Tsodikova S, Fridman M. 6″-Thioether tobramycin analogues: towards selective targeting of bacterial membranes. Angew. Chem., Int. Ed. 2012;51:5652–5656. doi: 10.1002/anie.201200761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrestha SK, Fosso MY, Green KD, Garneau-Tsodikova S. Amphiphilic Tobramycin Analogues as Antibacterial and Antifungal Agents. Antimicrob. Agents Chemother. 2015;59:4861–4869. doi: 10.1128/AAC.00229-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whalen JG, Mully TW, English JC., III Spontaneous Citrobacter freundii infection in an immunocompetent patient. Arch. Dermatol. 2007;143:124–125. doi: 10.1001/archderm.143.1.124. [DOI] [PubMed] [Google Scholar]
- 45.Berkov-Zrihen Y, Herzog IM, Benhamou RI, Feldman M, Steinbuch KB, Shaul P, Lerer S, Eldar A, Fridman M. Tobramycin and nebramine as pseudo-oligosaccharide scaffolds for the development of antimicrobial cationic amphiphiles. Chem.–Eur. J. 2015;21:4340–4349. doi: 10.1002/chem.201406404. [DOI] [PubMed] [Google Scholar]
- 46.Watkins D, Norris FA, Kumar S, Arya DP. A Fluorescence-based Screen for Ribosome Binding Antibiotics. Anal. Biochem. 2013;434:300–307. doi: 10.1016/j.ab.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green KD, Chen W, Houghton JL, Fridman M, Garneau-Tsodikova S. Exploring the substrate promiscuity of drug-modifying enzymes for the chemoenzymatic generation of N-acylated aminoglycosides. ChemBioChem. 2010;11:119–126. doi: 10.1002/cbic.200900584. [DOI] [PubMed] [Google Scholar]
- 48.Chen W, Biswas T, Porter VR, Tsodikov OV, Garneau-Tsodikova S. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9804–9808. doi: 10.1073/pnas.1105379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.