Abstract
The exocyst is an evolutionarily conserved octameric protein complex that mediates the tethering of post-Golgi secretory vesicles to the plasma membrane during exocytosis, and is implicated in many cellular processes such as cell polarization, cytokinesis, ciliogenesis and tumor invasion. Using cryo-electron microscopy (cryo-EM) and chemical cross-linking mass spectrometry (CXMS), we solved the structure of the Saccharomyces cerevisiae exocyst complex at an average resolution of 4.4 Å. Our model revealed the architecture of the exocyst, and led to the identification of the helical bundles that mediate the assembly of the complex at its core. Sequence analysis suggests that these regions are evolutionarily conserved across eukaryotic systems. Further cell biological studies suggest a mechanism for exocyst assembly that leads to vesicle tethering at the plasma membrane.
Vesicular trafficking in eukaryotic cells is mediated by an elaborate network of molecular interactions that ensure the orderly transport, docking and fusion of secretory vesicles to their cognate target membrane compartments. The initial contacts between the secretory vesicles and their target membrane is mediated by the tether family of proteins1–5. The Multi-subunit Tethering Complexes (MTCs) capture the vesicles to their specific target membranes prior to SNARE-mediated fusion at various stages of vesicular trafficking3,5,6. Elucidating the structure and assembly of the MTCs is essential to the understanding of the mechanisms of vesicle tethering and fusion.
The exocyst, first identified in the budding yeast Saccharomyces cerevisiae, consists of Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo847,8. The exocyst mediates the tethering of post-Golgi secretory vesicles to the plasma membrane and promotes the assembly of the SNARE complex for membrane fusion2,8–11. Besides exocytosis, the exocyst also plays pivotal roles in many cellular processes such as cell polarization, primary ciliogenesis, cytokinesis, pathogen invasion, tumorigenesis and metastasis10,12. The crystal structures of fragments of a few exocyst subunits have been solved; these regions were mostly helical-bundle repeats and GTPase-binding domains that assume certain levels of structural autonomy13. Previous quick-freeze deep-etch EM and negative staining EM showed that the exocyst has an elongated shape consisting of packed long rods14,15. The rod structure seems to be a shared feature for the Complex Associated with Tethering Containing Helical Rods (CATCHRs) proteins such as Conserved Oligomeric Golgi complex (COG) and Golgi-Associated Retrograde Protein complex (GARP)5,16,17. Recently, a light microscopic approach, based on fluorescence proteins tagging, provided new information on the exocyst complex in cells18. A topology regarding the connectivity of the exocyst subunits has also been depicted using biochemical analyses and negative-staining EM15. However, the structure model and mechanism of exocyst assembly remain unknown.
Using single particle cryo-EM, we have solved the structure of the fully assembled yeast exocyst complex at an average resolution of 4.4 Å. The cryo-EM, together with chemical cross-linking mass spectrometry (CXMS) and cell biological assays, provides insights to the hierarchical assembly of the exocyst complex and mechanism of vesicle tethering.
RESULTS
Structure determination of the exocyst complex
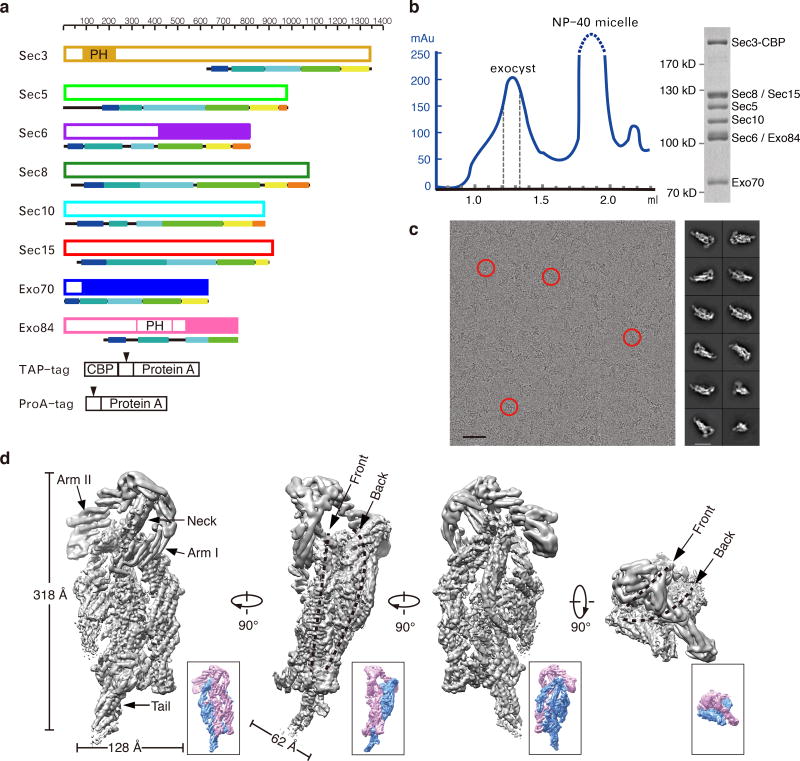
We tagged the chromosomal copy of individual exocyst subunits by TAP or ProA, and purified the exocyst complex from yeast cell lysates using affinity chromatography and size-exclusion chromatography (Fig. 1a and 1b). The purification yielded monodispersed intact exocyst complex comprising all of the eight subunits at equal stoichiometry (Fig. 1b). We next performed single-particle cryo-EM analysis of the complex and reconstituted the three-dimensional (3-D) structure of the intact complex at an average resolution of 4.4 Å (Fig. 1c–d, Supplementary Fig. 1–2). The holo-exocyst complex consists of long curved rods that pack against each other to form a hollow architecture measuring a length of 320 Å and a width of 130 Å (Fig. 1d). The central body appears as a double-layer oval disk with a thickness of 60 Å. The front layer nestles into the concave side of the back layer, forming a prolate cave between the two layers. From the top of the front layer, a neck-like structure stretches out, with two arms extending in opposite directions. One arm protrudes to the right edge of the front layer (henceforth termed “Arm I”), and the other extends backward over the back layer (termed “Arm II”). Stretching out from the bottom of the back layer is a tail-like structure (termed “Tail”). While the central portion of the reconstruction clearly shows bundles of α-helices with apparent helical pitches, Arm I, Arm II and Tail are less well resolved (Fig. 1d and Supplementary Fig. 2e), suggesting flexibility of these regions.
Figure 1. Cryo-EM structure of the exocyst complex.
a. Schematic diagram of the yeast exocyst subunits with their affinity tags. Each subunit is drawn following the scale above and color-coded as in later figures for consistency and referencing. Fragments with crystal structure solved are indicated in solid. Arrowheads indicate the TEV cleavage sites in TAP and ProA tag. b. Gel filtration of affinity purified exocyst complex. Peak fractions indicated within the dash lines were tested by SDS-PAGE (right panel, uncropped gel image is shown in Supplementary Data Set 4) and collected for cryo-EM sample preparation. c. A representative cryo-EM micrograph with particles of the exocyst complex indicated by red circles (left panel). Scale bar, 50 nm. Representative 2D class averages of the exocyst complex are shown to the right. Scale bar, 20 nm. d. The 4.4 Å map of the exocyst complex in different views. Dash lines indicate the two layers of the map. The front and back layers are also colored in pink and blue, respectively, in a lower resolution map (~6 Å, lower panel) shown in the lower right for better viewing.
The assignment and structure of the exocyst subunits
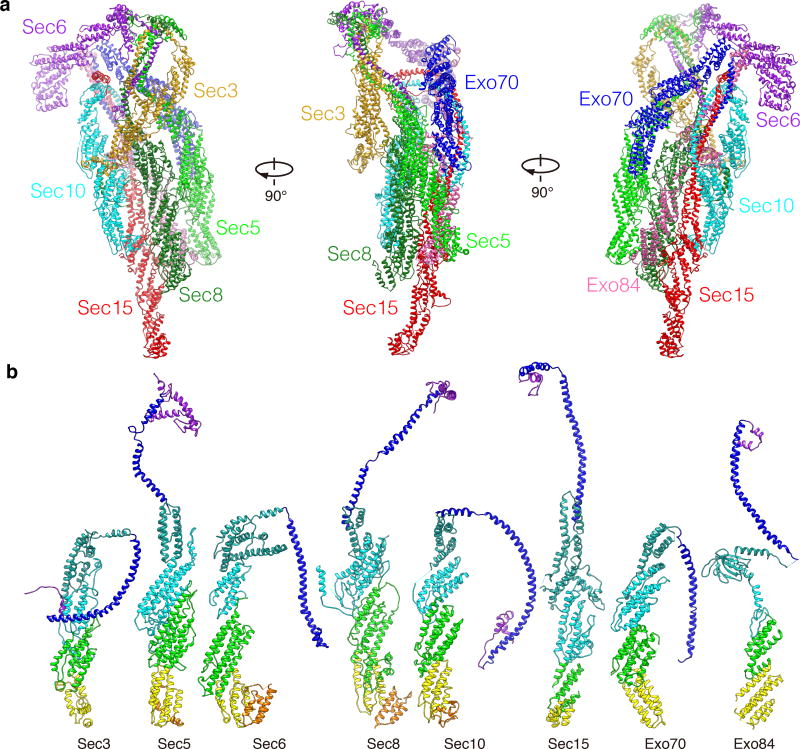
The 4.4 Å resolution does not allow clear resolution of the side chains. To gain better structure information of the exocyst, we performed extensive intra- and inter-molecular CXMS analyses. The combined approach allowed us to build a model of the exocyst complex consisting of the near full-length Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, Exo84, and the C-terminal half of Sec3 (Fig. 2a, Table 1, Supplementary Fig. 3–6, and Supplementary Table 1. Also see Method and Supplementary Video 1 for details in model building). When mapped onto the exocyst model, 86% of all crosslinks fall within 24 Å (Supplementary Data Set 1–3 and Supplementary Video 2), which is comparable with X-ray crystallography studies showing that ~70% of the crosslinks are compatible with such a constrain19. Even though the subunits share less than 10% sequence identity among themselves, all display an extended configuration with similar folds, consistent with previous prediction20 (Fig. 2b). All subunits contain an N-terminal long coiled-coil followed by a region of contiguous rod, which is composed of anti-parallel short helical bundle units (hereafter termed as “CAT domain” for its conservation in the CATCHR family proteins5). Each of the seven subunits (except for Exo84) has 4 CAT domains (henceforth termed as CAT-A to CAT-D) (Fig. 2b and Supplementary Table 1). For Exo84, there are only two CAT domains that follow its PH domain. The N-terminal half of Sec3 could not be modeled into the cryo-EM map, probably due to its flexibility. This is in agreement with the secondary structure prediction of this region, which contains a ~170 a.a. helix with flanking long unstructured sequences (Supplementary Fig. 7). The structure of the C-terminus of Sec3 was less well defined due to its poor density (Supplementary Fig. 5c). This is consistent with a conformational change observed at the rod region of Sec3 that accounts for 8.6% of the particles in the 3-D classifications (Supplementary Fig.1, Class 10).
Figure 2. The structure of the holo-exocyst complex and individual subunits.
a. The model of the exocyst complex. Subunits are color-coded as indicated. b. Structures of the exocyst subunits. All structures are color-coded based on domain arrangement (from N- to C-terminus: purple, N-terminal overhang; blue, the CorEx motif; sea green, CAT-A; cyan, CAT-B; green, CAT-C; yellow, CAT-D; Orange, C-terminus. In Exo84, sea green indicates the PH domain, and green and yellow indicate CAT-A and B, respectively).
Table 1.
Cryo-EM data collection, refinement and validation statistics
| The exocyst complex (PDB 5YFP) (EMD 6827) |
|
|---|---|
| Data collection | |
| Microscope | Titan Krios |
| Camera | Gatan K2 Summit |
| Magnification | 22,500 |
| Voltage (kV) | 300 |
| Electron dose (e–/Å2) | 4.8 |
| Defocus range (µm) | 2–3 |
| Pixel size (Å) | 1.30654 |
| Initial particles (no.) | 904,481 |
| Final particles (no.) | 343,342 |
| Model composition | |
| Nonhydrogen atoms | 33,148 |
| Protein residues | 5,982 |
| Ligands | 0 |
| Refinement | |
| Resolution range (Å) | 470-5.0 |
| FSC (or other resolution metric) | 0.8289 |
| Map sharpening B factor (Å2) | −200 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.331 |
| Validation | |
| MolProbity score | 2.50 |
| Clashscore | 16.80 |
| Poor rotamers (%) | 0.10 |
| Ramachandran plot | |
| Favored (%) | 81.49 |
| Allowed (%) | 17.71 |
| Disallowed (%) | 0.79 |
Hierarchical assembly of the exocyst complex
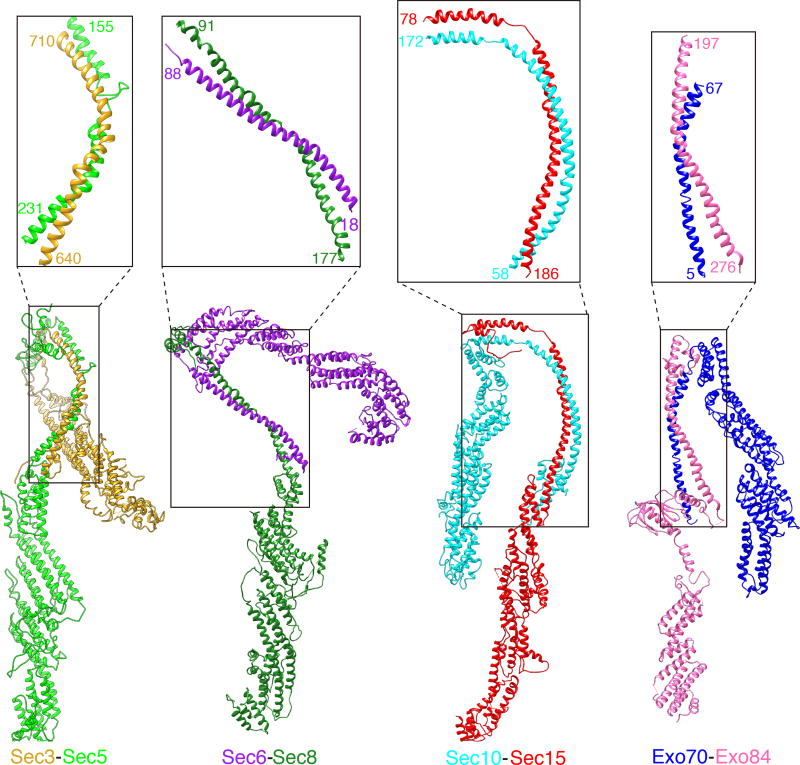
The eight subunits assemble into the exocyst complex in a hierarchical manner. Rather than tiling together as discrete units, the eight subunits knit elaborately with each other through intertwined long coiled-coils (Fig. 2a). The interactions can be divided into 4 pairs: Sec3-Sec5, Sec6-Sec8, Sec10-Sec15 and Exo70-Exo84 (Fig. 3, Supplementary Video 3). Within each pair, the coiled-coil region of each subunit participates in the central pair-wise interactions by forming an intact antiparallel “zipper” measuring 86~128 Å (63~115 residues) (Fig. 3). We term the core exocyst assembly region as “CorEx” (The Core of Exocyst).
Figure 3. Pair-wise interactions of the exocyst subunits through the CorEx motifs.
The four pairs of exocyst subunits, Sec3 and Sec5, Sec6 and Sec8, Sec10 and Sec15, Exo70 and Exo84, form zipper-like interactions through the CorEx motifs. The paring of the CorEx motifs is indicated in the boxes and close-up views, with the starting and ending residues of each CorEx motif labeled.
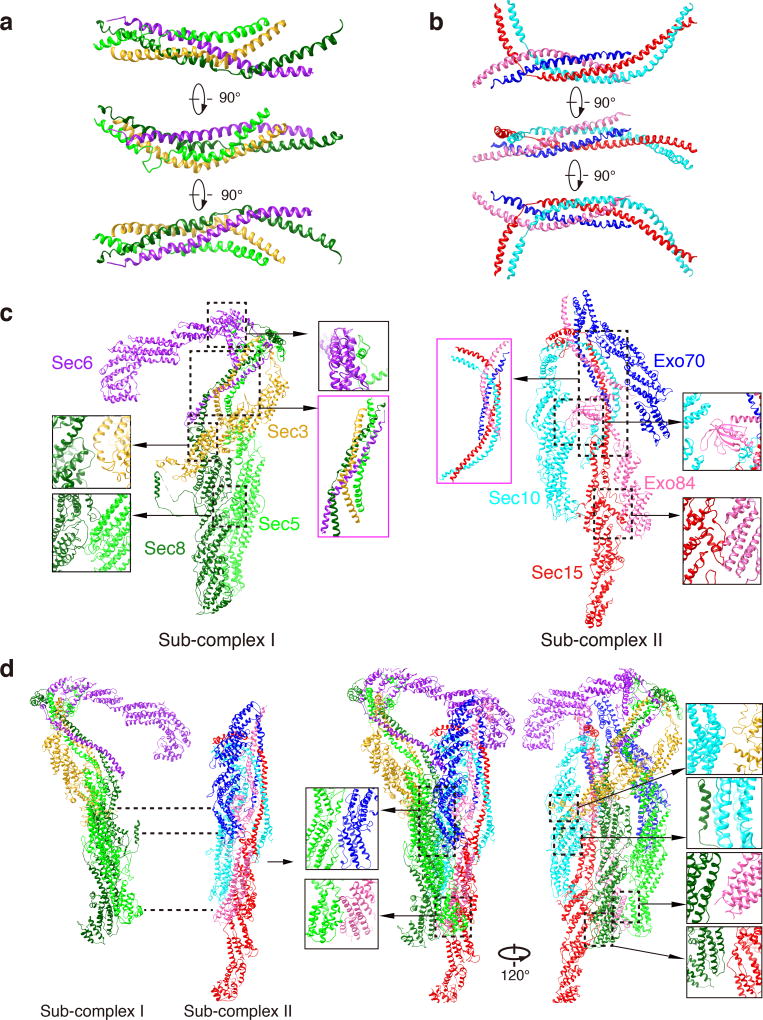
The four dimeric pairs further assemble into higher-order structures, in which the CorEx motifs of Sec3-Sec5 and Sec6-Sec8 form one four-helical bundle (named “CorEx I” (Fig. 4a), and those of Sec10-Sec15 and Exo70-Exo84 dimers form the other (named “CorEx II”) (Fig. 4b), resulting in the tetrameric “Sub-complex I” and “Sub-complex II”, respectively (Fig. 4c; Supplementary Video 3). Such molecular organization is consistent with previous speculation that Sec3-Sec5-Sec6-Sec8 and Sec10-Sec15-Exo70-Exo84 form two stable modules based on pulldown experiments15 and the prediction hinted from comparison of the exocyst with the Cog1–4 sub-complex21. In CorEx I, the two zippers of Sec3-Sec5 and Sec6-Sec8 are comparable in length, and they twist together at the N-terminal ends of Sec5 and Sec8 CorEx motifs, and C-terminal ends of Sec3 and Sec6 CorEx motifs, respectively. The four CorEx motifs intertwine as a four-helix bundle through about two-thirds of the helices, and then separate as two zippers (Fig. 4a). Thus, the shape of CorEx I resembles an upside-down letter “Y” (Fig. 4c, highlighted with magenta box). For CorEx II, the Sec10-Sec15 zipper is much longer than that of the Exo70-Exo84 pair (Fig. 4b). The intertwine of the four CorEx motifs is mainly located at the middle part of the Sec10-Sec15 zipper, and takes about two-thirds of the length of the Exo70-Exo84 zipper from the N-terminus of Exo70 CorEx motif and C-terminus of the Exo84 CorEx motif. The two CorExes are different from the SNARE complex, which is assembled in a parallel fashion22.
Figure 4. Assembly of the core sub-complexes and the holo-complex.
a. The four-helical bundle formed by the CorEx motifs of Sec3, Sec5, Sec 6 and Sec8. b. The four-helical bundle formed by the CorEx motifs of Sec10, Sec15, Exo70 and Exo84. c. Sec3, Sec5, Sec6 and Sec8 form the Sub-complex I; Sec10, Sec15, Exo70 and Exo84 form the Sub-complex II. Interactions among the subunits in the Sub-complex I and Sub-complex II are shown in close-up views, with the four-helical bundles highlighted with magenta boxes. d. Sub-complex I and Sub-complex II assemble into the holo-exocyst complex. Interactions between the Sub-complex I and Sub-complex II are indicated with dash lines in the left panel and close-up views in the right panel.
Beside the contacts among CorEx motifs, the interactions between CAT domains further help to stabilize the sub-complexes. In the Sub-complex I, the bottom of Sec5’s CAT-B domain protrudes into the cradle formed by a long loop (a.a. 771–820) from Sec8 CAT-C domain, and the N-terminal overhang (a.a. 1–30) of Sec5 clings to Sec6’s CAT-A domain. Sec3’s CAT-C domain inserts into the cleft formed between Sec8 CAT-A and CAT-B domain. In the Sub-complex II, the two CAT domains of Exo84 form an extensive interaction interface with Sec15’s CAT-A domain, and the PH domain of Exo84 berths between the CorEx II and CAT-B and CAT-C region of Sec10 (Fig. 4c).
At the highest hierarchy, Sub-complex I and Sub-complex II clap into the holo-exocyst complex as two layers (Fig. 4d, Supplementary Video 3), which coincide to the front layer and back layer of the 3-D cryo-EM assembly (Fig. 1d). Sec8 of Sub-complex I connects the two layers via its multiple interaction interfaces with the rod regions of Sec10, Sec15 and Exo84 in Sub-complex II (a.a. 460–550 of CAT-B in Sec8 with a.a. 570–780 of CAT-B and CAT-C in Sec10; a.a. 860–900 of CAT-D in Sec8 with a.a. 580–680 of CAT-B in Sec15; a.a. 870–970 of CAT-D in Sec8 with a.a. 620–720 of CAT-B in Exo84) (Fig. 4d). Sec5’s CAT-B and CAT-C domain from Sub-complex I form a large binding interface with CAT-C and CAT-D domains of Exo70 from Sub-complex II. The C-termini of Sec5 and Exo84 run very close to each other (Fig. 4d). Moreover, Sec3’s CAT-D domain binds to the joint of CAT-B and CAT-C domains from Sec10. Lastly, Sec6’s CAT-C and CAT-D domain may also interact with the four-helical bundle in Sub-complex II, which in turn helps stabilize the conformation of Sec6 in the exocyst complex (Fig. 4d). These inter-subunit interactions yield the complicated architecture of the holo-complex. Together, the structure reveals detailed binding interfaces amid the exocyst subunits, with the CorEx motifs as the central assembly determents.
The function of Sec3 CorEx motif in vesicle tethering
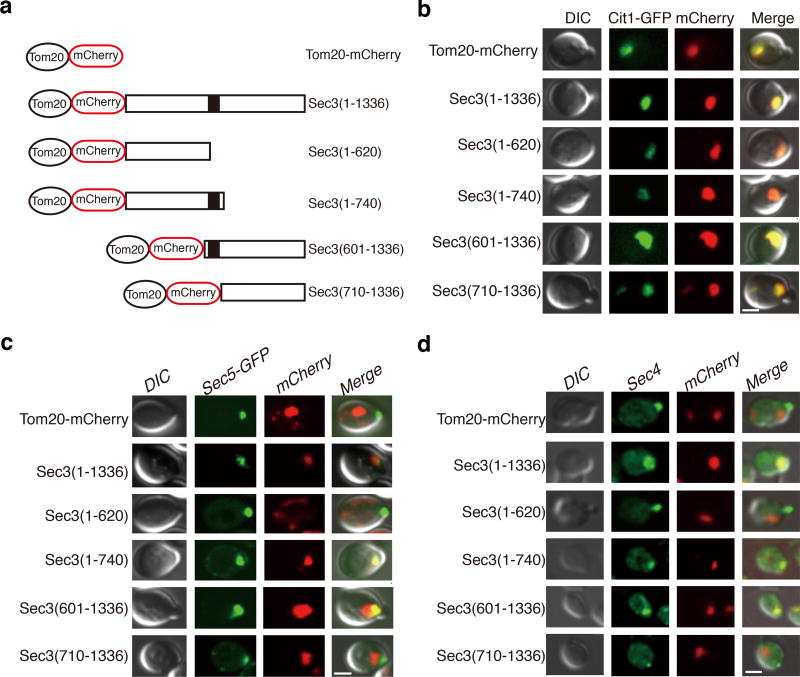
Among all of the exocyst subunits, Sec3 directly associates with the plasma membrane through its N-terminal PH domain23–27, while the other seven exocyst subunits are transported by the secretory vesicles to the plasma membrane marked by Sec328. Sec3 also plays a critical role in the recruitment of other exocyst subunits during vesicle tethering 23,25–27,29. Genetic manipulation suggests that the loss of Sec3 has the least destabilizing effect on the assembly of the exocyst complex15. Consistently, the lack of density for the N-terminal half, the conformational change of the rod region (Supplementary Fig. 1, Class 10) as well as poor density at the C-terminus of Sec3 (Fig. 1d, Supplementary Fig. 2e and 5c) all indicate its flexibility, and suggest that Sec3 is the last subunit assembled into the exocyst complex.
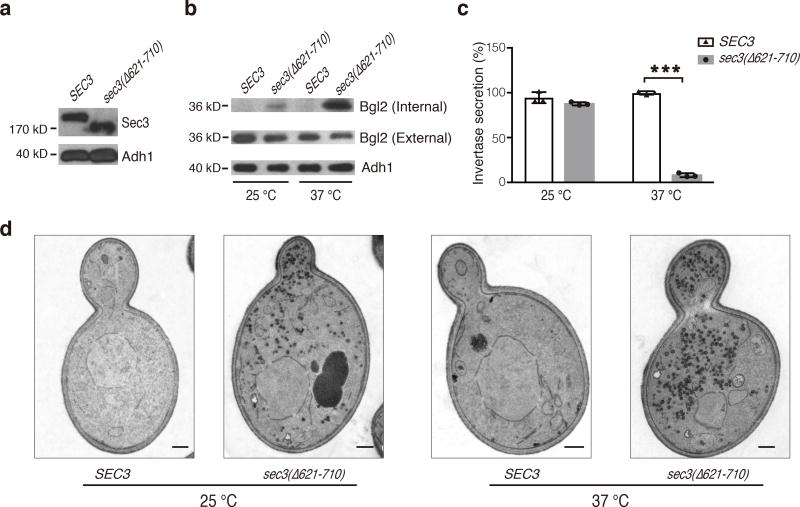
Structural analysis shows that the CorEx motif of Sec3 (a.a. 640–710) mediates its interactions with the remaining subunits (Fig. 4). We thus tested the role of Sec3 CorEx motif in subunits recruitment and vesicle tethering. Sec3, when tagged with Tom20, was targeted to mitochondria and was able to recruit the other exocyst subunits and tether secretory vesicles to the mitochondria29. We ectopically targeted various Sec3 truncation mutants to mitochondria using the same strategy (Fig. 5a–b). The exocyst subunit Sec5, and the Rab protein Sec4 (as a marker for post-Golgi secretory vesicles) were recruited to mitochondria clusters only in cells expressing the Sec3 variants containing the CorEx motif (Fig. 5c and 5d). This result suggests that the CorEx motif of Sec3 plays an important role in exocyst assembly and vesicle tethering. Next, we examined the effect of Sec3 CorEx motif deletion on exocytosis. The sec3(Δ621–710) cells accumulated Bgl2, a cell wall modification enzyme, inside the cells (Fig. 6a–b). In addition, the secretion of the periplasmic enzyme invertase was also decreased (Fig. 6c). Thin-section EM showed that the sec3(Δ621–710) cells accumulate electron-dense vesicles of diameters of 80–100 nm, which is characteristic of post-Golgi secretory vesicles (Fig. 6d). Taken together, the data suggest that disruption of the CorEx motif of Sec3 leads to defects in exocytosis.
Figure 5. The CorEx motif of Sec3 is crucial for exocyst complex assembly and vesicle tethering.
a. Schematic diagram of Sec3 full-length and truncation constructs for ectopic targeting to mitochondria through their fusion with Tom20-mCherry. The dark block indicates the CorEx motif. b. Co-localization of all of the Tom20-mCherry-tagged Sec3 proteins with Cit1-GFP, a marker protein of yeast mitochondria. Scale bar, 2 µm. c. Co-localization of Sec5-GFP and the Tom20-mCherry-tagged Sec3 proteins. Scale bar, 2 µm. d. Co-localization of Sec4, a marker of post-Golgi secretory vesicles, and the Tom20-mCherry-tagged Sec3 proteins. Scale bar, 2 µm. At least 5 images were taken for each sample, with all cells bearing both green and red fluorescence showing similar co-localization patterns as displayed here.
Figure 6. The Sec3 CorEx deletion mutant is defective in exocytosis.
a. Expression of Sec3-GFP and Sec3(Δ621–710)-GFP in yeast cell lysates was detected by Western blotting with an anti-GFP antibody. Alcohol dehydrogenase-1 (“Adh1”) was used as a loading control. b. Accumulation of Bgl2 in sec3(Δ621–710) cells at 25°C and 37°C. Internal and external Bgl2 was detected by Western blotting. Adh1 was used as a loading control. Uncropped blot images are shown in Supplementary Data Set 4. This experiment was independently performed twice with similar results. c. Accumulation of invertase in sec3(Δ621–710) cells at 37°C. The mean and s.d. value of the invertase secretion rate was shown as bars and error bars, respectively. n=3, as technical replicates (see METHOD); “***”, P<0.001. This experiment was independently performed twice with similar results. d. Accumulation of post-Golgi secretory vesicles in sec3(Δ621–710) cells as revealed by thin-section EM. Scale bar, 0.5 µm. At least five images were taken for each condition, showing similar results.
A model of exocyst mediated vesicle tethering
As elaborated above, a long linker region (a.a. 226–620) situates between the PH domain (a.a. 70–225) and the CorEx motif of Sec3 (Supplementary Fig. 7). The cross-linking of Sec3 (K119) to Sec15 (K278) suggests that the PH domain is located near the bottom of the back layer of the exocyst complex, far away from Sec3’s CorEx motif (Supplementary Fig. 8). The linker, spanning ~150 nm with high solvent accessibility, is sufficient to bridge the PH domain and the CorEx motif. Notably, the PI(4,5)P2-binding site of Exo70 is located on the same face of the exocyst complex (Supplementary Fig. 8)30, suggesting that the exocyst associates with the plasma membrane through its rear face of the back layer. It is possible that Sec3 berths at the secretion sites through its PH domain and extends far into the cytoplasm through the long flexible linker as a pre-assembly state. As the vesicle arrives, Sec3 binds to the remaining exocyst subunits through the CorEx motif to form a stable holo-complex and bring the vesicle to close proximity to the plasma membrane. Subsequently, the interaction between Exo70 and phospholipids30,31 completes the docking of the exocyst complex to the plasma membrane, allowing the exocyst to tether the secretory vesicle for subsequent SNARE-mediated membrane fusion.
DISCUSSION
Precise knowledge of the structure of MTCs is essential for the understanding of their assembly and vesicle tethering during various membrane trafficking processes. Our study, for the first time, provides a structural model of the fully assembled exocyst complex at high resolution. The model was built based on the 4.4 Å cryo-EM map, and 96% of cross-linked residue pairs have a Cα – Cα distance ≤ 35 Å when mapped onto the model (Supplementary Data Set 1–3). The only three intra-subunit cross-links with Cα – Cα space > 35 Å are all located in Sec3’s CAT-B and CAT-C domains. This is consistent with the conformational change of Sec3’s rod region found in our 3-D classifications (Supplementary Fig. 1). C-terminal GFP tagging further verified the subunit assignment in the model (Supplementary Fig. 2a).
Previous crystallographic studies have solved the structure of several fragments of the exocyst subunits from a few eukaryotic species26, 27, 32–40. These regions were mostly helical-bundle repeats and GTPase-binding domains that assume structural autonomy. Our study revealed the near full-length structures of the exocyst subunits. All subunits display an extended configuration with their CorEx motifs interacting with each other at the core of the complex. Cryo-EM provides us with the unique opportunity to determine the high-resolution structure of the intact exocyst complex. Moreover, it is only in the fully assembled state that we were able to identify the CorExes, and furthermore, the hierarchical assembly of the holo-complex. It is also important to note that the CorEx motif sequence in individual exocyst subunits is evolutionarily conserved (Supplementary Note 1). It is likely that the CorEx motifs mediate the core exocyst interactions in higher eukaryotes including humans. Notably, the CorEx motifs in the exocyst have also been previously noted to share structure similarities to the N-terminal sequences of subunits of the COG complex and GARP complex3,41, suggesting the conservation of this structure across the MTCs.
Sec3 directly associates with the plasma membrane, and the assembly of other exocyst components with Sec3 may serve as a mechanism for vesicle tethering. Using the ectopic targeting strategy, we demonstrated that the CorEx motif of Sec3 plays an important role in this process. It is also interesting to note that a long linker region connects Sec3’s PH domain and its CorEx motif. This is similar to the long coiled-coil tethers such as EEA1. Recently, it was reported that the conformational changes in EEA1 bring endosomal vesicles to close proximity for membrane fusion42. It would be interesting to examine whether this linker region of Sec3 undergoes similar conformational changes during vesicle tethering at the plasma membrane.
Since vesicle tethering precedes SNARE-mediated membrane fusion, subunits of the exocyst are direct targets of signaling proteins such as small GTPases and kinases, which spatially or kinetically regulate exocytosis9,10,43–50. Different regulators could promote or inhibit the assembly of the exocyst complex, thereby coordinating exocytosis with other cellular activities such as mitosis and tumor invasion46–50. For example, it was shown that mitotic phosphorylation of Exo84 by CDK1 disrupts the assembly of the exocyst complex and blocks exocytosis, thereby arresting cell surface expansion prior to the metaphase-anaphase transition48. Our structure data suggest that the phosphorylated residues are located at a region that mediates the interactions of Exo84 with other subunits. CDK1 phosphorylation of these residues may thus disrupt the assembly of the complex. Future studies based on the high-resolution structure may lead to better mechanistic understanding of such regulations.
To our knowledge, this is the first cryo-EM structure of a tethering complex. Although different MTCs function at different stages of vesicular trafficking, they often share similar features in vesicle tethering3,5. The exocyst cryo-EM structure may shed light to the assembly and function of other MTCs, which will ultimately help elucidate the molecular mechanisms of vesicular trafficking.
ONLINE MATERIALS AND METHODS
Yeast strains and plasmids
The budding yeast Saccharomyces cerevisiae strains and plasmids used in this study are listed in Supplementary Table 2. To integrate SEC3-GFP and sec3(Δ621–710)-GFP into the TRP1 locus with SEC3 promoter, pG215 and pG1980 were digested with SnaBI and transformed into the sec3Δ strain (GY2624).
Purification of the yeast exocyst complex
33 liters of yeast culture was grown in YPD medium at 30 °C for 10–14 hours to an OD600 of ~2.5. Cells were pelleted through centrifugation at 6,000 g for 8 min, and washed once with ddH2O. Normally, ~150 g of yeast cells were obtained from the culture. The yeast cells were resuspended in 40 ml (~1/4 (v/w) of the 5 × lysis buffer containing 50 mM HEPES pH 7.4, 750 mM NaCl, 0.5% NP-40, 5 mM EDTA and 50% glycerol and drop-frozen in liquid nitrogen. The cells were then disrupted into powders by SPEX 6870 Freezer Mill. The frozen powder was melted by stirring under cold water bath. Protease inhibitors and 2 mM DTT was added during the melting process. The cell lysate was then subjected to centrifugation at 35,000 g for 30 min, yielding ~100 ml supernatant. IgG Dynabeads were prepared as previously described15. 1 ml Dynabeads suspension was added to the supernatant and incubated at 4 °C for 3–6 hours. After extensive wash by a buffer containing 10 mM HEPES pH 7.4, 500 mM NaCl, 0.1% NP-40, 1 mM EDTA, 10% glycerol and 2 mM DTT, proteins were released by TEV protease cleavage for 1.5 hours at 4 °C in 0.5 ml wash buffer containing 150 mM NaCl. The eluent was then concentrated to a total volume of 45 µl and applied to a Superose 6 (3.2/300) column (GE Healthcare) with running buffer containing 10 mM HEPES pH 7.4, 150 mM NaCl, 2mM DTT. Peak fractions were collected for cross-linking mass spectrometry and Cryo-EM sample preparation.
Chemical Cross-linking mass spectroscopy (CXMS)
For CXMS, approximately 10 µg of purified exocyst complex was crosslinked at 25 °C with 0.25, 0.5, 1 mM DSS and BS3 for 1 hour, or with 2 mM EDC plus 3 mM sulfo-NHS for 2 hours. The reactions were quenched with 20 mM NH4HCO3 or 10 mM hydroxylamine, respectively. Proteins were precipitated with ice-cold acetone, resuspended in 8 M urea, 100 mM Tris pH 8.5, and then digested by trypsin (Promega, Inc.) in 2 M urea, 100 mM Tris (pH 8.5). The LC-MS/MS analysis was performed on an Easy-nLC 1000 II HPLC (Thermo Fisher Scientific) coupled to a Q-Exactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were loaded on a pre-column (75 µm ID, 6 cm long, packed with ODS-AQ 120 Å–10 µm beads from YMC Co., Ltd.) and further separated on an analytical column (75 µm ID, 12 cm long, packed with Luna C18 1.9 µm 100 Å resin from Welch Materials) with a linear reverse-phase gradient from 100% buffer A (0.1% formic acid in H2O) to 30% buffer B (0.1% formic acid in acetonitrile) in 56 min at a flow rate of 200 nl/min. The top 15 most intense precursor ions from each full scan (resolution 60,000) were isolated for HCD MS2 (resolution 15,000; normalized collision energy 27) with a dynamic exclusion time of 30s. Precursors with 1+, 2+, 7+ or above, or unassigned charge states were excluded. The pLink software was used to identify cross-linked peptides with precursor mass accuracy at 20 ppm, fragment ion mass accuracy at 20 ppm, and the results were filtered by applying a 5% FDR cutoff at the spectral level and then an E-value cutoff at 0.00151.
EM data acquisition and processing
For negative staining EM sample preparation, 4 µl of purified exocyst complex at a concentration of ~0.02 mg/ml was applied to a glow-discharged holy carbon EM grid covered with a thin layer of carbon film (Zhongjingkeyi Technology Co. Ltd). The grid was then stained by 2% (w/v) uranyl acetate solution for 1 min, dried in air and stored at room temperature. For EM data collection, the negative-stained grid was transferred into an FEI Tecnai Spirit Bio-TWIN electron microscope operated at an acceleration voltage of 120kV to manually collect micrographs with random conical tilt strategy52, using a Gantan US4000 4k×4k CCD camera.
Samples for cryo-EM were prepared with a Vitrobot Mark IV (FEI Company) under 100% humidity at 8 °C. 3.5 µl of 0.3 mg/ml untreated exocyst solution, or ~0.12 mg/ml of exocyst treated with freshly prepared glutaraldehyde at a final concentration of 0.0025% for 20 min on ice, was applied to a glow-discharged Quantifoil grid (Quantifoil Au R1.2/1.3), blotted for 2 seconds, and plunged into liquid ethane cooled by liquid nitrogen. To obtain enough particles with sufficient views for 3D reconstitution, we prepared cryo-EM specimens of exocyst complexes purified from several strains with different subunits fused to the TAP or ProA tag (Fig 1a and Supplementary Fig. 2a). The cryo-EM grids were transferred to a Titan Krios (FEI Company) electron microscope operated at 300kV and equipped with a direct electron counting device (Gatan K2 Summit). Using UCSF-Image4 interface53, we collected electron micrographs of the specimens with a nominal magnification of 22,500 in super-resolution mode on the K2 Summit camera as dose-fractionated movie stacks. Each movie stack contained 32 frames with a dose rate of about 8.25 counts per second per physical pixel (~6.25 e−/sec/Å2), resulting in a total exposure time of 8s and 50 e- per stack. A total of 6,466 cryo-EM movie stacks with defocus values ranging from 2~3 µm were collected, aligned and summed using whole-image motion correction54,55 with a binned pixel size of 1.30654 Å.
Image processing
30 pairs of un-tilt (0°) – tilt (50°) micrographs were collected for the negative-stained exocyst complexes, from which 4516 tilt-pair particles were picked by EMAN256 subroutine e2RCTboxer.py in an interactive RCT-boxing mode. A low-resolution 3D map was then generated by SPIDER57 and served as an initial model for cryo-EM 3D refinement.
For the cryo-EM dataset, the movie stacks were first corrected for motion into summed images using MotionCorr program54. The defocus value of each image was determined by CTFFIND358. From the summed micrographs, about 900,000 particles were semi-auto-picked by RELION1.459. After reference-free 2D classification and 3D classification, through which contaminants and defective particles were removed, 343,342 particles were used for further processing (Supplementary Fig. 1). The particles were classified into ten 3D classes to elucidate possible conformational variation; a density map with an overall resolution of 9.0 Å was obtained by auto-refinement, which was further improved to 7.35 Å by RELION post-processing and 6.75Å with body mask refinement. In parallel, we performed 5×5 fraction-image motion correction with MotionCor260 combined with dose weighting and re-extracted the 343,342 particles from the original stacks. This dataset was refined with post-processing to generate a reconstruction with an overall resolution of 5.5 Å. We also applied distortion magnification correction61,62 and a second round body mask to the body-masked 7.02 Å map and obtained a map at 4.35 Å resolution. We further performed local mask refinement for the head and tail parts of the complex to improve the local density quality at 6.2 Å, and 4.65 Å, respectively. These maps were integrated in Chimera63 for model building and figure presentation. Local resolution variations were estimated using ResMap64. Overall resolutions of the maps were based on the gold-standard FSC 0.143 criterion65.
Model building and refinement
We used a combination approach with the available crystal structures of some of the subunits and their homologues, the secondary and tertiary structural prediction, and the CXMS results to build models of the cryo-EM maps (Supplementary Fig. 3, Supplementary Video 1). Model building was initiated with the crystal structures of yeast exocyst subunits, Sec6 (a.a.411–805) (PDB code: 2FJI), Exo70 (a.a.67–623) (PDB code: 2B1E) and Exo84 (a.a.525–753) (PDB code: 2D2S). Exo84 (a.a.525–753) was fit into the map unambiguously as a rigid body by Situs66 (Supplementary Fig. 4a). Exo70 (a.a.67–623) is an extended helical bundle that couldn’t be well-docked into the map as a whole body. Since Exo70 helical bundle has the tendency to bend in the middle37, we split it into two halves: Exo70 (a.a.67–344) and Exo70 (a.a.345–623). The two halves fit into the map with high degree of agreement as rigid bodies, with the break sites close enough to form a peptide bond (Supplementary Fig. 4b). Based on the intermolecular crosslinking pairs between Exo70 and Sec6, we depicted Sec6 (a.a.411–805) proximal to the N-terminus of Exo70 (a.a.67–623) in the cryo-EM map (Supplementary Fig. 4c). The Sec6 model fits in the corresponding density very well. At this point, we have docked all of the known crystal structures of the yeast exocyst subunits into the map. Next, we fit the map with predicted tertiary structures obtained from homologue modelling. The tertiary structure of Sec10 (a.a.234–867) was predicted with Swiss-Model67 based on the atomic model of Zebrafish Sec10 (a.a.195–708) (PDB code: 5H11). After the loops of a.a.279–319 and a.a.456–568 were pruned, the protein was fit into the right edge of the back layer (Supplementary. 4d). For better fitting, it was split into two halves between Arg432 and Ser433 and docked into the map separately. The PH domain of Exo84 (a.a.346–470) and the C-terminal portion of Sec15 (a.a.482–896) were predicted by PHYRE268 based on the crystal structures of rat Exo84 (PDB code: 1ZC4) and Drosophila Sec15 (PDB code: 2A2F), and docked into the map (Supplementary Fig. 4e–f).
From the above docked atomic models, we performed manual extension of the C-alpha peptides of the corresponding proteins following the map density connectivity by Coot69. We started with helices assignment based on the secondary structure predictions (Supplementary Fig. 7) and then modeled the connecting loops guided by a lower threshold of the map showing the densities. Wherein a loop was too long and not enough density was visualized, the corresponding region was left un-modeled. Using such a strategy, near full-length Sec6, Sec10, Sec15 and Exo70, as well as Exo84 (a.a.170–753) were modeled in the map (Supplementary Fig. 4g–k).
To build the models of Sec5 and Sec8, we first identified their densities based on several cross-linked residue pairs. Pairs Exo84(725)-Sec5(961) and Exo84(731)-Sec5(961) revealed that the C-terminal residues of Sec5 were adjacent to the C-terminal end of Exo84. This information, together with the pair Exo70(167)–Sec5(228), assigned the left branch of the front layer to Sec5 (Supplementary. 5a). Similarly, pairs Exo84(707)-Sec8(893), Exo84(343)-Sec8(238) and Sec6(237)-Sec8(125) indicated that the long axle of the front layer is Sec8 (Supplementary. 5b). We thus manually modeled Sec5 and Sec8 from their C- to N-terminus.
After the above modeling, the remaining density corresponding to Arm I was assigned to Sec3 with one end of the density located near Exo70 and the other end reaching to the middle of Sec10. The crosslink pairs Exo70(228)-Sec3(613) and Sec10(515)-Sec3(1323) suggested that Sec3 extends from the first end with Lys613 to the second one with Lys1323. Based on the above information and the density shape, we manually modeled the Sec3(612–1332) into the map (Supplementary Fig. 5c). The very C-terminus of Sec3 starting from Phe1040 cannot be modeled precisely due to the poor density.
Since the resolution of 4.4 Å is not sufficient for clear side chain modeling, all of the docked tertiary structures derived from homologue structure prediction and manually built models consist of poly-alanines to trace the main chains. Only side chains derived from the crystal structures of yeast exocyst subunits were kept in the docked structures. To facilitate the CXMS analysis, all of the models are presented with their original sequences. The exocyst model was then validated with independent CXMS analyses, in which different crosslinking chemicals were used (Supplementary Tables 2–4, Supplementary Video 2). The Cα - Cα distance restraint of BS3/DSS as well as EDC cross-links is normally within a range of 24 Å19,51, and approximately 70% of the DSS or BS3 crosslinks are compatible with the X-ray structures19. As a common practice in protein structure modeling, the distance restraints are often relaxed to 30 Å or 35 Å70–72. When mapped onto the exocyst model, 96% of all crosslinks fall within 35 Å, 94% within 30 Å, 91% within 27 Å and 86% within 24 Å, confirming the overall accuracy of the assignment, fold and chain tracing of the exocyst model.
After the validation, the exocyst model was subject to 100 iterations of Phenix.real_space_refine73 against the 4.4 Å map with global geometry and secondary structure restraints. Sec6 was also auto-refined separately against the refined map of the head region (Supplementary Fig. 1). The quality of the models was evaluated with MolProbity74. Chimera63 was used for figure presentation.
Fluorescence microscopy
Yeast cells were grown to an OD600 of 1.0–1.5 in Leu- synthetic complete medium at 25 °C and harvested by centrifugation at 3000 rpm for 1 min. 2 µl of cell suspension was processed for fluorescence microscopy with a Nikon ECLIPSE Ti confocal fluorescence microscope equipped with an Apo TIRF 100×/1.49 oil immersion objective lens. Images were collected with a digital camera (CSU-X1, Nikon) operating with NIS-Elements AR 4.60.00 software (Nikon). Rabbit anti-Sec4p polyclonal antibody diluted to 1:2000 was used for immunofluorescence microscopy.
Bgl2 secretion assay
Yeast strains expressing SEC3-GFP and sec3(Δ621–710)-GFP under SEC3 promotor as the sole copy of Sec3 were grown to an early log phase in YPD medium at 25 °C, followed by another 2 hrs of culturing at 25 °C and 37 °C, respectively. 10 mM NaN3 and 10 mM NaF were added into the culture and a total of 10 O.D.s of cells were collected for each sample. Cells were washed by 50 mM Tris pH 7.5, 10 mM NaN3 and 10 mM NaF, and spheroplasted by Zymolase in spheroplast buffer containing 50 mM Tris pH 7.5, 1.4 M sorbitol, 10 mM NaN3 and 10 mM NaF. After pelleting, the supernatant was taken for external Bgl2 measurement. The spheroplast was washed once by spheroplast buffer and lysed by 0.5% Triton, and was used for the internal Bgl2 determination. The amounts of external and internal Bgl2 were determined by Western blotting, with Adh1 as the loading control.
Invertase secretion assay
Yeast strains with SEC3-GFP and sec3(Δ621–710)-GFP were grown to early log phase in YPD medium at 25 °C. 7 O.D.600 units of cells were collected for each strain and aliquoted into 7 tubes. 1 tube of the cells was immediately washed and resuspended in 10 mM NaN3 as un-induced control. The remaining 6 tubes was subject to 90 min culture in YP (1% yeast extract + 2% peptone) plus 0.1% glucose at 25 °C (3 tubes) or 37 °C (3 tubes) to induce the expression of invertase. After induction, cells were washed and resuspended in 10 mM NaN3. Half of the cells in each of the 7 tubes were used to measure the external invertase and to measure the O.D.600 for cell number normalization, while the other half was lysed for the measurement of the internal invertase. After normalization with cell number and subtracting the un-induced control, invertase secretion was calculated by external/ (external + internal). Student’s t-test (unpaired, two-tailed) was used for statistical analyses. Prism7 was used for data presentation. This assay was independently performed twice with similar results obtained.
Thin section electronic microscopy
Early log phase cells were harvested and prefixed with 0.1 M sodium cacodylate containing 3% glutaraldehyde, 5 mM CaCl2 and 5 mM MgCl2, pH 7.4 at room temperature for 1 hr. The cells were then embedded in 2% low melting point agarose in a 1:1 ratio and cut into small pieces (~1mm3). The samples were then fixed with 4% KMNO4 for 1 hr at 25 °C. After extensive wash with double-distilled water, the pieces were treated with 0.5% sodium meta-periodate for 15 min to increase cell penetrability and then placed into 2% uranyl acetate overnight at 25 °C after another wash. On the next day, the samples were dehydrated through a graded series of ethanol (50%–100%) wash at 4 °C and 100% ethanol incubation overnight at 25 °C, followed by two washes with propylene oxide (PO) and PO/Spurr resin (1:1) agitation overnight at room temperature. Finally, the samples were embedded in 100% Spurr resin and sectioned for electronic microscopy. Cells were observed using a JEM-1011 JEOL transmission microscopy at 15,000× magnification.
Statistics
For invertase secretion assay analysis, the online GraphPad software was used to do the statistics. Un-paired student’s t-test was used to analyze the difference between invertase secretion rate of SEC3-GFP and sec3(Δ621–710)-GFP. For data collected at 25 °C, the two-tailed P value is 0.1376, t value is 1.8526 and degree of freedom is 4; for data collected at 37 °C, the two-tailed P value is less than 0.0001, t value is 53.7907, and degree of freedom is 4.
Data availability and Accession Code Availability Statements
EM structures and associated atomic model have been deposited into the Electron Microscopy Data Bank and Protein Data Bank with the accession code EMD-6827 and 5YFP, respectively. Source data for Figure 6c is available with the paper online. Other data supporting this study are available upon reasonable request. A Life Sciences Reporting Summary for this article is available.
Supplementary Material
Acknowledgments
We are grateful to J. Lei, Y. Xu, Z. Yan, Q. Zhou, J. Wang and T. Yang for their help in cryo-EM experiments, structure determination, and model building. We thank the supports from Beijing National Protein Science Facility at Tsinghua University and Electron Microscopy Resource Laboratory at the University of Pennsylvania. This work is supported by National Institute of Health (R01 GM111128) to W.G., by National Science Foundation of China (Grant 31530018), Beijing Municipal Science & Technology Commission (Grant Z161100000116034), and the National Key Research and Development Program of MOST (Grant 2016YFA0501100) to H.-W.W.
Footnotes
AUTHOR CONTRIBUTIONS
W.G. initiated the project. W.G. and H.-W.W. designed and supervised the experiments. K.M., G.L. and P.Y. purified the exocyst complex. Y.L. and K.M. collected the EM data. Y.L., H.-W.W., J.W., K.M., and J.-J.L. analyzed the EM data and generated the EM map. G.S., Y.D. and M.-Q.D. performed the CXMS experiments. K.M., G.S., Y.L., M.-Q.D., W. G. and H.-W.W. analyzed the CXMS data. K.M., Y.L., H.-W.W. and W.G. built the atomic model. K.M., H.-W.W., W.G. and X.W. analyzed the structure. S.W. performed the microscopy of Sec3 mutations. S.W. and K.M. performed the secretion assays. K.M., S.W., and W.G. analyzed the cell biology data. W.G., K.M. and Y.L. wrote the draft. W.G., K.M., H.-W.W. and X.W. edited the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1999;1:E17–22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- 2.Guo W, Sacher M, Barrowman J, Ferro-Novick S, Novick P. Protein complexes in transport vesicle targeting. Trends Cell Biol. 2000;10:251–255. doi: 10.1016/s0962-8924(00)01754-2. [DOI] [PubMed] [Google Scholar]
- 3.Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- 4.Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am. J. Physiol. Cell Physiol. 2006;290:C11–26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- 5.Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 6.Brocker C, Engelbrecht-Vandre S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 2010;20:R943–952. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 7.TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TerBush DR, Maurice T, Roth D, Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 9.He B, Guo W. The exocyst complex in polarized exocytosis. Curr. Opin. Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu B, Guo W. The Exocyst at a Glance. J. Cell Sci. 2015;128:2957–2964. doi: 10.1242/jcs.156398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue P, et al. Sec3 promotes the initial binary t-SNARE complex assembly and membrane fusion. Nat. Commun. 2017;8:14236. doi: 10.1038/ncomms14236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat. Struct. Mol. Biol. 2006;13:577–81. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 14.Hsu SC, Hazuka CD, Foletti DL, Scheller RH. Targeting vesicles to specific sites on the plasma membrane: the role of the sec6/8 complex. Trends Cell Biol. 1999;9:150–153. doi: 10.1016/s0962-8924(99)01516-0. [DOI] [PubMed] [Google Scholar]
- 15.Heider MR, et al. Subunit connectivity, assembly determinants and architecture of the yeast exocyst complex. Nat. Struct. Mol. Biol. 2016;23:59–66. doi: 10.1038/nsmb.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lees JA, Yip CK, Walz T, Hughson FM. Molecular organization of the COG vesicle tethering complex. Nat. Struct. Mol. Biol. 2010;17:1292–1297. doi: 10.1038/nsmb.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou HT, Dukovski D, Chambers MG, Reinisch KM, Walz T. CATCHR, HOPS and CORVET tethering complexes share a similar architecture. Nat. Struct. Mol. Biol. 2016;23:761–763. doi: 10.1038/nsmb.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picco A, et al. The in vivo architecture of the exocyst provides structural basis for exocytosis. Cell. 2017;168:400–412. doi: 10.1016/j.cell.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Ding YH, et al. Increasing the Depth of Mass-Spectrometry-Based Structural Analysis of Protein Complexes through the Use of Multiple Cross-Linkers. Anal Chem. 2016;88:4461–4469. doi: 10.1021/acs.analchem.6b00281. [DOI] [PubMed] [Google Scholar]
- 20.Croteau NJ, Furgason ML, Devos D, Munson M. Conservation of helical bundle structure between the exocyst subunits. PLoS One. 2009;4:e4443. doi: 10.1371/journal.pone.0004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson LP, Kümmel D, Reinisch KM, Owen DJ. Structures and mechanisms of vesicle coat components and multisubunit tethering complexes. Curr Opin Cell Biol. 2012;24:475–483. doi: 10.1016/j.ceb.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 23.Finger FP, Hughes TE, Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- 24.Guo W, Tamanoi F, Novick P. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell. Biol. 2001;3:353–360. doi: 10.1038/35070029. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J. Cell Biol. 2008;180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita M, et al. Structural basis for the Rho- and phosphoinositide-dependent localization of the exocyst subunit Sec3. Nat. Struct. Mol. Biol. 2010;17:180–186. doi: 10.1038/nsmb.1722. [DOI] [PubMed] [Google Scholar]
- 27.Baek K, et al. Structure-function study of the N-terminal domain of exocyst subunit Sec3. J. Biol. Chem. 2010;285:10424–10433. doi: 10.1074/jbc.M109.096966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd C, Hughes T, Pypaert M, Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo G, Zhang J, Guo W. The role of Sec3p in secretory vesicle targeting and exocyst complex assembly. Mol. Biol. Cell. 2014;25:3813–3822. doi: 10.1091/mbc.E14-04-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He B, Xi F, Zhang X, Zhang J, Guo W. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 2007;26:4053–4065. doi: 10.1038/sj.emboj.7601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Zuo X, Yue P, Guo W. Phosphatidylinositol 4,5-bisphosphate mediates the targeting of the exocyst to the plasma membrane for exocytosis in mammalian cells. Mol. Biol. Cell. 2007;18:4483–4492. doi: 10.1091/mbc.E07-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat. Struct. Mol. Biol. 2006;13:555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 33.Jin R, et al. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 2005;24:2064–2074. doi: 10.1038/sj.emboj.7600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukai S, Matern HT, Jagath JR, Scheller RH, Brunger AT. Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. EMBO J. 2003;22:3267–3278. doi: 10.1093/emboj/cdg329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat. Struct. Mol. Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 36.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat. Struct. Mol. Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 37.Hamburger ZA, Hamburger AE, West AP, Jr, Weis WI. Crystal Structure of the S. cerevisiae Exocyst Component Exo70p. J. Mol. Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 38.Moore BA, Robinson HH, Xu Z. The Crystal Structure of Mouse Exo70 Reveals Unique Features of the Mammalian Exocyst. J. Mol. Biol. 2007;371:410–421. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, et al. Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E41–50. doi: 10.1073/pnas.1521248112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JX, et al. Crystal structure of Sec10, a subunit of the exocyst complex. Sci. Rep. 2017:7. doi: 10.1038/srep40909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- 42.Murray DH, et al. An endosomal tether undergoes an entropic collapse to bring vesicles together. Nature. 2016;537:107–111. doi: 10.1038/nature19326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novick P, Guo W. Ras family therapy: Rab, Rho and Ral talk to the exocyst. Trends Cell Biol. 2002;12:247–249. doi: 10.1016/s0962-8924(02)02293-6. [DOI] [PubMed] [Google Scholar]
- 44.Lipschutz JH, Mostov KE. Exocytosis: the many masters of the exocyst. Curr. Biol. 2002;12:R212–214. doi: 10.1016/s0960-9822(02)00753-4. [DOI] [PubMed] [Google Scholar]
- 45.Moskalenko S, et al. The exocyst is a Ral effector complex. Nat. Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 46.Moskalenko S, et al. Ral GTPases regulate exocyst assembly through dual subunit interactions. J. Biol. Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- 47.Sugihara K, et al. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat. Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- 48.Luo G, Zhang J, Luca FC, Guo W. Mitotic phosphorylation of Exo84 disrupts exocyst assembly and arrests cell growth. J. Cell Biol. 2013;202:97. doi: 10.1083/jcb.201211093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu H, et al. Oncogenic BRAF-Mediated Melanoma Cell Invasion. Cell Rep. 2016;15:2012–2024. doi: 10.1016/j.celrep.2016.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren J, Guo W. ERK1/2 regulate exocytosis through direct phosphorylation of the exocyst component Exo70. Dev. Cell. 2012;22:967–978. doi: 10.1016/j.devcel.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat. Meth. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 52.Liu X, Wang HW. Single Particle Electron Microscopy Reconstruction of the Exosome Complex Using the Random Conical Tilt Method. J. Vis. Exp. JoVE. 2011;49:2574. doi: 10.3791/2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Zheng S, Agard DA, Cheng Y. Aysnchronous data acquisition and on-the-fly analysis of dose fractionated cryo-EM images by UCSFImage. J. Biol. Chem. 2015;192:74–78. doi: 10.1016/j.jsb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 57.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 59.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant T, Grigorieff N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J. Struct. Biol. 2015;192:204–208. doi: 10.1016/j.jsb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu G, et al. An algorithm for estimation and correction of anisotropic magnification distortion of cryo-EM images without need of pre-calibration. J. Struct. Biol. 2016;195:207–215. doi: 10.1016/j.jsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 64.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 67.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 70.Ding YH, et al. Modeling protein excited-state structures from ‘over-length’ chemical cross-links. J. Biol. Chem. 2017;292:1187–1196. doi: 10.1074/jbc.M116.761841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herzog F, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 72.Erzberger JP, et al. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EM structures and associated atomic model have been deposited into the Electron Microscopy Data Bank and Protein Data Bank with the accession code EMD-6827 and 5YFP, respectively. Source data for Figure 6c is available with the paper online. Other data supporting this study are available upon reasonable request. A Life Sciences Reporting Summary for this article is available.