Abstract
Nucleo-cytoplasmic shuttling of class IIa histone deacetylases (i.e. HDAC4, -5, -7, and -9) is a synaptic activity- and nuclear calcium–dependent mechanism important for epigenetic regulation of signal-regulated gene expression in hippocampal neurons. HDAC4 in particular has been linked to the regulation of genes important for both synaptic structure and plasticity. Here, using a constitutively nuclear-localized, dominant-active variant of HDAC4 (HDAC4 3SA), we demonstrate that HDAC4 accumulation in the nucleus severely reduces both the length and complexity of dendrites of cultured mature hippocampal neurons, but does not affect the number of dendritic spines. This phenomenon appeared to be specific to HDAC4, as increasing the expression of HDAC3 or HDAC11, belonging to class I and class IV HDACs, respectively, did not alter dendritic architecture. We also show that HDAC4 3SA decreases the expression of vascular endothelial growth factor D (VEGFD), a key protein required for the maintenance of dendritic arbors. The expression of other members of the VEGF family and their receptors was not affected by the nuclear accumulation of HDAC4. VEGFD overexpression or administration of recombinant VEGFD, but not VEGFC, the closest VEGFD homologue, rescued the impaired dendritic architecture caused by the nuclear-localized HDAC4 variant. These results identify HDAC4 as an epigenetic regulator of neuronal morphology that controls dendritic arborization via the expression of VEGFD.
Keywords: histone deacetylase (HDAC), epigenetics, dendrite, hippocampus, calcium, neuronal morphology, nucleo-cytoplasmic shuttling, synaptic activity, VEGFD
Introduction
Lysine acetylation of histone and non-histone proteins is a major epigenetic mechanism regulating gene expression and a large spectrum of cellular functions (1). In neurons, changes in the balancing of histone acetylation are associated with induction or repression of the transcription of many genes implicated in physiological neuronal functions and in neurodegeneration (2, 3). Histone deacetylases (HDACs)3 generally act as transcriptional repressors by either directly interfering with DNA-binding proteins or by catalyzing the removal of acetyl groups from the N-terminal tails of histones, resulting in a denser chromatin structure (1, 4, 5). HDACs can be divided into four classes based on their structural, functional, and phylogenetic properties. Class I (HDAC1, -2, -3, and -8), class II (class IIa: HDAC4, -5, -7, and -9/class IIb: HDAC6 and -10), and class IV (HDAC11) are related to yeast Rpd3, Hda1, and Hos3 proteins, respectively, and constitute the classical family of zinc-dependent HDACs; class III consists of seven sirtuins, which are NAD+-dependent yeast Sir2 homologues (6). All members of class IIa can shuttle between the nucleus and cytoplasm in a signal-dependent manner, a complex process that is regulated by protein kinases and phosphatases, and takes place in cells of different types (7, 8). In neurons, the shuttling of class IIa HDACs is controlled by synaptic activity and nuclear calcium (9, 10).
In recent years, nuclear calcium has emerged as one of the main signals governing the expression of genes crucial for several adaptive processes in the nervous system such as plasticity, memory formation, acquired neuroprotection, and dynamics of neuronal morphology (11). Nuclear calcium also controls the expression of vascular endothelial growth factor D (VEGFD), a key factor for the maintenance of proper dendritic arborization (12–14). Mechanistically, nuclear calcium influences transcription both by regulating the activity of transcription factors and by modulating epigenetic processes, which includes induction of expression of DNA methyltransferases (15) and the nucleo-cytoplasmic shuttling of class IIa HDACs (10).
Among class IIa HDACs, HDAC4 is highly expressed in the rodent hippocampus and forebrain regions (16); under basal conditions, it is predominantly localized in the cytoplasm of neurons (17). Blockade of synaptic activity and/or nuclear calcium signaling causes an accumulation of HDAC4 in the nucleus (10), which, in turn, leads to changes in the rate of transcription of many genes (10, 18). HDAC4 activity can regulate the expression of synaptic proteins, alter the shape of synapses, and modulate neuronal survival (18). However, if and how the subcellular localization of HDAC4 affects dendritic architecture remains to be elucidated. Here, we used a constitutively nuclear-localized dominant-active mutant of HDAC4 (19) and analyzed its effects on dendrites in primary cultured mouse hippocampal neurons. The results established HDAC4 as a critical epigenetic regulator of neuronal architecture and identified VEGFD as a major mediator of the observed morphological changes.
Results
Nuclear HDAC4 affects neuronal morphology
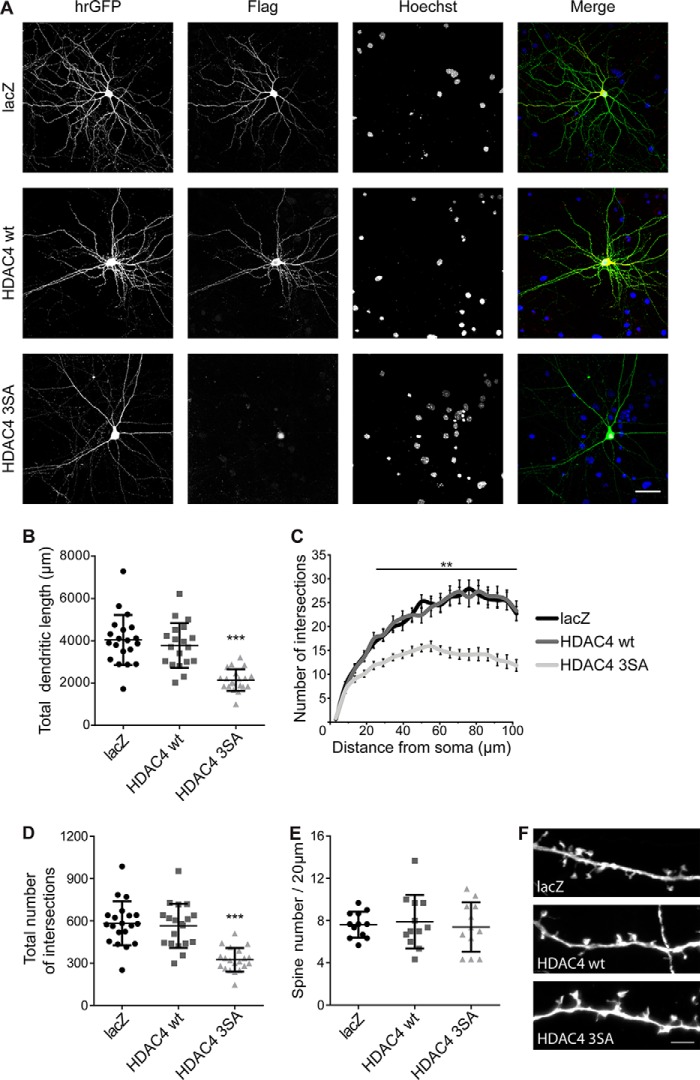
Given that interfering with nuclear calcium signaling results in both an alteration of neuronal morphology and a nuclear accumulation of class IIa HDACs (10, 13, 14), we sought to investigate if the two events are functionally linked. We focused our efforts on HDAC4, which is highly expressed in hippocampal neurons and we made use of the previously characterized dominant-active mutant of HDAC4 (HDAC4 3SA) (9, 10). This mutant carries three mutations in key phosphorylation sites necessary for nuclear export; as a result it is permanently localized in the nucleus. Neuronal architecture was visualized by transfecting a construct encoding for hrGFP (humanized Renilla reniformis GFP) together with constructs encoding for HDAC4 3SA, HDAC4 WT, and as an additional control, lacZ (Fig. 1A). HDAC4 3SA, HDAC4 WT, and lacZ constructs all carry an additional FLAG tag, which allows for detection and verification of subcellular localization. Immunolabeling of FLAG-tagged constructs showed a predominantly cytoplasmic distribution for HDAC4 WT (10), whereas HDAC4 3SA was localized in the nucleus as expected (Fig. 1A). We observed a distinct decline in the size and extent of the dendritic tree in HDAC4 3SA expressing neurons compared with neurons expressing HDAC4 WT or lacZ (Fig. 1A). Neurons expressing the nuclear localized HDAC4 mutant showed a significant reduction in total dendritic length compared with controls (lacZ, 3779 μm ± 243 μm; HDAC4 WT, 4037 μm ± 264 μm; HDAC4 3SA, 2138 μm ± 115 μm; Fig. 1B). This effect was accompanied by a severe reduction in the complexity of the dendritic tree determined by Sholl analysis (13) (Fig. 1C) and by computing the total number of intersections between the analyzed dendrites and the shells used for the Sholl analysis (lacZ, 583 ± 35; HDAC4 WT, 537 ± 44; HDAC4 3SA, 324 ± 19; Fig. 1D). In contrast, in agreement with previous reports (18), dendritic spine density was not affected by the nuclear HDAC4 mutant (Fig. 1, E and F). These results show that nuclear accumulation of HDAC4, a class IIa HDAC, has dramatic effects on the dendritic morphology of hippocampal neurons.
Figure 1.
Nuclear accumulation of HDAC4 results in a deterioration of neuronal morphology. A, representative images of cultured hippocampal neurons co-transfected with hrGFP and FLAG-tagged constructs HDAC4 WT, HDAC4 3SA, or lacZ. hrGFP fluorescence reveals neuronal architecture. FLAG tags were used to confirm the subcellular localization of lacZ, HDAC4 WT, and HDAC4 3SA. Nuclei were stained with Hoechst. Scale bar is 40 μm. B, quantification of the total dendritic length of hippocampal neurons transfected as in A. HDAC4 3SA versus HDAC4 WT, p = 1.65 × 10−5; HDAC4 3SA versus lacZ, p = 1.1 × 10−5; lacZ versus HDAC4 WT, p = 0.68. C, Sholl analysis of hippocampal neurons transfected as indicated. D, total number of intersections derived from the Sholl analysis shown in C. HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA versus lacZ, p = 1 × 10−4; lacZ versus HDAC4 WT, p = 0.61. E, quantification of spine density transfected as indicated. F, representative images of dendritic spines of hippocampal neurons transfected as in E. Scale bar is 5 μm. In total, 20 neurons (lacZ and HDAC4 3SA) and 19 neurons (HDAC4 WT) from 5 independent experiments were analyzed for each construct (B–D). For spine density analysis 12 neurons from 3 independent experiments were analyzed for each construct (E). Statistically significant differences were determined by one-way ANOVA (B, D, and E) and two-way ANOVA (C), followed by Tukey's post hoc test. ***, p < 0.001; **, p < 0.01. For scatter plots, each point represents a value derived from one neuron. Graphs represent mean ± S.D.
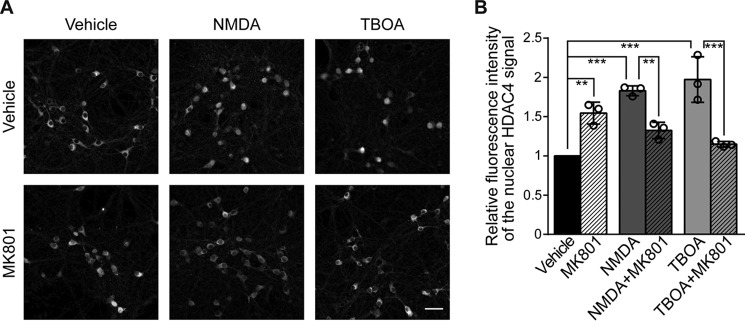
In neuronal cells, HDAC4 is primarily localized in the cytoplasm (10, 17, 18) but it can accumulate in the nucleus in conditions of deprived synaptic activity (10, 18) and in neuronal pathologies (20–24). In particular, extrasynaptic N-methyl-d-aspartate receptors (eNMDARs) that trigger excitotoxicity (25) have been associated with many neurodegenerative diseases (25–28). We therefore investigated the possibility that eNMDARs stimulation causes nuclear accumulation of HDAC4, which would impact neuronal morphology (Fig. 1). Hippocampal neurons were either exposed to bath applied NMDA, which directly activates eNMDARs, or treated with dl-threo-β-benzyloxyaspartic acid (TBOA). TBOA inhibits glutamate up-take systems and, as a result, leads to increased levels of glutamate in the extrasynaptic space and to stimulation of eNMDARs. Both treatments caused a significant increase in the nuclear content of endogenous HDAC4, which was prevented by the NMDA receptor blocker, MK801 (Fig. 2). In line with previous findings, which showed that reducing synaptic activity also causes an increase in nuclear content of HDAC4 (10, 18), hippocampal cultures treated with MK801 only, which blocks synaptic NMDARs at resting conditions, showed elevated HDAC4 nuclear levels (Fig. 2). These results indicate that activation of eNMDARs triggers a redistribution of HDAC4 from the cytosol toward the nucleus.
Figure 2.
Stimulation of extrasynaptic NMDA receptors induces nuclear accumulation of endogenous HDAC4. A, representative images of cultured hippocampal neurons immunostained for endogenous HDAC4 either left untreated (vehicle) or treated with NMDA (20 μm), TBOA (50 μm), and/or MK801 (10 μm) as indicated, for 1 h. Scale bar is 40 μm. B, quantitative measurements of the relative fluorescent intensity of the HDAC4 signal in the nucleus normalized to the respective untreated control. Vehicle versus NMDA, p = 1 × 10−4; vehicle versus NMDA + MK801, p = 0.12; vehicle versus TBOA, p = 1 × 10−4; vehicle versus TBOA + MK801, p = 0.79; vehicle versus MK801, p = 5.1 × 10−3; NMDA versus NMDA + MK801, p = 8.8 × 10−3; TBOA versus TBOA + MK801, p = 1 × 10−4. For each condition, 3 independent experiments were analyzed. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. ***, p < 0.001; **, p < 0.01; bars represent mean ± S.D.
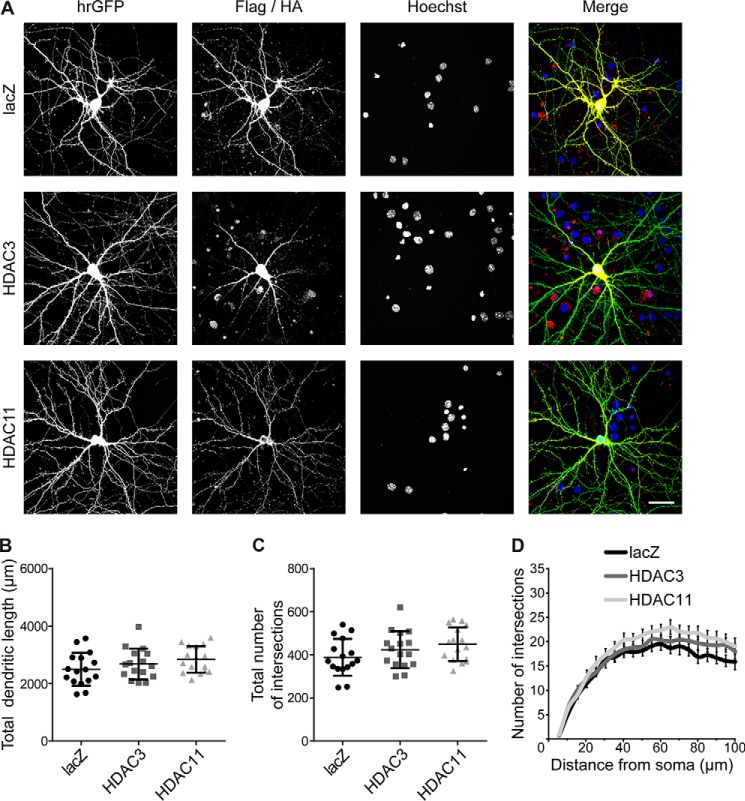
Next, we investigated if increasing the activity, by means of overexpression, of HDACs belonging to class I and IV would also influence dendritic architecture. We transfected neurons with constructs encoding for HDAC3 (class I), HDAC11 (class IV), or lacZ as control (Fig. 3). hrGFP was used for visualization of neuronal morphology. In contrast to HDAC4, morphometric analyses revealed that overexpression of HDAC3 or HDAC11 had no effect on the dendritic length (lacZ, 2499 μm ± 144 μm; HDAC3, 2681 ± 134 μm; HDAC11, 2838 ± 115 μm; Fig. 3B) or complexity (lacZ, 389 ± 21; HDAC3, 424 ± 21; HDAC11, 450 ± 20; Fig. 3, C and D) of mature neurons. These results indicate that among HDACs, class IIa HDAC4 specifically regulates the size and shape of mature dendritic arborization.
Figure 3.
HDACs of class I and IV do not affect dendritic architecture. A, representative images of cultured hippocampal neurons co-transfected with hrGFP and HA-tagged constructs HDAC3 and HDAC11, or lacZ. hrGFP fluorescence reveals neuronal architecture. Epitope tags were used to confirm the subcellular localization of lacZ, HDAC3, and HDAC11. Nuclei were stained with Hoechst. Scale bar is 40 μm. B, quantification of the total dendritic length of hippocampal neurons transfected as in A. lacZ versus HDAC3, p = 0.59; lacZ versus HDAC11, p = 0.17; HDAC3 versus HDAC11, p = 0.68. C, total number of intersections derived from the Sholl analysis shown in D. lacZ versus HDAC3, p = 0.46; lacZ versus HDAC11, p = 0.11; HDAC3 versus HDAC11, p = 0.66. D, Sholl analysis of hippocampal neurons transfected as indicated. In total, 16 neurons from 4 independent experiments were analyzed for each construct. Statistically significant differences were determined by one-way ANOVA (B and C) and two-way ANOVA (D), followed by Tukey's post hoc test. For scatter plots, each point represents a value derived from one neuron. Graphs represent mean ± S.D.
HDAC4 regulates the expression of the neuronal morphology regulator, VEGFD
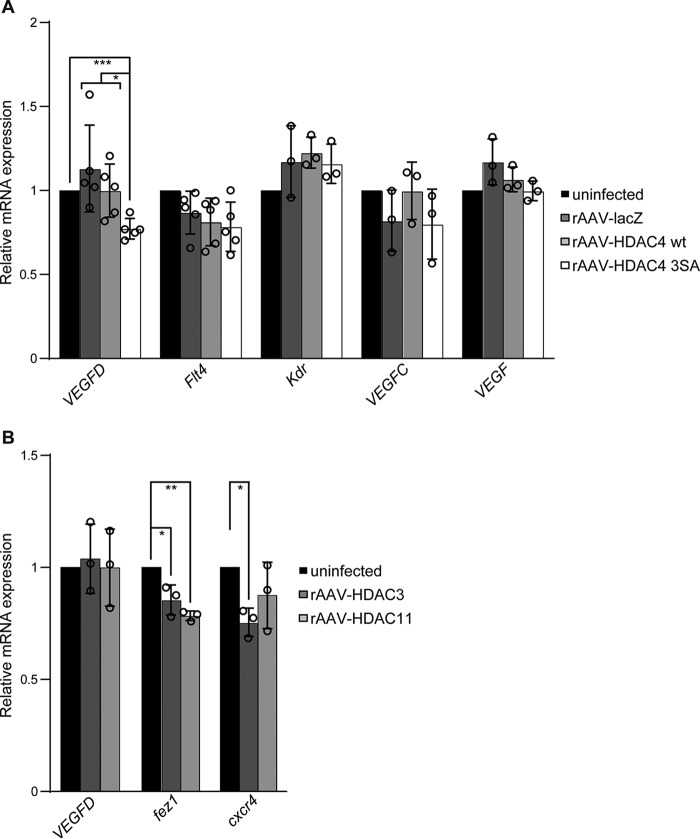
Our previous work identified vascular endothelial growth factor D (VEGFD) as a key regulator of neuronal morphology (12–14). Similar to class IIa HDACs shuttling, VEGFD expression is controlled by nuclear calcium signaling and affected by the calcium buffering capacity of the nucleus (13, 14). We therefore tested whether HDAC4 activity regulates VEGFD expression. We infected hippocampal neurons with rAAVs containing an expression cassette for either HDAC4 (rAAV-HDAC4 WT) or for the HDAC4 3SA mutant (rAAV-HDAC4 3SA). Uninfected cells and neurons infected with lacZ (rAAV-lacZ) served as controls (Fig. 4A). Quantitative reverse transcriptase PCR (qRT-PCR) revealed that expression of the constitutively nuclear and active HDAC4 3SA results in reduced mRNA levels of VEGFD, whereas the expression of other VEGF family members (VEGF, VEGFC) or their cognate receptors (Flt4, Kdr) was not affected by nuclear HDAC4 (VEGFD: rAAV-HDAC4 3SA, 0.77 ± 0.04; rAAV-HDAC4, WT 1 ± 0.04; rAAV-lacZ, 1.13 ± 0.13; Fig. 4A).
Figure 4.
HDAC4 regulates expression of VEGFD. A, quantitative RT-PCR analysis of VEGFD (n = 5), Flt4 (n = 5), Kdr (n = 3), VEGFC (n = 3), and VEGF (n = 3) mRNA levels in uninfected hippocampal neurons and in hippocampal neurons infected with rAAV-lacZ, rAAV-HDAC4 WT, or rAAV-HDAC4 3SA. VEGFD, rAAV-HDAC4 3SA versus rAAV-HDAC4 WT, p = 0.03; rAAV-HDAC4 3SA versus rAAV-lacZ, p = 0.04; rAAV-HDAC4 3SA versus uninfected, p = 4 × 10−4; rAAV-lacZ versus uninfected, p = 0.78; rAAV-HDAC4 wt versus uninfected, p = 0.99; rAAV-lacZ versus rAAV-HDAC4 WT, p = 0.6. B, quantitative RT-PCR analysis of VEGFD (n = 3), fez1 (n = 3), and cxcr4 (n = 3) mRNA levels in uninfected hippocampal neurons and in hippocampal neurons infected with rAAV-HDAC3 or rAAV-HDAC11. VEGFD, uninfected versus rAAV-HDAC3, p = 0.93; uninfected versus rAAV-HDAC11, p = 0.99. fez1, uninfected versus rAAV-HDAC3, p = 0.01; uninfected versus rAAV-HDAC11, p = 1.6 × 10−3. cxcr4, uninfected versus rAAV-HDAC3, p = 0.04; uninfected versus rAAV-HDAC11, p = 0.3. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. ***, p < 0.001; **, p < 0.01; *, p < 0.05. Bars represent mean ± S.D.
Next, we infected hippocampal neurons with rAAV-HDAC3 or rAAV-HDAC11. qRT-PCR revealed that overexpression of HDAC3 resulted in the expected reduced mRNA levels of its target gene cxcr4 (29), whose expression, compared with uninfected control cultures, was not affected by HDAC11 overexpression (Fig. 4B). Furthermore, HDAC11 overexpression caused a decrease of mRNA level of the previously identified HDAC11 target gene fez1 (30), which we also found was affected by HDAC3 overexpression (Fig. 4B). Overexpression of HDAC3 or HDAC11, which does not affect neuronal architecture (Fig. 3), had no effect on VEGFD expression (Fig. 4B). These results suggest that HDAC4 contributes to the regulation of VEGFD expression in hippocampal neurons.
VEGFD overexpression prevents the impairment of morphology caused by HDAC4 3SA expression
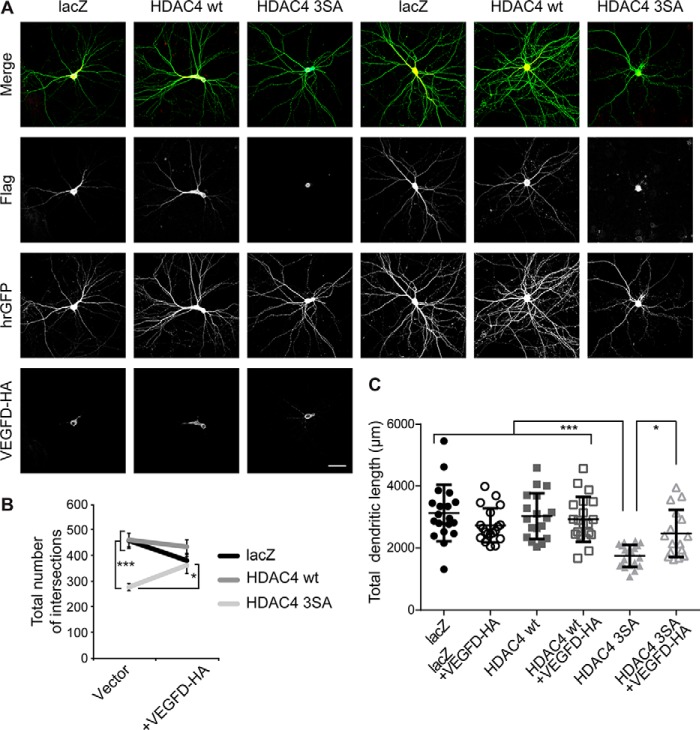
As nuclear accumulation of HDAC4 negatively affects the size and complexity of dendritic trees (Fig. 1), as well as the expression of VEGFD (Fig. 4), we next investigated if overexpression of VEGFD could rescue the impaired morphology of HDAC4 3SA expressing neurons. To this end, we made use of a construct encoding for VEGFD and carrying an HA tag (13). Morphometric analyses revealed that, as previously demonstrated (13), overexpression of VEGFD does not change the morphology of control hippocampal neurons (total dendritic length: lacZ, 3130 ± 209 μm; lacZ + VEGFD-HA, 2730 ± 127 μm; HDAC4 WT, 3026 ± 174 μm; HDAC4 WT + VEGFD-HA, 2925 ± 162 μm; Fig. 5, A and C and total number of intersections: lacZ, 460 ± 30; lacZ + VEGFD-HA, 382 ± 23; HDAC4 WT, 461 ± 27; HDAC4 WT + VEGFD-HA, 435 ± 27; Fig. 5, A and B). However, VEGFD overexpression can prevent the loss of structural integrity induced by HDAC4 3SA expression. Although total dendritic length of neurons expressing HDAC4 3SA was significantly reduced to an average value of 1746 ± 79 μm (Figs. 1 and 5, A and C), the dendrites of HDAC4 3SA + VEGFD-HA expressing neurons are similar in length (2466 ± 191 μm) to those of control groups analyzed (Fig. 5, A and C). Moreover, Sholl analysis showed that HDAC4 3SA + VEGFD-HA expressing cells displayed a higher degree of complexity than HDAC4 3SA expressing neurons, which is close to control values (HDAC4 3SA, 278 ± 14; HDAC4 3SA + VEGFD-HA, 363 ± 32; Fig. 5, A and B). These data show that overexpression of VEGFD can compensate for the down-regulation of endogenous VEGFD induced by nuclear-accumulated HDAC4 thereby preventing the loss of dendritic structures of hippocampal neurons.
Figure 5.
Overexpression of VEGFD can rescue impaired morphology of neurons caused by nuclear HDAC4. A, representative images of cultured hippocampal neurons co-transfected with hrGFP, FLAG-tagged constructs HDAC4 WT, HDAC4 3SA, lacZ with or without HA-tagged VEGFD as indicated. Epitope tags were detected with Alexa 594 (FLAG) and Alexa 633 (HA). Scale bar is 40 μm. B, total number of intersections derived from Sholl analysis of hippocampal neurons transfected as in A. lacZ versus lacZ + VEGFD-HA, p = 0.24; HDAC4 WT versus HDAC4 WT + VEGFD-HA, p = 0.98; HDAC4 3SA + VEGFD-HA versus HDAC4 3SA, p = 0.02; HDAC4 3SA + VEGFD-HA versus lacZ, p = 0.14; HDAC4 3SA + VEGFD-HA versus lacZ + VEGFD-HA, p = 0.99; HDAC4 3SA + VEGFD-HA versus HDAC4 WT, p = 0.13; HDAC4 3SA + VEGFD-HA versus HDAC4 WT + VEGFD-HA, p = 0.43. C, quantification of the total dendritic length of hippocampal neurons transfected as in A. lacZ versus lacZ + VEGFD-HA, p = 0.48; HDAC4 WT versus HDAC4 WT + VEGFD-HA, p = 0.99; HDAC4 3SA versus lacZ, p = 1 × 10−4; HDAC4 3SA versus lacZ + VEGFD-HA, p = 3 × 10−4; HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA versus HDAC4 WT + VEGFD-HA, p = 1 × 10−4; HDAC4 3SA versus HDAC4 3SA + VEGFD-HA, p = 0.03; HDAC4 3SA + VEGFD-HA versus lacZ, p = 0.06; HDAC4 3SA + VEGFD-HA versus lacZ + VEGFD-HA, p = 0.87; HDAC4 3SA + VEGFD-HA versus HDAC4 WT, p = 0.18; HDAC4 3SA + VEGFD-HA versus HDAC4 WT + VEGFD-HA, p = 0.36. In total, 19 neurons (lacZ and lacz + VEGFD-HA), 18 neurons (HDAC4 wt), 20 neurons (HDAC4 WT + VEGFD-HA and HDAC4 3SA), and 16 neurons (HDAC4 3SA + VEGFD-HA) from 5 independent experiments were analyzed for each condition. Statistically significant differences were determined by one-way ANOVA followed by Tukey's post hoc test. ***, p < 0.001; *, p < 0.05. For scatter plot, each point represents a value derived from one neuron. Graphs represent mean ± S.D.
VEGFD administration to HDAC4 3SA expressing hippocampal neurons with impaired dendritic architecture can restore structural integrity
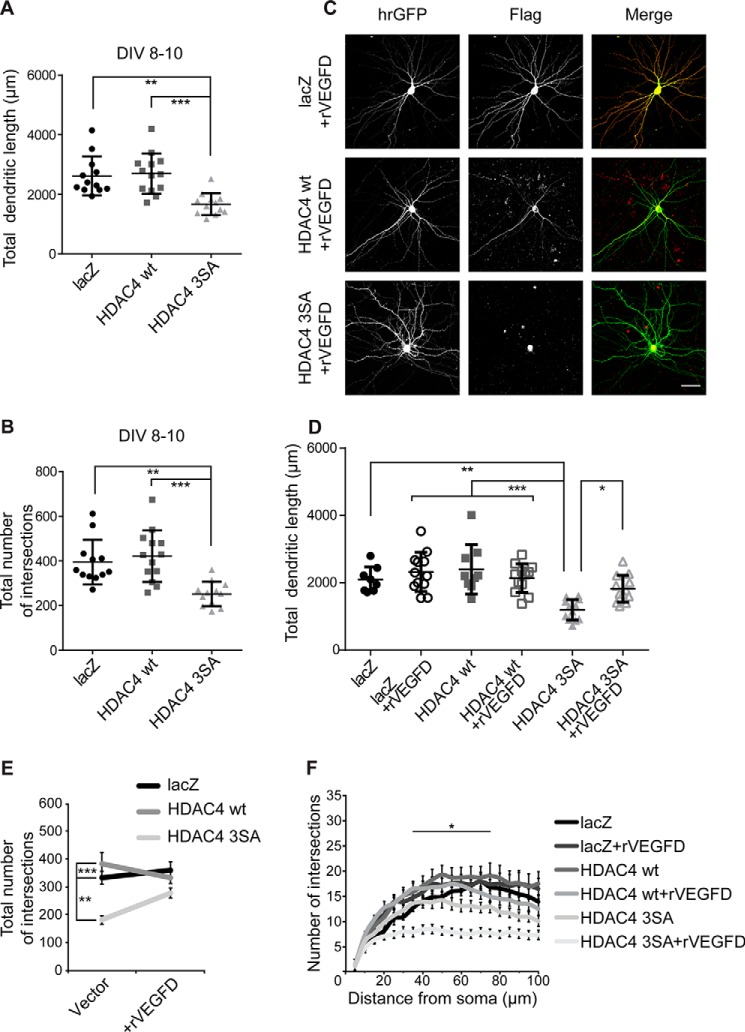
To investigate if VEGFD is capable to restore normal dendrite morphology when already impaired, we first performed morphometric analysis on neuronal cells expressing lacZ, HDAC4 WT, or HDAC4 3SA for 48 h. This is a shorter time period than our previous experiments where we allowed the transgenes to be expressed for 5 days before morphological assessment (Figs. 1 and 5). We observed that HDAC4 3SA expression impairs morphology even after a shorter expression time (total dendritic length: lacZ, 2617 ± 189 μm; HDAC4 WT, 2693 ± 188 μm; HDAC4 3SA, 1667 ± 106 μm; Fig. 6A and total number of intersections: lacZ, 394 ± 28; HDAC4 WT, 421 ± 32; HDAC4 3SA, 252 ± 16; Fig. 6B).
Figure 6.
VEGFD treatment prevents the simplification of dendritic morphology promoted by nuclear HDAC4. A, quantification of the total dendritic length of hippocampal neurons at DIV10. Cells were co-transfected with hrGFP and FLAG-tagged constructs lacZ, HDAC4 WT, and HDAC4 3SA at DIV8. HDAC4 3SA versus HDAC4 WT, p = 3 × 10−4; HDAC4 3SA versus lacZ, p = 1 × 10−3; lacZ versus HDAC4 WT, p = 0.94. B, total number of intersections derived from Sholl analysis at DIV10. HDAC4 3SA versus HDAC4 WT, p = 2 × 10−4; HDAC4 3SA versus lacZ, p = 2.1 × 10−3; lacZ versus HDAC4 WT, p = 0.76. C, representative images of cultured hippocampal neurons transfected as in A and treated or not with rVEGFD (100 ng/ml) for 3 days. FLAG tags were stained with Alexa 594. Scale bar is 40 μm. D, quantification of the total dendritic length of hippocampal neurons transfected as indicated, with or without treatment for 3 days with rVEGFD. HDAC4 3SA versus lacZ, p = 1.9 × 10−3; HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA versus HDAC4 3SA + rVEGFD, p = 0.03; HDAC4 3SA + rVEGFD versus lacZ + rVEGFD, p = 0.14; HDAC4 3SA + rVEGFD versus HDAC4 WT + rVEGFD, p = 0.61. E, total number of intersections derived from the Sholl analysis shown in F. HDAC4 3SA versus lacZ, p = 2.2 × 10−3; HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA + rVEGFD versus lacZ, p = 0.69; HDAC4 3SA + rVEGFD versus lacZ + rVEGFD, p = 0.15; HDAC4 3SA + rVEGFD versus HDAC4 WT, p = 0.06; HDAC4 3SA + rVEGFD versus HDAC4 WT + rVEGFD p = 0.57; lacZ versus lacZ + rVEGFD, p = 0.98; HDAC4 WT versus HDAC4 WT + rVEGFD, p = 0.76. F, Sholl analysis of hippocampal neurons transfected as indicated, with or without treatment for 3 days with rVEGFD. In total, 8 (lacZ), 9 (HDAC4 WT), and 12 neurons (lacZ + rVEGFD, HDAC4 WT + rVEGFD, HDAC4 3SA, HDAC4 3SA + rVEGFD) from 3 independent experiments were analyzed for each construct (D, E, and F). At DIV10, 12 (lacZ and HDAC4 3SA) and 13 (HDAC4 WT) neurons from 3 independent experiments were analyzed for each construct (A and B). Statistically significant differences were determined by one-way ANOVA (A, B, D, and E) and two-way ANOVA (F) followed by Tukey's post hoc test. ***, p < 0.001; **, p < 0.01; *, p < 0.05. For scatter plots, each point represents a value derived from one neuron. Graphs represent mean ± S.D.
For the rescue experiments we used recombinant VEGFD (rVEGFD). We first allowed the neurons to express lacZ, HDAC4 WT, or HDAC4 3SA for 48 h, which is a sufficient period of time to impair morphology (Fig. 6, A and B), and later we applied rVEGFD (100 ng/ml) to the culturing medium. After 3 days, morphometric analysis showed that, as expected (Figs. 1 and 5), HDAC4 3SA expressing neurons showed a lower total dendritic length compared with lacZ and HDAC4 WT expressing neurons (lacZ, 2095 ± 134 μm; HDAC4 WT, 2399 ± 245 μm; HDAC4 3SA, 1194 ± 88 μm; Fig. 6, C and D). rVEGFD administration was sufficient to restore the total dendritic length of HDAC4 3SA expressing neurons back to control levels. As shown for the overexpression experiments (Fig. 5) and as previously observed (13, 14), rVEGFD treatment did not have any effect on control neurons (HDAC4 3SA + rVEGFD, 1822 ± 116 μm; lacZ + rVEGFD, 2320 ± 168 μm; HDAC4 WT + rVEGFD, 2138 ± 123 μm; Fig. 6, C and D). Sholl analysis revealed that rVEGFD treatment also restores the complexity of the previously impaired dendritic arborization without affecting control neurons (total number of intersections: lacZ, 335 ± 24; HDAC4 WT, 384 ± 43; HDAC4 3SA, 185 ± 15; lacZ + rVEGFD, 363 ± 29; HDAC4 WT + rVEGFD, 336 ± 20; HDAC4 3SA + rVEGFD, 280 ± 18; Fig. 6, C, E, and F). Taken together, these results show that VEGFD can restore dendritic morphology when this is already impaired as a result of HDAC4 nuclear accumulation.
VEGFC fails to restore the morphology of HDAC4 3SA expressing neurons
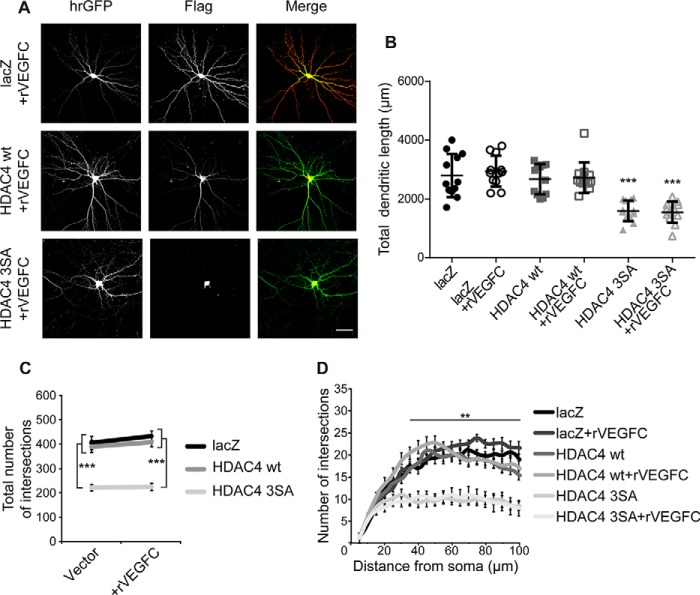
The closest homologue to VEGFD is VEGFC. The two factors belong to the same family of growth factors and have been reported to possibly activate the same tyrosine kinase receptors (31). Although expression of VEGFC in hippocampal neurons is not affected by HDAC4 3SA (see Fig. 4), we investigated the potential of VEGFC to counteract the loss of the dendritic structure induced by HDAC4 3SA. rVEGFC (100 ng/ml) was added to the culture medium over a period of 3 days, starting at 2 days after neurons had been transfected with hrGFP, lacZ, HDAC4 WT, or HDAC4 3SA. All neurons expressing HDAC4 3SA, with or without rVEGFC treatment, exhibited severely impaired morphological features compared with lacZ- or HDAC4 WT-transfected cells (total dendritic length: lacZ, 2797 ± 211 μm; HDAC4 WT, 2676 ± 150 μm; HDAC4 3SA, 1593 ± 101 μm; lacZ + rVEGFC, 2944 ± 150 μm; HDAC4 WT + rVEGFC, 2726 ± 151 μm; HDAC4 3SA + rVEGFC, 1546 ± 105 μm; Fig. 7, A and B). Likewise, VEGFC failed to restore dendrite complexity assessed by Sholl analysis (total number of intersections: lacZ, 421 ± 29; HDAC4 WT, 404 ± 24; HDAC4 3SA, 230 ± 15; lacZ + rVEGFC, 451 ± 22; HDAC4 WT + rVEGFC, 424 ± 22; HDAC4 3SA + rVEGFC, 233 ± 16; Fig. 7, A, C, and D). These results indicate that different from VEGFD, VEGFC cannot rescue the impairment of dendrite architecture caused by nuclear-localized HDAC4.
Figure 7.
VEGFC has no influence on the dendritic architecture of HDAC4 3SA expressing neurons. A, representative images of cultured hippocampal neurons co-transfected with hrGFP and FLAG-tagged constructs lacZ, HDAC4 WT, and HDAC4 3SA, treated or not with rVEGFC (100 ng/ml) for 3 days. FLAG tags were detected with Alexa 594. Scale bar is 40 μm. B, quantification of the total dendritic length of hippocampal neurons transfected as indicated, with or without treatment for 3 days with rVEGFC. HDAC4 3SA versus lacZ, p = 1 × 10−4; HDAC4 3SA versus lacZ + rVEGFC, p = 1 × 10−4; HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA versus HDAC4 WT + rVEGFC, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus lacZ, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus lacZ + rVEGFC, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus HDAC4 WT + rVEGFC, p = 1 × 10−4; lacZ versus lacZ + rVEGFC, p = 0.98; HDAC4 WT versus HDAC4 wt + rVEGFC, p = 0.99. C, total number of intersections derived from the Sholl analysis shown in D. HDAC4 3SA versus lacZ, p = 1 × 10−4; HDAC4 3SA versus lacZ + rVEGFC, p = 1 × 10−4; HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA versus HDAC4 WT + rVEGFC, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus lacZ, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus lacZ + rVEGFC, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA + rVEGFC versus HDAC4 WT + rVEGFC, p = 1 × 10−4; lacZ versus lacZ + rVEGFC, p = 0.94; HDAC4 WT versus HDAC4 WT + rVEGFC, p = 0.99. D, Sholl analysis of hippocampal neurons transfected as indicated, with or without treatment for 3 days with rVEGFC. In total, 12 neurons from 3 independent experiments were analyzed for each construct. Statistically significant differences were determined by one-way ANOVA (B and C) and two-way ANOVA (D) followed by Tukey's post hoc test. ***, p < 0.001; **, p < 0.01. For the scatter plot, each point represents a value derived from one neuron. Graphs represent mean ± S.D.
In conclusion, nuclear-localized HDAC4 leads to suppression of VEGFD expression and subsequent loss of structural integrity of the dendritic arbor. Supplementation of VEGFD prevents this impairment and, moreover, can restore a normal morphology even after neuronal structures have been lost.
Discussion
In this study, we demonstrated that nuclear accumulation of HDAC4, a class IIa HDAC, impairs dendritic architecture by down-regulating the expression of VEGFD, a key factor for dendrite maintenance.
Effects of nuclear localization of HDAC4 in neuronal cells
Our data show that nuclear accumulation of HDAC4 causes a simplification of dendritic arborization. In neurons, at resting conditions, HDAC4 is predominantly localized in the cytoplasm (17), where it appears to promote neuroprotective gene expression (32, 33). Indeed, synaptic activity-induced nuclear calcium-signaling, known for its survival-enhancing effects (34–37), has been shown to promote HDAC4 nuclear export (10). In contrast, toxic insults promote HDAC4 nuclear accumulation (9). Our data indicate that the eNMDAR, a key player in neurodegenerative processes (25–28), is responsible for excitotoxicity-associated nuclear accumulation of HDAC4 (Fig. 2). Increased levels of HDAC4 in the nucleus of neuronal cells have been observed in several neurodegenerative disorders including stroke, Parkinson's disease, Alzheimer's disease, and ataxia telangiectasia and are linked to disease progression and neuronal death (20–24). Thus, HDAC4 nuclear localization might represent an intermediate event on the road to neuronal death: downstream of the initiating signals but preceding and possibly facilitating progression toward full-blown degeneration. Our results demonstrate that HDAC4 nuclear localization is detrimental to the maintenance of dendrite integrity. Indeed, the neurodegenerative disorders mentioned above often exhibit aberrations in neuronal morphology in addition to a pathological HDAC4 nuclear accumulation (38, 39), suggesting that the two events may be causally linked. Future work needs to extend our cell culture experiments to in vivo studies of HDAC4-mediated regulation of neuronal morphology, considering in particular that depending on the brain region, transcriptional response to neuronal calcium signals can differentially regulate dendritic architecture (40).
Histone deacetylases and neuronal morphology
Different classes of HDACs have been implicated in the development of neuronal architecture both at the synaptic and dendritic levels. HDAC1 and HDAC2, which belong to class I, negatively regulate the formation and maturation of excitatory synapses in immature neurons (41, 42), and expression of HDAC4 mutants can lead to changes in the strength and internal structural organization of synapses without affecting their number (18). Here, we confirm that the number of spines of hippocampal neurons is not affected by the expression of HDAC4 3SA. HDAC6, HDAC5, and HDAC9 activity are thought to be involved in dendritogenesis specifically during the early stages of neurodevelopment (43–45). Our experiments uncovered that HDAC4 influences the structure of an established, complex dendritic tree at a developmental stage when most of dendritogenesis has taken place and the neurons are mature and have formed functional synaptically connected networks. Hippocampal neurons, cultured under our experimental settings, do not extend their dendritic connections in the analyzed timeframe (data not shown). Boosting either class I- or class IV-HDACs signaling by means of overexpression does not seem to influence mature dendritic arborization (see Fig. 3) or VEGFD expression (Fig. 4), suggesting that primarily the synaptic activity-regulated nucleo-cytoplasmic shuttling of class IIa HDACs controls dendrite architecture. Consistent with this hypothesis is the observation that blockade of synaptic activity causes accumulation of HDAC4 in the cell nucleus, but not of the other HDAC classes (10), and dendrite impairments (46). Expression of VEGFD, which is negatively regulated by nuclear HDAC4 (see Fig. 4), is also strongly decreased in conditions of lowered synaptic activity (13). Thus, basal synaptic activity ensures proper maintenance of dendritic arborization by promoting HDAC4 nuclear export thereby allowing for sufficiently high levels of VEGFD.
VEGFD is a key regulator of proper connectivity
VEGFD is a key factor for the maintenance of mature dendritic structure of pyramidal neurons (13, 14), and as such crucial for memory formation as well as consolidation and extinction of fear memory (12, 13). Several disorders of the nervous system are characterized by both aberrations of morphology and HDAC4 nuclear localization. It would be of interest to determine neuronal VEGFD expression levels in such pathological conditions since our present work links nuclear accumulation of HDAC4 to a decrease of neuronal complexity and VEGFD expression. However, HDAC4 nuclear localization might affect morphology by influencing additional genes besides VEGFD, and, moreover, the observed down-regulation of VEGFD expression may not be directly caused by HDAC4.
In this study we show that overexpression of VEGFD or treatment with recombinant VEGFD, but not with that of its homologue VEGFC, prevents the loss of dendrites caused by nuclear HDAC4. Moreover, using administration of recombinant VEGFD we succeeded in restoring dendritic architecture to control levels even at a stage where the loss of structural integrity had already taken place. Thus, VEGFD has the potential to prevent structural disintegration of neurons associated with neurodegenerative diseases and may even restore functional dendritic arbors.
Experimental procedures
Expression constructs
Expression vectors for HDAC3, HDAC11, HDAC4, HDAC4 3SA, VEGFD, and lacZ were used for co-transfection with hrGFP. All constructs were C-terminal epitope-tagged and previously described (10, 13). LacZ, HDAC4, and HDAC4 3SA carry a FLAG tag, whereas VEGFD, HDAC3, and HDAC11 constructs contain a HA tag.
Recombinant adeno-associated viruses
The method used to construct, package, and purify recombinant adeno-associated viruses (rAAVs) has been previously described (36). For viral infection, cultured hippocampal neurons were infected on day in vitro 3 (DIV) and harvested on DIV10.
Hippocampal neuronal cultures and treatments
Hippocampal neurons from newborn C57BL/6 mice were isolated and cultured as described before (47). DNA transfection was done after a culturing period of 8 days in vitro using Lipofectamine. Experiments were done at DIV10. Transfected cells were treated with 100 ng/ml of recombinant mouse VEGFD (R&D Systems GmbH) or 100 ng/ml of recombinant mouse VEGFC (Biocat) over a period of 3 days until analysis.
Immunocytochemistry
Following transfection and/or treatments, cells were fixed on DIV13 (or DIV10 when indicated in the text) for 20 min at room temperature with paraformaldehyde (4% paraformaldehyde, 4% sucrose in phosphate-buffered saline (PBS), pH 7.4). Antibodies were diluted in GDB (0.1% gelatin, 0.3% Triton X-100, 15 mm Na2HPO4, 400 mm NaCl) and cells were incubated overnight with primary antibodies and 45 min with secondary antibodies. Hoechst staining (1:6000) was used for visualization of nuclei. Coverslips were mounted with Mowiol 4-88 (Calbiochem).
Antibodies
Mouse monoclonal anti-FLAG-M2 (1:200, Sigma), rabbit monoclonal anti-HDAC4 (1:200, Cell Signaling), rabbit polyclonal anti-HA (1:200, Santa Cruz), and Alexa 594 goat anti-mouse and Alexa 633 goat anti-rabbit (1:400, Life Technologies) were used.
RNA extraction and cDNA synthesis
Total RNA was isolated at DIV10 from hippocampal primary neuron cultures with an RNeasy Mini Kit (Qiagen) including an optional DNase I treatment at room temperature for 15 min according to the manufacturer's instructions (Qiagen). 1.2 μg of extracted RNA was reverse transcribed into first strand cDNA using High Capacity cDNA Reverse Transcription kit (Applied Biosystems).
Real-time quantitative PCR
qRT-PCR was done on StepOne plus Real-Time PCR using TaqMan Gene Expression Assays for the indicated genes (Applied Biosystems). The following TaqMan Gene Expression Assays were used in this study: Gusb (Mm00446953_m1), VEGFD (Mm00438965_m1), VEGFC (Mm01202432_m1), VEGF (Mm01281449_m1), VEGFR3/Flt4 (Mm01292618_m1), VEGFR2/Kdr (Mm00440099_m1), fez1 (Mm00805945_m1), and cxcr4 (Mm01292123_m1). Expression of target genes was normalized against the expression of Gusb as the endogenous control gene. Data were derived from 3 to 7 independent experiments.
Morphometric analyses
For morphometric analyses, neurons were analyzed 2 or 5 days after transfection, as indicated under “Results,” using a Leica TCS SP2 confocal microscope or a Nikon A1R confocal microscope. Total dendritic length and complexity were calculated using Fiji (48). Briefly, a z-stack acquisition was imported, calibrated, and manually traced using the simple neurite tracer plugin (49). Total dendritic length was then computed. For three-dimensional Sholl analysis (50), the shell interval was set to 5 μm using a plugin available for Fiji. Total number of intersections was defined by the sum of all intersections between the traced dendrites and the shells used for Sholl analysis up to a radius of 185 μm. Dendritic spine density over randomly chosen 20-μm dendrite portions was manually computed. All analyses were performed blind. For each experimental condition, 8–20 neurons from 3 to 5 independent preparations were analyzed; details are always indicated in the respective figure legends.
Quantification of nuclear HDAC4
Cells were either treated with 20 μm NMDA, 50 μm dl-threo-β-benzyloxyaspartic acid, and/or 10 μm dizocilpine (MK 801) for 1 h. After fixation, cultures were immunostained with an antibody directed against HDAC4, nuclei were visualized using Hoechst. Up to 8 random pictures from each treated condition were acquired using a Nikon Ni-E upright fluorescence microscope. Fluorescent intensity of HDAC4 nuclear signal has been analyzed by defining the nuclei in the Hoechst channel as the region of interest in ImageJ. The average integrated density was measured, and the relative intensity of the nuclei compared with the untreated control cells was calculated.
Data analysis
Data are presented as mean ± S.D. Unless otherwise stated, statistical analysis has been done using one-way analysis of variance (ANOVA) with Tukey's post hoc test. Results were considered to be statistically significant for significance levels of p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***). Exact p values are displayed in the relative figure legends.
Author contributions
C. L. and D. M. formal analysis; C. L. and D. M. investigation; C. L. and D. M. methodology; C. L. and D. M. writing-original draft; C. L., H. B., and D. M. writing-review and editing; H. B. and D. M. conceptualization; H. B. resources; H. B. and D. M. funding acquisition; H. B. and D. M. project administration; D. M. supervision.
Acknowledgments
We thank I. Bünzli-Ehret for help with the preparation of hippocampal cultures and Markus Kardoff for technical help. Portions of the figures were acquired at the Nikon Imaging Center at Heidelberg University.
This work was supported in part by Deutsche Forschungsgemeinschaft Grants SFB1158 and FOR2325 (to D. M. and H. B.) and FRONTIER grant from the Heidelberg University (to D. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- HDAC
- histone deacetylase
- rVEGF
- recombinant vascular endothelial growth factor
- Flt4
- Fms-related tyrosine kinase 4
- Kdr
- kinase insert domain receptor
- fez1
- fasciculation and elongation protein ζ1
- cxcr4
- C-X-C motif chemokine receptor 4
- hrGFP
- humanized R. reniformis green fluorescent protein
- rAAV
- recombinant adeno-associated virus
- eNMDAR
- extrasynaptic N-methyl-d-aspartate receptor
- TBOA
- DL-threo-β-benzyloxyaspartic acid
- MK801
- dizocilpine
- DIV
- days in vitro
- ANOVA
- analysis of variance
- qRT-PCR
- quantitative reverse transcriptase-PCR.
References
- 1. Peserico A., and Simone C. (2011) Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011, 371832 10.1155/2011/371832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiang Y., Langley B., Lubin F. D., Renthal W., Wood M. A., Yasui D. H., Kumar A., Nestler E. J., Akbarian S., and Beckel-Mitchener A. C. (2008) Epigenetics in the nervous system. J. Neurosci. 28, 11753–11759 10.1523/JNEUROSCI.3797-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim M. S., Akhtar M. W., Adachi M., Mahgoub M., Bassel-Duby R., Kavalali E. T., Olson E. N., and Monteggia L. M. (2012) An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J. Neurosci. 32, 10879–10886 10.1523/JNEUROSCI.2089-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan J. K., Sun L., Yang X. J., Zhu G., and Wu Z. (2003) Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278, 23515–23521 10.1074/jbc.M301922200 [DOI] [PubMed] [Google Scholar]
- 5. Tse C., Sera T., Wolffe A. P., and Hansen J. C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18, 4629–4638 10.1128/MCB.18.8.4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang X. J., and Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haberland M., Montgomery R. L., and Olson E. N. (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao X., Ito A., Kane C. D., Liao T. S., Bolger T. A., Lemrow S. M., Means A. R., and Yao T. P. (2001) The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276, 35042–35048 10.1074/jbc.M105086200 [DOI] [PubMed] [Google Scholar]
- 9. Chawla S., Vanhoutte P., Arnold F. J., Huang C. L., and Bading H. (2003) Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85, 151–159 10.1046/j.1471-4159.2003.01648.x [DOI] [PubMed] [Google Scholar]
- 10. Schlumm F., Mauceri D., Freitag H. E., and Bading H. (2013) Nuclear calcium signaling regulates nuclear export of a subset of class IIa histone deacetylases following synaptic activity. J. Biol. Chem. 288, 8074–8084 10.1074/jbc.M112.432773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bading H. (2013) Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 14, 593–608 10.1038/nrn3531 [DOI] [PubMed] [Google Scholar]
- 12. Hemstedt T. J., Bengtson C. P., Ramírez O., Oliveira A. M. M., and Bading H. (2017) Reciprocal interaction of dendrite geometry and nuclear calcium-VEGFD signaling gates memory consolidation and extinction. J. Neurosci. 37, 6946–6955 10.1523/JNEUROSCI.2345-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mauceri D., Freitag H. E., Oliveira A. M., Bengtson C. P., and Bading H. (2011) Nuclear calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron 71, 117–130 10.1016/j.neuron.2011.04.022 [DOI] [PubMed] [Google Scholar]
- 14. Mauceri D., Hagenston A. M., Schramm K., Weiss U., and Bading H. (2015) Nuclear calcium buffering capacity shapes neuronal architecture. J. Biol. Chem. 290, 23039–23049 10.1074/jbc.M115.654962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oliveira A. M., Hemstedt T. J., and Bading H. (2012) Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 15, 1111–1113 10.1038/nn.3151 [DOI] [PubMed] [Google Scholar]
- 16. Broide R. S., Redwine J. M., Aftahi N., Young W., Bloom F. E., and Winrow C. J. (2007) Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci. 31, 47–58 10.1007/BF02686117 [DOI] [PubMed] [Google Scholar]
- 17. Darcy M. J., Calvin K., Cavnar K., and Ouimet C. C. (2010) Regional and subcellular distribution of HDAC4 in mouse brain. J. Comp. Neurol. 518, 722–740 10.1002/cne.22241 [DOI] [PubMed] [Google Scholar]
- 18. Sando R. 3rd, Gounko N., Pieraut S., Liao L., Yates J. 3rd, and Maximov A. (2012) HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151, 821–834 10.1016/j.cell.2012.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grozinger C. M., and Schreiber S. L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. U.S.A. 97, 7835–7840 10.1073/pnas.140199597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kassis H., Shehadah A., Chopp M., Roberts C., and Zhang Z. G. (2015) Stroke induces nuclear shuttling of histone deacetylase 4. Stroke 46, 1909–1915 10.1161/STROKEAHA.115.009046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen X., Chen J., Li J., Kofler J., and Herrup K. (2016) Neurons in vulnerable regions of the Alzheimer's disease brain display reduced ATM signaling. eNeuro 3, pii:ENEURO.0124–15.2016 10.1523/ENEURO.0124-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu Q., Yang X., Zhang L., Zhang Y., and Feng L. (2017) Nuclear accumulation of histone deacetylase 4 (HDAC4) exerts neurotoxicity in models of Parkinson's disease. Mol. Neurobiol. 54, 6970–6983 10.1007/s12035-016-0199-2 [DOI] [PubMed] [Google Scholar]
- 23. Yuan H., Denton K., Liu L., Li X. J., Benashski S., McCullough L., and Li J. (2016) Nuclear translocation of histone deacetylase 4 induces neuronal death in stroke. Neurobiol. Dis. 91, 182–193 10.1016/j.nbd.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 24. Li J., Chen J., Ricupero C. L., Hart R. P., Schwartz M. S., Kusnecov A., and Herrup K. (2012) Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat. Med. 18, 783–790 10.1038/nm.2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hardingham G. E., Fukunaga Y., and Bading H. (2002) Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat. Neurosci. 5, 405–414 10.1038/nn835 [DOI] [PubMed] [Google Scholar]
- 26. Parsons M. P., and Raymond L. A. (2014) Extrasynaptic NMDA receptor involvement in central nervous system disorders. Neuron 82, 279–293 10.1016/j.neuron.2014.03.030 [DOI] [PubMed] [Google Scholar]
- 27. Bading H. (2017) Therapeutic targeting of the pathological triad of extrasynaptic NMDA receptor signaling in neurodegenerations. J. Exp. Med. 214, 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hardingham G. E., and Bading H. (2010) Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat. Rev. Neurosci. 11, 682–696 10.1038/nrn2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim H. C., Choi K. C., Choi H. K., Kang H. B., Kim M. J., Lee Y. H., Lee O. H., Lee J., Kim Y. J., Jun W., Jeong J. W., and Yoon H. G. (2010) HDAC3 selectively represses CREB3-mediated transcription and migration of metastatic breast cancer cells. Cell Mol. Life Sci. 67, 3499–3510 10.1007/s00018-010-0388-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bryant D. T., Landles C., Papadopoulou A. S., Benjamin A. C., Duckworth J. K., Rosahl T., Benn C. L., and Bates G. P. (2017) Disruption to schizophrenia-associated gene Fez1 in the hippocampus of HDAC11 knockout mice. Sci. Rep. 7, 11900 10.1038/s41598-017-11630-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamazaki Y., and Morita T. (2006) Molecular and functional diversity of vascular endothelial growth factors. Mol. Divers 10, 515–527 10.1007/s11030-006-9027-3 [DOI] [PubMed] [Google Scholar]
- 32. Bolger T. A., and Yao T. P. (2005) Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 25, 9544–9553 10.1523/JNEUROSCI.1826-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen B., and Cepko C. L. (2009) HDAC4 regulates neuronal survival in normal and diseased retinas. Science 323, 256–259 10.1126/science.1166226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee B., Butcher G. Q., Hoyt K. R., Impey S., and Obrietan K. (2005) Activity-dependent neuroprotection and cAMP response element-binding protein (CREB): kinase coupling, stimulus intensity, and temporal regulation of CREB phosphorylation at serine 133. J. Neurosci. 25, 1137–1148 10.1523/JNEUROSCI.4288-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Papadia S., Stevenson P., Hardingham N. R., Bading H., and Hardingham G. E. (2005) Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J. Neurosci. 25, 4279–4287 10.1523/JNEUROSCI.5019-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang S. J., Steijaert M. N., Lau D., Schütz G., Delucinge-Vivier C., Descombes P., and Bading H. (2007) Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron 53, 549–562 10.1016/j.neuron.2007.01.025 [DOI] [PubMed] [Google Scholar]
- 37. Zhang S. J., Zou M., Lu L., Lau D., Ditzel D. A., Delucinge-Vivier C., Aso Y., Descombes P., and Bading H. (2009) Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 5, e1000604 10.1371/journal.pgen.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baloyannis S. J. (2009) Dendritic pathology in Alzheimer's disease. J. Neurolog. Sci. 283, 153–157 10.1016/j.jns.2009.02.370 [DOI] [PubMed] [Google Scholar]
- 39. Stephens B., Mueller A. J., Shering A. F., Hood S. H., Taggart P., Arbuthnott G. W., Bell J. E., Kilford L., Kingsbury A. E., Daniel S. E., and Ingham C. A. (2005) Evidence of a breakdown of corticostriatal connections in Parkinson's disease. Neuroscience 132, 741–754 10.1016/j.neuroscience.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 40. Mellström B., Kastanauskaite A., Knafo S., Gonzalez P., Dopazo X. M., Ruiz-Nuño A., Jefferys J. G., Zhuo M., Bliss T. V., Naranjo J. R., and DeFelipe J. (2016) Specific cytoarchitectureal changes in hippocampal subareas in daDREAM mice. Mol. Brain 9, 22 10.1186/s13041-016-0204-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Akhtar M. W., Raingo J., Nelson E. D., Montgomery R. L., Olson E. N., Kavalali E. T., and Monteggia L. M. (2009) Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J. Neurosci. 29, 8288–8297 10.1523/JNEUROSCI.0097-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guan J. S., Haggarty S. J., Giacometti E., Dannenberg J. H., Joseph N., Gao J., Nieland T. J., Zhou Y., Wang X., Mazitschek R., Bradner J. E., DePinho R. A., Jaenisch R., and Tsai L. H. (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459, 55–60 10.1038/nature07925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ageta-Ishihara N., Miyata T., Ohshima C., Watanabe M., Sato Y., Hamamura Y., Higashiyama T., Mazitschek R., Bito H., and Kinoshita M. (2013) Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun. 4, 2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gu X., Fu C., Lin L., Liu S., Su X., Li A., Wu Q., Jia C., Zhang P., Chen L., Zhu X., and Wang X. (2018) miR-124 and miR-9 mediated downregulation of HDAC5 promotes neurite development through activating MEF2C-GPM6A pathway. J. Cell. Physiol. 233, 673–687 10.1002/jcp.25927 [DOI] [PubMed] [Google Scholar]
- 45. Sugo N., Oshiro H., Takemura M., Kobayashi T., Kohno Y., Uesaka N., Song W. J., and Yamamoto N. (2010) Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur. J. Neurosci. 31, 1521–1532 [DOI] [PubMed] [Google Scholar]
- 46. Segal M. (2010) Dendritic spines, synaptic plasticity and neuronal survival: activity shapes dendritic spines to enhance neuronal viability. Eur. J. Neurosci. 31, 2178–2184 10.1111/j.1460-9568.2010.07270.x [DOI] [PubMed] [Google Scholar]
- 47. Bading H., and Greenberg M. E. (1991) Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science 253, 912–914 10.1126/science.1715095 [DOI] [PubMed] [Google Scholar]
- 48. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Longair M. H., Baker D. A., and Armstrong J. D. (2011) Simple neurite tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454 10.1093/bioinformatics/btr390 [DOI] [PubMed] [Google Scholar]
- 50. Sholl D. A. (1953) Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 87, 387–406 [PMC free article] [PubMed] [Google Scholar]