Abstract
Phytochromes are photoreceptors in plants, fungi, and various microorganisms and cycle between metastable red light–absorbing (Pr) and far-red light–absorbing (Pfr) states. Their light responses are thought to follow a conserved structural mechanism that is triggered by isomerization of the chromophore. Downstream structural changes involve refolding of the so-called tongue extension of the phytochrome-specific GAF-related (PHY) domain of the photoreceptor. The tongue is connected to the chromophore by conserved DIP and PRXSF motifs and a conserved tyrosine, but the role of these residues in signal transduction is not clear. Here, we examine the tongue interactions and their interplay with the chromophore by substituting the conserved tyrosine (Tyr263) in the phytochrome from the extremophile bacterium Deinococcus radiodurans with phenylalanine. Using optical and FTIR spectroscopy, X-ray solution scattering, and crystallography of chromophore-binding domain (CBD) and CBD–PHY fragments, we show that the absence of the Tyr263 hydroxyl destabilizes the β-sheet conformation of the tongue. This allowed the phytochrome to adopt an α-helical tongue conformation regardless of the chromophore state, hence distorting the activity state of the protein. Our crystal structures further revealed that water interactions are missing in the Y263F mutant, correlating with a decrease of the photoconversion yield and underpinning the functional role of Tyr263 in phytochrome conformational changes. We propose a model in which isomerization of the chromophore, refolding of the tongue, and globular conformational changes are represented as weakly coupled equilibria. The results also suggest that the phytochromes have several redundant signaling routes.
Keywords: photoreceptor, cell signaling, mutagenesis, protein structure, X-ray crystallography, structural biology, protein conformation, chromophore-binding domain, photoconversion, phytochrome
Introduction
Phytochromes are a family of red/far-red light–sensing photoreceptors found in plants, fungi, cyanobacteria, and eubacteria. They play a role for example in seed germination and shade avoidance in plants and chromatic adaptation in bacteria (1). They bind a linear bilin chromophore that is phytochromobilin in plants, phycocyanobilin, in cyanobacteria, and biliverdin in bacteria. In response to red light, the phytochromes cycle between two metastable states. In prototypical phytochromes, the resting state absorbs red light (Pr),3 and the activated state absorbs far-red light (Pfr). Some phytochromes, however, have Pfr as a resting state and are called bathy phytochromes. Phytochromes can be reversibly switched between the states with red light (Pr→Pfr) and far-red light (Pfr→Pr). In the dark, they thermally revert to their resting state.
Phytochromes are usually dimeric proteins that consist of two modules (2, 3). The photosensory module (PSM) contains a Per/Arnt/Sim (PAS), a cGMP phosphodiesterase, adenylyl cyclase, FhlA (GAF), and a phytochrome-specific GAF-related (PHY) domain (Fig. 1). In plants, the phytochromobilin chromophore is bound to a conserved cysteine in the GAF domain, whereas in bacteria, this cysteine resides N-terminal to the PAS domain (4). The chromophore itself, consisting of four rings (A–D), is nested in a chromophore-binding pocket that is formed by PAS and GAF domains (5). This PAS-GAF entity is therefore often called a chromophore-binding domain (CBD). The PSM is followed by an output module that is often a histidine kinase domain in bacteria, making these phytochromes act as sensors in two-component signaling systems (6). The output module is more variable in plants and includes, for example, additional PAS domains and a histidine kinase-related domain.
Figure 1.
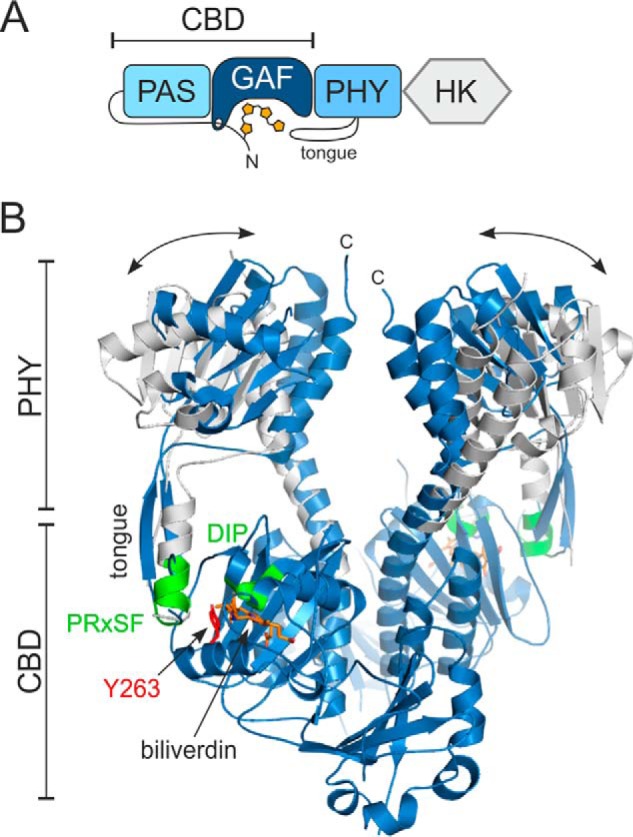
Overall structure of the bacterial phytochrome from D. radiodurans. A, simplified domain composition of the full-length phytochrome. The biliverdin bound to the N-terminal extension of the PAS domain and the PHY tongue are depicted. B, crystal structures of the photosensory module in closed (Pr, blue) and open (Pfr, white) conformations (9). Tongue, conserved DIP and PRXSF motifs (green), Tyr263 (red), and the chromophore (orange) are indicated. The double-headed arrows represent the changes in the PHY domain separation upon illumination. The CBD part has been shown only for the Pr-state structure for clarity. HK, histidine kinase.
Upon red light activation, the bilin chromophore isomerizes, which leads to changes in the rest of the phytochrome (Fig. 1). It has been shown that several residues surrounding the chromophore are important for the initial photoreactions. These include a conserved DIP motif in the GAF domain (3). The initial photoreactions in the immediate chromophore surroundings lead to changes in a so-called tongue extension of the PHY domain. In the Pr state, this PHY tongue adopts a β-hairpin conformation, whereas in the Pfr state, it adopts an α-helix and a coil (7–10). The tongue contains a conserved PRXSF motif that binds the DIP motif of the GAF domain. In the Pr state, a conserved Asp residue of the DIP motif, Asp207 in the Deinococcus radiodurans phytochrome (DrBphP), forms a salt bridge with an Arg residue (Arg466) of the tongue (3, 9, 11). In the Pr→Pfr transition, this salt bridge is broken, the entire PHY tongue refolds, and the Asp-interacting arginine is replaced by a serine (Ser468) of the tongue helix (9, 10, 12). Light-induced refolding of the PHY tongue affects the orientation of the PHY domain, opening the entire PSM dimer (9). In full-length phytochromes, this large-scale change is then relayed by the long connecting helices to the output module, causing changes in its structure and activity (13–15). The output activity is often a kinase activity but may vary depending on the species, and its changes will lead to biological responses.
The important DIP and PRXSF motifs are closely associated with a conserved tyrosine (Tyr263). The tyrosine resides close to the chromophore (Fig. 1B). A Y263F mutation obstructs the photocycle in the CBD fragment of DrBphP, leading to relatively high fluorescence quantum yield (16, 17). The same mutation has also been introduced to the CBD–PHY fragment of cyanobacterial phytochrome Cph1 and plant PhyB (18, 19). In this study, we show how Tyr263 in DrBphP plays an important role in the structural and spectroscopic fine-tuning of the phytochrome activity. By a Y263F mutation, we pinpoint the multiple roles that this tyrosine has for the photocycle and for stabilizing the tongue interactions. The results indicate that the coupling between the conformational states of the chromophore and protein is redundant and weaker than previously assumed and that the two entities may under certain conditions even be uncoupled.
Results
Absorption and fluorescence spectroscopy indicate a role of Tyr263 in the photocycle
First, we studied UV-visible absorption properties in the CBD and CBD–PHY fragments of DrBphP and their Y263F variants. The absorption spectra of these proteins (Fig. 2A) resemble the spectrum of the full-length DrBphP in the Pr form with absorption maxima at 698 nm (CBD), 700 nm (CBD–PHY), and 702 nm (CBD-Y263F and CBD–PHY-Y263F) (16, 18, 20, 21).
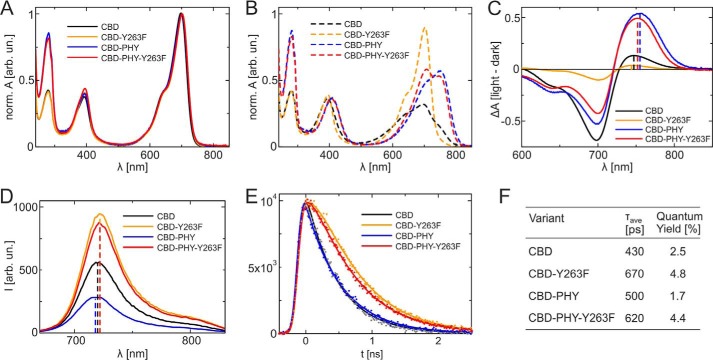
Figure 2.
Absorption and fluorescence spectra of the DrBphP CBD and CBD–PHY fragments and their Y263F variants. A, steady-state absorption spectra of CBD (black), Y263F (yellow), CBD–PHY (blue), and CBD–PHY-Y263F (red) in the dark-adapted state. The spectra were normalized to the absorption maximum. B, absorption spectra of the variants after irradiation with the 655-nm light. C, absorption difference spectra of the variants (red-irradiated spectra have been subtracted from dark-adapted spectra). Shown are fluorescence emission spectra (D) and emission decay of the same variants (E). The samples were excited at 660 nm, and the fluorescence signal was monitored at 720 nm. The solid lines show the multiexponential fits of the data. F, fluorescence quantum yield measurements and fluorescence lifetimes. The average lifetimes are calculated from the amplitude-weighted sum of the lifetimes observed in the single-photon counting experiments.
Generally, red illumination reversibly bleaches the 700-nm absorbance, and the absorption maximum shifts to the far-red region (Fig. 2C). In the CBD–PHY fragments, the absorbance maxima of the illuminated spectra appear at around 755 nm, but in the CBD fragments, this far-red peak is blue-shifted around 10 nm. The shift due to Tyr263 was hypsochromic in Pfr, as opposed to Pr. This indicates that the interaction of Tyr263 with the chromophore is different in Pfr compared with Pr. Five times longer illumination times were needed for CBD–PHY-Y263F to reach photoequilibrium compared with the WT CBD–PHY (Fig. S1A). Because the dark conversion rate from Pfr to Pr was hardly affected by the mutation (Fig. S1B), we conclude that the Y263F mutation reduces the photoconversion (Pr→Pfr) yield and possibly the absorption properties of the Pfr state.
As demonstrated earlier (16, 17, 22), the Y263F mutation influences the fluorescence properties of the DrBphP. We recorded fluorescence spectra after excitation at 630 nm (Fig. 2D). The spectra have maxima at 720 nm (CBD), 718 nm (CBD–PHY), and 722 nm (CBD-Y263F and CBD–PHY-Y263F). Again, a small bathochromic shift is seen in both Y263F variants, which is consistent with the steady-state absorption measurements for Pr. The fluorescence quantum yields increase from 2.5% (CBD) to 4.8% (CBD-Y263F) and from 1.7% (CBD–PHY) to 4.4% (CBD–PHY-Y263F) due to the Y263F mutation.
The fluorescence decay properties of the biliverdin molecule in the binding pocket were studied by time-correlated single-photon counting with an excitation wavelength of 660 nm and monitoring wavelength of 720 nm (Fig. 2E). The fluorescence decays were fitted with exponential functions (23). For Y263F variants, only a single decay component was needed. For the WT samples, multiexponential fitting curves were needed. The fluorescence decay of the CBD sample is described with lifetimes (and amplitudes) of 375 ps (77%) and 615 ps (23%), in line with values of (23). The CBD–PHY sample was best described with lifetimes of 173 ps (67%), 570 ps (31%), and 2 ns (2%). The fluorescence yields correlated with the averaged fluorescence lifetimes, which were 420 ps (CBD), 670 ps (CBD-Y263F), 500 ps (CBD–PHY), and 620 ps (CBD–PHY-Y263F) (Fig. 2F).
An increase in fluorescence yield implies a decrease in yield of forming the first ground state intermediate, Lumi-R. Thus, we conclude that the Y263F mutation hinders the Lumi-R formation. As demonstrated in the Fig. S1B, the thermal reversion rate from the Pfr state is equally slow or slower in the case of CBD–PHY-Y263F when compared with the WT CBD–PHY. This means that the CBD–PHY-Y263F sample can be populated to a similar photoequilibrium Pfr state with prolonged illumination of the red light as the WT CBD–PHY sample, although the Lumi-R formation is hampered due to mutation of the Tyr263.
FTIR spectroscopy reveals reduced PHY tongue refolding in the Y263F mutant
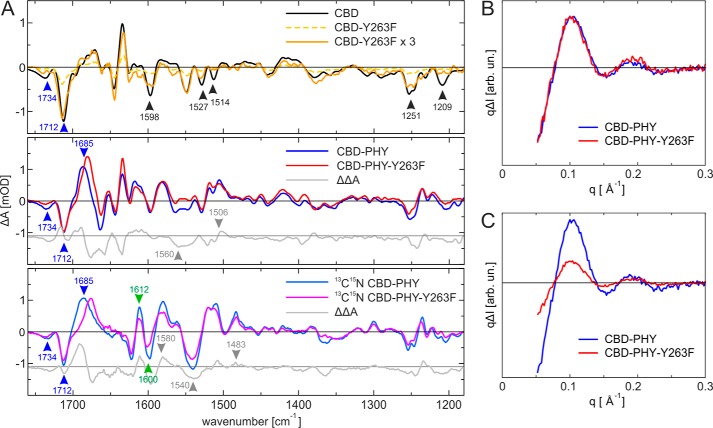
The FTIR spectroscopic analysis gives information from the light-induced changes in biliverdin and the protein. Fig. 3 shows the FTIR difference spectra between the illuminated and the dark states of WT CBD–PHY and CBD fragments and their Y263F mutants. The biliverdin-based transitions are distinguished from those originating from the protein by using uniformly 13C15N-apoprotein–labeled CBD–PHY and CBD–PHY-Y263F samples. The transitions from the biliverdin molecule remain unchanged in the 13C15N-labeled samples, whereas the protein peaks are shifted. As indicated in Fig. S1C, the negative bands at 1734 and 1712 cm−1 originate from the carbonyl vibration of the biliverdin rings A and D, respectively (24). The large positive band at 1688 cm−1 in the CBD–PHY is an induced absorption in the Pfr state. Most likely, this positive band originates largely from the carbonyl vibration of the D ring. In addition, changes in the biliverdin vibrations can be observed at 1250 and 1230 cm−1 (Fig. S1C).
Figure 3.
FTIR and X-ray scattering analysis of the studied phytochrome complexes. A, comparison of the CBD samples (top) indicates a very small signal for CBD-Y263F sample. The 3× scaled difference spectra of CBD-Y263F shows features similar to those of the CBD sample. The FTIR difference spectra (Pfr-Pr) of CBD–PHY and CBD–PHY-Y263F (middle) are similar except for the spectral regions around 1550 cm−1 and in range 1600–1660 cm−1, which show small deviations. The comparison of 13C15N–labeled CBD–PHY and CBD–PHY-Y263F (bottom) indicate that the β-sheet → α-helix transition takes place with a lower rate in the Y263F mutant. The spectra of each sample are normalized to the UV-visible absorbance value at 700 nm. The peak positions mentioned under “Results” are indicated as arrows, which are color-coded as changes in biliverdin ring (blue), changes in secondary structure (green), and other changes (black). B, difference X-ray scattering of the two CBD–PHY variants. Shown are the Pfr-Pr difference signals of WT (blue) and Y263F (red). The signals are scaled to the positive 0.1 Å−1 maximum, making the signal shapes fully comparable with each other. C, relative X-ray scattering signal amplitudes of the CBD–PHY variants. The signals are scaled to the absolute scattering of each sample, making the intensities fully comparable with each other. The signal amplitudes indicate a reduced photoconversion efficiency of the Y263F mutant.
The FTIR difference signal of the CBD-Y263F is small compared with the WT CBD (Fig. 3A, top), which is a result of the Pr state dominating the photoequilibrium after red light irradiation, as described in the UV-visible analysis (Fig. 2B). The 3-fold multiplication of the signal however still highlights a difference signal for CBD-Y263F variant. In particular, the negative band at around 1712 cm−1 shows that the D-ring environment is affected by illumination. On the other hand, the 1734 cm−1 band is missing in the CBD-Y263F variant, thus indicating no changes in the A-ring environment under illumination. Interestingly, many FTIR features originating from the protein environment of CBD are present and similar in CBD-Y263F, but a few missing bands in the variant can be assigned as potential signals from the Tyr263 in the CBD sample. Signals that are completely missing in the CBD-Y263F compared with the WT are found at 1514 and 1209 cm−1. Signals with reduced intensity were at 1251, 1527, and 1598 cm−1. These frequencies correspond well to Tyr-OH vibrations (25), except the 1527 cm−1, which may report changes in the Asp (COO−1) interactions.
In contrast to the CBD fragments, the FTIR difference spectrum of the CBD–PHY-Y263F variant has an amplitude similar to that of the WT CBD–PHY (Fig. 3A, middle). This reveals that after a prolonged irradiation time, these two samples have similar Pfr/Pr ratios, which is in line with the UV-visible analysis (Fig. 2B and Fig. S1A). The similar negative features at 1712 cm−1 in the CBD–PHY and CBD–PHY-Y263F variants indicate that their D-ring carbonyls experience similar interactions in the Pr state. On the other hand, the D-ring signal in the Pfr state is downshifted to 1685 cm−1 in the CBD–PHY-Y263F variant (Fig. 3A, middle and bottom). This indicates that the D-ring carbonyl experiences a different chemical environment in the mutant than in the WT, for example due to altered hydrogen bonding. The slight upshift of the negative 1734 cm−1 carbonyl vibration of the A ring, observed both in the nonlabeled and 13C15N-labeled samples, indicates that Tyr263 influences the biliverdin–protein interaction in the Pr state. Small changes are also seen in the region of 1527 cm−1 (−), 1514 cm−1 (−), and 1506 cm−1 (+), suggesting changes in the Tyr protonation state and interactions. The differences between the WT and Y263F samples at 1270–1230 cm−1 reflect to a large extent the light-induced changes in the biliverdin–protein interactions. Naturally, the Tyr263 hydrogen-bonding character between the Pr and Pfr states influences these changes as well (26).
The secondary structural changes in the CBD–PHY domains during the photoconversion are more clear in the 13C15N-isotope–labeled samples, as they separate better from the overlapping signal of the CBD fragment (Fig. 3A, bottom). In particular, the β-sheet to α-helix transition is now clearly pronounced as a positive-negative feature at 1612 cm−1 (+)/1600 cm−1 (−). This can be translated to the signal of the transition in the tongue region, similar to Kennis and co-workers with a signal pair at 1655 cm−1 (+)/1633 cm−1 (−) for the unlabeled samples of bacterial phytochromes from Rhodopseudomonas palustris (10, 12). The difference signal amplitude of CBD–PHY-Y263F is about 60% from that of the WT. This indicates fewer secondary structural changes, either due to a larger α-helix population in the Pr state or larger β-sheet population in the Pfr state in the mutant. The smaller amplitudes of CBD–PHY-Y263F in the changes of secondary structure are also observed in the amide II region, as indicated by the broad negative peak in the double difference spectrum at 1560 cm−1 (−) and positive signal at 1506 cm−1 (+) that have shifted due to isotope labeling to 1540 cm−1 (−) and 1483 cm−1 (+), respectively. An additional change at 1580 cm−1 (+) in labeled spectra may be related to the absorption of aspartate side chain, which would be likely, given the close proximity of Tyr263 to Asp207. In summary, the FTIR difference spectra demonstrate that the yield of the refolding of the tongue is lower in the Y263F mutant.
X-ray solution scattering reveals large-scale changes in all PSM variants
To study the role of Y263F mutation in large-scale structural changes in solution (9), we applied difference X-ray scattering for CBD–PHY and CBD–PHY-Y263F. The results shown in Fig. 3 (B and C) indicate that both variants give a highly similar Pfr-Pr difference signal, which has been assigned to the CBD–PHY dimer opening (9, 27). The signals were practically identical in shape (Fig. 3B), indicating that CBD–PHY-Y263F undergoes large-scale movements that are identical to those of the WT protein. The amplitudes of the differences signals, however, varied, which indicates a reduced yield of dimer opening when compared with the WT CBD–PHY (Fig. 3C). This reduction in yield may be due to the lower yield of forming Lumi-R or the reduced yield of tongue refolding. The effect was even more pronounced in the D207H mutant (Fig. S1D), which has a broken Asp207–Arg466 salt bridge and therefore impaired interactions between the PHY tongue and GAF domain.
Importantly, the X-ray scattering results, together with FTIR, indicate that in addition to the conserved Tyr263 (and Asp207 of the DIP motif), there should be other signaling routes that relay the structural signal from the chromophore to the PHY tongue. To reveal the structural details underlying the impaired photocycle and the alternative signaling routes to the tongue, we crystallized Y263F mutants of the CBD and CBD–PHY fragments, respectively.
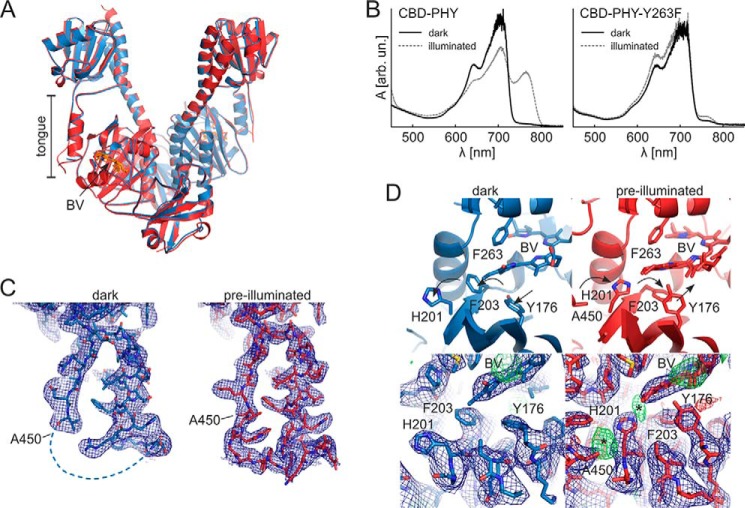
Crystal structure of CBD-Y263F reveals reduced water interactions
CBD-Y263F was crystallized in the same conditions as the WT CBD (28). Crystal data were processed up to 1.34 Å resolution, and the electron density appears completely comparable with the corresponding WT data. The overall CBD-Y263F structure was highly similar to the WT CBD (Fig. 4A) (28), with an overall root mean square deviation (RMSD) of 0.102 Å. The most notable difference between the structures is a missing water that resides between the pyrrole water and the Tyr263 phenyl ring. This “transient water” (W2) is present in some of the CBD structures (17, 22, 28) and interacts with so-called pyrrole water (W1), Asp207, and biliverdin ring A (Fig. 4B). The phenyl ring of the mutated Phe263 resides slightly closer to the pyrrole water than in the WT Tyr263, which may exclude the transient water from the binding pocket. The slight repositioning of the Phe263 side chain may also be a result of reduced steric effects and the lack of hydrogen bonding of the hydroxyl moiety of this residue. The hydroxyl appears to bind to one solvent water molecule in the WT (W3). This interaction is lost in the Y263F structure, and the water density is reduced (Fig. 4, C and D). Otherwise, no large changes between the CBD and CBD-Y263F structures were observed, and the pyrrole ring orientations seemed unchanged at this resolution.
Figure 4.
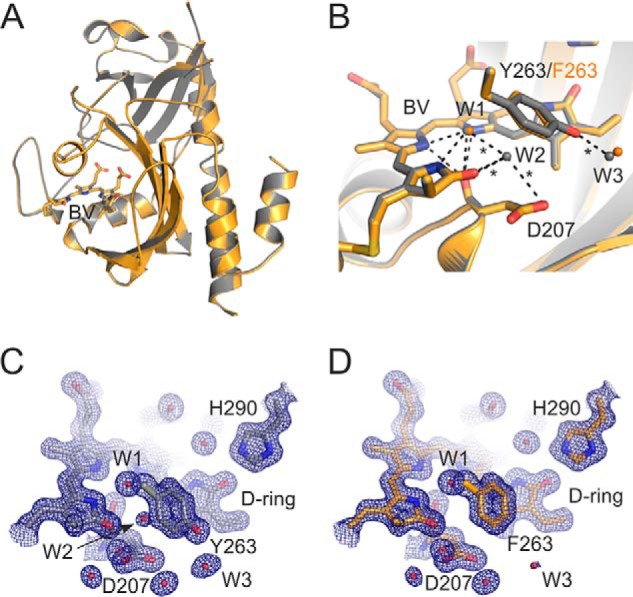
Crystal structure of CBD-Y263F and comparison with CBD. A, overall cartoon presentation of superimposed CBD-Y263F (orange) and the WT CBD structure (gray; PDB code 5K5B) (28). Biliverdin (BV) is presented as sticks. B, structural comparison of the CBD-Y263F and CBD in the biliverdin surroundings. Key residues are indicated, and the three waters are named accordingly: pyrrole water (W1), transient water (W2), and solvent water (W3). The hydrogen bonds of the WT structure are shown as dashed lines, and the interactions missing in the Y263F mutant structure are marked as asterisks. C and D, the electron densities of the chromophore surroundings in CBD (C) and CBD-Y263F (D). The 2Fo − Fc density (blue mesh) is plotted at 1.5 RMSD.
CBD–PHY-Y263F in Pr state crystallized exclusively in Pfr-like conformation
Although the WT CBD–PHY formed Pr-state crystals, as previously reported (14), the Y263F mutant did not (Fig. S2A). To obtain Pr structures from the CBD–PHY-Y263F variant, we screened for new crystallization conditions in the dark. Although CBD–PHY-Y263F could not be crystallized in Pr-like crystallization conditions, it crystallized readily in the previously reported Pfr-like crystallization conditions (9). Pre-illumination with red light greatly facilitated the crystal growth (Fig. S2A). These conditions yielded “pre-illuminated” crystals that diffracted up to 3.3 Å resolution. Surprisingly, we found out that both WT CBD–PHY and the Y263F variant could be crystallized in this Pfr-like crystallization condition even if they were kept in the Pr state. We found an additional condition in which the CBD–PHY-Y263F mutant crystallized especially well in complete darkness and without pre-illumination. The resulting “dark” crystals diffracted to 3.6 Å resolution and had an open PSM conformation, as in the previously reported Pfr structures (Fig. 5) (9, 29).
Figure 5.
The two CBD–PHY-Y263F crystal forms and the absorption spectra of CBD–PHY variant crystals. A, overall dimer structure of pre-illuminated (red) and dark (blue) crystal forms of CBD–PHY-Y263F. Biliverdin (BV) is shown in sticks. B, absorption spectra of CBD–PHY WT CBD–PHY and Y263F variant crystallized in the dark. The WT crystal shown here is crystallized in a published Pfr condition (9), and the Y263F variant was crystallized in the “dark” condition reported in this paper. The crystals had characteristic Pr spectra with a maximum at 700 nm (dark). The CBD–PHY crystals could be partially photoswitched with 660-nm light, whereas CBD–PHY-Y263F crystals responded only little to red light (illuminated). C, electron densities of the tongue region of both crystal structures. D, structure of the D-ring surroundings in CBD–PHY variants. Dark (left) and pre-illuminated (right) Y263F structures differ in the side-chain orientations of residues Tyr176, His201, and Phe203. These orientations are supported by the 2Fo − Fc electron density maps. However, weak positive electron density (*) at the sites where these residues would reside in the Pr state imply traces of Pr-state orientations in the pre-illuminated structure. See Fig. S2B for a comparison with published Pr and Pfr structures. The 2Fo − Fc maps (blue) are plotted at 1.0 RMSD, and Fo − Fc maps are plotted at 3.0 RMSD.
The relative tendency of the CBD–PHY variants to form the Pfr-like crystals in the dark varied, hinting at different propensities to adopt the open conformation in the dark (Fig. S2A). CBD–PHY-Y263F crystallized most readily in the “dark” crystallization condition reported here but also in a published Pfr crystallization condition (9). This propensity to crystallize implies that the CBD–PHY-Y263F adopts an open conformation more easily, probably due to the weakened interactions between the tongue and GAF domain. The WT CBD–PHY, on the other hand, crystallized faintly in Pfr-like crystallization conditions without pre-illumination. This implies that it rarely adopts a Pfr-like conformation in darkness due to stronger interactions between the GAF domain and the Pr-state tongue. On some occasions, however, even the WT CBD–PHY crystallized in these conditions. The absorption spectra of the dark crystals (Fig. 5B) verified that the PAS-GAF-PHY variants have indeed remained in the Pr state from the biliverdin point of view.
The pre-illuminated and dark CBD–PHY-Y263F crystals had highly similar packing and dimensions with each other and with previously reported WT pre-illuminated Pfr crystals (9, 29) (see Table 2). Indeed, both crystal structures closely resembled these published structures. The CBD–PHY mutant dimers seemed to adopt an “open” conformation with separated sister PHY domains and a helical PHY tongue conformation (Fig. 5, A and C).
Table 2.
X-ray diffraction data collection and refinement statistics for the DrBphP Y263F structures
| Parameters | CBD-Y263F | CBD-PHY-Y263F pre-illuminated | CBD-PHY-Y263F dark |
|---|---|---|---|
| Data collectiona | |||
| Space group | C121 | P 21 21 21 | P 21 21 21 |
| Cell dimensions | |||
| a, b, c (Å) | 94.24, 54.58, 70.56 | 85.7, 198.5, 223.6 | 84.1, 197.1, 214.1 |
| α, β, γ (degrees) | 90.00, 92.20, 90.00 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 19.93–1.34 (1.38–1.34) | 19.98–3.30 (3.38–3.30) | 19.94–3.60 (3.69–3.60) |
| Rmerge | 0.059 (1.333) | 0.157 (1.469) | 0.072 (1.414) |
| CC½ | 0.998 (0.311) | 0.994 (0.312) | 0.999 (0.343) |
| I/σ(I) | 10.51 (0.89) | 7.68 (0.96) | 11.59 (0.90) |
| Completeness (%) | 99.8 (99.7) | 99.1 (99.9) | 96.6 (98.8) |
| Redundancy | 4.13 (3.94) | 4.09 (4.31) | 4.39 (4.52) |
| Wilson B factor | 18.2 | 99.8 | 164.2 |
| Refinement | |||
| Resolution (Å) | 19.93–1.34 (1.38–1.34) | 19.98–3.30 (3.38–3.30) | 19.94–3.60 (3.69–3.60) |
| No. of reflections | 76,244 (5617) | 54,898 (3899) | 38,682 (2783) |
| Rwork/Rfree | 0.143/0.182b (0.528/0.530) | 0.234/0.267 (0.389/0.391) | 0.248/0.287 (0.373/0.359) |
| Overall B factor | 26.0 | 115.0 | 197.0 |
| No. of atoms | |||
| Proteinc | 2336 | 14,790 | 14,443 |
| Ligandd | 63 | 172 | 172 |
| Water | 318 | 5 | 0 |
| Geometry | |||
| RMSD | |||
| Bond lengths (Å) | 0.014 | 0.007 | 0.007 |
| Bond angles (degrees) | 1.754 | 1.172 | 1.175 |
| Ramachandran (%) | |||
| Favored | 99 | 96 | 95 |
| Allowed | 1 | 4 | 5 |
| Outliers | 0 | 0 | 0 |
| PDB code | 5NFX | 5NM3 | 5NWN |
a Outer shell values are in parentheses.
b Test set for Rfree calculation constitutes 5% of total reflections that were randomly chosen.
c Number of protein molecules in an asymmetric unit: one (CBD-Y263F) and four (CBD–PHY-Y263F dark and pre-illuminated).
d This includes atoms from a biliverdin, three acetates and a (4S)-2-methyl-2,4-pentanediol (CBD-Y263F), or four biliverdins (pre-illuminated and dark CBD–PHY-Y263F).
Closer inspection of the dark CBD–PHY-Y263F structure revealed that the chromophore-binding pocket resembles that of the Pr-state structure, but the rest of the protein adopts a Pfr state–like conformation. Notably, three residues (Tyr176, His201, and Phe203) along with Arg222 adopted exclusively a conformation that is characteristic of the Pr state (Fig. 5D). Although the resolution was not adequate to unequivocally confirm the details of the biliverdin conformation, the chromophore was modeled in the Pr-state (15Z) isomer. The electron density distribution (Fig. S2C) and the absorption spectrum (Fig. 5B) support this biliverdin state. The overall chromophore surroundings were highly similar to our WT Pfr structure, which has mixed Pr/Pfr photochromic state and Pr-like side-chain orientations (9).
In the pre-illuminated CBD–PHY-Y263F structure, the orientation of three residues (Tyr176, His201, and Phe203) was different from that of the dark CBD–PHY-Y263F structure. Whereas in the dark crystals, these residues reside in a Pr-like conformation, the Pfr-like conformations predominate in the pre-illuminated structure (Fig. 5D). It appears that these residues can adopt both Pr and Pfr orientations in this crystal form and that these orientations reflect the prevalent Pr/Pfr ratio within the crystals. The Arg222 residue close to biliverdin propionate B adopted a Pr-like conformation here, although it has been reported to have a different conformation in a saturated Pfr state (8, 29). The chromophore in the pre-illuminated structure is oriented in the pocket as an intermediate of Pr- and Pfr-state biliverdins, closely resembling the Pfr structure published by us (9). Because this crystal form is a result of pre-illumination with red light, the chromophores were modeled as a Pr/Pfr mixture. In addition, the propionate C of the biliverdin shows some flexibility in pre-illuminated crystals, as the electron density is diffuse in this site (see Fig. S2C), which is in accordance with the results with other Pfr structure of DrBphP PSM (29).
The orientation of the Tyr176-Phe203-His201 triplet in the Pr state led to a structural clash between His201 and the tongue residue Ala450 (Fig. 5D). This explains the unstructured region in the tongue near Ala450 that could not be modeled. The dark crystals therefore have substantially more unstructured PHY tongue region than the pre-illuminated crystals (Fig. 5C).
Discussion
The amino acids surrounding the chromophore are highly conserved in phytochromes, which reflects their importance for the function of the protein. Generally, the residues participate in coupling of the chromophore state to the conformation of the rest of the protein. Specifically, a triad of residues, Asp207 of the DIP motif, Ser466/Arg468 of the PRXSF motif, and Tyr263, is expected to couple chromophore isomerization with conformational changes in the PHY tongue. In this paper, we confirmed by mutational, spectroscopic, and structural analysis that Tyr263 plays a central role in the structural and spectral responses in D. radiodurans phytochrome. It influences the excited-state reactions of the biliverdin and stabilizes the tongue interactions to the GAF domain, thus affecting the structural equilibrium between Pr- and Pfr-like conformations. However, our data show that this coupling is weaker than commonly believed and that it can be modified. The Y263F mutation leads to reduced Pr→Pfr photoconversion of the biliverdin but also favors a constitutively active conformation of the photoreceptor.
The role of Tyr263 in early stages of the photocycle
The UV-visible absorption and fluorescence data indicate that Tyr263 plays an important role in the phytochrome photocycle. The role of this residue is most striking in the CBD-Y263F sample, which has a barely detectable photoconversion and an elevated fluorescence quantum yield (16) (Fig. 2). The increased fluorescence yield hints at a role of Tyr263 in the excited-state reactions of the biliverdin and therefore in the Lumi-R formation. Somewhat surprising is that the structural differences between the CBD-Y263F and the WT CBD are the missing or absent hydrogen bonds involving a few water molecules (Fig. 4). One missing water resides between Tyr263 and pyrrole water and potentially has an important role in the photocycle. Whether the changes in the hydrogen-bonding pattern and biliverdin electrostatics are the cause of the divergent photocycle of Y263F is a matter for future research.
The mutated PSM fragment (CBD–PHY-Y263F) also has a higher fluorescence yield compared with the WT PSM, suggesting an impaired Lumi-R formation (Fig. 2). This is in accordance with the fluorescence studies with the Y263F mutant of Cph1 (18). Due to the long thermal reversion times, prolonged illumination conditions can be used to populate the sample to a Pfr state similar to that of the WT PSM. This is consistent with a previous study with cyanobacterial Cph1, where it was concluded that the aromatic character, but not the hydroxyl group, of the conserved Tyr263 is critical for the Pfr formation (18). The hydroxyl of Tyr263 facilitates the photocycle and Pfr formation but is not indispensable for these events. The resulting Pfr spectrum of CBD–PHY-Y263F closely resembles that of CBD–PHY, speaking for a biliverdin end state similar to that of the WT protein. This is also apparent in the crystal structures reported here (Fig. 5 and Fig. S2). The ability of CBD–PHY-Y263F to reach the Pfr state indicates that other residues in the binding pocket and/or in the PHY tongue are likely to be engaged in the photoconversion.
The coupling of the PHY tongue to the chromophore
Phytochromes switch between the Pr and Pfr states in response to light, and this change in the photoactive state reflects their output activity state. We propose that the photochromic and conformational states of phytochromes do not necessarily go hand in hand. Although the Pr/Pfr ratio of the bilin chromophore can be shifted by light, the phytochrome activity may be uncoupled from this light regulation. By mutating the Tyr263 residue at the GAF/PHY interface, we were able to break this coupling. This resulted in a phytochrome that has its chromophore in the Pr state, but its PSM equilibrium is shifted toward a Pfr-like conformation.
This spectral and functional uncoupling has also been shown in other phytochrome species. The mutation of this conserved tyrosine in Arabidopsis thaliana PhyB (Y361F) led to greatly enhanced red light sensitivity (19). There is even evidence that the lack of the entire tongue favors the active state of the phytochrome, as an IsPadC tongue deletion mutant has high diguanylyl cyclase activity even in its dark state (30). The evidence speaks for the role of the conserved tyrosine in stabilizing the tongue interactions to the GAF domain. If this interaction is lost or weakened, the phytochrome more likely adopts a Pfr-like conformation that is in many cases physiologically active (1).
The absence of the Tyr263 hydroxyl group in the mutant presumably weakens the tongue interactions that involve a salt bridge between the conserved DIP and PRXSF motifs (Asp207–Arg466). This effect favors a Pfr-like tongue arrangement over the Pr-like conformation and causes a reduced light responsiveness of the phytochrome molecule. This may explain smaller signals of tongue refolding (Fig. 3A) and large-scale conformational changes of the Y263F mutant (Fig. 3, B and C). The importance of the tongue interactions in light responsiveness is further underlined by the greatly reduced difference X-ray scattering signal of CBD–PHY-D207H (Fig. S1D). This indicates that Asp207 plays an even more prominent role in tongue stabilization than Tyr263.
CBD–PHY-Y263F seemed to crystallize exclusively in an open Pfr-like conformation. This indicates that the mutation makes the PSM prefer this conformation over the Pr conformation in solution. During the crystal formation, packing effects may further favor a Pfr-like conformation by forcing the remaining Pr-state proteins to adopt this form. We note that even the WT PSM molecules in the Pr state can be forced to form Pfr-like crystals by the crystallization conditions (Fig. 5B), although they would be unlikely to adopt a Pfr-like conformation spontaneously. In contrast to CBD–PHY-Y263F in D. radiodurans, the Y263F mutant of cyanobacterial Cph1 has been successfully crystallized in the Pr-like conformation (18). This Cph1 mutant, however, has a slightly different tongue arrangement compared with the WT Cph1 and other Pr-state PSM structures (7, 9, 11, 14, 30–32). One explanation for this could be the crystal-packing effects; most phytochrome PSM structures in the Pr state, as in DrBphP CBD–PHY (9, 14), have crystal contacts between the tongues of the neighboring symmetry mates. If the native tongue conformation is disrupted, this packing interaction is compromised. Even the slightest changes in the tongue conformation and stability would therefore make the Pr-like crystal formation unfavorable.
The published Pfr-state crystal structures indicate that the Y263F hydroxyl may in theory interact with the biliverdin D ring (Fig. S3A). However, we did not detect any increase in the rate of dark reversion in the Y263F mutant (Fig. S1B), which speaks against the role of Tyr263 in stabilizing the Pfr state. Residue His201 in DrBphP is shown to stabilize the Pfr state by interacting with the biliverdin D ring (29). This histidine, however, is not conserved in most prototypical phytochromes (Fig. S3B). This additional interaction in DrBphP would explain its relatively stable Pfr state and slow dark reversion rate (9, 14, 20).
Redundant routes for coupling the PHY tongue to the chromophore
Upon photoconversion, three residues close to the D ring, Tyr176, Phe203, and His201, reorient (Fig. 5D). This change in orientation has been noted before (29), but its exact role in the phytochrome signaling remains unclear. Although the reorientation of His201 enables an interaction with the ring D and a more stable Pfr state in DrBphP, it may have a role different from that of leucine in other phytochromes. Residue Tyr176 and the aromatic character of Phe203 are conserved among all phytochromes (Fig. S3B), and their mutation obstructs the photoconversion of DrBphP (21, 29).
Although the residue Ala450 in the helical tongue may affect the reorienting of the Tyr176-Phe203-His201 triplet through steric packing interactions (arrows in Fig. 5D), this effect is too weak to affect the biliverdin spectrum in dark crystals. The three residues may allow more space for the D ring to rotate, which would enable the switching of these crystals between the Pr- and Pfr-like states without radically losing crystal integrity. Our data therefore indicate that both Pr and Pfr orientations of the triplet can co-exist in Pfr-like crystal form and that they mainly reflect the photochromic state of the biliverdin. This indicates that it is the chromophore, not the tongue conformation, that is the main determinant of the Tyr176-Phe203-His201 orientation. It is possible that this triplet plays a role in phytochrome signaling by affecting the tongue stability; in the Pr state, His201 points away from the chromophore-binding pocket and destabilizes the Pfr-like tongue conformation around residue Ala450. This is visible as missing electron density at this region in our dark Pr-state structure (Fig. 5C). In the Pfr state, on the other hand, His201 potentially clashes with the 451WGG453 region of the Pr-likes tongue, making this combination unlikely. This way, the Tyr176-Phe203-His201 route can act as a mediator by favoring the tongue conformation dictated by the chromophore. The residue triplet would facilitate the refolding process during the photoconversion and reduce the prospects of a misfolded PHY tongue. Indeed, mutating the PhyA and PhyB tyrosines that correspond to residues Tyr176 and Phe203 in DrBphP renders them constitutively active (33, 34).
We propose that there is a β-sheet/α-helix equilibrium for the tongue conformation and that it is affected by several means. The conserved Asp207-Arg466 salt bridge and Tyr263 play an important role in the stabilization of Pr conformation, whereas the Tyr176-Phe203-His201 route may facilitate the coupling of the chromophore state to the tongue conformation. In addition, a conserved proline in the PRXSF motif (Pro465 in DrBphP) has been proposed to play a role in the conformational trigger in the photoconversion through interaction with the biliverdin A ring (31). Together, these redundant interaction routes may couple the changes in chromophore to the tongue refolding. This coupling can be considered important for phytochromes, because the two states, dark reversion, and phototransition between them have to be supported by a single protein structure. Redundancy in the signal pathway is also consistent with the fact that these residues are highly conserved (Fig. S3B) but not entirely indispensable to the structural photoconversion.
Phytochromes function at several levels that affect each other
In light of the presented results, we propose a model in which light-induced responses in photoreceptors occur at several levels (Fig. 6). Phytochromes can be considered to switch conformation at the levels of the chromophore, the tongue, the PSM, and the entire protein. Each level resides in an equilibrium that is affected by external factors and the other levels. For example, light and tongue interactions affect the Pr/Pfr equilibrium of the biliverdin. On the other hand, the tongue is affected by the state of the chromophore and the quaternary PSM arrangement (35). The PSM arrangement finally affects the structure and hence the activity state of the output module (11, 13, 36).
Figure 6.
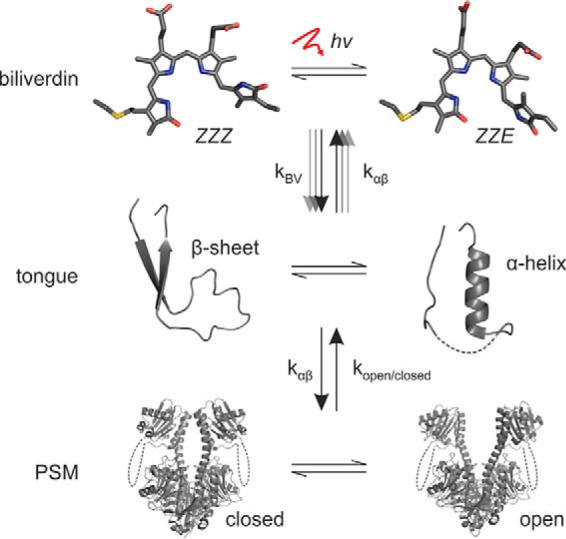
Schematic presentation of the three levels in phytochrome signaling and their relations. Each level resides in an equilibrium that is affected by the other levels or external factors (denoted by arrows and k-factors). In the case of CBD–PHY-Y263F, the Pr→Pfr transition is impaired, but the α-helical tongue is favored over the β-sheet, which increases the prospects of attaining an open PSM that has its biliverdin in the Pr (ZZZ) state. Multiple arrows between the “biliverdin” and “tongue” levels illustrate multiple, partly redundant signaling routes between the levels.
By introducing the Y263F mutation, we were able to shift the equilibria within each level. This caused deviant spectral and structural properties but did not totally obstruct the function of the PSM. This speaks for redundant signaling routes within this signaling network, which may be an evolutionarily conserved feature of phytochromes and other signaling enzymes. These properties would offer a photoreceptor sophisticated means to fine-tune the light-induced responses without compromising the function of the entire protein. A good example of this is a phytochrome from Stigmatella aurantiaca (SaBphP1), which has apparently defective photocycle (biliverdin level) but undergoes normal light-induced quaternary changes (PSM level) (27). The weak coupling and redundancy between the signaling levels has therefore enabled this phytochrome to adapt spectrally for novel purposes.
Experimental procedures
Molecular cloning and sample preparation
The constructs encoding the CBD and CBD–PHY fragments (amino acids 1–321 and 1–502) of the DrBphP bacteriophytochrome are described elsewhere (5, 21). The Y263F mutation was introduced to the WT fragments with QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies) and primer 5′–CCATGCACATGCAGTTCCTGCGGAACATGGGCGTC-3′ (nucleotides coding Phe263 underlined). Protein expression and purification was done as described previously (9, 12, 20, 23). The purified protein samples in final buffer (30 mm Tris/HCl, pH 8.0) were concentrated to 30 mg/ml and flash-frozen. Just before measurements, the frozen phytochrome samples were quickly thawed and filtered with 0.22-μm centrifugal filters (Amicon Ultrafree, Millipore) and diluted to the desired concentration with the final buffer. All measurements were carried out in the dark at room temperature.
UV-visible absorption and fluorescence spectroscopy
The steady-state absorption spectra were recorded as described previously (20). The fluorescence quantum yield was determined from the steady-state absorption and emission spectra, and the fluorescence decay was characterized using a time-correlated single-photon counting system (22). The fluorescence data were analyzed using Pygspec (37).
X-ray solution scattering
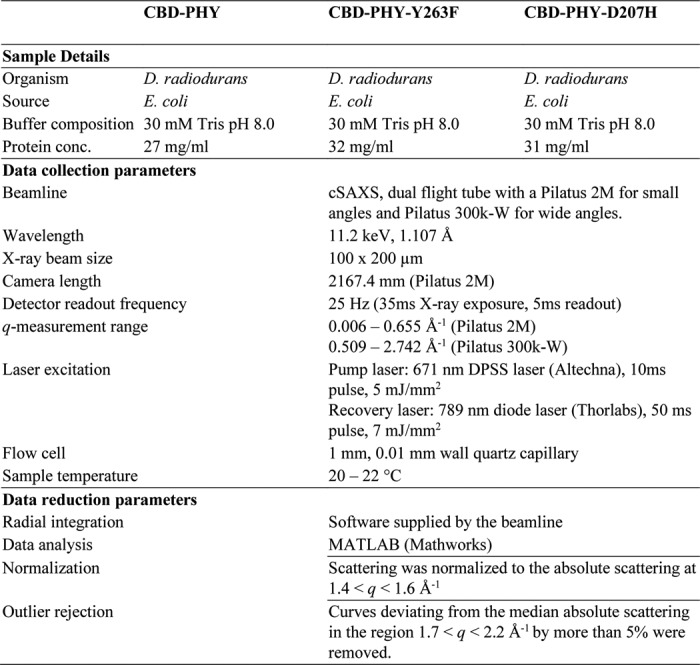
The X-ray scattering data acquisition was performed at the cSAXS beamline of the Swiss Light Source as described previously (13). In brief, the sample (∼30 mg/ml) was pumped through a 1.0-mm quartz capillary and switched between the Pr and Pfr states with a red diode-pumped solid-state laser (671 nm, 5 mJ/mm2; Altechna) and a far-red diode laser (789 nm, 7 mJ/mm2; Thorlabs). While switching the sample, an overlapping X-ray beam (11.2 keV) was used to probe the difference scattering signal. More details can be found in Table 1 and in the supporting information.
Table 1.
X-ray solution scattering data collection
FTIR spectroscopy
The samples were concentrated to approximately 2.5 mm. A sample volume of 2 μl was pipetted on a CaF2 window and sandwiched without any spacer between two window plates, resulting in ∼10-μm sample thickness. The hydration was assessed with an absolute IR absorption spectrum in each sample, which resulted in a ratio of 0.9/0.35 between amide I (at 1645 cm−1) and amide II region (at 1580 cm−1). The difference spectra were recorded with an FTIR spectrometer (Nicolet) by utilizing red (λ = 655 nm) and far-red (λ = 785 nm) light–emitting diodes in a consecutive manner, similar to a method described previously (12). The baseline was recorded during each switch before changing the illumination conditions. About 20 scans of both light-activated states (Pr and Pfr) were acquired.
Protein crystallography
The CBD-Y263F crystals were grown as described (28). The hanging-drop vapor diffusion method was applied, in which 20 mg/ml protein was mixed at a 1:1 ratio with reservoir solution (67 mm sodium acetate, pH 4.95, 3.3% PEG 400, 30% 2-methyl-2,4-pentanediol, 1 mm DTT). Large crystals formed after 24–48-h incubation at room temperature. For crystallographic data collection, the crystals were flash-frozen with liquid nitrogen in cryoloops under green light. Diffraction data were collected at beamline ID23-2 of the European Synchrotron Radiation Facility (ESRF), under a 100 K cryostream and an X-ray wavelength of 0.873 Å. The data were processed with the XDS program package (38) and cut at 1.34 Å resolution, which corresponds to a correlation coefficient (CC½) value of 0.31 (39). The crystal belonged to the C121 space group with cell parameters highly similar to those of the published WT CBD structure (28). The CBD-Y263F crystal structure was solved by molecular replacement with Phaser, and structure 2O9C (40) was used as a search model. The structure was refined using REFMAC5 (41). In the refinement, the geometry was restrained with an X-ray matrix-weighting term of 0.5 and anisotropic temperature factors, resulting in Rwork/Rfree values of 0.143/0.182. The statistics of data collection, processing, structure determination, and refinement of all data sets are summarized in Table 2.
The pre-illuminated CBD–PHY-Y263F crystals were acquired by hanging-drop vapor diffusion by mixing (1:1) 20 mg/ml protein with reservoir solution (100 mm Tris/HCl, pH 8.5, 200 mm NaCl, 25% PEG 3350) in conditions identical to those described previously (9). Right after setting up the crystallization, drops were pre-illuminated with a red light–emitting diode (655 nm, 5 milliwatts/cm2). The crystals were flash-frozen after a week without cryoprotection. Diffraction data were collected like the CBD-Y263F data at beamline ID23-2 of ESRF. The data were cut at 3.3 Å, which corresponded to a CC½ value of 31.2. The pre-illuminated CBD–PHY-Y263F crystal structure was solved by molecular replacement with Phaser. A modified D. radiodurans CBD–PHY structure 5C5K (29) was used as a search model. The side chains of Tyr176, His201, Phe203, and Arg222 and the biliverdin were removed from the search model and modeled only at later stages of the refinement. Like the WT Pfr crystals (9), the pre-illuminated CBD–PHY-Y263F crystals belonged to P212121 space group with two dimers in an asymmetric unit. In the refinement, automatically generated local noncrystallographic symmetry restraints were applied, and the geometry was restrained tightly with an X-ray matrix-weighting term of 0.005, resulting in Rwork/Rfree values of 0.234/0.267.
The dark CBD–PHY-Y263F crystals were grown by the hanging-drop vapor diffusion method against reservoir conditions (100 mm Tris/HCl, pH 8.5, 200 mm sodium acetate trihydrate, 30% PEG 4000). The Pr state of the protein was ensured by illuminating the crystallization drops briefly with a 785-nm LED (30.7 milliwatts) and keeping the drops in complete darkness at room temperature. Crystals were flash-frozen without additional cryoprotection under green-filtered light. Diffraction data were collected at beamline ID23-1 of ESRF. The data were cut at 3.6 Å (CC½ = 34.3). The cell parameters and packing indicated a crystal form highly similar to that of the pre-illuminated CBD–PHY-Y263F, and the molecular replacement was therefore conducted as above. To minimize the bias caused by the initial search model, the biliverdin and the neighboring residues that differ between Pr and Pfr states (Tyr176, His201, Phe203, and Arg222) were removed and added only at the later stages of the refinement. The final refinement steps were conducted with tight noncrystallographic symmetry restraints between the chains and an X-ray matrix-weighting term of 0.003, which gave final Rwork/Rfree values of 0.248/0.287. All electron density maps were prepared from the final structure factor files with FFT of the CCP4 interface, and the crystallography figures were created with the PyMOL Molecular Graphics System version 2.0 (Schrödinger, LLC). The RMSD between WT CBD and CBD-Y263F was calculated with PyMOL after outlier rejection (13 of 315 atoms rejected).
Absorption spectroscopy of crystals
The crystal absorption spectra were measured using a home-built microfocusing device connected by optical fiber to a standard deuterium lamp and a diode array spectrometer (Ocean Optics), as in Ref. 9. The crystals were mounted in cryoloops in reservoir solution with 15% glycerol and flash-frozen with liquid nitrogen. The absorption measurements were conducted under a 100 K cryostream, which impairs photoconversion. To test the photoswitching of the crystals, the focused detection light was replaced with a red light–emitting diode (660 nm, 13 milliwatts; Thorlabs) for 5–15 s after the cryostream (and other light sources) was temporarily blocked. Visual inspection showed that the crystals remained intact through the brief illumination treatments.
Author contributions
H. T. cloned, purified, and crystallized the proteins, solved the crystal structures, and measured the UV-visible spectra. H. L. measured and analyzed the UV-visible and FTIR spectra. R. N. measured the labeled FTIR spectra. O. B., A. H., S. N., M. P., L. H., and A. M. measured and O. B. analyzed the X-ray scattering data. H. T., H. L., S. W., and J. A. I. designed and wrote the paper with input from all other authors.
Supplementary Material
Acknowledgments
We acknowledge beamline access and personnel at the cSAXS in the Swiss Light Source and at ESRF and thank A. Liukkonen for assistance in laboratory work.
This work was supported by Academy of Finland Grants 285461 (to H. T.), 277194 (to H. L.), and 296135 (to J. A. I.); the Foundation of Strategic Research, Sweden, Grant FFL09-0106, and European Research Council, Agreement 279944 (to S. W.); the Emil Aaltonen Foundation (to H. L.); and Jane and Aatos Erkko Foundation (to J. A. I.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
The atomic coordinates and structure factors (codes 5NFX, 5NM3, and 5NWN) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Pr
- red light–absorbing
- Pfr
- far-red light–absorbing
- CBD
- chromophore-binding domain
- PHY
- phytochrome-specific GAF-related
- PSM
- photosensory module
- PDB
- Protein Data Bank
- DrBphP
- D. radiodurans bacteriophytochrome
- PAS
- Per/Arnt/Sim
- GAF
- cGMP phosphodiesterase, adenylyl cyclase, FhlA
- ESRF
- European Synchrotron Radiation Facility
- RMSD
- root mean square deviation.
References
- 1. Quail P. H. (2002) Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell. Biol. 3, 85–93 10.1038/nrm728 [DOI] [PubMed] [Google Scholar]
- 2. Rockwell N. C., Su Y. S., and Lagarias J. C. (2006) Phytochrome structure and signaling mechanisms. Annu. Rev. Plant. Biol. 57, 837–858 10.1146/annurev.arplant.56.032604.144208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burgie E. S., and Vierstra R. D. (2014) Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell 26, 4568–4583 10.1105/tpc.114.131623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamparter T., Michael N., Caspani O., Miyata T., Shirai K., and Inomata K. (2003) Biliverdin binds covalently to agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 278, 33786–33792 10.1074/jbc.M305563200 [DOI] [PubMed] [Google Scholar]
- 5. Wagner J. R., Brunzelle J. S., Forest K. T., and Vierstra R. D. (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 10.1038/nature04118 [DOI] [PubMed] [Google Scholar]
- 6. Casino P., Rubio V., and Marina A. (2010) The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol. 20, 763–771 10.1016/j.sbi.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 7. Essen L. O., Mailliet J., and Hughes J. (2008) The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 10.1073/pnas.0806477105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang X., Kuk J., and Moffat K. (2008) Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 10.1073/pnas.0806718105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J. A., and Westenhoff S. (2014) Signal amplification and transduction in phytochrome photosensors. Nature. 509, 245–248 10.1038/nature13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stojković E. A., Toh K. C., Alexandre M. T., Baclayon M., Moffat K., and Kennis J. T. (2014) FTIR spectroscopy revealing light-dependent refolding of the conserved tongue region of bacteriophytochrome. J. Phys. Chem. Lett. 5, 2512–2515 10.1021/jz501189t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burgie E. S., Bussell A. N., Walker J. M., Dubiel K., and Vierstra R. D. (2014) Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184 10.1073/pnas.1403096111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takala H., Niebling S., Berntsson O., Björling A., Lehtivuori H., Häkkänen H., Panman M., Gustavsson E., Hoernke M., Newby G., Zontone F., Wulff M., Menzel A., Ihalainen J. A., and Westenhoff S. (2016) Light-induced structural changes in a monomeric bacteriophytochrome. Struct. Dyn. 3, 054701 10.1063/1.4961911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Björling A., Berntsson O., Lehtivuori H., Takala H., Hughes A. J., Panman M., Hoernke M., Niebling S., Henry L., Henning R., Kosheleva I., Chukharev V., Tkachenko N. V., Menzel A., Newby G., et al. (2016) Structural photoactivation of a full-length bacterial phytochrome. Sci. Adv. 2, e1600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgie E. S., Wang T., Bussell A. N., Walker J. M., Li H., and Vierstra R. D. (2014) Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J. Biol. Chem. 289, 24573–24587 10.1074/jbc.M114.571661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kacprzak S., Njimona I., Renz A., Feng J., Reijerse E., Lubitz W., Krauss N., Scheerer P., Nagano S., Lamparter T., and Weber S. (2017) Intersubunit distances in full-length, dimeric, bacterial phytochrome Agp1, as measured by pulsed electron-electron double resonance (PELDOR) between different spin label positions, remain unchanged upon photoconversion. J. Biol. Chem. 292, 7598–7606 10.1074/jbc.M116.761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Auldridge M. E., Satyshur K. A., Anstrom D. M., and Forest K. T. (2012) Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 287, 7000–7009 10.1074/jbc.M111.295121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhattacharya S., Auldridge M. E., Lehtivuori H., Ihalainen J. A., and Forest K. T. (2014) Origins of fluorescence in evolved bacteriophytochromes. J. Biol. Chem. 289, 32144–32152 10.1074/jbc.M114.589739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mailliet J., Psakis G., Feilke K., Sineshchekov V., Essen L. O., and Hughes J. (2011) Spectroscopy and a high-resolution crystal structure of Tyr263 mutants of cyanobacterial phytochrome Cph1. J. Mol. Biol. 413, 115–127 10.1016/j.jmb.2011.08.023 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J., Stankey R. J., and Vierstra R. D. (2013) Structure-guided engineering of plant phytochrome B with altered photochemistry and light signaling. Plant Physiol. 161, 1445–1457 10.1104/pp.112.208892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takala H., Lehtivuori H., Hammarén H., Hytönen V. P., and Ihalainen J. A. (2014) Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome. Biochemistry 53, 7076–7085 10.1021/bi501180s [DOI] [PubMed] [Google Scholar]
- 21. Wagner J. R., Zhang J., von Stetten D., Günther M., Murgida D. H., Mroginski M. A., Walker J. M., Forest K. T., Hildebrandt P., and Vierstra R. D. (2008) Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 283, 12212–12226 10.1074/jbc.M709355200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lehtivuori H., Bhattacharya S., Angenent-Mari N. M., Satyshur K. A., and Forest K. T. (2015) Removal of chromophore-proximal polar atoms decreases water content and increases fluorescence in a near infrared phytofluor. Front. Mol. Biosci. 2, 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lehtivuori H., Rissanen I., Takala H., Bamford J., Tkachenko N. V., and Ihalainen J. A. (2013) Fluorescence properties of the chromophore-binding domain of bacteriophytochrome from Deinococcus radiodurans. J. Phys. Chem. B 117, 11049–11057 10.1021/jp312061b [DOI] [PubMed] [Google Scholar]
- 24. Foerstendorf H., Benda C., Gärtner W., Storf M., Scheer H., and Siebert F. (2001) FTIR studies of phytochrome photoreactions reveal the C=O bands of the chromophore: consequences for its protonation states, conformation, and protein interaction. Biochemistry 40, 14952–14959 10.1021/bi0156916 [DOI] [PubMed] [Google Scholar]
- 25. Barth A., and Zscherp C. (2002) What vibrations tell us about proteins. Q. Rev. Biophys. 35, 369–430 10.1017/S0033583502003815 [DOI] [PubMed] [Google Scholar]
- 26. Takahashi R., Okajima K., Suzuki H., Nakamura H., Ikeuchi M., and Noguchi T. (2007) FTIR study on the hydrogen bond structure of a key tyrosine residue in the flavin-binding blue light sensor TePixD from Thermosynechococcus elongatus. Biochemistry 46, 6459–6467 10.1021/bi7004653 [DOI] [PubMed] [Google Scholar]
- 27. Björling A., Berntsson O., Takala H., Gallagher K. D., Patel H., Gustavsson E., St Peter R., Duong P., Nugent A., Zhang F., Berntsen P., Appio R., Rajkovic I., Lehtivuori H., Panman M. R., et al. (2015) Ubiquitous structural signaling in bacterial phytochromes. J. Phys. Chem. Lett. 6, 3379–3383 10.1021/acs.jpclett.5b01629 [DOI] [PubMed] [Google Scholar]
- 28. Edlund P., Takala H., Claesson E., Henry L., Dods R., Lehtivuori H., Panman M., Pande K., White T., Nakane T., Berntsson O., Gustavsson E., Båth P., Modi V., Roy-Chowdhury S., et al. (2016) The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography. Sci. Rep. 6, 35279 10.1038/srep35279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgie E. S., Zhang J., and Vierstra R. D. (2016) Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 10.1016/j.str.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 30. Gourinchas G., Etzl S., Göbl C., Vide U., Madl T., and Winkler A. (2017) Long-range allosteric signaling in red light-regulated diguanylyl cyclases. Sci. Adv. 3, e1602498 10.1126/sciadv.1602498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagano S., Scheerer P., Zubow K., Michael N., Inomata K., Lamparter T., and Krauss N. (2016) The crystal structures of the N-terminal photosensory core module of Agrobacterium phytochrome Agp1 as parallel and anti-parallel dimers. J. Biol. Chem. 291, 20674–20691 10.1074/jbc.M116.739136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang X., Stojković E. A., Ozarowski W. B., Kuk J., Davydova E., and Moffat K. (2015) Light signaling mechanism of two tandem bacteriophytochromes. Structure 23, 1179–1189 10.1016/j.str.2015.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Su Y. S., and Lagarias J. C. (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19, 2124–2139 10.1105/tpc.107.051516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jeong A. R., Lee S. S., Han Y. J., Shin A. Y., Baek A., Ahn T., Kim M. G., Kim Y. S., Lee K. W., Nagatani A., and Kim J. I. (2016) New constitutively active phytochromes exhibit light-independent signaling activity. Plant Physiol. 171, 2826–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takala H., Björling A., Linna M., Westenhoff S., and Ihalainen J. A. (2015) Light-induced changes in the dimerization interface of bacteriophytochromes. J. Biol. Chem. 290, 16383–16392 10.1074/jbc.M115.650127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berntsson O., Diensthuber R. P., Panman M. R., Björling A., Gustavsson E., Hoernke M., Hughes A. J., Henry L., Niebling S., Takala H., Ihalainen J. A., Newby G., Kerruth S., Heberle J., Liebi M., et al. (2017) Sequential conformational transitions and α-helical supercoiling regulate a sensor histidine kinase. Nat. Commun. 8, 284 10.1038/s41467-017-00300-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tkachenko N. V. (2006) Optical Spectroscopy: Methods and Instrumentations, Elsevier, Amsterdam [Google Scholar]
- 38. Kabsch W. J. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 10.1107/S0021889893005588 [DOI] [Google Scholar]
- 39. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wagner J. R., Zhang J., Brunzelle J. S., Vierstra R. D., and Forest K. T. (2007) High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 282, 12298–12309 10.1074/jbc.M611824200 [DOI] [PubMed] [Google Scholar]
- 41. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.