Abstract
The conserved serine/threonine protein kinase target of rapamycin (TOR) is a major regulator of eukaryotic cellular and organismal growth and a valuable target for drug therapy. TOR forms the core of two evolutionary conserved complexes, TOR complex 1 (TORC1) and TORC2. In the fission yeast Schizosaccharomyces pombe, TORC2 responds to glucose levels and, by activating the protein kinase Gad8 (an orthologue of human AKT), is required for well-regulated cell cycle progression, starvation responses, and cell survival. Here, we report that TORC2–Gad8 is also required for gene silencing and the formation of heterochromatin at the S. pombe mating-type locus and at subtelomeric regions. Deletion of TORC2–Gad8 resulted in loss of the heterochromatic modification of histone 3 lysine 9 dimethylation (H3K9me2) and an increase in euchromatic modifications, including histone 3 lysine 4 trimethylation (H3K4me3) and histone 4 lysine 16 acetylation (H4K16Ac). Accumulation of RNA polymerase II (Pol II) at subtelomeric genes in TORC2–Gad8 mutant cells indicated a defect in silencing at the transcriptional level. Moreover, a concurrent decrease in histone 4 lysine 20 dimethylation (H4K20me2) suggested elevated histone turnover. Loss of gene silencing in cells lacking TORC2–Gad8 is partially suppressed by loss of the anti-silencer Epe1 and fully suppressed by loss of the Pol II–associated Paf1 complex, two chromatin regulators that have been implicated in heterochromatin stability and spreading. Taken together, our findings suggest that TORC2–Gad8 signaling contributes to epigenetic stability at subtelomeric regions and the mating-type locus in S. pombe.
Keywords: Akt PKB, heterochromatin, gene silencing, gene expression, Schizosaccharomyces pombe, Epe1, mating-type locus, Paf1 complex, subtelomeres, TORC2
Introduction
The target of rapamycin (TOR)3 pathway is a central regulator of metabolic pathways, cell growth, and proliferation in response to nutritional and stress signals, and its deregulation is implicated in cancer, diabetes, neurodegenerative disease, and aging (1, 2). TOR is a serine/threonine protein kinase that is found in two structurally and functionally distinct complexes, TORC1 and TORC2. In mammalian cells, a single catalytic subunit, mTOR, is present in both TORC1 and TORC2 (mTORC1 and mTORC2). Many features of the two TOR complexes are conserved in evolution, and a substantial amount of research has suggested multiple conserved pathways by which TOR proteins control cellular growth, including regulation of transcription, translation, enzymatic metabolism, and autophagy (3, 4). TORC1 is inhibited by rapamycin and its derivatives and is well-known for its conserved role as a positive regulator of cellular growth and a negative regulator of starvation responses. TORC2 has been implicated in growth, metabolism, and survival; however, our understanding of TORC2 lags behind that of TORC1. This is partly due to the lack of TORC2-specific inhibitors, a disadvantage that is mitigated in genetically tractable organisms such as yeasts (5, 6).
Schizosaccharomyces pombe contains two TOR homologues, Tor1 and Tor2, which were numbered based on the order of their discovery (7). Tor2 interacts with the Raptor-like subunit, Mip1, to form TORC1, whereas Tor1 interacts with the Rictor-like subunit Ste20 and with Sin1 (mSin1 in humans) to form TORC2 (8, 9). S. pombe TORC1 (SpTORC1) promotes cellular growth in response to nitrogen and amino acid availability while inhibiting starvation responses (8, 10, 11). SpTORC2 is critical for several responses to starvation, including entrance into the sexual development pathway or stationary (quiescence) phase and for survival under osmotic or oxidative stress (7, 12), DNA-damaging, or DNA replication stress conditions (13, 14). More recently, it was demonstrated that SpTORC2 controls the sexual development pathway in coordination with SpTORC1 via a cross-talk mechanism that involves the PP2A-B55 phosphatase (15) and the SAGA (Spt–Ada–Gcn5 acetyltransferase) co-activation transcription complex (16). Under normal growth conditions, SpTORC2 plays a role in regulating entrance into mitosis (13, 17, 18), cytokinesis (19), and nutrient assimilation (11, 20). SpTORC2 mediates its known functions via phosphorylation and activation of the Gad8 kinase, the homologue of mammalian AKT or SGK1 kinases that lie downstream of mTORC2 (21, 22). Unlike SpTORC1, glucose, but not nitrogen, is required for activation of SpTORC2–Gad8 (23, 24).
Transcriptional profile analyses of cells lacking the catalytic subunit of SpTORC2 (Δtor1) revealed an extensive similarity with the transcriptomes of chromatin structure or function mutant cells, such as similarity with mutants of histone deacetylases (clr6-1 or Δclr3 clr6-1) or mutants of the SWI/SNF chromatin-remodeling complexes RSC (remodeling the structure of chromatin) (Δrsc58) (13). This finding led us to discover several defects in nuclear functions in TORC2–Gad8 mutant cells, including elongated telomeres, increased chromosomal loss, and elevated levels of Rad52 foci (13). Moreover, Tor1 and Gad8 are found in the nucleus in association with the chromatin fraction, physically interact with the MluI cell cycle box–binding factor (MBF) transcription complex, and affect MBF-dependent gene transcription (25). These findings suggest the possibility that TORC2–Gad8 has direct effects on nuclear and chromatin functions.
Regulation of chromatin states is an efficient way to rapidly and reversibly control cellular growth in response to changing environmental conditions. Chromatin states primarily consist of silent and compact heterochromatic regions and less condensed, active euchromatic regions. Reversible transition between heterochromatic and euchromatic states is characteristic of facultative heterochromatin, which provides regulatory mechanisms for gene expression (26, 27). The dynamics between heterochromatic and euchromatic states plays an important role during early development and stem cell differentiation in higher eukaryotes (28), and its regulation is crucial for epigenetic stability, the process that ensures that the epigenetic state is faithfully transferred and maintained in daughter cells.
In S. pombe, constitutive heterochromatin is formed at highly repetitive sequences residing at the centromeres, telomeres, and the silent mating-type locus and is characterized by H3K9 methylation, hypo-acetylation, and low histone turnover (27). Facultative heterochromatin, enriched in stress-regulated and meiotic genes, is found at small blocks of heterochromatic islands dispersed along the chromosomes (29, 30). The establishment of heterochromatin at centromeres and repetitive elements is primarily regulated by the RNAi machinery that includes the argonaute, dicer, and RNA-dependent RNA polymerase gene homologs (31, 32). The RNAi machinery recruits the Clr4–Rik1–Cul4 complex containing the histone methyl transferase Clr4 (Suv39h in humans) (33). Clr4 is the sole H3K9 methylase responsible for di- and tri-H3K9 methylation (34, 35), and its activity provides a docking site for chromodomain proteins, including Swi6 (heterochromatin protein 1, HP1 in humans), Chp1, Chp2, and Clr4 itself (36). Transcription factors (37) and RNA processing and surveillance mechanisms (38, 39) also contribute to heterochromatin formation.
The spreading of the chromatin is restricted by several mechanisms. At the centromeres and mating-type locus, heterochromatin is confined by sequence-specific boundaries, such as the IRC and IR elements, respectively (26, 29). These boundaries are responsible for a sharp decrease of the H3K9me signature outside the heterochromatic region. At the telomeres, the borders between the heterochromatin and the euchromatin are not well-defined, and the H3K9me2 signature gradually declines as the heterochromatin spreads away from the telomeric repeats (29). The Jumonji C protein Epe1, which acts to antagonize heterochromatin spreading, accumulates at the well-defined boundaries through the binding to Swi6 (40–43) and is selectively abolished from the interior parts of the heterochromatin by the Cul4-Ddb1Cdt2 ubiquitin ligase (44). In the absence of Epe1, the heterochromatin spreads outside the boundaries of the mating-type locus (40) or the centromeres (43), leading to silencing of nearby genes, whereas the overexpression of Epe1 leads to desilencing at subtelomeric regions (42). The mechanism by which Epe1 contributes to regulation of heterochromatin spreading is not clear. The sequence of Epe1 predicts that it may act as a histone de-methylase, but no such activity has been observed in vitro (45). Epe1 contributes to restricting the spreading of heterochromatin by recruitment of the bromodomain protein Bdf2, which antagonizes the deacytelation of H4K16 (46) or by competition with the Clr3–histone deacetylase complex for binding to Swi6 (47). More recently it was suggested that Epe1 may act in coordination with the RNA polymerase-associated factor 1 complex (Paf1C) (48, 49). The Paf1C complex, a conserved multifunctional regulator of eukaryotic gene expression that is involved in transcriptional elongation and termination and RNA 3′-end formation, has more recently been shown to affect histone modifications and chromatin states (50). In S. pombe, Paf1C antagonizes heterochromatin spreading by promoting H4K16 acetylation (48), histone turnover (49) or efficient transcription termination (51). The Paf1C complex is required to regulate gene repression at the subtelomeric regions and its loss results in spreading of the subtelomeric heterochromatin into the neighboring euchromatic region (48).
Here we show that TORC2–Gad8 is required for gene silencing of a reporter gene embedded at the mating-type locus and genes located at subtelomeric regions, specifically those characterized by intermediate levels of H3K9 methylation. Loss of TORC2–Gad8 results in unstable maintenance of heterochromatin-mediated gene silencing, loss of heterochromatic markers and gain of euchromatic markers. We show that the defects in gene silencing in TORC2–Gad8 mutant cells are partially suppressed by Epe1 and fully suppressed by the Paf1C complex. Our findings indicate that TORC2–Gad8 plays a role in gene silencing at the transcriptional level and is involved in determining specific epigenetic states.
Results
TORC2–Gad8 is required for silencing of a reporter gene embedded in the silent mating-type locus
The mating-type region in S. pombe contains the mat1, mat2, and mat3 genes. The mat1+ gene is transcriptionally active and determines the mating type, whereas mat2+ and mat3+ lie within the constitutive heterochromatin, flanked by the two inverted IR boundary elements. The cenH element, which lies in between the two silent mating-type cassettes, acts as a heterochromatin nucleation center (see schematic presentation in Fig. 1A). We previously showed that loss of the catalytic subunit of TORC2 (Δtor1) results in de-silencing of the ade6+ reporter gene inserted at the SacI site (13) that is located at the left IR boundary, IR-L (Fig. 1A). The mat2::SacI::ade6+ reporter gene is only weakly silenced in WT cells (40); thus, the de-silencing observed in Δtor1 cells is not striking (13). Here, we employed the mat2::BamHI::ade6+ reporter gene that resides within the mat2 gene (40). This reporter gene is completely silenced in WT cells, resulting in cells that cannot grow in medium lacking adenine and accumulate a red pigment under adenine-limiting conditions (Fig. 1B). Remarkably, mutant cells carrying deletion mutations in TORC2–Gad8 (Δtor1, Δsin1, Δste20, or Δgad8) carrying mat2::BamHI::ade6+ grew on minimal medium lacking adenine, and they produced white colonies under limiting adenine conditions, suggesting that TORC2–Gad8 plays a prominent role in gene silencing of the reporter gene (Fig. 1B). Consistently, quantitative RT-PCR (qRT-PCR) analysis in Δtor1 cells revealed a strong expression of the ade6+ transcript (∼120 fold increase compared with WT), similar to the de-repression observed in Δclr4 cells, lacking the sole H3K9 methyl transferase in S. pombe (34) (Fig. 1C). Similar de-repression of the ade6+ gene is observed in Δgad8 mutant cells (Fig. S1A).
Figure 1.
The TORC2–Gad8 pathway promotes silencing at the mating-type region. A, schematic representation of the mating-type locus in S. pombe. The mating-type region contains three genes: mat1, mat2, and mat3. mat1 is transcriptionally active and determines the mating type. mat2-P and mat3-M reside at the heterochromatic region shown within the gray box. This region is flanked by the two inverted repeats boundary elements IR-L and IR-R. The ade6+ cassettes inserted at the SacI or BamHI sites are indicated. B, mutations in TORC2–Gad8 result in de-silencing at the mating-type locus. WT strain (ade6+) and strains containing ade6-M210 in addition to the indicated mutations, with or without the mat2:BamHI::ade6+ reporter gene, were spotted onto rich medium plates, supplemented with a standard amount of adenine or low adenine (YE+Ade, YE−Ade) or minimal medium plates containing a standard amount of adenine or no adenine (ADE+, ADE−). White colonies growing on ADE− plates indicate de-silencing of the ade6+ gene. C, expression levels of the ade6+ gene inserted at the mat2 locus (mat2:BamHI::ade6+); qRT-PCR analysis. The level of act1+ mRNA was used as a reference. Each value is the mean of at least three independent assays. D, stability of ectopic heterochromatin in mitotic cells. Five independent colonies of each strain were isolated on nondifferential medium (YE+Ade) and replated on differential medium (YE−Ade), and their color was scored. n, number of scored colonies. E, hereditability of the repressed or active chromatin state in Δtor1 cells. Five independent red or white Δtor1 colonies were isolated on limiting adenine (YE−Ade) plates and replated on YE-Ade plates, and their color was scored. n, number of scored colonies.
The repression of a euchromatic gene upon its insertion into a heterochromatic region is a universally conserved epigenetic phenomenon termed position effect variegation (PEV), which has been used to identify key regulators of heterochromatin-induced silencing (reviewed in Ref. 27). However, the sensitivity to silencing of the reporter gene is not identical to that of the endogenous mating-type cassettes. Thus, for example, disruption of clr4+ results in de-silencing of reporter genes at the mating-type locus but has little or no effect on the expression level of the silent mat2+ gene (52). Similarly, the Δtor1 or Δgad8 mutations had only a very limited effect on the expression of the endogenous mat2+ gene (Fig. S1B). Thus, TORC2–Gad8, similar to Clr4, strongly affects the spreading of heterochromatin across the euchromatic reporter gene but is incapable of overriding the heterochromatic-dependent silencing at the endogenous mating-type gene. This probably reflects the existence of redundant repressive mechanisms at the endogenous locus.
Instability of chromatin states in TORC2–Gad8 mutant cells
We noted that Δtor1 mat2::BamHI::ade6+ cells that grew under rich, nondefined media (YE+Ade plates) produced a majority of white colonies but could also give rise to variegated/pink (35%) or red (10%) colonies under limiting adenine conditions (Fig. 1D). In comparison, Δclr4 (lacking the H3K9 methylase) or Δsir2 (lacking the sirtuin-type histone deacetylase) cells carrying the mat2::BamHI::ade6+ reporter gene produced only white colonies, whereas WT cells carrying the reporter gene produced only red colonies (Fig. 1D). We compared the morphology and stress sensitivity of white versus red Δtor1 mat2::BamHI::ade6+ cells. Both displayed the characteristic phenotypes for Δtor1 cells (7), including sensitivity to high temperature or osmotic stress and the typical aberrant cell morphology (data not shown). Thus, the red colonies that repressed expression of mat2::BamHI::ade6+ did not rise from accumulation of general suppressors of the Δtor1 mutation but rather from the instability of the transcriptional state of the reporter gene. When we followed the fate of isolated white or red Δtor1 colonies by replating on limiting adenine plates, we observed that both the repressive and the active states of the reporter gene were relatively stable over 30 generations (Fig. 1E).
Similar to Δtor1, Δste20, or Δsin1, mutant cells that were isolated on rich, nondefined media and then plated under limiting adenine conditions gave rise mainly to white colonies but also produced a small proportion of red or pink colonies (Fig. 1D). Δgad8 mutant cells exhibited a slightly different pattern of inheritance of the epigenetic state as they gave rise to a larger proportion of pink and red colonies and a high proportion of sectored colonies (Fig. 1D and Fig. S1C), suggesting that lack of Gad8 results in further instability of the epigenetic state of mat2::BamHI::ade6+. The reason for the differences observed for the stability of the expression of mat2::BamHI::ade6+ in the different mutant cells of the TORC2–Gad8 module is not clear and may reflect the highly stochastic nature of the process of heterochromatin spreading. In any case, the unstable expression state of mat2::BamHI::ade6+ in TORC2–Gad8 mutant cells is in contrast with the stable expressed state of the reporter gene in Δclr4 or Δsir2 cells. These findings suggest that TORC2–Gad8 affects the stability of the heterochromatin but is not absolutely required for its formation.
TORC2–Gad8 is required for gene silencing at the subtelomeric region
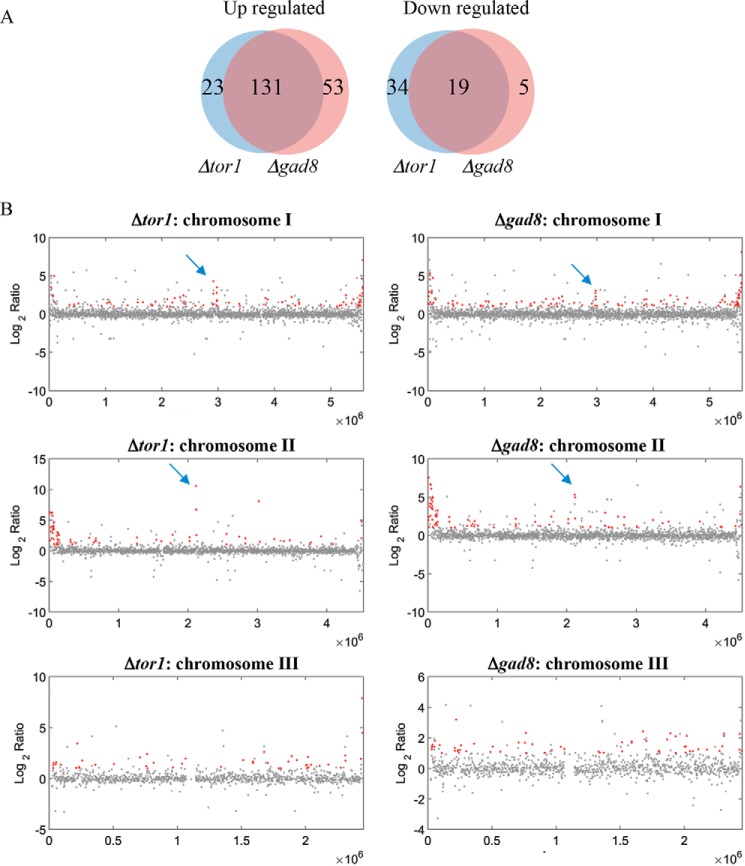
We previously showed an extensive similarity with the transcriptional profiles of Δtor1 cells and the profiles of histone deacetylase mutant cells (Dclr3 or clr6-1) and nucleosome-remodeling complex mutant cells (Drsc8) but not RNAi mutant cells (such as Δdcr1 or Δago1) (13). Here, we have extended this analysis by performing RNA-Seq of Δtor1 and Δgad8 cells (Table S3). In agreement with the activation of Gad8 by Tor1, there is a significant overlap between the expression profiles of Δtor1 or Δgad8 mutant cells (Fig. 2A). The RNA-Seq analysis recapitulated several of our previous results for gene expression in Δtor1 cells (13). For example, the majority of the genes that are abnormally expressed are up-regulated (154 and 184 genes in Δtor1 and Δgad8, respectively), whereas a minority of the genes are down-regulated (53 and 24 genes in Δtor1 and Δgad8, respectively). As reported previously for Δtor1 cells (13), among the genes that are down-regulated, there is an enrichment for membrane transporters (Table S3). No enrichment for specific functional groups is detected within the abnormally up-regulated genes. However, when plotted along the chromosomes, we noted that genes located close to the telomeres of chromosome I and II are up-regulated in Δtor1 or Δgad8 mutants (Fig. 2B). In contrast, the Δtor1 or Δgad8 mutations do not affect genes at the ends of chromosome III that contain ribosomal DNA repeats. The subtelomeric region of chromosomes I and II consists of 20–40 kb of heterochromatin that is characterized by a gradual decline in H3K9me2 at the telomere-distal end (29). We also noted two clusters of up-regulated genes in Δtor1 or Δgad8 cells that reside in the middle of chromosomal arms I and II (see blue arrows in Fig. 2B). These clusters are enriched in noncoding RNA transcripts (Fig. S2), but no enrichment in H3K9me2 has been reported in these regions or was detected in our ChIP-Seq analysis (described below).
Figure 2.
TORC2–Gad8 represses transcription of genes located close to the telomeres of chromosome I and II. A, RNA-Seq analysis shows extensive similarity between the transcriptional profiles of Δgad8 and Δtor1. The genes that are up-regulated or down-regulated by at least log2-fold are presented in Venn diagrams. B, up-regulated genes in Δgad8 and Δtor1 cells are clustered at the subtelomeric regions of chromosome I and II. The log2 ratio of the distribution of the Δtor1 versus WT (left) or Δgad8 versus WT (right) is plotted as a function of the location of the chromosome. The chromosome number is indicated above each plot. Genes that had a ratio >2, adjusted p value <0.05, and a count of at least 20 in at least one of the samples are shown in red. The arrows indicate clusters of genes along the arms of chromosomes I or II that are found to be abnormally up-regulated in Dgad8 and Δtor1 cells. These clusters are not associated with heterochromatic structure.
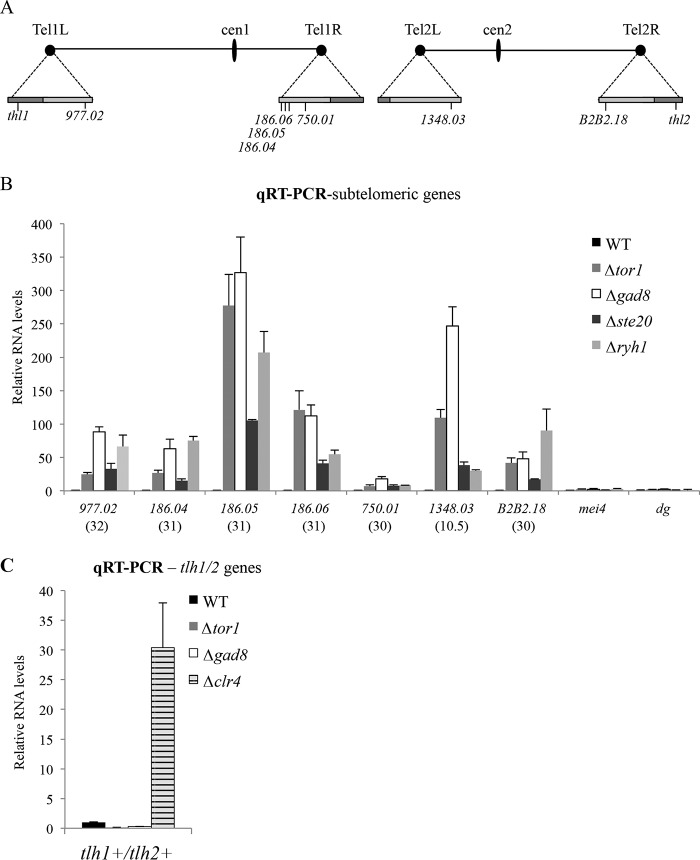
Using qRT-PCR with specific primers, we confirmed that the subtelomeric genes spac186.04+, spac186.05+, spac186.06+, spac750.01+ (chromosome I, right arm), spac977.02+ (chromosome I, left arm), spbc1348.03+ (chromosome II, left arm), and spbpb2b2.18+ (chromosome II, right arm) are up-regulated in cells carrying mutations in Δtor1, Δste20, Δgad8 or in Δryh1 cells, lacking the small Ryh1 GTPase that activates TORC2 (53) (Fig. 3, A and B). All of these subtelomeric genes are located at the regions characterized by intermediate levels of H3K9me2 (see schematic presentation in Fig. S3 and Refs. 29 and 54). Unlike Δclr4 cells, mutations in Δtor1 or Δgad8 did not result in de-silencing of tlh1+/tlh2+, the most telomere-proximal genes located at the high-level H3K9me2 subtelomeric region (Fig. 3C). The tlh1+/tlh2+ genes encode RecQ-like helicases and contain repeat elements in their coding regions, similar to the repeats at the cenH element at the mating-type locus or the dg/dh centromeric repeats (26). The de-silencing of subtelomeric genes but not tlh1+/tlh2+ is similar to the differential effect observed for cohesin mutant cells, the cohesin loader Mis4, or the cohesin core component Rad21 (54). Thus, similar to mutations in cohesin, TORC2 affects gene expression only at the subtelomeric region that is characterized by an intermediate level of H3K9me2.
Figure 3.
TORC2–Gad8 is required for silencing at a specialized subtelomeric region but not for silencing of centromeric repeats or meiotic genes. A, schematic presentations of chromosomes I and II. The triangles on the chromosomal edges are magnifications of the subtelomeric regions. Dark and light gray boxes indicate high- or low-H3K9me2 regions, respectively. B, expression levels of subtelomeric genes on chromosome I (ChI) or chromosome II (ChII), mei4+, and the centromeric dg repeat were examined in WT or cells carrying mutations in Δtor1, Δgad8, Δste20, or Δryh1 cells. The numbers in parenthesis indicate the distance of each subtelomeric gene from the nearby telomeric repeats. Expression levels were determined by qRT-PCR. The level of act1+ mRNA was used as a reference. Each value is the mean of at least three independent assays, and the error bars indicate S.D. C, the levels of the tlh1+/tlh2+ genes that are located next to the telomeric repeats are unchanged or slightly reduced in TORC2–Gad8 mutant cells.
Disruption of TORC2–Gad8 did not affect the level of expression of centromeric dg repeats (Fig. 3B) or the silencing of reporter genes inserted into the centromeric innermost repeats (imr1L) or outermost repeats (otr1L) (Fig. S4). Thus, our data indicate that TORC2–Gad8 is not required for silencing at the centromeric constitutive heterochromatin. We also found no effects of Δtor1 or Δgad8 cells on the expression levels of facultative heterochromatic islands encoding meiotic genes (our RNA-Seq analysis; see qRT-PCR for mei4+ in Fig. 3B), ruling out the involvement of complexes that are dedicated to meiotic mRNA decay, such as the Erh1-Mmi (EMC) and Mtl1–Red1 core (MTREC) complexes (55).
Loss of TORC2–Gad8 is accompanied by loss of heterochromatic markers at subtelomeric regions
H3K9me2 is a key histone modification that characterizes heterochromatic regions. As our RNA-Seq experiments suggest that TORC2–Gad8 signaling has a global effect on gene expression that is associated with chromosomal location, we investigated the effects of Δtor1 or Δgad8 on genome-wide H3K9me2 levels by ChIP-Seq analysis. We found that the strongest effect of the Δtor1 or Δgad8 mutations was at the subtelomeric region of the right telomere of chromosome 1 (tel1R), where a 33-kb region that resides 30 kb away from the telomeric repeats became almost completely devoid of H3K9me2 in the mutant cells (Fig. 4A). Using ChIP–qRT-PCR analyses, we confirmed that the level of H3K9me2 is reduced at the subtelomeric genes that reside within the 33-kb region of tel1R but is unchanged for the centromeric dg repeats (Fig. 4B). The subtelomeric genes that reside at the other chromosomal ends and exhibited elevated levels of transcription in Δtor1 mutant cells (Fig. 3B) also showed a significant reduction in H3K9me2 levels (Fig. 4B). We examined the binding levels of Swi6, the main HP1 protein in S. pombe that binds H3K9me2. We detected enrichment for Swi6 in WT cells in five of the seven examined subtelomeric genes. Of these, four genes lost Swi6 binding in Δtor1 cells (Fig. 4C). The exception was the most tel1R-proximal gene, spac750.01+, which retained Swi6 binding in Δtor1 cells. The level of Swi6 binding at the dg repeats was not affected by Δtor1 (Fig. 4C), consistent with the lack of effect on H3K9me2 or transcription at the centromeric repeats.
Figure 4.
Loss of TORC2–Gad8 leads to a decrease in heterochromatic markers at the subtelomeric region. A, genome browser view showing ChIP-Seq analysis of H3K9me2 levels in WT (green), Δtor1 (red), and Δgad8 (orange) in log2 scale. B, levels of H3K9me2 by ChIP-quantitative PCR at subtelomeric genes. C, levels of Swi6 by ChIP–qRT-PCR at subtelomeric genes. Δclr4 cells, used as control for no Swi6 binding, showed no significant signal. Each value is the mean of at least three independent assays, and the error bars indicate S.D.
Loss of TORC2–Gad8 elevates chromatin modifications associated with transcriptional active chromatin
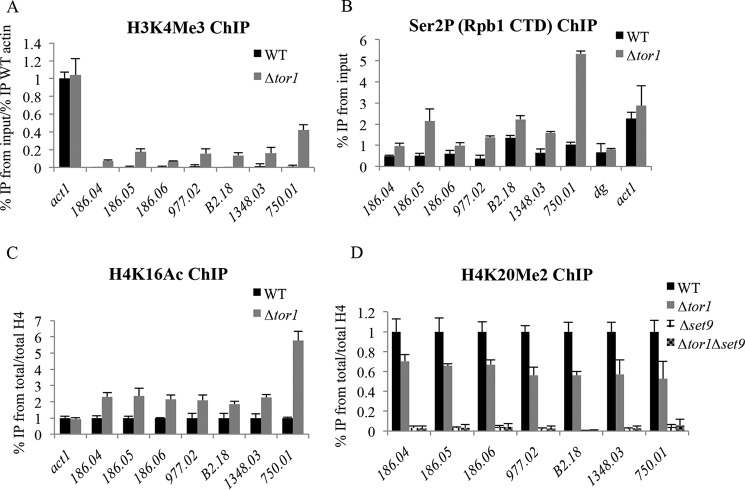
We next examined changes in trimethylation of H3K4 (H3K4me3), a modification that is associated with transcriptionally active chromatin (56). We found an increase in H3K4me3 in all subtelomeric genes examined in Δtor1 mutant cells (Fig. 5A). The histone methyltransferase Set1, a component of the set1C–COMPSS complex, is the sole methyltransferase responsible for mono-, di-, or tri-methylation of H3K4 in S. pombe (57). Although Set1 is responsible for a histone modification that is associated with active euchromatin, Set1 acts as a transcriptional repressor at many chromosomal loci, including the subtelomeric and mating-type regions (57). Indeed, we found that deletion of set1+ resulted in up-regulation of subtelomeric genes, very similar to that observed in Δtor1 cells (Fig. S5A). The double mutant Δtor1 Δset1 cells showed no enhancement of the up-regulation of gene expression at the subtelomeric genes (Fig. S5A), suggesting that Tor1 and the repressive function of Set1 may lie on the same pathway. Because we observed an increase in the level of H3K4me3 in Δtor1 cells, it is unlikely that the elevation in H3K4me3 is responsible for the increase in gene transcription in Δtor1 cells. Rather, the increase in H3K4me3 is the result of elevated levels of transcription in this region.
Figure 5.
Changes in histone modification patterns at the subtelomeric region in tor1 mutant cells. Levels of H3K4me3 (A), Ser2P (the phosphorylated form of the C-terminal domain of Rpb1) (B), H4K16Ac (C), or H4K20me2 (D) were measured at the indicated genes by ChIP–qRT-PCR analysis. Each value is the mean of at least three independent assays, and the error bars indicate S.D.
The reduction in heterochromatic markers and the increase in euchromatic markers at the subtelomeric genes in Δtor1 mutant cells suggest a higher level of gene transcription. We therefore investigated the accessibility of RNA Pol II to the subtelomeric regions in WT compared with mutant cells. As RNA Pol II moves from the initiation site, the Rbp1 C-terminal domain becomes phosphorylated at Ser-2. We observed an accumulation of the elongating form of RNA Pol II in Δtor1 cells at the subtelomeric genes, consistent with elevated transcription at this region (Figs. 3B and 5B). There is no change in RNA Pol II occupancy at the dg repeats, consistent with the lack of effect on transcription at this site (Figs. 3B and 5B). We also detected an increase in transcription-associated H4K16 acetylation (H4K16Ac) at the subtelomeric genes in Δtor1 cells (Fig. 5C). The H4K20me2 modification is carried out by Set9 and can be used as a marker for “old” histones, due to the progressive nature of Set9 and the absence of an H4K20me2 demethylase (58). We found a decrease of H4K20me2 at the subtelomeric genes, which suggests an increase in histone turnover (Fig. 5D). This increase may be the result of an elevated level of transcription at the subtelomeric region.
Finally, we considered the possibility that TORC2–Gad8 may affect gene silencing via degradation of the RNAs emerging from heterochromatic regions, a process that involves the exosome (38). Deletion of the core subunit of the exosome, rrp6+, resulted in de-silencing of the subtelomeric genes, but the effect was generally very different in its strength compared with that of the Δtor1 mutation (Fig. S5B). Moreover, combining the Δtor1 and Δrrp6 mutations resulted in an additive effect on gene de-silencing for three subtelomeric genes (spac186.04+, spac186.05+, and spac750.01+; Fig. S5B), suggesting that Rrp6 and Tor1 act in different pathways.
The Leo1–Paf1 subcomplex and the anti-silencer Epe1 antagonize the silencing activity of TORC2
The unstable chromatin states that we observed for TORC2–Gad8 mutant cells (Fig. 1D) are reminiscent of the phenotypes observed in Δepe1 (40) or mutations in the Leo1–Paf1 subcomplex of Paf1C (49). However, whereas TORC2–Gad8 functions to maintain the repressed state of the chromatin, Epe1 or the Paf1C complex functions to antagonize heterochromatin spreading. Accordingly, overexpression of Epe1 resulted in up-regulation of subtelomeric genes (42), whereas Δleo1 resulted in spreading of the heterochromatin at the subtelomeric regions (48). Therefore, we tested for genetic interactions between TORC2–Gad8 and Epe1 or the Leo1–Paf1 subcomplex. Deletion of epe1+ partially suppressed the de-silencing defect of Δtor1 cells, as evidenced by the pink color of Δtor1 Δepe1 double mutant cells carrying the mat2::BamHI::ade6+ construct (Fig. 6A). In contrast, Δleo1 completely suppressed the de-silencing defect of Δtor1 or Δgad8, resulting in fully red colonies (Fig. 6B). Similarly, qRT-PCR analyses show that the de-silencing defect in Δtor1 mutant cells at the subtelomeric region is partially suppressed by Δepe1 and fully suppressed by Δpaf1 (Fig. 6, C and D).
Figure 6.
The silencing defects in Δtor1 or Δgad8 mutant cells are partially suppressed by disruption of epe1+ and fully suppressed by disruption of the Paf1 complex. A and B, suppression at the mat2 locus. Strains containing the indicated mutations were spotted onto low-adenine plates. WT cells that are prototrophic or auxotrophic to adenine were used as a control. C and D, suppression at subtelomeres. Expression levels of the indicated genes were determined by qRT-PCR as described in the legend to Fig. 3.
The partial suppression of the de-silencing defects in TORC2–Gad8 by Δepe1 could be explained if the global levels of Epe1 or the efficiency by which Epe1 is bound to the chromatin is altered in TORC2–Gad8 mutant cells. However, we did not detect changes in the protein level of Epe1-FLAG in Δtor1 or Δgad8 cells or abnormal accumulation of Epe1-FLAG in Δtor1 mutant cells at the mating-type locus (data not shown). TORC2–Gad8 may suppress Leo1–Paf1 activity or act via a distinct pathway that antagonizes Leo1–Paf1 activity. Distinguishing between these two possibilities requires further investigation.
Discussion
The S. pombe TORC2 complex is composed of well-conserved subunits: the catalytic subunit Tor1 and the auxiliary subunits Ste20 (Rictor in human) and Sin1 (mSin1 in humans). A highly conserved feature of TORC2 signaling is the phosphorylation and activation of members of the family of AGC kinases (protein A protein G protein C kinases), Gad8 in S. pombe and AKT in human cells. Similar to mTORC2-AKT, TORC2–Gad8 is implicated in different aspects of growth and proliferation and plays critical roles in starvation responses, cell survival, and aging.
TOR signaling and the regulation of epigenetic information
TOR-dependent signaling often exerts its effects via transcriptional regulation. In particular, TORC1 was shown to regulate gene transcription via multiple mechanisms, including nuclear localization of transcription factors, the regulation of their activity, and regulation of the activity of RNA polymerases (59). TORC1 has also been implicated in chromatin-mediated transcriptional regulation. In S. cerevisiae, TORC1 has been linked to histone H4 acetylation (60, 61) and affects the activity of the sirtuin deacetylases Hst3 and Hst4 (62). Functional links between mTORC1 and SIRT4 have also been reported (63). mTORC1 has also been shown to phosphorylate H2B, thereby mediating histone modifications required for early adipogenesis (64). As in many other aspects, our understanding of the roles of TORC2 in regulating gene expression lags behind that of TORC1. Our studies reveal for the first time that TORC2–Gad8 regulates chromatin-mediated gene expression, epigenetic states, and their stability.
We demonstrate that TORC2–Gad8 is required to silence a reporter gene embedded at the hetrochromatic region of the mating-type locus (mat2::BamHI::ade6+) and affects heterochromatic spreading and gene silencing at subtelomeres. The use of mat2::BamHI::ade6+ as a reporter for chromatin-dependent gene silencing revealed the unstable nature of the epigenetic states in TORC2–Gad8 mutant cells. Accordingly, the epigenetic state of mat2::BamHI::ade6+ in TORC2–Gad8 mutant cells can be found either in its expressed or repressed state, whereas WT cells or mutations in major histone modifiers, such as the Clr4 or Sir2, stably maintain the reporter gene in either its repressed or expressed state, respectively. Epigenetic instability is characteristic of mutations in epe1, a heterochromatin boundary element or mutations in the RNA Pol II–associated complex, Paf1C (48, 49, 51). However, whereas loss of Epe1 or Paf1C results in an unstable extension of the heterochromatin, the deletion of TORC2–Gad8 results in contraction of the heterochromatin. Loss of function of Epe1 or Leo1–Paf1 suppresses the de-silencing defect in TORC2–Gad8 mutant cells, indicating that these chromatin factors act in an antagonistic manner to TORC2–Gad8.
Roles of TORC2–Gad8 in the formation of specific heterochromatic regions at subtelomeres
Our genome-wide transcription (RNA-Seq) and H3K9me2 (ChIP-Seq) analyses, followed by validations of specific subtelomeric genes, indicate that TORC2–Gad8 has a role in regulating heterochromatic spreading and gene silencing at subtelomeres of chromosomes I and II. S. pombe contains three chromosomes, of which chromosomes I and II contain telomeric repeats followed by species-specific homologous DNA sequences (SH sequences) and a variety of genes encoding for proteins. In contrast, arrays of ribosomal DNA repeats, which are typically transcribed by RNA polymerase I, lie next to the telomeres of chromosome III (29, 65). The up-regulation of genes in Δtor1 or Δgad8 at the subtelomeric regions of chromosome I and II, but not chromosome III, is in agreement with previous reports that suggest that chromosomes I and II contain a distinct form of heterochromatin state at their edges (29, 54, 65, 66).
The subtelomeric regions of chromosomes I and II can be divided into to a telomere-proximal high-level H3K9me2 region and a telomere-distal low level H3K9me2 region (29, 54). We detected elevated levels of gene expression only at the low-level H3K9me2 regions. The high- and low-H3K9me2 regions may serve different purposes. The high-level H3K9me2 regions may provide repressive structures, guarding the chromosomal ends from rearrangements that could result from illegitimate recombination between nearly identical sequences. The low-level H3K9me2 region may provide a way to regulate gene expression via chromatin-mediated mechanisms. Indeed, many genes that reside at the low H3K9me2 region become expressed in response to environmental changes, such as starvation and other stress conditions (67). The seven subtelomeric genes that we monitored by qRT-PCR and ChIP–qRT-PCR in TORC2–Gad8 mutant cells encode for putative plasma membrane and transporter proteins (spac186.04+, spac186.05+, spbpb2b2.18+) or metabolic enzymes (spac186.06+, spac750.01+). According to global gene expression databases, these genes are up-regulated in meiosis and/or in response to oxidative DNA damage or cadmium stress (see the Bähler Expression viewer) (67). The specific effect of TORC2–Gad8 at relatively low methylated H3K9me2 regions is remarkably similar to the effects of mutations in cohesin loader mis4 or cohesin core component rad21 mutations (54). Cohesin is recruited to silent chromatin via Swi6 (68, 69) but may also affect Swi6 binding, leading to a cooperative mechanism by which Swi6 levels are maintained at the low-H3K9me2 subtelomeric regions (54). Whether the effects of TORC2–Gad8 and cohesin at subtelomeric region are related remains to be investigated.
TORC2–Gad8 affects gene silencing at the transcriptional level
Loss of TORC2–Gad8 results in up-regulation of gene expression at the subtelomeric regions that is accompanied by an increase in histone modifications associated with elevated levels of transcription, including elevated levels of H3K4me3 and H4K16Ac and accumulation of the elongating form of RNA Pol II. Thus, our data suggest that Δtor1 mutant cells are defective in gene silencing at the transcriptional level. In contrast, mutations in the exosome produced an additive effect when combined with the Δtor1 mutation.
We observed an increase in H4K20me2 in subtelomeric genes in TORC2–Gad8 mutant cells, a modification change that indicates an increased level of histone turnover (58). Suppression of histone turnover is characteristic of heterochromatic regions and accompanies histone modifications that result in suppression of transcription (70). Both Epe1 and the Leo1–Paf1 subcomplex were suggested to mediate their effects on chromatin states through histone turnover (49). Because deletion of the Leo1–Paf1 subcomplex completely abolished gene de-silencing in TORC2–Gad8 mutant cells, it is possible that TORC2–Gad8 is also involved in the regulation of histone turnover. Alternatively, the elevated levels of transcription in TORC2–Gad8 mutant cells bring about an increase in histone turnover. These two possibilities are not mutually exclusive, as histone turnover and elevated levels of transcriptions are two closely related processes. Interestingly, phosphosite mutations of Ser-2 or Ser-7 at the C-terminal domain region of RNA Pol II resulted in up-regulation of S. pombe subtelomeric genes of chromosome I and II (66). Thus, the phosphorylation of these sites may provide a mechanism that links transcription and chromatin modifications. Moreover, phosphorylation of the CDT region of RNA Pol II plays a role in recruitment of the Set1–Compass complex and co-transcriptional methylation of H3K4 in S. cerevisiae or S. pombe (71, 72). The Paf1C complex has also been shown to play a role in recruitment of Set1–Compass in S. cerevisiae (73). Because we observed elevation of H3K4me3 in Δtor1 mutant cells, it is possible that the absence of TORC2 allows the access of RNA Pol II to the subtelomeric region, bringing with it accessory factors, such as the Paf1C complex and Set1–Compass. Alternatively, but not mutually exclusive, TORC2–Gad8 may affect directly RNA Pol II or its associated factors, leading to maintenance of the subtelomeric region in its heterochromatic form. The detailed molecular mechanism by which TORC2–Gad8 regulates gene silencing awaits the isolation of the relevant direct substrate(s).
Experimental procedures
Strains and growth conditions
S. pombe strains used in this study are listed in Table S1. Standard procedures were used for growth and genetic manipulation. Yeast cells were cultured in rich YE medium supplemented with adenine and uracil at 30 °C or in Edinburgh minimal medium (5 g/liter NH4Cl) as described (7). Gene deletions and tagging were performed by standard PCR-based methods (74).
ChIP
50 ml of each strain was grown to A600 ∼1 in YE. 1.5 ml of formaldehyde (37% solution) was added for 15 min, and the formaldehyde was quenched with 2.5 ml of 2.5 m glycine for 5 min. Cells were harvested, washed once with 15 ml of cold PBS, and broken down for 10 min with glass beads in 600 μl of lysis buffer (50 mm HEPES-KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholic acid). The supernatant was removed to a new tube (the lysate). The glass beads were washed with 500 μl of lysis buffer and centrifuged, and the supernatant was added to the lysate. The lysate was sonicated six times for 10 s at 80% amplitude with 1 min on ice between each time. The sonicated material was centrifuged for 30 min at 2500 rpm. The supernatant was used for immunoprecipitations. The sonicated proteins were precleared with 25 μl of protein A–Sepharose and protein G–Sepharose bead mixture (GE Healthcare), and the appropriate antibodies were added to the cleared extract and incubated overnight at 4 °C. Swi6-FLAG, H3K9me2, H3K4me3, Ser2P RPD1, H4K16Ac, H3, H4, and H4K20me2 were immunoprecipitated with 2–5 μg of anti-FLAG antibody (Sigma, F3165), anti-H3K9me2 (Abcam, ab1220), anti-H4K16Ac (Active Motif, 39167-AM), anti-H4 (Abcam, ab10158), anti-H4K20me2 (Abcam, ab9052), anti-H3 (Abcam, ab1791), anti-H3K4me3 (Abcam, ab8580), and anti-Ser2P RPD1 (Millipore, MABE953). A total of 10% of the extract was saved as input. The beads after the immunoprecipitations were washed once with lysis buffer, once with lysis buffer with 360 mm NaCl, once with washing buffer (10 mm Tris/HCl, pH 8, 0.25 m LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholic acid, 1 mm EDTA) and once with TE (10 mm Tris/HCl, pH 8, and 10 mm EDTA). The washed beads and the input were treated with elution buffer (50 mm Tris/HCl, pH 8, 10 mm EDTA, 1% SDS) overnight at 65 °C. The DNA was precipitated, resuspended in water, and used for PCR real-time analysis. All experiments are plotted as the average of at least three independent biological repeats, and each biological repeat is the average of three technical PCR repeats.
qRT-PCR
RNA extractions and qRT-PCR analysis were performed as described by Laor et al. (75), followed by treatment of DNase (RQ1, Promega). For RT-PCR, complementary DNA was synthesized using the ImPromII Reverse transcription system (Promega) with poly(dT) and analyzed by quantitative PCR using the StepOne real-time PCR system. Data were normalized against actin and investigated using the C(t) method. Sequences of the primers used for qRT-PCR analyses are listed in Table S2.
RNA-Seq
RNA was extracted from 50 ml of logarithmic cells using the RNeasy minikit (Qiagen, catalog no. 74104). 1000 ng of total RNA from nine samples was processed using the TruSeq RNA sample preparation kit version 2 (Illumina) (RS-122-2001). Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of nine samples on one lane. Single-end 60-bp reads were sequenced on an Illumina HiSeq 2500 V4 instrument. The sequence yield was between 26.6 and 30.5 million reads per sample.
ChIP-Seq
9–20 ng of ChIP DNA was processed as described previously (76). Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of 30 samples on one lane. Single-end 60-bp reads were sequenced to a median depth of ∼9 million reads/sample on an Illumina HiSeq 2500 version 4 instrument. The reads were aligned uniquely to the S. pombe genome, version ASM294v2 (Ensembl), using Bowtie (version 1.0.0) (77). Differential binding was performed using csaw (78).
Author contributions
A. C., A. H., D. L., and S. Y. investigation; A. C. writing-original draft; M. K. and R. W. supervision; R. W. conceptualization; R. W. writing-review and editing.
Supplementary Material
Acknowledgments
We thank M. Yamamoto, A. Cohen, H. D. Madhani, R. Allshire, K. Shiozaki, S. Whitehall, T. C. Humphery, and B. Xhemalce for strains and Takeshi Urano for the anti-H3K9me2 antibodies. We thank Gilgi Friedlander and Gil Hornung and members of the Mantoux Bioinformatics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine at the Weizmann Institute of Science for help in the analyses of the RNA-Seq and ChIP-Seq. The library preparation and sequencing were performed at the genomics unit, the Crown Genomics Institute. We are very thankful to Mickaël Durand-Dubief and Karl Ekwall for helpful advice on and analysis of the ChIP-Seq experiments and for comments on the manuscript.
This work was supported by Israel Science Foundation Grant 688/14 and Open University of Israel Grant 31040 (to R. W.). The authors declare that they have no conflicts of interest with the contents of this article.
RNA-seq and ChIP-seq data are available using the BioProject ID PRJNA429430.
This article contains Tables S1–S3 and Figs. S1–S5.
- TOR
- target of rapamycin
- mTOR
- mechanical target of rapamycin
- TORC1 and -2
- TOR complex 1 and 2, respectively
- Paf1C
- Paf1 complex
- MBF
- MluI cell cycle box–binding factor
- H3 and H4
- histone H3 and H4, respectively
- K4
- K9, K16, and K20, lysine 4, 9, 16, and 20, respectively
- me
- me2, and me3, methyl, dimethylation, and trimethylation, respectively
- HP1
- heterochromatin protein 1
- kb
- kilobase(s)
- Pol
- polymerase
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Shimobayashi M., and Hall M. N. (2014) Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15, 155–162 10.1038/nrm3757 [DOI] [PubMed] [Google Scholar]
- 2. Saxton R. A., and Sabatini D. M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dazert E., and Hall M. N. (2011) mTOR signaling in disease. Curr. Opin. Cell Biol. 23, 744–755 10.1016/j.ceb.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 4. Zoncu R., Efeyan A., and Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loewith R., and Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weisman R. (2016) Target of rapamycin (TOR) regulates growth in response to nutritional signals. Microbiol. Spectr. 4 10.1128/microbiolspec.FUNK-0006-2016 [DOI] [PubMed] [Google Scholar]
- 7. Weisman R., and Choder M. (2001) The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276, 7027–7032 10.1074/jbc.M010446200 [DOI] [PubMed] [Google Scholar]
- 8. Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., Kokubu A., Ebe M., and Yanagida M. (2007) Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 12, 1357–1370 10.1111/j.1365-2443.2007.01141.x [DOI] [PubMed] [Google Scholar]
- 9. Matsuo T., Otsubo Y., Urano J., Tamanoi F., and Yamamoto M. (2007) Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 27, 3154–3164 10.1128/MCB.01039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alvarez B., and Moreno S. (2006) Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 119, 4475–4485 10.1242/jcs.03241 [DOI] [PubMed] [Google Scholar]
- 11. Weisman R., Roitburg I., Schonbrun M., Harari R., and Kupiec M. (2007) Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics 175, 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawai M., Nakashima A., Ueno M., Ushimaru T., Aiba K., Doi H., and Uritani M. (2001) Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39, 166–174 10.1007/s002940100198 [DOI] [PubMed] [Google Scholar]
- 13. Schonbrun M., Laor D., López-Maury L., Bähler J., Kupiec M., and Weisman R. (2009) TOR complex 2 controls gene silencing, telomere length maintenance, and survival under DNA-damaging conditions. Mol. Cell. Biol. 29, 4584–4594 10.1128/MCB.01879-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schonbrun M., Kolesnikov M., Kupiec M., and Weisman R. (2013) TORC2 is required to maintain genome stability during S phase in fission yeast. J. Biol. Chem. 288, 19649–19660 10.1074/jbc.M113.464974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martín R., Portantier M., Chica N., Nyquist-Andersen M., Mata J., and Lopez-Aviles S. (2017) A PP2A-B55-mediated crosstalk between TORC1 and TORC2 regulates the differentiation response in fission yeast. Curr. Biol. 27, 175–188 10.1016/j.cub.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laboucarié T., Detilleux D., Rodriguez-Mias R. A., Faux C., Romeo Y., Franz-Wachtel M., Krug K., Maček B., Villén J., Petersen J., and Helmlinger D. (2017) TORC1 and TORC2 converge to regulate the SAGA co-activator in response to nutrient availability. EMBO Rep. 18, 2197–2218 10.15252/embr.201744942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Petersen J., and Nurse P. (2007) TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat. Cell Biol. 9, 1263–1272 10.1038/ncb1646 [DOI] [PubMed] [Google Scholar]
- 18. Petersen J. (2009) TOR signalling regulates mitotic commitment through stress-activated MAPK and Polo kinase in response to nutrient stress. Biochem. Soc. Trans. 37, 273–277 10.1042/BST0370273 [DOI] [PubMed] [Google Scholar]
- 19. Baker K., Kirkham S., Halova L., Atkin J., Franz-Wachtel M., Cobley D., Krug K., Maček B., Mulvihill D. P., and Petersen J. (2016) TOR complex 2 localises to the cytokinetic actomyosin ring and controls the fidelity of cytokinesis. J. Cell Sci. 129, 2613–2624 10.1242/jcs.190124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weisman R., Roitburg I., Nahari T., and Kupiec M. (2005) Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics 169, 539–550 10.1534/genetics.104.034983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsuo T., Kubo Y., Watanabe Y., and Yamamoto M. (2003) Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22, 3073–3083 10.1093/emboj/cdg298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otsubo Y., and Yamamato M. (2008) TOR signaling in fission yeast. Crit. Rev. Biochem. Mol. Biol. 43, 277–283 10.1080/10409230802254911 [DOI] [PubMed] [Google Scholar]
- 23. Cohen A., Kupiec M., and Weisman R. (2014) Glucose activates TORC2–Gad8 protein via positive regulation of the cAMP/cAMP-dependent protein kinase A (PKA) pathway and negative regulation of the Pmk1 protein-mitogen-activated protein kinase pathway. J. Biol. Chem. 289, 21727–21737 10.1074/jbc.M114.573824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hatano T., Morigasaki S., Tatebe H., Ikeda K., and Shiozaki K. (2015) Fission yeast Ryh1 GTPase activates TOR Complex 2 in response to glucose. Cell Cycle 14, 848–856 10.1080/15384101.2014.1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cohen A., Kupiec M., and Weisman R. (2016) Gad8 protein is found in the nucleus where it interacts with the MluI cell cycle box-binding factor (MBF) transcriptional complex to regulate the response to DNA replication stress. J. Biol. Chem. 291, 9371–9381 10.1074/jbc.M115.705251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grewal S. I., and Jia S. (2007) Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- 27. Allshire R. C., and Ekwall K. (2015) Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 7, a018770 10.1101/cshperspect.a018770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Atlasi Y., and Stunnenberg H. G. (2017) The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 18, 643–658 10.1038/nrg.2017.57 [DOI] [PubMed] [Google Scholar]
- 29. Cam H. P., Sugiyama T., Chen E. S., Chen X., FitzGerald P. C., and Grewal S. I. (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37, 809–819 10.1038/ng1602 [DOI] [PubMed] [Google Scholar]
- 30. Zofall M., Yamanaka S., Reyes-Turcu F. E., Zhang K., Rubin C., and Grewal S. I. (2012) RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335, 96–100 10.1126/science.1211651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., and Martienssen R. A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 32. Shimada Y., Mohn F., and Bühler M. (2016) The RNA-induced transcriptional silencing complex targets chromatin exclusively via interacting with nascent transcripts. Genes Dev. 30, 2571–2580 10.1101/gad.292599.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bayne E. H., White S. A., Kagansky A., Bijos D. A., Sanchez-Pulido L., Hoe K. L., Kim D. U., Park H. O., Ponting C. P., Rappsilber J., and Allshire R. C. (2010) Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140, 666–677 10.1016/j.cell.2010.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakayama J., Rice J. C., Strahl B. D., Allis C. D., and Grewal S. I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 10.1126/science.1060118 [DOI] [PubMed] [Google Scholar]
- 35. Jih G., Iglesias N., Currie M. A., Bhanu N. V., Paulo J. A., Gygi S. P., Garcia B. A., and Moazed D. (2017) Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature 547, 463–467 10.1038/nature23267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thon G., and Verhein-Hansen J. (2000) Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155, 551–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jia S., Noma K., and Grewal S. I. (2004) RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971–1976 10.1126/science.1099035 [DOI] [PubMed] [Google Scholar]
- 38. Reyes-Turcu F. E., Zhang K., Zofall M., Chen E., and Grewal S. I. (2011) Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat. Struct. Mol. Biol. 18, 1132–1138 10.1038/nsmb.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee N. N., Chalamcharla V. R., Reyes-Turcu F., Mehta S., Zofall M., Balachandran V., Dhakshnamoorthy J., Taneja N., Yamanaka S., Zhou M., and Grewal S. I. (2013) Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell 155, 1061–1074 10.1016/j.cell.2013.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ayoub N., Noma K., Isaac S., Kahan T., Grewal S. I., and Cohen A. (2003) A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol. Cell. Biol. 23, 4356–4370 10.1128/MCB.23.12.4356-4370.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zofall M., and Grewal S. I. (2006) Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22, 681–692 10.1016/j.molcel.2006.05.010 [DOI] [PubMed] [Google Scholar]
- 42. Isaac S., Walfridsson J., Zohar T., Lazar D., Kahan T., Ekwall K., and Cohen A. (2007) Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 175, 1549–1560 10.1534/genetics.106.068684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trewick S. C., Minc E., Antonelli R., Urano T., and Allshire R. C. (2007) The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin. EMBO J. 26, 4670–4682 10.1038/sj.emboj.7601892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Braun S., Garcia J. F., Rowley M., Rougemaille M., Shankar S., and Madhani H. D. (2011) The Cul4-Ddb1(Cdt)(2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144, 41–54 10.1016/j.cell.2010.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trewick S. C., McLaughlin P. J., and Allshire R. C. (2005) Methylation: lost in hydroxylation? EMBO Rep. 6, 315–320 10.1038/sj.embor.7400379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J., Tadeo X., Hou H., Tu P. G., Thompson J., Yates J. R. 3rd, and Jia S. (2013) Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes Dev. 27, 1886–1902 10.1101/gad.221010.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shimada A., Dohke K., Sadaie M., Shinmyozu K., Nakayama J., Urano T., and Murakami Y. (2009) Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 23, 18–23 10.1101/gad.1708009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Verrier L., Taglini F., Barrales R. R., Webb S., Urano T., Braun S., and Bayne E. H. (2015) Global regulation of heterochromatin spreading by Leo1. Open Biol. 5, 150045 10.1098/rsob.150045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sadeghi L., Prasad P., Ekwall K., Cohen A., and Svensson J. P. (2015) The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast. EMBO Rep. 16, 1673–1687 10.15252/embr.201541214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Van Oss S. B., Cucinotta C. E., and Arndt K. M. (2017) Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem. Sci. 42, 788–798 10.1016/j.tibs.2017.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kowalik K. M., Shimada Y., Flury V., Stadler M. B., Batki J., and Bühler M. (2015) The Paf1 complex represses small-RNA-mediated epigenetic gene silencing. Nature 520, 248–252 10.1038/nature14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hansen K. R., Hazan I., Shanker S., Watt S., Verhein-Hansen J., Bähler J., Martienssen R. A., Partridge J. F., Cohen A., and Thon G. (2011) H3K9me-independent gene silencing in fission yeast heterochromatin by Clr5 and histone deacetylases. PLoS Genet. 7, e1001268 10.1371/journal.pgen.1001268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tatebe H., Morigasaki S., Murayama S., Zeng C. T., and Shiozaki K. (2010) Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr. Biol. 20, 1975–1982 10.1016/j.cub.2010.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dheur S., Saupe S. J., Genier S., Vazquez S., and Javerzat J. P. (2011) Role for cohesin in the formation of a heterochromatic domain at fission yeast subtelomeres. Mol. Cell. Biol. 31, 1088–1097 10.1128/MCB.01290-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sugiyama T., Thillainadesan G., Chalamcharla V. R., Meng Z., Balachandran V., Dhakshnamoorthy J., Zhou M., and Grewal S. I. (2016) Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol. Cell 61, 747–759 10.1016/j.molcel.2016.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roguev A., Schaft D., Shevchenko A., Aasland R., Shevchenko A., and Stewart A. F. (2003) High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 278, 8487–8493 10.1074/jbc.M209562200 [DOI] [PubMed] [Google Scholar]
- 57. Mikheyeva I. V., Grady P. J., Tamburini F. B., Lorenz D. R., and Cam H. P. (2014) Multifaceted genome control by Set1 Dependent and independent of H3K4 methylation and the Set1C/COMPASS complex. PLoS Genet. 10, e1004740 10.1371/journal.pgen.1004740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Svensson J. P., Shukla M., Menendez-Benito V., Norman-Axelsson U., Audergon P., Sinha I., Tanny J. C., Allshire R. C., and Ekwall K. (2015) A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin. Genome Res. 25, 872–883 10.1101/gr.188870.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Workman J. J., Chen H., and Laribee R. N. (2014) Environmental signaling through the mechanistic target of rapamycin complex 1: mTORC1 goes nuclear. Cell Cycle 13, 714–725 10.4161/cc.28112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rohde J. R., and Cardenas M. E. (2003) The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23, 629–635 10.1128/MCB.23.2.629-635.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen H., Workman J. J., Tenga A., and Laribee R. N. (2013) Target of rapamycin signaling regulates high mobility group protein association to chromatin, which functions to suppress necrotic cell death. Epigenetics Chromatin 6, 29 10.1186/1756-8935-6-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Workman J. J., Chen H., and Laribee R. N. (2016) Saccharomyces cerevisiae TORC1 controls histone acetylation by signaling through the Sit4/PP6 phosphatase to regulate sirtuin deacetylase nuclear accumulation. Genetics 203, 1733–1746 10.1534/genetics.116.188458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Csibi A., Fendt S. M., Li C., Poulogiannis G., Choo A. Y., Chapski D. J., Jeong S. M., Dempsey J. M., Parkhitko A., Morrison T., Henske E. P., Haigis M. C., Cantley L. C., Stephanopoulos G., Yu J., and Blenis J. (2013) The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854 10.1016/j.cell.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yi S. A., Um S. H., Lee J., Yoo J. H., Bang S. Y., Park E. K., Lee M. G., Nam K. H., Jeon Y. J., Park J. W., You J. S., Lee S. J., Bae G. U., Rhie J. W., Kozma S. C., et al. (2016) S6K1 phosphorylation of H2B mediates EZH2 trimethylation of H3: a determinant of early adipogenesis. Mol. Cell 62, 443–452 10.1016/j.molcel.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tashiro S., Nishihara Y., Kugou K., Ohta K., and Kanoh J. (2017) Subtelomeres constitute a safeguard for gene expression and chromosome homeostasis. Nucleic Acids Res. 45, 10333–10349 10.1093/nar/gkx780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Inada M., Nichols R. J., Parsa J. Y., Homer C. M., Benn R. A., Hoxie R. S., Madhani H. D., Shuman S., Schwer B., and Pleiss J. A. (2016) Phospho-site mutants of the RNA polymerase II C-terminal domain alter subtelomeric gene expression and chromatin modification state in fission yeast. Nucleic Acids Res. 44, 9180–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mata J., Lyne R., Burns G., and Bähler J. (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32, 143–147 10.1038/ng951 [DOI] [PubMed] [Google Scholar]
- 68. Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., and Allshire R. C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 10.1126/science.1064027 [DOI] [PubMed] [Google Scholar]
- 69. Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S. I., and Watanabe Y. (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4, 89–93 10.1038/ncb739 [DOI] [PubMed] [Google Scholar]
- 70. Aygün O., Mehta S., and Grewal S. I. (2013) HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol. 20, 547–554 10.1038/nsmb.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ng H. H., Robert F., Young R. A., and Struhl K. (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719 10.1016/S1097-2765(03)00092-3 [DOI] [PubMed] [Google Scholar]
- 72. Mbogning J., Pagé V., Burston J., Schwenger E., Fisher R. P., Schwer B., Shuman S., and Tanny J. C. (2015) Functional interaction of Rpb1 and Spt5 C-terminal domains in co-transcriptional histone modification. Nucleic Acids Res. 43, 9766–9775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., and Shilatifard A. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 10.1016/S1097-2765(03)00091-1 [DOI] [PubMed] [Google Scholar]
- 74. Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A. M. 3rd, Steever A. B., Wach A., Philippsen P., and Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 10.1002/(SICI)1097-0061(199807)14:10%3C943::AID-YEA292%3E3.0.CO%3B2-Y [DOI] [PubMed] [Google Scholar]
- 75. Laor D., Cohen A., Kupiec M., and Weisman R. (2015) TORC1 regulates developmental responses to nitrogen stress via regulation of the GATA transcription factor Gaf1. MBio 6, e00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., and Amit I. (2013) High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 8, 539–554 10.1038/nprot.2013.023 [DOI] [PubMed] [Google Scholar]
- 77. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lun A. T. L., and Smyth G. K. (2016) csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 44, e45 10.1093/nar/gkv1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.